Abstract
Copper homoeostasis was investigated in the Drosophila melanogaster S2 cell line to develop an insect model for the study of copper regulation. Real-time PCR studies have demonstrated expression in S2 cells of putative orthologues of human Cu regulatory genes involved in the uptake, transport, sequestration and efflux of Cu. Drosophila orthologues of the mammalian Cu chaperones, ATOX1 (a human orthologue of yeast ATX1), CCS (copper chaperone for superoxide dismutase), COX17 (a human orthologue of yeast COX17), and SCO1 and SCO2, did not significantly respond transcriptionally to increased Cu levels, whereas MtnA, MtnB and MtnD (Drosophila orthologues of human metallothioneins) were up-regulated by Cu in a time- and dose-dependent manner. To examine the effect on Cu homoeostasis, expression of several key copper homoeostasis genes was suppressed using double-stranded RNA interference. Suppression of the MTF-1 (metal-regulatory transcription factor 1), reduced both basal and Cu-induced gene expressions of MtnA, MtnB and MtnD, significantly reducing the tolerance of these cells to increased Cu. Suppression of either Ctr1A (a Drosophila orthologue of yeast CTR1) or Ctr1B significantly reduced Cu uptake from media, demonstrating that both these proteins function to transport Cu into S2 cells. Significantly, Cu induced Ctr1B gene expression, and this could be prevented by suppressing MTF-1, suggesting that Ctr1B might be involved in Cu detoxification. Suppression of DmATP7, the putative homologue of human Cu transporter genes ATP7A and ATP7B, significantly increased Cu accumulation, demonstrating that DmATP7 is essential for efflux of excess Cu. This work is consistent with previous studies in mammalian cells, validating S2 cells as a model system for studying Cu transport and identifying novel Cu regulatory mechanisms.
Keywords: copper, copper homoeostasis, Drosophila, gene expression, S2 cell, viability
Abbreviations: ATOX1, a human orthologue of yeast ATX1; ATP7A, human copper-transporting ATPase; CCS, copper chaperone for superoxide dismutase; CHO cells, Chinese-hamster ovary cells; dsRNAi, double-stranded RNA interference; MBS, metal-binding site; MTF-1, metal-regulatory transcription factor 1; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; SFM, serum-free media
INTRODUCTION
The maintenance of Cu homoeostasis at the cellular level is crucial for aerobic organisms. The redox properties of Cu ensure that it is an essential cofactor for several key enzymes including cytochrome c oxidase (mitochondrial electron transport chain), CuZn superoxide dismutase (antioxidant defences), tyrosinase (pigmentation) and dopamine β-hydroxylase (catecholamine synthesis) [1]. However, an excess of Cu is toxic since these redox properties also allow Cu ions to generate reactive oxygen species [2].
Studies in yeast and mammalian systems have identified well-conserved mechanisms for maintaining Cu homoeostasis, including regulation of uptake, distribution, sequestration and efflux [3]. In humans, several proteins have been identified as having an important role in Cu homoeostasis: Cu enters the cell via the high-affinity uptake protein, hCTR1 (a human orthologue of yeast CTR1) [4]; the specific chaperones CCS (copper chaperone for superoxide dismutase), COX17 and ATOX1 (human orthologues of yeast COX17 and ATX1 respectively) are involved in the distribution of Cu to superoxide dismutase, cytochrome c oxidase and the secretory pathway respectively [5–7]; sequestration of excess Cu by metallothioneins prevents cellular damage; metallothioneins are encoded by several genes that are transcriptionally regulated by MTF-1 (metal-regulatory transcription factor 1) [8]; and the heavy metal P-type ATPase ATP7A (human copper-transporting ATPase), expressed in extrahepatic tissues, and ATP7B (human copper-transporting ATPase), expressed predominantly in the liver, allow efflux of excess Cu from the cell [9].
Orthologues of several key Cu homoeostasis genes have been described in Drosophila melanogaster. Cu uptake genes Ctr1A, Ctr1B and Ctr1C (Drosophila orthologues of yeast CTR1) are classified as members of the Ctr1 family on the basis of conservation of sequence motifs present in mammalian and yeast Ctr1 genes [10,11]. Studies with these Drosophila Ctr genes transfected into S2 cells have shown that Ctr1A and Ctr1B could stimulate Cu uptake, whereas Ctr1C could not [10]. The human genome contains ten functional metallothionein genes, as well as seven pseudogenes [12]. In contrast, only four metallothioneins have been identified in Drosophila, designated MtnA, MtnB, MtnC and MtnD, all containing the metal response element TGCRCNC (R=A or G and N=any nucleotide) [13–15]. MtnB, MtnC and MtnD share a high level of sequence similarity [15]. MTF-1 has been identified as the Drosophila transcription factor capable of inducing expression of these Mtn genes in response to Cu [15]. In addition, the present study has identified a Drosophila orthologue of ATP7A and ATP7B, designated DmATP7. The Drosophila cell line S2 has been used to elucidate the function of the Drosophila Cu uptake proteins [10]; however, little else is known about Cu homoeostasis in S2 cells.
The specific aims of the present study were to identify orthologues of mammalian Cu homoeostasis genes, and use dsRNAi (double-stranded RNA interference) to suppress their gene expression in S2 cells. We suppressed Ctr1A, Ctr1B, MTF-1 and DmATP7 and examined Cu accumulation and retention, cell viability and gene expression. The results demonstrated the similarity between Drosophila S2 cells and mammalian systems and establishes S2 cells as a model system for exploring Cu homoeostatic mechanisms.
EXPERIMENTAL
Cell culture
Drosophila S2 cells were propagated in Drosophila SFM (serum-free media Invitrogen); supplemented with 367 μM L-glutamine and penicillin/streptomycin (Sigma). Cells were maintained at 27 °C and passaged every 7 days. S2 cells were loosely attached to the surface of tissue culture flasks and could be dislodged by pipetting. For all experiments, Cu was added to SFM as CuCl2 at the concentration specified in the Figure legends.
Real-time PCR
Total RNA was extracted using the RNeasy kit, including DNase treatment, according to the manufacturer's instructions (Qiagen, Chartsworth, CA, U.S.A.). cDNA was transcribed from 1 μg of total RNA using AMV Reverse Transcriptase in a 20 μl reaction according to the manufacturer's instructions (Promega, Charbonnières, France). Primers for real-time PCR were designed using Primer3 software [16] available at http://www.broad.mit.edu/genome_software/other/primer3.html.
Forward and reverse primer sequences were: Ctr1A: GCTGGAATATCGACCTGTGA, ATCGAAGGTGGTTGCTTGT; Ctr1B: AGCAGCGTAGGAAGAACGA, CAGGGACTGGACGATGTG; Ctr1C: GGCGATTCAGAGACCATTC, GAGCACAGCAACCATGAAA; CG32444: GGCGATAAGGTCGAGAAAGT, ACGTAGGTGGTGCTCTTGC; CCS: TGGACAAGACACCAATCCAG, ACAACCACTCCAGGCTTCTT; CG9065: GCAGCTCCAAGTGTTTCG, ATGCGGTGGTTGTGGAT; CG8885: GGCAGTTGGGAATTAGTTGAT, CTCTAGCTCATCGGGACAAA; DmATP7: CCCACTGACCTTCTTCGATAC, GGTCTTTCCCTTGGCTATGT; MtnA: CCTGCAACTGCGGATCT, CGCAGGCGGATTTCTT; MtnB: ATGGTTTGCAAGGGTTGTG, TTGCAGGCGCAGTTGT; MtnC: CAAAGGCTGCGGAACAA, GCACTTGCAGTCCTGATTACAG; MtnD: AGTGCTCCGCCACCAA, TGTCCTTGGGTCCGTTCT; MTF-1: ACAAGTGGCGAGCAACAG, GGCTGTAGGAAGTGAGGAATG; and Actin42A: GCTTCGCTGTCTACTTTCCA, CAGCCCGACTACTGCTTAGA.
Real-time PCR was performed using the Rotor Gene 3000 (Corbett Research, Mortlake, NSW, Australia). Reverse-transcribed total RNA (20 ng) was amplified in a 25 μl reaction containing 1 μM of each primer and 12.5 μl of 2×QuantiTect SYBR Green PCR Master Mix (Qiagen).
The amount of gene product in each sample was determined using two different methods, the established ΔCT method [17] and the comparative quantification method using the Rotor Gene 5.0 software (Corbett Research). These two methods provided very similar results, and all data shown in this paper were obtained using the comparative quantification method.
Actin42A, a gene that does not transcriptionally respond to copper, was used as a housekeeping gene. The amount of gene product for the gene of interest was expressed relative to that of Actin42A to normalize for differences in total cDNA between samples.
dsRNA production and RNA interference
Production of dsRNA was conducted as reported previously [18]. The gene of interest was PCR-amplified using forward and reverse primers, each containing the T7 RNA polymerase promoter sequence at the 5′-end and pSK(bluescript)-specific sequence at the 3′-end. Forward and reverse primer gene-specific sequences were TGTAATACGACTCACTATAGGGCAATTAACCCTCACTAAAGGGAA and TGTAATACGACTCACTATAGGGC respectively. PCR products were purified using Ultra-Clean PCR Clean Up (Mo Bio Laboratories, Carlsbad, CA, U.S.A.). RNA was transcribed in vitro from 1 μg of DNA template using MEGAscript T7 (Ambion, Austin, TX, U.S.A.). RNA was precipitated, dissolved in water, incubated at 65 °C for 30 min and slowly cooled to 25 °C to allow annealing of the complementary RNA strands.
The method described by Worby et al. [18] to introduce dsRNA into S2 cells for suppressing native RNA was adapted for use with SFM. S2 cells were seeded at a concentration of 1×106 cells/ml in SFM. dsRNA (15 μg) was added and the media were swirled vigorously for 10 s. Cells were incubated at 27 °C for 48 h before subsequent experiments.
Cell viability
Cell viability following Cu exposure was determined colorimetrically using MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; Sigma] by a method adapted from La Fontaine et al. [19]. Cells were seeded in a 96-well plate at a concentration of 2×105 cells (0.2 ml per well) and exposed to basal media or media supplemented with Cu for 48 h. The media were replaced with 0.3 ml SFM containing 2.4 mM MTT for 4 h. Media were removed and cells were solubilized in 0.3 ml DMSO (Sigma). The absorbance was read at 600 nm using an EIA plate reader (model no. 2550, Bio-Rad) and the percentage viability was determined by expressing the average absorbance from a given Cu treatment relative to the average absorbance from a reference sample treated with basal media.
64Cu accumulation and retention
64Cu accumulation and retention experiments were conducted using a method adapted from Camakaris et al. [20]. Cells (1 ×106 ml−1·well−1) were seeded in a 24-well plate and incubated with approx. 0.4 MBq of 64Cu (Australian Radioisotopes) and various concentrations of unlabelled Cu for 1 or 24 h. Cu accumulation was stopped by washing cells four times with 1 ml of cold Hanks balanced salt solution containing 1 mM histidine (Sigma). Cells were then lysed in 100 μl of 0.1% SDS, containing 2 mM EDTA. For Cu-retention experiments, cells were incubated with various concentrations of Cu for 24 h, washed as described above, and incubated for an additional 24 h in 1 ml of basal media. Cells were then washed and lysed as described above. Radioactivity was measured with a γ-counter (1282 CompuGamma, LKB Wallac) and 64Cu content was determined as described previously [20]. Cu levels were standardized to total cellular protein, which was determined using a Bio-Rad protein reagent according to the manufacturer's instructions (Bio-Rad).
Statistics
Statistical analysis was conducted using SPSS v11 (SPSS). A one-sample Kolomogorov–Smirinov test was used to assess whether data were normally distributed. Statistical analyses are described in the Figure legends. P<0.05 was deemed statistically significant.
RESULTS AND DISCUSSION
S2 cells are highly Cu tolerant
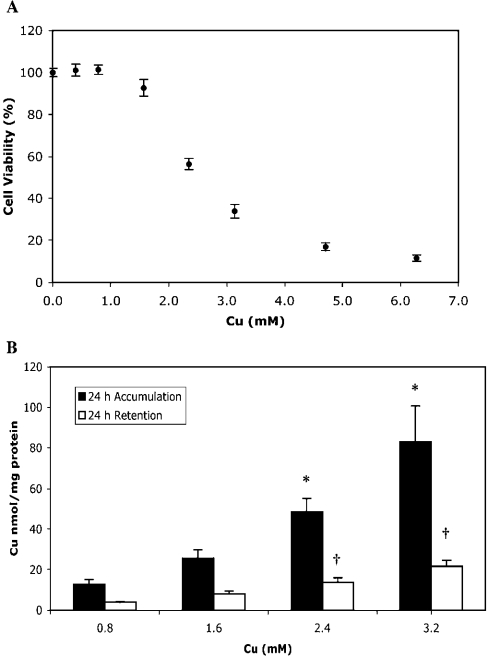
S2 cells were exposed to Cu concentrations ranging from basal to 6.3 mM and viability was measured 48 h later (Figure 1A). The Cu concentration needed to reduce cell viability to 50% of control (EC50) was 2.8 mM, which is consistent with a previous study that found that viability of S2 cells decreased to 70% of control after 48 h exposure to 2 mM CuSO4 [21]. Cu tolerance of S2 cells was much higher than that seen in CHO cells (Chinese-hamster ovary cells), which have a 24 h EC50 of approx. 0.33 mM CuCl2 [19]. However, comparisons between different cell types are difficult due to different measures of viability, different cell-culture media and the bioavailability of Cu. Both CuCl2 and CuSO4 are used for such studies and Cu is sometimes added as a complex. Comparable levels of Cu tolerance have been found with other cell lines: HL-60, a human leukaemia cell line, has an approximate EC50 of 1 mM when exposed to copper sulphate/nitrilotriacetate for 14 h [22], and the rat hepatoma cell line HAC has an EC50 of 0.4 mM when exposed to Cu(II) acetate for 72 h [23].
Figure 1. Cell viability and Cu accumulation and retention in S2 cells.
(A) Cell viability was measured with an MTT assay after 48 h exposure to 0.2, 0.4. 0.8, 1.6, 2.4, 3.2, 4.8 and 6.4 mM Cu, and is expressed as a percentage of viable cells compared with cells exposed to basal media. Values are means±S.E.M. of six replicates from two independent experiments. (B) Intracellular Cu accumulation and retention in cells exposed to 0.8, 1.6, 2.4 and 3.2 mM Cu labelled with 64Cu. Accumulation was measured after 24 h Cu exposure. Retention was measured after 24 h Cu exposure and an additional 24 h exposure to basal media. Intracellular Cu was normalized to total cellular protein as described in the Experimental section. Values are means±S.E.M. of six replicates from two independent experiments. A one-way ANOVA with Games–Howell post-hoc test was used to determine statistically significant differences between groups. *P<0.05 compared with 0.8 mM Cu accumulation; †P<0.05 compared with 0.8 mM Cu retention. A Pearson correlation showed Cu exposure was positively correlated with Cu accumulation (r=0.98, P<0.05) and Cu retention (r=0.99, P<0.05).
Variations in Cu tolerance may be due to regulation of Cu uptake, sequestration of free Cu or regulating efflux of excess Cu. Both CHO and HAC cell lines can be adapted to tolerate increased Cu levels. The increased tolerance of CHO-Cur3 cells occurs as a result of ATP7A gene amplification, resulting in increased Cu efflux [20]. In contrast, HAC800 cells have increased Cu tolerance as a result of metallothionein gene amplification. Increased metallothionein mRNA levels in these cells were positively correlated with intracellular Cu concentrations, demonstrating that tolerance is due to efficient Cu sequestration [24].
S2 cells accumulate and retain excess Cu
Cu accumulation was examined in S2 cells exposed to 0.8, 1.6, 2.4 or 3.2 mM Cu, labelled with 64Cu (Figure 1B). A 24 h exposure resulted in a dose-dependent increase in Cu accumulation. Cells exposed to 3.2 mM Cu accumulated five times as much Cu as those exposed to 0.8 mM. These results demonstrate that S2 cells do not appear to regulate intracellular Cu via changes in uptake, since exposure to high levels is sufficient to cause cells to accumulate Cu in excess of cellular requirements. Similar results were obtained with CHO cells for 6, 20 and 72 h [25,26].
Cu retention was measured in S2 cells exposed to Cu for 24 h, and then returned to basal media for another 24 h (Figure 1B). Cu ‘efflux’, the amount of Cu exported from the cell over that period, could be calculated from the difference between accumulation and retention. Cu retention was positively correlated with the initial Cu exposure. Cells were able to export some Cu at all exposure concentrations; however, cells initially exposed to 3.2 mM Cu retained 5-fold more Cu than those exposed to 0.8 mM. These results demonstrate that S2 cells have a mechanism for exporting excess Cu; however, this mechanism is not sufficient to maintain optimal intracellular Cu levels when exposed to a very high Cu concentration. The high levels of Cu accumulation and retention of S2 cells exposed to increased Cu concentrations suggests that the relatively high Cu tolerance of these cells is probably a result of efficient intracellular Cu sequestration than decreased uptake or increased efflux.
S2 cells express orthologues of Cu homoeostasis genes
The sequencing of the D. melanogaster genome has facilitated the identification of orthologues of mammalian Cu homoeostasis genes involved in uptake, distribution, sequestration and efflux of Cu [27]. These Drosophila orthologues show significant protein sequence identity and similarity (Table 1). Three copper uptake genes, Ctr1A, Ctr1B and Ctr1C, have been identified [10,11]. Four metallothioneins, designated MtnA, MtnB, MtnC and MtnD [13–15], and MTF-1, a transcription factor capable of inducing the expression of these Mtn genes, have also been identified [15].
Table 1. D. melanogaster Cu homoeostasis genes are expressed in S2 cells.
Putative D. melanogaster orthologues of human copper homoeostasis genes were identified with a default BLASTn search of the D. melanogaster genome (http://flybase.bio.indiana.edu). Percentage protein identity and similarity were determined using a default Gap alignment of human and Drosophila sequences (http://www.accelrys.com).
| Human gene (GenBank® accession ID) | Protein function | Drosophila orthologue | Protein sequence identity/similarity (%) | Fold induction by Cu (means±S.E.M.) |
|---|---|---|---|---|
| hCTR1 (Al831843) | Cu uptake | CG3977 Ctr1A | 47.6/57.8 | 1.2±0.1 |
| CG15551 | PA 38.1/48.4 | Not detected | ||
| Ctr1C* | PB 37.5/47.2 | |||
| CG7459 Ctr1B | 38.4/46.5 | 4.3±1.6 | ||
| ATOX1 (AY165037) | Cu chaperone | CG32444 | 22.4/35.8 | 1.2±0.1 |
| CCS (AF002210) | Cu chaperone | CG17753 CCS | 51.0/64.1 | 1.1±0.1 |
| COX17 (L77701) | Cu chaperone | CG9065 | 57.1/60.3 | 0.9±0.1 |
| SCO1 (AF026852) | Cu chaperone | CG8885 | 51.0/62.4 | 1.1±0.1 |
| SCO2 (AL021683) | Cu chaperone | CG8885 | 44.7/51.6 | |
| ATP7A (L06133) | Cu efflux | CG1886 | 49.9/58.9 | 1.7±0.6 |
| ATP7B (U11700) | Cu efflux | CG1886 | 49.9/59.1 | |
| MT† | Cu sequestration | CG9470 MtnA | N/A | 46±12‡ |
| Cu sequestration | CG4312 MtnB | N/A | 1443±517*‡ | |
| Cu sequestration | CG5097 MtnC | N/A | Not detected | |
| Cu sequestration | CG33192 MtnD | N/A | 474.4±149.3*‡ | |
| MTF-1 (X78710) | MT transcription | CG3743 | PA 32.0/38.5 | 1.4±0.2 |
| factor | MTF-1* | PB 27.0/32.4 |
* FlyBase identified two protein sequences, designated PA and PB, for both Ctr1C and MTF-1. Gene expression was measured with real-time PCR and normalized to Actin42A expression as described in the Experimental section. Gene expression was measured in S2 cells exposed to basal media and 0.8 mM Cu for 24 h. Values are gene expression in cells exposed to Cu relative to cells exposed to basal media and are the means±S.E.M. of four replicates from two independent experiments. A one-sample t test was used to determine if gene expression was significantly different with Cu exposure.
† Drosophila Mtn sequences were similar to multiple members of the human MT family and were not assigned (N/A) to a single putative orthologue.
‡ P<0.05.
A default BLASTn search of the Drosophila genome identified putative orthologues of the human Cu chaperones ATOX1, CCS, COX17 and SCO1 and SCO2. One putative Cu-transporting P-type ATPase was also identified, designated DmATP7, which shows strong sequence similarity to both ATP7A and ATP7B of humans (Table 1). The sequence similarity between human ATP7A and ATP7B was recently reviewed, and across the entire protein sequence, ATP7A and ATP7B are 54% identical; however, sequence similarity is much higher for several key motifs [9,28]. These include six N-terminal MBSs (metal-binding sites) characterized by a CXXC motif, eight transmembrane domains and the TGES/A, DKTG, TGDN and AMXGDGVND motifs in the major cytosolic loop. An alignment of the human ATP7A and ATP7B and the putative DmATP7 protein sequences using CLUSTAL W [29] demonstrated that all these key motifs were conserved in DmATP7 (results not shown). The conservation of these key motifs in Drosophila suggests that this protein is probably a functional Cu-transporting ATPase. Interestingly, the protein sequence alignment showed that DmATP7 has only four N-terminal MBS, corresponding to MBS 3–6 in human ATP7A and ATP7B.
S2 cells are of embryonic origin, and it was important to establish whether these putative Drosophila Cu genes were expressed in these cells. Real-time PCR was used to examine gene expression in cells under basal conditions and after exposure to 0.8 mM Cu. All the genes in Table 1, with the exception of Ctr1C and MtnC, were expressed in S2 cells. PCR failure was unlikely to be responsible for the inability to detect expression of these genes since expression was detected using adult D. melanogaster cDNA under the same PCR conditions (results not shown). A previous study using an RNA blot also failed to detect either Ctr1B or Ctr1C expression in S2 cells [10]. However, using real-time PCR, Ctr1B was detectable and expression was also induced by Cu exposure (Table 1).
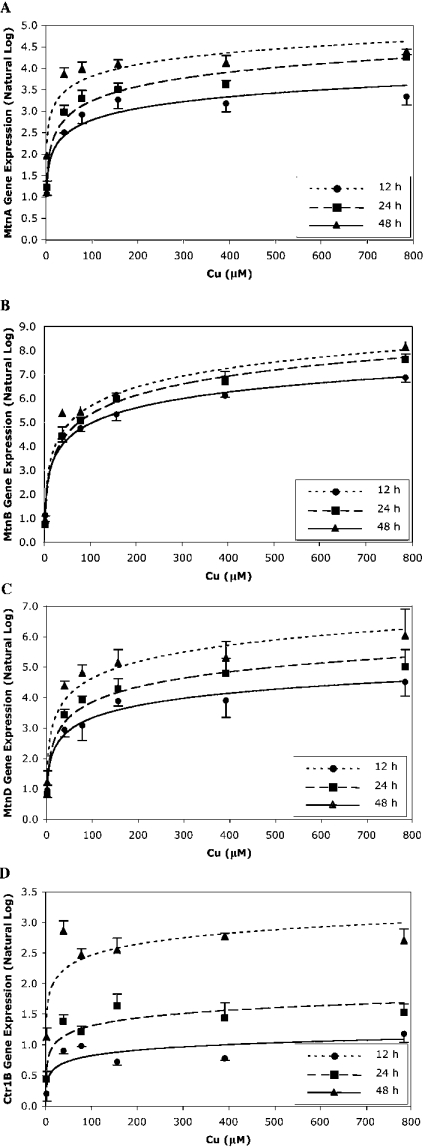
Orthologues of the mammalian Cu chaperones ATOX1, CCS, COX17 and SCO1 and SCO2 did not significantly respond transcriptionally to increased Cu levels. As expected, the expression of MtnA, MtnB and MtnD genes in S2 cells was significantly increased after Cu exposure (Table 1). Drosophila Mtn expression has been shown to be temporally and tissue-specifically induced by Cu [13–15]. To explore the Mtn gene expression profile further, we exposed S2 cells to various Cu concentrations for 12, 24 and 48 h and measured gene expression with real-time PCR (Figures 2A–2C). Gene expression increased in a time- and dose-dependent manner for all three Mtn genes, although the expression of MtnB and MtnD increased more than that of MtnA at all time points. Interestingly, under these Cu conditions, Ctr1B gene expression also increased in a time- and dose-dependent manner (Figure 2D). In contrast, Ctr1A gene expression did not change after Cu exposure (results not shown). To the best of our knowledge, this is the first time a Ctr1 gene has been shown to be up-regulated by Cu. Studies using Drosophila larvae found that Ctr1B gene expression increased after Cu depletion and decreased with excess Cu [10] and similar results are seen in yeast [30].
Figure 2. Metallothionein gene expression in S2 cells after Cu exposure.
S2 cells were exposed to basal media and 2, 39, 79, 157, 393 and 785 μM Cu for 12, 24 and 48 h, and then MtnA (A), MtnB (B), MtnD (C) and Ctr1B (D) gene expressions were measured with real-time PCR. Expression was normalized to Actin42A and expressed relative to cells exposed to basal media. Data were loge-transformed and values are the means±S.E.M. of three replicates. The gene expression profiles of MtnA, MtnB, MtnD and Ctr1B were significantly related to Cu exposure concentration and time of Cu exposure, as determined from a general linear model univariate analysis, P<0.05.
To investigate the role of Cu homoeostasis genes in S2 cells, we utilized dsRNAi to suppress gene expression of Ctr1A, Ctr1B, MTF-1 and DmATP7. S2 cells were treated with dsRNA for 48 h before analysis. Real-time PCR was used to measure the gene expression, which was expressed as a percentage of that in control cells treated with non-specific dsRNA. Mean Ctr1A gene expression was reduced to 9.9% of control (S.E.M.=2.8%), Ctr1B was reduced to 15.0% (S.E.M.=4.6%), MTF-1 was reduced to 24.1% (S.E.M.=2.6%) and DmATP7 was reduced to 6.9% (S.E.M.=2.0%). Suppression was maintained at these levels for at least 4 days and often up to 7 days or more (results not shown). Similar dsRNAi experiments in S2 cells have also shown gene suppression that lasted over 7 days [21].
Ctr1A and Ctr1B transport Cu into S2 cells
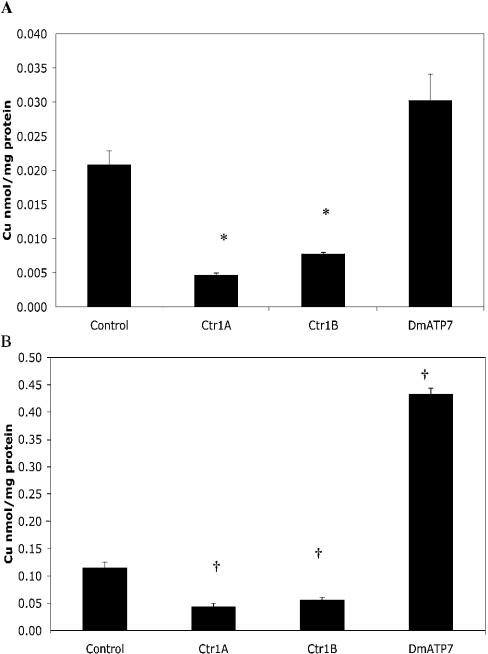
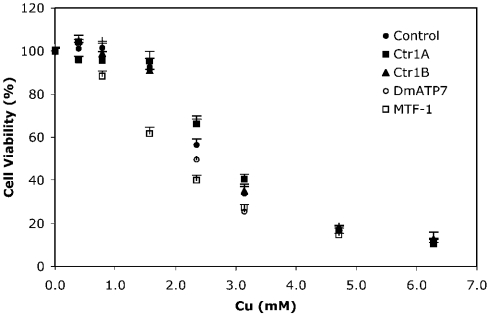
The expression of Ctr1A and Ctr1B was suppressed using dsRNAi, and Cu accumulation in S2 cells was then determined after exposure to 2 μM Cu. As expected, both Ctr1A- and Ctr1B-dsRNAi-treated cells took up significantly less Cu than control cells over 1 h (Figure 3A), and when exposed to 2 μM Cu for 24 h, only accumulated half as much Cu as control cells (Figure 3B). Suppression of each Ctr1 gene did not affect the gene expression of the other (results not shown), suggesting that the unaffected Ctr1 could function to transport Cu into the cell. However, it appears that both Ctr1A and Ctr1B are necessary for maximal Cu accumulation, under physiological Cu conditions. Cu tolerance was not significantly affected by suppression of either Ctr1A or Ctr1B (Figure 4). Unfortunately, attempts to use dsRNAi to suppress both Ctr1A and Ctr1B simultaneously were unsuccessful; hence it was not possible to determine if there are other mechanisms of Cu uptake active in these cells.
Figure 3. Cu accumulation in S2 cells after dsRNAi treatment.
S2 cells were pretreated with Ctr1A, Ctr1B, DmATP7 or control dsRNA. (A) Cells were exposed to 2 μM Cu-labelled with 64Cu, and intracellular Cu was measured after 1 h. Cu was normalized to total cellular protein. Values are means±S.E.M. of nine replicates from three independent experiments. Data were normally distributed and a one-way ANOVA with Games–Howell post-hoc test was used to determine significant differences. *P<0.05 compared with control. (B) Cells were exposed to 2 μM Cu labelled with 64Cu, and intracellular Cu was measured after 24 h. Cu was normalized to total cellular protein. Values are means±S.E.M. of nine replicates from three independent experiments. Data were not normally distributed and a Mann–Whitney test was used to determine significant differences. †P<0.05 compared with control.
Figure 4. S2 cell viability after dsRNAi treatment.
S2 cells were pretreated with MTF-1, DmATP7, Ctr1A, Ctr1B or control dsRNA. Cell viability was measured with an MTT assay after 48 h exposure to 0.2, 0.4. 0.8, 1.6, 2.4, 3.2, 4.8 and 6.4 mM Cu and expressed as a percentage of viable cells compared with those exposed to basal media. Values are means±S.E.M. of six replicates from two independent experiments. Viability was significantly dependent on Cu exposure concentration and different between groups as determined from a general linear model univariate analysis; P<0.05. A Dunnett post-hoc test found that viability of MTF-1-treated cells was significantly different from control cells; P<0.05.
MTF-1 suppression reduces Cu tolerance
Cu tolerance was examined after suppression of MTF-1. The viability of treated cells was significantly lower than that of control when exposed to Cu (Figure 4), probably due to an inhibition of Cu-induced Mtn expression. To test this hypothesis, we exposed MTF-1-dsRNAi-treated cells to basal media and 0.8 mM Cu and measured the expression of MTF-1 and the Mtn genes. Relative to basal conditions, Cu significantly increased expression of MtnA, MtnB and MtnD in control cells; however, suppression of MTF-1 significantly reduced this response (Table 2). After MTF-1 suppression, MtnA, MtnB and MtnD expressions were also reduced under basal conditions, demonstrating an additional role for this transcription factor in the absence of increased Cu. Expression of the Mtn genes tended to increase slightly in MTF-1-dsRNAi-treated cells after Cu exposure, which is probably due to the ability of residual MTF-1 protein to induce some Mtn expression. Alternatively, there may be another, as yet unidentified mechanism capable of inducing a limited amount of Mtn expression in response to Cu.
Table 2. Gene expression in S2 cells after MTF-1 dsRNAi treatment.
S2 cells were pretreated with MTF-1 or control dsRNA and then exposed to either basal media or 0.8 mM Cu for 24 h. MTF-1, MtnA, MtnB and MtnD gene expressions were measured with real-time PCR and normalized to Actin42A expression. Values were expressed relative to control cells exposed to basal media and are the means±S.E.M. of four replicates from two independent experiments. MTF-1, MtnA and Ctr1B gene expression were normally distributed and a one-way ANOVA with Games–Howell post-hoc test was used to determine significant differences. *P<0.05 compared with control cells exposed to basal media. †P<0.05 compared with MTF-1 cells exposed to basal media. MtnB and MtnD gene expression were not normally distributed and a Mann–Whitney test was used to determine significant differences. ‡P<0.05 compared with control cells exposed to basal media. §P<0.05 compared with MTF-1 cells exposed to basal media.
| dsRNA | Cu (mM) | MTF-1 | MtnA | MtnB | MtnD | Ctr1B |
|---|---|---|---|---|---|---|
| Control | Basal | 1.00±0.05 | 1.00±0.04 | 1.00±0.04 | 1.00±0.10 | 1.00±0.03 |
| Control | 0.8 | 1.87±0.05* | 5.79±0.23* | 402±44.6† | 1326±156† | 2.71±0.11* |
| MTF-1 | Basal | 0.24±0.01* | 0.20±0.01* | 0.18±0.02† | 0.34±0.05† | 0.32±0.05* |
| MTF-1 | 0.8 | 0.26±0.04* | 1.51±0.14*‡ | 3.43±0.27†§ | 6.54±1.29†§ | 0.60±0.08* |
Interestingly, MTF-1 suppression also reduced the basal and Cu-induced expressions of Ctr1B (Table 2). Ctr1B contains several copies of the metal-response elements that are bound by MTF-1, suggesting that this transcription factor may regulate Ctr1B as well as the Mtns [10]. Studies in Drosophila have shown that Ctr1B [10] and MTF-1 [15] knockout flies are sensitive to both increased and decreased Cu levels. Zhou et al. [10] proposed that sensitivity of MTF-1 null flies to low Cu levels may be due to loss of Ctr1B regulation and that sensitivity of Ctr1B null flies to high Cu levels may be due a potential role of this protein in Cu detoxification. This is supported by studies with yeast Ctr1 and human Ctr1 that show that excess Cu stimulates the endocytosis of these proteins [31,32]. If Ctr1B translocates in response to Cu it would be very interesting to explore whether this occurs to prevent excess Cu uptake or to sequester and detoxify intracellular Cu actively. The MTF-1-mediated up-regulation of Ctr1B expression found in S2 cells exposed to Cu supports an active detoxification role for Ctr1B, where this protein may mediate a sequestration of excess Cu.
DmATP7 is required for Cu efflux from S2 cells
Cu accumulation and retention studies with wild-type S2 cells showed that these cells were capable of effluxing Cu (Figure 1A). Given that bioinformatic analysis suggests that Drosophila has only one putative Cu translocating P-type ATPase (Table 1), it seemed likely that this protein is responsible for Cu efflux from S2 cells, similar to the role of ATP7A and ATP7B in mammalian cells [20]. Cu accumulation was examined in DmATP7-dsRNAi-treated cells. After exposure to 2 μM Cu for 1 h, cells did not show a significant increase in Cu accumulation (Figure 3A); however, 24 h exposure to Cu resulted in a 4-fold increase in Cu accumulation (Figure 3B). These results demonstrate that DmATP7 is required for efflux of Cu from S2 cells.
Suppressing DmATP7 gene expression did not significantly affect S2 cell viability in response to Cu (Figure 4), suggesting that it may not be the major copper detoxification mechanism. It is possible that a complete knockout of DmATP7 would cause reduced tolerance to increases in copper; however, the level of DmATP7 gene suppression in the present study was sufficient to increase intracellular Cu levels significantly without any significant effect on cell viability. Since wild-type S2 cells continue to accumulate Cu when exposed to increased concentrations, it is probable that the excess intracellular Cu must be efficiently sequestered to prevent cellular damage from reactive oxygen species. Unfortunately, we were not able to suppress the expression of both DmATP7 and MTF-1 with dsRNAi simultaneously. We predict that S2 cells lacking significant levels of DmATP7 and MTF-1 would be extremely sensitive to Cu exposure, as there does not appear to be another P-type ATPase for Cu efflux, and Mtn gene expression could not be significantly induced by Cu in S2 cells with reduced MTF-1 expression (Table 2).
We have examined mechanisms of Cu homoeostasis in Drosophila S2 cells and found that they not only accumulate and export Cu in a comparable way to mammalian cells, but also there is an expression of orthologues of mammalian genes involved in Cu uptake, transport, sequestration and efflux. Although both Ctr1A and Ctr1B are involved in Cu uptake, Cu-induced expression of Ctr1B is mediated by MTF-1, suggesting a dual role in Cu uptake and detoxification. Sequestration appears to be the primary method of Cu detoxification in S2 cells, as preventing induction of Mtn expression by suppressing MTF-1 reduces copper tolerance. However, DmATP7 is also important, as it is essential for efflux of excess Cu. This work is consistent with previous studies in mammalian cells and validates S2 cells as an efficient and malleable system for further exploring mechanisms of Cu homoeostasis.
Acknowledgments
This work was supported by grants from the International Copper Association and The Australian Institute of Nuclear Science and Engineering. R. B. was a recipient of a Peter Doherty Fellowship from the National Health and Medical Research Council and currently receives a J. N. Peters Bequest Fellowship from the University of Melbourne. MTF-1 cDNA was a gift from Professor Walter Schaffner (Institute of Molecular Biology, University of Zürich, Zürich, Switzerland). Ian Gordon from the Statistical Consulting Centre of the University of Melbourne is acknowledged for statistical assistance.
References
- 1.Linder M. C., Hazegh-Azam M. Copper biochemistry and molecular biology. Am. J. Clin. Nutr. 1996;63:797S–811S. doi: 10.1093/ajcn/63.5.797. [DOI] [PubMed] [Google Scholar]
- 2.Halliwell B., Gutteridge J. M. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camakaris J., Voskoboinik I., Mercer J. F. Molecular mechanisms of copper homeostasis. Biochem. Biophys. Res. Commun. 1999;261:225–232. doi: 10.1006/bbrc.1999.1073. [DOI] [PubMed] [Google Scholar]
- 4.Zhou B., Gitschier J. hCTR1: a human gene for copper uptake identified by complementation in yeast. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7481–7486. doi: 10.1073/pnas.94.14.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Culotta V. C., Klomp L. W., Strain J., Casareno R. L., Krems B., Gitlin J. D. The copper chaperone for superoxide dismutase. J. Biol. Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 6.Amaravadi R., Glerum D. M., Tzagoloff A. Isolation of a cDNA encoding the human homolog of COX17, a yeast gene essential for mitochondrial copper recruitment. Hum. Genet. 1997;99:329–333. doi: 10.1007/s004390050367. [DOI] [PubMed] [Google Scholar]
- 7.Klomp L. W., Lin S. J., Yuan D. S., Klausner R. D., Culotta V. C., Gitlin J. D. Identification and functional expression of HAH1, a novel human gene involved in copper homeostasis. J. Biol. Chem. 1997;272:9221–9226. doi: 10.1074/jbc.272.14.9221. [DOI] [PubMed] [Google Scholar]
- 8.Ghoshal K., Jacob S. T. Regulation of metallothionein gene expression. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:357–384. doi: 10.1016/s0079-6603(00)66034-8. [DOI] [PubMed] [Google Scholar]
- 9.Voskoboinik I., Camakaris J., Mercer J. F. Understanding the mechanism and function of copper P-type ATPases. Adv. Protein Chem. 2002;60:123–150. doi: 10.1016/s0065-3233(02)60053-1. [DOI] [PubMed] [Google Scholar]
- 10.Zhou H., Cadigan K. M., Thiele D. J. A copper-regulated transporter required for copper acquisition, pigmentation, and specific stages of development in Drosophila melanogaster. J. Biol. Chem. 2003;278:48210–48218. doi: 10.1074/jbc.M309820200. [DOI] [PubMed] [Google Scholar]
- 11.Lee J., Petris M. J., Thiele D. J. Characterization of mouse embryonic cells deficient in the ctr1 high affinity copper transporter. Identification of a Ctr1-independent copper transport system. J. Biol. Chem. 2002;277:40253–40259. doi: 10.1074/jbc.M208002200. [DOI] [PubMed] [Google Scholar]
- 12.Garrett S. H., Somji S., Todd J. H., Sens M. A., Sens D. A. Differential expression of human metallothionein isoform I mRNA in human proximal tubule cells exposed to metals. Environ. Health Perspect. 1998;106:825–832. doi: 10.1289/ehp.98106825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mokdad R., Debec A., Wegnez M. Metallothionein genes in Drosophila melanogaster constitute a dual system. Proc. Natl. Acad. Sci. U.S.A. 1987;84:2658–2662. doi: 10.1073/pnas.84.9.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lastowski-Perry D., Otto E., Maroni G. Nucleotide sequence and expression of a Drosophila metallothionein. J. Biol. Chem. 1985;260:1527–1530. [PubMed] [Google Scholar]
- 15.Egli D., Selvaraj A., Yepiskoposyan H., Zhang B., Hafen E., Georgiev O., Schaffner W. Knockout of ‘metal-responsive transcription factor’ MTF-1 in Drosophila by homologous recombination reveals its central role in heavy metal homeostasis. EMBO J. 2003;22:100–108. doi: 10.1093/emboj/cdg012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozen S., Skaletsky H. J. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S., Misener S., editors. Totowa: Humana Press; 2000. pp. 365–386. [Google Scholar]
- 17.Lorentzos P., Kaiser T., Kennerson M. L., Nicholson G. A. A rapid and definitive test for Charcot-Marie-Tooth 1A and hereditary neuropathy with liability to pressure palsies using multiplexed real-time PCR. Genet. Test. 2003;7:135–138. doi: 10.1089/109065703322146821. [DOI] [PubMed] [Google Scholar]
- 18.Worby C. A., Simonson-Leff N., Dixon J. E. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Science STKE 2001(95) 2001:PL1. doi: 10.1126/stke.2001.95.pl1. [DOI] [PubMed] [Google Scholar]
- 19.La Fontaine S., Firth S. D., Lockhart P. J., Brooks H., Parton R. G., Camakaris J., Mercer J. F. Functional analysis and intracellular localization of the human Menkes protein (MNK) stably expressed from a cDNA construct in Chinese hamster ovary cells (CHO-K1) Hum. Mol. Genet. 1998;7:1293–1300. doi: 10.1093/hmg/7.8.1293. [DOI] [PubMed] [Google Scholar]
- 20.Camakaris J., Petris M. J., Bailey L., Shen P., Lockhart P., Glover T. W., Barcroft C., Patton J., Mercer J. F. Gene amplification of the Menkes (MNK; ATP7A) P-type ATPase gene of CHO cells is associated with copper resistance and enhanced copper efflux. Hum. Mol. Genet. 1995;4:2117–2123. doi: 10.1093/hmg/4.11.2117. [DOI] [PubMed] [Google Scholar]
- 21.Radyuk S. N., Sohal R. S., Orr W. C. Thioredoxin peroxidases can foster cytoprotection or cell death in response to different stressors: over- and under-expression of thioredoxin peroxidase in Drosophila cells. Biochem. J. 2003;371:743–752. doi: 10.1042/BJ20021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawai K., Liu S. X., Tyurin V. A., Tyurina Y. Y., Borisenko G. G., Jiang J. F., St Croix C. M., Fabisiak J. P., Pitt B. R., Kagan V. E. Antioxidant and antiapoptotic function of metallothioneins in HL-60 cells challenged with copper nitrilotriacetate. Chem. Res. Toxicol. 2000;13:1275–1286. doi: 10.1021/tx000119l. [DOI] [PubMed] [Google Scholar]
- 23.Freedman J. H., Weiner R. J., Peisach J. Resistance to copper toxicity of cultured hepatoma cells. Characterization of resistant cell lines. J. Biol. Chem. 1986;261:11840–11848. [PubMed] [Google Scholar]
- 24.Czaja M. J., Weiner F. R., Freedman J. H. Amplification of the metallothionein-1 and metallothionein-2 genes in copper-resistant hepatoma cells. J. Cell. Physiol. 1991;147:434–438. doi: 10.1002/jcp.1041470308. [DOI] [PubMed] [Google Scholar]
- 25.Petris M. J., Camakaris J., Greenough M., LaFontaine S., Mercer J. F. A C-terminal di-leucine is required for localization of the Menkes protein in the trans-Golgi network. Hum. Mol. Genet. 1998;7:2063–2071. doi: 10.1093/hmg/7.13.2063. [DOI] [PubMed] [Google Scholar]
- 26.Voskoboinik I., Strausak D., Greenough M., Brooks H., Petris M., Smith S., Mercer J. F., Camakaris J. Functional analysis of the N-terminal CXXC metal-binding motifs in the human Menkes copper-transporting P-type ATPase expressed in cultured mammalian cells. J. Biol. Chem. 1999;274:22008–22012. doi: 10.1074/jbc.274.31.22008. [DOI] [PubMed] [Google Scholar]
- 27.FlyBase Consortium. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 2003;31:172–175. doi: 10.1093/nar/gkg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutsenko S., Petris M. J. Function and regulation of the mammalian copper-transporting ATPases: insights from biochemical and cell biological approaches. J. Membr. Biol. 2002;191:1–12. doi: 10.1007/s00232-002-1040-6. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labbe S., Zhu Z., Thiele D. J. Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J. Biol. Chem. 1997;272:15951–15958. doi: 10.1074/jbc.272.25.15951. [DOI] [PubMed] [Google Scholar]
- 31.Ooi C. E., Rabinovich E., Dancis A., Bonifacino J. S., Klausner R. D. Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 1996;15:3515–3523. [PMC free article] [PubMed] [Google Scholar]
- 32.Petris M. J., Smith K., Lee J., Thiele D. J. Copper-stimulated endocytosis and degradation of the human copper transporter, hCtr1. J. Biol. Chem. 2003;278:9639–9646. doi: 10.1074/jbc.M209455200. [DOI] [PubMed] [Google Scholar]