Abstract
The histone-like HU protein is ubiquitous in the eubacteria. A role for Escherichia coli HU in compaction of the bacterial genome has been reported, along with regulatory roles in DNA replication, transposition, repair and transcription. We show here that HU from the human pathogen Helicobacter pylori, which has been implicated in the development of ulcers and gastric cancer, exhibits enhanced thermal stability and distinct DNA substrate specificity. Thermal denaturation of HpyHU (H. pylori HU) measured by CD spectroscopy yields a melting temperature (Tm) of 56.4±0.1 °C. HpyHU binds linear duplex DNA with a site size of ∼19 bp and with low affinity, but in striking contrast to E. coli HU, HpyHU has only modest preference for DNA with mismatches, nicks or gaps. Instead, HpyHU binds stably to four-way DNA junctions with half-maximal saturation of 5 nM. Substitution of two residues adjacent to the DNA-intercalating prolines attenuates both the preference for flexible DNA and the ability to bend and supercoil DNA. These observations suggest that proline intercalation generates hinges that must be stabilized by adjacent residues; insufficient stabilization leads to reduced bending and a failure to bind preferably to DNA with flexure points, such as gaps and mismatches.
Keywords: bacterial chromatin, Helicobacter pylori, histone-like HU protein, Holliday junction, thermal denaturation, type II DNA-binding protein
Abbreviations: EMSA, electrophoretic mobility-shift assay; HBsu, Bacillus subtilis HU; HpyHU, Helicobacter pylori HU; HpyHU-RN, HpyHU with Lys62→Arg and Val63→Asn; IHF, integration host factor; Tm, melting temperature
INTRODUCTION
Helicobacter pylori infection is associated with gastritis, peptic ulcer disease or gastric cancer [1]. H. pylori, which colonizes the human gastrointestinal tract, shows significant genetic diversity, reflected in sequence variations within otherwise well-conserved genes and by the presence of non-conserved genes, mobile genetic elements and chromosomal rearrangements [2–4]. Specialized to live in a single environment, H. pylori has a small genome (1.67 megabases), encoding a minimal set of metabolic genes, including specialized factors required for colonization and survival [5–7].
Bacterial genomes are compacted by association with histone-like proteins in a complex known as bacterial chromatin [8]. Most thoroughly characterized in Escherichia coli, the proteins primarily associated with the DNA are H-NS, Fis, HU and the HU homologue IHF (integration host factor), all of which are present at concentrations up to or even exceeding 10 μM [9]. HU homologues are ubiquitous, but proteins with homology to E. coli Fis or H-NS are absent in many eubacteria, including H. pylori. HU may therefore be primarily responsible for genomic compaction and for specific regulatory functions in such organisms. Consistent with this notion, inactivation of the HU genes in Bacillus subtilis and in Pseudomonas putida was shown to be lethal [10,11].
E. coli HU binds non-specifically and with micromolar affinity to ∼9 bp sites in duplex DNA, and with >100-fold higher affinity to cruciform DNA, specific DNA structures induced by supercoiling, and DNA with nicks and gaps [12–16]. Although it is known that E. coli HU acts as an accessory factor in many processes, such as regulation of DNA supercoiling, recombination and repair [17,18], the molecular mechanisms whereby it executes these functions remain incompletely understood. HU proteins are dimeric, usually comprising 90–99 amino acid subunits forming a compact core of intertwined monomers from which two β-strands extend to embrace a DNA helix [19–21]. The structures of E. coli IHF or Anabaena HU in complex with DNA show that highly conserved proline residues at the tips of these β-strands mediate two sharp DNA kinks. In the IHF–DNA structure, an ∼160° DNA bend is generated by the two proline residues intercalating between specific DNA base pairs, whereas Anabaena HU introduces a range of bend angles [22,23]. Binding properties of HU homologues vary, with binding sites of 9–37 bp and affinities for duplex DNA between 5 nM and 2.5 μM [14,24–29].
H. pylori experiences changes in the internal milieu that include transient pH fluctuations [4,6,7]. As it is predicted to play essential roles in regulation of nucleoprotein-complex formation, HpyHU (H. pylori HU) would be expected to tolerate such changing conditions. Curiously, HpyHU was also shown to be among 13 proteins that are released from H. pylori by mechanisms other than non-specific lysis, and presumed to contribute to gastric inflammation and epithelial damage [30]. We show here that HpyHU is folded under pH conditions encountered in vivo and exhibits greater thermal stability compared with orthologues from other mesophiles. Whereas HpyHU shares certain properties with E. coli HU, such as the ability to introduce negative DNA supercoils, its DNA substrate specificity is distinct: it engages a longer DNA duplex, and it does not bind with significant preference to DNA with nicks, gaps and mismatches, but only to four-way DNA junctions. Our results suggest that the proline-mediated DNA bends must be stabilized by adjacent residues, and that insufficient stabilization correlates with reduced bending and a failure to bind preferably to DNA with flexure points such as gaps and mismatches.
EXPERIMENTAL
Cloning, overexpression and purification of HpyHU and HpyHU-RN (HpyHU with Lys62→Arg and Val63→Asn)
The gene encoding HpyHU was amplified from H. pylori genomic DNA (reference type strain N.C.T.C. 11637). Primers, designed according to the genomic DNA sequence for H. pylori strain J99, were used to amplify a 512 bp product using a mixture of Taq and Pfu polymerases. The sequence, confirmed using two independently obtained PCR products, is available from GenBank® under accession number AY255799. The 512 bp PCR product was used as template to amplify a 355 bp product, using primers modified to introduce NdeI sites at both ends of the PCR product (primer sequences available on request). The PCR product was digested with NdeI and cloned into pET5a, generating plasmid pET-HpyHU. Integrity of the construct was confirmed by sequencing. Plasmid pET-HpyHU was transformed into E. coli strain BL21(DE3)pLysS, and overexpression was initiated by addition of IPTG (isopropyl β-D-thiogalactoside) to a final concentration of 1 mM. Cells were harvested 2 h after induction.
HpyHU-RN was generated by PCR amplification of plasmid pET-HpyHU, using a forward primer (5′-CAAAGAAGGTAGA-AACCCAGGAAGCG-3′) designed to introduce the Lys→Arg and Val→Asn substitutions at positions 62 and 63 of HpyHU (converting the original AAA and GTG codons to AGA and AAC respectively; underlined) and a reverse primer positioned to abut the forward primer (5′-CCTTTTTGCTCTGCGGTTTCAAATTTG-3′). The PCR reaction, carried out with a mixture of Taq and Pfu polymerase, generated full-length plasmid with the mutated HpyHU gene. The original pET-HpyHU template was removed by DpnI digestion, and the plasmid harbouring the mutated HpyHU gene was used to transform E. coli BL21(DE3)pLysS. The sequence of the mutated HpyHU gene was confirmed by sequencing. Overexpression was accomplished as described for wild-type HpyHU.
Both proteins were purified as follows: cells were lysed, and lysates were fractionated by ammonium sulphate precipitations, as described previously [31]. The precipitate formed after the final (75%) ammonium sulphate precipitation was dissolved in buffer A (20 mM potassium phosphate, pH 7.0, 50 mM KCl, 5% (v/v) glycerol, 1 mM EDTA, 0.2 mM PMSF and 3.5 mM 2-mercaptoethanol), dialysed against buffer A, and applied to a CM–Sepharose column equilibrated in buffer A. The proteins were eluted with a linear gradient of 50 mM to 1 M KCl in buffer A. Peak fractions were adjusted to 50% saturation with (NH4)2SO4 while stirring and applied to a phenyl-Sepharose column equilibrated in buffer A containing 50% (NH4)2SO4. HU proteins were eluted with a linear gradient of 50% to 0% (NH4)2SO4 and dialysed against buffer A. Purity was ascertained by Coomassie Blue staining of SDS/polyacrylamide gels. Protein concentrations were determined by quantitation of Coomassie Blue-stained SDS/polyacrylamide gels, using the HU homologue TF1 as a standard. Cloning and purification of HBsu (B. subtilis HU) will be described elsewhere.
Glutaraldehyde-mediated crosslinking
HU proteins were diluted in buffer with 10–500 mM NaCl and incubated for 30 min at room temperature (24 °C) in the presence of 0.1% glutaraldehyde; buffers were 20 mM bicine, pH 8.5, phosphate, pH 8.0, 7.0 or 6.0, or acetate, pH 5.2. Reactions were terminated by addition of 2× Laemmli sample buffer and separated by SDS/PAGE (17% gel). Proteins were visualized by Coomassie Blue staining.
CD spectroscopy
CD spectra were recorded on an AVIV Model 202 CD spectrometer. For wavelength scans, the protein concentration was 0.05 mg/ml in 10 mM sodium phosphate buffer, pH 7.0, with 50 mM NaCl. For thermal denaturation, the protein concentration was 0.1 mg/ml in the same buffer, and spectra were recorded from 190 to 250 nm in 1 nm steps at each temperature. Data were collected from 5 °C to 90 °C in 1.5–5° intervals, with the smallest temperature steps (1.5°) in the transition region. Protein was incubated for 6 min with stirring at each temperature in a 1-cm path-length rectangular cuvette with a screw top seal. Reversibility of denaturation was measured to ensure that the system was at thermodynamic equilibrium, with the fraction of native protein recovered calculated from the CD values by linearly extrapolating the pre-transition and post-transition baselines. CD signals at 218, 219, 220 and 221 nm were used for analysis of the thermal denaturation curves which were fitted to a modified form of the van't Hoff equation that simultaneously fits the native and denatured baselines and the transition region to obtain the Tm (melting temperature of DNA) and ΔH° values for denaturation [32]:
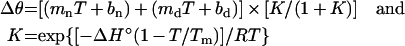 |
where Δθ is the ellipticity, mn and bn are the slope and intercept of the native state baseline, md and bd are the slope and intercept of the denatured state baseline, T is the temperature, ΔH° is the van't Hoff enthalpy, and R is the gas constant. Tm and ΔH° values are reported as the mean of fits to four different wavelengths as a function of temperature. Data were fitted using the program KaleidaGraph.
EMSAs (electrophoretic mobility-shift assays) using agarose gels
Supercoiled or EcoRI-linearized pET5a (50 ng) was mixed with HpyHU in 10 μl of binding buffer (20 mM Tris/HCl, pH 8.0, 50 mM KCl, 0.1 mM EDTA, 0.1 mM dithiothreitol, 0.05% Brij58, 10 μg/ml BSA and 5% glycerol). The entire reaction was loaded on to a 0.5% agarose gel, using either TBE [45 mM Tris/borate, (pH 8.3)/1 mM EDTA] or TAE [40 mM Tris/acetate (pH 8.0)/1 mM EDTA] as the running buffer. DNA was visualized by ethidium bromide staining following electrophoresis.
EMSA and quantitation of protein–DNA complexes
Oligonucleotides used for preparation of 80 bp duplex or 37 bp duplex or duplex with loops, bulge-loops, nicks or gaps and oligonucleotides used for generation of four-way junction DNA were purchased and purified by denaturing PAGE. To generate nicked DNA, two oligonucleotides (3′-GGATCCGATGTGGATGAG-5′ and 3′-AAACATTCTTAATTCGAAG-5′, terminating with free hydroxy groups) were annealed to the 37-nt top strand. To generate DNA with a 1-nt gap, one nucleotide was omitted from the 3′-end of the second complementary strand, while two nucleotides were omitted to generate DNA with a 2-nt gap. Integrity of the nicked/gapped DNA constructs was readily confirmed, as incompletely annealed constructs had distinct electrophoretic mobilities under the conditions used. DNA with two bulge-loops was generated by annealing the 37-nt top strand to 3′-GGATCCGATGTGGACCCTGAGAAACACCCTTCTTAATTCGAAG-5′, in which bulged-out nucleotides are shown in bold. The sequence of oligonucleotides used to generate four-way junctions were as reported in [29]. The sequence of oligonucleotides used to generate the 80 bp duplex are available on request from the authors. DNA was 32P-labelled at the 5′-end with T4 polynucleotide kinase, and equimolar amounts of complementary oligonucleotides were mixed, heated to 90 °C and slowly cooled to 4 °C to form duplex DNA. The four-way junction was prepared by annealing strands 1–4, followed by purification of the junctions on native polyacrylamide gels. A 148 bp duplex was obtained by PCR amplification of a fragment of plasmid pUC18 (primer sequences are available on request from the authors).
EMSAs were performed using 10% (w/v) polyacrylamide gels (39:1, acrylamide/bisacrylamide) in TBE [45 mM Tris/borate (pH 8.3)/1 mM EDTA]. Gels were pre-run for 30 min at 20 mA at room temperature before loading the samples with the power on, except for experiments with bulged, nicked, gapped or four-way junction DNA for which electrophoresis was performed at 4 °C. DNA and protein were mixed in binding buffer, and each sample contained 50–100 fmol of DNA in a total reaction volume of 10 μl, unless specified otherwise. After electrophoresis, gels were dried and protein–DNA complexes were visualized and quantified by phosphorimaging, using software supplied by the manufacturer (ImageQuant 1.1). The region on the gel between complex and free DNA was considered as complex. Complex dissociation during electrophoresis was measured as described previously [31], and the observed fraction of complex F(t) was corrected for dissociation during electrophoresis according to the equation Fcorr=F(t)/[exp(−kdisst)], where kdiss is the exponential decay constant for complex dissociation during electrophoresis and t is time of electrophoresis.
Data were fitted to the equation y=ymax×[P]/([P]1/2+[P]) where [P] is the total protein concentration, [P]1/2 is the protein concentration at half-maximal saturation, and ymax corresponds to maximal saturation. A modified version of the McGhee–von Hippel binding isotherm for non-specific binding that takes into account binding of protein to a finite DNA lattice was also used to evaluate the data, following determination of the non-specific occluded site size by stoichiometric titrations (where [DNA]>Kd; [33,34]). For non-cooperative binding of ligand to a finite DNA lattice of N monomer units (bp), the modified McGhee–von Hippel isotherm is:
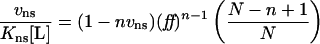 |
(1) |
where Kns is the apparent equilibrium binding constant for non-specific binding to any site, [L] is the free ligand concentration, n is the non-specific site size, vns is the binding density in units of mol bound ligand per mole lattice unit and ff=(1−nvns)/[1−(n−1)vns]. For cooperative binding, the modified isotherm becomes:
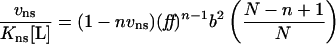 |
(2) |
where
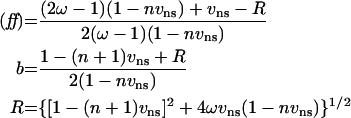 |
and ω is the cooperativity parameter specifying the relative affinity of ligand for a contiguous site vesus an isolated binding site. The measured fractional saturation of DNA lattice θ=nvns and the total ligand concentration LT=L+vnsN. Fits were performed using the program KaleidaGraph, and quality of the fits was evaluated by visual inspection, χ2 values and correlation coefficients. Reported Kd values are apparent values. All experiments were carried out at least in triplicate. The errors are calculated from non-linear fits.
Supercoiling assays
Negatively supercoiled pGEM5 was relaxed by incubation at 37 °C with 4 units of vaccinia topoisomerase I (Epicentre) in 50 mM Tris, pH 8.0, 50 mM NaCl and 0.1 mM EDTA. Increasing concentration of HU protein was added, and the reaction was allowed to continue for 60 min at 37 °C. Reactions were terminated by addition of proteinase K to a final concentration of 0.17 mg/ml and incubation at 37 °C for 1 h. DNA topoisomers were resolved by electrophoresis on 1% agarose gels in TBE at approx. 3 V/cm for 17 h and visualized by ethidium bromide staining. To distinguish positive and negative supercoiling, gels were soaked in chloroquine (3 μg/ml) after electrophoresis in the first dimension, turned 90°, and the DNA resolved by electrophoresis in the second dimension. Gels were stained with ethidium bromide following electrophoresis.
DNA cyclization assays
Plasmid pET5a was digested with BspHI to yield a 105 bp fragment, which was 32P-labelled at the 5′-end with T4 polynucleotide kinase. Ligation experiments with increasing concentrations of HpyHU were performed. Reactions were initiated by addition of 80 units of T4 DNA ligase to a final volume of 10 μl. Reactions containing 100 fmol of DNA and the desired concentration of HU were incubated in 1× binding buffer [20 mM Tris/HCl, pH 8.0, 10 mM MgCl2, 50 mM NaCl, 0.1 mM Na2EDTA, 0.1 mM dithiothreitol, 0.05% (w/v) Brij58 and 100 μg/ml BSA] with 1× ligase buffer (New England Biolabs) and 0.5 mM ATP at room temperature for 60 min and terminated with 5 μl of 75 mM EDTA, 3 mg/ml proteinase K, 15% glycerol, bromophenol blue and xylene cyanol, followed by a 15 min incubation at 55 °C. Cyclized DNA was identified by its resistance to digestion by exonuclease III. Reactions were analysed on 8% polyacrylamide gels [39:1 (w/w), acrylamide/bisacrylamide] in TBE. After electrophoresis, gels were dried and ligation products visualized and quantified by phosphorimaging.
RESULTS
Sequence characteristics of HpyHU
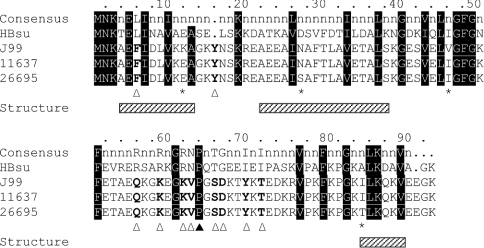
The gene encoding the H. pylori HU homologue, HpyHU, was amplified from genomic DNA (N.C.T.C. number 11637). Comparison with the sequence of HU genes from H. pylori strains J99 and 26695 reveals 18 positions of nucleotide polymorphism that translates into only four positions of amino acid variation (Figure 1). These variable amino acid residues are all found at positions where no overall consensus is present, as defined by alignment of 60 HU homologues [27]. This level of variation is equivalent to the overall level of strain-specific genetic diversity reported based on comparison of the J99 and 26695 complete genomic sequences [35].
Figure 1. Amino acid sequence alignment of HU from H. pylori J99, 11637 and 26695.
The consensus sequence, derived from alignment of 60 HU homologues (with positions of >80% homology identified; [27]), is shown at the top, followed by the sequence of HBsu. Residues are numbered based on the H. pylori HU sequences. Residues that vary between HpyHU J99, 11637 and 26695 sequences are identified with an asterisk. Residues that correspond to the overall consensus sequence are highlighted in black, and HpyHU-specific divergent residues are identified with an open triangle. The DNA-intercalating Pro64 is identified by the closed triangle. Helical segments, based on the structure of B. stearothermophilus HU, are indicated by hatched boxes below the alignment.
In contrast, a comparison of the HpyHU amino acid sequences with the consensus sequence reveals ten positions at which HpyHU sequences differ; at all these positions, the three HU orthologues feature the same divergent residue, signifying a potential adaptation of HpyHU thermodynamic or DNA-binding properties to the unique needs of this organism (Figure 1). Overall similarity of structure is predicted by the conservation of hydrophobic residues in the protein core, and conservation of the DNA-intercalating Pro64 predicts DNA bending by the same mechanism as described for IHF and Anabaena HU [22,23]. The conservative substitution of Phe for Leu in position 6, where helix-1 packs against helix-2 in the body of the protein, may not affect structure or DNA binding significantly. This substitution is also found in the sequence of HU from the closely related pathogen, Campylobacter jejuni [36,37]. In position 16, a tyrosine is inserted in the loop between helices 1 and 2. Increasing the length of this connecting loop may increase its flexibility and thereby optimize helix packing. Other HU homologues have amino acid insertions in this loop, however, it is absent from C. jejuni HU.
Eight substitutions are found between positions 56 and 72 in the β-ribbon arms that engage the DNA duplex (Figure 1). Except for Val63, none of these substitutions are found in C. jejuni HU. Gln56 corresponds to a position where replacement of a conserved arginine with glutamine significantly impairs DNA binding by B. stearothermophilus HU [38]. Substitution of several residues at the tip of the DNA-binding arms, surrounding the DNA-intercalating Pro64, is likely to generate distinct structural and dynamic properties and hence affect DNA binding.
Purification and characterization of HpyHU and HpyHU-RN
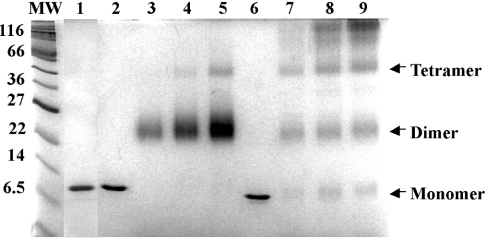
We overexpressed and purified wild-type HpyHU and the HpyHU-RN mutant protein in which Lys62 and Val63 were replaced with Arg and Asn respectively; these residues were chosen as the initial target for mutagenesis as Arg and Asn are almost completely conserved among HU homologues [27]. Our original expectation was, therefore, for the HpyHU-RN mutant protein to exhibit properties akin to those described for other homologues, with wild-type HpyHU possibly exhibiting distinct DNA-binding characteristics. As shown below, this expectation turned out not to be fulfilled. Both proteins were purified to apparent homogeneity, as determined by Coomassie Blue staining of SDS/PAGE gels (Figure 2). Consistent with expectations, HpyHU exists predominantly as a dimer in solution, as shown by glutaraldehyde-mediated crosslinking. However, higher-order oligomeric assemblies, predominant at higher concentrations of HBsu, are not formed by HpyHU, and no residual monomeric protein is seen (Figure 2). This pattern is observed in a pH range of 5.2–8.5, and it is not affected by [NaCl] between 10 and 500 mM (results not shown). By contrast, HBsu is only partially folded at pH 5.2, as shown both by the presence of significant amounts of monomeric protein after treatment with glutaraldehyde and by its mass spectrometry profile [39]. The cross-linking pattern of HpyHU-RN is indistinguishable from that of wild-type HpyHU (results not shown).
Figure 2. HpyHU exists as a dimer in solution.
SDS/PAGE of HpyHU-RN (lane 1), HpyHU (lanes 2–5) and HBsu (lanes 6–9). Lanes 1, 2 and 6 contain 1 μg of unmodified proteins, and lanes 3–5 and 7–9 contain 1, 2 and 3 μg respectively of HpyHU (lanes 3–5) or HBsu (lanes 7–9) cross-linked with glutaraldehyde in bicine (pH 8.5)/50 mM NaCl. With a theoretical pI of 9.05, HpyHU migrates slightly slower than predicted from the calculated molecular mass of 10422 Da. Molecular-mass markers are indicated at the left (in kDa).
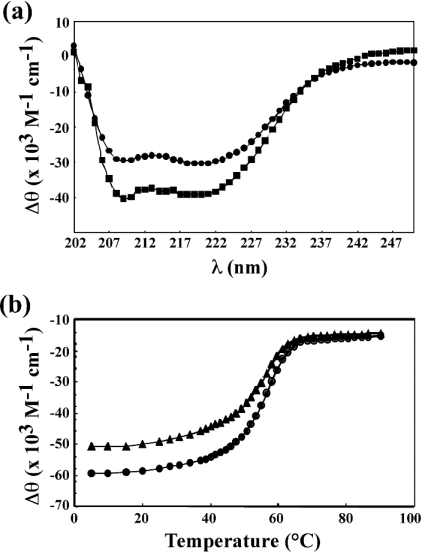
The secondary structure of HpyHU was examined by CD spectroscopy (Figure 3a). Consistent with secondary structure predictions, significant α-helicity is evident. Spectra obtained in 10 mM sodium acetate, pH 5.2, with 50 mM NaCl show that the protein remains folded in the acidic medium. Thermal denaturation was determined at pH 7.0 by recording the ellipticity at 218–221 nm at increasing temperatures, showing a gradual disruption of secondary structure (Figure 3b). HpyHU exhibited 85% reversible denaturation with two successive denaturations yielding a melting temperature Tm of 56.4±0.1 °C and a van't Hoff enthalpy change at Tm of 55.6 kcal/mol−1. Evidently, HpyHU is more thermally stable than HBsu (Tm=48.6 °C in sodium phosphate buffer, pH 7.0, containing 200 mM NaCl; [40]).
Figure 3. Thermal stability of HpyHU.
(a) CD spectrum of HpyHU at room temperature, recorded in phosphate buffer, pH 7.0, (•) or in acetate, pH 5.2, (▪). (b) The ellipticity as a function of temperature. The midpoint of the temperature transition corresponds to the melting temperature Tm. Denaturation curve (•), renaturation curve (▴).
Interaction with plasmid DNA
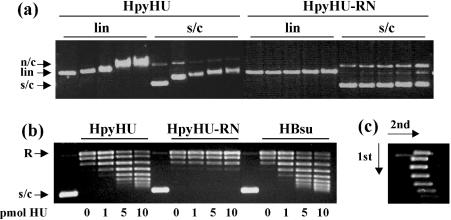
HpyHU binds with modest preference to supercoiled DNA compared with linear DNA (Figure 4a), consistent with reported properties of other HU homologues [15]. Surprisingly, HpyHU-RN binds supercoiled DNA only at higher protein concentrations; evidently, the Lys62→Arg and Val63→Asn substitutions result in a lower DNA-binding affinity, and they attenuate the modest preference for supercoiled DNA seen with wild-type HpyHU. The ability of HpyHU to constrain DNA supercoils was examined (Figure 4b). As expected, HpyHU introduces supercoils into plasmid DNA relaxed with topoisomerase I (left panel). Equivalent concentrations are required compared with HBsu (right panel). However, consistent with its diminished binding to supercoiled DNA, HpyHU-RN is effectively inactive in this assay (middle panel). Even when incubated for 5–30 min with 10-fold higher concentrations of vaccinia topoisomerase I, no supercoiling by HpyHU-RN is detected, while the distribution of supercoiled topoisomers generated by wild-type HpyHU is unaltered (results not shown). These data suggest that the failure of HpyHU-RN to retain negative supercoils is not simply due to a substantially reduced half-life of its complex with DNA. As determined by two-dimensional electrophoresis, HpyHU introduces negative DNA supercoiling (Figure 4c).
Figure 4. Interaction of HpyHU with plasmid DNA.
(a) Agarose gel electrophoresis of HpyHU variants binding to supercoiled (s/c) or linear (lin) DNA. n/c, nicked circular DNA. HpyHU variants are identified at the top, and protein concentrations, identical for all panels, are 0, 50, 100, 200 and 250 nM from left to right. (b) HpyHU introduces DNA supercoiling. Plasmid relaxed with topoisomerase I is supercoiled in the presence of HpyHU (left panel) or HBsu (right panel). HpyHU-RN is nearly inactive (middle panel). Protein concentrations are indicated below the panels. Relaxed (R) and supercoiled (s/c) DNA is indicated on the left. Lanes 1, 6 and 11 (numbered from the left) contain plasmid DNA not treated with topoisomerase I. (c) Two-dimensional electrophoresis of DNA topoisomers generated in the presence of 10 pmol of HpyHU. Negatively supercoiled DNA loses superhelicity in the presence of chloroquine, forming the left branch of the arc. The directions of the first and second dimensions are indicated by arrows.
Affinity for duplex DNA
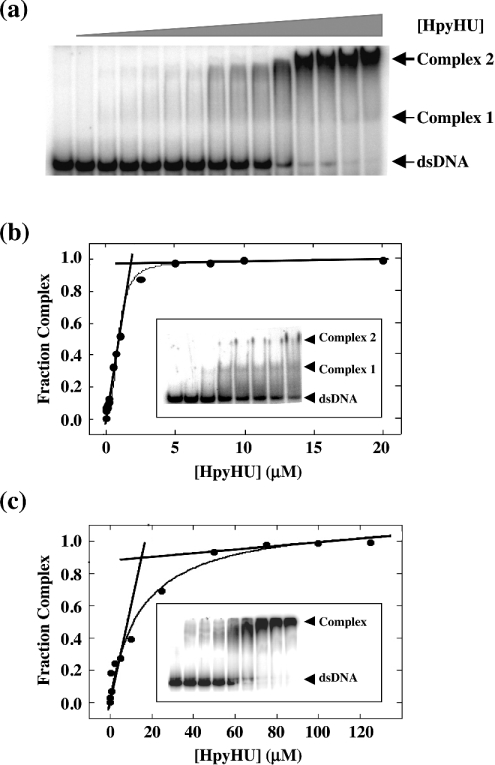
In analogy with HU from E. coli and B. subtilis, HpyHU is expected to bind DNA non-specifically, implying the existence of numerous overlapping sites on the DNA. The modified version of the McGhee–von Hippel binding isotherm for non-specific binding to a finite DNA lattice would therefore be required to evaluate the data [33,34]. Although it is theoretically possible to determine the non-specific site size, n, the observed equilibrium association constant, Ka, and the cooperativity parameter, ω, simultaneously from a three-parameter fit, experimental uncertainty may render such a fit ambiguous. Therefore we determined the occluded site size first by stoichiometric titrations (where [DNA]>Kd). We used both a 37 bp duplex, which represents the longest site size reported for HU homologues and an 80 bp duplex. Using the equation Ka≈1/(n×[P1/2]) to estimate the apparent dissociation constant [41], a site size of 10–20 bp, equivalent to that seen for E. coli or Anabaena HU, would yield a Kd of 350–700 nM for binding to the 37 bp duplex, and [DNA]>1 μM were used for stoichiometric titrations. Under these conditions, a break is seen at the equivalence point, where the molar ratio of lattice residues (base pairs) to ligand concentration is equal to site size (Figure 5). Only two distinct complexes may be seen on 37 bp DNA, immediately suggesting a longer site size compared with E. coli HU which was shown to form three complexes with 29 bp DNA [24]. An occluded site size of 20±1 bp was measured at 50 mM KCl on 37 bp DNA and 19±1 bp on 80 bp DNA (Figure 5c), larger than the site sizes of non-specific binding for E. coli HU (∼9 bp, measured by counting complexes on 21–42 bp duplexes or by fluorescence anisotropy and analytical ultracentrifugation using 13 or 34 bp DNA; [12,42]) or IHF (9–16 bp, depending on salt concentration, measured by isothermal titration calorimetry using 14 bp and 160 bp DNA; [43]), but equivalent to the suggested site size for Anabaena HU (19 bp, estimated from crystallographic data; [23]). At lower salt concentrations (10 mM KCl, the conditions under which E. coli HU forms three distinct complexes on 29 bp DNA), the 37 bp duplex also supports only two complexes with HpyHU (Figure 5b, inset).
Figure 5. Determination of the non-specific site size for HpyHU.
(a) Titration of HpyHU with 1.0 μM perfect duplex 37 bp DNA [25]. Complex and free DNA is identified at the right. [HpyHU], 0–20 μM. (b) Binding isotherm corresponding to the titration shown in (a). The breakpoint corresponds to the molar equivalence between ligand concentration and lattice residues, defining the occluded non-specific site size. The inset shows formation of two distinct complexes with increasing concentration of HpyHU binding to 37 bp duplex at 10 mM KCl. Protein concentrations are 0, 5, 10, 25, 50, 100, 250 and 500 nM from left to right respectively. (c) Binding isotherm for HpyHU binding to 3.5 μM 80 bp DNA. Inset shows representative titration of HpyHU with 80 bp DNA.
The measured value of n=19 bp was used in the modified McGhee–von Hippel equations. Because of overlap binding, available sites become less accessible with increasing saturation of the DNA. Overlap binding therefore is effectively negatively cooperative as it becomes increasingly difficult to bind additional ligands as saturation is approached, and such negative cooperativity may render this binding model less accurate for the shorter duplexes. Binding to a 148 bp duplex was therefore measured. Only complex corresponding to complete saturation of the DNA is sufficiently stable to electrophoresis to yield a discrete band, and multiple bands corresponding to individual HpyHU dimers binding to the DNA probe are not seen (Figure 6a). Whereas fits to the non-cooperative McGhee–von Hippel equation failed to converge (r=0.6875), fits to the cooperative McGhee–von Hippel equation showed modest cooperativity of binding (ω=64±6) and Kd=2.1±0.5 μM (r=0.9947; Figure 6b). This value of Kd is comparable with the 2.5 μM affinity reported for E. coli HU at 200 mM NaCl [14].
Figure 6. Affinity for duplex DNA.
(a) Electrophoretic analysis of HpyHU binding to 148 bp DNA. Protein concentrations are 0–1500 nM. (b) Binding isotherm for HpyHU binding to 148 bp DNA.
HpyHU bends DNA
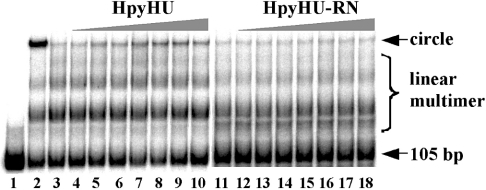
To assess the ability of HpyHU to bend DNA, ligase-mediated cyclization assays were performed. This assay measures the efficiency with which T4 DNA ligase mediates ring closure of DNA fragments that are shorter than the persistence length. As shown in Figure 7, HpyHU mediates cyclization of a 105 bp DNA fragment, although not nearly as effectively as HBsu (lane 2). The HpyHU-RN mutant protein does not mediate DNA cyclization above that seen in absence of HU protein, consistent with its reduced affinity and very limited ability to supercoil DNA. While DNA bending by HpyHU is expected based on properties of other HU proteins, the failure to bend DNA that is characteristic of HpyHU-RN suggests that residues surrounding the DNA-intercalating proline are important for stabilizing the DNA kinks.
Figure 7. HpyHU bends DNA.
105 bp DNA was incubated with T4 DNA ligase for 60 min in the presence of 2 pmol of HBsu (lane 2) or 0–7 pmol of HpyHU respectively (lanes 3–10). The reaction mixture in lane 1 contained no ligase. Less than 10% of total DNA is cyclized at the highest [HpyHU]. HpyHU-RN (1–7 pmol) does not cyclize the 105 bp DNA (lanes 12–18) above that seen in absence of HU (lane 11). DNA was resolved on an 8% polyacrylamide gel.
HpyHU binds preferentially to four-way junction DNA
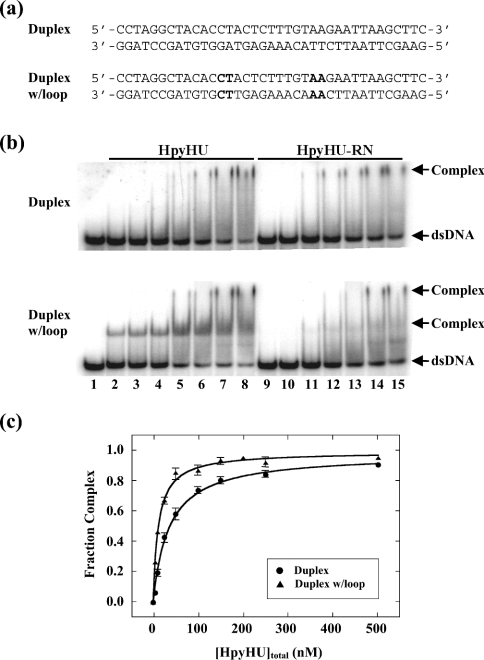
Binding to 37 bp perfect duplex DNA was compared with loop-containing DNA in which two 4-nt loops are placed in the DNA, symmetrically disposed about the centre and with a spacing of 9 bp (Figure 8a); this DNA construct was shown to serve as a preferred substrate for other HU homologues, including Thermotoga maritima HU (which has an ∼37 bp site size; [27]) and Anabaena HU (∼19 bp site size; [31]). Whereas complex 1 is very unstable, complex 2, which corresponds to saturation of the probe, migrates as a distinct species (Figure 8b). Introduction of the set of 4-nt loops results in the formation of two complexes with distinct mobility, both of which are unstable during electrophoresis. The midpoint of the binding isotherm for HpyHU binding to perfect duplex DNA (35±3 nM) is only approx. 3-fold higher than for binding to looped DNA (12±1 nM; Figure 8c), indicating that HpyHU binds DNA with increased flexure with only a modest preference. The slower-migrating complex seen on perfect duplex DNA is present at higher protein concentrations, most likely corresponding to non-specific binding of HpyHU on the DNA; for a competition between preferred binding (to DNA loops) and non-specific binding, preferred binding would predominate at [DNA]/[protein]>1, whereas non-specific binding would be expected at [DNA]/[protein]<1. The modestly increased affinity for looped DNA is also seen with 37 bp DNA containing two bulge-loops separated by 9 bp of duplex (results not shown). Bulge-loops, which confer predisposed DNA bends, evidently do not provide optimal complementarity with the HpyHU binding interface to yield stable complex formation. Introduction of a central nick or a 1–2 nt gap (terminating with free hydroxy groups) into the 37 bp duplex does not result in enhanced complex formation compared with perfect duplex, again suggesting a limited ability of HpyHU to engage DNA with greater flexibility stably (results not shown).
Figure 8. EMSA of HpyHU binding to 37 bp DNA.
(a) Sequence of the DNA probes. The sequence corresponds to the thymine-containing version of a preferred binding site for the B. subtilis bacteriophage SPO1-encoded HU homologue TF1. Loops are generated by substituting the sequence of the bottom strand to generate tandem mismatches of identical opposing bases. Sequences generating loops are in bold. (b) Titrations of HpyHU (left panel) and HpyHU-RN (right panel) with perfect duplex DNA (upper panel) and loop-containing DNA (lower panel). Complex and free DNA is identified at the right. HpyHU variants are identified at the top, and protein concentrations (identical for both panels) are as follows: Lane 1, no protein; lanes 2–8 and 9–15, reactions with 5, 10, 25, 50, 100, 250, 500 nM protein respectively (from left to right). (c) Binding isotherm for HpyHU binding to 37 bp duplex DNA (•) or duplex with loops (▴), corrected for complex dissociation during electrophoresis.
HpyHU-RN which binds the short duplex DNA comparably with wild-type HpyHU (half-maximal saturation of 50±6 nM; Figure 8b), does not bind markedly better to the looped DNA compared with perfect duplex, indicating that increased flexure at potential sites of kinking fails to stabilize complex formation significantly. The almost complete lack of preference for distorted DNA is consistent with the limited ability of HpyHU-RN to bend and supercoil DNA (Figures 4b and 7). Complex formation measured at pH 6.0 is equivalent to that seen at pH 8.0 (Figure 8 and results not shown).
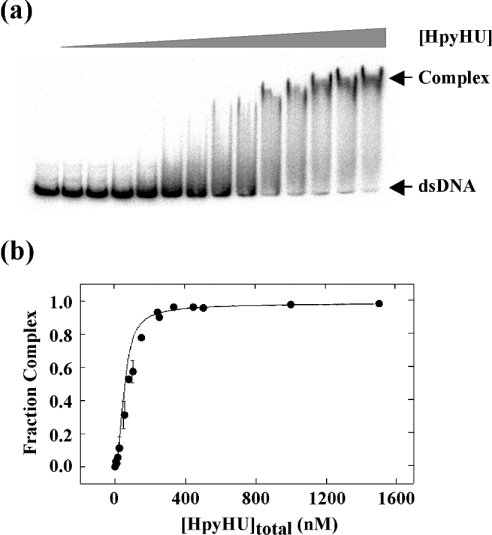
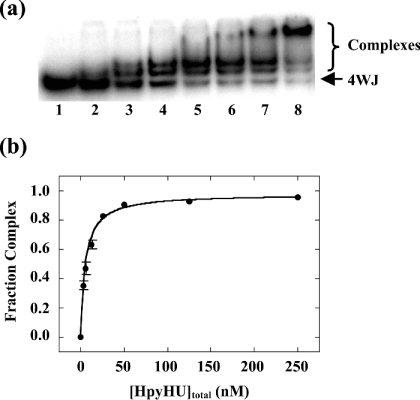
Four-way junction DNA was generated based on the sequence used for analysis of E. coli HU [12]. Two stable HpyHU complexes, whose high mobility suggest a compacted structure, are formed at low protein concentration, and an additional complex is seen at higher concentrations (Figure 9). No evidence of cooperativity of binding was observed. This pattern of complexes is similar to that observed with E. coli HU, where two HU dimers bind to opposite angles of the four-way junction, leading to two complexes at low protein concentrations, with higher-order complexes corresponding to HU binding to the linear branches [12]. HpyHU binds the four-way junction with half-maximal saturation of 5.3±0.5 nM. HpyHU-RN also binds preferentially to the four-way junction, the higher half-maximal saturation of 13.7±2.2 nM, reflecting the already observed reduced binding affinity of the mutant protein (results not shown). The significant preference for the pre-bent DNA construct suggests that only DNA in which the energetic cost of bending has been significantly lessened serves as an optimal substrate for HpyHU.
Figure 9. HpyHU binds preferentially to four-way junction DNA.
Titration of HpyHU with four-way junction DNA. Binding was assayed in the stacked X conformation preferred in the presence of MgCl2 [53]. Complex and free DNA is identified at the right. Protein concentrations are 0, 2.5, 5, 12.5, 25, 50, 125, and 250 nM respectively.
DISCUSSION
Thermal stability of HpyHU
Comparison of HpyHU sequences from three strains reveals several amino acids that differ from the overall consensus (Figure 1). One of these substitutions is an insertion in the loop connecting helices 1 and 2, and this connecting loop also features a glycine in position 14. It was previously shown that loop flexibility correlates with optimal helix packing and thermal stability: substituting Gly15 with glutamic acid in B. stearothermophilus HU reduces the loop flexibility and causes a reduction in Tm from 64 °C to 54 °C, and increasing the loop flexibility through replacement of Glu15 with Gly in HBsu or in the B. subtilis bacteriophage SPO1-encoded HU homologue, TF1, in both cases increases the Tm by 11 °C [40,44,45]. In general, Gly15 is found in HU encoded by thermophilic organisms, such as B. stearothermophilus and Thermus aquaticus, and insertions in this connecting loop are found for instance in HU from Aquifex aeolicus [27]. Kawamura et al. [40] reported a Tm for HBsu of 48.6 °C, measured by CD spectroscopy in phosphate buffer, pH 7.0, with 200 mM NaCl, a Tm that is significantly lower than the 56.4 °C measured for HpyHU under somewhat less stringent conditions (phosphate buffer, pH 7.0, with 50 mM NaCl; Figure 3). Additional reports have been published presenting a Tm for HBsu {33 °C, 39.7 °C and 43 °C respectively [46–48], based on CD spectra recorded in sodium cacodylate, pH 7.5, with 100 mM KF (Tm=33 °C) or under unspecified solution conditions}. On the basis of the correlation between flexibility of the loop connecting helices 1 and 2 and thermal stability, optimized packing mediated by flexibility of this connecting loop is likely to be the reason for the enhanced stability of HpyHU [40,44,45]. We note that the CD spectrum at pH 5.2 shows that HpyHU remains folded at lower pH (Figure 2); under these conditions, HBsu was shown to be partially unfolded and the unfolded monomer unable to dimerize [39]. It may be that the enhanced thermal stability is an accidental consequence of evolutionary pressures to remain folded when exposed to transient increases in intracellular acidity.
Four-way junction DNA is an optimal substrate for HpyHU
HpyHU binds non-specifically and with low affinity to an ∼19 bp DNA site, and its preference for DNA with greater than average flexure is modest (Figure 8). This is in remarkable contrast to E. coli HU which was reported to bind ∼9 bp of linear duplex DNA with at least 100-fold lower affinity compared with DNA with nicks and gaps (Kd=2–8 nM in 200 mM KCl; [13]). Loops or discontinuities in the DNA backbone are considered to increase local flexure, thereby facilitating formation of the protein-mediated DNA kinks [49–51]. However, HpyHU has only modest preference for such flexible sites (Figure 8), suggesting an inability to stabilize the proline-mediated DNA kinks significantly, even when DNA flexure is greater than average. The biological corollary may be that HpyHU in vivo does not discriminate significantly between perfect duplex DNA and DNA with breaks or mismatches, arguing against a role in recognition of substrates for the DNA repair systems. In contrast, HpyHU has significant binding preference for DNA in which the energetic cost of bending has been reduced by pre-bending the DNA (Figure 9). Such properties would be consistent with in vivo roles in stabilization of four-way junction structures or other severely bent DNA conformations. HpyHU introduces negative DNA supercoiling (Figure 4), consistent with a role in compaction of the bacterial genome.
Substrate specificity of HU proteins is determined by their ability to stabilize DNA bends
Upon substitution of Lys62-Val preceding the DNA-intercalating proline with Arg-Asn, DNA binding and bending is attenuated. Notably, preference for DNA with flexure points, such as gaps or mismatches, is also lost, whereas binding to prebent DNA is only modestly reduced. It is conceivable that the HpyHU-specific substitutions within the DNA-binding β-arms render a conformation that is incompatible with a spatial disposition of arginine that permits DNA contacts, and that lysine is specifically required for electrostatic contacts to the DNA. The significant effect of the Lys62-Val→Arg-Asn substitutions on DNA bending and on preference for DNA with imposed flexure suggests that a stabilization of the proline-mediated DNA kinks by adjacent residues is essential. We propose that insufficient stabilization of the DNA kinks leads to a failure to bend duplex DNA and an inability to utilize flexible DNA as a preferred substrate, while retaining a significant preference for pre-bent DNA, as seen for HpyHU-RN. Such properties were recently shown also to characterize the Deinococcus radiodurans-encoded HU homologue, which binds preferably only to four-way junction DNA and which fails to bend DNA [29]. A modest stabilization of the flexible DNA hinges would correlate with preferred binding to more flexible DNA sites, as seen for E. coli and Anabaena HU [12–14,23,31]. The distribution of charged residues in the region surrounding the DNA kink has been previously shown to have a marked effect on affinity and substrate specificity; for example, variants of the SPO1-encoded HU homologue TF1 with serine or glutamine in the position corresponding to Arg62 of the consensus sequence fail to bind DNA unless it is pre-bent [52]. Accordingly, substrate specificity of HU proteins appears to vary as a function of the ability of individual proteins to stabilize a severely bent DNA conformation.
Acknowledgments
We are grateful to C.-C. Liu for assistance with CD measurements and to V. LiCata and members of the Grove laboratory for helpful discussions.
References
- 1.McGowan C. C., Cover T. L., Blaser M. J. Helicobacter pylori and gastric acid: biological and therapeutic implications. Gastroenterology. 1996;110:926–938. doi: 10.1053/gast.1996.v110.pm8608904. [DOI] [PubMed] [Google Scholar]
- 2.Blaser M. J. Genetic basis for Helicobacter pylori diversity. In: Hunt R. E., Tytgat G. N. J., editors. Helicobacter pylori: Basic Mechanisms to Clinical Use. Boston: Kluwer Academic Publishers; 1996. pp. 33–39. [Google Scholar]
- 3.Jiang Q., Hiratsuka K., Taylor D. E. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol. Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 4.Sachs G., Weeks D. L., Melchers K., Scott D. R. The gastric biology of Helicobacter pylori. Annu. Rev. Physiol. 2003;65:349–369. doi: 10.1146/annurev.physiol.65.092101.142156. [DOI] [PubMed] [Google Scholar]
- 5.Tomb J. F., White O., Kerlavage A. R., Clayton R. A., Sutton G. G., Fleischmann R. D., Ketchum K. A., Klenk H. P., Gill S., Dougherty B. A., et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 6.Scott D. R., Weeks D., Hong C., Postius S., Melchers K., Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998;114:58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 7.Wen Y., Marcus E. A., Matrubutahm U., Gleeson M. A., Scott D. R., Sachs G. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 2003;71:5921–5939. doi: 10.1128/IAI.71.10.5921-5939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kellenberger E., Arnold-Schultz-Gahmen B. Chromatins of low-protein-content: special features of their compaction and condensation. FEMS Microbiol. Lett. 1992;100:361–370. doi: 10.1111/j.1574-6968.1992.tb14064.x. [DOI] [PubMed] [Google Scholar]
- 9.Azam T. A., Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. J. Biol. Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 10.Micka B., Marahiel M. A. The DNA-binding protein HBsu is essential for normal growth and development in Bacillus subtilis. Biochimie. 1992;74:641–650. doi: 10.1016/0300-9084(92)90136-3. [DOI] [PubMed] [Google Scholar]
- 11.Bartels F., Fernandez S., Holtel A., Timmis K. N., de Lorenzo V. The essential HupB and HupN proteins of Pseudomonas putida provide redundant and nonspecific DNA-bending functions. J. Biol. Chem. 2001;276:16641–16648. doi: 10.1074/jbc.M011295200. [DOI] [PubMed] [Google Scholar]
- 12.Bonnefoy E., Takahashi M., Rouvière-Yaniv J. DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J. Mol. Biol. 1994;242:116–129. doi: 10.1006/jmbi.1994.1563. [DOI] [PubMed] [Google Scholar]
- 13.Castaing B., Zelwer C., Laval J., Boiteux S. HU protein of Escherichia coli binds specifically to DNA that contains single-strand breaks or gaps. J. Biol. Chem. 1995;270:10291–10296. doi: 10.1074/jbc.270.17.10291. [DOI] [PubMed] [Google Scholar]
- 14.Pinson V., Takahashi M., Rouvière-Yaniv J. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J. Mol. Biol. 1999;287:485–497. doi: 10.1006/jmbi.1999.2631. [DOI] [PubMed] [Google Scholar]
- 15.Kobryn K., Lavoie B. D., Chaconas G. Supercoiling-dependent site-specific binding of HU to naked Mu DNA. J. Mol. Biol. 1999;289:777–784. doi: 10.1006/jmbi.1999.2805. [DOI] [PubMed] [Google Scholar]
- 16.Kamashev D., Balandina A., Rouvière-Yaniv J. The binding motif recognized by HU on both nicked and cruciform DNA. EMBO J. 1999;18:5434–5444. doi: 10.1093/emboj/18.19.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huisman O., Faelen M., Girard D., Jaffe A., Toussaint A., Rouvière-Yaniv J. Multiple defects in Escherichia coli mutants lacking HU protein. J. Bacteriol. 1989;171:3704–3712. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boubrik F., Rouvière-Yaniv J. Increased sensitivity to gamma irradiation in bacteria lacking protein HU. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3958–3962. doi: 10.1073/pnas.92.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka I., Appelt K., Dijk J., White S. W., Wilson S. 3-Å resolution structure of a protein with histone-like properties in prokaryotes. Nature (London) 1984;310:376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- 20.Vis H., Mariani M., Vorgias C. E., Wilson K. S., Kaptein R., Boelens R. Solution structure of the HU protein from Bacillus stearothermophilus. J. Mol. Biol. 1995;254:692–703. doi: 10.1006/jmbi.1995.0648. [DOI] [PubMed] [Google Scholar]
- 21.Jia X., Grove A., Ivancic M., Hsu V. L., Geiduschek E. P., Kearns D. R. Structure of the Bacillus subtilis phage SPO1-encoded type II DNA-binding protein TF1 in solution. J. Mol. Biol. 1996;263:259–268. doi: 10.1006/jmbi.1996.0573. [DOI] [PubMed] [Google Scholar]
- 22.Rice P. A., Yang S. W., Mizuuchi K., Nash H. A. Crystal structure of an IHF–DNA complex: A protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 23.Swinger K. K., Lemberg K. M., Zhang Y., Rice P. A. Flexible DNA bending in HU–DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonnefoy E., Rouvière-Yaniv J. HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein–DNA complexes with short DNA fragments. EMBO J. 1991;10:687–696. doi: 10.1002/j.1460-2075.1991.tb07998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grove A., Galeone A., Mayol L., Geiduschek E. P. Localized DNA flexibility contributes to target site selection by DNA-bending proteins. J. Mol. Biol. 1996;260:120–125. doi: 10.1006/jmbi.1996.0386. [DOI] [PubMed] [Google Scholar]
- 26.Kobryn K., Naigamwall D. Z., Chaconas G. Site-specific DNA binding and bending by the Borrelia burgdorferi Hbb protein. Mol. Microbiol. 2000;37:145–155. doi: 10.1046/j.1365-2958.2000.01981.x. [DOI] [PubMed] [Google Scholar]
- 27.Grove A., Lim L. High-affinity DNA binding of HU protein from the hyperthermophile Thermotoga maritima. J. Mol. Biol. 2001;311:491–502. doi: 10.1006/jmbi.2001.4763. [DOI] [PubMed] [Google Scholar]
- 28.Grove A. Surface salt bridges modulate DNA wrapping by the type II DNA binding protein TF1. Biochemistry. 2003;42:8739–8747. doi: 10.1021/bi034551o. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh S., Grove A. Histone-like protein HU from Deinococcus radiodurans binds preferentially to four-way DNA junctions. J. Mol. Biol. 2004;337:561–571. doi: 10.1016/j.jmb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Kim N., Weeks D. L., Shin J. M., Scott D. R., Young M. K., Sachs G. Proteins released by Helicobacter pylori in vitro. J. Bacteriol. 2002;184:6155–6162. doi: 10.1128/JB.184.22.6155-6162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grove A., Galeone A., Mayol L., Geiduschek E. P. On the connection between inherent DNA flexure and preferred binding of hydroxymethyluracil-containing DNA by the type II DNA-binding protein TF1. J. Mol. Biol. 1996;206:196–206. doi: 10.1006/jmbi.1996.0392. [DOI] [PubMed] [Google Scholar]
- 32.Eftink M. R., Ramsay G. D. Analysis of multidimensional spectroscopic data to monitor unfolding of proteins. Methods Enzymol. 1994;240:615–645. doi: 10.1016/s0076-6879(94)40066-0. [DOI] [PubMed] [Google Scholar]
- 33.McGhee J. D., von Hippel P. H. Theoretical aspects of DNA–protein interactions: cooperative and noncooperative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 34.Tsodikov O. V., Holbrook J. A., Shkel I. A., Record M. T., Jr Analytic binding isotherms describing competitive interactions of a protein ligand with specific and nonspecific sites on the same DNA oligomer. Biophys. J. 2001;81:1960–1969. doi: 10.1016/S0006-3495(01)75847-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alm R. A., Ling L.-S. L., Moir D. T., King B. L., Brown E. D., Doig P. C., Smith D. R., Noonan B., Guild B. C., deJonge B. L., et al. Genome-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature (London) 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 36.Konkel M. E., Marconi R. T., Mead D. J., Cieplak W., Jr Cloning and expression of the hup encoding a histone-like protein of Campylobacter jejuni. Gene. 1994;146:83–86. doi: 10.1016/0378-1119(94)90837-0. [DOI] [PubMed] [Google Scholar]
- 37.Parkhill J., Wren B. W., Mungall K., Ketley J. M., Churcher C., Basham D., Chillingworth T., Davies R. M., Feltwell T., Holroyd S., et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature (London) 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 38.Saitoh S. F., Kawamura S., Yamasaki N., Tanaka I., Kimura M. Arginine-55 in the β-arm is essential for the activity of DNA-binding protein HU from Bacillus stearothermophilus. Biosci. Biotechnol. Biochem. 1999;63:2232–2235. doi: 10.1271/bbb.63.2232. [DOI] [PubMed] [Google Scholar]
- 39.Vis H., Heinemann U., Dobson C. M., Robinson C. V. Detection of a monomeric intermediate associated with dimerization of protein Hu by mass spectrometry. J. Am. Chem. Soc. 1998;120:6427–6428. [Google Scholar]
- 40.Kawamura S., Kakuta Y., Tanaka I., Hikichi K., Kuhara S., Yamasaki N., Kimura M. Glycine-15 in the bend between two α-helices can explain the thermostability of DNA binding protein HU from Bacillus stearothermophilus. Biochemistry. 1996;35:1195–1200. doi: 10.1021/bi951581l. [DOI] [PubMed] [Google Scholar]
- 41.Kowalczykowski S. C., Paul L. S., Lonberg N., Newport J. W., McSwiggen J. A., von Hippel P. H. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- 42.Wojtuszewski K., Hawkins M. E., Cole J. L., Mukerji I. HU binding to DNA: evidence for multiple complex formation and DNA bending. Biochemistry. 2001;40:2588–2598. doi: 10.1021/bi002382r. [DOI] [PubMed] [Google Scholar]
- 43.Holbrook J. A., Tsodikov O. V., Saecker R. M., Record M. T., Jr Specific and non-specific interactions of integration host factor with DNA: thermodynamic evidence for disruption of multiple IHF surface salt-bridges coupled to DNA binding. J. Mol. Biol. 2001;310:379–401. doi: 10.1006/jmbi.2001.4768. [DOI] [PubMed] [Google Scholar]
- 44.Andera L., Spangler C. J., Galeone A., Mayol L., Geiduschek E. P. Interrelations of secondary structure stability and DNA-binding affinity in the bacteriophage SPO1-encoded type II DNA-binding protein TF1. J. Mol. Biol. 1994;236:139–150. doi: 10.1006/jmbi.1994.1124. [DOI] [PubMed] [Google Scholar]
- 45.Liu W., Vu H. M., Geiduschek E. P., Kearns D. R. Solution structure of a mutant of transcription factor 1: implications for enhanced DNA binding. J. Mol. Biol. 2000;302:821–830. doi: 10.1006/jmbi.2000.4084. [DOI] [PubMed] [Google Scholar]
- 46.Welfle H., Misselwitz R., Welfle K., Groch N., Heinemann U. Salt-dependent and protein-concentration-dependent changes in the solution structure of the DNA-binding histone-like protein, HBsu, from Bacillus subtilis. Eur. J. Biochem. 1992;204:1049–1055. doi: 10.1111/j.1432-1033.1992.tb16727.x. [DOI] [PubMed] [Google Scholar]
- 47.Wilson K. S., Vorgias C. E., Tanaka I., White S. W., Kimura M. The thermostability of DNA-binding protein HU from bacilli. Prot. Eng. 1990;4:11–22. doi: 10.1093/protein/4.1.11. [DOI] [PubMed] [Google Scholar]
- 48.Christodoulou E., Vorgias C. E. The thermostability of DNA-binding protein HU from mesophilic, thermophilic and extreme thermophilic bacteria. Extremophiles. 2002;6:21–31. doi: 10.1007/s007920100235. [DOI] [PubMed] [Google Scholar]
- 49.Kahn J. D., Yun E., Crothers D. M. Detection of localized DNA flexibility. Nature (London) 1994;368:163–166. doi: 10.1038/368163a0. [DOI] [PubMed] [Google Scholar]
- 50.Mills J. B., Cooper J. P., Hagerman P. J. Electrophoretic evidence that single-stranded regions of one or more nucleotides dramatically increase the flexibility of DNA. Biochemistry. 1994;33:1797–1803. doi: 10.1021/bi00173a024. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y., Crothers D. M. High-throughput approach for detection of DNA bending and flexibility based on cyclization. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3161–3166. doi: 10.1073/pnas.0530189100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grove A., Figueiredo M. L., Galeone A., Mayol L., Geiduschek E. P. Twin hydroxymethyluracil-A basepair steps define the binding site for the DNA-bending protein TF1. J. Biol. Chem. 1997;272:13084–13087. doi: 10.1074/jbc.272.20.13084. [DOI] [PubMed] [Google Scholar]
- 53.Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]