Abstract
Abnormal deposition of Aβ (amyloid-β peptide) is one of the hallmarks of AD (Alzheimer's disease). This peptide results from the processing and cleavage of its precursor protein, APP (amyloid-β precursor protein). We have demonstrated previously that TGF-β (transforming growth factor-β), which is overexpressed in AD patients, is capable of enhancing the synthesis of APP by astrocytes by a transcriptional mechanism leading to the accumulation of Aβ. In the present study, we aimed at further characterization of the molecular mechanisms sustaining this TGF-β-dependent transcriptional activity. We report the following findings: first, TGF-β is capable of inducing the transcriptional activity of a reporter gene construct corresponding to the +54/+74 region of the APP promoter, named APPTRE (APP TGF-β-responsive element); secondly, although this effect is mediated by a transduction pathway involving Smad3 (signalling mother against decapentaplegic peptide 3) and Smad4, Smad2 or other Smads failed to induce the activity of APPTRE. We also observed that the APPTRE sequence not only responds to the Smad3 transcription factor, but also the Sp1 (signal protein 1) transcription factor co-operates with Smads to potentiate the TGF-β-dependent activation of APP. TGF-β signalling induces the formation of nuclear complexes composed of Sp1, Smad3 and Smad4. Overall, the present study gives new insights for a better understanding of the fine molecular mechanisms occurring at the transcriptional level and regulating TGF-β-dependent transcription. In the context of AD, our results provide additional evidence for a key role for TGF-β in the regulation of Aβ production.
Keywords: Alzheimer's disease, cytokine, luciferase, promoter, Smad, transforming growth factor-β
Abbreviations: AD, Alzheimer's disease; APP, amyloid-β precursor protein; APPTRE, APP TGF-β-responsive element; Aβ, amyloid-β peptide; EMSA, electrophoretic mobility-shift assay; MLP, major late promoter; Mv1Lu, mink lung epithelial cells; PAI-1, plasminogen activator inhibitor-1; Smad, signalling mother against decapentaplegic peptide; Sp1, signal protein 1; TGF-β, transforming growth factor-β; UTR, untranslated region
INTRODUCTION
AD (Alzheimer's disease) is a neurodegenerative disease associated with progressive memory loss and leading to dementia. Two histological characteristics are observed in AD patients after autopsy: extracellular plaques and intracellular tangles. Filamentous tangles are composed of paired helical filaments composed of neurofilament and hyperphosphorylated tau protein, named microtubule-associated protein. Extracellular plaques are mostly composed of the Aβ (amyloid-β peptide), a peptide derived from the APP (amyloid-β precursor protein). In addition to genetic influences, environmental factors such as cytokines may play important roles in the development and progression of AD and thus explain late-onset/sporadic AD. Several cytokines have been associated with AD progression, such as interleukin-1, interleukin-6, tumour necrosis factor-α and TGF-β (transforming growth factor-β) [1–6].
APP is a protein that is present in three major isoforms in the brain, called APP695, APP751 and APP770 based on their number of amino acid residues. The proteolytic activity of three proteases, denoted as α, β and γ secretases, lead to the production of several APP cleavage products, including Aβ.
In a previous study, we have demonstrated that TGF-β1, a cytokine overexpressed in the brain of AD patients [5,7], induced APP synthesis in vivo and in vitro and promoted Aβ formation by a transcriptional mechanism involving the Smad3 (signalling mother against decapentaplegic peptide 3) signalling pathway in astrocytes [8]. Accordingly, other groups have reported an overexpression of mRNA for the APP751 and APP770 isoforms of APP by TGF-β in cultured astrocytes [9,10]. In addition, we have observed that the TGF-β-transcriptional activity involved the activation of a TGF-β-responsive element at the position +54/+74 of APP promoter. Moreover, it was reported that the overexpression of TGF-β1 in the brain parenchyma was sufficient to induce Aβ deposition in cerebral blood vessels and meninges of aged mice [6]. Finally, as demonstrated in aged mice overexpressing human APP and TGF-β1, it was suggested that TGF-β1 could increase Aβ deposits by activating perivascular astrocytes, but could also reduce Aβ deposition in brain parenchyma by activating microglia [11].
However, the mechanisms sustaining these effects of TGF-β need to be investigated further. TGF-βs belong to a family of peptides that play pivotal roles in intercellular communication [12]. TGF-β1 is the prototype of three different isoforms (TGF-β1,-β2 and -β3) in mammalian species that transduce their biological signal through the activation of a set of serine–threonine kinase receptors (TβR-I and TβR-II) [13]. Subsequent activation of the Smad transcription factor cascade regulates the transcription of TGF-β target genes [14]. Smads are the only substrate and signalling transducers of the activated TGF-β-receptors known so far. Nevertheless, the positive and negative changes in the gene expression induced by TGF-β signalling cannot occur with the Smad proteins only. Thus Smad-dependent regulation of gene transcription is modulated by the interaction with transcriptional co-activators or co-repressors [13]. Briefly, ubiquitous or cell-specific transcription factors are capable of modulating the transcriptional responses driven by the Smad complexes.
In the present study, we have further investigated the mechanisms by which TGF-β could influence the transcriptional activity of APP promoter and thus participate in the AD progression.
EXPERIMENTAL
Cell transfection
Mv1Lu (mink lung epithelial cells) were transiently transfected with the indicated constructs using the Transfast® Transfection Reagent (Promega, Charbonnières, France) according to the manufacturer's instructions.
Reporter gene assay
Cells were treated, 24 h after transfection, and luciferase activities (firefly luciferase and Renilla luciferase) were evaluated after 1 day using the Dual Luciferase® Reporter Assay System (Promega). Values were normalized to the Renilla luciferase activity (Promega). The Dual-Luciferase® Reporter Assay System refers to the simultaneous expression and measurement of two individual reporter enzymes within a single system. Typically, the ‘experimental’ reporter (firefly luciferase) is correlated with the effect of specific experimental conditions (e.g. TGF-β1 treatment), whereas the activity of the co-transfected ‘control’ (Renilla luciferase) reporter provides an internal control, which serves as the baseline of the response. Indeed, the pRL-TK control vector contains the herpes simplex virus thymidine kinase promoter region upstream of Renilla luciferase. It provides low level and constitutive expression in transfected cells. Normalizing the activity of the experimental reporter to the activity of the internal control minimizes experimental variability caused by differences in cell toxicity and transfection efficiency, and enables the anti-proliferative effect of TGF-β to be taken into account.
APP promoter constructs
pGL3-APPTRE-Luc constructs (where APPTRE is APP TGF-β-responsive element) were prepared on the pGL3-basic vector (Promega) containing a major late promoter sequence (pGL3-MLP) [15]. Wild-type or mutated luciferase reporter vectors corresponding to the +54/+74 of Rhesus monkey APP promoter were obtained by inserting into pGL3-MLP a double-stranded oligonucleotide corresponding to the respective sequences (see Figure 3A) and flanked by two XhoI restriction sites.
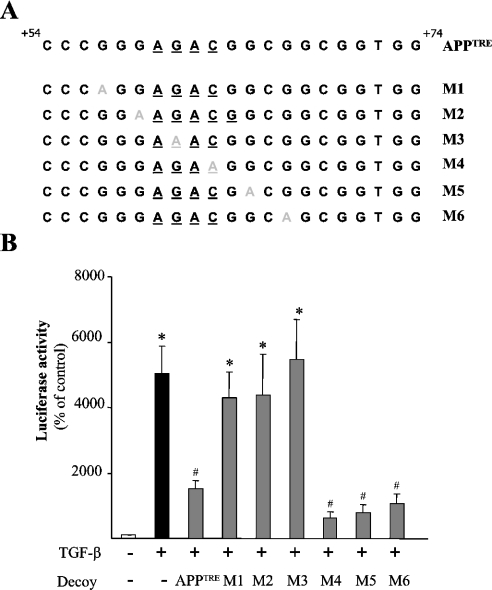
Figure 3. Point mutations of the APPTRE block the TGF-β-dependent transcription of APPTRE.
(A) Sequences of the wild-type (APPTRE) and mutated (M1–M6) double-stranded decoy oligonucleotides used in the study. The mutations performed in the APPTRE sequence (M1–M6) are shown in grey. The critical TGF-β-responsive element AGAC has been underlined in each sequence. (B) Means±S.E.M. of the luciferase activity of Mv1Lu cell line transiently co-transfected with (grey bars) or without (black bars and white bars) the APPTREMLP-Luc and with double-stranded decoy oligonucleotides corresponding to wild-type or point-mutated versions of the APPTRE sequence (n=12). For each condition, transfected cells were treated with (black bars and grey bars) or without (white bar) 1 ng/ml TGF-β1 for 24 h. *P<0.01, significantly superior to control; #, P<0.01, significantly inferior to TGF-β treatment condition, ANOVA followed by the Bonferroni–Dunn test.
Decoy oligonucleotide assays
Decoy double-stranded oligonucleotides were transfected to Mv1Lu cells in an attempt to interfere with the binding of TGF-β-activated transcription factors to their cis-acting elements within the APPTRE sequence. The sequences of the decoy oligonucleotides used are presented in Figure 4(A). These oligonucleotides were co-transfected in the Mv1Lu cells together with the reporter constructs. After incubation for 24 h, cell lysates were prepared and assayed for dual-luciferase assay.
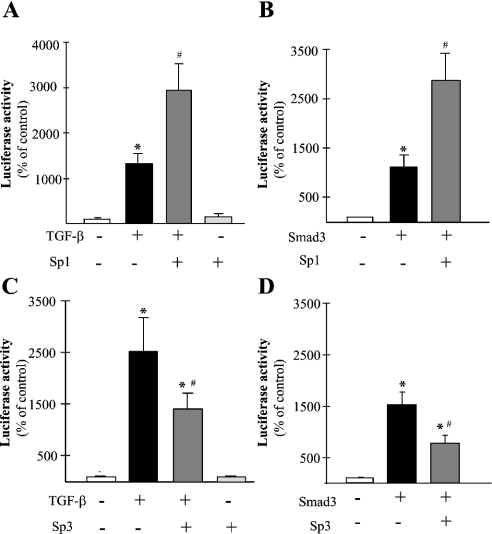
Figure 4. Effects of Sp1 and Sp3 transcription factors on the activation of APPTRE by TGF-β and Smad3.
(A) Means±S.E.M. of the luciferase activity of Mv1Lu cell line transiently co-transfected with the APPTREMLP-Luc and with an expression vector encoding Sp1 (n=12). For each condition, transfected cells were treated with or without 1 ng/ml TGF-β1 for 24 h. *Significantly (P<0.01) superior to control; #, significantly (P<0.01) superior to TGF-β treatment condition, ANOVA followed by the Bonferroni–Dunn test. (B) Means±S.E.M. of the luciferase activity of Mv1Lu cell line transiently co-transfected with the APPTREMLP-Luc and with expression vectors encoding Sp1 and Smad 3 (n=12). *Significantly (P<0.01) superior to control; #, significantly (P<0.01) superior to Smad3-transfection condition, ANOVA followed by the Bonferroni–Dunn test. (C) Means±S.E.M. of the luciferase activity of Mv1Lu cell line transiently co-transfected with the APPTREMLP-Luc and with an expression vector encoding Sp3 (n=12). For each condition, transfected cells were treated with or without 1 ng/ml TGF-β1 for 24 h. *Significantly (P<0.01) superior to control; #, significantly (P<0.01) inferior to TGF-β treatment condition, ANOVA followed by the Bonferroni–Dunn test. (D) Means±S.E.M. of the luciferase activity of Mv1Lu cell line transiently co-transfected with the APPTREMLP-Luc and with expression vectors encoding Sp3 and Smad 3 (n=12). *Significantly (P<0.01) superior to control; #, significantly (P<0.01) inferior to Smad3-transfection condition, ANOVA followed by the Bonferroni–Dunn test.
Nuclear extracts
Nuclear extracts were prepared from control and TGF-β1-treated Mv1Lu cells (1 ng/ml). Cells were harvested for 30 min after treatment and processed as described previously [15].
Western-blot analyses
Cells were harvested in a lysis solution containing 50 mM Tris/HCl (pH 7.6), 1% Nonidet P40 (Sigma), 150 mM NaCl, 2 mM EDTA with 100 μM PMSF in the presence of a protease inhibitor mixture (Sigma). Electrophoreses were performed on 8% SDS/polyacrylamide Tris/glycine gels. Gels were transferred on to a PVDF membrane (PolyScreen®; PerkinElmer Life Sciences, Great Shelford, Cambridge, U.K.); the membranes were blocked in non-fat milk and probed with an appropriate antiserum. Blots were finally developed with an enhanced chemiluminescence Western-blotting detection system (PerkinElmer Life Sciences).
Immunoprecipitation
Mv1Lu cells were solubilized in lysis buffer (20 mM Tris/HCl, pH 7.4, 150 mM NaCl and 0.5% Triton X-100) in the presence of protease inhibitors at 4 °C for 30 min. Insoluble debris was removed by microcentrifugation at 11000 g for 5 min. Proteins were immunoprecipitated by incubation in the presence of specific antibodies for 1 h at 4 °C, followed by adsorption on Protein G–Sepharose (Amersham Biosciences), and subjected to further Western-blot analyses.
EMSA (electrophoretic mobility-shift assay)
Oligonucleotides were end-labelled with [α-32P]dCTP using the Klenow fragment of DNA polymerase. Binding reactions, using 10 μg of nuclear extracts and 2 ng of labelled oligonucleotides, were performed for 20 min at 37 °C in an appropriate binding buffer [15]. The sequence of the double-stranded oligonucleotide used as a probe was 5′-CCCGGGAGACGGCGGCGGTGG-3′. Protein–DNA complexes were resolved on 5% (w/v) polyacrylamide gels containing 0.5×TBE (Tris/borate/EDTA). For antibody interference assays, 1 μl of anti-Sp1 (where Sp1 stands for signal protein 1), anti-Smad3 or anti-smad4 antibodies (Santa Cruz Biotechnology, Heidelberg, Germany) were added to each reaction mixture for 120 min at room temperature (20 °C), then for 15 min at 4 °C. The oligonucleotide probe was finally added to the binding reaction and further incubated for 15 min at room temperature, before resolution on polyacrylamide gels.
Statistical analysis
Results are expressed as means±S.D. Statistical analyses were performed with StatView (Abacus, Berkeley, CA, U.S.A.) by one-way ANOVA followed by the Bonferroni–Dunn test.
RESULTS
The Smad3-dependent TGF-β signalling pathway activates the APPTRE
We identified previously a highly conserved region of the APP promoter, common to human, Rhesus monkey, mouse and rat, located between base numbers +56 and +71 of the 5′-UTR (where UTR stands for untranslated region) as a TGF-β-responsive element [8]. To investigate further the molecular mechanisms controlling the transcription of APP under TGF-β exposure, we have generated a luciferase reporter gene construct by inserting the +54/+74 region of the Rhesus monkey APP promoter on the pGL3-basic luciferase reporter containing an MLP sequence. When transfected in the Mv1Lu, characterized previously as a TGF-β-responsive cell line, this reporter construct is highly activated after TGF-β treatment (Figure 1A), as was shown previously in cultured astrocytes [8] These observations allowed us to term this +54/+74 sequence of the APP promoter as APPTRE.
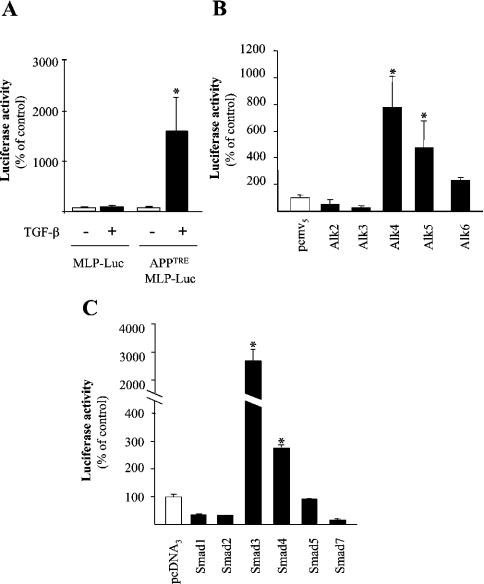
Figure 1. TGF-β induces transcriptional activation of the +54/+74 region of the APP promoter via TGF-β receptor activation and the Smad3-dependent transduction pathway.
(A) Means±S.E.M. of the luciferase activity of Mv1Lu cell line transiently transfected with a luciferase reporter construct containing five copies of the +54/+74 sequence of the APP promoter inserted 5′ of the pGL3-Basic luciferase reporter vector containing the MLP sequence (APPTREMLP-luc). For each condition, transfected cells were treated with (black bars) or without (open bars) 1 ng/ml TGF-β1 for 24 h (n=12). (B) Means±S.E.M. of the luciferase activity of Mv1Lu cell line transiently co-transfected with the APPTREMLP-Luc and with expression vectors encoding autoactivated versions of receptors of the TGF-β family (black bars) or with the corresponding empty vector pCMV5 (open bar) (n=12). (C) Means±S.E.M. of the luciferase activity of Mv1Lu cell line transiently co-transfected with the APPTREMLP-Luc and with expression vectors encoding Smad transcription factors (black bars) or with the corresponding empty vector pcDNA3 (open bar) (n=12). (A–C) *P<0.01, ANOVA followed by the Bonferroni–Dunn test.
To characterize further the transduction pathway involved in the activation of APPTRE by TGF-β, we have transiently transfected Mv1Lu cells with expression vectors encoding constitutively activated versions of receptors for members of the TGF-β family (Alks). Expression of either Alk4 or Alk5, both related to TGF-β signalling, led to enhanced activity of the co-transfected APPTRE luciferase reporter vector (Figure 1B), whereas the other Alks, related to bone morphogenetic protein signalling, failed to modify the activity of this reporter construct. These results showed that the transcriptional activity of APPTRE is restricted to TGF-β1 and potentially Activin, among the TGF-β-related peptides.
TGF-β is known to transduce intracellularly its signal through activation of members of the Smad family of transcription factors. To investigate the role of these factors in the activation of APPTRE by TGF-β, we have transiently transfected Mv1Lu cells with expression vectors encoding different members of the Smad family. Among the different factors, only Smad3, known as an intracellular mediator of TGF-β signalling and, with a lower amplitude, Smad4, described as a common mediator for all members of the TGF-β family, were capable of inducing the activation of APPTRE promoter activity (Figure 1C). Smad1 and Smad5, known to mediate bone morphogenetic protein signalling and, more surprisingly, Smad2, related to TGF-β signalling, failed to promote the transcription of the APPTRE promoter. The inhibitory Smad, Smad7, also had no effect on the activity of the APPTRE reporter construct (Figure 1C).
Taken together, these results suggest that TGF-β, through its specific receptors, is capable of activating Smad3 but not Smad2, a transcription factor that, together with Smad4 transcription factor, targets the +54/+74 region of the APP promoter (APPTRE).
The APPTRE sequence contains critical response elements, in addition to the TGF-β-responsive element AGAC box
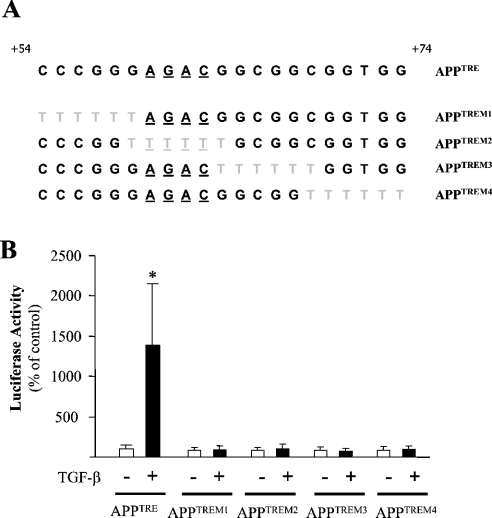
We next aimed at studying the response elements contained in the APPTRE sequence. It has been reported that for binding to DNA, Smad3/Smad4 complexes require the presence, in promoter sequences, of TGF-β target genes of the critical AGAC motif [15], also contained in the APPTRE sequence. We proceeded to perform several mutations of the APPTRE sequence contained in the APPTRE-MLP-Luc reporter vector (APPTREM1−4; Figure 2A). Four mutants were generated by turning the +54/+59, the +59/+64, the +64/+69, and the +69/+74 sequences respectively on a T6 sequence (Figure 2A). In doing so, we targeted not only the AGAC box contained in this sequence but also different portions of this sequence located upstream and downstream of the AGAC box. Among these mutations, only the TREM2 mutation overlapped the AGAC sequence contained in APPTRE. However, the four mutations we performed abolished the TGF-β-induced transcription activity of APP (Figure 2B). These results suggest that, in addition to the AGAC box, other elements contained in the APPTRE sequence are critical for mediating the response to TGF-β.
Figure 2. Mutations of the APPTRE block the transcriptional activation of APPTRE by TGF-β.
(A) Sequences of the wild-type (APPTRE) and mutated (APPTREM1 to APPTREM4) double-stranded decoy oligonucleotides used in the present study. The mutations performed in the APPTRE sequence are shown in grey. The critical TGF-β-responsive element AGAC has been underlined in each sequence. (B) Means±S.E.M. of the luciferase activity of Mv1Lu cell line transiently transfected with a luciferase reporter construct containing the wild-type APPTRE sequence or with mutated versions of this sequence (APPTREM1 to APPTREM4). For each condition, transfected cells were treated with (black bars) or without (open bars) 1 ng/ml TGF-β1 for 24 h (n=12). *P<0.01, ANOVA followed by the Bonferroni–Dunn test.
However, the loss of response induced by the TREM mutations could be explained by a modification of the close environment of the AGAC sequence, and does not reflect the actual presence of additional TGF-β-responsive elements within the APPTRE sequence. To consider this possibility, we performed decoy oligonucleotide assays to identify further the critical TGF-β-responsive elements contained in the APPTRE sequence. A decoy double-stranded oligonucleotide, whose sequence corresponds to the APPTRE, was generated and transfected to Mv1Lu cells in an attempt to interfere with the binding of TGF-β-activated transcription factors to their cis-acting elements within the APPTRE sequence. The use of this decoy oligonucleotide partially blocked the TGF-β-induced activation of the co-transfected APPTRE-MLP-Luc reporter vector (Figure 3B). Six point mutations of this initial decoy oligonucleotide were then generated: guanidines located at positions +57, +59, +61, +65 and +67 as well as a cytosine located at position +63 were respectively mutated to adenines (M1–M6, Figure 3A). Transfection of decoy oligonucleotides M4, M5 and M6 was efficient in reversing the TGF-β-induced activation of the co-transfected APPTRE-MLP-Luc reporter vector. However, decoy oligonucleotides M1, M2 and M3 failed to influence the response to TGF-β (Figure 3B).
These results indicate that the mutations of bases +57, +59 and +61 affected the ability of the decoy oligonucleotides to interfere with TGF-β-induced activation of APPTRE. Thus these results suggest that, in addition to the AGAC sequence, the region of APPTRE located between bases +54 and +59 plays a role in the APPTRE responsiveness to TGF-β signal.
The Sp1 transcription factor co-operates with Smad3 and Smad4 to activate the transcriptional activity of APPTRE
Our next step was to identify potential transcription factors that are capable of co-operating with Smad3 and Smad4 and controlling the activation of APP. Among the multiple cofactors of Smad identified so far, the Sp1 transcription factor appeared to be a good candidate as a cofactor of Smad3 on APPTRE promoter sequence, since it is potentially capable of binding to GC-rich sequences, such as the C3G3 motive located at positions +54 to +59.
To investigate the effect of Sp1 transcription factors on the promoter activity of APPTRE, we have transiently transfected Mv1Lu cells with an expression vector encoding Sp1. The activity of the co-transfected APPTRE-MLP-Luc reporter vector was not affected by the transfection of Sp1 alone (Figure 4A). However, the effect of TGF-β1 treatment on this reporter construct was strongly potentiated by the expression of Sp1 (Figure 4A). When co-transfected with an expression vector encoding Smad3, the Sp1 expression vector also potentiated the effect of the Smad3 expression on the activity of APPTRE-MLP-Luc reporter vector (Figure 4B). To confirm the implication of Sp1 in the transduction of TGF-β signal, we investigated whether TGF-β transcriptional effects could be reversed by antagonizing Sp1. Therefore, we tested the effect of another transcription factor related to Sp1, termed Sp3, on the TGF-β- and Smad3-induced activation of APPTRE-MLP-Luc reporter vector. Sp3 transcription factor is structurally related to Sp1 and these two proteins have been shown to act on the same GC-rich binding sites on DNA, potentially providing an Sp1 antagonist effect for Sp3. We transiently transfected Mv1Lu cells with an expression vector encoding Sp3. Although Sp3 expression alone did not modify the activity of APPTRE-MLP-Luc reporter vector, it could inhibit the effects of both TGF-β1 treatment (Figure 4C) and Smad3 expression (Figure 4D) on the activity of APPTRE-MLP-Luc reporter vector.
The above results suggest that Sp1 could be considered as a cofactor of Smad, without intrinsic effect on the transcriptional activity of the APPTRE sequence, but capable of positively modulating the TGF-β-dependent transcriptional activity of APP. Sp1 would then act as a co-activator of the Smad-dependent transduction pathway.
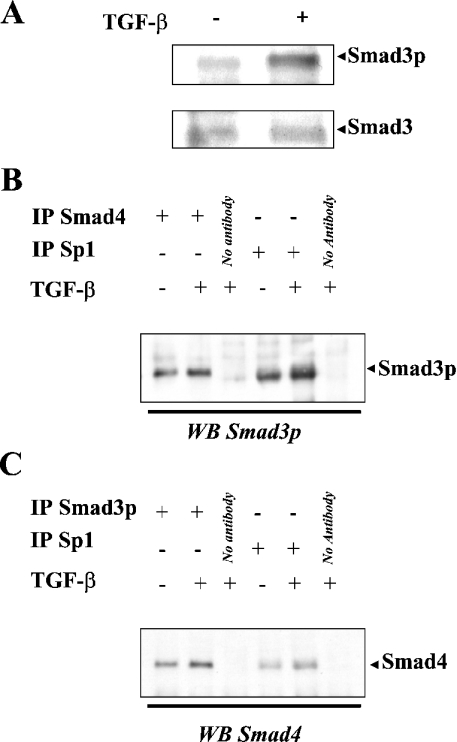
To study further the molecular mechanisms leading to the co-operation between Smad3 and Sp1, we extracted total proteins or proteins contained in nuclei of Mv1Lu cells, under control conditions or after treatment in the presence of 1 ng/ml TGF-β1 for 30 min. Total proteins were subjected to Western blotting, revealed with antibodies raised against either Smad3 or Smad3p (phosphorylated active form of Smad3). Although TGF-β1 did not modify the total amount of Smad3 protein, a larger amount of active Smad3 (Smad3p) was detected in TGF-β1-treated cells (Figure 5A). This result indicates that TGF-β induces the phosphorylation of pre-existing Smad3 proteins, not requiring de novo Smad3 protein synthesis. We then subjected proteins from nuclei to immunoprecipitation with an antibody raised against Smad4, followed by Western blotting revealed with an antibody raised against the phosphorylation-activated form of Smad3 to identify active Smad3/Smad4 complexes. As expected, we observed that such complexes were present in the nuclei of Mv1Lu cells under control conditions and that treatment with TGF-β enhanced the quantity of these complexes (Figure 5B). Conversely, immunoprecipitations using an antibody raised against active Smad3, followed by Western blotting revealed with an antibody raised against Smad4, led to the same observation (Figure 5C). Additional experiments were performed, using Sp1 immunoprecipitation, followed by Western blotting against activated Smad3. These experiments revealed the presence of active Smad3/Sp1 complexes, potentiated by TGF-β1 treatment (Figure 5B). Sp1-immunoprecipitated material was also subjected to Western blotting revealed with an anti-Smad4 antibody. The presence of Sp1/Smad4 complexes under control conditions and enhancement of the quantity of these complexes after TGF-β1 treatment were noticed (Figure 5C).
Figure 5. TGF-β induces the formation of Smad3–Smad4–Sp1 complexes.
(A) Total proteins were extracted from Mv1Lu cells under control conditions or after treatment with 1 ng/ml TGF-β1 for 24 h, and subjected to Western blotting, revealed with antibodies raised against total Smad3 or Smad3p. (B) Proteins were extracted from nuclei of Mv1Lu cells under control conditions or after treatment with 1 ng/ml TGF-β1 for 24 h and subjected to immunoprecipitation using antibodies raised against either Smad4 or Sp1 transcription factors. Smad4- and Sp1-immunoprecipitated material were then subjected to Western blotting revealed with an antibody raised against Smad3p. (C) Proteins were extracted from nuclei of Mv1Lu cells under control conditions or after treatment with 1 ng/ml TGF-β1 for 24 h and subjected to immunoprecipitation using antibodies raised either against Smad3p or Sp1 transcription factors. Smad3p- and Sp1-immunoprecipitated material were then subjected to Western blotting revealed with an antibody raised against Smad4. No antibody, immunoprecipitation control experiment performed in the absence of IgG.
We were able to show that Sp1 transcription factor is capable of interacting physically with active Smad3 and Smad4 transcription factors. Taken together, the above results indicate that TGF-β is capable of inducing the formation of complexes containing the active form of Smad3, Smad4 and Sp1.
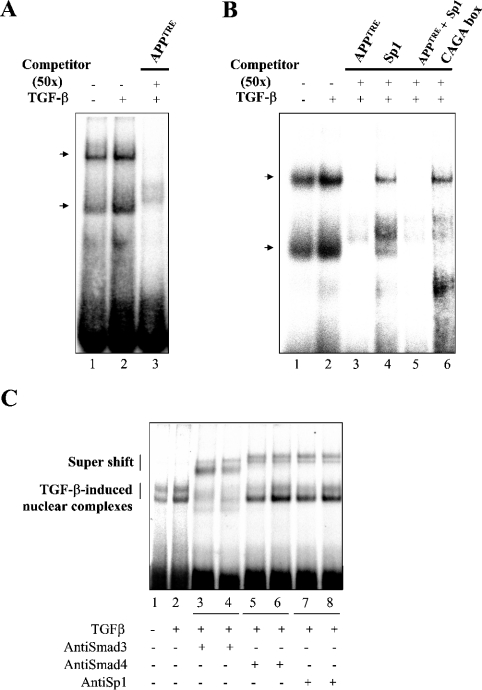
In a next step, we attempted to characterize the DNA-binding activity of these complexes on the APPTRE sequence. We performed EMSA from nuclear extracts of Mv1Lu treated in the presence of 1 ng/ml TGF-β1 for a time period of 30 min, using a 32P-radiolabelled probe corresponding to the APPTRE sequence. As described previously for the CAGA box [15], two bands were observed, corresponding to two binding complexes (Figure 6A). These complexes were detected under control conditions, and clearly increased after TGF-β exposure (Figure 6A, lanes 1, 2; Figure 6B, lanes 1, 2). The specificity of this binding was confirmed by the observation that the bands corresponding to these complexes were displaced by an excess of unlabelled APPTRE oligonucleotide (Figure 6A, lane 3; Figure 6B, lane 3). Furthermore, we observed that an unlabelled oligonucleotide corresponding to the Sp1 consensus binding site was also capable of partly displacing the bands corresponding to binding complexes to APPTRE (Figure 6B, lane 4). Although unlabelled oligonucleotides corresponding to the Sp1-binding site and to the APPTRE sequence, when applied alone, were only able to displace partially the bands corresponding to APPTRE-binding complexes described above (Figure 6B, lanes 3 and 4), these bands were completely displaced when the two unlabelled oligonucleotides were used in combination (Figure 6B, lane 5). Finally, to identify further the elements contained in the APPTRE-binding complexes, we also applied an excess of an unlabelled oligonucleotide corresponding to the previously characterized TGF-β-responsive element CAGA-box [15] (Figure 6B, lane 6). This sequence is contained in the promoter of the PAI-1 (plasminogen activator inhibitor-1) gene and has been shown to bind Smad3/Smad4 but not Smad2/Smad4 complexes of transcription factors [15]. We observed that an unlabelled oligonucleotide corresponding to this sequence was capable of partially displacing the APPTRE-binding complexes (Figure 6B, lane 6), indicating that these complexes are, at least in part, composed of Smad3 and Smad4 proteins. Antibody interference assays were finally performed to identify the transcription factors binding to the APPTRE-sequence. TGF-β-induced nuclear complexes were supershifted with antibodies against Smad3 (Figure 6C, lanes 3 and 4), Smad 4 (Figure 6C, lanes 5 and 6) and Sp1 (Figure 6C, lanes 7 and 8), indicating that these proteins are capable of binding the APPTRE sequence.
Figure 6. Binding properties of the APPTRE sequence.
Nuclear extracts from Mv1Lu cells under control conditions (A, lane 1; B, lane 1) after treatement with TGF-β1 (1 ng/ml, 30 min; A, lanes 2, 3; B, lanes 2–6) were subjected to EMSA using 32P-labelled oligonucleotides corresponding to the APPTRE sequence. A 50-fold excess of unlabelled oligonucleotides corresponding to the APPTRE sequence (A, lane 3; B, lane 3), to the consensus binding site for Sp1 transcription factor (B, lane 4) to the APPTRE sequence and to the consensus binding site for Sp1 transcription factor (B, lane 5) or to the consensus binding site for Smad3 (CAGA box; B, lane 6) were added as competitors. Arrows indicate the TGF-β-induced nuclear complexes. (C) Antibody interference assays using specific antibodies were performed in duplicate (lanes 3 and 4, anti Smad3; lanes 5 and 6, anti Smad4; lanes 7 and 8, anti Sp1) as indicated in the Experimental section. TGF-β-induced nuclear complexes and supershifted complexes are indicated in the Figure.
Altogether, these results indicate that TGF-β1 induces the formation of nuclear complexes containing Sp1, Smad3 and Smad4, capable of binding to the APPTRE sequence, and that these complexes participate in the regulation of transcriptional activity of the APP promoter.
DISCUSSION
Aβ peptide accumulation is considered as one of the critical events leading to the development of AD, and it is assumed that a better understanding of the mechanisms leading to the production of this peptide would help to develop new treatments for AD. One critical point concerns the activity of the APP secretases that lead to the production of different APP fragments, including Aβ, from the APP peptide. Another field of research concerns the study of the mechanisms leading to the synthesis of APP peptide. We report in the present study new elements concerning the molecular events sustaining the effect of the cytokine TGF-β on the transcription of the APP gene. These results are of particular interest since inflammation has been reported in numerous neurodegenerative disorders such as Parkinson's disease, stroke and AD as a key element in the progression of the disease. In AD, the inflammatory response is mainly located at the vicinity of amyloid plaques. Cytokines, such as interleukins-1-, -6-, tumour necrosis factor-α and TGF-β have been found to be involved in this inflammatory process. Although their expression is induced by the presence of Aβ, these cytokines are also capable of promoting the accumulation of Aβ, thus leading to a vicious circle in the progression of the disease.
We document here that a short DNA sequence, referred to as APPTRE, contained in the 5′-UTR region of the APP promoter, is capable of responding positively to TGF-β signal through the recruitment of Smad3 and Smad4 transcription factors, which would co-operate with Sp1. Our observation of the presence of a TGF-β-responsive element within the APP promoter is consistent with previous studies concerning the effects of TGF-β on the transcription of various genes including PAI-1, Muc5ac mucin and α2(I) procollagen [15–17]. As we have also observed in the APPTRE sequence, all these promoters share the AGAC motif, which is now considered as a critical TGF-β-responsive element in the promoters of TGF-β-activated genes. This sequence is considered to be necessary for binding of TGF-β-induced Smad2/Smad4 and Smad3/Smad4 complexes of transcription factors to the promoters of TGF-β target genes. However, the affinity of Smad proteins for DNA is relatively low, so that additional transcription factors, complexing to Smads, appear to be necessary for an efficient transcriptional response to TGF-β. These cofactors have been classified into different subtypes. Some of them, such as FAST and Mixer proteins, function as adaptators of Smad and lack intrinsic transcriptional activity. Others, such as JUNB or TFE3, can co-operate with Smad proteins on target promoters but also display their own transcriptional activity, independent of Smads, in other contexts [13]. We have noted, in the present study, that Sp1 transcription factor is capable of potentiating TGF-β-induced promoter activity of APPTRE, probably through a physical interaction with Smad3/Smad4 complexes. These results are in accordance with previous studies reporting a co-operation of Sp1 with Smad proteins in mediating the TGF-β-induced activation of various genes including PAI-1, p21Waf1, α2(I) collagen or the β5 integrin subunit [18–21]. In our work, Sp1 did not show any transcriptional activity by itself on the APPTRE sequence, which would define Sp1 as a co-activator adaptator of Smad3/Smad4 complexes on the APP promoter.
We also report that Smad3, but not Smad2, is capable of inducing the transcriptional activity of the APPTRE. This discrepancy between Smad2 and Smad3 has already been observed by other groups on different TGF-β- and activin-responsive elements [15,22]. This specificity of action of Smad3 could be explained by its structural differences from Smad2. If these two factors have very similar structures, it appears that Smad3 lacks an insert that is present in Smad2, close to the DNA-binding site of this protein, and that would prevent direct binding of Smad2 to DNA [23,24].
By using EMSA, we showed that TGF-β1 induces the binding of nuclear factors to the APPTRE sequence. Two distinct TGF-β-induced binding complexes could be observed, which is in accordance with previous reports [15] concerning another TGF-β-responsive element, CAGA box. In this previous study [15], these two CAGA-box-binding complexes had been demonstrated to contain both Smad3 and Smad4 transcription factors. In the present study, an oligonucleotide corresponding to this CAGA-box sequence was able to compete with the binding of TGF-β-induced complexes to APPTRE, suggesting that Smad3 and Smad4 are present in the APPTRE binding complexes. Similarly, an oligonucleotide corresponding to the Sp1 consensus binding site was also able to displace these complexes. In conclusion, our present results strongly support the idea that TGF-β induces complexes of Smad3 and Smad4 transcription factors, associated with the Sp1 protein.
Overall, this study increases our understanding of how TGF-β discriminates between its various target genes depending on the context. Alternative pathways involving Smad2 or Smad3, as well as recruitment of different cofactors of Smads, such as Sp1, participate in this discrimination. This better understanding appears to be critical for designing new, efficient therapeutic strategies that would be based on action at the promoter level. Considering the multiple targets of TGF-β, this would enable them to act more specifically on genes of therapeutic interest. In the particular context of AD, strategies aiming at interfering with transduction pathways of cytokines such as TGF-β to down-regulate the synthesis of APP at the transcriptional level could prove efficient in the future. Moreover, these results suggest that the overexpression of TGF-β may initiate or promote amyloidogenesis in AD and may thus be a risk factor for developing AD, even though no polymorphism has so far been detected in its gene at this time.
Acknowledgments
We thank Professor Philippe Galéra for his useful comments. MLP-Luc reporter vector, Alks and Smad plasmids were provided by Dr Joan Massagué (The Sloane Kettering Cancer Center, New York, NY, U.S.A.), Dr Peter ten Dijke and Dr Sylvianne Dennler, both from The Netherlands Cancer Institute (Amsterdam, The Netherlands). This work was supported by grants from the Regional Council of Lower Normandy (to F. D. and S. L.) and the French National Center for Scientific Research CNRS (to C. G.).
References
- 1.Griffin W. S., Stanley L. C., Ling C., White L., MacLeod V., Perrot L. J., White C. L., III, Araoz C. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1989;86:7611–7615. doi: 10.1073/pnas.86.19.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fillit H., Ding W. H., Buee L., Kalman J., Altstiel L., Lawlor B., Wolf-Klein G. Elevated circulating tumor necrosis factor levels in Alzheimer's disease. Neurosci. Lett. 1991;129:318–320. doi: 10.1016/0304-3940(91)90490-k. [DOI] [PubMed] [Google Scholar]
- 3.van der Wal E. A., Gomez-Pinilla F., Cotman C. W. Transforming growth factor-beta 1 is in plaques in Alzheimer and Down pathologies. Neuroreport. 1993;4:69–72. doi: 10.1097/00001756-199301000-00018. [DOI] [PubMed] [Google Scholar]
- 4.Huell M., Strauss S., Volk B., Berger M., Bauer J. Interleukin-6 is present in early stages of plaque formation and is restricted to the brains of Alzheimer's disease patients. Acta Neuropathol. 1995;89:544–551. doi: 10.1007/BF00571510. [DOI] [PubMed] [Google Scholar]
- 5.Flanders K. C., Lippa C. F., Smith T. W., Pollen D. A., Sporn M. B. Altered expression of transforming growth factor-beta in Alzheimer's disease. Neurology. 1995;45:1561–1569. doi: 10.1212/wnl.45.8.1561. [DOI] [PubMed] [Google Scholar]
- 6.Wyss-Coray T., Masliah E., Mallory M., McConlogue L., Johnson-Wood K., Lin C., Mucke L. Amyloidogenic role of cytokine TGF-beta1 in transgenic mice and in Alzheimer's disease. Nature (London) 1997;389:603–606. doi: 10.1038/39321. [DOI] [PubMed] [Google Scholar]
- 7.Chao C. C., Ala T. A., Hu S., Crossley K. B., Sherman R. E., Peterson P. K., Frey W. H., 2nd Serum cytokine levels in patients with Alzheimer's disease. Clin. Diagn. Lab. Immunol. 1994;1:433–436. doi: 10.1128/cdli.1.4.433-436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesné S., Docagne F., Gabriel C., Liot G., Lahiri D. K., Buée L., Plawinski L., Delacourte A., MacKenzie E. T., Buisson A., Vivien D. Transforming growth factor-beta 1 potentiates amyloid-beta generation in astrocytes and in transgenic mice. J. Biol. Chem. 2003;278:18408–18418. doi: 10.1074/jbc.M300819200. [DOI] [PubMed] [Google Scholar]
- 9.Gray C. W., Patel A. J. Regulation of beta-amyloid precursor protein isoform mRNAs by transforming growth factor-beta 1 and interleukin-1 beta in astrocytes. Brain Res. Mol. Brain Res. 1993;19:251–256. doi: 10.1016/0169-328x(93)90037-p. [DOI] [PubMed] [Google Scholar]
- 10.Amara F. M., Junaid A., Clough R. R., Liang B. TGF-beta(1), regulation of alzheimer amyloid precursor protein mRNA expression in a normal human astrocyte cell line: mRNA stabilization. Brain Res. Mol. Brain Res. 1999;71:42–49. doi: 10.1016/s0169-328x(99)00158-8. [DOI] [PubMed] [Google Scholar]
- 11.Wyss-Coray T., Lin C., Yan F., Yu G. Q., Rohde M., McConlogue L., Masliah E., Mucke L. TGF-beta1 promotes microglial amyloid-beta clearance and reduces plaque burden in transgenic mice. Nat. Med. 2001;7:612–618. doi: 10.1038/87945. [DOI] [PubMed] [Google Scholar]
- 12.Massagué J. The transforming growth factor-beta family. Annu. Rev. Cell. Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 13.Massagué J. How cells read TGF-beta signals. Nat. Rev. Mol. Cell. Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 14.Massagué J., Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonckheere N., Van Der Sluis M., Velghe A., Buisine M. P., Sutmuller M., Ducourouble M. P., Pigny P., Buller H. A., Aubert J. P., Einerhand A. W., et al. Transcriptional activation of the murine Muc5ac mucin gene in epithelial cancer cells by TGF-beta/Smad4 signalling pathway is potentiated by Sp1. Biochem. J. 2003;377:797–808. doi: 10.1042/BJ20030948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S. J., Yuan W., Lo S., Trojanowska M., Varga J. J. Interaction of smad3 with a proximal smad-binding element of the human alpha2(I) procollagen gene promoter required for transcriptional activation by TGF-beta. Cell Physiol. 2000;3:381–392. doi: 10.1002/(SICI)1097-4652(200006)183:3<381::AID-JCP11>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 18.Datta P. K., Blake M. C., Moses H. L. Regulation of plasminogen activator inhibitor-1 expression by transforming growth factor-beta-induced physical and functional interactions between smads and Sp1. J. Biol. Chem. 2000;275:40014–40019. doi: 10.1074/jbc.C000508200. [DOI] [PubMed] [Google Scholar]
- 19.Pardali K., Kurisaki A., Morén A., ten Dijke P., Kardassis D., Moustakas A. Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-beta. J. Biol. Chem. 2000;275:29244–29256. doi: 10.1074/jbc.M909467199. [DOI] [PubMed] [Google Scholar]
- 20.Poncelet A. C., Schnaper H. W. Sp1 and Smad proteins cooperate to mediate transforming growth factor-beta 1-induced alpha 2(I) collagen expression in human glomerular mesangial cells. J. Biol. Chem. 2001;276:6983–6992. doi: 10.1074/jbc.M006442200. [DOI] [PubMed] [Google Scholar]
- 21.Lai C.-F., Feng X., Nishimura R., Teitelbaum S. L., Avioli L. V., Ross F. P., Cheng S.-L. Transforming growth factor-beta up-regulates the beta 5 integrin subunit expression via Sp1 and Smad signaling. J. Biol. Chem. 2000;275:36400–36406. doi: 10.1074/jbc.M002131200. [DOI] [PubMed] [Google Scholar]
- 22.Nagarajan R. P., Chen Y. Structural basis for the functional difference between Smad2 and Smad3 in FAST-2 (forkhead activin signal transducer-2)-mediated transcription. Biochem. J. 2000;350:253–259. [PMC free article] [PubMed] [Google Scholar]
- 23.Yagi K., Goto D., Hamamoto T., Takenoshita S., Katom M., Miyazono K. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J. Biol. Chem. 1999;274:703–709. doi: 10.1074/jbc.274.2.703. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y., Wang Y. F., Jayaraman L., Yang H., Massagué J., Pavletich N. P. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell (Cambridge, Mass.) 1998;5:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]