Abstract
The Escherichia coli multi-promoter region of the gapA gene ensures a high level of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) production under various growth conditions. In the exponential phase of growth, gapA mRNAs are mainly initiated at the highly efficient gapA P1 promoter. In the present study, by using site-directed mutagenesis and chemical probing of the RPo (open complex) formed by Eσ70 (holoenzyme associated with σ70) RNAP (RNA polymerase) at promoter gapA P1, we show that this promoter is an extended −10 promoter that needs a −35 sequence for activity. The −35 sequence compensates for the presence of a suboptimal −10 hexamer. A tract of thymine residues in the spacer region, which is responsible for a DNA distortion, is also required for efficient activity. We present the first chemical probing of an RPo formed at a promoter needing both a −10 extension and a −35 sequence. It reveals a complex array of RNAP–DNA interactions. In agreement with the fact that residue A-11 in the non-template strand is flipped out in a protein pocket in previously studied RPos, the corresponding A residue in gapA P1 promoter is protected in RPo and is essential for activity. However, in contrast with some of the previous findings on RPos formed at other promoters, the −12 A:T pair is opened. Strong contacts with RNAP occur both with the −35 sequence and the TG extension, so that the σ4 and σ2 domains may simultaneously contact the promoter DNA. RNAP–DNA interactions were also detected immediately downstream of the −35 hexamer and in a more distal upstream segment, reflecting a wrapping of RNAP by the core and upstream promoter DNA. Altogether, the data reveal that promoter gapA P1 is a very efficient promoter sharing common properties with both extended −10 and non-extended −10 promoters.
Keywords: DNA distortion, DNA–RNA polymerase interaction, Escherichia coli, extended −10 promoter, gapA gene, open complex chemical probing
Abbreviations: α-CTD, C-terminal domain of the α subunit; DMS, dimethylsulphate; DTT, dithiothreitol; Eσ70/Eσ32, holoenzyme associated with σ70 or σ32; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ORF, open reading frame; PGK, phosphoglycerate kinase; RNAP, RNA polymerase; RPo, open complex; Taq, Thermus aquaticus; WT, wild-type
INTRODUCTION
GAPDH (D-glyceraldehyde-3-phosphate dehydrogenase) with phosphorylating activity is a key enzyme for glucose metabolism. It catalyses the reversible oxidation of D-glyceraldehyde-3-phosphate to 1,3-diphosphoglycerate. In many bacteria species, the gene encoding the phosphorylating GAPDH (gap gene) precedes the pgk gene encoding the PGK (phosphoglycerate kinase) [1,2]. However, these genes have a different organization in γ-proteobacteria compared with other bacteria. The best-studied example is Escherichia coli, where an epd ORF (open reading frame) is located upstream of the pgk ORF. It encodes a protein showing homology with GAPDH, but having a non-phosphorylating erythrose-4-phosphate dehydrogenase activity [3,4]. The E. coli phosphorylating GAPDH is encoded by the gapA gene that is located at another locus on the chromosome [5,6]. Nevertheless, GAPDH and PGK are co-ordinately expressed in E. coli.
In an initial study [7], we showed that gapA transcription is governed by a multi-promoter region that responds to several of the general regulatory mechanisms used by E. coli. Promoters P1, P3 and P4 are recognized by the general Eσ70 (holoenzyme associated with σ70) RNAP (RNA polymerase), whereas promoter P2 is recognized by the Eσ32 RNAP. Promoter P3 is subjected to catabolic repression. An interplay between the different gapA promoters ensures the expression of GAPDH under various environmental conditions. However, of the four gapA promoters, promoter P1 plays an essential role, since transcripts initiated at promoter P1 are the most abundant ones throughout the growth period and at the beginning of the stationary phase [7]. The strong activity of promoter P1 explains the very high level of expression of the cloned E. coli gapA gene (80% of soluble proteins) [5]. Promoter gapA P1 probably also plays an important role in the co-ordinated expression of the gapA and pgk ORFs. Indeed, GAPDH and PGK production is increased in the presence of glucose in the medium and these increases are explained by an augmentation of the amounts of transcripts initiated at the gapA P1 promoter and gapB P0 promoter, which are present upstream of the epd and pgk ORF tandem [8]. Such increases in GAPDH and PGK production are not observed in a ptsG− strain, suggesting that the amounts of transcripts initiated at both the gapA P1 and gapB P0 promoters depend on the uptake of glucose through its specific transporter, EIIBCGlc, encoded by the ptsG gene [8]. In addition, promoter P1 is also very efficiently used in vitro by the Eσ70 RNAP, which is an indication of strong intrinsic activity [7]. On the basis of all these observations, an in-depth study of the gapA P1 promoter was of high interest.
Despite having a high intrinsic efficiency [5], the promoter structure is not well defined. Indeed, the gapA P1 promoter has a clear −35 element (TTGACG) and two possible −10 sequences: either a canonical −10 sequence spaced 16 bp from the −35 hexamer or an extended −10 sequence with a non-canonical −10 hexamer, which is spaced 17 bp from the −35 sequence. When they were first discovered, the extended −10 promoters of E. coli were considered to be constitutive promoters [9]. Indeed, the TG dinucleotide at positions −14 and −15 is recognized by domain 3.0 (2.5) of factor σ70 [9–13] and this interaction can compensate for the absence of a −35 sequence [14–18]. However, a recent, in-depth inspection of E. coli promoters revealed that TG-containing promoters are more frequent than previously expected (20% of the promoters) [19,20] and that 74% of them have a −35 hexamer with 3/6 or more matches to the −35 consensus sequence [20]. The functional importance of the TG dinucleotide upstream of the −10 hexamer was experimentally demonstrated for seven of them [20]. However, the requirement of the −35 element was not tested in parallel. The optimal length of the spacer region in E. coli promoters recognized by the Eσ70 RNAP is 17 bp, allowing interactions of the −10 and −35 hexamers with two distinct domains of factor σ70 (regions 2.4 and 4.2 respectively) [21–25] (see [26,27] for reviews). On the basis of this knowledge, an experimental analysis of the E. coli gapA P1 promoter was needed to identify its functional sequences.
In the present study, by using site-directed mutagenesis, we demonstrate that promoter gapA P1 belongs to the class of extended −10 promoters and needs both a TG dinucleotide and a −35 sequence for activity. We also show the importance of the sequence of the spacer region for promoter conformation and activity. Chemical probing of the RPo (open complex) structure revealed a complex array of DNA–protein interactions. This is the first complete chemical probing of a natural promoter from the extended −10 class that needs a −35 hexamer for activity. The results we obtained are discussed taking into account the three-dimensional model of the complex formed by the Taq (Thermus aquaticus) RNAP and a fork-junction promoter DNA containing an extended −10 sequence (RF complex) [12].
EXPERIMENTAL
Bacterial strains and growth conditions
Propagation of recombinant BSIISK+ phagemids (Stratagene, La Jolla, CA, U.S.A.) was performed in E. coli DH5α. Epicurian Coli™ XL1-Blue cells were used to study the in vivo GAPDH production and the transcriptional activity of the gapA P1 variant promoters. Bacteria were grown at 37 °C in LB (Luria–Bertani) broth medium, and 100 mg/l ampicillin was added for the growth of transformed cells.
Genetic constructions and site-directed mutagenesis
Plasmid pBS::EcogapA [7] was used as a reference for GAPDH and mRNA production by the WT (wild-type) gapA gene. In this construct, the gapA ORF is preceded by a 483 bp upstream non-coding sequence containing the multi-promoter region. To create variants (mt-9, -TG, -12, −17T to C, −17T to G, −19T to C, −19T to G, −19T to A, -35, -sp3′), site-directed mutageneses were performed on plasmid pBS::EcogapA. Other variants (mt-16T to G, −16T to C, −18T to C, −18T to G, −18T to A, -cons-10, -cons-10/mt-35, mt-20G to C, mt-20G to T) were obtained by site-directed mutagenesis of plasmid pBS::EcogapAmut62 with a Quik Change™ Site-Directed Mutagenesis kit (Stratagene). This plasmid carries the same promoter region as plasmid pBS::EcogapA, except that an NdeI restriction site was introduced at the ATG codon. The presence of this restriction site does not change the in vivo levels of gapA mRNAs and GAPDH when compared with the WT sequence (results not shown). After mutagenesis, a HindIII–NdeI DNA fragment carrying the mutated sequence was cloned between the HindIII and NdeI sites of plasmid pBS::EcogapAmut62.
Derivatives of the pRLG770 plasmid [28] were built for in vitro run-off transcription assays. To this end, DNA fragments containing the WT or variant gapA P1 promoters were PCR-amplified from position +1 to −84 (+1 corresponds to the transcription-start site) using two primers (1280 and 1278), which introduced an EcoRI site at the 5′-end and a HindIII site at the 3′-end respectively. To obtain the series of pRLG::gapAP1 plasmids, the EcoRI–HindIII fragments were ligated into the pRLG770 plasmid [28] that was digested by these two enzymes.
Protein extract preparation and enzymic assay
Enzymic activities were measured on the soluble fraction obtained after sonication of the cells harvested at an absorbance A600 0.5. GAPDH activity was measured as described previously [8]. The β-lactamase activity measurements were performed as described by Sargent [29].
Extraction of total RNA and mRNA analysis
Total RNA was isolated as described previously by the hot phenol extraction procedure [7]. RNA extractions were performed on a fixed number of cells (5×108) grown at 37 °C in LB/ampicillin medium until the A600 reached 0.5. The gapA mRNAs, as well as the truncated yeaA mRNAs also encoded by plasmid pBS::EcogapA, were analysed by primer extension, using conditions described previously that allow quantitative measurement of mRNA levels [7]. Briefly, 5 μg of total RNA isolated from transformed cells was annealed with 5 ng of 5′-end-labelled gapA primer 1043 (5′-TTTCTGAGCAGCACGGAA-3′) or yeaA primer 437 (5′-GATTCTGCGTCACGT-3′) at 65 °C for 10 min. Primer extensions were performed for 30 min at 45 °C with 1 unit of AMV (avian myeloblastosis virus) Reverse Transcriptase (Life Science) in 50 mM Tris/HCl (pH 8.3), 40 mM KCl and 6 mM MgCl2. The synthesized cDNAs were fractionated by electrophoresis on a 6% sequencing gel and the fractionation pattern was revealed by autoradiography. Since the WT and variant gapA mRNAs initiated at the P1 promoter had identical lengths and sequences, they were expected to have identical half-lives under similar growth conditions. Hence, we assumed that the steady-state levels of gapA P1 mRNA in cells carrying the variant genes were directly correlated to gapA P1 promoter efficiency. As a prerequisite for a quantitative measurement of the gapA P1 mRNA level, we verified that the contribution of the chromosomal gapA gene to gapA P1 mRNA level was negligible. To take into account possible variations of the plasmid copy number in the different cultures, the steady-state level of the yeaA transcript was used as an internal reference. The part of the yeaA gene that is present in plasmid pBS::EcogapA directs transcription initiation on the opposite strand compared with promoter gapA P1 (B. Charpentier, unpublished work). The ratio between gapA P1 and yeaA mRNA levels was estimated for each experiment by measuring the radioactivity contained in the cDNA bands using a PhosphorImager. The variations of gapA P1 mRNA levels in cells containing the variant genes were expressed as a percentage of the gapA P1/yeaA P1 mRNA level found in cells transformed with the WT pBS::EcogapA plasmid. For each variant gapA P1 gene, the experiment was repeated three times on independent cell cultures and the values given are the mean values for the three experiments. The range error on this mean value was estimated using Excel software.
In vitro run-off transcription
Plasmid pRLG::gapAP1 and its derivatives were used as supercoiled DNA templates for in vitro run-off transcription assays. Plasmids (0.8–1 nM) were incubated for 15 min at 22 °C in 25 μl of 10 mM Tris/HCl (pH 8), 150 mM NaCl, 10 mM MgCl2, 1 mM DTT (dithiothreitol) and 100 μg/ml BSA. Then, heparin (10 μg/ml) was added and incubation was continued for 5 min before the addition of a mixture of 500 μM ATP, 200 μM CTP and GTP, 10 μM UTP, 5 μCi of [α-32P]UTP and 0.2 unit of RNAP (Eσ70; Roche). The reaction was stopped after 20 min by the addition of 5 μl of a stop solution [10 mM EDTA, 1% SDS, 2×TBE (180 mM Tris/borate, pH 8.3, and 4 mM EDTA) and 8 M urea, 0.05% Bromophenol and 0.025% xylene cyanol]. Samples were loaded on to 5% polyacrylamide denaturing gels and the radioactivities contained in the DNA bands corresponding to the gapA P1-rrnB and RNAI transcripts were measured using a PhosphorImager with the Image Quant software (Molecular Dynamics). Variations of the levels of the gapA P1 transcript in assays performed with the variant promoters were expressed as a percentage of the ratio of gapA P1 transcript/RNAI transcript found for the WT promoter. The experiment was repeated three times for each variant promoter and the values given are the mean values for three independent experiments. The range error on the mean value for three different experiments was estimated using Excel software.
Analysis of DNA conformation
The following method was used for the PCR amplification of DNA fragments carrying the WT or variant gapA P1 promoters. The supercoiled plasmid pBS::EcogapA (50 ng) was denatured for 3 min at 94 °C in 40 μl of 10 mM Tris/HCl (pH 9), 50 mM KCl, 1.5 mM MgCl2, 0.01% (w/v) gelatin and 0.1% Triton X-100, in the presence of 100 ng of each primer 110 and 351 (5′-TATTTCAGCACATATGCCATG-3′ and 5′-ATTGCCCTTTAAAATTCGGG-3′ respectively). Then, 0.2 mM of each dNTP and 0.25 unit of Taq DNA polymerase were added to a final volume of 50 μl. A series of 30 cycles was performed in a PerkinElmer Thermal Cycler with incubations at 94 °C for 30 s for denaturation, 55 °C for 30 s for hybridization and 72 °C for 1 min for polymerization. The amplified DNAs were extracted with a phenol/chloroform/isoamylalcohol (24:24:1, by vol.) mixture, precipitated with ethanol, dissolved in TE buffer (1 mM EDTA and 1 mM Tris/HCl, pH 8.0) and fractionated by electrophoresis on a 6% polyacrylamide non-denaturing gel. A 29:1 ratio of acrylamide/bisacrylamide was used. Electrophoresis was performed at 4 °C in Tris/borate buffer (2 mM EDTA and 90 mM Tris/boric acid, pH 8.5) at 7 V/cm. The Sau3AI digestion products of pBR322 were used as molecular-mass standards. After electrophoresis, the gels were stained with ethidium bromide (0.5 μg/ml) and DNA was visualized by UV illumination.
The DNA bending model of WT and variant gapA P1 promoters were made by using the plot.it server (http://www.icgeb.trieste.it/dna/) [30]. WebLab Viewer Pro was used to visualize and represent the different models.
Chemical probing of DNA
The reactivity of promoter DNA towards DMS (dimethylsulphate) or potassium permanganate (KMnO4) in the absence or presence of purified Eσ70 RNAP was studied by the method of Borowiec et al. [31]. A supercoiled template (5 nM) was incubated for 30 min at 37 °C with the purified commercial Eσ70 holoenzyme (100 nM) in 50 μl of buffer I [40 mM Tris/HCl (pH 8), 30 mM KCl, 10 mM MgCl2, 1 mM DTT and 300 μg/ml BSA] for plasmid pBS::EcogapA or in 50 μl of buffer II [10 mM Tris/HCl (pH 8), 150 mM NaCl, 10 mM MgCl2 and 100 μg/ml BSA] for plasmid pRLGgapAP1. Heparin was added to a final concentration of 10 μg/ml before adding the chemical reagent. For DMS modification, DMS was added to a 5 mM concentration. After 5 min of incubation at 37 °C, the reaction was stopped by the addition of 12.5 μl of a G-stop solution [1.5 M sodium acetate (pH 7) and 1 M 2-mercaptoethanol], and the DNA was ethanol-precipitated, washed with 70% (v/v) ethanol, dried and dissolved in 100 μl of 1 M piperidine. After a 30 min incubation at 90 °C, the DNA was again precipitated with ethanol. For KMnO4 modification, KMnO4 (8 mM) was added for 2 min. The reaction was stopped by the addition of 1.2 M 2-mercaptoethanol followed by ethanol precipitation. The pellets from DMS and KMnO4 modifications were washed with 70% ethanol, dried and dissolved in 35 μl of MilliQ water.
Positions modified by KMnO4 and DMS were identified by primer extension by the method of Borowiec et al. [31]. Alkaline denaturation of the double-strand plasmid was performed with NaOH (1 mM) and 5 ng of 5′-end-labelled oligonucleotide [1043 (sequence shown above) or 352 (5′-GGAAGAGTGAGGCG-3′) for plasmid pBS::EcogapA or 2559 (5′-ACCATCGGCGCTACGGCG-3′) for plasmid pRLGgapAP1] were added. The DNA was denatured at 80 °C for 2 min and cooled on ice for 5 min. Samples were then neutralized with 5 μl of 10×buffer [500 mM Tris/HCl (pH 7.2), 100 mM MgSO4 and 2 mM DTT] and heated for 3 min at 45 °C for annealing. Samples were cooled again on ice for 5 min before the addition of 5 μl of a dNTP mixture (5 mM each). Primer extensions were performed for 10 min at 50 °C with 1 unit of Klenow fragment enzyme. The reaction products were precipitated, and the pellet was dissolved in formamide dye buffer and analysed by electrophoresis on 7% sequencing gels. Dideoxy-sequencing reaction products, obtained with the same primers, were run in parallel.
RESULTS
Activity of the gapA P1 promoter depends on an extended −10 hexamer
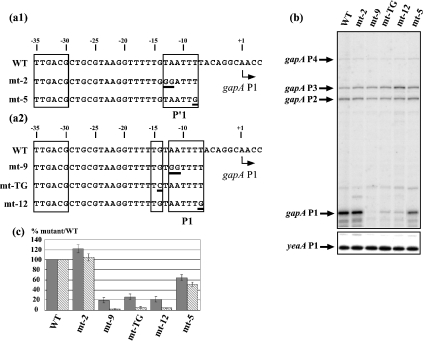
The two possible architectures (P′1 and P1) of promoter gapA P1 are shown in Figure 1(a). To test for the activity of each putative −10 element, their 5′-terminal dinucleotide and 3′-terminal residues, which are known to play an important role in the initiation process [12,20,22], were substituted for a GG dinucleotide (mt-2 to test for the P′1 −10 sequence and mt-9 to test for the P1 −10 sequence) and a G residue (mt-5 for the P′1 −10 sequence and mt-12 for the P1 −10 sequence) respectively (Figure 1a). For an in vivo analysis of their activities, the mutated promoters were cloned into plasmid pBS::EcogapA [7]. The amounts of gapA transcripts were estimated by primer-extension analysis with reverse transcriptase under the conditions described in the Experimental section (Figures 1a and 1b), using an oligonucleotide primer complementary to the gapA ORF. To take into account possible variations of the plasmid copy number, the steady-state level of transcripts from the 5′ part of the yeaA gene, which is located upstream of the gapA gene and is transcribed from the opposite strand (B. Charpentier, unpublished work), was used as an internal reference. GAPDH production in recombinant cells was also monitored by enzymic activity measurement of cell extracts, as described in the Experimental section (Figure 1c). The amount of mRNAs initiated at the gapA P1 promoter was significantly reduced by mutations at the 5′ or 3′ extremities of the P1 −10 sequence (AATTTT). In contrast, no decrease was observed when the TA dinucleotide at the 5′-end of the P′1 −10 hexamer (TAATTT) was mutated (Figure 1c, cf. lanes mt-9 and mt-12 with lanes mt-2 and mt-5). The activity was divided by a factor of 2 for the mt-5 mutation. Similar variations were reproducibly obtained in several independent series of experiments. In agreement with these observed variations of the gapA P1 mRNA levels, the rates of GAPDH production were decreased by factors of between 4 and 6 for the variant mt-12 and mt-9 genes. The lower decreases in GAPDH activity compared with gapA P1 mRNA levels are explained by the contribution of promoters P2 and P3 to GAPDH production. Taken together, these results suggested that the AATTTT sequence, in spite of its divergent sequence compared with the consensus −10 sequence, is the functional −10 hexamer sequence of promoter gapA P1. The functionality of the extended −10 architecture was confirmed by the strong decrease in promoter gapA P1 efficiency after substitution of the TG dinucleotide upstream of the AATTTT sequence for a TC dinucleotide (Figure 1b, lane mt-TG). As a result, GAPDH production was low (Figure 1c). Taken together, these results demonstrated that promoter gapA P1 belongs to the class of extended −10 promoters.
Figure 1. Promoter gapA P1 belongs to the class of extended −10 promoters.
(a1, a2) Nucleotide sequences of the two alternative −10/−35 P′1 (a1) and P1 (a2) promoters present in the gapA P1 promoter region are shown. The −10 hexamers, the TG extension and the −35 hexamer are boxed. A broken arrow marks the transcription start site (+1). Sequences of the variant gapA P1 promoter regions are shown below the WT sequence [variants with mutations in the P′1 −10 sequence (a1) and variants with mutations in the P1 extended sequence (a2)]. Positions of the base substitutions are underlined. (b) Analysis of the effects of mutations on the in vivo gapA mRNA levels by reverse transcription. Epicurian Coli™ XL1-Blue cells transformed with plasmid pBS::EcogapA carrying the WT or variant gapA P1 promoter were grown to an absorbance A600 0.5. The gapA mRNAs were analysed by primer extension under conditions allowing quantitative measurements (see the Experimental section). cDNAs corresponding to transcripts initiated at the P1, P2, P3 and P4 promoters are indicated. The yeaA mRNA primer-extension analysis (yeaA P1) was used for quantifying the effects of mutations on gapA P1 activity (see the Experimental section). (c) A histogram representing the effects of the mutations on GAPDH production and the in vivo gapA P1 mRNA level. GAPDH-specific activities and the plasmid-encoded β-lactamase-specific activities were measured as described in the Experimental section and their relative ratios in strains transformed with plasmids carrying the variant gapA genes were compared with that measured in the strain transformed with the WT pBS::EcogapA plasmid. In (c), variations of GAPDH-specific activities were represented as percentages of the GAPDH/β-lactamase ratio found for the WT plasmid (grey bars). The cDNA signals corresponding to gapA P1 and yeaA P1 mRNAs were quantified using a PhosphorImager. Variations of the gapA P1/yeaA P1 ratios (stippled bars) in strains transformed with plasmids carrying the variant genes were expressed as a percentage of the gapA P1/yeaA P1 ratio found in the strain transformed with the WT pBS::EcogapA plasmid. The values given in (c) correspond to the mean values for three different experiments and the estimated errors are indicated by vertical bars.
Structural analysis of the RPo formed at promoter gapA P1
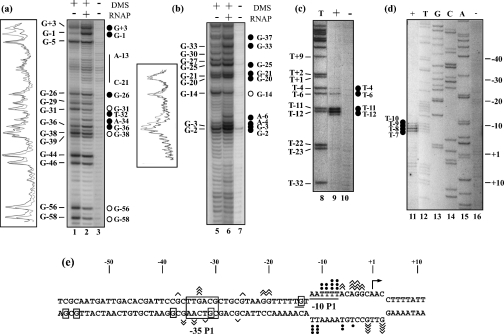
For a complete demonstration that promoter gapA P1 was an extended −10 promoter, RPos were formed with purified Eσ70 RNAP and the supercoiled pBS::EcogapA plasmid, as described in the Experimental section. DMS was used to probe the single-stranded adenine residues and to test for guanine methylation at position N-7 in single- and double-stranded regions (Figures 2a and 2b). KMnO4 was used to probe the single-stranded thymine residues (Figures 2c and 2d). Positions of the modifications were identified by primer extension analysis using the Klenow fragment of the E. coli DNA polymerase. In the RPo formed with promoter gapA P1, a large number of hypersensitive sites were detected on both strands of the −10 hexamer. This allowed us to map the DNA single-stranded region from position −12 (included) to +3 (included), taking position +1 as the transcription initiation site (Figure 2e).
Figure 2. Numerous DNA–protein contacts take place in the gapA P1 RPo.
RPos were formed with the commercial Eσ70 RNAP and the supercoiled pBS::EcogapA plasmid under conditions described in the Experimental section. Probing experiments were performed on the RPo (RNAP+) and on the naked DNA (RNAP–) with DMS (a, b) or KMnO4 (c, d) (see the Experimental section). DMS modifications were followed by piperidine treatment for strand cleavage at modified G and A residues. A control elongation was performed without chemical treatment [lane 3 in (a) and lane 7 in (b)]. Positions of DMS and KMnO4 modifications were identified by primer extension using primer 352 for the template strands (a) and (c) and primer 1043 for the non-template strands (b) and (d). The T sequence ladder in (c) and A, C, G and T sequence ladders in (d) were obtained with primer 1043. Positions of the modified residues in the gapA P1 sequence are given on the left of the panels. The transcription start site was at position +1. Nucleotides that become protected in RPos or nucleotides with an enhanced reactivity compared with DNA are indicated by open and full circles respectively. Densitograms of portions of lanes 1, 2, 5 and 6 were made with the Molecular Analysis software (Bio-Rad). Black lines correspond to lane 1 or 5 (RNAP–) and grey lines to lane 2 or 6 (RNAP+). (e) Compilation of the results of the probing experiments. Angular brackets and closed circles indicate positions of nucleotides whose reactivity to the probe (DMS or KMnO4) is enhanced in the presence of RNAP. Their number is proportional to the level of increase. Boxed nucleotides were less sensitive to the chemical probe in the RPo than in the naked DNA. The −10 sequence and the TG extension are underlined and the −35 sequence is boxed. The opened DNA region is shown as well as the transcription initiation site (broken arrow).
In favour of the proposal that promoter gapA P1 belongs to the class of extended −10 promoters, the TG dinucleotide extension was protected against DMS methylation in the presence of RNAP (residue G-14, Figures 2b and 2e). Our probing experiments revealed strong variations in the accessibility of the putative gapA P1 −35 hexamer in the RPo compared with naked DNA. In the RPo, residue G-31 in the template strand was protected in the presence of RNAP (Figures 2a and 2e), whereas residues A-34 and T-32 showed an increased reactivity to DMS and, in the non-template strand, residue G-33 had a significantly increased sensitivity to DMS (Figures 2a and 2b). Moreover, upstream of the −35 hexamer sequence, residues G-36 and G-38 in the template strand had modified reactivities in the RPo (increased and decreased respectively; Figure 2a). Altogether, the strong variations of DMS modifications found within and upstream of the putative −35 hexamer strongly suggested a functional role for this hexamer, and we tested this hypothesis as described below.
Our RPo probing experiments also revealed variations of DMS methylation in the spacer region. Altogether, in the two strands of the spacer region, four of the six G residues had an increased sensitivity to DMS in RPo (residues G-20 and G-21 and, to a lesser extent, residue G-25 in the non-template strand and residue G-26 in the opposite strand; Figures 2a and 2b). Finally, our probing data also suggested an interaction of the RNAP holoenzyme with an upstream DNA sequence of promoter gapA P1, since residues G-56 and G-58 were strongly protected against DMS methylation in RPo (Figure 2a). In conclusion, our probing experiments revealed a tight and complex interaction of the Eσ70 RNAP holoenzyme with promoter gapA P1.
The −35 sequence is required for promoter activity
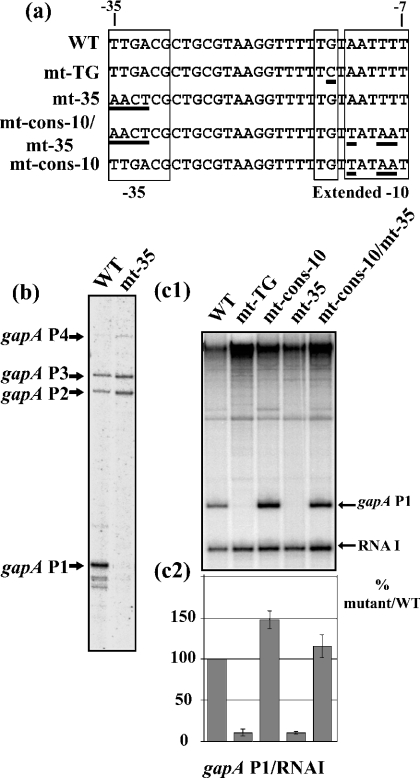
To test for the functional importance of the putative −35 hexamer of promoter P1, we substituted the 5′-terminal TTGA sequence for an AACT sequence (mt-35; Figure 3a). No transcripts initiated at the mt-35 gapA P1 promoter were detected in vivo by primer-extension analysis (Figure 3b). Only initiation at the gapA P2 and P3 promoters was detected. To test whether the mutation influenced the binding of cellular factor(s) or had a direct negative effect on promoter P1 efficiency, in vitro transcription assays were performed with purified Eσ70 RNAP. To this end, we produced a series of pRLG::gapAP1 plasmid derivatives that carried the sequence coding the transcription terminator of the E. coli rrnB operon, under the control of WT or variant gapA P1 promoters (Figures 3c1 and 3c2). Since plasmid pRLG::gapAP1 derivatives also encode RNAI [28], the ratio between the amounts of gapA P1-rrnB and RNAI transcripts was used for the normalization of the data. To compare the effects of mutations in the −35 hexamer and the TG dinucleotide extension on the in vitro efficiency of promoter P1, the mt-TG gapA P1 variant promoter was included in the experiment. We reproducibly found that alteration of the −35 sequence has the same strong negative effect as mutation of the TG dinucleotide (compare lane WT with lanes mt-TG and mt-35 in the in vitro transcription experiment shown in Figure 3c1). Transcripts initiated at the mt-TG and mt-35 promoters were nearly undetectable by in vitro transcription (Figure 3c2). We concluded that the high efficiency of the gapA P1 promoter depends on the presence of both a −35 hexamer and a TG extension. One possible reason could be the sequence divergence of the −10 hexamer compared with the consensus sequence (AATTTT versus TATAAT). Thus we analysed the effect of the conversion of the WT −10 sequence into a consensus −10 sequence in the presence (variant mt-cons-10) or the absence (variant mt-cons-10/mt-35) of an active −35 hexamer (Figure 3a). The in vitro efficiency of the variant promoter (mt-cons-10 P1), containing a −10 consensus hexamer and the WT −35 hexamer, was approx. 1.5-fold superior to that of the WT promoter (Figure 3c). In the absence of an active −35 hexamer, conversion of the WT sequence into a consensus sequence restored the WT activity (Figure 3c). Taken together, these results demonstrated that a −35 hexamer is needed for the efficient activity of the gapA P1 promoter because of a suboptimal −10 hexamer.
Figure 3. The −35 sequence is required for gapA P1 activity.
(a) Sequences of the variant gapA P1 promoters produced to test for the importance of the −10 and −35 sequences are shown. Positions of base substitutions are underlined. (b) Effect of absence of the −35 sequence on the in vivo level of gapA mRNAs. The RNA extracts were prepared and analysed as described in the legend to Figure 1. (c1) Plasmid pRLG::gapAP1 carrying the WT gapA P1 promoter and its derivatives carrying the mutated promoters were used as templates for the in vitro run-off transcriptions [28]. After transcription, the products were fractionated on a 5% polyacrylamide denaturing gel as described in the Experimental section. The RNA I transcript, encoded by the plasmid, was used for the normalization of the amount of gapAP1 transcript recovered in each transcription assay. (c2) For each variant promoter, the gapA P1-rrnB transcript/RNAI transcript ratios were estimated using a PhosphorImager. The gapA P1 promoter activity of the variant promoters is represented as a percentage of the gapA P1-rrnB transcript/RNAI transcript ratio of the WT promoter. The values given in (c) are mean values for three different experiments and the estimated error is indicated by vertical bars.
Role of the spacer sequence in promoter P1 efficiency
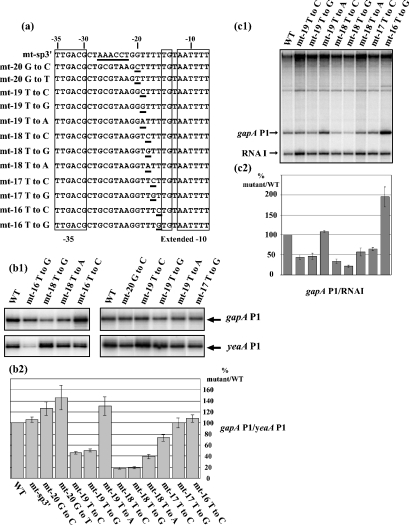
The appearance of additional DMS modifications in the gapA P1 spacer region upon RPo formation (Figure 2) suggested its distortion upon RNAP binding. DNA distortions of the spacer region were found by X-ray analysis of the complex formed between the Taq RNAP and a DNA promoter segment: DNA is bent around the RNAP and this is due to kinks centred at base-pair positions −35 (36°) and −25 (8°) and due to a sharp bend (37°) at base-pair position −16 [12]. Interestingly, the gapA P1 promoter contains a 5-T tract and a 4-T tract spaced by 8 bp (Figure 4a). Formation of intrinsic kinks and bends in double-stranded DNA is known to be sequence-dependent. In particular, the presence of multiple dAn and dTn (n=4–6) runs aligned in phase with the B-DNA helix repeat are known to induce DNA bends [32]. We tested whether the 5-T tract, which is located in the spacer region, may influence promoter activity. To this end, each of the 4-T residues located from positions −16 to −19 were substituted by C and G residues (Figure 4a). The effect of a T to A substitution was tested at positions −18 and −19, as well as G to C and G to T substitutions at position −20, immediately upstream of the T tract (Figure 4a). Finally, the entire sequence from positions −22 to −27 was substituted for an AAACCT sequence (variant mt-sp3′) (Figure 4a). The effects of the mutations were tested in vivo using pBS::EcogapA plasmid derivatives. As above, the efficiency of the variant gapA P1 promoters was estimated by primer-extension analysis and we used the amount of yeaA P1 transcript for normalization of the results. Examples of cDNA fractionations are presented in Figure 4(b1). The histogram in Figure 4(b2) compares the mean values for the ratios of gapA P1/yeaA P1 transcripts that were established for the WT and variant genes in three independent experiments. For the variants showing a marked variation of gapA P1 activity, run-off experiments were also performed. To this end, a series of pRLG::gapAP1 variant plasmids was built. An example of a run-off experiment is shown in Figure 4(c1). In Figure 4(c2), we compared the mean values for the normalized amounts of gapA P1-rrnB transcript obtained for the WT and variant promoters in three independent experiments.
Figure 4. The sequence of the spacer region is important for gapA P1 activity.
(a) The variant gapA P1 promoters with mutations in the spacer region that were produced are represented. The −35 and the extended −10 regions are boxed. The mutated residues are underlined. Names of the mutant promoters are given on the left. (b1, b2) Activities of the variant promoters were measured in vivo as described in Figure 1. Results are expressed as a percentage of the gapA P1/yeaA mRNA level in cells containing the WT gene. The given values are mean values for three independent experiments. The estimated error is indicated by a thin vertical bar. (c1) Plasmid pRLG::gapAP1 carrying the WT promoter and its derivatives, which were carrying gapA P1 promoters mutated in the spacer region, were used as templates for in vitro run-off transcription assays [28]. The transcription products were fractionated on a denaturing gel. (c2) As in Figure 3, the RNA I transcript was used for normalization of the amount of gapA P1 transcript recovered after each transcription experiment. Variations of the gapA P1 activity for the variant promoter are expressed as a percentage of the gapA P1/RNAI ratio found for the WT promoter (100%).
Not only positions but also identities of base substitutions in the T tract were found to influence promoter gapA P1 activity. For instance, the T to G substitution at position −16 had a strong positive effect in vitro (factor of 2), whereas the T to C substitution at the same position only had a modest effect. Interestingly, for variant −16T to G, in agreement with the increase in gapA P1 activity observed in vitro, an increased value of the gapA P1/yeaA P1 mRNA ratio was found in vivo. However, the total amount of gapA P1 mRNA in the transformed cells was low compared with that for the WT gene. This was explained by a marked decrease in the plasmid copy number. Our interpretation is that the amount of GAPDH produced from a gapA gene with a T to G substitution at position −16 is so high that it is deleterious for cell growth. Only the cells that were carrying plasmids with mutations decreasing their copy number were viable. In line with this observation, a G is positioned 5′ to the TG dinucleotide in variant −16T to G and the presence of a G residue at this position was already found to increase the activity of promoters from the extended −10 class [19,20]. Thus, here again, the gapA P1 promoter behaved as an extended −10 promoter.
For the three other T residues in the T tract, the strongest effect of base substitution was detected at position −18 (second T residue in the tract): a T to A substitution decreased the efficiency by a factor of approx. 2.5, and T to C and T to G substitutions had more severe negative effects (factor of approx. 5.5). T to C and T to G substitutions at position −19 divided the activity by a factor of approx. 2, whereas a T to A substitution at this position increased the activity. The G to C and G to T substitutions at position −20, immediately upstream of the T tract, slightly increased the promoter activity and the complete substitution of the AACCT sequence (positions −22 to −27) had a very limited effect on the activity.
Taken together, these results suggested a functional importance for the T tract sequence located in the 3′ half of the spacer region, whereas the sequence of the 5′ half of the spacer region did not seem to have any functional importance.
The presence of a DNA distortion in the gapA P1 promoter
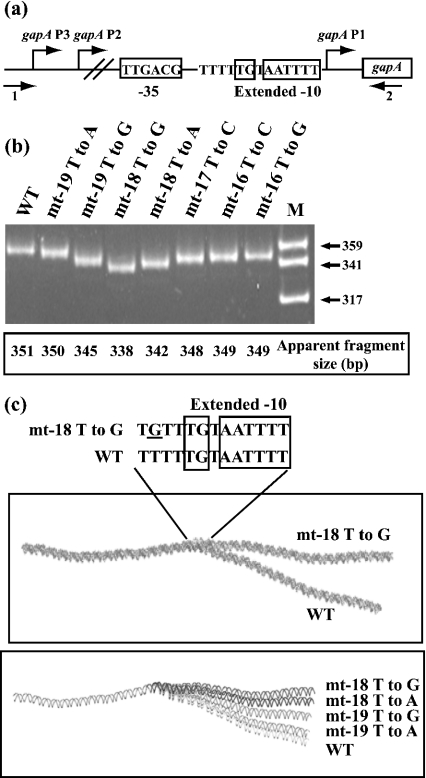
One possible explanation for the functional importance of the 3′ half of the spacer sequence was the induction of a DNA distortion by the 5-T tract that is in phase with the 4-T tract of the −10 hexamer. As DNA distortions modify DNA electrophoretic mobility, we studied the electrophoretic mobilities of DNA fragments of equal length that contained gapA P1 promoters. They were amplified from the WT and variant gapA genes by using the same pair of primers (Figure 5a). As shown in Figure 5(b), the WT DNA had an unusually low electrophoretic mobility compared with that predicted from its length (apparent mobility of 351 bp for a length of 333 bp). In addition, fragments with higher electrophoretic mobilities contained less efficient promoters, as is the case for promoter −18T to G. In contrast, the WT and mt-19T to A promoters, which have similar efficiencies, gave amplification fragments with similar low electrophoretic mobilities. These observations reinforced the idea of a DNA distortion located in the gapA P1 spacer that is important for promoter activity. For a better delineation of this distortion, we used the plot.it server established by Munteanu et al. [30], which allows prediction of bending of a DNA helix. As illustrated in Figure 5(c), a bend of the WT DNA is predicted in the region that contains the two series of T residues and encompasses the extended −10 sequence. In agreement with the observed electrophoretic mobilities, promoter mt-18T to G is predicted to have only a very modest bend (Figure 5c). We concluded that bending of the region, including the 3′ part of the spacer region and the extended −10 element, is probably important for promoter activity.
Figure 5. A DNA distortion is present in the core gapA P1 promoter.
(a) Plasmid pBS::EcogapA and its derivatives were used as the templates for PCR amplifications with oligonucleotides 1 (5′-ATTGCCCTTTAAAATTCGGGG-3′) and 2 (5′-TATTTCAGCACATATGCCATG-3′) of fragments (+155 to −140) that carry the WT or mutated promoter sequences (see the Experimental section). (b) The amplified fragments, generated from the promoter DNA indicated above the lanes, were fractionated on a 6% polyacrylamide non-denaturing gel. The molecular-mass standard (lane M) was produced by Sau3AI digestion of plasmid pBR322. It was used to determine the apparent lengths of the amplified DNA fragments. (c) Models of the DNA bending of the variant gapA P1 promoters generated by using the plot.it server [30]. Upper panel: differential bending between WT and variant mt-18T to G gapA P1 promoters; lower panel: effect of a mutation at positions −18 and −19 on DNA bending.
Role of the cis-acting sequences for RPo formation
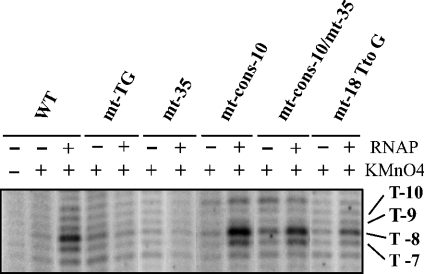
The question was whether DNA bending was required for RPo formation or a step further in the transcription initiation process. To answer this question, we used the KMnO4 probing technique for the estimation of the amount of RPo formed with the variant gapA P1 promoters. The single-stranded thymine T-8 on the non-template strand is highly modified by KMnO4 in the WT promoter (Figures 2 and 6) and is present in all the variant promoters. The intensity of the band corresponding to its modification by KMnO4 was used as an indicator of the amount of RPo formed. As seen in Figure 6, no RPo formation was detected for variant mt-TG and mt-35. This demonstrated the requirement of both the TG dinucleotide and the −35 sequence for RPo formation. The amounts of RPo detected for the WT and variant mt-18T to G, mt-cons-10 and mt-cons-10/mt-35 promoters were proportional to the amounts of in vitro transcripts produced by these promoters (Figures 3 and 4): compared with the WT promoter, very low, similar and larger amounts of RPo were detected for the variants mt-18T to G, mt-cons-10/mt-35 and mt-cons-10 respectively. We concluded that the effects of the studied mutations took place before or at RPo formation.
Figure 6. The TG dinucleotide, the −35 sequence and the spacer sequence of gapA P1 promoter are required for RPo formation.
RPos were formed with the commercial Eσ70 RNAP and the supercoiled plasmid pRLG::gapAP1 carrying the WT promoter or with the pRLG::gapAP1 derivatives containing the gapA P1 promoter variant, under conditions described in the Experimental section. RPos were treated with KMnO4 and analysed by primer extension with oligonucleotide 2559 (5′-ACCATCGGCGCTACGGCG-3′) under conditions described in Figure 2. A control elongation was performed without chemical treatment (lane 1). Nucleotides with an enhanced reactivity in the RPo are indicated on the right side of the autoradiogram.
DISCUSSION
Sequence comparison of E. coli promoters with an extended −10 sequence suggested that some of these promoters might require a −35 sequence for activity. We bring experimental evidence for this hypothesis by an in-depth study of the gapA P1 promoter and performed the first chemical probing of an RPo formed at a promoter displaying this property.
Eσ70 RNAP selects the extended −10 non-canonical P1 instead of the canonical gapA P′1 promoter
The presence in the gapA P1 promoter region of a putative promoter (P′1) with canonical hexamers and a short spacer region (16 bp), together with a putative promoter (P1) with an extended −10 region containing a non-canonical −10 hexamer and a −35 sequence located 17 bp upstream, was an opportunity to compare the relative efficiencies of these two promoter organizations. The strong deleterious effects of the mutations at positions −14, −11 and −7 in the P1 extended −10 hexamer, as well as RPo probing data, revealed an almost unique selection of promoter P1 by Eσ70 RNAP (Figures 1 and 2). Both in vivo and in vitro, the putative P′1 promoter was not active or had a very low activity. Although we cannot exclude promoter P′1 utilization under different growth conditions, we concluded that the strong transcriptional activity of the gapA P1 region depends on a promoter that contains an extended −10 hexamer with a non-canonical −10 sequence (3/6 matches).
It was recently proposed that promoters that have a poor match to the −10 hexamer compensate by having both a TG dinucleotide and a −35 hexamer [20]. In the present study, we demonstrate that this is the case for the E. coli gapA P1 promoter. Indeed, the strong gapA P1 promoter, which plays an important role in exponentially growing cells [8], displays this kind of organization. We could, however, reinforce its activity by optimization of the −10 hexamer or by a T to G substitution at position −16. A compilation of E. coli promoters with putative extended −10 sequences was recently made [20]. However, owing to the occurrence of the overlapping canonical P′1 promoter, promoter gapA P1 was not included in the list. Thus the occurrence of a functional TG extension among E. coli promoters might be more frequent than established recently.
The −12 A:T pair is opened in the gapA P1 RPo
Strand separation in promoter DNA induced by E. coli RNAP is considered to be initiated at the conserved A residue located at position −11 in the −10 hexamer. The adenine at position −11 in the non-template strand is supposed to be flipped out from the helix and tightly bound in a hydrophobic pocket of the 2.3 region of σ70 [33–35]; in the three-dimensional structure established for the complex formed between the Taq RNAP and a fork-junction DNA promoter, the T:A base-pair at position −12 is not melted [12]. A highly conserved tryptophan residue (Trp-433 in E. coli) of the σ 2.3 region is positioned to stack on the exposed faces of the bases at position −12. This structural arrangement is expected to delineate the upstream edge of the transcription bubble [12]. Numerous other previous experiments have strongly suggested that, for extended −10 as well as non-extended −10 promoters, the −12 T:A base-pair located at the extremity of the −10 hexamer element is not disrupted in the RPo [33,34,36,37]. In discordance with all these observations, our chemical probing of the gapA P1 RPo reveals an opening of the A:T base-pair, which is present at position −12, instead of the canonical T:A pair. Since only trace amounts of RPo were formed with the mt-TG variant promoter, we can exclude the possibility that a mixture of RPos formed at the P1 and P′1 promoter sequences was present in our probing experiments (Figure 6). However, owing to the original utilization of the promoter active sequences, the base-pair at position −12 may have a lesser importance in promoter gapA P1 compared with other promoters studied previously. Indeed, the TA (−13 to −12) to GG (variant mt-2) substitution was found to have a limited effect, whereas the AA (−12 to −11) to GG (variant mt-9) substitution had a strong deleterious effect on gapA P1 activity (Figure 1). This indicates that the identity of the residue at the 5′ extremity of the P1 −10 functional hexamer is not important for gapA P1 activity, whereas that of the residue at position −11 is essential. This functional importance of residue A-11 is in agreement with its flipping out in an RNAP pocket [33–35]. Accordingly, the second residue, which is considered to be a key recognition determinant for non-template strand binding (T at position −7) [38], is also required for efficient gapA P1 activity. Hence, we conclude that, as for other promoters, nucleotides −11 and −7 in the −10 hexamer are probably essential for the interaction with domain 2 of σ70. However, the A:T base-pair at position −12 does not constitute the edge of the open bubble. The fact that the T−13 to G substitution (variant mt-2, Figure 1) had a very limited effect on gapA P1 activity excludes the possibility that the T:A base-pair at position −13 functionally replaces the T:A generally found at position −12. It should be pointed out that, in almost all RPos previously probed by KMnO4, there was a T residue at position −12 in the upper strand and this part of the strand is protected from permanganate attack by direct interaction with RNAP even if it is unwound. Thus the −12 bp opening may have escaped detection in some of the previously studied RPos.
In gapA P1 RPo, the −10 element establishes numerous distinct contacts with RNAP
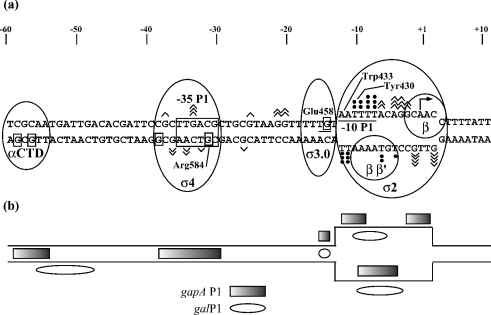
Under the conditions we used for RPo modification by DMS, both single-stranded A and G residues were expected to be modified, this being with a greater reactivity for G residues compared with A residues [31]. The observed pattern of modification brings information about the strong contact established between RNAP and the two single-stranded regions of the gapA P1 −10 hexamer, and the putative RNAP partners of the various DNA regions are indicated in Figure 7. In the non-template strand, whereas residue A-4 was strongly reactive, residue A-6 was less modified, and residues A-11 and A-12 were completely protected. Thus, in addition to residue A-11, residue A-12 is probably in close contact with factor σ70 (Figure 7a). The strong protection of residues A-11 and A-12 in the non-template strand is in contrast with the strong accessibility to KMnO4 of the two opposite T residues in the template strand (Figure 2). Compared with residues T-8 and T-7, residue T-10 in the non-template strand was poorly sensitive to KMnO4. In the RF three-dimensional model, residues −9/−10 are within reach of a Tyr-283 residue (430 in E. coli; Figure 7). Altogether, protection of the 5′ part of the −10 hexamer in the non-template strand is less extended in gapA P1 RPo as compared with galP1 RPo (Figures 2 and 7b) [36]. It is more similar to that found for the RPo formed at the conventional Pilv promoter of Bacillus subtilis [39] and fits perfectly with crystallographic data [12]. In gapA P1 RPo, residues A+1 and A+2 in the non-template strand were also protected against DMS modification and their partners in the template strand were not accessible to KMnO4. A greater sensitivity to KMnO4 of this region was observed in the galP1 RPo [36] (Figure 7b) and, here again, the sensitivity found for promoter gapA P1 is more similar to that found for the conventional B. subtilis Pilv promoter [39]. According to the RF three-dimensional model, the −2 to +4 part of the non-template strand is held in a groove between two lobes of the β subunit [12] (Figure 7a). This may explain the observed protection. Finally, the limited accessibility of the template strand from position −5 to −10 in the gapA P1 RPo (Figures 2 and 7a) is in agreement with strong contacts of this DNA strand with the tunnel formed by the β′ and β subunit and factor σ70 in the RPo [40]. Taken together, our probing data are indicative of three sites of contact between RNAP and the −10 element (Figure 7). In addition, the observed protection of residue G-14 in the non-template strand is indicative of a direct contact of RNAP with the TG extension. Whereas residues His-455 and Glu-458 of σ3 were found to be involved in the recognition of the −10 extension [20], no direct contact between amino acid side chains and bases at positions −14 and −15 was detected in the RF complex containing an extended −10 sequence [12]. As promoters with extended and non-extended −10 hexamers were found to form different contacts with RNAP [36,41], it will be interesting to determine which kind of interaction is responsible for protection of residue G-14 in the gapA P1 RPo.
Figure 7. A complex array of DNA–protein interactions in the gapA P1 RPo.
(a) The possible interactions of RNAP components with the upstream and core promoter regions in gapA P1 RPos. Interactions of the σ2 domain with the −10 region are in agreement with Murakami et al. [12] and Young et al. [40]. The putative interactions of residues A-11 and A-12 with Trp-433 and residues T-9 and T-10 with Tyr-430 are based on the established RF three-dimensional structure [12]. Interactions of the σ3.0 domain with the TG extension (Glu-458) are in agreement with Barne et al. [10] and interactions between σ4 (Arg-581) and the −35 hexamer have been described by Campbell et al. [42]. The potential interaction between α-CTD and the −60 DNA region is represented according to Ross et al. [45]. (b) Comparison of the protected areas in the gapA P1 RPo (the present study; rectangle) and the galP1 RPo (ellipse) [36].
In gapA P1 RPo, the −35 element contacts RNAP
In the gapA P1 RPo, RNAP probably contacts both the −35 sequence and the TG dinucleotide, since residue G-31 in the template strand was also protected against DMS modification (Figures 2 and 7a). This protection is in perfect agreement with the interaction of residue G-31 with the Arg-409 of σ4 (counterpart of E. coli Arg-584) found by X-ray analysis in the complex between Taq σ4 domain and −35 DNA [42] (Figure 7a). Protection in gapA P1 RPo extends upstream of the −35 hexamer in the template strand (protection of residue G-38). No DNA–protein interaction was detected at this position in the complex between Taq σ4 domain and −35 DNA, and the interactions with the template strand were restricted to position −31 to −33 [42]. In this complex, the non-template strand from position −35 to −38 was in contact with the σ4 domain through protein–phosphate interactions. No protection of the corresponding segment was detected in the gapA P1 RPo. Hence, in gapA P1 RPo, RNAP may develop different contacts with the −35 region compared with those identified in the σ4–DNA complex [42] (Figure 7a). The increased reactivities to DMS of residues G-33 and G-37 in the non-template strand and residues G-36, A-34 and T-32 in the template strand, which are found in gapA P1 RPo, are indicative of a profound distortion occurring upon RNAP binding, which is in agreement with the strong dependence of promoter gapA P1 activity on the −35 sequence. One important question to address is whether the interactions of σ4 with the −35 element and σ3 with the TG extension may occur simultaneously in the gapA P1 RPo. This question is important to address, since the three-dimensional model of the replication fork structure gave no information on these interactions [12]. As the function of the −35 sequence is to provide an anchor for the initial binding of RNAP [26], our results suggest that the −35 sequence plays such a role in the extended −10 gapA P1 promoter. The TG motif might (i) also serve as an anchor by interacting with region 3.0 of σ70 [10,11,13] and/or (ii) facilitate RPo formation as reported for other extended promoters [19,43] and/or (iii) stabilize the RPo as found for Gram-positive bacterial promoters [44].
The protection at positions −56 and −58 in the template strand probably represents supplementary binding sites for the RNAP (Figure 7). The bend in the −35 element, which is induced by RNAP binding, was proposed to alter the trajectory of the upstream DNA, bringing it closer to RNAP and facilitating interaction between the α-CTD (C-terminal domain of the α subunit) and upstream DNA [12]. One question that remains to be addressed is whether the α-CTD is implicated in the distal contacts detected in gapA P1 RPo, as found for other promoters from the non-extended class [43,45,46]. Interestingly, an upstream interaction with the RNAP holoenzyme was also detected by chemical probing of the galP1 RPo [36] and the corresponding sequence was found to be important for promoter activity [14]. As the −35 sequence of promoter gapA P1 becomes dispensable when the consensus −10 sequence is present (variant mt-cons–10/mt-35 in Figure 3), the −10 hexamer, the TG motif and the upstream sequence should be sufficient for promoter recognition and stable RNAP binding.
A natural DNA distortion is important for promoter gapA P1 activity
Another interesting feature of promoter gapA P1 is the important role played by the spacer region. On the basis of the electrophoretic behaviour of a PCR-amplification product, we concluded that there is some kind of helix distortion in the core region of gapA P1 (Figure 5). The presence of two T tracts in phase with the B-DNA helix strongly suggests that the distortion consists of DNA bending. In addition, we found a correlation between the amplitude of the distortion and the efficiency of promoter gapA P1 (Figure 5), which suggests a functional importance of this natural distortion. DNA distortions were often found to be involved in the correct positioning of the −10 and −35 hexamers within RPos [26,47,48]. Residues −20 and −21, which are located immediately upstream of the T tract in the spacer region, had an increased reactivity towards DMS in the gapA P1 RPo. This may reflect a reinforcement of the natural DNA bending upon RNAP binding. Hence, the pre-existence of a natural DNA distortion may favour RPo formation at promoter gapA P1. The spacer sequence is also important for the −10 extended galP1 promoter activity. Interestingly, the important sequence was also found to correspond to a series of T residues [14].
In conclusion, promoter gapA P1 is an interesting promoter that shares common properties with extended −10 promoters that do not need a −35 sequence, as well as with conventional promoters with a non-extended −10 sequence and a functional −35 sequence. For further identification of the numerous RNAP–DNA contacts we detected in the gapA P1 RPo, it will be interesting to test the possibility of forming the RPo with variant RNAPs carrying mutations in the α-CTD and different regions of factor σ and of solving the three-dimensional structure of an RNAP–gapA P1 DNA complex. The numerous contacts established between DNA and RNAP, together with the natural capacity of bending of promoter gapA P1, favour a tight wrapping of the Eσ70 RNAP by the gapA P1 DNA, which probably facilitates RPo formation and explains the high efficiency of the E. coli gapA P1 promoter.
Acknowledgments
We are grateful to R. Gourse (University of Wisconsin) for providing us with plasmid pRLG770. M. Buckle and B. Sclavi (UMR 8113 CNRS, ENS Cachan, Cachan, France) are thanked for helpful discussions and a critical reading of this paper. G. Branlant (UMR 7567 CNRS-UHP), P. Fourneret (INIST CNRS, Nancy, France) and M. Eghbali are acknowledged for their participation in the initiation of this work. B. T. was supported by a fellowship from the French Ministère délégué à la Recherche et aux Nouvelles Technologies. This work was financially supported by the Centre National de la Recherche Scientifique (CNRS), by the French Ministère délégué à la Recherche et aux Nouvelles Technologies and by funds from the Region Lorraine (PRST Bioingénierie).
References
- 1.Conway T., Ingram L. O. Phosphoglycerate kinase gene from Zymomonas mobilis: cloning, sequencing, and localization within the gap operon. J. Bacteriol. 1988;170:1926–1933. doi: 10.1128/jb.170.4.1926-1933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branlant C., Oster T., Branlant G. Nucleotide sequence determination of the DNA region coding for Bacillus stearothermophilus glyceraldehyde-3-phosphate dehydrogenase and of the flanking DNA regions required for its expression in Escherichia coli. Gene. 1989;75:145–155. doi: 10.1016/0378-1119(89)90391-0. [DOI] [PubMed] [Google Scholar]
- 3.Boschi-Muller S., Azza S., Pollastro D., Corbier C., Branlant G. Comparative enzymatic properties of GapB-encoded erythrose-4-phosphate dehydrogenase of Escherichia coli and phosphorylating glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 1997;272:15106–15112. doi: 10.1074/jbc.272.24.15106. [DOI] [PubMed] [Google Scholar]
- 4.Zhao G., Pease A. J., Bharani N., Winkler M. E. Biochemical characterization of gapB-encoded erythrose 4-phosphate dehydrogenase of Escherichia coli K-12 and its possible role in pyridoxal 5′-phosphate biosynthesis. J. Bacteriol. 1995;177:2804–2812. doi: 10.1128/jb.177.10.2804-2812.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branlant G., Flesch G., Branlant C. Molecular cloning of the glyceraldehyde-3-phosphate dehydrogenase genes of Bacillus stearothermophilus and Escherichia coli, and their expression in Escherichia coli. Gene. 1983;25:1–7. doi: 10.1016/0378-1119(83)90161-0. [DOI] [PubMed] [Google Scholar]
- 6.Branlant G., Branlant C. Nucleotide sequence of the Escherichia coli gap gene. Different evolutionary behavior of the NAD+-binding domain and of the catalytic domain of D-glyceraldehyde-3-phosphate dehydrogenase. Eur. J. Biochem. 1985;150:61–66. doi: 10.1111/j.1432-1033.1985.tb08988.x. [DOI] [PubMed] [Google Scholar]
- 7.Charpentier B., Branlant C. The Escherichia coli gapA gene is transcribed by the vegetative RNA polymerase holoenzyme Eσ70 and by the heat shock RNA polymerase Eσ32. J. Bacteriol. 1994;176:830–839. doi: 10.1128/jb.176.3.830-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charpentier B., Bardey V., Robas N., Branlant C. The EIIGlc protein is involved in glucose-mediated activation of Escherichia coli gapA and gapB-pgk transcription. J. Bacteriol. 1998;180:6476–6483. doi: 10.1128/jb.180.24.6476-6483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bown J. A., Barne K. A., Minchin S. D., Busby S. J. W. Extended −10 promoters. In: Eckstein F., Lilley D. M. J., editors. Nucleic Acids and Molecular Biology, vol. 11. Berlin: Springer; 1997. pp. 41–52. [Google Scholar]
- 10.Barne K. A., Bown J. A., Busby S. J., Minchin S. D. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the ‘extended–10’ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bown J. A., Owens J. T., Meares C. F., Fujita N., Ishihama A., Busby S. J., Minchin S. D. Organization of open complexes at Escherichia coli promoters. Location of promoter DNA sites close to region 2.5 of the σ70 subunit of RNA polymerase. J. Biol. Chem. 1999;274:2263–2270. doi: 10.1074/jbc.274.4.2263. [DOI] [PubMed] [Google Scholar]
- 12.Murakami K. S., Masuda S., Campbell E. A., Muzzin O., Darst S. A. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 13.Sanderson A., Mitchell J., Minchin S., Busby S. Substitutions in the Escherichia coli RNA polymerase σ70 factor that affect recognition of extended −10 elements at promoters. FEBS Lett. 2003;544:199–205. doi: 10.1016/s0014-5793(03)00500-3. [DOI] [PubMed] [Google Scholar]
- 14.Chan B., Busby S. Recognition of nucleotide sequences at the Escherichia coli galactose operon P1 promoter by RNA polymerase. Gene. 1989;84:227–236. doi: 10.1016/0378-1119(89)90496-4. [DOI] [PubMed] [Google Scholar]
- 15.Keilty S., Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J. Biol. Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 16.Kumar A., Malloch R. A., Fujita N., Smillie D. A., Ishihama A., Hayward R. S. The minus 35-recognition region of Escherichia coli σ70 is inessential for initiation of transcription at an ‘extended minus 10’ promoter. J. Mol. Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 17.Ponnambalam S., Webster C., Bingham A., Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal −35 region sequences. J. Biol. Chem. 1986;261:16043–16048. [PubMed] [Google Scholar]
- 18.Belyaeva T., Griffiths L., Minchin S., Cole J., Busby S. The Escherichia coli cysG promoter belongs to the ‘extended–10′ class of bacterial promoters. Biochem. J. 1993;296:851–857. doi: 10.1042/bj2960851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burr T., Mitchell J., Kolb A., Minchin S., Busby S. DNA sequence elements located immediately upstream of the −10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res. 2000;28:1864–1870. doi: 10.1093/nar/28.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell J. E., Zheng D., Busby S. J., Minchin S. D. Identification and analysis of ‘extended −10’ promoters in Escherichia coli. Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lisser S., Margalit H. Compilation of E coli. mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auble D. T., deHaseth P. L. Promoter recognition by Escherichia coli RNA polymerase. Influence of DNA structure in the spacer separating the −10 and −35 regions. J. Mol. Biol. 1988;202:471–482. doi: 10.1016/0022-2836(88)90279-3. [DOI] [PubMed] [Google Scholar]
- 24.Spassky A., Kirkegaard K., Buc H. Changes in the DNA structure of the lac UV5 promoter during formation of an open complex with Escherichia coli RNA polymerase. Biochemistry. 1985;24:2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]
- 25.Kuznedelov K., Minakhin L., Niedziela-Majka A., Dove S. L., Rogulja D., Nickels B. E., Hochschild A., Heyduk T., Severinov K. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 26.Record M. T., Reznikoff W. S., Craig M. L., McQuade K. L., Schlax P. J. Escherichia coli RNA polymerase (Eσ70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhart F., Curtiss R., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd edn. Washington, DC: American Society for Microbiology Press; 1996. pp. 792–820. [Google Scholar]
- 27.Murakami K. S., Darst S. A. Bacterial RNA polymerases: the whole story. Curr. Opin. Struct. Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 28.Ross W., Thompson J. F., Newlands J. T., Gourse R. L. E coli. Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sargent M. G. Rapid fixed-time assay for penicillinase. J. Bacteriol. 1968;95:1493–1494. doi: 10.1128/jb.95.4.1493-1494.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munteanu M. G., Vlahovicek K., Parthasarathy S., Simon I., Pongor S. Rod models of DNA: sequence-dependent anisotropic elastic modelling of local bending phenomena. Trends Biochem. Sci. 1998;23:341–347. doi: 10.1016/s0968-0004(98)01265-1. [DOI] [PubMed] [Google Scholar]
- 31.Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J. Mol. Biol. 1987;196:101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- 32.Koo H. S., Wu H. M., Crothers D. M. DNA bending at adeninethymine tracts. Nature (London) 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 33.Helmann J. D., deHaseth P. L. Protein-nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry. 1999;38:5959–5967. doi: 10.1021/bi990206g. [DOI] [PubMed] [Google Scholar]
- 34.Fenton M. S., Gralla J. D. Function of the bacterial TATAAT −10 element as single-stranded DNA during RNA polymerase isomerization. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9020–9025. doi: 10.1073/pnas.161085798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsujikawa L., Strainic M. G., Watrob H., Barkley M. D., DeHaseth P. L. RNA polymerase alters the mobility of an A-residue crucial to polymerase-induced melting of promoter DNA. Biochemistry. 2002;41:15334–15341. doi: 10.1021/bi026539m. [DOI] [PubMed] [Google Scholar]
- 36.Chan B., Spassky A., Busby S. The organization of open complexes between Escherichia coli RNA polymerase and DNA fragments carrying promoters either with or without consensus −35 region sequences. Biochem. J. 1990;270:141–148. doi: 10.1042/bj2700141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burns H. D., Belyaeva T. A., Busby S. J., Minchin S. D. Temperature-dependence of open-complex formation at two Escherichia coli promoters with extended −10 sequences. Biochem. J. 1996;317:305–311. doi: 10.1042/bj3170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marr M. T., Roberts J. W. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- 39.Juang Y. L., Helmann J. D. A promoter melting region in the primary sigma factor of Bacillus subtilis. Identification of functionally important aromatic amino acids. J. Mol. Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- 40.Young B. A., Gruber T. M., Gross C. A. Views of transcription initiation. Cell (Cambridge, Mass.) 2002;109:417–420. doi: 10.1016/s0092-8674(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 41.Minchin S., Busby S. Location of close contacts between Escherichia coli RNA polymerase and guanine residues at promoters either with or without consensus −35 region sequences. Biochem. J. 1993;289:771–775. doi: 10.1042/bj2890771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell E. A., Muzzin O., Chlenov M., Sun J. L., Olson C. A., Weinman O., Trester-Zedlitz M. L., Darst S. A. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 43.Burns H. D., Ishihama A., Minchin S. D. Open complex formation during transcription initiation at the Escherichia coli galP1 promoter: the role of the RNA polymerase alpha subunit at promoters lacking an UP-element. Nucleic Acids Res. 1999;27:2051–2056. doi: 10.1093/nar/27.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voskuil M. I., Chambliss G. H. The TRTGn motif stabilizes the transcription initiation open complex. J. Mol. Biol. 2002;322:521–532. doi: 10.1016/s0022-2836(02)00802-1. [DOI] [PubMed] [Google Scholar]
- 45.Ross W., Gosink K. K., Salomon J., Igarashi K., Zou C., Ishihama A., Severinov K., Gourse R. L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 46.Minakhin L., Severinov K. On the role of the Escherichia coli RNA polymerase σ70 region 4.2 and a subunit C-terminal domain in promoter complex formation on the extended −10 galP1 promoter. J. Biol. Chem. 2003;278:29710–29718. doi: 10.1074/jbc.M304906200. [DOI] [PubMed] [Google Scholar]
- 47.Lozinski T., Adrych-Rozek K., Markiewicz W. T., Wierzchowski K. Effect of DNA bending in various regions of a consensus-like Escherichia coli promoter on its strength in vivo and structure of the open complex in vitro. Nucleic Acids Res. 1991;19:2947–2953. doi: 10.1093/nar/19.11.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.deHaseth P. L., Helmann J. D. Open complex formation by Escherichia coli RNA polymerase: the mechanism of polymerase-induced strand separation of double helical DNA. Mol. Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]