ABSTRACT
The inflammatory tumor microenvironment (TME) is a key driver for tumor-promoting processes. Tumor-associated macrophages are one of the main immune cell types in the TME and their increased density is related to poor prognosis in prostate cancer. Here, we investigated the influence of pro-inflammatory (M1) and immunosuppressive (M2) macrophages on prostate cancer lineage plasticity. Our findings reveal that M1 macrophage secreted factors upregulate genes related to stemness while downregulating genes associated with androgen response in prostate cancer cells. The expression of cancer stem cell (CSC) plasticity markers NANOG, KLF4, SOX2, OCT4, and CD44 was stimulated by the secreted factors from M1 macrophages. Moreover, AR and its target gene PSA were observed to be suppressed in LNCaP cells treated with secreted factors from M1 macrophages. Inhibition of NFκB signaling using the IKK16 inhibitor resulted in downregulation of NANOG, SOX2, and CD44 and CSC plasticity. Our study highlights that the secreted factors from M1 macrophages drive prostate cancer cell plasticity by upregulating the expression of CSC plasticity markers through NFκB signaling pathway.
KEYWORDS: Cancer cell plasticity, prostate cancer, tumor-associated macrophages
1. Introduction
Tumor microenvironment (TME) exhibits high heterogeneity with the composition of different immune cells, cancer-associated fibroblasts, tumor cells, and extracellular matrix changing during tumor progression.1 An inflammatory TME provides favorable conditions for several pro-tumor processes, such as angiogenesis2 and epithelial–mesenchymal transition (EMT).3
Tumor-associated macrophages (TAMs) are an abundant immune cell population in the inflammatory TME, where they have distinct anti- and protumor functions. They are heterogeneous group of macrophages that have both pro-inflammatory (M1) and immunosuppressive (M2) properties. TAMs are originated from tissue-resident macrophages or circulating monocytes, with their specific phenotype determined by the prevailing cytokines and other mediators in TME.4 M1 macrophages are primarily known to have tumor-suppressive properties, and they secrete a diverse pattern of pro-inflammatory cytokines, including TNFα, IL-1β, IL-6, and CXCL8, which have diverse effects in the TME. Conversely, M2 macrophages release immunosuppressive mediators, such as IL-10 and TGF-β, and are known to have a tumor-promoting role in TME.5 The phenotypic TAM composition within the TME varies during tumor progression and their increased number correlates with angiogenesis and metastasis and thus with poor prognosis in prostate cancer.6
The cellular plasticity of prostate cancer cells plays a central role in the progression of castration-resistant prostate cancer (CRPC) during androgen deprivation therapy (ADT).7 In CRPC, cancer cells are no longer dependent on androgen receptor (AR) signaling and the cells are resistant to androgen inhibitor therapies.8 Additionally, AR downregulation is a key characteristic in the prostate cancer cell plasticity.9 Cancer stem cells (CSC) exhibit increased ability for self-renewal, tumor initiation, and drug resistance.10 Prostate cancer stem-like cells are acknowledged for expressing several stem cell markers, such as transcription factors NANOG, KLF4, SOX2, and OCT411,12 in addition to the cell surface protein CD44.13 These factors are involved in several tumor cell functions, including cell proliferation, pluripotency, tumorigenesis, and therapeutic resistance12,14,15 collectively driving cancer progression.
Several studies have suggested a positive correlation between TAM infiltration and functions promoting prostate cancer progression,16,17 such as tumor angiogenesis, negative response to ADT6 as well as prostate cancer cell migration and invasion.18 While the majority of studies investigating the tumor promoting effects of TAMs in prostate cancer are mainly focused on M2-polarized TAMs, it is noteworthy that part of these TAMs in localized and metastatic prostate tumors exhibit a pro-inflammatory M1 phenotype.19 The influence of these M1 macrophages on tumor progression has gained less attention. In the present study, we investigated the role of pro-inflammatory (M1) and immunosuppressive (M2) macrophages on prostate cancer cell plasticity by studying the alterations in the expression of CSC markers (NANOG, KLF4, SOX2, OCT4, and CD44) and AR.
2. Materials and methods
2.1. Reagents used
The reagents used are described in the Supplementary Materials and Methods
2.2. Cell culture
THP-1 human monocyte, LNCaP, LNCaP-C42B (C42B), 22Rv1, and VCaP prostate cancer cells lines were cultured as described in the Supplementary Materials and Methods.
2.3. THP-1 monocyte differentiation
THP-1 cells were differentiated and polarized to M0, M1, and M2 macrophages and macrophage culture media were collected as described previously.20 Briefly, THP-1 cells were seeded on 10/15 cm culture dish (6.25 × 105 cells/ml) with 10 ng/ml 12-O-tetradecanoylphorbol-13-acetate (PMA) for 24 h. Thereafter, cells were treated with a fresh culture medium for 1 h. M0 macrophages were polarized to M1 macrophages using 15 ng/ml LPS and 20 ng/ml IFNγ or M2 macrophages using 25 ng/ml IL-4 and IL-13 for 48 h. M0 macrophages were cultured with fresh THP-1 culture medium. Media from polarized macrophages were collected and sterile-filtered (0.22 µm, FPE-204-030, Jet-Biofil, Guandong, China or 16532-K, Sartorius, Goettingen, Germany) (=conditioned medium, CM). The polarization of macrophages was verified with qPCR.
2.4. Prostate cancer cell treatments
LNCaP or C42B cells were seeded in culture medium on 6 cm dish, 6-well plate, 8-well µ-slide (Ibidi GmbH, Martinsried, Germany) or 96-well plate (Ibidi GmbH, Martinsried, Germany) (2 × 106, 3 × 105, 4 × 104, and 7.5 × 103 cells/well, respectively). Thereafter, the cells were pre-treated (RPMI containing 1% heat-inactivated FBS, P/S and L-glutamine) for 3 h. The medium was substituted with CM diluted with RPMI (10% heat-inactivated FBS, 1% P/S, L-glutamine) at 1:4 ratio. RPMI containing 10% heat-inactivated FBS, 1% P/S and L-glutamine served as the control treatment. LNCaP cells were exposed to the respective treatment media for 0–72 h.
22Rv1 cells were seeded on 6-well plate, 8-well µ-slide, 96-well plate (3 × 105, 3 × 104, and 7.5 × 103 cells/well, respectively), and VCaP cells were seeded on 8-well µ-slide (3 × 104 cells/well). After 2 days, the treatments were performed similarly as with LNCaP cells. 22Rv1 and C42B cells were treated with 100% CM.
Treatments for inhibitor and AR activation assays, RNA sequencing (RNA-seq), RNA extraction, cDNA synthesis, qPCR, SDS-PAGE, western blotting, proliferation and apoptosis assays, immunofluorescence stainings and microscopy are described in the Supplementary Materials and Methods.
2.5. Statistical analysis
Statistical analyses were performed using GraphPad Prism 9 (GraphPad software, Boston, MA, USA). Before statistical tests, the data were tested for normal distribution. Treatment comparisons were analyzed with Mixed-effect model with Tukey’s multiple comparison, nonparametric Kruskal-Wallis, or Mann–Whitney tests. Significance was considered significant when p < 0.05.
3. Results
3.1. Secreted factors from M1 macrophages promote lineage plasticity and the expression of CSC markers in prostate cancer cells
We hypothesized that secreted factors from pro-inflammatory or immunosuppressive macrophages could affect prostate cancer lineage plasticity. To determine this, macrophages were polarized to M1 type using LPS and IFNγ, and M2 type using IL-4 and IL-13, and AR-positive prostate cancer cells were exposed to CM from the M0, M1, or M2 macrophages. Cytokine composition of CM of M0, M1, and M2 macrophages was analyzed previously.20
First, to determine which pathways are affected by the secreted factors from M1 or M2 type macrophages, RNA-seq and Gene set enrichment analysis (GSEA) from LNCaP and C42B cells was utilized. M1 CM treatment in both cell lines resulted in significantly different gene expression pattern compared to control, M0 CM and M2 CM treatments (Supplementary Figure S1A-D). Moreover, M1 CM treatment induced morphological changes with elongated cell protrusions (Supplementary Figure S2A), and reduced proliferation and induced apoptosis in LNCaP cells (Supplementary Figure S2B-C). This was also supported by the GSEA, which showed enrichment in gene sets related to apoptosis, while downregulation in genes related to proliferation and cell cycle in M1 CM -treated cells (Figure 1a, Supplementary Figure S1E). Moreover, inflammatory response, EMT, complement system, and NFκB signaling-associated genes were enriched in M1 CM -exposed LNCaP and C42B cells (Figure 1a, Supplementary Figure S1E). Surprisingly, the secreted factors from M0, M1, and M2 macrophages downregulated androgen response-related genes (Figure 1a). M2 CM upregulated genes associated with DNA repair, E2F targets, and oxidative phosphorylation. GSEA also revealed enrichment of gene sets associated with cell stemness in M1 CM treatment in both LNCaP and C42B cells (Figure 1b–c). M1 CM treatment upregulated genes associated with CSC plasticity (ALCAM, CXCR4, KLF4, TACSTD2, SOX9, CDH1, SOX4, PLXNB2, and LIF) in LNCaP cells (Supplementary Figure S2D).
Figure 1.
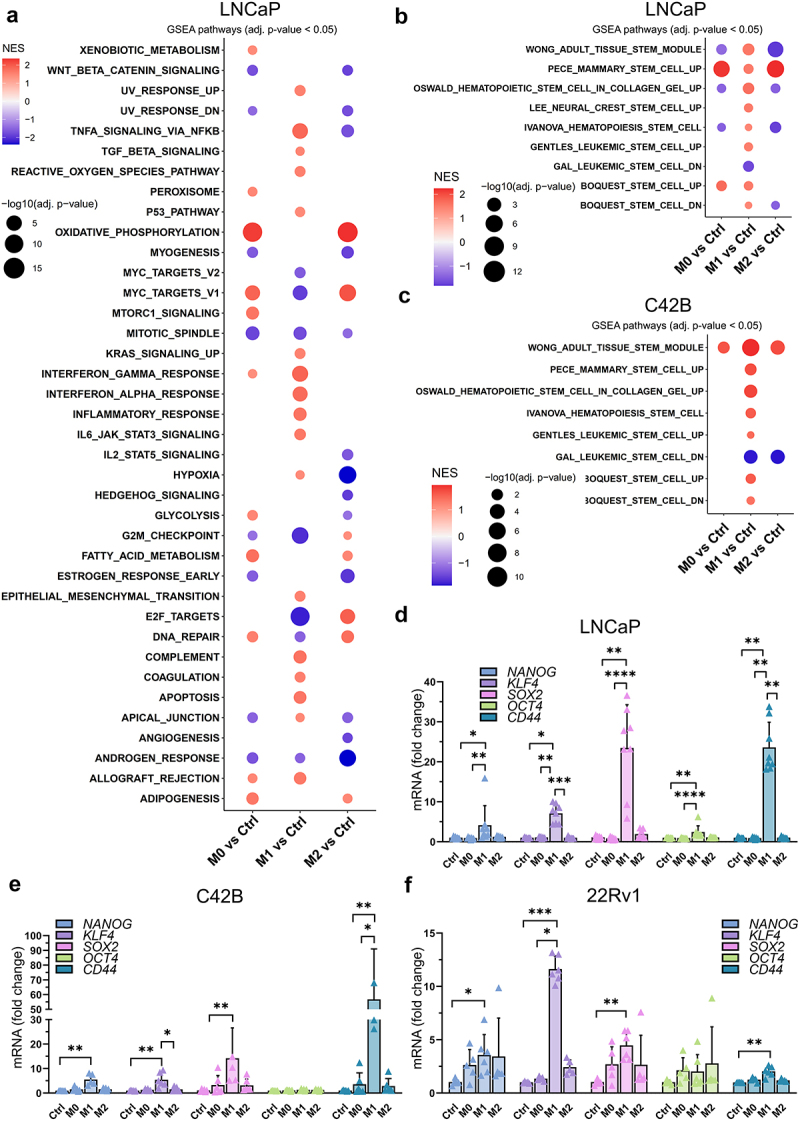
Effects of secreted factors from differently polarized macrophages on the CSC plasticity marker expression. (a) GSEA from hallmark gene sets in pairwise comparisons of LNCaP cells treated with control (ctrl) medium or M0, M1 or M2 CM. NES: normalized enrichment score. Adjusted p-value <0.05. (b) LNCaP and (c) C42B RNA-seq GSEA data about stemness-associated gene sets with pairwise comparisons listed above. The expression of CD44, NANOG, SOX2, KLF4 and OCT4 in (d) LNCaP, (e) C42B and (f) 22Rv1 cells after 48-hour treatment with control media (ctrl), M0, M1 or M2 CM (n = 3). The data represents mean ± SD. Statistical significances were tested with the Kruskal-Wallis nonparametric test. *p < 0.05; *p < 0.01; ***p < 0.001; ****p < 0.0001.
To further validate differential expression of the pluripotency genes in M1 CM -exposed cells, qPCR was performed revealing a significant upregulation of NANOG, KLF4, SOX2, OCT4, and CD44 in M1 CM -treated LNCaP, C42B, and 22Rv1 cells (Figure 1d–f). Similar upregulation of NANOG, SOX2, and CD44 was also seen in SCC9 (squamous cell carcinoma) cells (Supplementary Figure S2E). Next, the association between the expression of stem cell plasticity genes to M1 macrophage infiltration in prostate cancer patient samples was studied using TIMER2.0 database.21 The TIMER2.0 demonstrated a significant positive correlation of KLF4, SOX2, and CD44 expression with M1 macrophage infiltration. These results suggest that the expression of stem cell plasticity genes correlate with TME infiltration of M1 macrophages (Figure 2).
Figure 2.
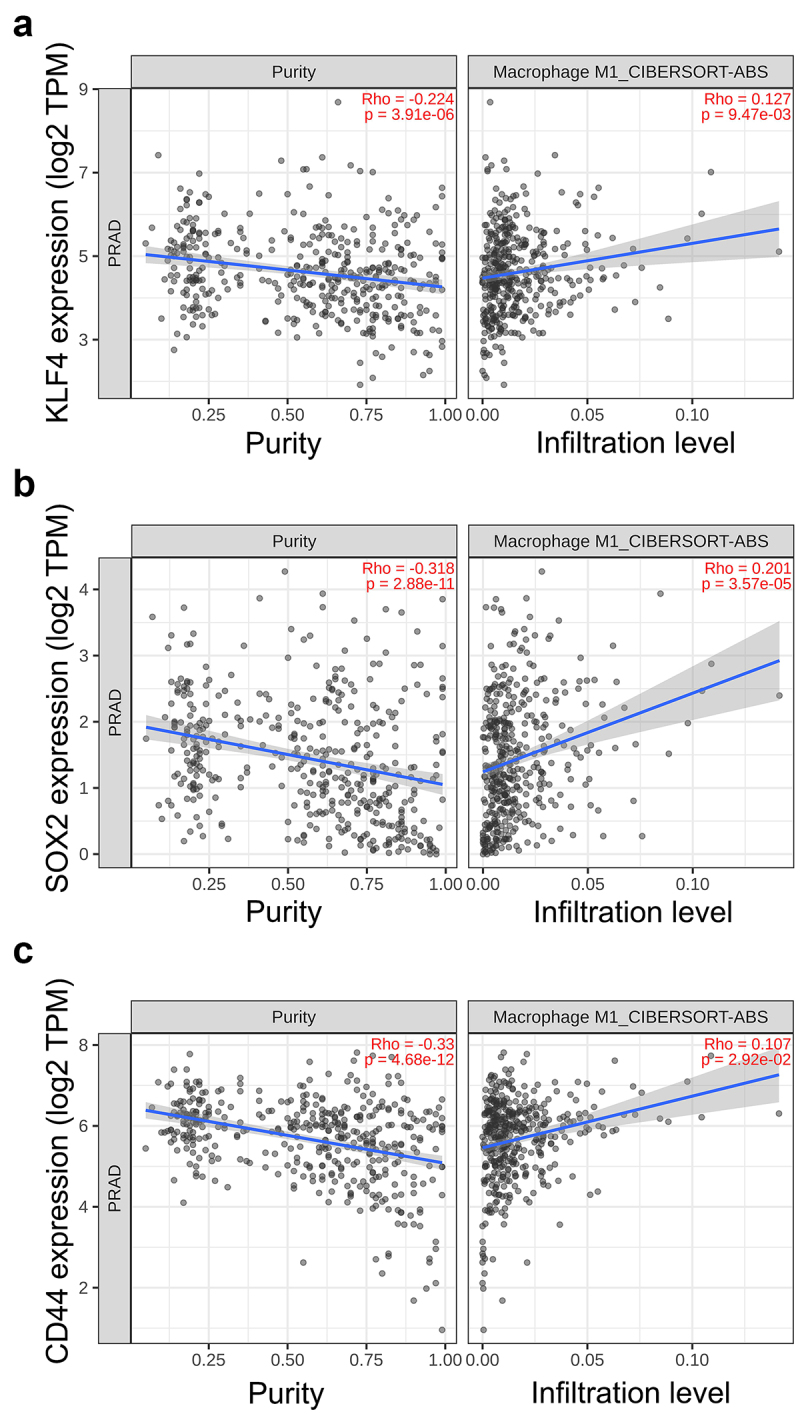
Association of CSC plasticity markers with M1 macrophage infiltration in prostate cancer patient samples. Scatter plots representing the correlation of (a) KLF4, (b) SOX2 and (c) CD44 expression with tumor purity (left) and M1 macrophage infiltration level (right) estimated by TIMER2.0 in adenocarcinoma.
Since secreted factors from M1 macrophages induced the CSC plasticity marker gene expression, we further studied protein level effects on prostate cancer cells. The secreted factors from M1 macrophages induced the expression of NANOG (70 kDa) and KLF4 (64 kDa) proteins, as identified from LNCaP cell lysates (Figure 3a–b). 38 kDa NANOG expression was detected with weak intensity. Immunofluorescent staining revealed that NANOG was localized to cytosol in LNCaP cells (Figure 3c). Ctrl, M0 CM, and M2 CM -treated cells showed faint cytoplasmic staining, while M1 CM -treated cells showed more intense staining, confirming that the secreted factors from M1 macrophages upregulate NANOG expression (Figure 3f). M1 CM -induced KLF4 and CD44 expression were detectable in VCaP and 22Rv1 cells (Figure 3d–e,g–j). In summary, the secreted factors from M1 macrophages stimulate the expression of CSC markers NANOG, KLF4, SOX2, OCT4, and CD44 in prostate cancer cells.
Figure 3.
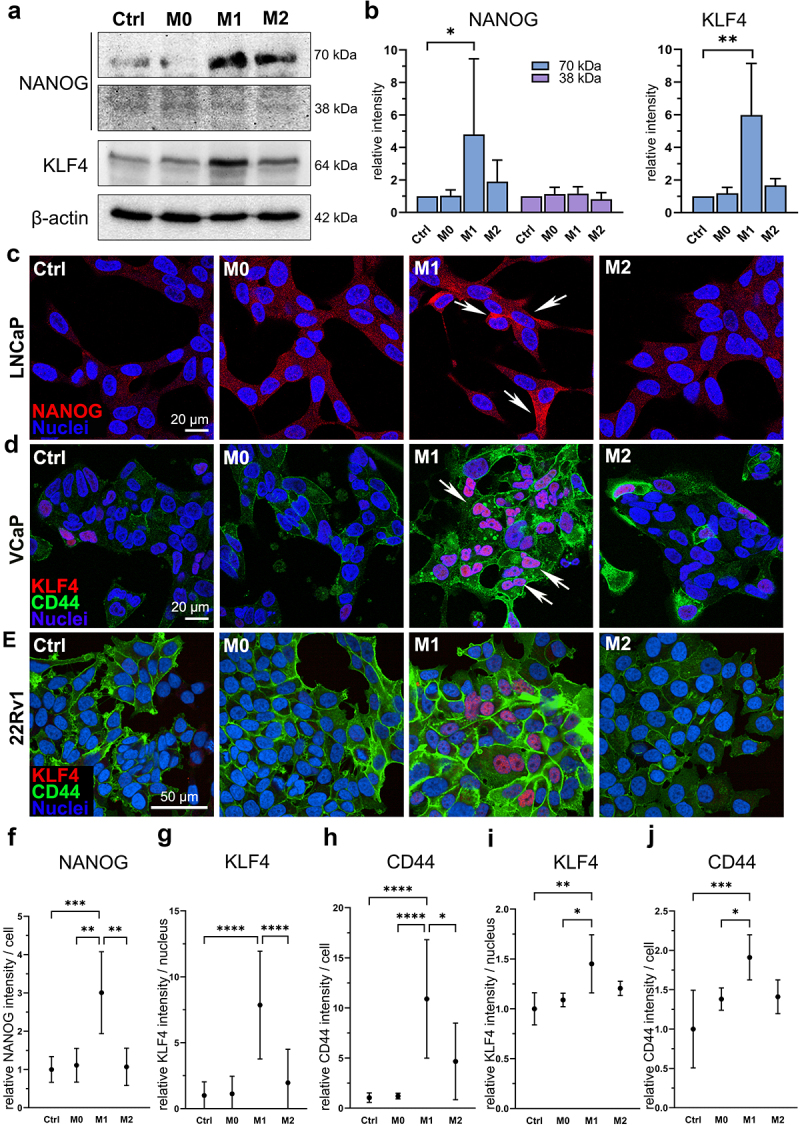
M1 macrophage secretome stimulate NANOG, KLF4 and CD44 protein expression in prostate cancer cells. (a) NANOG (70 and 38 kDa), KLF4 and β-actin western blot images and (b) quantified band densities from LNCaP cell lysates after 72-hour ctrl, M0, M1 or M2 CM treatment (n = 3). The data represents mean ± SD. (c) fixed LNCaP cells stained for NANOG (red, arrows) and nuclei (blue). Fixed (d) VCaP and (e) 22Rv1 cells stained for KLF4 (red, arrows), CD44 (green) and nuclei (blue). Quantified normalized (f) NANOG intensities from LNCaP cells, and KLF4 and CD44 intensities from (g-h) VCaP cells and (i-j) 22Rv1 cells. Cells in (c) and (d) were imaged using Zeiss LSM 800 microscope (40× objective) and cells in (e) were imaged using Opera Phenix Plus microscope (40× objective). Statistical significances were tested with the Kruskal–Wallis nonparametric test. *p < 0.05; *p < 0.01; ***p < 0.001; ****p < 0.0001.
3.2. M1 macrophages suppress AR signaling
Since downregulation of AR signaling is associated with induced pluripotency and prostate cancer cell plasticity, the effects of secreted factors from pro-inflammatory macrophages on AR signaling were studied. AR signaling was induced by synthetic androgen R1881 to investigate if secreted factors from M1 macrophages suppress the induced AR signaling (Figure 4a). The secreted factors from M1 macrophages suppressed AR expression under the R1881 activation in LNCaP cells (Figure 4b, Supplementary Figure S3A). Immunofluorescent staining revealed nuclear localization of AR in R1881-preinduced M0 CM -treated LNCaP and C42B cells, which was inhibited in M1 CM -exposed cells (Figure 4c–d). In VCaP cells, this effect was modest (Supplementary Figure S3B). qPCR also revealed the downregulation of AR (Figure 5a) and the AR target gene PSA (KLK3) (Figure 5b) in R1881-induced M1 CM-treated LNCaP cells. Moreover, AR downstream target genes including KLK2, KLK3, FKPB5, and MAF were downregulated in M1 CM -exposed LNCaP cells (Figure 5c). In summary, the secreted factors from M1 macrophages attenuate AR signaling in prostate cancer cells.
Figure 4.
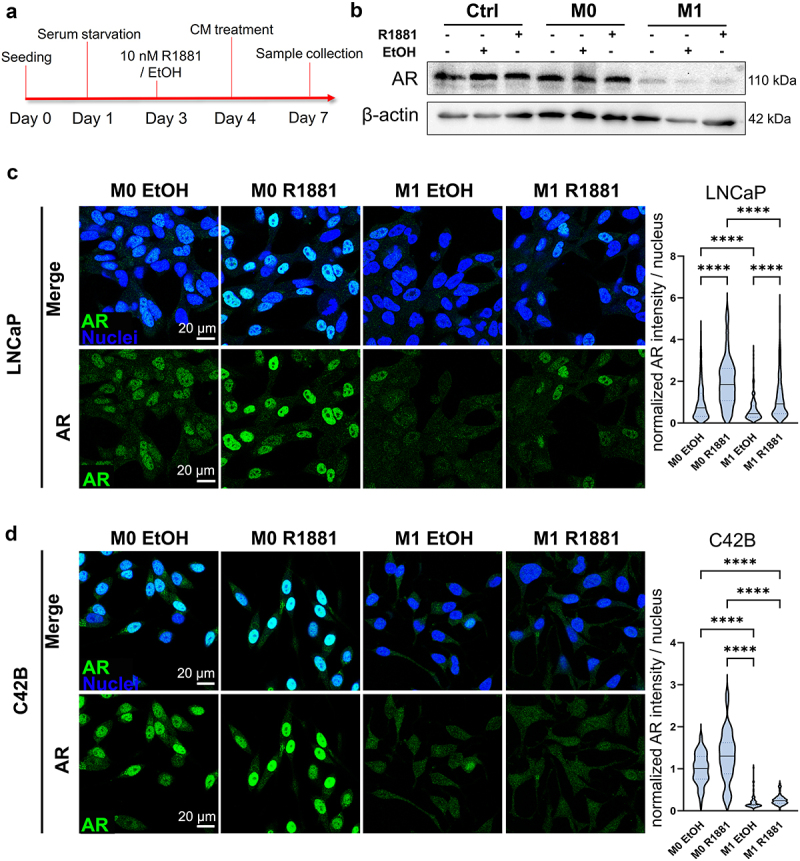
Secreted factors from M1 macrophages suppress AR signaling. (a) Timeline illustrating treatment for AR induction with AR agonist R1881 followed by CM treatments. (b) Western blot images representing AR and β-actin expression in control media (ctrl), M0 or M1 CM -treated LNCaP cells. The cells were treated with (+) or without (-) R1881 in advance. Equal amount of EtOH served as a control. Confocal microscopy images from fixed (c) LNCaP and (d) C42B cells treated with R1881 or EtOH followed by M0 or M1 CM treatment for 48 hours. The cells were labelled against AR (green) and nuclei (blue). Violin blots represents normalized AR intensities/nucleus. The data represents mean ± SD. Statistical significances were tested with the Kruskal–Wallis nonparametric test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Figure 5.
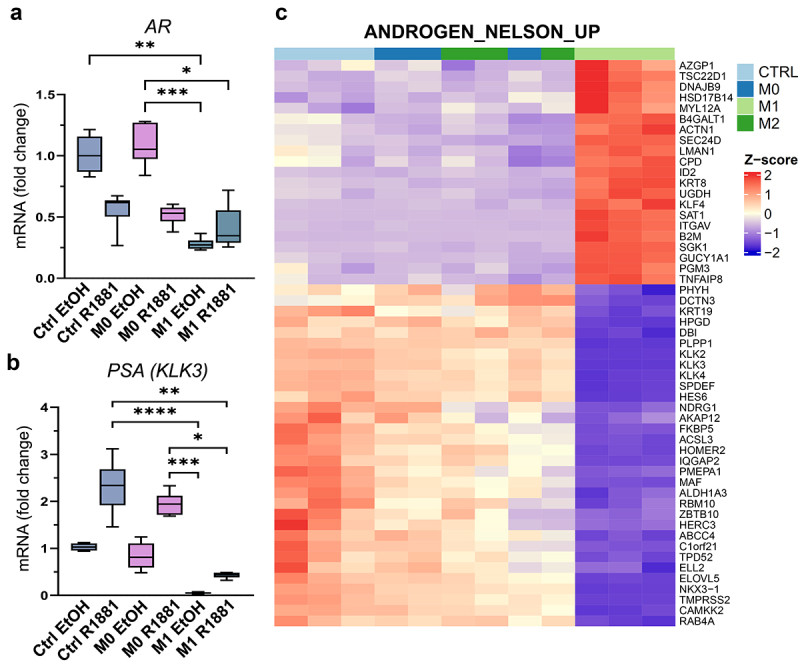
M1 macrophage -secreted factors suppress the expression of AR target genes. (a) AR and (b) PSA (KLK3) expression after pre-treatment with R1881 followed by control (ctrl), M0 or M1 CM exposure. (c) Heatmap showing DEGs related to androgen response (ANDROGEN_NELSON_UP gene set) in control (ctrl), M0, M1 or M2 CM treatment. p <0.01, |logFC|>0.5, transcripts per million (TPM)>1.05. The data represents mean ± SD. Statistical significances were tested with the Kruskal–Wallis nonparametric test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
To investigate the association of AR suppression with pluripotency gene expression in M1 CM -induced LNCaP prostate cancer cells, AR signaling was suppressed with enzalutamide or induced with R1881 in M1 CM -treated cells (Supplementary Figure S3C). R1881 treatment resulted in downregulation of NANOG, SOX2, OCT4, and CD44 whereas suppression of AR with enzalutamide showed slight upregulation of NANOG, SOX2, and OCT4 (Supplementary Figure S3D). The KLF4 expression was not altered with either enzalutamide or R1881. These results suggest that AR inhibition is associated with elevated expression of NANOG, SOX2, OCT4, and CD44 in M1 CM -treated cells.
3.3. Secreted factors from M1 macrophages induce the expression of NANOG, SOX2, and CD44 via NFκB pathway
Since factors secreted by pro-inflammatory M1 macrophages induced prostate cancer cell plasticity -associated gene expression and enriched Hallmark pathway TNFα signaling via NFκB, it was reasonable to study whether the upregulation of genes is due to induced NFκB signaling. GSEA from RNA-seq data showed reversal effect in the expression of Hallmark gene sets when LNCaP cells were treated with M1 CM in combination with NFκB signaling inhibitor IKK16 (NFκBi) compared to M1 CM with DMSO. M1 CM together with NFκBi resulted in the downregulation of genes associated with TNFα signaling via NFκB, inflammatory response and EMT, concomitant with the upregulation of oxidative phosphorylation and the cell cycle -related genes (Figure 6a). Remarkably, the expression of these gene sets exhibited an opposite profile when comparing M1 CM to M0 CM. Similar outcome was observed in gene sets related to cell stemness (Figure 6b). The insights from GSEA suggest the pivotal role of the NFκB pathway in orchestrating the interaction between M1 macrophages and prostate cancer cells, thereby directing prostate cancer cell plasticity under the influence of the secreted factors of M1 macrophages.
Figure 6.
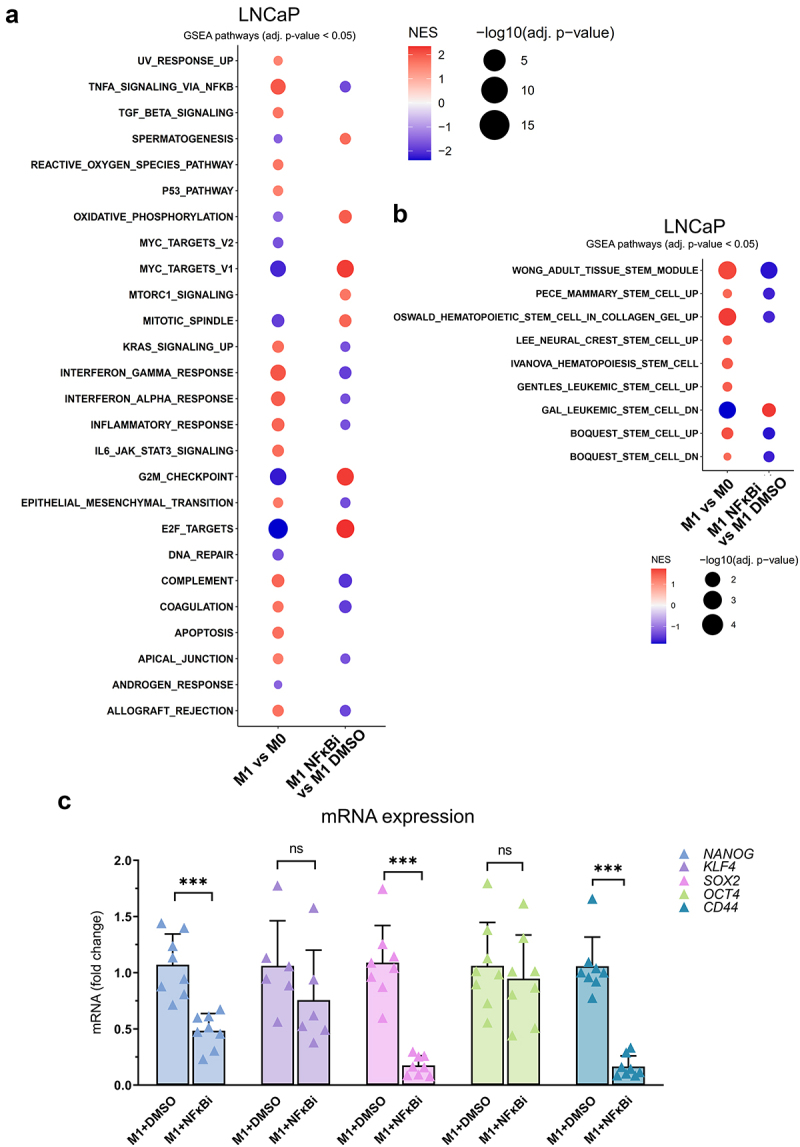
M1 macrophage -secreted factors simulate stemness gene expression pattern through NFκB signaling. (a) Enrichment of hallmark gene sets in pairwise comparisons in LNCaP cells treated with M0 or M1 CM, or M1 CM with IKK16 inhibitor (NFκBi) or DMSO. NES: normalized enrichment score, adjusted p-value <0.05. (b) GSEA from gene sets related to stem cell characteristics. (c) The effects of M1 CM NFκBi or DMSO on the expression of NANOG, KLF4, OCT4, SOX2 and CD44. The data represents mean ± SD. Statistical significances were tested with the Mann–Whitney test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Next, the role of NFκB signaling in M1 macrophage -induced pluripotency gene expression in LNCaP cells was further validated. Inhibition of NFκB signaling reversed the M1 macrophage -induced NANOG, SOX2, and CD44 expression, but not KLF4 or OCT4 (Figure 6c). Similar effects were also observed in NANOG and KLF4 protein expression (Figure 7a–b). The downregulation of NANOG in M1 CM and NFκBi -treated cells was observed both in cell lysates and immunofluorescent stainings. Significant suppression in NANOG intensity was observed in M1 CM -induced LNCaP cells treated with NFκBi (Figure 7c–d). To further verify the association of pro-inflammatory factors, stem cell markers, AR, and its target protein PSA (KLK3) we used STRING protein network interaction analysis tool.22 STRING analysis suggested that M1 macrophage -secreted pro-inflammatory mediators (that M1 CM contains), especially IL-6 and TNFα are linked with OCT4 (POU5F1), NANOG, SOX2, KLF4, CD44 as well as AR and PSA (KLK3) (Figure 7e). In conclusion, the secreted factors from M1 macrophages promote the expression of NANOG, CD44, and SOX2 via NFκB signaling.
Figure 7.
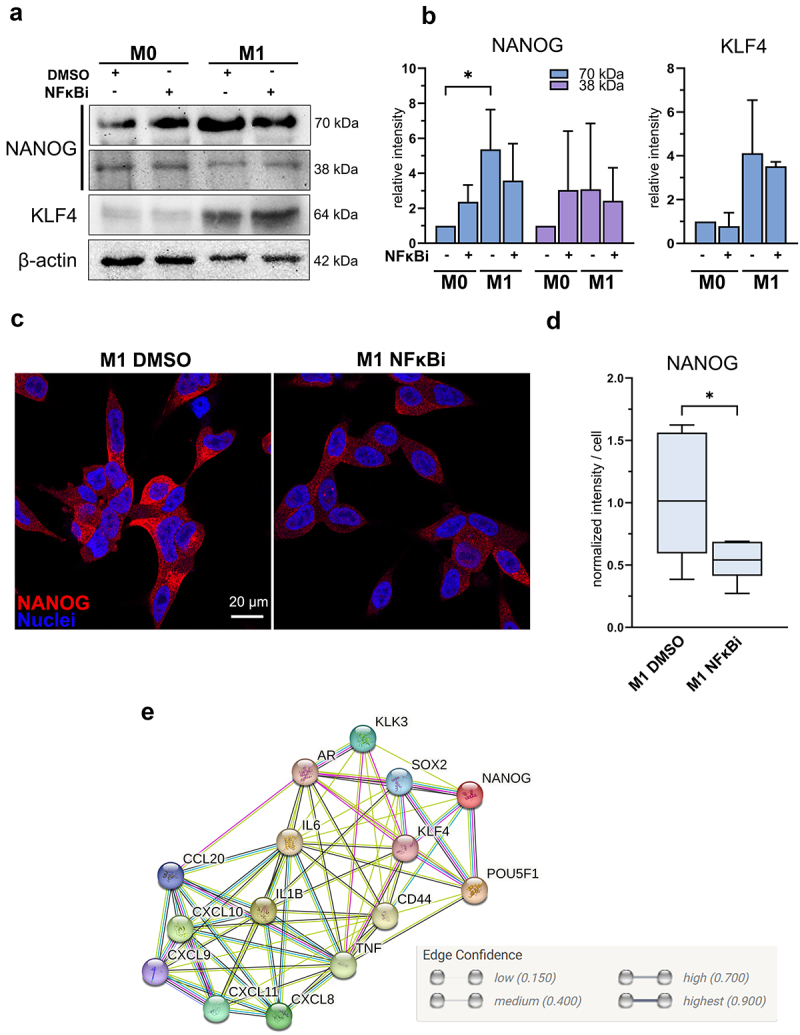
M1 macrophage -induced NANOG expression is suppressed by inhibition of NFκB signaling (a) NANOG, KLF4 and β-actin western blot images and (b) quantified band densities from LNCaP lysates after 72-hour exposure with M0 or M1 CM together with IKK16 inhibitor (NFκBi) normalized to β-actin (n = 3). Statistical significances were tested with the Kruskal-Wallis nonparametric test. (c) Confocal microscopy images from fixed LNCaP cells stained against NANOG (red) and nuclei (blue) in M1 DMSO and M1 NFκBi treatments. (d) Quantified normalized intensities of NANOG per cell. Statistical significance was tested with the Mann–Whitney nonparametric test. (e) Protein network interactions performed by STRING network analysis (version 12) with medium confidence (0.4000) threshold. The data represents mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
4. Discussion
In this study, we showed that pro-inflammatory M1 macrophages secrete factors stimulating prostate cancer stem-like phenotype by promoting the expression of CSC markers NANOG, KLF4, SOX2, OCT4, and CD44 and suppressing AR signaling. Moreover, our results indicated that the upregulation of NANOG, SOX2, and CD44 was due to induced NFκB signaling.
Immune cells play an important role in the formation of favorable environment for tumor progression. The number of TAMs is elevated in advanced cancers23 and both M1 and M2 macrophage characteristics exist in prostate cancer tumors.19,24,25 We and others have showed tumor-promoting role of pro-inflammatory M1 macrophages on melanoma cells,20 in oral squamous cell carcinoma26,27 and breast cancer.28,29 Here, we demonstrate that the secretome of M1-polarized macrophages induce a highly different gene expression pattern in LNCaP and C42B prostate cancer cells including the upregulation of inflammatory response, TNFα signaling via NFκB and EMT-associated genes compared to secretome from M0 or M2 macrophages, while secreted factors from M2 macrophages upregulated genes related to DNA repair, E2F targets, and oxidative phosphorylation. M1 macrophages secrete various pro-inflammatory cytokines, such as TNFα, IL-1β, IL-6, and CXCL830,31 that drive inflammatory response in target cells inducing tumor-promoting effects in cancer cells. Previous studies have shown specific cytokines, such as TNFα,32 IL-1β,33,34 and IL-635,36 to promote EMT in several cancer cell types. These results are in line with our findings that M1 macrophage -secreted factors upregulate genes associated with EMT in melanoma20 and prostate cancer cells. Moreover, our results suggest that M1 macrophage infiltration correlates with the expression of stem cell plasticity markers (KLF4, SOX2, and CD44) which may promote tumor progression.
Since EMT and CSC plasticity are strongly linked,37 we discovered the M1 CM-induced upregulation of stemness -associated genes from GSEA data. There is increasing evidence that the diverse TME actively promotes and sustains CSC phenotype. For instance, TAMs contribute to increased sphere formation, and higher number of TAMs correlate with the number of CD44+ hepatocellular carcinoma cells.38 Moreover, M2-TAMs promote the expression of Sox2, Oct4, and Nanog in murine breast cancer coculture model.39 While the relationship between TAMs and CSC plasticity is shown previously, the impact of M1 macrophage phenotype on CSC plasticity is less studied. In this study, we observed upregulation of stem cell markers NANOG, KLF4, SOX2, OCT4, and CD44 in prostate cancer cells induced by the secreted factors from M1 macrophages. In LNCaP cells, the M1 CM -induced NANOG expression was also assessed at the protein level revealing an increased expression of high molecular weight (70 kDa) isoform as NANOG is also expressed.40 Moreover, M1 CM -treated cells showed elevated cytoplasmic NANOG immunostaining. Cytoplasmic NANOG is associated with high-grade in oral squamous cell carcinoma41 and cancer progression in laryngeal carcinoma42 and cervical cancer.43 Additionally, cytoplasmic NANOG is detected in prostate cancer tumors, and its expression is increased in prostate adenocarcinoma.44 These results suggest a potential role of pro-inflammatory macrophage-secreted factors in prostate cancer tumor progression through the upregulation of NANOG.
Previous studies have shown that NANOG and OCT4 are upregulated by secreted factors from inflammation-induced monocytes,45 while KLF4 in breast cancer cells46 and OCT4 in oral squamous carcinoma cells47 are induced by TNFα. Moreover, pro-inflammatory cytokine IL-1β induces the expression of NANOG, SOX2, and OCT4 in squamous cell carcinoma and melanoma cells48 and IL-6 elevates CD44 levels in prostate cancer.49 The stimulated expression of stem cell markers NANOG, KLF4, SOX2, OCT4, and CD44 confirmed our GSEA findings from LNCaP and C42B cells that the secreted factors from M1 macrophages promote the features of CSC plasticity in prostate cancer cells.
It is widely acknowledged that the depletion of AR signaling is associated with prostate cancer cell reprogramming, driving them toward cancer stem-like stage,50,51 as evidenced by increased levels of NANOG and OCT4.52 Our study revealed that the secreted factors from M1 macrophages suppressed the AR and its target gene KLK3 expression in androgen-induced LNCaP cells. Moreover, decreased nuclear expression of AR was also detected in M1 CM -exposed C42B cells. DiNatale et al. have shown a negative correlation between IL-1β and AR as well as AR target genes, such as KLK3 in metastatic CRPC.53 In LNCaP cells, AR levels are also diminished by TNFα,54 and TNFα alone restrains the AR cistrome, but together with androgen stimulation it reprograms LNCaP cell transcription.55 M1 macrophage secretome contains both IL-1β and TNFα which might be the potential factors suppressing AR expression in M1 CM -treated LNCaP cells. Additionally, activation of AR signaling with dihydrotestosterone suppresses NFκB signaling whereas AR depletion leads to NFκB activation in androgen-sensitive prostate cancer cell lines.56 These molecular interactions highlight the role of TAMs and inflammatory monocytes in driving resistance to anti-androgen therapies in murine prostate cancer model.57 M1 macrophages release several mediators that suppress AR signaling while enhancing NFκB activation to stimulate the expression of cancer stem-like factors.
Since M1 macrophages are primarily known to have antitumor effects, reprogramming of pro-inflammatory macrophages is suggested as cancer immunotherapy in several cancer types58 including prostate cancer.59,60 However, the overall impact of pro-inflammatory macrophages on tumor progression is still unclear. The current study presents the effects of differently polarized macrophages on prostate cancer cell gene and protein expression, revealing the stimulatory role of pro-inflammatory M1 macrophages on the expression of CSC plasticity markers (NANOG, KLF4, SOX2, OCT4, CD44). The stimulatory effect on the expression of NANOG, SOX2, and CD44 was due to the activation of NFκB pathway. Moreover, we found that factors secreted from M1 macrophages suppress AR signaling. These outcomes indicate that inflammatory TME has a role on tumor progression and pro-inflammatory macrophages highly influence on prostate cancer cell transcriptome. Our novel findings suggest that M1 macrophages also play a role in tumor promotion, which should be taken into account when considering reprogramming therapy. However, further studies are needed to investigate effects of our findings in vivo.
Supplementary Material
Acknowledgments
The authors acknowledge Janne Capra, Heidi Kaljunen, Roosa Kaarijärvi, Noora Leppänen, Merja Räsänen, and Taija Hukkanen. Financial support for this work was provided by the Sigrid Juselius Foundation, Research Council of Finland, Cancer Foundation Finland, Paavo Koistinen foundation, the Northern Savo Cancer Foundation, Finnish Cultural Foundation North Savo Regional Fund, Finnish Cultural Foundation, and Kuopio University Foundation. This work was carried out with the support of the Cell and Tissue Imaging Unit, University of Eastern Finland; Biocenter Kuopio and Biocenter Finland.
Funding Statement
The work was supported by the Research Council of Finland [324238, 324009, 328928, 352964, 356947, 331847, 340927], Cancer Foundation Finland, Sigrid Jusélius foundation, Finnish Cultural foundation, Finnish Cultural foundation North Savo Regional Fund, the Cancer Society of North Savo, Kuopio University foundation and Paavo Koistinen foundation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Abbreviations
- CSC
cancer stem cell
- CM
conditioned medium
- TME
tumor microenvironment
- AR
androgen receptor
- TAM
tumor associated macrophages
- CRPC
castration-resistant prostate cancer
- ADT
androgen deprivation therapy
Author contributions
Conceptualization: KKa, KKe, SP-S; Data Curation: EAN; Formal Analysis: KKa, EAN, KKe; Funding Acquisition: SP-S, KKe; Investigation: KKa, EAN, JK, KH, KM-M, VP, MM, KKe, SP-S Methodology: KKa, EAN, KKe, SP-S; Project Administration: KKe, SP-S; Supervision: KKe, SP-S; Validation: KKa, Visualization: KKa; Writing – Original Manuscript Preparation: KKa; Writing – Review and Editing: KKa, EAN, JK, KH, KM-M, VP, MM, KKe, SP-S
Data availability statement
Raw data from RNA-seq data are available with GEO accession GSE264191 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE264191). The data that support the findings of this study are available from the corresponding author, [KK], upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2024.2393442.
References
- 1.Ge R, Wang Z, Cheng L.. Tumor microenvironment heterogeneity an important mediator of prostate cancer progression and therapeutic resistance. npj precis. Onc. 2022;6(1):1–12. doi: 10.1038/s41698-022-00272-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer M, Dogra N, Kyprianou N. Inflammation as a Driver of prostate cancer metastasis and therapeutic resistance. Cancers (Basel). 2020;12(10):2984. doi: 10.3390/cancers12102984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suarez‐Carmona M, Lesage J, Cataldo D, Gilles C. EMT and inflammation: inseparable actors of cancer progression. Mol Oncol. 2017;11(7):805–823. doi: 10.1002/1878-0261.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larionova I, Tuguzbaeva G, Ponomaryova A, Stakheyeva M, Cherdyntseva N, Pavlov V, Choinzonov E, Kzhyshkowska J. Tumor-associated Macrophages in human breast, colorectal, lung, ovarian and prostate cancers. Front Oncol. 2020;10:10. doi: 10.3389/fonc.2020.566511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C, Xu X, Wei S, Jiang P, Xue L, Wang J. Tumor-associated macrophages: potential therapeutic strategies and future prospects in cancer. J Immunother Cancer. 2021;9(1):e001341. doi: 10.1136/jitc-2020-001341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuri P, Shigemura K, Kitagawa K, Hadibrata E, Risan M, Zulfiqqar A, Soeroharjo I, Hendri AZ, Danarto R, Ishii A. et al. Increased tumor-associated macrophages in the prostate cancer microenvironment predicted patients’ survival and responses to androgen deprivation therapies in Indonesian patients cohort. Prostate Int. 2020;8(2):62–69. doi: 10.1016/j.prnil.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kushwaha PP, Verma S, Kumar S, Gupta S. Role of prostate cancer stem-like cells in the development of antiandrogen resistance. Cancer Drug Resist. 2022;5(2):459–471. doi: 10.20517/cdr.2022.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Formaggio N, Rubin MA, Theurillat J-P. Loss and revival of androgen receptor signaling in advanced prostate cancer. Oncogene. 2021;40(7):1205–1216. doi: 10.1038/s41388-020-01598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop JL, Davies A, Ketola K, Zoubeidi A. Regulation of tumor cell plasticity by the androgen receptor in prostate cancer. Endocr-Relat Cancer. 2015;22(3):R165–R182. doi: 10.1530/ERC-15-0137. [DOI] [PubMed] [Google Scholar]
- 10.Lytle NK, Barber AG, Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat Rev Cancer. 2018;18(11):669–680. doi: 10.1038/s41568-018-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf I, Gratzke C, Wolf P. Prostate cancer stem cells: clinical aspects and targeted therapies. Front Oncol. 2022;12:12. doi: 10.3389/fonc.2022.935715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Wet L, Williams A, Gillard M, Kregel S, Lamperis S, Gutgesell LC, Vellky JE, Brown R, Conger K. et al. SOX2 mediates metabolic reprogramming of prostate cancer cells. Oncogene. 2022;41(8):1190–1202. doi: 10.1038/s41388-021-02157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurt EM, Kawasaki BT, Klarmann GJ, Thomas SB, Farrar WL. CD44+CD24− prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer. 2008;98(4):756–765. doi: 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Sheng M, Lin L, Li H, Guo S, Zhang J, Chen G, Chen H. NANOG regulates the proliferation of PCSCs via the tgf-β1/SMAD pathway. Open Med (Wars). 2020;15(1):841–849. doi: 10.1515/med-2020-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y-D, Cai N, Wu X-L, Cao H-Z, Xie L-L, Zheng P-S. OCT4 promotes tumorigenesis and inhibits apoptosis of cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis. 2013;4(8):e760–e760. doi: 10.1038/cddis.2013.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nonomura N, Takayama H, Nakayama M, Nakai Y, Kawashima A, Mukai M, Nagahara A, Aozasa K, Tsujimura A. Infiltration of tumour-associated macrophages in prostate biopsy specimens is predictive of disease progression after hormonal therapy for prostate cancer. BJU Int. 2011;107(12):1918–1922. doi: 10.1111/j.1464-410X.2010.09804.x. [DOI] [PubMed] [Google Scholar]
- 17.Comito G, Giannoni E, Segura CP, Barcellos-de-Souza P, Raspollini MR, Baroni G, Lanciotti M, Serni S, Chiarugi P. Cancer-associated fibroblasts and M2-polarized macrophages synergize during prostate carcinoma progression. Oncogene. 2014;33(19):2423–2431. doi: 10.1038/onc.2013.191. [DOI] [PubMed] [Google Scholar]
- 18.Maolake A, Izumi K, Shigehara K, Natsagdorj A, Iwamoto H, Kadomoto S, Takezawa Y, Machioka K, Narimoto K, Namiki M. et al. Tumor-associated macrophages promote prostate cancer migration through activation of the CCL22–CCR4 axis. Oncotarget. 2016;8(6):9739–9751. doi: 10.18632/oncotarget.14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deichaite I, Sears TJ, Sutton L, Rebibo D, Morgan K, Nelson T, Rose B, Tamayo P, Ferrara N, Asimakopoulos F. et al. Differential regulation of TNFα and IL-6 expression contributes to immune evasion in prostate cancer. J Transl Med. 2022;20(1):527. doi: 10.1186/s12967-022-03731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kainulainen K, Takabe P, Heikkinen S, Aaltonen N, de la Motte C, Rauhala L, Durst FC, Oikari S, Hukkanen T, Rahunen E. et al. M1 macrophages induce protumor inflammation in melanoma cells through tnfr–NF-κB signaling. J Invest Dermatol. 2022;142(11):3041–3051.e10. doi: 10.1016/j.jid.2022.04.024. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva N, Pyysalo S. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salmi S, Siiskonen H, Sironen R, Tyynelä-Korhonen K, Hirschovits-Gerz B, Valkonen M, Auvinen P, Pasonen-Seppänen S. The number and localization of CD68+ and CD163+ macrophages in different stages of cutaneous melanoma. Melanoma Res. 2019;29(3):237–247. doi: 10.1097/CMR.0000000000000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maynard JP, Godwin TN, Lu J, Vidal I, Lotan TL, De Marzo AM, Joshu CE, Sfanos KS. Localization of macrophage subtypes and neutrophils in the prostate tumor microenvironment and their association with prostate cancer racial disparities. The Prostate. 2022;82(16):1505–1519. doi: 10.1002/pros.24424. [DOI] [PubMed] [Google Scholar]
- 25.Siefert JC, Cioni B, Muraro MJ, Alshalalfa M, Vivié J, van der Poel HG, Schoots IG, Bekers E, Feng FY, Wessels LFA. et al. The prognostic potential of human prostate cancer-associated macrophage subtypes as revealed by single-cell transcriptomics. Mol Cancer Res. 2021;19(10):1778. doi: 10.1158/1541-7786.MCR-20-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lv C, Li S, Zhao J, Yang P, Yang C. M1 macrophages enhance survival and invasion of oral squamous cell carcinoma by inducing GDF15-mediated ErbB2 phosphorylation. ACS Omega. 2022;7(13):11405–11414. doi: 10.1021/acsomega.2c00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You Y, Tian Z, Du Z, Wu K, Xu G, Dai M, Wang Y, Xiao M. M1-like tumor-associated macrophages cascade a mesenchymal/stem-like phenotype of oral squamous cell carcinoma via the IL6/Stat3/THBS1 feedback loop. J Exp Clin Cancer Res. 2022;41(1):10. doi: 10.1186/s13046-021-02222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshi M, Tokumaru Y, Asaoka M, Yan L, Satyananda V, Matsuyama R, Matsuhashi N, Futamura M, Ishikawa T, Yoshida K. et al. M1 macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci Rep. 2020;10(1):16554. doi: 10.1038/s41598-020-73624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suárez-Arriaga MC, Méndez-Tenorio A, Pérez-Koldenkova V, Fuentes-Pananá EM. Claudin-low breast cancer inflammatory signatures support polarization of M1-like macrophages with protumoral activity. Cancers (Basel). 2021;13(9):2248. doi: 10.3390/cancers13092248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickman E, Smyth T, Cobos-Uribe C, Immormino R, Rebuli ME, Moran T, Alexis NE, Jaspers I. Expanded characterization of in vitro polarized M0, M1, and M2 human monocyte-derived macrophages: bioenergetic and secreted mediator profiles. PLoS One. 2023;18(3):e0279037. doi: 10.1371/journal.pone.0279037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reales-Calderón JA, Aguilera‐Montilla N, Corbí ÁL, Molero G, Gil C. Proteomic characterization of human proinflammatory M1 and anti-inflammatory M2 macrophages and their response to Candida albicans. Proteomics. 2014;14(12):1503–1518. doi: 10.1002/pmic.201300508. [DOI] [PubMed] [Google Scholar]
- 32.Li C-W, Xia W, Huo L, Lim S-O, Wu Y, Hsu JL, Chao C-H, Yamaguchi H, Yang N-K, Ding Q. et al. Epithelial–mesenchymal transition induced by tnf-α requires nf-κB–Mediated transcriptional Upregulation of Twist1. Cancer Res. 2012;72(5):1290–1300. doi: 10.1158/0008-5472.CAN-11-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li R, Ong SL, Tran LM, Jing Z, Liu B, Park SJ, Huang ZL, Walser TC, Heinrich EL, Lee G. et al. Chronic IL-1β-induced inflammation regulates epithelial-to-mesenchymal transition memory phenotypes via epigenetic modifications in non-small cell lung cancer. Sci Rep. 2020;10(1):377. doi: 10.1038/s41598-019-57285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Wang L, Pappan L, Galliher-Beckley A, Shi J. IL-1β promotes stemness and invasiveness of colon cancer cells through Zeb1 activation. Mol Cancer. 2012;11(1):87. doi: 10.1186/1476-4598-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan NJ, Sasser AK, Axel AE, Vesuna F, Raman V, Ramirez N, Oberyszyn TM, Hall BM. Interleukin-6 induces an epithelial–mesenchymal transition phenotype in human breast cancer cells. Oncogene. 2009;28(33):2940–2947. doi: 10.1038/onc.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Q, Shang Y, Sun F, Dong X, Niu J, Li F. Interleukin-6 promotes epithelial-mesenchymal transition and cell invasion through integrin β6 upregulation in colorectal cancer. Oxid Med Cell Longev. 2020;2020:1–13. doi: 10.1155/2020/8032187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng X, Dai F, Feng L, Zou H, Feng L, Xu M. Communication between epithelial–mesenchymal plasticity and cancer stem cells: new insights into cancer progression. Front Oncol. 2021;11:617597. doi: 10.3389/fonc.2021.617597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan S, Zhao E, Kryczek I, Vatan L, Sadovskaya A, Ludema G, Simeone DM, Zou W, Welling TH. Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology. 2014;147(6):1393–1404. doi: 10.1053/j.gastro.2014.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Liao D, Chen C, Liu Y, Chuang T-H, Xiang R, Markowitz D, Reisfeld RA, Luo Y. Tumor-associated Macrophages regulate murine breast cancer stem cells through a novel paracrine EGFR/Stat3/Sox-2 signaling pathway. STEM Cells. 2013;31(2):248–258. doi: 10.1002/stem.1281. [DOI] [PubMed] [Google Scholar]
- 40.Liu B, Badeaux MD, Choy G, Chandra D, Shen I, Jeter CR, Rycaj K, Lee C-F, Person MD, Liu C. et al. Nanog1 in NTERA-2 and recombinant NanogP8 from somatic cancer cells adopt multiple protein conformations and migrate at multiple M.W species. PLoS One. 2014;9(3):e90615. doi: 10.1371/journal.pone.0090615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H-J, Kang Y-H, Lee J-S, Byun J-H, Kim U-K, Jang S-J, Rho G-J, Park B-W. Positive expression of NANOG, mutant p53, and CD44 is directly associated with clinicopathological features and poor prognosis of oral squamous cell carcinoma. BMC Oral Health. 2015;15(1):153. doi: 10.1186/s12903-015-0120-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigo JP, Villaronga MÁ, Menéndez ST, Hermida-Prado F, Quer M, Vilaseca I, Allonca E, Pedregal Mallo D, Astudillo A, García-Pedrero JM. et al. A novel role for nanog as an early cancer risk Marker in patients with laryngeal precancerous lesions. Sci Rep. 2017;7(1):11110. doi: 10.1038/s41598-017-11709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu T-T, Liu S-Y, Zheng P-S. Cytoplasmic nanog-positive stromal cells promote human cervical cancer progression. Am J Pathol. 2012;181(2):652–661. doi: 10.1016/j.ajpath.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Miyazawa K, Tanaka T, Nakai D, Morita N, Suzuki K. Immunohistochemical expression of four different stem cell markers in prostate cancer: high expression of NANOG in conjunction with hypoxia-inducible factor-1α expression is involved in prostate epithelial malignancy. Oncol Lett. 2014;8(3):985–992. doi: 10.3892/ol.2014.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang T-S, Chen C-L, Wu Y-C, Liu J-J, Kuo YC, Lee K-F, Lin S-Y, Lin S-E, Tung S-Y, Kuo L-M. et al. Inflammation promotes expression of stemness-related properties in HBV-Related hepatocellular carcinoma. PLoS One. 2016;11(2):e0149897. doi: 10.1371/journal.pone.0149897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang B, Zhao M-Z, Cui N-P, Lin D-D, Zhang A-Y, Qin Y, Liu C-Y, Yan W-T, Shi J-H, Chen B-P. et al. Krüppel-like factor 4 induces apoptosis and inhibits tumorigenic progression in SK-BR-3 breast cancer cells. FEBS Open Bio. 2015;5(1):147–154. doi: 10.1016/j.fob.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun S, Yang, el Al H, Wang F, Zhao S. Oct4 downregulation-induced inflammation increases the migration and invasion rate of oral squamous cell carcinoma. ABBS. 2021. Nov. 10(11):1440–1449. doi: 10.1093/abbs/gmab127. [DOI] [PubMed] [Google Scholar]
- 48.Lu L, Wang P, Zou Y, Zha Z, Huang H, Guan M, Wu Y, Liu G. IL-1β promotes stemness of tumor cells by activating Smad/ID1 signaling pathway. Int J Med Sci. 2020;17(9):1257. doi: 10.7150/ijms.44285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu C-T, Huang Y-C, Chen W-C, Chen M-F. Effect of tumor burden on tumor aggressiveness and immune modulation in prostate cancer: association with IL-6 signaling. Cancers. 2019;11(7):992. doi: 10.3390/cancers11070992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaarijärvi R, Kaljunen H, Nappi L, Fazli L, Kung SHY, Hartikainen JM, Paakinaho V, Capra J, Rilla K, Malinen M. et al. DPYSL5 is highly expressed in treatment-induced neuroendocrine prostate cancer and promotes lineage plasticity via EZH2/PRC2. Commun Biol. 2024;7(1):108. doi: 10.1038/s42003-023-05741-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leppänen N, Kaljunen H, Kaarijärvi R, Paatero I, Paakinaho V, Ketola K. SIX2 promotes cell plasticity via Wnt/ß-catenin signalling in androgen receptor independent prostate cancer. Nucleic Acids Res. 2024. Mar. 30:gkae206. doi: 10.1093/nar/gkae206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramesh S, Selvakumar P, Ameer MY, Lian S, Abdullah Alzarooni AIM, Ojha S, Mishra A, Tiwari A, Kaushik A, Jung YD. et al. State-of-the-art therapeutic strategies for targeting cancer stem cells in prostate cancer. Front Oncol. 2023;13:1059441. doi: 10.3389/fonc.2023.1059441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiNatale A, Worrede A, Iqbal W, Marchioli M, Toth A, Sjöström M, Zhu X, Corey E, Feng FY, Zhou W. et al. IL1β expression driven by androgen receptor absence or inactivation promotes prostate cancer bone metastasis. Cancer Res Commun. 2022;2(12):1545–1557. doi: 10.1158/2767-9764.CRC-22-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizokami A, Gotoh A, Yamada H, Keller ET, Matsumoto T. Tumor necrosis factor-α repressess androgen sensitivityin the LNCaP prostate cancer cell line. J Urol. 2000;164(3 Pt 1):800–805. doi: 10.1016/S0022-5347(05)67318-1. [DOI] [PubMed] [Google Scholar]
- 55.Malinen M, Niskanen EA, Kaikkonen MU, Palvimo JJ. Crosstalk between androgen and pro-inflammatory signaling remodels androgen receptor and nf-κB cistrome to reprogram the prostate cancer cell transcriptome. Nucleic Acids Res. 2017;45(2):619–630. doi: 10.1093/nar/gkw855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Basílio J, Hochreiter B, Hoesel B, Sheshori E, Mussbacher M, Hanel R, Schmid JA. Antagonistic functions of androgen receptor and nf-κB in prostate cancer—experimental and computational analyses. Cancers. 2022;14(24):6164. doi: 10.3390/cancers14246164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X-F, Selli C, Zhou H-L, Cao J, Wu S, Ma R-Y, Lu Y, Zhang C-B, Xun B, Lam AD. et al. Macrophages promote anti-androgen resistance in prostate cancer bone disease. J Exp Med. 2023;220(4):e20221007. doi: 10.1084/jem.20221007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mantovani A, Allavena P, Marchesi F, Garlanda C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov. 2022;21(11):799–820. doi: 10.1038/s41573-022-00520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Mitri D, Mirenda M, Vasilevska J, Calcinotto A, Delaleu N, Revandkar A, Gil V, Boysen G, Losa M, Mosole S. et al. Re-education of tumor-associated Macrophages by CXCR2 blockade drives senescence and tumor inhibition in advanced prostate cancer. Cell Rep. 2019;28(8):2156–2168.e5. doi: 10.1016/j.celrep.2019.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Praharaj M, Shen F, Lee AJ, Zhao L, Nirschl TR, Theodros D, Singh AK, Wang X, Adusei KM, Lombardo KA. et al. Metabolic reprogramming of tumor-associated macrophages using glutamine antagonist JHU083 drives tumor immunity in myeloid-rich prostate and bladder cancers. Cancer Immunol Res. [2024 June 4]. 12(7):854–875. doi: 10.1158/2326-6066.CIR-23-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data from RNA-seq data are available with GEO accession GSE264191 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE264191). The data that support the findings of this study are available from the corresponding author, [KK], upon reasonable request.


