Abstract
Phosphorylation of the human AR (androgen receptor) is directly correlated with the appearance of at least three AR isoforms on an SDS/polyacrylamide gel. However, it is still not clear to what extent phosphorylation is involved in the occurrence of isoforms, which sites are phosphorylated and what are the functions of these phosphosites. The human AR was expressed in COS-1 cells and AR phosphorylation was studied further by mutational analyses and by using reversed-phase HPLC and MS. The reversed-phase HPLC elution pattern of the three isoforms revealed that Ser-650 was phosphorylated constitutively. After de novo synthesis, only Ser-650 was phosphorylated in the smallest isoform of 110 kDa and both Ser-650 and Ser-94 were phosphorylated in the second isoform of 112 kDa. The hormone-induced 114 kDa isoform shows an overall increase in phosphorylation of all the isolated peptides. The activities of the Ser–Ala substitution mutant S650A (Ser-650→Ala) was found to be identical with wild-type AR activation in four different cell lines and three different functional analyses, e.g. transactivation, N- and C-terminal-domain interaction and co-activation by transcriptional intermediary factor 2. This was also found for mutants S94A and S515A with respect to transactivation. However, the S515A mutation, which should eliminate phosphorylation of the potential mitogen-activated protein kinase site, Ser-515, resulted in an unphosphorylated form of the peptide containing Ser-650. This suggests that Ser-515 can modulate phosphorylation at another site. The present study shows that the AR isoform pattern from AR de novo synthesis is directly linked to differential phosphorylation of a distinct set of sites. After mutagenesis of these sites, no major change in functional activity of the AR was observed.
Keywords: androgen receptor, isoform, MS, phosphorylation, reversed-phase HPLC, transactivation
Abbreviations: AR, androgen receptor; CHO cell line, Chinese-hamster ovary cell line; dcc, dextran-coated charcoal; DMEM, Dulbecco's modified Eagle's medium; FCS, fetal calf serum; MAPK, mitogen-activated protein kinase; MMTV, murine-mammary-tumour virus; RP, reversed phase; TIF2, transcriptional intermediary factor 2; WT, wild-type
INTRODUCTION
The AR (androgen receptor) is a ligand-dependent transcription factor belonging to the family of steroid hormone receptors. Similar to other members of the steroid hormone receptor family including the oestrogen receptor, progesterone receptor, glucocorticoid receptor and mineralocorticoid receptor, the AR becomes activated after ligand binding. This results in stabilization, a conformational change and tight nuclear binding of the receptor and, eventually, in a positive and/or negative transcription regulation of target genes.
Ligand binding is not the only regulatory event in the functions of steroid hormone receptors. Post-translational modification of steroid hormone receptors by proteins participating in other signal-transduction pathways plays a role in the regulation as well. Phosphorylation of oestrogen receptor α influences transactivation and association with the co-activators p160 and p300/CBP [CREB (cAMP-response-element-binding protein)-binding protein] [1–3] or with the co-repressors N-CoR (nuclear receptor co-repressor) and SMRT (silencing mediator for retinoic acid receptor and thyroid hormone receptor) [4]. Transactivation of the glucocorticoid receptor is also regulated by phosphorylation [5–7]. A hormone-dependent phosphosite in the progesterone receptors A and B plays a role in receptor degradation [8], transactivation [9] and nuclear export [10,11]. Thus phosphorylation of specific sites in steroid hormone receptors has been shown to play a role in various processes.
Post-translational modifications such as acetylation and sumoylation have been shown to influence the transactivation potential of the AR [12,13]. However, it is not clear whether phosphorylation has an effect on the properties and activity of the AR. It has been shown that the AR is a phosphoprotein [14,15] and extra phosphorylation of the AR is induced when cells are exposed to androgens, in addition to the so-called basal AR phosphorylation observed in the absence of androgens [15,16]. Phosphorylation occurs predominantly at serine residues [16,17], which are mainly located in the N-terminal domain [18]. Furthermore, phosphorylation is correlated with the three AR isoforms that appear on an SDS/polyacrylamide gel [19]. Within minutes after the start of de novo synthesis, the AR appears as a 110 kDa isoform, whereas generation of the second (112 kDa) isoform follows within 15 min as shown by radioactive methionine-labelling studies [20,21]. Only after hormone binding does the third (114 kDa) isoform appear [19]. The AR isoform pattern is correlated with AR phosphorylation as was shown in previous studies by using phosphatases. The dephosphorylation of AR by phosphatases resulted in the loss of one isoform either in the presence or absence of hormone [19]. This effect was also observed when AR phosphosites were mutated [19]. Furthermore, several phosphorylation sites have been identified. The first identified phosphosites, Ser-81, Ser-94 and Ser-650, were found by mutagenesis analyses in combination with SDS/PAGE [19,22]. Ser-308 was the first phosphosite identified by mutagenesis and MS [23]. Ser-16, Ser-81, Ser-94, Ser-256, Ser-308, Ser-424 and Ser-650 were all identified and confirmed as phosphosites by mutagenesis, peptide mapping and MS [16]. However, cell-free in vitro phosphorylation reaction studies on AR mutants also demonstrated Ser-213, Ser-515 and Ser-791 to be phosphosites [24–26].
It is still not clear to what extent phosphorylation is involved in the appearance of isoforms and which sites in the different isoforms are phosphorylated. In the present study, phosphorylation of the isoforms was further explored by studying the differential phosphorylation of the three AR isoforms. An attempt was made to identify all the phosphosites. Furthermore, the consequences of transcriptional activation of the identified and potential phosphosites were evaluated using functional assays, which tested the transactivation, N- and C-terminal-domain interaction and co-activation by TIF2 (transcriptional intermediary factor 2), and the functional assays were performed in COS-1 and CHO (Chinese-hamster ovary) cell lines and two prostate cancer cell lines (PC-3 and DU-145). In contrast with a previous study, which showed that Ser-94 was constitutively phosphorylated [16], our study showed that the Ser-650 is constitutively phosphorylated.
MATERIALS AND METHODS
Materials
Phosphate-free DMEM (Dulbecco's modified Eagle's medium) and goat anti-mouse agarose beads were purchased from Sigma–Aldrich (St. Louis, MO, U.S.A.). Media (GlutaMAX I-supplemented DMEM/F12 and RPMI 1640) and filtertop culture flasks were purchased from Invitrogen (Paisley, U.K.). Multiwell tissue culture plates were obtained from Nunc (Roskilde, Denmark). FCS (fetal calf serum) was obtained from Greiner (Frickenhausen, Germany) and the mixture of penicillin and streptomycin was from BioWhittaker (Walkersville, MD, U.S.A.). The oligonucleotides used in plasmid construction and sequencing were synthesized by Eurogentec (Liege, Belgium). FuGENE 6, Complete inhibitor EDTA-free, shrimp alkaline phosphatase and Rapid DNA Ligation kits were obtained from Roche (Basel, Switzerland) and Quik Change Site-Directed Mutagenesis kit was from Stratagene (La Jolla, CA, U.S.A.). Steady-Glo Luciferase assay system and sequencing-grade modified trypsin (specific activity, 16000 units/mg) were obtained from Promega (Madison, WI, U.S.A.). [32P]Pi was from Amersham Biosciences (Uppsala, Sweden). F39.4.1 is a mouse monoclonal antibody raised against amino acids 301–320 of the human AR [27] and SP197 is a rabbit polyclonal antibody [18]. The 10× Tris/glycine/SDS electrophoresis buffer and N,N,N′,N′-tetramethylethylenediamine were obtained from Bio-Rad Laboratories (Hercules, CA, U.S.A.). HPLC reagents were of sequencing grade and obtained from Merck (Darmstadt, Germany). Deltapack C18 column was purchased from Waters and C-18 Z-tips were from Millipore (Milford, MA, U.S.A.). NEN Life Science Products (Boston, MA, U.S.A.) supplied R1881 (methyltrienolone). MMTV-Luc reporter plasmid (where MMTV stands for murine-mammary-tumour virus) was kindly provided by Dr R. Dijkema (Organon, Oss, The Netherlands) and has been described previously [28]. SpeedVac concentrator was obtained from Thermo Savant (Division of Thermo Electron Corporation, Waltham, MA, U.S.A.).
Plasmid construction
Plasmid construction was performed according to standard methods [29] and, where indicated, the plasmids were rendered blunt-ended with Klenow. Constructs including a PCR-amplification step for preparation were sequenced to verify the correct reading frame and the absence of random mutations. All the AR amino acid numbers used in the present study are based on the National Center for Biotechnology Information accession number AAA51729, which refers to the AR consisting of 919 amino acids [30]. The mutants AR S650A (Ser-650→Ala) and AR S515A were constructed by site-directed mutagenesis using PCR DNA amplification techniques. The following sense oligos were used for introducing the substitution with a Quik Change: AR S650A, 5′-CCAGCACCACCGCCCCCACTGAG-3′; and AR S515A, 5′-CCTATCCCGCTCCCACTTGT-3′. The mutations are indicated in italics and underlined. AR104 was described previously as pSVAR-104, which is the AR construct containing sequences encoding the C-terminal amino acids 537–919 [31]. The N-terminal construct, pSVAR(TAD1–494), consists of the AR amino acids 1–503 [32]. AR104/S650A was prepared by the digestion of BHEXARS650A with SacI and ligating the fragment into AR104 by using the Rapid DNA Ligation kit. TIF2 is described as a co-activator in [33,34].
COS-1 cell culture, transfection, metabolic labelling with [32P]Pi and immunoprecipitation
COS-1 cells were cultured in DMEM/F12 supplemented with GlutaMAX I and 5% (v/v) FCS treated with dextran-coated charcoal (dcc-FCS). For steady-state labelling, 1.5×106 COS-1 cells were grown overnight in an 80 cm2 flask with 8 ml of DMEM/F12, followed by transfection with the following mix: 4 μg of the indicated plasmid with 12 μl of FuGENE 6 transfection reagent in 200 μl of serum- and antibiotics-free DMEM/F12. Transfections were performed according to the manufacturer's instructions for FuGENE 6. After 30 h, cells were washed twice with 0.9% (w/v) NaCl and incubated in phosphate-free DMEM, supplemented with 50 mM Hepes buffer and 5% dcc-FCS, which had been dialysed for 24 h against 0.9% NaCl. Subsequently, cells were incubated with R1881 as indicated and [32P]Pi (0.333 mCi/ml) for 16 h. Cells were harvested and lysed at 4 °C in immunoprecipitation buffer A [40 mM Tris/HCl (pH 7.4), 5 mM EDTA (pH 8.0), 10% (v/v) glycerol, 10 mM sodium phosphate, 10 mM sodium molybdate, 50 mM sodium fluoride, 0.5 mM sodium orthovanadate, 0.6 mM PMSF, 0.5 mM Bacitracin, Complete inhibitor EDTA-free and 10 mM dithiothreitol], supplemented with 1% (v/v) Triton X-100, 0.5% (w/v) deoxycholic acid and 0.08% (w/v) SDS. Subsequently, the lysate was centrifuged at 100000 g for 30 min at 4 °C. The supernatant was then incubated at 4 °C with the antibody F39.4.1, which was linked to goat anti-mouse agarose. After 2 h, the agarose beads were washed as follows: three times with buffer A supplemented with 1% Triton X-100, 0.5% deoxycholic acid and 0.08% SDS, three times with buffer A supplemented with 0.2% Triton X-100 and 0.4 M NaCl and three times with buffer A without any additions. The immunoprecipitated AR was separated by SDS/PAGE (7% gel). After fixing the gel in 10% (v/v) acetic acid and 50% (v/v) methanol, the gel was subjected to Coomassie Blue staining and destaining. Subsequently, the AR band was excised from the gel and digested with sequencing-grade modified trypsin.
In-gel digestion and RP (reversed-phase) HPLC analysis
The excised AR spot was in-gel-digested as described by Shevchenko et al. [35] with 20 units of sequencing-grade modified trypsin for 16 h at 37 °C. The amount of trypsin necessary to secure full digestion of the higher amount of AR protein in the presence of hormone was verified by varying the amount. The peptides were extracted as described by Shevchenko et al. [35] and dried in a SpeedVac for 1.5 h. The peptides were dissolved in 0.1% (v/v) trifluoroacetic acid. Then, 25 μl of this solution was applied to a 2 mm×150 mm Waters Deltapack C18 column. The flow was set to 0.18 ml/min and fractions were collected every 1.5 min until a gradient of 18% (v/v) acetonitrile in 0.1% trifluoroacetic acid was reached.
Peptide gel
A 40% (w/v) acrylamide alkaline peptide gel was cast and run as described by West et al. [36].
Characterization of the HPLC fractions with MS
AR tryptic peptides were separated by RP-HPLC. The fractions with retention times corresponding to 32P-labelled tryptic phosphopeptides were collected and, after drying in a SpeedVac, they were dissolved in 20 μl of 50% acetonitrile. With electrospray ionization MS and MS/MS, data were collected from the individual fractions using a Q-TOF (Micromass, Wythenshaw, Manchester, U.K.). The peptides were directly infused in the Q-TOF with a nanospray needle (Micromass) containing a 3 μl sample plus 0.3 μl of 10% (v/v) formic acid. Low-energy collision-induced dissociation of selected precursor ions was used to obtain fragmentation spectra. These were deconvoluted (Masslynx software; Micromass) and used to identify the corresponding tryptic peptides, including modifications, of the AR.
CHO, PC-3, DU-145 and COS-1 cell culture, transfection and luciferase assay
CHO and COS-1 cells were maintained in DMEM/F12 culture medium, supplemented with 5% dcc-FCS. PC-3 and DU-145 were cultured in RPMI 1640 medium, supplemented with 5% FCS. For transcription activation experiments, the cells were plated in 24-well plates at a density of 2×104 cells/well (1.9 cm2) in 500 μl of either DMEM/F12 or RPMI 1640 and grown overnight. Cells were transfected using 100 μl of either serum- and antibiotics-free DMEM/F12 or RPMI 1640 containing FuGENE 6 (FuGENE/DNA ratio of 3:1) with AR expression plasmids and, where indicated, with TIF2 reporter plasmids (50 ng/well) and pTZ19 carrier plasmid to a total DNA concentration of 250 ng/well. After 5 h, R1881 was added, followed by an overnight incubation; at the end of this incubation, the cells were harvested for a luciferase assay. Then, 50 μl of lysis buffer [25 mM Tris/phosphate (pH 7.8), 15% glycerol, 1% Triton X-100, 8 mM MgCl and 1 mM dithiothreitol] was added to the cells. After incubation for 10 min, 25 μl of the supernatants were transferred to white non-transparent 96-well assay plates, and 25 μl of 16 mg Steady-Glo Luciferase assay substrate per ml of Steady-Glo luciferase assay buffer was added. Luciferase activity was measured with a TopCount luminometer (Packard Bioscience, PerkinElmer Life Sciences, Zaventem, Belgium).
Western blotting
COS-1 cells were plated at a density of 1×106 cells/80 cm2 flask and transfected with 4 μg of AR expression plasmid and 12 μl of FuGENE. After an overnight incubation with hormones, the cells were washed once with PBS, and immunoprecipitation buffer (see the COS-1 cell culture subsection), supplemented with protease inhibitors, was added. Lysates were centrifuged for 10 min at 400000 g and AR was immunoprecipitated with the monoclonal antibody F39.4.1. Next, samples were subjected to SDS/PAGE and blotted on to a nitrocellulose membrane. AR was immunoblotted with the AR polyclonal antibody SP197 and visualized by chemiluminescence detection or by using an alkaline phosphatase-conjugated secondary antibody.
RESULTS
AR isoform expression
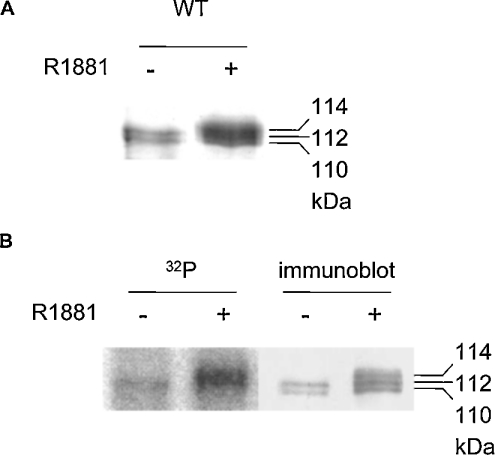
To verify the AR isoform pattern on an SDS/polyacrylamide gel, WT (wild-type) human AR was expressed by transient transfection into COS-1 cells and metabolically labelled with [32P]Pi either in the absence or presence of the synthetic androgen R1881. The same amount of total lysate of the two conditions was used for AR immunoprecipitation. After immunopurification, the gel was stained with Coomassie Blue. In the absence of R1881, the two isoforms (110 and 112 kDa) were clearly visible and the presence of R1881 resulted in the appearance of the third isoform of 114 kDa (Figure 1A).
Figure 1. AR isoforms on a Coomassie Blue-stained SDS/polyacrylamide gel.
WT AR was expressed in COS-1 cells and labelled with [32P]Pi for 16 h in the absence or presence of 10 nM R1881 for 16 h. The same amount of total lysate of the two conditions was used for AR immunoprecipitation with the monoclonal antibody F39.4.1. The immunoprecipitated AR was separated by SDS/PAGE (7% gel) and Coomassie Blue-stained (A) or blotted and immunostained with the polyclonal antibody SP197 (B). The corresponding autoradiogram is also shown in (B).
Lysates obtained by the same procedure as the previous experiment were also immunoprecipitated and blotted. The immunoblot showed the same isoform pattern (Figure 1B) as the Coomassie Blue-stained gel. An autoradiogram of the corresponding immunoblot showed an upshift of a phosphorylated band and an increase in phosphorylation (Figure 1B). This increase is partly due to stabilization of the AR. These results confirm that the AR is phosphorylated in the absence of hormone and that phosphorylation of the AR is increased in the presence of hormone [16]. Furthermore, this suggests that the upshift of a phosphorylated band corresponds to the 114 kDa isoform.
Changes in AR phosphorylation induced by R1881
To investigate whether androgenic activation increases the phosphorylation of existing phosphosites and/or induces phosphorylation of new sites, the AR was expressed by transient transfection into COS-1 cells and metabolically labelled with [32P]Pi either in the absence or presence of R1881. After immunopurification and subsequent digestion with trypsin, the resulting peptides were separated on an RP HPLC column. The RP HPLC elution pattern of AR in absence of R1881 showed that several fractions contained 32P-labelled peptides (Figure 2A, fractions A, B, D and E). After stimulating the cells with 10 nM R1881, an increase in phosphorylation of the peptides in fractions A, B, D and E was observed (Figure 2B). In addition, the relative phosphorylation level of two peptides in fractions C and F was slightly increased (Figure 2B). It is important to note that the overall phosphorylation pattern was highly reproducible in ten independent experiments. These results are in agreement with a previous report that hormone binding results in increased phosphorylation of existing phosphorylated sites [16].
Figure 2. R1881-induced changes in AR phosphorylation pattern.
AR was expressed in COS-1 cells and labelled with [32P]Pi for 16 h in the absence (A) or presence (B) of 10 nM R1881. The same amount of total lysate of the two conditions was used for AR immunoprecipitation (with anti-AR monoclonal antibody F39.4.1). The precipitated AR was digested with trypsin and the peptides obtained were separated on an RP HPLC C18 column. HPLC fractions were collected and the amount of [32P]Pi incorporated was determined. The different radioactive fractions are indicated with A–F. Note that the ordinates have a different scale.
Differential phosphorylation of AR isoforms
To study differential phosphorylation of the three isoforms of 110, 112 and 114 kDa in more detail, COS-1 cells were transfected with AR and stimulated for 16 h with R1881 and labelled with [32P]Pi. Each 32P-labelled AR isoform was isolated separately from an SDS/polyacrylamide gel. The phosphorylation pattern of tryptic fragments of the individual isoforms revealed that the 110 kDa isoform is predominantly phosphorylated on the peptide in fraction D (Figure 3A). The phosphorylation level of fraction D in the 112 kDa isoform was similar to that in the 110 kDa isoform (Figures 3A and 3B) and slightly increased further in the 114 kDa isoform (Figure 3C). Since fraction D contains the peptide with the phosphorylated Ser-650 (see below), these results indicate that Ser-650 is constitutively phosphorylated.
Figure 3. Differential phosphorylation of AR isoforms.
AR was expressed in COS-1 cells and labelled with [32P]Pi for 16 h in the presence of 10 nM R1881. AR was immunoprecipitated (with anti-AR monoclonal antibody F39.4.1) and the isoforms of 110 kDa (A), 112 kDa (B) and 114 kDa (C) were separately cut out from the same lane on an SDS/7% polyacrylamide gel followed by trypsin digestion. The peptides obtained were separated on an RP HPLC column. HPLC fractions were collected and the amount of [32P]Pi incorporated was determined. The different radioactive fractions are indicated with A–F.
The phosphorylation level of the different RP HPLC fractions for each isoform was found to be different and to get an impression of quantitative changes, the fold increase has been calculated. First, the phosphorylation level of each peptide is based on the sum of the radioactivities in three consecutive fractions. Since phosphorylation of fraction D was shown to be more or less constant in the different isoforms, the level of phosphorylation in the other fractions was calculated relative to the level of phosphorylation in fraction D. Furthermore, the change in ratios for the 112 and 114 kDa isoforms were calculated relative to the ratios for the 110 kDa isoform, resulting in fold increase (Table 1). The fold increase of the fractions A, B, C, E and F obtained from the 110 kDa isoform were low. For the 112 kDa isoform, the fold increase of fractions A and B was 2–3-fold higher (Table 1). The fold increase of fractions C, E and F was the same in the 112 kDa isoform compared with the 110 kDa isoform.
Table 1. Phosphorylation ratios of AR peptides.
The phosphorylation ratio of different fractions is calculated relative to the intensity of phosphorylation of fraction D (1.0, see also text).
| Phosphorylation | |||||
|---|---|---|---|---|---|
| HPLC fractions | Fraction X/D (110 kDa isoform) | Fraction X/D (112 kDa isoform) | Fold increase (relative to 110 kDa) | Fraction X/D (114 kDa isoform) | Fold increase (relative to 110 kDa) |
| A | 0.3 | 0.8 | 2.9 | 1.4 | 5.1 |
| B | 0.2 | 0.4 | 1.9 | 0.9 | 4.3 |
| C | 0.1 | 0.1 | 0.7 | 0.2 | 1.3 |
| D | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| E | 0.3 | 0.3 | 1.0 | 0.4 | 1.2 |
| F | 0.1 | 0.1 | 1.1 | 0.2 | 2.9 |
Moreover, analysis of the 114 kDa isoform showed that, in the presence of R1881, the fold increase of fractions C and E was rather small, but the fold increase of phosphorylation in fractions A, B and F were higher (Table 1). These results confirm that a correlation exists between phosphorylation status and the SDS/PAGE migration pattern for the three AR isoforms [19]. In conclusion, increase in phosphorylation is correlated with a decreased migration rate.
Phosphopeptide analysis
To characterize the HPLC fractions in more detail, the most intensely phosphorylated fractions (A, B, D and E) were subjected to further analysis on a 40% acrylamide alkaline peptide gel. Peptide analysis revealed that fractions A, D and E each contained a single phosphorylated peptide and that each peptide was different from the others (Figure 4). However, fraction B contained two phosphorylated peptides. The peptide in fraction B, which migrated into the gel as far as the peptide in fraction A, is most probably identical with the phosphorylated peptide from fraction A and is present in fraction B due to incomplete resolution during HPLC separation. These data show that at least four different peptides derived from the AR are phosphorylated.
Figure 4. Separation of individual HPLC fractions on a 40% acrylamide alkaline peptide gel.
Peptides from HPLC fractions were separated on a 40% acrylamide alkaline peptide gel. Note that the separation is according to size and charge. Phosphopeptides were visualized by using a PhosphorImager (Molecular Dynamics). The different radioactive fractions are indicated with A, B, D and E.
Peptide analysis by MS
To identify the phosphorylated amino acid residues, MS analysis was used to characterize first the phosphorylated peptides in the HPLC fractions. Out of the six HPLC fractions collected (fractions A–F), five contained tryptic peptides from the AR (Table 2). The identified peptides in fractions B, D and E were phosphorylated. The theoretical mass of these peptides was increased by the mass of one phosphate group (79.9799 Da) and, moreover, MS/MS data confirmed the presence of this modification, probably on a serine residue. The MS/MS data from fraction D could not discriminate between two possible serine phosphosites (Ser-647 and Ser-650). Both of the identified tryptic AR peptides in fractions A and F contained a serine residue, but were not phosphorylated. The results are summarized in Table 2 together with information on possible putative phosphosites. A small number of the fractions displayed a variable low level of phosphorylation, most probably due to contaminations, and were excluded from further analyses.
Table 2. MS data of phosphorylated peptides in RP HPLC fractions.
| Fraction | Measured mass | Tryptic peptide fragment* | AR peptide | Putative phosphosite | Prediction† |
|---|---|---|---|---|---|
| A | 962.4 | T70–T71 Cys-852 with acrylamide adduct | 847–854 | Ser-851 | 0.962 |
| B | 2056.8 | T7–T8 plus phosphate probably on Ser-94 | 84–100 | Ser-94 | 0.572 |
| C | n.d.‡ | ||||
| D | 2232.0 | T50 plus phosphate probably on Ser-647 or Ser-650 | 639–658 | Ser-647 | 0.760 |
| Ser-650 | 0.997 | ||||
| E | 1226.6 | T28 plus phosphate probably on Ser-515; Cys-518 with carbamidomethyl | 511–520 | Ser-515 | 0.967 |
| F | 1554.7 | T15 | 221–235 | § |
* Peptide identification is based on comparison of the detected mass and the corresponding MS/MS information of the measured peptides with all possible tryptic fragments (T1–T79).
† NetPhos 2.0 [40] was used to search for possible phosphorylation sites. A prediction of 0.5 and higher was considered as representing a potential phosphorylation site.
‡ n.d., not determined.
§ No potential phospho-serine residue was present in the peptide.
Phosphosite identification by site-directed mutagenesis
The peptide in fraction D (amino acid residues 639–658) contains two potential phosphorylation sites, Ser-647 and Ser-650 (Table 2). Substitution of Ser-650 with an alanine residue resulted in the disappearance of peptide phosphorylation in fraction D (Figure 5A). In contrast, substitution of Ser-647 to Ala-647 did not result in any change in phosphorylation (results not shown). This substantiates the observation that fraction D contains the peptide consisting of amino acids 639–658 and shows that Ser-650, but not Ser-647, is a phosphorylation site. Another peptide identified by MS and consisting of amino acid residues 84–100 (fraction B) contains the potential phosphosite Ser-94 (Table 2). Substitution of Ser-94 to Ala-94 resulted in a 40% loss of radioactive phosphate incorporation in both fractions A and B (Figure 5B and Table 3). A third phosphorylated peptide identified in these experiments is the peptide from fraction E consisting of amino acid residues 511–520 and containing the potential phosphosite Ser-515. Remarkably, substitution of Ser-515 to Ala-515 did not result in disappearance of radioactivity from fraction E. In contrast, fraction D, which contained the peptide with phosphosite Ser-650, was no longer radioactive after the Ser-515 to Ala-515 substitution (Figure 5C). This result indicates that changes in Ser-515 might exert an influence on the phosphorylation status of Ser-650. In fraction A, the peptide consisting of amino acids 847–854 has the predicted phosphosite Ser-851. Substitution of Ser-851 with an alanine residue did not result in a change in the phosphorylation pattern (results not shown). This indicates that the identified peptide consisting of amino acids 847–854 present in fraction A does not contain the predicted Ser-851 phosphosite. No phosphosites were predicted for the identified peptide present in fraction F and no peptides could be detected in fraction C.
Figure 5. AR phosphorylation on Ser-650 and Ser-94 and the phosphorylation influenced by Ser-515.
The AR mutant S650A (A), S94A (B) or S515A (C) was expressed in COS-1 cells and labelled with [32P]Pi for 16 h in the presence of 10 nM R1881. The AR mutant was immunoprecipitated (with anti-AR monoclonal antibody F39.4.1) and digested with trypsin. The peptides obtained were separated on an RP HPLC C18 column. HPLC fractions were collected and the amount of [32P]Pi incorporated was determined.
Table 3. Phosphorylation of S94A mutant.
The phosphorylation of peptides in fractions A and B from the WT AR (Figure 2B) and the S94A mutant (Figure 5B) were normalized to their corresponding fractions D. Percentage change in phosphorylation between the mutant and WT for fractions A and B was calculated from the ratios.
| Phosphorylation | |||
|---|---|---|---|
| Ratio | WT AR | Mutant S94A | Change in phosphorylation (%) |
| A/D | 0.65 | 0.38 | −42 |
| B/D | 0.42 | 0.23 | −44 |
Phosphorylation and AR transcriptional activity
Since the phosphorylation level of the AR was changed in the presence of R1881, AR phosphorylation might regulate the functional activity of the AR. The transactivation activity of AR mutants S515A, S650A and S94A was tested using an MMTV-Luciferase reporter assay in CHO cells. Substitution of Ser-650 to Ala-650 did not result in a change in transactivation of the MMTV-Luc reporter gene when compared with the WT AR (Figure 6A). The same neutral effect was found for the S515A and S94A mutants (results not shown). To exclude the possibility that CHO cells exert AR phosphorylation in a peculiar cell-type-specific manner, the same experiment was performed in COS-1 as well as PC-3 and DU-145 cells (both prostate cancer cell lines). No differences in AR transactivation activity for the three AR mutants could be detected in any of these three cell lines (results not shown). These results indicate that loss of AR phosphorylation on Ser-650, Ser-515 and Ser-94 does not have an effect on AR transactivation activity.
Figure 6. Functional studies on WT AR and the AR mutant S650A.
CHO cells were transfected with 50 ng of reporter construct MMTV-Luc, different amounts of DNA plasmids from either the AR or AR mutant S650A (A), C-terminal construct AR104 or C-terminal mutant construct AR104/S650A together with 10 ng of N-terminal construct pSVAR(TAD1–494) (B) or 100 ng of TIF2 cDNA (C). The cells were harvested 16 h after treatment with ethanol or 1 nM R1881. Fold induction is shown at the top of each bar and represents the ratio of activity determined after incubation in the presence or absence of R1881.
The AR protein can undergo conformational changes resulting in intra-molecular interaction between the N- and C-terminal domains [32,37] and this N- and C-terminal interaction can be influenced by several mutations in the AR, resulting in altered transcriptional activation activity of the AR [38,39]. To examine whether phosphorylation exerts an influence on this interaction, the C-terminal-domain construct containing the S650A mutant and the N-terminal-domain construct were co-transfected in CHO cells. As shown in Figure 6(B), the S650A mutant displayed a similar functional N- and C-terminal interaction as the WT AR C-terminal construct.
To investigate whether phosphorylation has an influence on TIF2 co-activation, CHO cells were co-transfected with constructs of AR C-terminal domain with or without the S650A substitution and TIF2. The mutant showed a similar functional interaction with TIF2 as the WT AR C-terminal domain (Figure 6C).
AR isoforms on SDS/PAGE
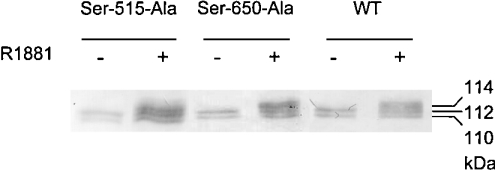
It has been shown that substitution of the phosphosite Ser-94 by an alanine residue results in a loss of isoforms both in the absence and presence of hormone [19]. To determine whether there is a change in isoform pattern of the AR mutants S650A and S515A, these mutants were expressed in COS-1 cells and immunoprecipitated. The precipitated AR mutants were separated by SDS/PAGE. The isoform patterns of both mutants were similar to the WT AR (Figure 7). Both mutants expressed two isoforms in the absence of hormone and three isoforms in the presence of R1881. This indicates that Ser-515 and Ser-650 are not essential for the migration of 112 and 114 kDa isoforms in the absence or presence of R1881. Furthermore, these results demonstrate that loss of phosphorylation on certain sites does not always result in a change in isoform pattern.
Figure 7. Immunoblot of S650A and S515A AR mutant isoforms.
AR S650A and S515A mutants were expressed in COS-1 cells in the absence or presence of 10 nM R1881 for 16 h. The AR mutants were immunoprecipitated with the monoclonal antibody F39.4.1, separated by SDS/PAGE (7% gel), blotted and immunostained with the polyclonal antibody SP197.
DISCUSSION
The present study shows that the AR isoform pattern after AR de novo synthesis is directly linked to differential phosphorylation. It appeared that, after synthesis, the AR 110 kDa isoform is predominantly phosphorylated at Ser-650, and there is a higher phosphorylation level for other existing sites in the 112 and 114 kDa isoforms. These results reveal that Ser-650 is constitutively phosphorylated. Moreover, loss of phosphorylation on certain sites does not always result in a change in isoform pattern. Functional analysis of the identified phosphosites Ser-94, Ser-650 and Ser-515 revealed that substitution of these sites with alanine does not influence AR function.
Recently, several AR phosphosites were identified after transient expression in COS-1 cells, by sequencing AR peptides using tandem MS and Edmann degradation [16]. Our approach was different, i.e. RP HPLC was used instead of two-dimensional thin-layer electrophoresis and ascending chromatography. With the purified fractions of the tryptic peptides containing possible phosphosites, tandem MS can be a good tool for identification. However, it is not always possible to identify the exact position of the phosphate group. Fragmentation data may lack the specific fragment ions containing the phosphate group owing to the individual fragmentation behaviour of the peptide or by loss of their positive charges during the collision-induced dissociation. Although all fractions contained radioactivity, only three phosphopeptides could be identified. A possible explanation is that the concentration of the peptides containing the phosphate group is too low or the peptides containing the phosphate group are not easily ionized with electrospray. MS is a valuable tool, but additional information, such as site-directed mutagenesis in this case, is necessary.
The autoradiogram and the HPLC elution pattern revealed that the phosphorylation was increased approx. ten times. A previous study showed that hormone binding results in an increased phosphorylation of existing phosphorylated sites by approx. 1.8 times [15]. This indicates that the apparent extra phosphorylation of approx. five times results from stabilization by hormone binding.
The phosphorylation pattern of the isoforms revealed that Ser-650 is already phosphorylated in the 110 kDa isoform and Ser-94 in the 112 kDa isoform. However, these phosphorylation patterns were studied in the presence of R1881, which raises the question whether Ser-650 is phosphorylated in the 110 kDa isoform and Ser-94 in the 112 kDa isoform in the absence of R1881 as well. However, the RP HPLC elution pattern of the AR in the absence of R1881 also showed the presence of phosphorylation in fractions A, B and D (Ser-94 and Ser-650; Figure 2A). It is therefore probable that the phosphorylation pattern of the 110 and 112 kDa isoforms in absence of R1881 is the same as that in the presence of R1881. Moreover, this suggests that phosphorylation of fraction D in the 110 kDa isoform and phosphorylation of fractions A and B in the 112 kDa isoform are not hormone-induced.
Surprisingly, site-directed mutagenesis of the Ser-515 to an alanine residue revealed dephosphorylation of a totally different site, Ser-650. This type of distal influence on phosphorylation caused by the substitution of another site has been observed by others [16]. Gioeli et al. [16] suggested that substitution of Ser-424 with an alanine residue resulted either in the mobility shift of a phosphorylated peptide or in phosphorylation of new sites. In the present study, the dephosphorylation of Ser-650 induced by the S515A substitution might be due to a conformational change, which resulted in a surface more accessible for phosphatases or less accessible to kinases. Interestingly, both sites are flanking the DNA-binding domain and are located in flexible regions of the AR protein.
The reason why substitution of Ser-515 with an alanine residue does not result in the disappearance of phosphorylation in fraction E might be that other serine residues in this peptide are phosphorylated as well. Although these serine sites are not predicted as potential phosphosites by NetPhos [40], it predicts that Tyr-513 is a phosphorylation consensus site [40]; however, two-dimensional phosphopeptide mapping studies have shown that only serine residues in the AR are phosphorylated [16,17]. Substitution of Ser-94 with an alanine residue resulted in a partial decrease in phosphorylation level (40%) in fractions A and B. This suggests that one phosphopeptide of the two in fraction B as seen on the peptide gel was identical with a peptide with the same mass and charge in fraction A. In addition, this also suggests that there might be another peptide present in both fractions. Alternatively, the presence of a partially digested peptide containing Ser-94 is excluded. The partially digested peptide would consist of amino acids 41–99 and contains the identified phosphosite Ser-81 [16]. However, the peptide will be very large and, therefore, difficult to elute from an SDS/polyacrylamide gel. Furthermore, the presence of two phosphopeptides only in fraction B and not in fraction A is in favour of a complete digestion.
Substitution of Ser-851 with an alanine residue did not result in the disappearance of radioactive phosphate in fraction A. Partial digestion of AR, resulting in a peptide containing Ser-851 and another potential phosphorylation site, is unlikely, since this would result in a very large peptide that cannot be eluted from a gel. Substitution of Ser-851 did not result in a change of phosphorylation in fraction A or other fractions (results not shown), which indicates that Ser-851 is not a phosphosite at all. The presence of another phosphopeptide cannot be excluded.
A change in AR phosphorylation appears to have no prominent function in AR transactivation. Similar findings were reported by Gioeli et al. [16] by using the PSA-Luc reporter construct in CV1 cells. In contrast, Zhou et al. [22] showed a decreased transactivation of the mutant S650A of 10–30% on the MMTV-Luc reporter also in CV1 cells.
It appears that phosphorylation does not play a major role in hormone-induced AR N- and C-terminal interaction or TIF2 co-activation, although it cannot be excluded that the cellular context as well as the reporter construct could influence this activity and that other so far unidentified phosphosites are involved [41].
After AR de novo synthesis, the 110 kDa isoform became immediately and predominantly phosphorylated on Ser-650. The 112 kDa isoform displayed an additional phosphorylation of Ser-94 and another peptide in fractions A and B only. The relationship between phosphorylation of Ser-94 and the appearance of the 112 kDa isoform corresponds to an immunoblot study in which S94A caused disappearance of the 112 kDa isoform in the absence of hormone and disappearance of the 114 kDa isoform in the presence of hormone [19].
In contrast with S94A AR mutant, substitution of Ser-650 with an alanine residue does not influence the isoform pattern on SDS/PAGE, which is understandable because Ser-650 phosphorylation occurs already in the 110 kDa isoform and is unchanged in the 112 kDa isoform and slightly changed in the 114 kDa isoform. The Ser-650 site represents basal phosphorylation of the AR and only changes in phosphorylation of other sites could perhaps contribute to the appearance of the isoforms. Substitution of the phosphosite Ser-81 to a glycine residue resulted in the loss of the largest isoform irrespective of the presence of ligand [19]. Similar results were obtained for the double-mutant S81/94A [22]. However, substitution of Ser-81 with an alanine residue resulted in the loss of one isoform only in the presence of hormone (results not shown). The RP HPLC elution pattern of the three isoforms illustrated the correlation between the phosphorylation status of sites other than Ser-650 and the migration pattern of the three AR isoforms during SDS/PAGE. Phosphorylation does not necessarily contribute to the appearance of isoform as shown by the mutant S650A.
The appearance of the 114 kDa isoform induced by R1881 is directly linked to an overall increase in phosphorylation of several sites as compared with the 110 and 112 kDa isoforms. This overall increase in phosphorylation was shown previously by van Laar et al. [15] and Gioeli et al. [16]. However, newly phosphorylated sites could not be identified. It is quite probable that these sites are important for the hormone-regulated transactivation of the AR, because their phosphorylation is linked to DNA-binding and transcription activation. Furthermore, these phosphorylation sites could be target sites for AR activation via crosstalk with other signal transduction pathways as well. Thus identifying the kinase(s) involved in the phosphorylation of these sites and the possible signalling pathway will be useful to elucidate the mechanisms of ligand-independent activation of the AR.
There are several kinases predicted to be involved in the phosphorylation of AR. The identified phosphosites Ser-94 and Ser-650 are so-called Ser-Pro sites, which can be phosphorylated by serine-proline-directed kinases (Ser/Thr-Pro), MAPK (mitogen-activated protein kinase) and cyclin-dependent kinases such as Cdc2 and Cdks. In addition, Ser-650 is a specific consensus site for casein kinases 1 and 2. There are conflicting data concerning which kinases phosphorylate the AR. It has been shown that protein kinase C [16,28], MAPK and AKT kinase [16] have no influence on the phosphorylation of the AR. However, other in vitro kinase studies showed that AKT [24,25] is capable of phosphorylating the AR. Furthermore, MAPK was also a candidate protein kinase of the AR [26]. However, in that particular study, the phosphorylation status of the MAPK site S515A AR mutant was not investigated [26].
Although basal phosphorylation of the AR does not seem to have a function, it is quite probable that a hormone-regulated phosphorylation of the AR is associated with transcriptional activation. Identification of the hormone-induced AR phosphorylation sites and subsequent elucidation of their possible function by mutational analysis in vivo could contribute significantly to our understanding of the mechanism of androgen action. Furthermore, it could reveal new targets for intervention in androgen action in prostate cancer.
Acknowledgments
This work was partially supported by the Dutch Cancer Society (NKB/KWF; grant no. EUR 98-1776) and the European Commission (QLRT-2000-00602). We thank the Nijbakker-Morra Foundation for providing financial support to purchase the SpeedVac equipment. The Q-TOF mass spectrometer was largely funded by grants from the Council for Medical Sciences of The Netherlands Organization for Scientific Research (NWO). We also thank Dr H. Gronemeyer and Dr P. Chambon of the Institut de Genetique et de Biologie Moleculaire et Cellulaire (CNRS/INSERM/ULP, College de France, Illkirch, France) for providing the TIF2 construct and Dr R. Dijkema for the MMTV-Luc construct.
References
- 1.Lannigan D. A. Estrogen receptor phosphorylation. Steroids. 2003;68:1–9. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- 2.Endoh H., Maruyama K., Masuhiro Y., Kobayashi Y., Goto M., Tai H., Yanagisawa J., Metzger D., Hashimoto S., Kato S. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol. Cell. Biol. 1999;19:5363–5372. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Dutertre M., Smith C. L. Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen receptor-alpha: regulation by phosphorylation sites in the A/B region depends on other receptor domains. Mol. Endocrinol. 2003;17:1296–1314. doi: 10.1210/me.2001-0316. [DOI] [PubMed] [Google Scholar]
- 4.Lavinsky R. M., Jepsen K., Heinzel T., Torchia J., Mullen T. M., Schiff R., Del-Rio A. L., Ricote M., Ngo S., Gemsch J., et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodwell J. E., Webster J. C., Jewell C. M., Cidlowski J. A., Hu J. M., Munck A. Glucocorticoid receptor phosphorylation: overview, function and cell cycle-dependence. J. Steroid Biochem. Mol. Biol. 1998;65:91–99. doi: 10.1016/s0960-0760(97)00185-4. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z., Garabedian M. J. Modulation of glucocorticoid receptor transcriptional activation, phosphorylation, and growth inhibition by p27Kip1. J. Biol. Chem. 2003;278:50897–50901. doi: 10.1074/jbc.M310297200. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z., Frederick J., Garabedian M. J. Deciphering the phosphorylation ‘code’ of the glucocorticoid receptor in vivo. J. Biol. Chem. 2002;277:26573–26580. doi: 10.1074/jbc.M110530200. [DOI] [PubMed] [Google Scholar]
- 8.Lange C. A., Shen T., Horwitz K. B. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1032–1037. doi: 10.1073/pnas.97.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen T., Horwitz K. B., Lange C. A. Transcriptional hyperactivity of human progesterone receptors is coupled to their ligand-dependent down-regulation by mitogen-activated protein kinase-dependent phosphorylation of serine 294. Mol. Cell. Biol. 2001;21:6122–6131. doi: 10.1128/MCB.21.18.6122-6131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu M., Olsen A., Faivre E., Horwitz K. B., Lange C. A. Mitogen-activated protein kinase regulates nuclear association of human progesterone receptors. Mol. Endocrinol. 2003;17:628–642. doi: 10.1210/me.2002-0378. [DOI] [PubMed] [Google Scholar]
- 11.Lange C. A. Making sense of cross-talk between steroid hormone receptors and intracellular signaling pathways: who will have the last word? Mol. Endocrinol. 2004;18:269–278. doi: 10.1210/me.2003-0331. [DOI] [PubMed] [Google Scholar]
- 12.Fu M., Wang C., Wang J., Zhang X., Sakamaki T., Yeung Y. G., Chang C., Hopp T., Fuqua S. A., Jaffray E., et al. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol. Cell. Biol. 2002;22:3373–3388. doi: 10.1128/MCB.22.10.3373-3388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poukka H., Karvonen U., Janne O. A., Palvimo J. J. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc. Natl. Acad. Sci. U.S.A. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Laar J. H., Bolt-de Vries J., Zegers N. D., Trapman J., Brinkmann A. O. Androgen receptor heterogeneity and phosphorylation in human LNCaP cells. Biochem. Biophys. Res. Commun. 1990;166:193–200. doi: 10.1016/0006-291x(90)91930-q. [DOI] [PubMed] [Google Scholar]
- 15.van Laar J. H., Berrevoets C. A., Trapman J., Zegers N. D., Brinkmann A. O. Hormone-dependent androgen receptor phosphorylation is accompanied by receptor transformation in human lymph node carcinoma of the prostate cells. J. Biol. Chem. 1991;266:3734–3738. [PubMed] [Google Scholar]
- 16.Gioeli D., Ficarro S. B., Kwiek J. J., Aaronson D., Hancock M., Catling A. D., White F. M., Christian R. E., Settlage R. E., Shabanowitz J., et al. Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. J. Biol. Chem. 2002;277:29304–29314. doi: 10.1074/jbc.M204131200. [DOI] [PubMed] [Google Scholar]
- 17.Kuiper G. G., Brinkmann A. O. Phosphotryptic peptide analysis of the human androgen receptor: detection of a hormone-induced phosphopeptide. Biochemistry. 1995;34:1851–1857. doi: 10.1021/bi00006a005. [DOI] [PubMed] [Google Scholar]
- 18.Kuiper G. G., de Ruiter P. E., Trapman J., Boersma W. J., Grootegoed J. A., Brinkmann A. O. Localization and hormonal stimulation of phosphorylation sites in the LNCaP-cell androgen receptor. Biochem. J. 1993;291:95–101. doi: 10.1042/bj2910095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenster G., de Ruiter P. E., van der Korput H. A., Kuiper G. G., Trapman J., Brinkmann A. O. Changes in the abundance of androgen receptor isotypes: effects of ligand treatment, glutamine-stretch variation, and mutation of putative phosphorylation sites. Biochemistry. 1994;33:14064–14072. doi: 10.1021/bi00251a015. [DOI] [PubMed] [Google Scholar]
- 20.Kuiper G. G., de Ruiter P. E., Grootegoed J. A., Brinkmann A. O. Synthesis and post-translational modification of the androgen receptor in LNCaP cells. Mol. Cell. Endocrinol. 1991;80:65–73. doi: 10.1016/0303-7207(91)90143-g. [DOI] [PubMed] [Google Scholar]
- 21.Kuiper G. G., de Ruiter P. E., Brinkmann A. O. Androgen receptor heterogeneity in LNCaP cells is caused by a hormone independent phosphorylation step. J. Steroid Biochem. Mol. Biol. 1992;41:697–700. doi: 10.1016/0960-0760(92)90407-a. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z. X., Kemppainen J. A., Wilson E. M. Identification of three proline-directed phosphorylation sites in the human androgen receptor. Mol. Endocrinol. 1995;9:605–615. doi: 10.1210/mend.9.5.7565807. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Z., Becklin R. R., Desiderio D. M., Dalton J. T. Identification of a novel phosphorylation site in human androgen receptor by mass spectrometry. Biochem. Biophys. Res. Commun. 2001;284:836–844. doi: 10.1006/bbrc.2001.5030. [DOI] [PubMed] [Google Scholar]
- 24.Lin H. K., Yeh S., Kang H. Y., Chang C. Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7200–7205. doi: 10.1073/pnas.121173298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen Y., Hu M. C., Makino K., Spohn B., Bartholomeusz G., Yan D. H., Hung M. C. HER-2/neu promotes androgen-independent survival and growth of prostate cancer cells through the Akt pathway. Cancer Res. 2000;60:6841–6845. [PubMed] [Google Scholar]
- 26.Yeh S., Lin H. K., Kang H. Y., Thin T. H., Lin M. F., Chang C. From HER2/Neu signal cascade to androgen receptor and its coactivators: a novel pathway by induction of androgen target genes through MAP kinase in prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5458–5463. doi: 10.1073/pnas.96.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zegers N. D., Claassen E., Neelen C., Mulder E., van Laar J. H., Voorhorst M. M., Berrevoets C. A., Brinkmann A. O., van der Kwast T. H., Ruizeveld de Winter J. A., et al. Epitope prediction and confirmation for the human androgen receptor: generation of monoclonal antibodies for multi-assay performance following the synthetic peptide strategy. Biochim. Biophys. Acta. 1991;1073:23–32. doi: 10.1016/0304-4165(91)90178-j. [DOI] [PubMed] [Google Scholar]
- 28.de Ruiter P. E., Teuwen R., Trapman J., Dijkema R., Brinkmann A. O. Synergism between androgens and protein kinase-C on androgen-regulated gene expression. Mol. Cell. Endocrinol. 1995;110:R1–R6. doi: 10.1016/0303-7207(95)03534-e. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J., Fritsch E. F., Maniatis T. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 30.Lubahn D. B., Joseph D. R., Sar M., Tan J., Higgs H. N., Larson R. E., French F. S., Wilson E. M. The human androgen receptor: complementary deoxyribonucleic acid cloning, sequence analysis and gene expression in prostate. Mol. Endocrinol. 1988;2:1265–1275. doi: 10.1210/mend-2-12-1265. [DOI] [PubMed] [Google Scholar]
- 31.Jenster G., van der Korput H. A., Trapman J., Brinkmann A. O. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J. Biol. Chem. 1995;270:7341–7346. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- 32.Doesburg P., Kuil C. W., Berrevoets C. A., Steketee K., Faber P. W., Mulder E., Brinkmann A. O., Trapman J. Functional in vivo interaction between the amino-terminal, transactivation domain and the ligand binding domain of the androgen receptor. Biochemistry. 1997;36:1052–1064. doi: 10.1021/bi961775g. [DOI] [PubMed] [Google Scholar]
- 33.Berrevoets C. A., Doesburg P., Steketee K., Trapman J., Brinkmann A. O. Functional interactions of the AF-2 activation domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor 2) Mol. Endocrinol. 1998;12:1172–1183. doi: 10.1210/mend.12.8.0153. [DOI] [PubMed] [Google Scholar]
- 34.Voegel J. J., Heine M. J., Zechel C., Chambon P., Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 35.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 36.West M. H. P., Wu R. S., Bonner W. M. Polyacrylamide gel electrophoresis of small peptides. Electrophoresis. 1984;5:133–138. [Google Scholar]
- 37.Langley E., Zhou Z. X., Wilson E. M. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J. Biol. Chem. 1995;270:29983–29990. doi: 10.1074/jbc.270.50.29983. [DOI] [PubMed] [Google Scholar]
- 38.Ghali S. A., Gottlieb B., Lumbroso R., Beitel L. K., Elhaji Y., Wu J., Pinsky L., Trifiro M. A. The use of androgen receptor amino/carboxyl-terminal interaction assays to investigate androgen receptor gene mutations in subjects with varying degrees of androgen insensitivity. J. Clin. Endocrinol. Metab. 2003;88:2185–2193. doi: 10.1210/jc.2002-021324. [DOI] [PubMed] [Google Scholar]
- 39.Langley E., Kemppainen J. A., Wilson E. M. Intermolecular NH2-/carboxyl-terminal interactions in androgen receptor dimerization revealed by mutations that cause androgen insensitivity. J. Biol. Chem. 1998;273:92–101. doi: 10.1074/jbc.273.1.92. [DOI] [PubMed] [Google Scholar]
- 40.Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 41.Takimoto G. S., Hovland A. R., Tasset D. M., Melville M. Y., Tung L., Horwitz K. B. Role of phosphorylation on DNA binding and transcriptional functions of human progesterone receptors. J. Biol. Chem. 1996;271:13308–13316. doi: 10.1074/jbc.271.23.13308. [DOI] [PubMed] [Google Scholar]