Abstract
Arginine methylation of proteins affects major processes in the cell, including transcriptional regulation, mRNA metabolism, signal transduction and protein sorting. Arginine methylation of Ad (adenovirus) E1B 55-kDa-associated protein E1B-AP5 was recently described by us [Kzhyshkowska, Schutt, Liss, Kremmer, Stauber, Wolf and Dobner (2001) Biochem. J. 358, 305–314]. In this first example of protein arginine methylation analysis in Ad-infected cells, we investigated methylation of the E1B-AP5 and the viral L4-100 kDa protein. We demonstrate that E1B-AP5 methylation is enhanced during the course of infection in a cell-type-specific manner. We also show that L4-100 kDa is efficiently methylated in Ad-infected cells. L4-100 kDa formed complex with methyltransferase in vivo during productive infection, and can be methylated by HRMT1L2 (human protein arginine methyltransferase 1) in vitro. Comparative analysis of E1B-AP5 and L4-100 kDa protein methylation in Ad-infected HeLa, MCF-7 and H1299 cells revealed that the profile of protein arginine methylation correlates with the efficiency of Ad proteins production. Our results suggest that protein arginine methylation is an important host-cell function required for efficient Ad replication.
Keywords: adenovirus, E1B-AP5, L4-100 kDa, protein arginine methyltransferase, S-adenosyl-L-methionine
Abbreviations: Ad, adenovirus; GST, glutathione S-transferase; hnRNP, heterogeneous nuclear ribonucleoprotein; hpi, hours post-infection; IP, immunoprecipitation; mAb, monoclonal antibody; NP40, Nonidet P40; NTM, no-translation mix; PRMT, protein arginine methyltransferase
INTRODUCTION
Ads (adenoviruses) are active molecular parasites exploiting host-cell biosynthetic systems during productive infection. The lytic cycle of Ads is organized into early and late phases separated by the onset of viral DNA replication [1,2]. The early phase serves to establish an optimal environment for viral particle production and is characterized by an expression of viral regulatory proteins. In the late phase of infection, large quantities of the viral genome and structural proteins are produced. In addition, the late phase is associated with significant changes in cellular gene expression at the level of nucleocytoplasmic mRNA transport and protein translation. These changes favour preferential synthesis of viral proteins and efficient assembly of virions [3,4].
One of the major mechanisms for a rapid host-cell response to viral demands is a rapid change in post-translational modifications of cellular proteins. In other words, the virus actively modifies the host-cell molecular context by influencing the post-translational modifications of cellular proteins used by the viral replication machinery [3,5–7]. Arginine methylation, a post-translation protein modification identified more than 30 years ago, has recently stimulated significant interest. Arginine methylation in proteins is apparently involved in fundamental processes of gene expression, such as transcriptional regulation and post-transcriptional control [8–10]. Post-translational methylation of arginine contributes to the regulation of protein–protein and protein–nucleic acid interactions [11–13]. Biochemically, arginine methylation represents the formation of ω-NG,NG-dimethylarginine residues present in RGG or RXR sequence motifs [14]. The enzymes catalysing this modification were recently cloned and comprise the PRMT (protein arginine methyltransferase) family. Several PRMTs are involved in ligand-induced signal-transduction pathways, as well as transcriptional and post-transcriptional regulation [15,16].
The major group of substrates for this class of enzymes includes the hnRNPs (heterogeneous nuclear ribonucleoproteins). In mammalian cells, hnRNPs contain approx. 65% of the total NG,NG-dimethylarginine found in the cell nucleus [14]. The hnRNP family includes a broad range of proteins acting at all stages of mRNA metabolism: transcriptional regulation, mRNA processing, nucleocytoplasmic transport and commitment to translation [17–21]. One of the hnRNP family members, E1B-AP5, was identified by our group as a target of the early Ad E1B 55 kDa protein [22]. This interaction was suggested to have a dual function: first, E1B 55-kDa/E1B-AP5 complex formation contributes to efficient accumulation of viral late mRNA in the cytoplasm [22]; secondly, E1B-AP5 might be a cofactor required for E1B 55-kDa-dependent transcriptional repression [23]. Arginine methylation of E1B-AP5, described recently in detail in [24], targets its RGG-box domain, which presumably mediates protein–RNA and/or protein–protein interactions. Human PRMT2 (HRMT1L1) but not the predominant hPRMT1 was found to interact with E1B-AP5, and was suggested to be responsible for specific E1B-AP5 methylation.
The initial aim of the present study was to analyse arginine methylation of E1B-AP5 in Ad-infected cells. We demonstrate that the level of E1B-AP5 methylation is modified during the late phase of infection in cell-type-specific manner. Surprisingly, we also observed a highly methylated viral late product, which we identified as the L4-100 kDa protein. We show that L4-100 kDa is a substrate for the hPRMT1 (human protein arginine methyltransferase 1). Comparative analysis of the dynamics of L4-100 kDa and E1B-AP5 methylation and viral late protein synthesis established a correlation between late-stage viral protein synthesis efficiency and the ability of the host cell to provide increased protein arginine methylation.
EXPERIMENTAL
Cells, viruses and infections
MCF-7 [25], H1299 and HeLa (A.T.C.C. CCL 2, Rockville, MD, U.S.A.) cells were maintained as monolayers in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal calf serum, 100 units of penicillin and 100 μg of streptomycin/ml. H5wt300 served as the wild-type Ad5 virus in these studies. For infection with H5wt300, cells were passaged 24 h before infection to achieve approx. 30% confluence at the time of infection. Cells were washed twice with PBS, and the final wash was replaced with H5wt300 virus (15–20 plaque-forming units/cell) in Dulbecco's modified Eagle's medium. The virus was added at one-fourth of the normal culture volume, and the cells were gently rocked for 60 min at 37 °C. The virus suspension was then replaced with normal growth medium.
Plasmids and purification of GST (glutathione S-transferase) fusion proteins
The plasmids pGST-AP5-N/RG and pGST-AP5-RGG express fragments of E1B-AP5 from amino acid positions 171 to 193 and 611 to 688 respectively fused to GST [24]. pGEX-HRMT1L2 was a gift from M. F. Henry (University of Medicine and Dentistry of New Jersey, NJ, U.S.A.). It produces the human arginine methyltransferase HRMT1L2 (hPRMT1) protein fused to GST [26]. Expression and purification of GST fusion proteins were exactly as described previously [24,27]. The amount of purified GST fusion proteins was determined by comparison with BSA standards using SDS/PAGE followed by protein staining with gelcode blue stain reagent (Pierce).
Generation of anti L4-100 kDa mAb (monoclonal antibody)
N-terminal 255 amino acid fragment of L4-100 kDa protein was expressed in GST-fusion form, purified out of Escherichia coli and used for rat immunization. Positive clone selection was performed using purified immunogen in ELISA. Specificity of L4-6B10 (rat IgG2a) mAb was confirmed using HeLa and A549 cells transiently transfected with pcDNA3-FLAG L4100 (a gift from R. Schneider, New York University, U.S.A.) and Ad5 wt300-infected HeLa and A549 cells.
Analyses of polypeptides and IPs (immunoprecipitations)
The following mAbs were used in the present study: the rat mAb 4A11 is specific for the E1B-AP5 protein [24]. Anti-L4-100 kDa protein 2100K-1 and anti-hexon mouse mAbs were kindly provided by S. J. Flint (Princeton University). The anti-E2A 72 kDa DNA-binding protein mouse mAb B6-8 [28] was a gift from A. Levine (Princeton University).
For the analysis of proteins by Western blotting or IP, total cell extracts were prepared in NP40 (Nonidet P40)/1% lysis buffer (50 mM Tris/HCl, pH 8.0, 150 mM NaCl and 1% NP40) supplemented with a protease inhibitor cocktail (Roche). For IPs, 250 μg of samples of total cell extracts were precleared with Protein G–Sepharose and incubated overnight with mAbs bound to Protein G–Sepharose in a total volume of 1.5 ml. The immunocomplexes were washed 5× with NP40/1% lysis buffer, resuspended in Laemmli loading buffer and heated at 95 °C for 5 min. After separation on 10% (w/v) SDS/polyacrylamide gels, proteins were detected by immunoblotting using the appropriate mAbs. For radiolabelled proteins, gels were incubated for 1 h with Enlight solution (EnerGene), dried and exposed to Kodak BioMax film at −80 °C.
In vivo and in vitro methylation reactions
Before in vivo cell labelling, normal growth medium was replaced by methionine-deficient MEM (minimal essential medium) supplemented with 100 μg of cycloheximide and 40 μg of chloramphenicol/ml (NTM, no-translation mix). After 1 h at 37 °C, L-[methyl-3H]methionine (Amersham Biosciences) was added to a final concentration of 30 μCi/ml to detect methylated proteins. Alternatively, protein translation inhibition was monitored by adding 20 μCi/ml [35S]methionine (ICN, Cleveland, OH, U.S.A.) to the medium. After 3.5 h, cells were washed 2× with PBS, harvested and lysed in NP40/1% lysis buffer. Total cell extracts were either used directly for SDS/PAGE or subjected to IP.
For in vitro methylation of Ad5 L4-100 kDa, H5wt300-infected cells were grown in the presence of methylation inhibitor AdOx (periodate-oxidized adenosine; Sigma) at a final concentration of 20 μM in the culture medium. Cells (1×106) were lysed 16 h after infection in NP40/1% lysis buffer and the L4-100 kDa protein was immunoprecipitated with mAb 2100K-1. One-tenth of the precipitate was added to a final volume of 35 μl of methylation buffer containing 50 mM Na-Mops, 0.3 M NaCl, 2 mM EDTA, 3 μCi of S-adenosyl[methyl-3H]-L-methionine (15 Ci/mmol) and protease inhibitors. Samples were incubated for 35 min at 37 °C. The reaction was stopped by adding 2× SDS-loading buffer and heating for 5 min at 95 °C. Aliquots of each reaction were analysed by SDS/PAGE, followed by incubation with Enlight solution (EnerGene) for 1 h, drying and autoradiography at −80 °C. For the activity-reconstitution experiments, samples were heated for 6 min at 70 °C, and 0.2 μg of purified GST-hPRMT1 was added to each reaction.
RESULTS
Analysis of E1B-AP5 arginine methylation during the time course of infection
To analyse arginine methylation of E1B-AP5 in Ad-infected cells, we performed experiments as outlined in Figure 1. HeLa, MCF-7 and H1299 cells were infected with Ad5 wild-type (H5wt300). At various time points after infection, cells were labelled with L-[methyl-3H]methionine for 3.5 h in the presence of protein synthesis inhibitors (+NTM). Total cell extracts were prepared and subjected to IP by using the E1B-AP5-specific mAb 4A11 (Figure 2A). In non-infected HeLa, MCF-7 and H1299 cells, a 120 kDa methylated protein was precipitated corresponding to E1B-AP5 as described by us [24]. Additional methylated protein migrating at a molecular mass of approx. 100 kDa was co-precipitated during viral time course of infection in all three cell lines analysed. Analysis of E1B-AP5 steady-state level with 4A11 antibody in total cell extracts revealed that the amount of E1B-AP5 120 kDa form is not changed during viral infection course except for a slight decrease at 22 hpi (hours post-infection) in MCF-7 cells (Figure 3B). Additionally, 4A11 antibody detected a second form of E1B-AP5 at 22 hpi in HeLa and H1299 cells, but not in MCF-7 cells. This form migrated with a molecular mass of approx. 110 kDa and did not correspond to co-precipitated methylated protein.
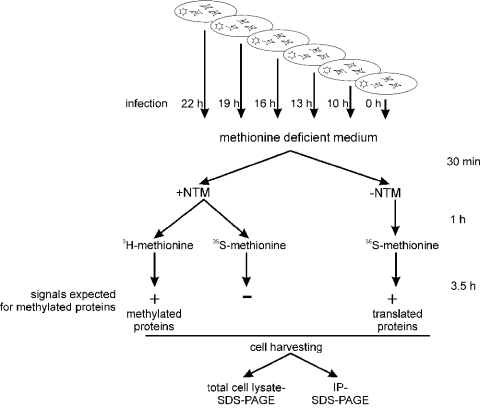
Figure 1. Experimental scheme for analysing protein arginine methylation in productive Ad-infected cells.
At various time points after infection (22–10 h) and control, non-infected (0 h) cells were transferred to a methionine-deficient medium in the presence or absence of protein synthesis inhibitors (+NTM or −NTM) as described in the Experimental section. To monitor protein methylation, the methyl-group donor L-[methyl-3H]methionine was added to the NTM-containing medium. Under these conditions, incorporation of 3H into proteins can only occur through post-translational methylation. Inhibition of protein synthesis was confirmed by parallel cell labelling with [35S]methionine in the presence of NTM. In contrast, total protein synthesis was monitored by labelling with [35S]methionine in the absence of NTM. Samples were subsequently analysed by SDS/PAGE of total cell lysates or immunoprecipitates.
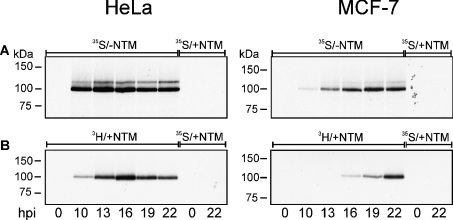
Figure 2. Protein methylation in H5wt300-infected HeLa, MCF-7 and H1299 cells.
At the indicated time points after infection (hpi), cells were incubated with MEM-lacking methionine and labelled either with L-[methyl-3H]methionine in the presence of +NTM (A, C) or with [35S]methionine but without −NTM (B). Inhibition of protein synthesis was confirmed by parallel cell labelling with [35S]methionine +NTM (lanes C in A–C) and cells were harvested 22 h post-infection. (A) Methylation of E1B-AP5 during the time course of infection. E1B-AP5 was immunoprecipitated from total cell extracts using anti-E1B-AP5 mAb 4A11. Samples were analysed by SDS/PAGE followed by autoradiography. The signal was analysed after 7 days of exposure. (B) Total protein synthesis during the time course of infection. Proteins (20 μg of samples) from each time point were separated by SDS/PAGE followed by autoradiography (6 h exposure). (C) Total protein methylation during the time course of infection. Proteins (20 μg of samples) from each time point were separated by SDS/PAGE followed by autoradiography (3 days exposure). Arrows indicate E1B-AP5 120 kDa (A) and methylated L4-100 kDa proteins (C).
Figure 3. Protein steady-state level in HeLa, MCF-7 and H1299 cells infected with H5wt300.
Aliquots of lysates (20 μg of samples) used in Figure 2 were separated by PAGE and analysed by Western blotting using the following antibodies: (A) (B6-8) mouse hybridoma supernatant recognizing Ad5 E2A 72 kDa to control the infection efficiency; (B) rat hybridoma supernatant 4A11 anti-E1B-AP5; (C) rat hybridoma supernatant L4-6B10 anti-L4-100 kDa.
Significant increase in the methylation of the 120 kDa E1B-AP5 form was observed in HeLa cells between 10 and 16 hpi with maximum at 13 hpi, followed by a sequential decrease at later time points. In contrast, the level of methylation of E1B-AP5 remained almost constant throughout the infection in MCF-7 and H1299 cells, except for a decrease at 22 hpi in H1299 cells (Figure 2A). These results demonstrated that methylation of E1B-AP5 is enhanced in the late phase of adenoviral infection in a cell-type-specific manner.
To ensure that inhibition of protein synthesis is complete and only methyl-group incorporation contributed to the protein labelling during the in vivo methylation procedure, we monitored the efficiency of protein translation inhibition by labelling cells with [35S]methionine in the presence of NTM. Analyses of total protein lysates by SDS/PAGE revealed no radioactive signals in the samples containing cells treated with NTM (Figure 2B, lanes C). Intense labelling in samples corresponding to control cell lysates propagated without NTM throughout the experiment represents the level of protein synthesis at the indicated time points after infection (Figure 2B). We also observed that virus-induced host-cell protein synthesis shut-off started earlier and was much more efficient in HeLa cells when compared with MCF-7 or H1299 cells (Figure 2B). Furthermore, synthesis of viral L4-100 kDa protein started earlier in HeLa cells (Figure 3C). The infection efficiency of Ad5 wild-type in all three cell types was evaluated by analysis of E2A-72 kDa protein expression using indirect immunofluorescence (results not shown) and Western blotting (Figure 3A). In all cell types, cells were infected 100% but we observed a lower amount of E2A-72 kDa protein in MCF-7 when compared with HeLa cells.
Interestingly, analysis of 3H-labelled total cell lysates during viral infection revealed the appearance of a highly methylated protein with a molecular mass of approx. 100 kDa. The signal was detected in HeLa, MCF-7 and H1299 cells, although the time point when the methylated protein appeared was cell-type-specific (Figure 2C). A strong signal was already observed in HeLa cells 10 h post-infection, whereas in MCF-7 and H1299 cells the first signal was detected 16 h after infection. Several facts indicated that a highly methylated protein has a viral origin. First, methylation signal in total cell lysates (20 μg of protein/sample) was extremely strong and was already detectable after 1 day of membrane exposure to the X-ray film (the presented result shows signal after 3 days exposure), whereas a 7 day exposure was necessary to visualize the methylated host-cell protein E1B-AP5, which was concentrated by IP out of 250 μg of cell lysate. Secondly, the new methylated protein appeared in cell lysates after the onset of viral replication, and the rate of methylated protein accumulation corresponded to the rate of viral protein synthesis (Figure 2B). The next step was to identify this protein.
Methylation of the L4-100 kDa protein during the lytic cycle of Ad infection
Two viral proteins of 100 and 110 kDa respectively are synthesized in the late phase of Ad-infected cells: L4-100 kDa, a non-structural polypeptide implicated in preferential synthesis of viral late proteins [4] and L3-110 kDa (hexon), a major structural unit of the capsid [2]. To identify if either of these two candidates undergoes methylation, we used our in vivo methylation/IP approach. HeLa cells were infected with Ad5 wild-type for 16 h before commencing in vivo methylation, performed according to our standard scheme (Figure 1). Protein lysates were divided equally and used for IPs with mAbs recognizing L4-100 kDa protein (2100K-1) and hexon. Methylation signals were only observed in samples containing immunoprecipitated L4-100 kDa protein (Figure 4). Both L4-100 kDa and hexon, analysed in the absence of protein inhibition, were efficiently synthesized during the late phase of infection, moreover, 2100K-1 IP out of 35S-labelled lysates revealed co-precipitation of hexon together with L4-100 kDa protein as described previously [29,30].
Figure 4. Ad5 L4-100 kDa is methylated in H5wt300-infected cells.
Cells were labelled with L-[methyl-3H]methionine or [35S]methionine in the presence (+NTM) or absence (−NTM) of NTM as indicated. Total cell extracts were prepared from non-infected cells (0 h) and 16 h post-infection and subjected to IP using anti-L4-100 kDa mAb 2100K-1 (A) or anti-hexon mAb (B). The immunocomplexes were separated by SDS/PAGE and proteins were visualized by autoradiography.
L4-100 kDa protein is methylated by hPRMT1 in vitro
L4-100 kDa protein exhibits RNA-binding activity and apparently provides functions that increase the translation efficiency of viral late transcripts [30,31]. Protein methylation on arginine residues is a characteristic feature of many cellular and viral proteins involved in mRNA metabolism. Since PRMT1 was clearly demonstrated to be the predominant cellular arginine methyltransferase [32], we examined whether this enzyme can methylate the L4-100 kDa protein. A combined IP/in vitro methylation assay was used to obtain substrates under optimal conditions, i.e. corresponding to complexes existing in vivo, in contrast with bacterially purified proteins [24]. L4-100 kDa protein was immunoprecipitated out of Ad5 wild-type-infected HeLa cells 16 h post-infection. To obtain non-methylated substrates, the cells were grown in the presence of methylation inhibitor AdOx. Incubation of immunoprecipitated L4-100 kDa with methyl group donor [35S]adenosylmethionine in vitro resulted in intense 3H incorporation in the protein without addition of any methyltransferase, indicating that the active enzyme forms a complex with L4-100 kDa in vivo and is co-precipitated (Figure 5, lane 1). No activity was precipitated when using non-specific mAb for IP (results not shown). Addition of bacterially produced GST-hPRMT1 slightly increased the 3H incorporation in L4-100 kDa (Figure 5, lane 2). To confirm whether GST-hPRMT1 is using L4-100 kDa as a substrate, the co-precipitated activity was inactivated by heating the whole complex to 70 °C for 6 min, followed by addition of bacterially produced GST-hPRMT1 to the reaction. Heat inactivation of co-precipitated activity was complete, and addition of the enzyme resulted in intense 3H incorporation in L4-100 kDa, identical with the non-inactivated substrate complexed with hPRMT1 (Figure 5, cf. lanes 1 and 4). The intensity of 3H incorporation was also comparable with the recombinant bacterially produced E1B-AP5 RGG box. In contrast with full-length E1B-AP5, its RGG box taken separately serves as a substrate for PRMT1 and can be used as positive control in vitro [24]. We concluded that L4-100 kDa is a specific substrate for hPRMT1, and that L4-100 kDa undergoes arginine methylation during lytic Ad infection.
Figure 5. Ad5 L4-100 kDa is methylated by hPRMT1/HRMT1L2 in vitro.
HeLa cells, grown in the presence of the methylation inhibitor AdOx, were infected with H5wt300 at a multiplicity of 20 plaque-forming units/cell. Total cell extracts were prepared 16 h post-infection and subjected to IP using anti-L4-100 kDa mAb 2100K-1 (IP α-L4-100 kDa). To inactivate co-precipitated methyltransferase activity half of the precipitate was incubated at 70 °C for 6 min (lanes 3 and 4). GST-hPRMT1 (0.2 μg) was added to the reactions (lanes 2 and 4) and samples were analysed by SDS/PAGE followed by autoradiography. Lanes 5–8 show control reactions where two different fragments of E1B-AP5 fused to GST were used as substrates for GST-hPRMT1 (lanes 6 and 8). GST-AP5-RGG (lanes 7 and 8) contains the RGG domain of E1B-AP5 previously shown to be methylated by GST-hPRMT1 in vitro [24].
Comparison of L4-100 kDa synthesis and methylation during viral infection
We compared the profile of L4-100 kDa protein methylation and the accumulation of its total amount during viral infection course. Western-blot analysis with L4-6B10 antibody revealed the detectable level of L4-100 kDa in HeLa cells at 13 hpi, and the protein accumulated gradually with the virus cycle progression (Figure 3C). The highest intensity of L4-100 kDa methylation was observed in HeLa cells at 13 hpi. In contrast with total protein amount, L4-100 kDa methylation signal was not accumulating, but was decreasing during the viral-cycle progression (Figure 2C). In MCF-7 and H1299 cells, accumulation of total amount of L4-100 kDa protein and its methylation had similar profiles and increased during the viral-cycle progression. In all three cell lines methylation signal was observed at earlier time points, compared with the signal from Western blotting due to higher sensitivity of radioactive detection (cf. Figures 2C and 3C).
To analyse whether the increase in L4-100 kDa methylation in HeLa cells is determined by the increase in its synthesis at the same time point, we compared the level of synthesis and methylation using methods of similar sensitivity. The level of L4-100 kDa synthesis was estimated by labelling cells with [35S]methionine in methionine-deficient medium. In HeLa cells, efficient synthesis of L4-100 kDa protein was already detected 10 h post-infection, and the amount of newly synthesized protein remained at a similar level during the entire late phase (Figure 6A). In parallel, methylation of L4-100 kDa protein analysed by labelling cells with [3H]methionine showed significant methylation in HeLa cells 10 h post-infection, indicating that post-translational modification occurs immediately after protein synthesis. The level of methylation reached a maximum between 13 and 16 h post-infection, and decreased at the end of late phase (Figures 2C and 6B). As a control, we used MCF-7 cells in which the methylation signal increase was paralleled by the increase in L4-100 kDa synthesis (Figure 6). We concluded that increase in L4-100 kDa methylation is not due to the accumulation of the total amount of protein, and is specific for HeLa cells.
Figure 6. Ad5 L4-100 kDa protein synthesis (A) and methylation (B) during the time course of infection.
H5wt300-infected HeLa and MCF-7 cells were labelled with L-[methyl-3H]methionine or [35S]methionine in the presence (+NTM) or absence (−NTM) of NTM. At the indicated time points after infection (hpi), total cell extracts were prepared and subjected to IP using anti-L4-100 kDa mAb 2100K-1. The immunocomplexes were separated by SDS/PAGE and L4-100 kDa was visualized by autoradiography [6 h exposure for (A) and 2 days exposure for (B)].
DISCUSSION
The present study is the first example of arginine methylation analysis during lytic Ad infection. Attention has been refocused on this post-translational modification due to several recent findings exposing the broad range of basic cellular processes that depend on protein arginine methylation. Among these are transcriptional regulation and mRNA metabolism. However, this post-translational protein modification has never been analysed in Ad-infected cells. The predominant population of arginine-methylated proteins includes hnRNP family members involved in different stages of mRNA metabolism. Recently, we demonstrated that hnRNP E1B-AP5, identified by our group as a protein probably used by Ad to increase the efficiency of viral late mRNA nucleocytoplasmic export, undergoes post-translational arginine methylation [24]. We supposed that changing its post-translational modification status alters normal E1B-AP5 cellular function. Indeed, in the present study, we demonstrated the cell-type-specific increase in the level of E1B-AP5 methylation between 10 and 16 h post-infection, suggesting that this parameter may contribute to the efficiency of viral mRNA metabolism and as a consequence to the efficiency of viral protein production.
Unexpectedly, we observed a highly methylated protein species synthesized during the late phase of Ad infection. We identified the protein as L4-100 kDa. In vitro studies revealed that the predominant mammalian methyltransferase PRMT1 catalyses arginine methylation of native L4-100 kDa protein immunoprecipitated from viral-infected cells. Recently, we demonstrated by the same approach, using native immunoprecipitated E1B-AP5, that a distinct member of the protein arginine methyltransferase family, presumably hPRMT2 (HRMT1L1), is responsible for E1B-AP5 methylation [24]. L4-100 kDa protein is a non-structural Ad protein abundant in the late phase of productive infection. A large proportion of the protein was found to bind viral and cellular mRNA species in the cytoplasm [30,31,33]. L4-100 kDa protein is required for efficient and selective initiation of viral late mRNA translation. The selectivity is achieved by displacing kinase Mnk1 from the translation initiation complex eIF4E in the host cell [34]. Under these conditions, only viral mRNA containing a tripartite leader sequence can initiate translation [35,36]. The structural background for this L4-100 kDa protein function is its ability to form complexes with specific mRNA sequences and protein components of the translation initiation complex. Since protein arginine methylation contributes to protein–RNA binding and is a critical factor for certain specific protein–protein interactions, further investigation of L4-100 kDa arginine methylation will lead to a better understanding of molecular mechanisms of L4-100 kDa function.
The fact that L4-100 kDa and E1B-AP5 are methylated by different protein arginine methyltransferases indicates that enhancement of arginine methylation during lytic Ad infection is not achieved by up-regulation of a single enzyme activity. An important question to be addressed in future studies is whether the host-cell enzymic machinery responsible for this post-translational modification is specifically targeted by the virus or represents a beneficial, but passive consequence of viral entry into the host cell.
The methylation profile of L4-100 kDa protein was cell-type-specific, and did not simply correspond to the total amount of L4-100 kDa protein accumulating during viral-infection course progression. Increase in methylation of L4-100 kDa was detected in HeLa cells between 13 and 16 hpi of late stage of infection and correlated with the increase in E1B-AP5 methylation. Since both proteins are involved in viral mRNA metabolism and arginine methylation is known to affect protein–RNA interaction for many hnRNPs, we suggest that the ability of the host cell to provide efficient protein arginine methylation for cellular and viral proteins could represent an important host-cell factor, which is utilized by virus and contribute to the viral mRNA metabolism.
Acknowledgments
We are grateful to S.J. Flint for providing the mAbs against viral proteins, M.F. Henry for the HRMT1L1 expression construct and R. Schneider for L4-100 kDa expression construct. We also thank D. Büchner for the excellent technical assistance and A. Gratchev for a critical reading of the paper. This work was supported by grants from Deutsche Forschungsgemeinschaft (Do343/4-3 to T. D.).
References
- 1.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B. N., Knipe D. M., Howley P. M., editors. Virology. New York: Lippincott-Raven; 1996. pp. 2111–2148. [Google Scholar]
- 2.Flint S. J. Nature Publishing Group; 2001. Adenoviruses, Encyclopedia of Life Sciences. Electronic Citation. [Google Scholar]
- 3.Dobner T., Kzhyshkowska J. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 2001;259:25–54. doi: 10.1007/978-3-642-56597-7_2. [DOI] [PubMed] [Google Scholar]
- 4.Flint S. J., Gonzalez R. A. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 2003;272:287–330. doi: 10.1007/978-3-662-05597-7_10. [DOI] [PubMed] [Google Scholar]
- 5.Kleinberger T. Induction of apoptosis by adenovirus E4orf4 protein. Apoptosis. 2000;5:211–215. doi: 10.1023/a:1009644210581. [DOI] [PubMed] [Google Scholar]
- 6.Branton P. E., Roopchand D. E. The role of adenovirus E4orf4 protein in viral replication and cell killing. Oncogene. 2001;20:7855–7865. doi: 10.1038/sj.onc.1204862. [DOI] [PubMed] [Google Scholar]
- 7.Ben Israel H., Kleinberger T. Adenovirus and cell cycle control. Front. Biosci. 2002;7:d1369–d1395. doi: 10.2741/ben. [DOI] [PubMed] [Google Scholar]
- 8.Boisvert F. M., Cote J., Boulanger M. C., Richard S. A proteomic analysis of arginine methylated protein complexes. Mol. Cell Proteom. 2003;2:1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Kwak Y. T., Guo J., Prajapati S., Park K. J., Surabhi R. M., Miller B., Gehrig P., Gaynor R. B. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell. 2003;11:1055–1066. doi: 10.1016/s1097-2765(03)00101-1. [DOI] [PubMed] [Google Scholar]
- 10.Yadav N., Lee J., Kim J., Shen J., Hu M. C., Aldaz C. M., Bedford M. T. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mears W. E., Rice S. A. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 1996;70:7445–7453. doi: 10.1128/jvi.70.11.7445-7453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedford M. T., Frankel A., Yaffe M. B., Clarke S., Leder P., Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 2000;275:16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- 13.Raman B., Guarnaccia C., Nadassy K., Zakhariev S., Pintar A., Zanuttin F., Frigyes D., Acatrinei C., Vindigni A., Pongor G., et al. N(omega)-arginine dimethylation modulates the interaction between a Gly/Arg-rich peptide from human nucleolin and nucleic acids. Nucleic Acids Res. 2001;29:3377–3384. doi: 10.1093/nar/29.16.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gary J. D., Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 15.McBride A. E., Silver P. A. State of the arg: protein methylation at arginine comes of age. Cell (Cambridge, Mass.) 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 16.Stallcup M. R. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene. 2001;20:3014–3020. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- 17.Krecic A. M., Swanson M. S. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 18.Hirose Y., Manley J. L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 19.Mili S., Shu H. J., Zhao Y., Pinol-Roma S. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol. Cell Biol. 2001;21:7307–7319. doi: 10.1128/MCB.21.21.7307-7319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed R., Magni K. A new view of mRNA export: separating the wheat from the chaff. Nat. Cell Biol. 2001;3:E201–E204. doi: 10.1038/ncb0901-e201. [DOI] [PubMed] [Google Scholar]
- 21.Reed R., Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell (Cambridge, Mass.) 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- 22.Gabler S., Schutt H., Groitl P., Wolf H., Shenk T., Dobner T. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 1998;72:7960–7971. doi: 10.1128/jvi.72.10.7960-7971.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kzhyshkowska J., Rusch A., Wolf H., Dobner T. Regulation of transcription by the heterogeneous nuclear ribonucleoprotein E1B-AP5 is mediated by complex formation with the novel bromodomain-containing protein BRD7. Biochem. J. 2003;371:385–393. doi: 10.1042/BJ20021281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kzhyshkowska J., Schutt H., Liss M., Kremmer E., Stauber R., Wolf H., Dobner T. Heterogeneous nuclear ribonucleoprotein E1B-AP5 is methylated in its Arg-Gly-Gly (RGG) box and interacts with human arginine methyltransferase HRMT1L1. Biochem. J. 2001;358:305–314. doi: 10.1042/0264-6021:3580305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soule H. D., Vazguez J., Long A., Albert S., Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 26.Scott H. S., Antonarakis S. E., Lalioti M. D., Rossier C., Silver P. A., Henry M. F. Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2) Genomics. 1998;48:330–340. doi: 10.1006/geno.1997.5190. [DOI] [PubMed] [Google Scholar]
- 27.Rosorius O., Heger P., Stelz G., Hirschmann N., Hauber J., Stauber R. H. Direct observation of nucleocytoplasmic transport by microinjection of GFP-tagged proteins in living cells. Biotechniques. 1999;27:350–355. doi: 10.2144/99272rr02. [DOI] [PubMed] [Google Scholar]
- 28.Reich N. C., Sarnow P., Duprey E., Levine A. J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology. 1983;128:480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- 29.Cepko C. L., Sharp P. A. Analysis of Ad5 hexon and 100K ts mutants using conformation-specific monoclonal antibodies. Virology. 1983;129:137–154. doi: 10.1016/0042-6822(83)90402-6. [DOI] [PubMed] [Google Scholar]
- 30.Riley D., Flint S. J. RNA-binding properties of a translational activator, the adenovirus L4 100-kilodalton protein. J. Virol. 1993;67:3586–3595. doi: 10.1128/jvi.67.6.3586-3595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayes B. W., Telling G. C., Myat M. M., Williams J. F., Flint S. J. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 1990;64:2732–2742. doi: 10.1128/jvi.64.6.2732-2742.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang J., Kao P. N., Herschman H. R. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J. Biol. Chem. 2000;275:19866–19876. doi: 10.1074/jbc.M000023200. [DOI] [PubMed] [Google Scholar]
- 33.Adam S. A., Dreyfuss G. Adenovirus proteins associated with mRNA and hnRNA in infected HeLa cells. J. Virol. 1987;61:3276–3283. doi: 10.1128/jvi.61.10.3276-3283.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuesta R., Xi Q., Schneider R. J. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 2000;19:3465–3474. doi: 10.1093/emboj/19.13.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yueh A., Schneider R. J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 36.Yueh A., Schneider R. J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]