Abstract
Vitamin A homoeostasis requires the gene encoding cellular retinol-binding protein-1 (Crbp1) which stimulates conversion of retinol into retinyl esters that serve as a storage form of vitamin A. The gene encoding alcohol dehydrogenase-1 (Adh1) greatly facilitates degradative metabolism of excess retinol into retinoic acid to protect against toxic effects of high dietary vitamin A. Crbp1−/−/Adh1−/− double mutant mice were generated to explore whether the stimulatory effect of CRBP1 on retinyl ester formation is due to limitation of retinol oxidation by ADH1, and whether ADH1 limits retinyl ester formation by opposing CRBP1. Compared with wild-type mice, liver retinyl ester levels were greatly reduced in Crbp1−/− mice, but Adh1−/− mice exhibited a significant increase in liver retinyl esters. Importantly, relatively normal liver retinyl ester levels were restored in Crbp1−/−/Adh1−/− mice. During vitamin A deficiency, the additional loss of Adh1 completely prevented the excessive loss of liver retinyl esters observed in Crbp1−/− mice for the first 5 weeks of deficiency and greatly minimized this loss for up to 13 weeks. Crbp1−/− mice also exhibited increased metabolism of a dose of retinol into retinoic acid, and this increased metabolism was not observed in Crbp1−/−/Adh1−/− mice. Our findings suggest that opposing actions of CRBP1 and ADH1 enable a large fraction of liver retinol to remain esterified due to CRBP1 action, while continuously allowing some retinol to be oxidized to retinoic acid by ADH1 for degradative retinoid turnover under any dietary vitamin A conditions.
Keywords: alcohol dehydrogenase (ADH), cellular retinol-binding protein (CRBP), retinoic acid, retinol, vitamin A
Abbreviations: ADH, alcohol dehydrogenase; CRBP1, cellular retinol-binding protein type 1; LRAT, lecithin:retinol acyltransferase; RA, retinoic acid; RAR, RA receptor; RALDH, retinaldehyde dehydrogenase; SDR, short-chain dehydrogenase/reductase; VAD, vitamin A deficiency
INTRODUCTION
The function of vitamin A (retinol) in chordate animals is unique in that it acts not only as a precursor for retinaldehyde used in the visual cycle [1], but also as a precursor for RA (retinoic acid) which serves as a ligand for RARs (RA receptors) controlling gene regulation during embryonic development and adult tissue regeneration [2–4]. The isomers all-trans-RA (a ligand for RARs) and 9-cis-RA (a ligand for retinoid X receptors; RXR) were both originally hypothesized to be important for gene regulation [5], but recent studies indicate that an RAR ligand is sufficient to rescue an embryonic lethal genetic defect in RA synthesis [6]. Also, as all-trans-RA can be easily detected in many adult and embryonic tissues, whereas 9-cis-RA cannot be detected in these same tissues [6–10], it is quite possible that all-trans-RA is the only ligand needed physiologically for RA signalling. RA is not produced by all cells of the body at all stages of development, but is instead produced in a unique spatiotemporal pattern as observed in mouse embryos carrying an RA-reporter transgene [11]. Retinol is transported in the blood via serum retinol-binding protein which makes it available to essentially all cells for potential conversion into RA [12–14]. Many cells (including hepatocytes and liver stellate cells) express CRBP1 (cellular retinol-binding protein-1) [15]. CRBP1 facilitates esterification of retinol in stellate cells by LRAT (lecithin:retinol acyltransferase) to produce retinyl esters for storage, and retinyl ester hydrolysis in stellate cells then regenerates retinol which can either be re-esterified, exported to hepatocytes, transported into the blood or metabolized into RA [16,17]. The stellate cells of the adult liver are the main site of retinyl ester storage in the body and this source of retinol is severely depleted in Crbp1−/− mice [18].
Conversion of retinol into RA is a two-step enzymic pathway with retinaldehyde as the intermediate. Oxidation of retinol into retinaldehyde (a reversible reaction) is ubiquitous, as it is catalysed by ubiquitously expressed ADH3 (alcohol dehydrogenase class III) [19], as well as by several overlapping tissue-specific enzymes including ADH1 and ADH4 [20–23] and numerous microsomal SDR (short-chain dehydrogenase/reductase) family members [24–26]. In addition, enzymes have been identified from the SDR family and AKR (aldo-keto reductase) family that act preferentially as retinaldehyde reductases to convert retinaldehyde into retinol [27–29]. Oxidation of retinaldehyde into RA (an irreversible reaction) is controlled by the tissue-specific retinaldehyde dehydrogenases RALDH1 (ALDH1A1), RALDH2 (ALDH1A2) and RALDH3 (ALDH1A3) which synthesize RA in distinct spatiotemporal patterns [30–37]. Some RA becomes bound to RA receptors for signalling, but some is subjected to oxidative degradation by various P450 enzymes (including CYP26A1, CYP26B1 and CYP26C1) that metabolize RA to more easily excretable forms [38–40]. The various retinol metabolic pathways are summarized in Scheme 1.
Scheme 1. Retinol levels in serum during VAD.
Intracellular retinol can bind CRBP with the equilibrium greatly in favour of the bound form. Retinol can be covalently linked to a fatty acid by LRAT to produce retinyl esters for storage, and a retinyl ester hydrolase can regenerate retinol. Several forms of ADH and SDR catalyse oxidation of retinol into retinaldehyde, and several forms of aldehyde dehydrogenase (ALDH) catalyse oxidation of retinaldehyde into RA. RA can enter the nucleus and function as a ligand for RARs, or it can be metabolized by several members of the P450 (CYP) enzyme family into 4-oxo-RA, which constitutes the first step in a degradative pathway ultimately resulting in retinoid excretion. RA can also bind cellular RA-binding protein (CRABP) which may affect the degradation pathway.
Previous studies on Adh1−/− mice demonstrated that ADH1 is not required for survival and growth on normal or vitamin A-deficient diets [23]. However, ADH1 is present at very high levels in adult liver where it plays a role in oxidative metabolism of excess dietary retinol to provide protection against retinol toxicity [23]. Further mouse genetic studies have demonstrated that the dominant pathway of degradative retinol turnover in the liver proceeds through oxidation of retinol into retinaldehyde by ADH1, followed by irreversible oxidation of retinaldehyde into RA by RALDH1 [41]. As ADH1 and RALDH1 are expressed at high levels in both hepatocytes and stellate cells [42], their retinoid activity may effect the ability of CRBP1 to sequester retinol for retinyl ester synthesis. CRBP1 is expressed at high levels in adult liver (particularly high in stellate cells), where it has been hypothesized to bind most of the retinol, leaving very little free retinol available [43,44]. These findings suggest that exposure to high dietary retinol intake leads to an increase in free retinol unbound to CRBP1, which is then oxidized into RA by ADH1 and RALDH1. Analysis of Crbp1−/− mice supports the hypothesis that liver CRBP1 sequesters retinol primarily for the purpose of retinyl ester synthesis rather than RA synthesis [18]. In the present study we test the hypothesis that liver CRBP1 limits oxidation of retinol by ADH1, thus increasing the ability of esterifying enzymes to produce retinyl esters in stellate cells. We also examine whether ADH1 plays a constant role in retinol turnover during vitamin A sufficiency, deficiency or excess. Results from genetic crosses between Crbp1−/− and Adh1−/− mice indicate that CRBP1 and ADH1, indeed, have opposing actions on liver retinol metabolism that control the balance between retinol storage and retinol turnover under any dietary vitamin A conditions.
EXPERIMENTAL
Generation of Crbp1−/−/Adh1−/− double mutant mice
Crbp1−/− mice have been described previously [18], as have Adh1−/− mice [23,45]. Both strains are viable and fertile. Crbp1−/− and Adh1−/− mice were mated to generate Crbp1−/+/Adh1−/+ double heterozygous adult mice, and then further matings of these mice were performed to generate Crbp1−/−/Adh1−/− double homozygous adult mice identified by Southern blot analysis of tail DNA. The Crbp1−/−, Adh1−/− and wild-type littermates obtained from these double heterozygote matings were used in all studies for comparison with Crbp1−/−/Adh1−/− mice. Mice were on a mixed 129/Sv-C57BL/6-Black Swiss genetic background.
Generation of vitamin A-deficient mice
Gestational VAD (vitamin A deficiency) was induced as described previously [18,19]. For each mouse strain, mice were placed on Purina VAD diet 5822 (vitamin A<0.22 unit/g) at 4 weeks of age and maintained on this diet until analysis. Prior to placement on the VAD diet, mice were maintained on normal Purina Formulab chow diet 5008 (vitamin A=15.0 units/g).
Retinol treatment of mice
All-trans-retinol (Sigma) was dissolved in acetone/Tween 20/water (0.25:5:4.75, by vol.) as described previously [46], and was administered at a dose of 50 mg/kg by oral injection to adult female mice (age- and weight-matched). After 2 h, liver and blood were collected and processed for HPLC analysis of retinoids as described below.
Quantification of serum RA and retinol
Serum (0.2 ml) was mixed with 0.2 ml of methanol and extracted twice with 1 ml of hexane. All retinoid extraction and analytical procedures were carried out in a darkened room to protect retinoids from exposure to light. The hexane layers were collected, combined and evaporated under vacuum. The residue was dissolved in 150 μl of HPLC mobile phase, and 100 μl of this was analysed by HPLC as described previously [35].
Quantification of retinoids in liver
Hepatic levels of all-trans-RA, all-trans-retinol and retinyl esters were quantified by HPLC as described previously [35]. Briefly, liver (0.5 g) was homogenized in 1 ml of PBS (0.01 M, pH 7.4) and 150 μl (10%) was removed for analysis of retinyl esters, whereas the remaining homogenate was used for analysis of all-trans-RA and all-trans-retinol. Retinyl esters were quantified as described previously using liver samples subjected to saponification to convert retinyl esters into retinol [47]. Liver retinyl ester content was obtained from the difference of the total retinol measurement (from saponified liver) and the unesterified retinol measurement (from non-saponified liver).
All samples were subjected to reversed-phase HPLC analysis performed on a Waters 2695 HPLC system using a SUPLEX pkB-100 analytical column (250 mm×4.6 mm) (SUPELCO). Detection of retinoids was performed using a photodiode array detector (Waters model 2996) which collected spectra between 200 and 450 nm. Standard solutions of all-trans-retinol and all-trans-RA (Sigma) were used to obtain the calibration curves. Characteristic peak spectra and retention times were used to identify each retinoid, and quantification of peak areas was calculated at λmax using Waters Millennium Chromatography Manager software. The detection limit for the all-trans-RA standard was 0.2 ng per sample. Statistical significance was determined for raw data using the unpaired Student's t test (Statistica version 5.0).
RESULTS
Generation of Crbp1−/−/Adh1−/− mice
Matings of Crbp1−/+/Adh1−/+ double heterozygous mice resulted in generation of double homozygous Crbp1−/−/Adh1−/− mice (7 out of 90 pups, or 1/13) at approximately the expected Mendelian ratio (1/16). Matings of Crbp1−/−/Adh1−/− mice produced normal litter sizes with viable offspring which were used for all subsequent studies. Wild-type, Crbp1−/− and Adh1−/− littermates generated from the double heterozygote matings were also used for the studies reported below.
Crbp1−/−/Adh1−/− genotype restores liver retinyl ester levels to normal
Retinoid levels in mice maintained on normal mouse chow (vitamin A sufficiency) were quantified by HPLC (Table 1). Compared with wild-type mice, liver retinyl ester levels were reduced by 3.5-fold in Crbp1−/− mice and increased by 1.6-fold in Adh1−/− mice, but Crbp1−/−/Adh1−/− mice exhibited levels that were not significantly different from wild-type. Levels of retinol in serum and RA in liver were not significantly different among the four strains (Table 1). Liver retinol levels were moderately lower in Crbp1−/−, as well as Crbp1−/−/Adh1−/− mice, and moderately higher in Adh1−/− mice. As the concentration of CRBP1 in liver has been reported to be approximately the same as the total liver retinol concentration, and as the dissociation constant of CRBP1 for retinol is 0.1 nM, it has been estimated that the amount of free retinol in wild-type liver is only 0.25 nM [43,44]. In Crbp1−/−, as well as Crbp1−/−/Adh1−/−, mice the amount of free retinol must be substantially higher than this, as the total retinol concentration in the absence of CRBP1 dropped only 2-fold. Overall, the biggest effect of CRBP1 or ADH1 null mutations was on liver retinyl ester levels. These findings suggest that oxidation of retinol by ADH1 under conditions of dietary vitamin A sufficiency is responsible for the loss of retinyl esters observed when CRBP1 is missing. As ADH1 activity had previously been shown to be essential for clearance of excess dietary retinol [23], this now expands the role of ADH1 in retinol oxidation to physiological vitamin A conditions. The lack of a major effect on liver steady-state RA levels when ADH1 activity is lost indicates that other enzymes (including ADH3) also contribute to liver RA synthesis in vivo as suggested previously [19]. Furthermore, continuous degradation of RA by P450s [39] may ultimately be responsible for setting a low steady-state level of liver RA that can still be maintained when RA synthesis by ADH1 is lost, but RA synthesis by other enzymes continues.
Table 1. Steady-state levels of retinoids in serum and liver of mice on normal diet.
The results are expressed as the means±S.E.M. (n=3). All mice examined were males at 10 weeks of age. Note that RA values are in ng/g. *P<0.05 compared with wild-type mice. †P<0.05 compared with Crbp1−/− mice.
| Serum | Liver | |||
|---|---|---|---|---|
| Genotype | All-trans-retinol (μg/ml) | All-trans-RA (ng/g) | All-trans-retinol (μg/g) | Retinyl esters (μg/g) |
| Wild-type | 0.28±0.02 | 2.2±0.1 | 7.32±0.88 | 416±41 |
| Crbp1−/− | 0.27±0.02 | 1.8±0.1 | 3.66±0.22* | 120±2* |
| Crbp1−/−/Adh1−/− | 0.34±0.03 | 1.9±0.4 | 4.34±0.27* | 318±38† |
| Adh1−/− | 0.36±0.06 | 1.4±0.3 | 11.7±2.7 | 685±59* |
Loss of retinoids observed in Crbp1−/− mice during VAD is delayed in Crbp1−/−/Adh1−/− mice
Mice from all four strains were placed on a VAD diet at 4 weeks of age, and retinoids were periodically quantified by HPLC. The degree of VAD obtained in our experiments (i.e. the rate of liver retinyl ester loss) was not as severe as that previously published for wild-type and Crbp1−/− mice [18], perhaps due to differing sources of VAD diet and differing genetic backgrounds. However, an effect of the Adh1−/− genotype was still observed.
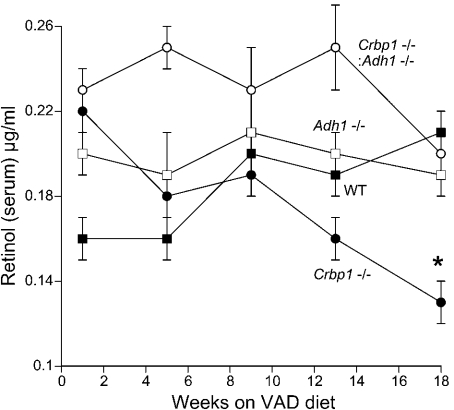
From 1–13 weeks on the VAD diet there was not a significant difference in serum retinol among the four strains (Figure 1). However, after 18 weeks of VAD, Crbp1−/− mice exhibited significantly lower serum retinol than wild-type mice, whereas levels in Crbp1−/−/Adh1−/− mice remained similar to wild-type (Figure 1). This indicates that ADH1 is involved in the loss of serum retinol observed during VAD when CRBP1 is missing. This finding suggests that ADH1 plays a role in retinol oxidation even during VAD when dietary retinol is no longer entering the body.
Figure 1. Retinol levels in serum during VAD.
Wild-type (WT), Crbp1−/−, Crbp1−/−/Adh1−/− and Adh1−/− female mice were placed on a VAD diet at 4 weeks of age and serum all-trans-retinol was periodically quantified by HPLC. The results are expressed as the means±S.E.M. (n=3). *P<0.05 compared with wild-type mice.
From 5–13 weeks of VAD there was a significant reduction in liver retinol observed in Crbp1−/− mice relative to wild-type mice and an increase in Adh1−/− mice, but levels were restored to wild-type in Crbp1−/−/Adh1−/− mice (Figure 2). At 18 weeks of VAD, liver retinol levels in both Crbp1−/− and Crbp1−/−/Adh1−/− mice were significantly lower than wild-type, probably due to excessive depletion of retinyl ester stores in the absence of CRBP1 (see below).
Figure 2. Retinol levels in liver during VAD.
Liver all-trans-retinol values determined by HPLC are shown for the same mice described in Figure 2. The results are expressed as the means±S.E.M. (n=3). *P<0.05 compared with wild-type (WT) mice.
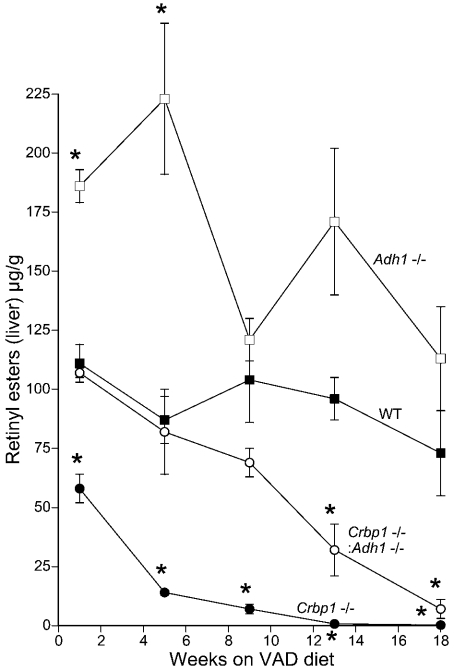
After 1–5 weeks of VAD, retinyl ester levels were significantly lower in Crbp1−/− mice and significantly higher in Adh1−/− mice relative to wild-type mice, but levels were restored to wild-type in Crbp1−/−/Adh1−/− mice (Figure 3). At 9–13 weeks of VAD, retinyl ester levels began to drop in Crbp1−/−/Adh1−/− mice relative to wild-type mice, but remained approx. 10-fold higher than that observed in Crbp1−/− mice (Figure 3). By 18 weeks of VAD, retinyl ester values were 0.2±0.1 μg/g for Crbp1−/− and 7±4 μg/g for Crbp1−/−/Adh1−/− mice, a 35-fold difference. Thus the deficiency status is significantly delayed in double null mutants relative to Crbp1−/− mice. This provides further evidence that metabolism of retinol by ADH1 is primarily responsible for the loss of retinyl esters observed when CRBP1 is missing. This conclusion is further supported by our observation that retinyl ester levels were higher in Adh1−/− mice relative to wild-type mice during VAD (Figure 3).
Figure 3. Retinyl ester levels in liver during VAD.
Liver retinyl ester values determined by HPLC are shown for the same mice described in Figure 2. The results are expressed as the means±S.E.M. (n=3). *P<0.05 compared with wild-type (WT) mice.
Liver RA levels are maintained during VAD
In contrast to the large decreases in liver retinol and retinyl ester levels observed in Crbp1−/− mice during VAD, liver RA levels were not significantly different among the four mouse strains during VAD. Liver RA values after 1 week of VAD were not significantly different than those observed after 36 weeks of VAD for all four strains (Figure 4). However, between 9–13 weeks of VAD, we observed a temporary reduction in liver RA for all four strains, but by 36 weeks of VAD all four strains exhibited liver RA levels similar to those observed at the beginning of VAD (Figure 4). These results suggest that during VAD there may be a mechanism in play which restores liver RA levels following a temporary drop, and that neither CRBP1 nor ADH1 are needed for this mechanism, as it occurred in Crbp1−/−/Adh1−/− mice.
Figure 4. RA levels in liver during VAD.
Liver all-trans-RA values determined by HPLC are shown for the same mice described in Figure 2. Data are also included for additional female mice examined for RA after 36 weeks on the VAD diet. Due to the large inter-mouse variation in all-trans-RA values in wild-type (WT) and Crbp1−/− mice after 1 week on the VAD diet, the values for these mice were not significantly different than those for Crbp1−/−/Adh1−/− and Adh1−/− mice at this time point. Such large variations were not observed in mice examined from 5 to 36 weeks on the VAD diet, but overall there was not a significant difference among the four strains during the 36 week period. The results are expressed as the means±S.E.M. (n=3).
After 36 weeks of VAD, retinol and retinyl ester levels were very low in all four strains (Table 2). At this stage of VAD nearly all liver retinyl esters had been depleted and serum retinol levels were reduced by 10-fold compared with mice on a normal diet. Also, by 36 weeks of VAD liver retinol levels (6–11 ng/g) were similar to liver RA levels (2.5–4.1 ng/g), rather than being about 1000-fold higher as observed in mice on a normal diet. These findings demonstrate that there is a remarkable ability to maintain liver RA levels during VAD, even after serum and liver retinol levels have dropped drastically.
Table 2. Retinoid levels in serum and liver of mice on VAD diet for 36 weeks.
The results are expressed as the means±S.E.M. (n=3). All mice examined were females. Note that values are in ng/g (or ml).
| Serum | Liver | |||
|---|---|---|---|---|
| Genotype | All-trans-retinol (ng/ml) | All-trans-RA (ng/g) | All-trans-retinol (ng/g) | Retinyl esters (ng/g) |
| Wild-type | 28±9 | 2.5±0.3 | 6±1 | 39±13 |
| Crbp1−/− | 51±1 | 3.6±0.1 | 11±2 | 33±4 |
| Crbp1−/−/Adh1−/− | 38±2 | 2.7±0.5 | 6±1 | 23±3 |
| Adh1−/− | 34±1 | 4.1±0.3 | 9±1 | 63±3 |
Increased metabolism of a dose of retinol to RA by Crbp1−/− mice is eliminated in Crbp1−/−/Adh1−/− mice
We have previously shown that ADH1 is responsible for most of the serum and liver RA produced following a 50 mg/kg oral dose of retinol [23]. In order to further examine the effect of CRBP1 and ADH1 mutations on metabolism of retinol into RA, we quantified retinoids at 2 h after oral injection of retinol (50 mg/kg). In the absence of CRBP1 and/or ADH1, more of the injected retinol accumulated in the serum (approx. 2-fold higher serum retinol levels for all three mutant strains relative to wild-type), indicating that both proteins normally function to clear excess retinol from the blood (Table 3). Liver retinol levels following a retinol dose were not significantly affected by a loss of CRBP1, but there was a slight increase in liver retinol when ADH1 was missing. Compared with wild-type mice, Crbp1−/− mice exhibited 3.4-fold higher serum RA levels and 3-fold higher liver RA levels following an oral dose of retinol (Table 3). These increases were not observed in Crbp1−/−/Adh1−/− mice which exhibited values similar to Adh1−/− mice, i.e. approx. 5-fold lower serum and liver RA compared with wild-type. After retinol treatment, liver retinyl ester values were lower in Crbp1−/− mice, higher in Adh1−/− mice, whereas Crbp1−/−/Adh1−/− mice accumulated more retinyl esters than Crbp1−/− mice (Table 3). These findings indicate that the absence of CRBP1 makes more retinol available to ADH1 for metabolism to RA. Conversely, the absence of ADH1 makes more retinol available to CRBP1 for conversion to retinyl esters.
Table 3. Dynamic levels of retinoids in serum and liver of mice at 2 h after injection of retinol (50 mg/kg).
The results are expressed as the means±S.E.M. (n=3). All mice examined were females at 10 weeks of age. Note that values are in μg/g (or ml). *P<0.05 compared with wild-type mice. †P<0.05 compared with Crbp1−/− mice.
| Serum | Liver | ||||
|---|---|---|---|---|---|
| Genotype | All-trans-RA (μg/ml) | All-trans-retinol (μg/ml) | All-trans-RA (μg/g) | All-trans-retinol (μg/g) | Retinyl esters (μg/g) |
| Wild-type | 0.67±0.11 | 0.92±0.06 | 1.20±0.18 | 11.2±0.21 | 612±57 |
| Crbp1−/− | 2.26±0.34* | 1.80±0.04* | 3.58±0.53* | 12.1±0.65 | 209±9* |
| Crbp1−/−/Adh1−/− | 0.11±0.02† | 1.85±0.07* | 0.15±0.04† | 15.2±1.49* | 462±45† |
| Adh1−/− | 0.12±0.02* | 1.65±0.11* | 0.27±0.06* | 18.2±2.17* | 925±33 |
DISCUSSION
Opposing actions of CRBP1 and ADH1 maintain retinyl ester homoeostasis
Previous studies have demonstrated that CRBP1 is expressed in both hepatocytes and liver stellate cells where it contributes to vitamin A homoeostasis by facilitating uptake of serum retinol into hepatocytes and transport to stellate cells for conversion into retinyl esters for long-term storage of vitamin A [15]. Crbp1−/− mice have much lower liver retinyl ester stores than wild-type mice and experience a much quicker loss of retinyl esters during VAD [18]. ADH1 is also expressed in both hepatocytes and stellate cells [42]. Our analysis of Crbp1−/−/Adh1−/− double mutant mice has demonstrated that the loss of liver retinyl esters observed in the absence of CRBP1 can be prevented by the additional loss of ADH1. We further demonstrate that ADH1 is involved in a continuous oxidative turnover of retinol into RA irrespective of dietary vitamin A conditions (i.e. vitamin A sufficiency, deficiency or excess) and that CRBP1 limits this turnover.
Based upon these findings, we hypothesize that a major function of CRBP1 in liver (hepatocytes and stellate cells) is to protect free retinol from ADH1. ADH1 is very active in metabolizing free retinol [19–21] and is present at very high levels in hepatocytes and stellate cells, accounting for 0.9% of total mouse liver protein [48]. Whereas ADH1 activity may be very useful to clear excess retinol to prevent toxicity [23], too much activity by ADH1 could severely reduce the amount of retinol available for conversion into retinyl esters for storage. Thus CRBP1 provides a pool of retinol for retinyl ester synthesis that is mostly protected from ADH1 activity, and retinyl esters themselves are protected from ADH1 oxidation by the fatty acid moiety covalently bound to the alcohol group. Most vitamin A is thus normally protected from ADH1 as it is present as retinyl esters, and when retinyl esters are hydrolysed to produce free retinol, most of this is presumably bound once again to CRBP1 which precludes ADH1 oxidation. Thus during cyclic retinyl ester formation, retinyl ester hydrolysis and re-esterification, only a small fraction of total vitamin A exists as retinol bound to CRBP1 at any given point in time, and an even smaller fraction exists as free retinol upon dissociation from CRBP1. Based upon the observation that the amount of CRBP1 present in liver exceeds the amount of retinol, it has been calculated that most intracellular retinol is bound non-covalently to CRBP1 [43,44], but a small fraction of free retinol may exist. Our observation that Adh1−/− mice accumulate more retinyl esters than wild-type mice even during VAD (when no new retinol is entering the body) provides evidence that a small amount of free retinol does in fact normally exist in liver which is continuously oxidized into RA by ADH1 for retinol turnover.
We propose that opposing actions of CRBP1 and ADH1 on liver retinol metabolism continuously regulate the amount of retinyl ester storage. This mechanism provides a long-term source of stored vitamin A while ensuring that retinoid turnover still occurs under any dietary vitamin A conditions. As all-trans-RA is the only retinoid known to be physiologically required for gene regulation [6], continuous retinol oxidation by ADH1 activity may be useful for turnover of retinyl esters that have suffered oxidation during their long-term storage; i.e. retinoids containing an oxidized β-ionine ring such as 4-hydroxy or 4-oxo have less biological activity [49]. Retinol turnover may also be particularly critical during ingestion of food sources containing high vitamin A content (i.e. liver or milk) to ensure that toxic amounts of vitamin A do not accumulate. Our previous studies on Adh1−/− mice have demonstrated that in order to minimize retinol toxicity in adult mice it is most beneficial to clear retinol by ADH1-mediated oxidation into RA [23]. Although RA is a teratogen [50], RA is less toxic to adult tissues than retinol due to the fact that RA is very quickly further metabolized by P450s to more easily excretable forms such as 4-oxo-RA [38]. Alternative disposition mechanisms for retinol lead to excessive toxicity in adult animals; for example, when retinol builds up, this results in excessive retinol glucuronidation that leads to depletion of uridine diphosphoglucuronic acid needed to perform other essential glucuronidations [51].
Serum retinol homoeostasis by CRBP1 and ADH1
As part of the vitamin A homoeostatic mechanism, serum retinol levels are normally maintained within a very narrow range, so that peripheral tissues receive a constant amount of retinol to ensure uninterrupted production of RA and visual pigment despite varying amounts of dietary vitamin A. Serum retinol-binding protein plays an important role in this mechanism [13], as does liver CRBP1 and liver ADH1 as shown in the present study. Previous studies have shown that wild-type mice maintain a constant amount of serum retinol for at least 23 weeks of VAD, but that Crbp1−/− mice do not maintain normal serum retinol levels after 13 weeks of VAD [18]; this was confirmed in the present study. In the present studies, we found that Crbp1−/−/Adh1−/− mice maintain serum retinol levels during VAD similar to wild-type, presumably due to higher liver retinyl ester stores in the double mutant where ADH1 is not present to oxidize retinol. Additionally, we show that ADH1 and CRBP1 both function to prevent excessive accumulation of retinol in serum following ingestion of excess retinol. We suggest that CRBP1 does this by stimulating accumulation of retinol in the liver for conversion into retinyl esters, and that ADH1 does this by oxidizing any retinol in the liver that is not bound to CRBP1. Our findings thus demonstrate that CRBP1 and ADH1 both contribute significantly to the disposition of dietary retinol to maintain a minimum level of serum retinol and to prevent excessive serum retinol accumulation.
Liver RA homoeostasis
We have demonstrated that vitamin A turnover in the liver occurs primarily through oxidation of retinol to RA by ADH1 and RALDH1 [23,41]. However, the steady-state amount of RA in liver is very low and fairly constant among all the strains tested here (2–4 ng/g in mice not treated with retinol). This may be due to very efficient further metabolism by P450 enzymes [39] and binding to cellular RA-binding proteins which may protect some RA from degradation [44]. This mechanism is temporarily overwhelmed following treatment with excess retinol, revealing very high levels of liver RA. This is a dynamic situation as RA levels return to the steady-state level at approx. 8 h after retinol treatment of wild-type mice [46]. In the absence of CRBP1 we now show that treatment with retinol leads to even higher levels of liver RA, and we demonstrated that this is due to greater access of retinol to ADH1. Our observation that steady-state liver RA levels are not significantly reduced when ADH1 activity is lost may be explained by the existence of additional enzymes that oxidize retinol for RA synthesis. Indeed, analysis of Adh3−/− mice has previously shown that class III ADH (ADH3) also contributes to liver retinol oxidation in vivo [52]. In addition, several microsomal SDRs may contribute to retinol oxidation in liver [24–26].
Mechanism of CRBP1 action
For cells that express CRBP1, most intracellular retinol is bound non-covalently to CRBP1, leaving very little free retinol [43,44]. Our findings suggest that CRBP1 greatly reduces the access of retinol to ADH1, but that a small fraction of free retinol always exists which can be oxidized by ADH1 for retinol degradation. The ability of ADH1 to use the free retinol available in the presence of CRBP1 may be the result of its very high expression in the liver which allows utilization of free retinol at very low concentrations. Based upon kinetic studies, microsomal LRAT and some forms of microsomal SDR have been reported to utilize the CRBP1–retinol complex as a substrate to produce either retinyl esters [16] or retinaldehyde for RA synthesis [53] respectively. This implies a physical interaction between these microsomal enzymes and the CRBP1–retinol complex to facilitate entry of retinol into the enzyme active site. A cross-linking study using a UV-activated reagent indicated that such an interaction occurs between rat RoDH1 (microsomal retinol dehydrogenase) and CRBP1–retinol [53]. As we have shown that liver RA levels are maintained in Crbp1−/− mice grown on normal or VAD diets, physical interaction of enzymes with CRBP1 is not absolutely required for oxidation of retinol in the liver which may be designed primarily for retinol degradation. However, outside the liver, CRBP1 or related binding proteins may facilitate retinol oxidation by enzymes that can physically bind CRBP1 to enhance synthesis of low levels of RA needed for signalling processes. Thus two categories of retinol-oxidizing enzymes may exist: CRBP1-binding forms designed to synthesize RA for signalling, and forms unable to bind CRBP1 designed to degrade free retinol.
Acknowledgments
We thank the Burnham Institute Animal Resources Facility for long-term maintenance of mice on a VAD diet. This work was supported by National Institutes of Health Grant AA09731 (to G.D.).
References
- 1.Wald G. The chemistry of rod vision. Science. 1951;113:287–291. doi: 10.1126/science.113.2933.287. [DOI] [PubMed] [Google Scholar]
- 2.Kastner P., Mark M., Chambon P. Nonsteroid nuclear receptors: What are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 3.Zile M. H. Function of vitamin A in vertebrate embryonic development. J. Nutr. 2001;131:705–708. doi: 10.1093/jn/131.3.705. [DOI] [PubMed] [Google Scholar]
- 4.Clagett-Dame M., DeLuca H. F. The role of vitamin A in mammalian reproduction and embryonic development. Annu. Rev. Nutr. 2002;22:347–381. doi: 10.1146/annurev.nutr.22.010402.102745E. [DOI] [PubMed] [Google Scholar]
- 5.Heyman R. A., Mangelsdorf D. J., Dyck J. A., Stein R. B., Eichele G., Evans R. M., Thaller C. 9-Cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 6.Mic F. A., Molotkov A., Benbrook D. M., Duester G. Retinoid activation of retinoic acid receptor but not retinoid X receptor is sufficient to rescue lethal defect in retinoic acid synthesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7135–7140. doi: 10.1073/pnas.1231422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton C., Maden M. Endogenous distribution of retinoids during normal development and teratogenesis in the mouse embryo. Dev. Dyn. 1995;202:312–323. doi: 10.1002/aja.1002020310. [DOI] [PubMed] [Google Scholar]
- 8.Kurlandsky S. B., Gamble M. V., Ramakrishnan R., Blaner W. S. Plasma delivery of retinoic acid to tissues in the rat. J. Biol. Chem. 1995;270:17850–17857. doi: 10.1074/jbc.270.30.17850. [DOI] [PubMed] [Google Scholar]
- 9.Ulven S. M., Gundersen T. E., Weedon M. S., Landaas V. O., Sakhi A. K., Fromm S. H., Geronimo B. A., Moskaug J. O., Blomhoff R. Identification of endogenous retinoids, enzymes, binding proteins, and receptors during early postimplantation development in mouse: Important role of retinal dehydrogenase type 2 in synthesis of all-trans-retinoic acid. Dev. Biol. 2000;220:379–391. doi: 10.1006/dbio.2000.9634. [DOI] [PubMed] [Google Scholar]
- 10.Werner E. A., DeLuca H. F. Metabolism of a physiological amount of all-trans-retinol in the vitamin A-deficient rat. Arch. Biochem. Biophys. 2001;393:262–270. doi: 10.1006/abbi.2001.2495. [DOI] [PubMed] [Google Scholar]
- 11.Rossant J., Zirngibl R., Cado D., Shago M., Giguère V. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- 12.Soprano D. R., Blaner W. S. Plasma retinol-binding protein. In: Sporn M. B., Roberts A. B., Goodman D. S., editors. The Retinoids: Biology, Chemistry, and Medicine. 2nd edition. New York: Raven Press Ltd; 1994. pp. 257–281. [Google Scholar]
- 13.Quadro L., Blaner W. S., Salchow D. J., Vogel S., Piantedosi R., Gouras P., Freeman S., Cosma M. P., Colantuoni V., Gottesman M. E. Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. EMBO J. 1999;18:4633–4644. doi: 10.1093/emboj/18.17.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wendler C. C., Schmoldt A., Flentke G. R., Case L. C., Quadro L., Blaner W. S., Lough J., Smith S. M. Increased fibronectin deposition in embryonic hearts of retinol-binding protein-null mice. Circ. Res. 2003;92:920–928. doi: 10.1161/01.RES.0000069030.30886.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem. J. 2000;348:481–495. [PMC free article] [PubMed] [Google Scholar]
- 16.Yost R. W., Harrison E. H., Ross A. C. Esterification by rat liver microsomes of retinol bound to cellular retinol-binding protein. J. Biol. Chem. 1988;263:18693–18701. [PubMed] [Google Scholar]
- 17.Ong D. E., Newcomer M. E., Chytil F. Cellular retinoid-binding proteins. In: Sporn M. B., Roberts A. B., Goodman D. S., editors. The Retinoids: Biology, Chemistry, and Medicine. 2nd edition. New York: Raven Press Ltd; 1994. pp. 283–317. [Google Scholar]
- 18.Ghyselinck N. B., Båvik C., Sapin V., Mark M., Bonnier D., Hindelang C., Dierich A., Nilsson C. B., Håkansson H., Sauvant P., et al. Cellular retinol-binding protein I is essential for vitamin A homeostasis. EMBO J. 1999;18:4903–4914. doi: 10.1093/emboj/18.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molotkov A., Fan X., Deltour L., Foglio M. H., Martras S., Farrés J., Parés X., Duester G. Stimulation of retinoic acid production and growth by ubiquitously-expressed alcohol dehydrogenase Adh3. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5337–5342. doi: 10.1073/pnas.082093299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boleda M. D., Saubi N., Farrés J., Parés X. Physiological substrates for rat alcohol dehydrogenase classes: Aldehydes of lipid peroxidation, omega-hydroxyfatty acids, and retinoids. Arch. Biochem. Biophys. 1993;307:85–90. doi: 10.1006/abbi.1993.1564. [DOI] [PubMed] [Google Scholar]
- 21.Han C. L., Liao C. S., Wu C. W., Hwong C. L., Lee A. R., Yin S. J. Contribution to first-pass metabolism of ethanol and inhibition by ethanol for retinol oxidation in human alcohol dehydrogenase family: Implications for etiology of fetal alcohol syndrome and alcohol-related diseases. Eur. J. Biochem. 1998;254:25–31. doi: 10.1046/j.1432-1327.1998.2540025.x. [DOI] [PubMed] [Google Scholar]
- 22.Kedishvili N. Y., Gough W. H., Davis W. I., Parsons S., Li T. K., Bosron W. F. Effect of cellular retinol-binding protein on retinol oxidation by human class IV retinol alcohol dehydrogenase and inhibition by ethanol. Biochem. Biophys. Res. Commun. 1998;249:191–196. doi: 10.1006/bbrc.1998.9105. [DOI] [PubMed] [Google Scholar]
- 23.Molotkov A., Deltour L., Foglio M. H., Cuenca A. E., Duester G. Distinct retinoid metabolic functions for alcohol dehydrogenase genes Adh1 and Adh4 in protection against vitamin A toxicity or deficiency revealed in double null mutant mice. J. Biol. Chem. 2002;277:13804–13811. doi: 10.1074/jbc.M112039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M., Chen W. G., Smith S. M., Napoli J. L. Molecular characterization of a mouse short chain dehydrogenase/reductase active with all-trans-retinol in intact cells, mRDH1. J. Biol. Chem. 2001;276:44083–44090. doi: 10.1074/jbc.M105748200. [DOI] [PubMed] [Google Scholar]
- 25.Shang E. Y., Lai K., Packer A. I., Paik J., Blaner W. S., Vieira M. D., Gouras P., Wolgemuth D. J. Targeted disruption of the mouse cis-retinol dehydrogenase gene: visual and nonvisual functions. J. Lipid Res. 2002;43:590–597. [PubMed] [Google Scholar]
- 26.Lapshina E. A., Belyaeva O. V., Chumakova O. V., Kedishvili N. Y. Differential recognition of the free versus bound retinol by human microsomal retinol/sterol dehydrogenases: Characterization of the Holo-CRBP dehydrogenase activity of RoDH-4. Biochemistry. 2003;42:776–784. doi: 10.1021/bi026836r. [DOI] [PubMed] [Google Scholar]
- 27.Crosas B., Cederlund E., Torres D., Jörnvall H., Farrés J., Parés X. A vertebrate aldo-keto reductase active with retinoids and ethanol. J. Biol. Chem. 2001;276:19132–19140. doi: 10.1074/jbc.M010478200. [DOI] [PubMed] [Google Scholar]
- 28.Kedishvili N. Y., Chumakova O. V., Chetyrkin S. V., Belyaeva O. V., Lapshina E. A., Lin D. W., Matsumura M., Nelson P. S. Evidence that the human gene for prostate short-chain dehydrogenase/reductase (PSDR1) encodes a novel retinal reductase (RalR1) J. Biol. Chem. 2002;277:28909–28915. doi: 10.1074/jbc.M202588200. [DOI] [PubMed] [Google Scholar]
- 29.Crosas B., Hyndman D. J., Gallego D., Martras S., Parés X., Flynn T. G., Farrés J. Human aldose reductase and human small intestine aldose reductase are efficient retinal reductases: consequences for retinoid metabolism. Biochem. J. 2003;373:973–979. doi: 10.1042/BJ20021818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu L. C., Chang W. C., Hoffmann I., Duester G. Molecular analysis of two closely related mouse aldehyde dehydrogenase genes: identification of a role for Aldh1, but not Aldh-pb, in the biosynthesis of retinoic acid. Biochem. J. 1999;339:387–395. [PMC free article] [PubMed] [Google Scholar]
- 31.Niederreither K., Subbarayan V., Dollé P., Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 32.Mic F. A., Haselbeck R. J., Cuenca A. E., Duester G. Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice. Development. 2002;129:2271–2282. doi: 10.1242/dev.129.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCaffery P., Posch K. C., Napoli J. L., Gudas L., Dräger U. C. Changing patterns of the retinoic acid system in the developing retina. Dev. Biol. 1993;158:390–399. doi: 10.1006/dbio.1993.1197. [DOI] [PubMed] [Google Scholar]
- 34.Grün F., Hirose Y., Kawauchi S., Ogura T., Umesono K. Aldehyde dehydrogenase 6, a cytosolic retinaldehyde dehydrogenase prominently expressed in sensory neuroepithelia during development. J. Biol. Chem. 2000;275:41210–41218. doi: 10.1074/jbc.M007376200. [DOI] [PubMed] [Google Scholar]
- 35.Fan X., Molotkov A., Manabe S.-I., Donmoyer C. M., Deltour L., Foglio M. H., Cuenca A. E., Blaner W. S., Lipton S. A., Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol. Cell. Biol. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blentic A., Gale E., Maden M. Retinoic acid signalling centres in the avian embryo identified by sites of expression of synthesising and catabolising enzymes. Dev. Dyn. 2003;227:114–127. doi: 10.1002/dvdy.10292. [DOI] [PubMed] [Google Scholar]
- 37.Dupé V., Matt N., Garnier J.-M., Chambon P., Mark M., Ghyselinck N. B. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts E. S., Vaz A. D. N., Coon M. J. Role of isozymes of rabbit microsomal cytochrome P-450 in the metabolism of retinoic acid, retinol, and retinal. Mol. Pharmacol. 1992;41:427–433. [PubMed] [Google Scholar]
- 39.White J. A., Guo Y. D., Baetz K., Beckett-Jones B., Bonasoro J., Hsu K. E., Dilworth F. J., Jones G., Petkovich M. Identification of the retinoic acid-inducible all-trans-retinoic acid 4-hydroxylase. J. Biol. Chem. 1996;271:29922–29927. doi: 10.1074/jbc.271.47.29922. [DOI] [PubMed] [Google Scholar]
- 40.Niederreither K., Abu-Abed S., Schuhbaur B., Petkovich M., Chambon P., Dollé P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat. Genet. 2002;31:84–88. doi: 10.1038/ng876. [DOI] [PubMed] [Google Scholar]
- 41.Molotkov A., Duester G. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J. Biol. Chem. 2003;278:36085–36090. doi: 10.1074/jbc.M303709200. [DOI] [PubMed] [Google Scholar]
- 42.Casini A., Pellegrini G., Ceni E., Salzano R., Parola M., Robino G., Milani S., Dianzani M. U., Surrenti C. Human hepatic stellate cells express class I alcohol dehydrogenase and aldehyde dehydrogenase but not cytochrome P4502E1. J. Hepatol. 1998;28:40–45. doi: 10.1016/s0168-8278(98)80200-6. [DOI] [PubMed] [Google Scholar]
- 43.Ross A. C. Cellular metabolism and activation of retinoids: Roles of cellular retinoid-binding proteins. FASEB J. 1993;7:317–327. doi: 10.1096/fasebj.7.2.8440409. [DOI] [PubMed] [Google Scholar]
- 44.Napoli J. L. Interactions of retinoid binding proteins and enzymes in retinoid metabolism. Biochim. Biophys. Acta. 1999;1440:139–162. doi: 10.1016/s1388-1981(99)00117-1. [DOI] [PubMed] [Google Scholar]
- 45.Deltour L., Foglio M. H., Duester G. Metabolic deficiencies in alcohol dehydrogenase Adh1, Adh3, and Adh4 null mutant mice: Overlapping roles of Adh1 and Adh4 in ethanol clearance and metabolism of retinol to retinoic acid. J. Biol. Chem. 1999;274:16796–16801. doi: 10.1074/jbc.274.24.16796. [DOI] [PubMed] [Google Scholar]
- 46.Collins M. D., Eckhoff C., Chahoud I., Bochert G., Nau H. 4-Methylpyrazole partially ameliorated the teratogenicity of retinol and reduced the metabolic formation of all-trans-retinoic acid in the mouse. Arch. Toxicol. 1992;66:652–659. doi: 10.1007/BF01981505. [DOI] [PubMed] [Google Scholar]
- 47.Satre M. A., Ugen K. E., Kochhar D. M. Developmental changes in endogenous retinoids during pregnancy and embryogenesis in the mouse. Biol. Reprod. 1992;46:802–810. doi: 10.1095/biolreprod46.5.802. [DOI] [PubMed] [Google Scholar]
- 48.Algar E. M., Seeley T.-L., Holmes R. S. Purification and molecular properties of mouse alcohol dehydrogenase isozymes. Eur. J. Biochem. 1983;137:139–147. doi: 10.1111/j.1432-1033.1983.tb07807.x. [DOI] [PubMed] [Google Scholar]
- 49.Frolik C. A. Metabolism of retinoids. In: Sporn M. B., Roberts A. B., Goodman D. S., editors. The Retinoids, vol. 2. Orlando: Academic; 1984. pp. 177–208. [Google Scholar]
- 50.Armstrong R. B., Ashenfelter K. O., Eckhoff C., Levin A. A., Shapiro S. S. General and reproductive toxicology of retinoids. In: Sporn M. B., Roberts A. B., Goodman D. S., editors. The Retinoids: Biology, Chemistry, and Medicine. 2nd edition. New York: Raven Press Ltd.; 1994. pp. 545–572. [Google Scholar]
- 51.Bray B. J., Rosengren R. J. Retinol potentiates acetaminophen-induced hepatotoxicity in the mouse: Mechanistic studies. Toxicol. Appl. Pharmacol. 2001;173:129–136. doi: 10.1006/taap.2001.9170. [DOI] [PubMed] [Google Scholar]
- 52.Molotkov A., Fan X., Duester G. Excessive vitamin A toxicity in mice genetically deficient in either alcohol dehydrogenase Adh1 or Adh3. Eur. J. Biochem. 2002;269:2607–2612. doi: 10.1046/j.1432-1033.2002.02935.x. [DOI] [PubMed] [Google Scholar]
- 53.Boerman M. H. E. M., Napoli J. L. Characterization of a microsomal retinol dehydrogenase: A short-chain alcohol dehydrogenase with integral and peripheral membrane forms that interacts with holo-CRBP (type I) Biochemistry. 1995;34:7027–7037. doi: 10.1021/bi00021a014. [DOI] [PubMed] [Google Scholar]