Abstract
Secondary contact between closely related taxa represents a “moment of truth” for speciation—an opportunity to test the efficacy of reproductive isolation that evolved in allopatry and to identify the genetic, behavioral, and/or ecological barriers that separate species in sympatry. Sex chromosomes are known to rapidly accumulate differences between species, an effect that may be exacerbated for neo-sex chromosomes that are transitioning from autosomal to sex-specific inheritance. Here we report that, in the Solomon Islands, two closely related bird species in the honeyeater family—Myzomela cardinalis and Myzomela tristrami—carry neo-sex chromosomes and have come into recent secondary contact after ~1.1 my of geographic isolation. Hybrids of the two species were first observed in sympatry ~100 years ago. To determine the genetic consequences of hybridization, we use population genomic analyses of individuals sampled in allopatry and in sympatry to characterize gene flow in the contact zone. Using genome-wide estimates of diversity, differentiation, and divergence, we find that the degree and direction of introgression varies dramatically across the genome. For sympatric birds, autosomal introgression is bidirectional, with phenotypic hybrids and phenotypic parentals of both species showing admixed ancestry. In other regions of the genome, however, the story is different. While introgression on the Z/neo-Z-linked sequence is limited, introgression of W/neo-W regions and mitochondrial sequence (mtDNA) is highly asymmetric, moving only from the invading M. cardinalis to the resident M. tristrami. The recent hybridization between these species has thus enabled gene flow in some genomic regions but the interaction of admixture, asymmetric mate choice, and/or natural selection has led to the variation in the amount and direction of gene flow at sex-linked regions of the genome.
Author summary
When an invasive species colonizes an island and interacts with a closely related resident species, we are provided with a rare opportunity to study the consequences of interbreeding and the factors that keep species distinct. Regions of the genome that evolve rapidly and/or influence mate choice may be especially likely to act as incidental barriers to gene flow. The red Myzomela cardinalis, birds in the honeyeater family, have invaded Makira in the Solomon Islands, coming into contact with the endemic, all black Myzomela tristrami. We used population genomic analyses of individuals in geographic isolation, and those in geographic contact, to understand the history of these two species and the consequences of their recent range overlap. We found that sex-specific regions of the genome (i.e., sex chromosomes) were either limited in their ability to move between species, or only moved in one direction, from the invading M. cardinalis to the resident M. tristrami. This work highlights how certain regions of the genome may be especially important in defining species boundaries and the generation and maintenance of biodiversity.
Introduction
When taxa are geographically isolated, it is difficult to know whether or not they are “good” biological species that are no longer reproductively compatible with each other [1–3]. Secondary geographic contact, therefore, provides a kind of “moment of truth” for speciation—an opportunity to test the efficacy of reproductive isolation in sympatry [1,4]. The presence or absence of interspecific pairings and/or hybrid offspring serve as phenotypic proxies for reproductive isolation, but with genetic data we now know that hybridization between seemingly good biological species is not uncommon (reviewed by [5]). A genic view of speciation allows us to distinguish compatible regions of the genome from those that maintain species divergence [6]. Typically, secondary contact is studied in long-standing hybrid or tension zones [7,8], in which the interaction of gene flow, selection, and recombination occurring over hundreds to thousands of generations allows specific loci to be identified that either move between species or are refractory to introgression due to selection and linkage [9–13]. The consequences of hybridization at the initiation of secondary contact may be transient and difficult to observe. Studying systems in which sympatry is hypothesized to have occurred relatively recently can therefore be especially important to understanding the genetic and/or phenotypic factors which either facilitate or prevent gene flow in the earliest stages of secondary contact [14–16].
Interspecific introgression can occur if traits under sexual or natural selection are globally adaptive, increasing fitness in the genomic background of either species [17,18]. Neutral alleles can introgress due to the demographic dynamics of range expansion precipitating the contact event [19]. However, introgression can be limited for locally adaptive alleles involved in sexual, ecological, and/or intrinsic genetic incompatibilities. Sexual incompatibilities may result from differences in courtship signals or mate preferences [1,20,21]. Ecological incompatibilities can result if hybrids possess intermediate phenotypes poorly suited to either parental habitat [22,23]. Intrinsic genetic incompatibilities can reduce the fertility or viability of hybrids [24–26].
Incompatibilities may be especially likely to arise on sex chromosomes, as these regions of the genome are expected to diverge rapidly. Sex chromosomes have lower effective population sizes compared to autosomes and selection in the heterogametic sex can, under some conditions, lead to “faster-X” (or -Z) evolution [27–30]. Although demography, mating system, and dosage compensation may mediate the strength of faster-X evolution [31], empirical evidence confirms that sex-linked regions show elevated substitution rates and rapid divergence of gene expression for a wide range of taxa [29,32,33]. The rapid evolution on sex chromosomes may explain their disproportionately large role in speciation [34,35]. Species recognition [36] and mating behaviors [37] have been mapped to sex-linked loci, suggesting these regions can be important in maintaining species boundaries via sexual incompatibilities [38]. Sex chromosomes are also known to limit gene flow between taxa through genetic incompatibilities that contribute to Haldane’s rule [34,39] and/or large X/Z effect [34,40–44].
Neo-sex chromosomes—often formed by the fusion of an autosome to an existing sex chromosome—experience a shift to sex-specific transmission, becoming heterogametic in one sex and homogametic in the other [45–48]. The rapid evolutionary transition from autosomal to sex-linked inheritance could enrich neo-sex chromosomes for sexual, ecological, and/or genetic incompatibilities, which reduce gene flow between taxa [49–52]. Indeed, neo-sex chromosomes have been implicated in speciation in plants, insects, and fish [47,49,52]. In birds, karyotype evolution was thought to be largely conservative, with sex chromosomes being syntenic [53,54]. However, neo-sex chromosomes have now been discovered in several bird lineages [45,48,55–61]. Despite their potential importance, few studies have considered the role of neo-sex chromosomes in reproductive isolation in avian taxa [62,63].
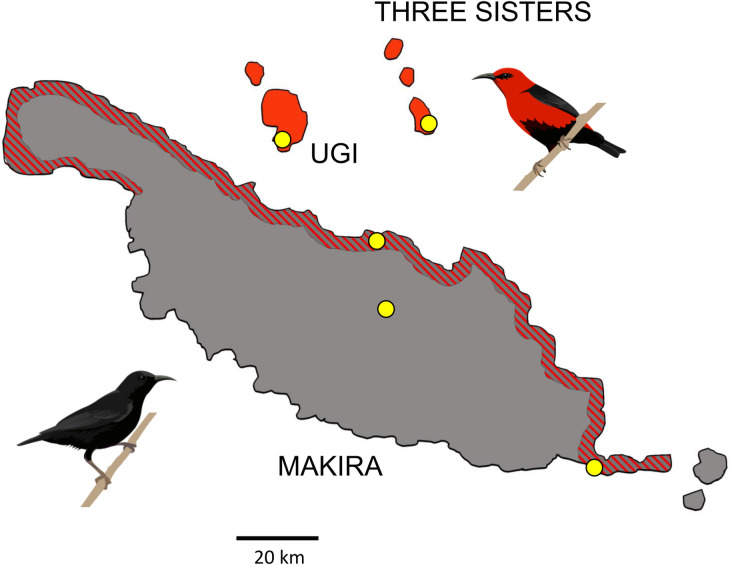
Myzomela honeyeaters of the Solomon Islands present an opportunity to study the genomics of reproductive isolation in a system with neo-sex chromosomes [4,64,65]. The sexually monomorphic, all black Sooty honeyeater (M. tristrami) is endemic to the large island of Makira (Fig 1). The sexually dimorphic, red Cardinal honeyeater (M. cardinalis), which shared a common ancestor with M. tristrami <3 mya [66], is distributed across many islands of the South Pacific, including the small satellite islands of Ugi and Three Sisters, 8 km and 20 km from Makira, respectively (Fig 1). Secondary contact occurred recently when M. cardinalis expanded its range and established a narrow, coastal region of sympatry with M. tristrami. Birds with intermediate plumage collected on Makira in 1908 were identified as putative hybrids [67] and, in 1927, Ernst Mayr collected phenotypically pure M. cardinalis as well as putative hybrids as part of the Whitney South Seas Expedition [68]. In subsequent expeditions no phenotypic hybrids were collected, and M. cardinalis was found to be more abundant than the native M. tristrami in sympatry [67], leading Mayr and Diamond [4] to propose that hybridization occurred only when conspecific mates were scarce. Preliminary genetic investigations of the system using six nuclear and two mitochondrial loci uncovered evidence for gene flow and for potential neo-sex chromosomes by aligning ddRADseq data to the Zebra Finch reference genome [64,65].
Fig 1.
Sampling sites (yellow dots) for allopatric Mtris (black bird, gray regions), allopatric Mcard (red bird, red regions), and sympatric individuals of both species and phenotypic hybrids (red and gray striped regions). Map downloaded and modified from https://download.geofabrik.de/australia-oceania/solomon-islands.html, illustrations by Emily Powell.
We revisit this classic case of recent secondary contact to address three major questions. (1) What is the history of divergence and secondary contact between M. cardinalis and M. tristrami? (2) What is the extent of admixture in the newly established region of sympatry? (3) How does the amount and direction of introgression vary across the genome and, in particular, on sex and neo-sex chromosome sequence? To answer these questions, we use a new, high quality reference genome for M. tristrami, and whole-genome resequencing for 143 individuals, including samples from both allopatric and sympatric populations of each species. We find evidence for bidirectional introgression on autosomes, limited introgression on the ancestral Z and neo-Z regions, and strong asymmetric introgression of W, neo-W, and mitochondria from M. cardinalis into M. tristrami.
Results and discussion
Neo-sex chromosomes formed by fusion of sex chromosomes with chromosome 5
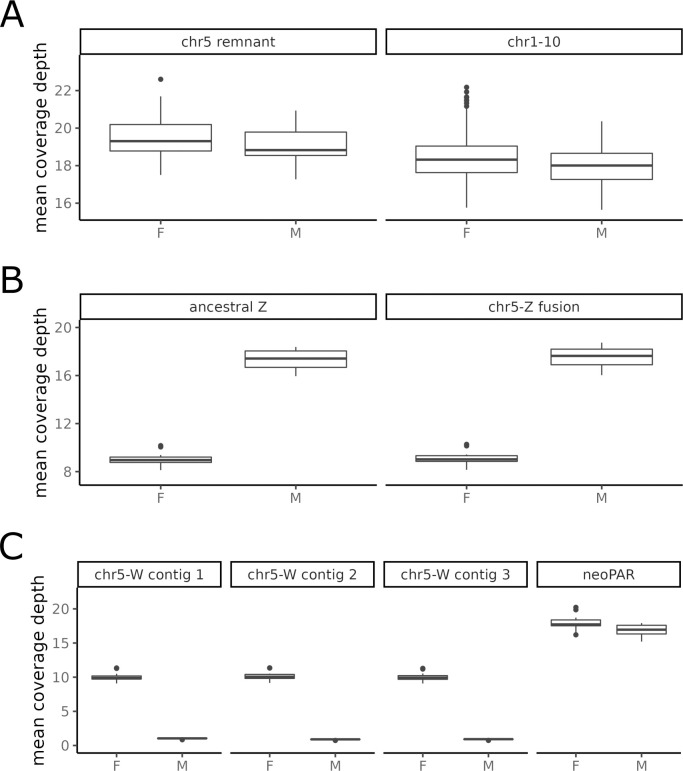
We generated a highly contiguous, chromosome-level reference assembly for a Myzomela tristrami (Mtris) female using PacBio HiFi long-read data at approximately 70X autosomal coverage. The primary raw assembly had an N50 of 25.7 Mb and a total length of 1505.7 Mb (S1 Table). After scaffolding and removal of autosomal haplotigs, we conducted a quantitative assessment of conserved avian single-copy orthologs using BUSCO [69], finding an overall BUSCO completeness score of 96.6%. The completed reference genome has a length of 1257.8 Mb and consists of 31 standard autosomes, 11 tentative microchromosomes, a mitochondrial genome, and partially scaffolded Z and W chromosomes. For the Z chromosome we overall find broad collinearity between our assembly and the published assembly of the Z chromosome from a closely related species, Lichenostomus melanops cassidix [70], with a major difference: we detect fusion between the sex chromosomes and an autosome (S1 Fig). We find, in particular, that ~86% of chromosome 5 (chr5) is now fused to Z- and W-linked contigs, while the remaining 14% of chr5 (hereafter “chr5 remnant”) assembles separately (S1 Fig). A recently published long-read reference assembly for another honeyeater, Entomyzon cyanotis, identified the same fusion between chr5 and ancestral Z sequence and two contigs from an independently assembling region of former chr5 corresponding to our chr5 remnant (their scaffold 13 and contig 14, S1 Fig) [62]. Raw coverage of short-read data from 10 male and 10 female Mtris birds (allopatric sample; see below) shows that chr5 remnant has similar coverage in the two sexes, comparable with that of autosomes (Fig 2A). In striking contrast, reads mapping to the chr5 region fused to chromosome Z show a sex difference in mean coverage—~2-fold lower in females than in males—that is inconsistent with being an autosome but consistent with Z-linkage (Fig 2B). We hereafter refer to the Z-fused chr5 region as the neo-Z region. Reads mapping to the chr5 region fused to three of the chromosome W contigs are nearly exclusive to females, inconsistent with being an autosome but consistent with W-linkage (Fig 2C). We refer to this W-fused chr5 region hereafter as neo-W sequence. Although neo-Z and neo-W chr5 derived segments are homologous, our ability to map reads specifically to neo-Z versus to neo-W chromosome arms implies appreciable sequence differentiation that could only accrue over time in the absence of recombination. Finally, the remaining ~18Mb of chr5 sequence not captured by either the chr5 remnant or the rearranged neo-Z or neo-W segments is fused to a small segment of ancestral W sequence in our assembly; in the E. cyanotis assembly this region is part of the “added-Z” contig 4 (S1 Fig). The region shows comparable coverage in both sexes (Fig 2C). We conjecture that this region may be acting as a pseudo-autosomal region (PAR), mediating meiotic recombination on the fused chromosomes, and refer to it as the neo-PAR. We have kept the sex chromosome contigs partially unscaffolded to minimize assumptions of organization. However, we expect the W chromosome to be a contiguous sequence composed of the four contigs identified, while the Z chromosome is expected to be composed of the ancestral Z, the Z1-Z2 fusion contig, and the neo-PAR portion of the fourth W contig. Further in-depth analyses of the origins, structure, and evolution of the neo-sex chromosomes in Myzomela will be discussed elsewhere.
Fig 2.
Raw short read sequence coverage from male (M) and female (F) Mtris individuals in the allopatric dataset mapped to Mtris reference assembly for chromosome 5 remnant contig and representative autosomes (chr1-10; A), Z-linked contigs (B) and W-linked contigs (C).
Allopatric populations show distinct demographic histories
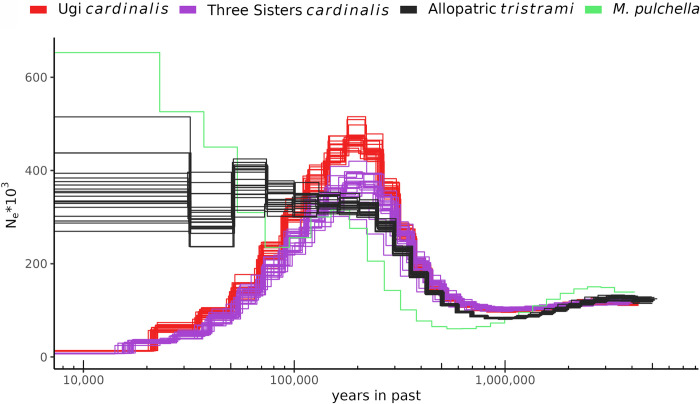
For population genomics analyses, we generated short-read sequence data for a total of 143 individuals: 60 sampled from allopatric and 70 from sympatric regions of the Mtris and M. cardinalis (Mcard) ranges; 12 phenotypic hybrids; and 1 individual from an outgroup species, M. pulchella (Mpulc; S2 Table and Fig 1). After alignment to the Mtris reference genome and filtering for quality and depth (see Methods), our dataset consisted of 30,283,937 single nucleotide polymorphisms (S3 Table). To infer the speciation and demographic histories of the allopatric populations of Mtris, Mcard (Ugi and Three Sisters), and the outgroup Mpulc, we used pairwise sequential Markov coalescent (PSMC) analyses of the autosomes [71]. These analyses suggest that Mtris and Mcard diverged ~1.1 mya and, then, both later experienced a period of sustained expansion (Fig 3). During the past ~200Ky, however, the two exhibit starkly different demographic histories: the inferred effective population size (Ne) of Mtris has been relatively stable, whereas that for Mcard shows evidence of steady decline (Fig 3). The scattered geographic distribution of Mcard subspecies across south Pacific islands suggests Ugi and Three Sisters populations may be at the leading edge of the species’ range [4]. We therefore infer that the reduction in Ne reflects its history of serial founder events during geographic expansion via repeated island colonization.
Fig 3. PSMC analyses of allopatric Myzomela.
PSMC inferred demographic history using autosomes of allopatric individuals show similar, declining effective populations sizes for Mcard on Ugi and Three Sisters, while Mtris and the outgroup species Mpulc have maintained or increased effective population size in the recent past.
The smaller Ne of allopatric Mcard versus Mtris is consistent with the lower average nucleotide diversities across all genomic compartments, including autosomes, neo-PAR, Z, neo-Z, W, neo-W, and mitochondria (Tables 1 and S4). Tajima’s D for Mtris autosomes show an excess of rare SNPs consistent with modest recent population growth, whereas Mcard autosomes show an excess of intermediate-frequency SNPs consistent with a recent reduction in Ne (Table 2). In particular, the Three Sisters population of Mcard shows lower nucleotide diversity and more positive Tajima’s D values for W and neo-W compared to Ugi Mcard, potentially indicating a more recent population reduction and/or founder event; alternatively, the smaller geographic area of Three Sisters (1/6th that of Ugi) may support fewer individuals. Sex chromosome diversity is considerably lower than autosomal diversity in both species. The Z/A ratio of diversity is, for example, much lower than the 0.75 expected on the assumptions of equal sex ratios, random mating, and equal mutation rates in the two sexes: for allopatric Mtris, Z/A = 0.646, whereas for Ugi Mcard, Z/A = 0.452 and for Three Sisters Mcard, Z/A = 0.298 (S5 Table). The lower Z/A ratio in allopatric Mcard is also consistent with a population bottleneck in its recent history. The difference between the Ugi versus Three Sisters populations of Mcard in Z/A nucleotide diversity may reflect population-specific demographic histories and/or sweeps in the Z/neo-Z region [72]. Together, these data suggest that the allopatric Mcard populations are relatively new arrivals to Ugi and Three Sisters. Their most recent dispersal event was to Makira, where they encountered the historically stable population of the endemic Mtris. A recent invasion of Makira by Mcard is consistent with the phenotypic observations of expeditions in the 20th century and the hypothesized history proposed in the literature [4]. We next turn to the genomic consequences of secondary contact.
Table 1. Nucleotide diversity, π.
| population | n | autosome | neo-PAR | Z | neo-Z | W | neo-W | mtDNA |
|---|---|---|---|---|---|---|---|---|
| Myzomela cardinalis | ||||||||
| Allopatry | 40 | 0.0023 (7 x 10−6) |
0.0025 (3.8 x 10−5) | 0.0009 (1.9 x 10−5) | 0.0008 (2 x 10−5) |
5 x 10−6 (1 x 10−6) |
4 x 10−6 (0) |
0.0004 (NA) |
| Sympatry | 40 | 0.0025 (7 x 10−6) |
0.0026 (4.1 x 10−5) | 0.0010 (1.9 x 10−5) | 0.0007 (2.1 x 10−5) | 5 x 10−6 (1 x 10−6) |
4 x 10−6 (0) |
0.0002 (NA) |
| Myzomela tristrami | ||||||||
| Allopatry | 20 | 0.0028 (9 x 10−6) |
0.0034 (4.2 x 10−5) | 0.0017 (2 x 10−5) |
0.0015 (1.7 x 10−5) | 2.3 x 10−5 (2 x 10−6) |
2 x 10−5 (0) |
0.0013 (NA) |
| Sympatry | 30 | 0.0029 (9 x 10−6) |
0.0034 (4.1 x 10−5) | 0.0018 (2.1 x 10−5) | 0.0015 (1.7 x 10−5) |
0.0006 (1.5 x 10−5) | 0.0006 (4 x 10−6) |
0.0138 (NA) |
Nucleotide diversity (π) averaged across 50 kb windows, standard error in parentheses for each sampled population of phenotypic parental Myzomela cardinalis and M. tristrami. Number of individuals for each species/sampling region shown in column n.
Table 2. Tajima’s D.
| population | autosome | neo-PAR | Z | neo-Z | W | neo-W | mtDNA |
|---|---|---|---|---|---|---|---|
| Myzomela cardinalis | |||||||
| Ugi | 0.6466 (0.0059) | 0.5342 (0.0336) | 0.3706 (0.0402) | 0.4694 (0.059) | -0.2637 (0.067) | -0.2402 (0.0263) | -1.4813 (NA) |
| Three Sisters | 0.7613 (0.008) | 0.5563 (0.0468) | 0.1301 (0.042) | -0.0289 (0.0644) | 0.0804 (0.0698) | 0.0779 (0.0263) | 0.9942 (NA) |
| Sympatry | 0.2976 (0.0056) | 0.3412 (0.0366) | -0.3043 (0.0435) | -0.9828 (0.061) | -0.2246 (0.0749) | -0.3755 (0.0246) | -1.3924 (NA) |
| Myzomela tristrami | |||||||
| Allopatry | -1.1067 (0.0026) | -1.0639 (0.0125) | -0.8908 (0.0094) | -0.7560 (0.0149) | -0.3088 (0.046) | -0.3165 (0.0194) | -0.8481 (NA) |
| Sympatry | -1.2067 (0.0024) | -1.1609 (0.0118) | -1.0246 (0.0092) | -0.9158 (0.0141) | 1.7281 (0.0222) | 2.0505 (0.0042) | 1.5222 (NA) |
Tajima’s D, averaged across 50 kb windows, standard error in parentheses for each sampled population of phenotypic parental Myzomela cardinalis and M. tristrami.
Autosomal loci introgress in both directions at secondary contact
We captured 187 birds in sympatry. Of these, 68 were phenotypically Mtris, 107 were phenotypically Mcard, and 12 were identified as “phenotypic hybrids”—individuals with plumage characteristics clearly intermediate between Mtris and Mcard (e.g., mostly black with some red feathers). These individuals raise the possibility that our sample includes backcross or advanced generation hybrids that are indistinguishable from the parental species. To test for the possibility of cryptic hybrids, we start by focusing on analyses of autosomal regions of the genome. We used five approaches to characterize autosomal admixture. First, simple summaries of differentiation and divergence restricted to phenotypically parental individuals (excluding phenotypic hybrids) support admixture between sympatric populations (Tables 3, S7 and S8). Autosomal differentiation (FST) between allopatric Mcard and Mtris is 0.282, whereas that for sympatric Mcard and Mtris drops to FST = 0.197 (Table 3). The difference in absolute divergence between species is less dramatic but still larger in allopatry (dxy ≈ 0.0035) than in sympatry (dxy = 0.0033; S8 Table). The observed shifts in estimated differentiation and divergence in sympatry when looking solely at phenotypically pure Mcard versus Mtris are consistent with introgression.
Table 3. Population differentiation, FST.
| population comparison | autosome | neo-PAR | Z | neo-Z | W | neo-W | mtDNA |
|---|---|---|---|---|---|---|---|
| Myzomela cardinalis | |||||||
| Allopatry vs. Sympatry |
0.0323 (2 x 10−4) |
0.0326 (0.0013) | 0.0508 (0.0016) | 0.0668 (0.0033) | -0.0067 (0.0061) | -0.0032 (0.002) | 0.0019 (NA) |
| Myzomela tristrami | |||||||
| Allopatry vs. Sympatry |
0.0061 (1 x 10−4) |
0.0047 (2 x 10−4) |
0.0054 (3 x 10−4) |
0.0051 (5 x 10−4) |
0.1548 (8 x 10−4) |
0.1557 (2 x 10−4) |
0.1233 (NA) |
| Heterospecific | |||||||
| Allo. Mcard vs. Allo. Mtris |
0.2818 (0.001) | 0.2757 (0.0061) | 0.5921 (0.0048) | 0.6606 (0.0052 | 0.9917 (7 x 10−4) |
0.9924 (2 x 10−4) | 0.9831 (NA) |
| Allo. Mcard vs. Sym. Mtris |
0.2423 (9 x 10−4) |
0.2426 (0.0054) | 0.5704 (0.0048) | 0.644 (0.0049) | 0.7409 (8 x 10−4) |
0.7411 (2 x 10−4) | 0.8017 (NA) |
| Allo. Mtris vs. Sym. Mcard |
0.2354 (9 x 10−4) |
0.2509 (0.0066) | 0.5717 (0.005) | 0.6630 (0.0067) | 0.9920 (6 x 10−4) |
0.9927 (2 x 10−4) | 0.9855 (NA) |
| Sym. Mcard vs. Sym. Mtris |
0.1972 (8 x 10−4) |
0.2178 (0.0059) | 0.5511 (0.005) | 0.6433 (0.0064) | 0.7411 (7 x 10−4) |
0.7413 (2 x 10−4) | 0.8041 (NA) |
FST averaged across 50kb windows for each region of the genome, for pairwise comparisons of phenotypic parental populations of Myzomela cardinalis (Mcard) and M. tristrami (Mtris). Allopatry and sympatry abbreviated as Allo. and Sym., respectively. Standard errors in parentheses.
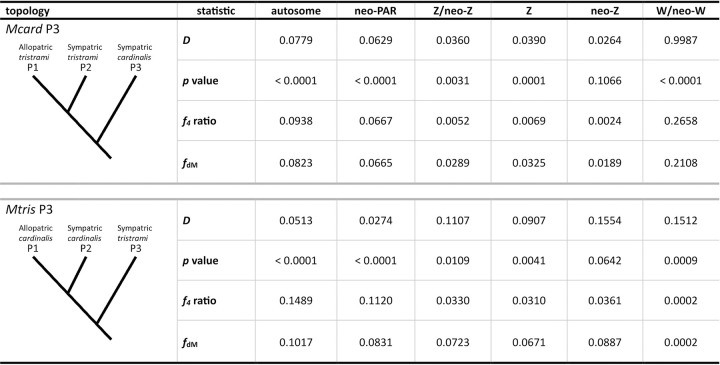
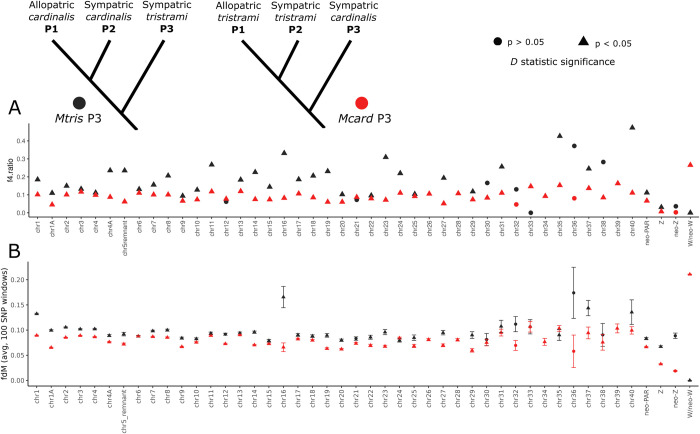
Second, to infer the amount and direction of introgression between species we conducted ABBA-BABA analyses using the program Dsuite [73] to calculate the D-statistic, f4, and fdM admixture statistics (Fig 4). ABBA-BABA uses absolute counts of the distribution of derived alleles shared among four taxa using the topology (((P1,P2)P3)P4) to determine if gene flow has occurred among the three ingroup taxa [74]. We used Mpulc as the P4 outgroup and analyzed two different topologies: “Mcard P3” with allopatric Mtris (P1), sympatric Mtris (P2), and sympatric Mcard (P3) as the ingroup; and “Mtris P3” with allopatric Mcard (P1), sympatric Mcard (P2), and sympatric Mtris (P3) as the ingroup. For both topologies, D statistics were positive and significant, indicating excess sharing of derived alleles between sympatric Mcard and Mtris consistent with autosomal gene flow. The f4 statistic calculates a proportion of admixture assuming unidirectional gene flow from P3 into P2. A slightly higher admixture proportion (f4 ratio) calculated for the Mtris P3 topology (0.15) suggests more gene flow from sympatric Mtris (P3) into sympatric Mcard (P2) than the reverse (0.09; Fig 4 and Fig 5A). The fdM statistic also estimates admixture [75] and does not assume unidirectional gene flow but nevertheless echoes the f4 ratio, showing slightly higher and more variable admixture in the Mtris P3 topology (Fig 5B). Parsing signals of introgression by chromosome, D statistics are significant for both topologies for most autosomes (74% for Mtris P3, 95% for Mcard P3; Figs 5 and S2). The relatively lower overall level of Mcard→Mtris introgression on autosomes (f4 statistic) may be attributable to relative abundance of the two species, to asymmetric mate preferences, and/or to more efficient selection against foreign alleles in the Mtris population owing to its larger effective population size.
Fig 4. Test of introgression, ABBA-BABA results.
A positive D statistic indicates gene flow between P2 and P3, and the p value specifies whether D is significantly different from zero, using block-jackknife procedure. The f4 ratio is calculated by splitting P3 population into two subsets to calculate the admixture proportion, assuming unidirectional introgression P3 → P2. The average fdM statistic is calculated using 100 SNP non-overlapping windows (W/neo-W calculated in 50 SNP non-overlapping windows) where fdM ≥ 0, indicating either no gene flow (fdM = 0) or gene flow between P2 and P3 (fdM > 0). Allopatric M. cardinalis includes samples from both Ugi and Three Sisters. M. pulchella is the P4 outgroup in both topologies.
Fig 5. Admixture statistics (f4, A; fdM, B) for autosomes and sex chromosome regions.
Color of the point indicates which topology the statistic was calculated for (Mtris P3 or Mcard P3), and shape of the point indicates whether the D statistic for that chromosome/region was significantly different from zero, determined using the block-jackknife procedure. fdM statistics were calculated across 100 SNP windows, restricted to windows where fdM ≥ 0 (potential gene flow between sympatric populations) and averaged, with bars showing standard error across windows for that chromosome/region. Admixture f4 ratios calculated for Mtris P3 taxa on chr 26, 28, 34, and 39 used an alternative topology indicating nonsignificant introgression from sympatric Mtris into allopatric Mcard, shown in S2 Fig.
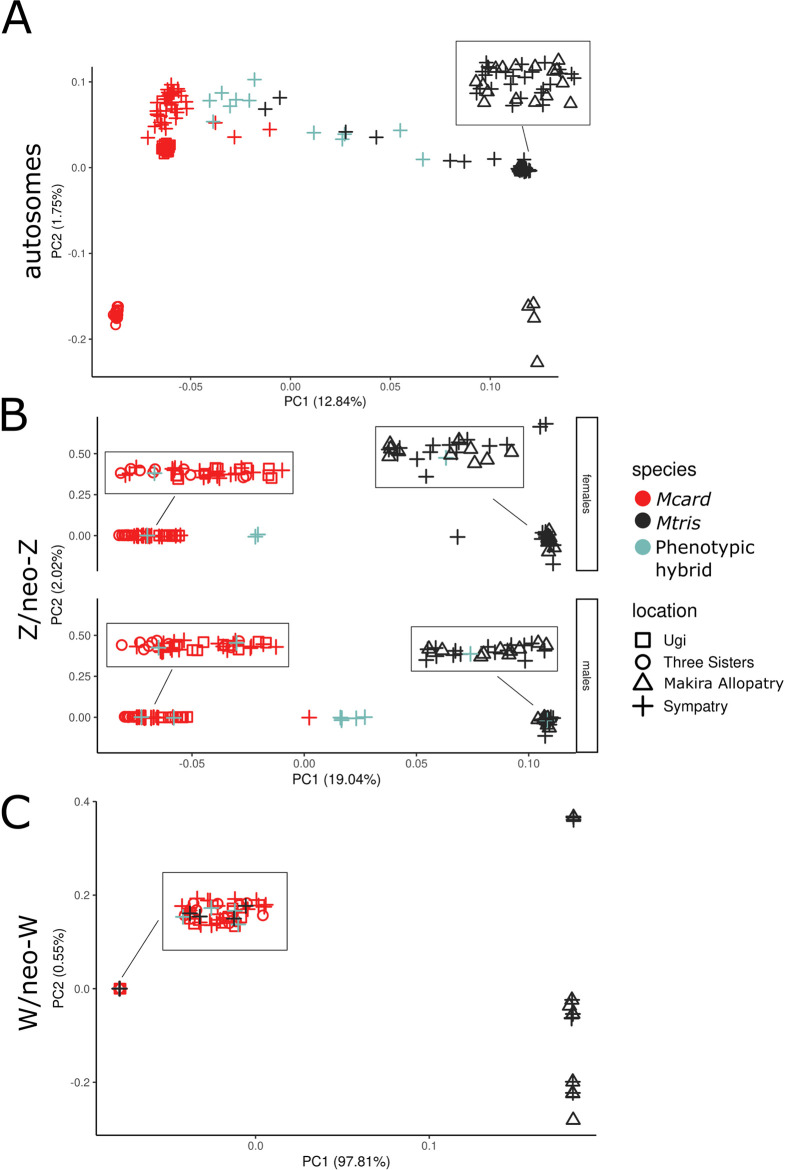
Third, we used principal component analyses (PCA) to identify admixed individuals. PCA revealed clear separation of allopatric Mcard and Mtris along principal component 1 (PC1, Fig 6A). As expected, phenotypic hybrids were intermediate in PC1 values (Fig 6A). Notably, several phenotypic Mcard and Mtris individuals also fell between the two clusters of parental species. These “cryptic hybrids” are likely backcross or advanced generation hybrids.
Fig 6.
Principal components analysis of autosomal (A), Z/neo-Z (B) and W/neo-W (C) sequence. Symbol color represents phenotypic species assignment while symbol shape indicates sampling locality. Inset boxes show points jittered for visualization. Plot for Z/neo-Z is separated by sex to distinguish homogametic males and heterogametic females.
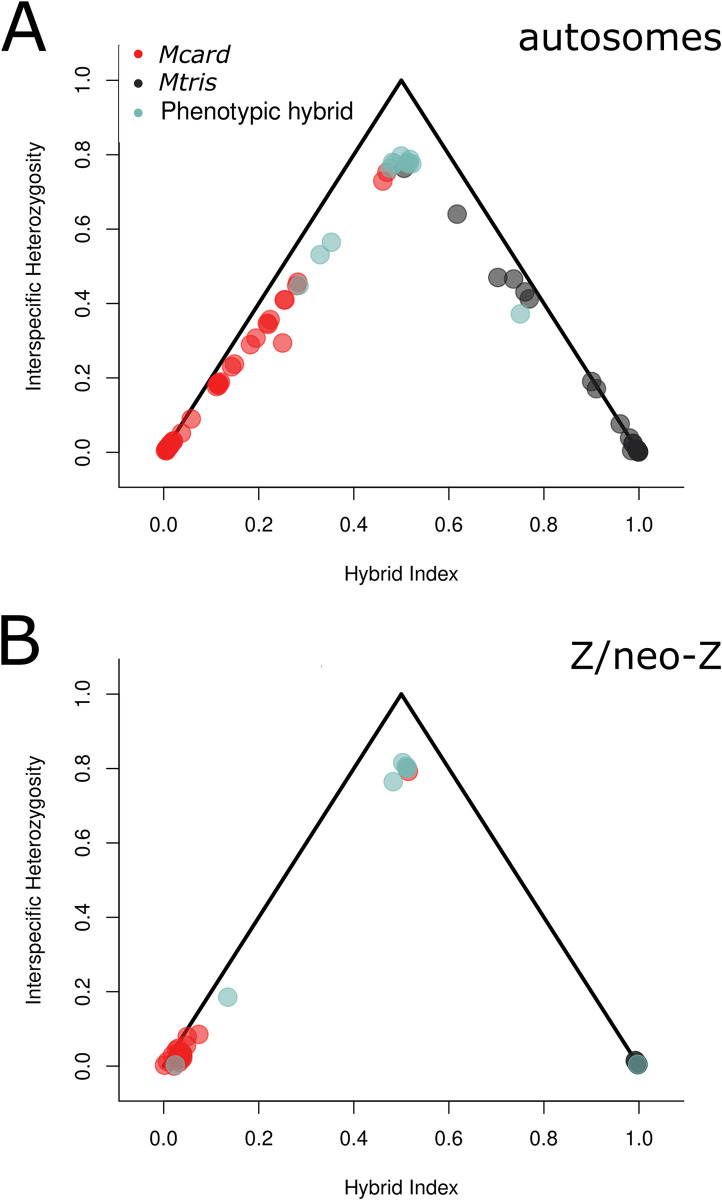
To explicitly test for production of F1s and advanced generation backcrosses we used autosomal SNPs fixed between allopatric Mcard and Mtris to estimate the interspecific heterozygosity and the hybrid index for each sympatric individual (Fig 7A). We see clear evidence of F1s with near-maximum interspecific heterozygosity and hybrid indices ≈ 0.5. Most are phenotypic hybrids (n = 8), but phenotypic Mcard (n = 2) and Mtris (n = 1) also appear to be F1 individuals. We detect 28 backcross hybrids in both crossing directions but no F2 individuals (Fig 7A). The absence of F2s may be due to limited sampling, low frequency of F1s limiting opportunities for F1 x F1 matings, the mating behavior of hybrids, or fitness breakdown in F2s.
Fig 7.
Triangle plots showing the relationship between interspecific heterozygosity and hybrid index calculated using 2,449 autosomal (A) and 37,613 Z/neo-Z (B) SNPs fixed between species in allopatry. Circles are sympatric individuals of both sexes (A) or sympatric males (B), colored by phenotype (see legend). F1 individuals fall at the maximum heterozygosity and hybrid index ≈ 0.5, while advanced generation backcrosses fall along the legs of the triangle as interspecific heterozygosity declines and hybrid index is closer to 0 (Mcard ancestry) or increases to 1 (Mtris ancestry).
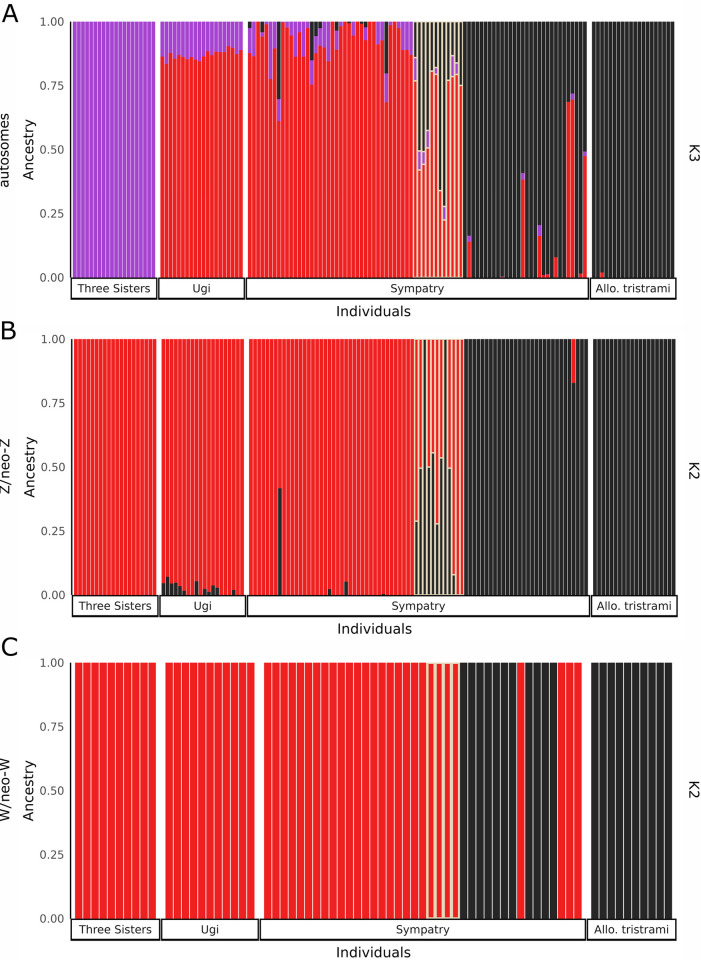
Finally, we used ADMIXTURE to estimate individual ancestry proportions [76]. Cross-validation error was minimized when the number of groups (K) was equal to two (S3 Fig). However, K = 3 was similarly well-supported for autosomal sites and informative about structure between the Ugi and Three Sisters Mcard populations (Fig 8A; see below for further discussion of Mcard allopatric populations). All phenotypic hybrids, as well as several individuals in sympatry (phenotypically Mtris and phenotypically Mcard) show autosomal ancestry from both species. In summary, autosomal sequence indicates bidirectional admixture between Mcard and Mtris in sympatry. The presence of advanced-generation backcrosses in our sample confirms that some F1 hybrids are fertile. The relative abundance of Mcard in sympatry (107 of 187 individuals captured in 2008–2015) and the continued production of phenotypic hybrids suggests ecological incompatibilities do not strongly influence reproductive isolation in the region of sympatry, although additional work is necessary to confirm this.
Fig 8.
Proportion ancestry calculated in ADMIXTURE for autosomes (A), Z/neo-Z (B), and W/neo-W (C). Autosomes are shown at K = 3 (see S3 Fig for K = 2), while Z/neo-Z and W/neo-W shown at K = 2. Individuals are grouped by sampling location. Phenotypic hybrids in sympatry are outlined in yellow, with phenotypic Mcard in sympatry to the left and phenotypic Mtris in sympatry to the right of phenotypic hybrids.
New pseudo-autosomal region shares patterns of introgression with autosomes
The new pseudo-autosomal region, or neo-PAR, is a region of the former autosome, chr5, that is now linked to the neo-sex chromosomes but has continued to recombine in both sexes. The neo-PAR therefore serves as an important point of comparison with the neo-sex chromosome regions that have limited recombination. The neo-PAR has nucleotide diversity similar to autosomes, likely preserved by a history of uninterrupted recombination since the initial formation of the neo-sex chromosome fusion (Table 1). Differentiation within and between species on the neo-PAR is also in line with that of autosomes (Table 3). Finally, signals of introgression on the neo-PAR parallel those of the autosomes, with significant D statistics for both topologies. Admixture statistics were only slightly lower for the neo-PAR than for autosomes as a whole, and tracked values of individual autosomes of similar size (Fig 4 and S4 Fig). Thus, despite transitioning from autosomal sequence to a part of the sex chromosomes, the neo-PAR region appears to have a history similar to the autosomes. We now compare the bidirectional pattern of autosomal and neo-PAR gene flow to introgression patterns observed on sex and neo-sex chromosome sequence, which we expect to differ due to variable ploidy and recombination (S8 Table).
Z /neo-Z refractory to introgression
The Z/neo-Z linked sequence shows evidence of admixture, but the degree of introgression is markedly reduced compared to autosomes. Differentiation on the Z and neo-Z regions between Mcard and Mtris is slightly lower in sympatry (Z: FST = 0.551, neo-Z: FST = 0.643) than in allopatry (Z: FST = 0.592, neo-Z: FST = 0.661), but is overall much higher than autosomes (Table 3). Divergence between Mcard and Mtris in allopatry is similar to that in sympatry (S8 Table). In the ABBA-BABA analysis, D statistics for the Z/neo-Z region were significant, consistent with gene flow between sympatric Mcard and Mtris, but f4 and fdM admixture statistics were lower than those estimated for autosomal sites (Fig 4 and Fig 5). We also estimated admixture separately for ancestral Z and neo-Z regions of the Z chromosome. Admixture statistics (D, f4 and fdM) for the ancestral Z are consistent with those estimated from the combined Z/neo-Z region (Fig 4). However, the neo-Z region yielded more complex results. First, D statistics for the neo-Z are not statistically significant in either the Mcard P3 or the Mtris P3 topologies (Fig 4), consistent with limited or no introgression. The Mcard P3 f4 and fdM admixture statistics for neo-Z sites are lower than those for the autosomes and the Z, suggesting Mcard→Mtris neo-Z introgression may be negligible. For Mtris P3, however, interpretation is more difficult: f4 admixture ratios are similar between Z and neo-Z while fdM statistics were on average slightly higher in neo-Z than Z. However, we note again that D statistics for the neo-Z region were non-significant and admixture statistics should therefore be interpreted with caution.
Analyses of individual genotypes revealed F1, backcross, and advanced generation backcross individuals in our sample. For PCA of the Z/neo-Z, we separated homogametic males and heterogametic females for visualization (Fig 6B). Mcard and Mtris clearly separated along PC1. Five of eight phenotypic hybrid males and one phenotypic Mcard male were intermediate in PC1, potentially F1 individuals heterozygous for species’ Z/neo-Z haplotypes. However, there were also phenotypic hybrid males which fell within the species’ clusters at either end of PC1, indicating those individuals may be advanced generation backcross hybrids homozygous for Z/neo-Z haplotypes from either Mcard (n = 2) or Mtris (n = 1). Although females only have one Z haplotype, three individuals had intermediate values of PC1 (Fig 6B). These females are likely backcross hybrids carrying recombinant Z/neo-Z chromosomes with both Mcard and Mtris sequence. Plotting hybrid index against interspecific heterozygosity for diploid males confirmed five of eight phenotypic hybrid males and one phenotypic Mcard male were heterozygous for their Z/neo-Z haplotype (Fig 7B). Individual-level estimates of ancestry proportions calculated in ADMIXTURE showed some but not all phenotypic hybrids were admixed for their Z/neo-Z haplotype, while only two phenotypically parental individuals had appreciable ancestry from the other species (Fig 8B).
Together, these results show introgression of Z/neo-Z in sympatry is limited relative to autosomal sequence. Aggregated, population-level estimates of differentiation are similar in allopatric and sympatric species comparisons, while estimates of introgression and admixture proportion are very low, and in the case of the neo-Z, not significant. Although sympatric individuals span a range of autosomal admixture, only a few carry Z/neo-Z genetic material from both species (S5A Fig). The absence of sex chromosome recombination in female hybrids limits production of individuals with admixed ancestry on the Z/neo-Z chromosome, in contrast to autosomes which assort independently and have the potential to recombine in both male and female hybrids. However, admixture statistics for Z and neo-Z are lower than those calculated for single autosomes of similar size and nucleotide diversity (S4 and S6 Figs). Thus, it seems likely that selection, perhaps in combination with reduced recombination, limits introgression of Z/neo-Z.
Strong, asymmetric introgression of W/neo-W and mitochondria
The W, neo-W, and mitochondrial sequence are all maternally co-transmitted to females, while males receive only mitochondrial sequence from their mother. Mcard and Mtris carry distinct sets of W, neo-W, and mitochondrial haplotypes and in our sampling of sympatric individuals all three regions show strong, asymmetric introgression from Mcard into Mtris. Interspecific differentiation between Mtris and Mcard is lower in sympatry (FST = 0.74) than in allopatry for W and neo-W regions (FST = 0.99; Table 3), consistent with introgression. As a result, Mtris now shows within-species differentiation between allopatric and sympatric individuals at W/neo-W (FST = 0.15) and mitochondrial sites (FST = 0.12) but not autosomal ones (FST < 0.01; Table 3). The introgression of Mcard W/neo-W and mitochondrial haplotypes has produced a striking positive shift in Tajima’s D for sympatric Mtris birds (Table 2). ABBA-BABA analyses for W/neo-W haplotypes further support asymmetric introgression from Mcard into Mtris. The Mcard P3 topology showed a significant D statistic and an f4 admixture proportion of 0.27; Mtris P3 topology also had a significant D statistic, but the f4 admixture proportion indicating gene flow Mtris → Mcard was ≈ 0 (Fig 4 and Fig 5). The extreme W/neo-W introgression from Mcard into Mtris is particularly apparent when plotting Mcard P3 fdM values against chromosome/region nucleotide diversity calculated in the allopatric Mtris population (S6 Fig). For other genomic regions a slightly positive relationship between nucleotide diversity and introgression is expected and observed, but despite negligible nucleotide diversity on W/neo-W, we observe strong introgression indicated by a high fdM statistic.
Movement of Mcard W/neo-W haplotypes into Mtris is also evident in analysis of individual genotypes. All phenotypic hybrid females and four of 15 phenotypic Mtris females cluster with Mcard individuals in PCA of W/neo-W sites, whereas none of the sympatric Mcard females cluster with Mtris (Fig 6C). Females carrying the Mcard W/neo-W haplotype also exhibit a wide range of autosomal ancestry, while those with Mtris W/neo-W are exclusively Mtris in autosomal background (S5B Fig). All sympatric Mcard, phenotypic hybrid, and four of 15 sympatric Mtris females have only Mcard ancestry in ADMIXTURE analyses (Fig 8C). Reassuringly, the proportion of Mtris females carrying W/neo-W haplotypes (4/15 = 0.267) matches the Mcard→Mtris f4 admixture ratio calculated using the Mcard P3 topology (Fig 4). The haplotype network for the full mitochondrial genome shows the same asymmetric pattern of introgression and provides insight to the maternal contribution to hybrid males. Mitochondrial haplotypes for Mtris are restricted to phenotypic Mtris, whereas Mcard mitochondrial haplotypes are carried by phenotypic hybrids and by sympatric Mcard and Mtris (S7 Fig). These results extend and confirm previous analyses of mitochondrial markers [65]. Consistent with asymmetric introgression, our sample has four phenotypic hybrid females, four phenotypic Mtris females, and 12 admixed males carrying Mcard W/neo-W and/or mitochondrial haplotypes. These individuals are therefore ultimately the product of crosses between Mcard females and Mtris males; we observe no admixed individuals produced by the reciprocal cross (i.e., carrying Mtris W/neo-W and/or mitochondrial haplotypes). Notably, the Mcard→Mtris introgression we observe for W/neo-W and mtDNA is counter to the expected direction of local→invading species resulting from initial introgression of local alleles and subsequent dilution of invading alleles [19,77]. The absence of admixed birds with Mtris W/neo-W or mtDNA in our dataset may reflect our limited sample size and/or limited rate of successful hybridization involving Mtris females. However, even with modest sample sizes we find that W/neo-W/mitochondrial introgression is significantly asymmetric: Mtris individuals are more likely than Mcard individuals to carry heterospecific W/neo-W (Fisher’s exact test, p = 0.026, females only, S9 Table) and/or mitochondria (Fisher’s exact test, p = 0.005, males and females, S9 Table).
We propose three potential explanations for asymmetric introgression of the Mcard W/neo-W and mitochondria. First, if matings between Mtris females and Mcard males occur, Haldane’s rule for lethality predicts a dearth of F1 hybrid daughters [39], which is often manifest in only one direction of the species cross due to genetic incompatibilities involving sex chromosomes or mitochondria [25,78,79]. There are, however, several reasons to consider alternative explanations. For one, Haldane’s rule for hybrid sterility in both directions tends to evolve before hybrid lethality [80]. In addition, hybrid lethality in birds tends to occur between much older species pairs [3,81]. Because mitochondrial sequence is co-inherited with W/neo-W and shares the pattern of asymmetric introgression, mitonuclear interactions or incompatibilities may also or instead be the target of selection [82,83]. It remains possible, however, that rapid evolution of the neo-W has accelerated the evolution of hybrid lethality or mitonuclear incompatibilities [50].
Sexual incompatibilities provide a second explanation for asymmetric introgression. Asymmetry in mate choice or mate availability may limit crosses between Mtris females and Mcard males [38]. Indeed, Mayr and Diamond [4] proposed that hybridization in sympatry initially occurred due to a lack of conspecific mates for recently arrived Mcard. Since then, however, the relative abundance of Mcard has surpassed that of the endemic Mtris. It therefore seems unlikely that Mcard females are constrained to pair with heterospecific males [65]. In fact, for a sample of molecularly sexed individuals captured foraging at flowering trees, sympatric Mcard shows a ratio of 1.1 males per female (n = 81), whereas Mtris has an even higher ratio of 2.1 males per female (n = 56; S10 Table). Nevertheless, phenotypic hybrids and admixed individuals continue to be observed decades after invasion. It is therefore possible that introgression of maternally coinherited sequences from Mcard into Mtris genomic backgrounds occurs via asymmetric mate choice or natural selection [84,85].
Positive selection which favors Mcard W/neo-W and/or mitochondria in the Mtris population is a third potential explanation for the asymmetric introgression we observe. The Mcard W/neo-W/mitochondria could, for instance: carry some globally adaptive mutation(s) absent from Mtris [86]; have a smaller load of unconditionally deleterious mutations [87]; and/or possess a transmission advantage (meiotic drive) in an Mtris genetic background [88]. Future work is necessary to distinguish among the three possible drivers of W/neo-W and mitochondrial introgression from Mcard into Mtris.
More than one source for sympatric Mcard population
To determine if the Mcard population(s) that invaded Makira originated from Ugi, from Three Sisters, or from both, we assessed relationships between the two allopatric Mcard and the sympatric Mcard populations. We identified very few alleles private to sympatric Mcard or fixed between sympatric Mcard and either allopatric Mcard population, while many alleles were shared among all three Mcard populations (S11 and S12 Tables). Three Sisters Mcard show distinct ancestry from Ugi Mcard in ADMIXTURE analyses allowing for three ancestral groups (K = 3; Fig 8A). Across all genomic sites, differentiation between Ugi and Three Sisters populations of Mcard is higher than that between either allopatric population and sympatric Mcard (S6 Table). Ugi Mcard does appear to have been a stronger contributor to the sympatric Mcard population, based on differentiation between the populations and clustering in autosomal PCA (S6 Table and Fig 6A). The mitochondrial network, however, shows that haplotypes otherwise unique to Ugi or to Three Sisters are both present in sympatric Mcard, sympatric Mtris, and phenotypic hybrids (S7 Fig). Thus, it is likely both Ugi and Three Sisters individuals contributed to the founding population of Mcard on Makira.
Conclusions
Our population genomic study of Myzomela honeyeaters in the Solomon Islands sheds light on a system with complex and ongoing introgression following very recent secondary contact of closely related taxa carrying neo-sex chromosomes. Sex chromosomes are known to play a large role in speciation [34]. Neo-sex chromosomes transitioning from autosomal to sex-specific inheritance undergo rapid structural and molecular evolution, which may incidentally contribute to incompatibilities between species [50,51,89]. Consistent with genetic and/or sexual incompatibilities accumulating on sex and neo-sex chromosomes, we see limited introgression of Z/neo-Z. Modest autosomal introgression into both species occurs. Surprisingly, and in contrast to other chromosomes, we observe asymmetric introgression of the W/neo-W and mitochondria. The patterns of introgression that we see highlight the potential for gene flow to vary across the genome, and the importance of sex chromosomes in maintaining species boundaries. Importantly, gene flow is reduced for Z/neo-Z sequence relative to autosomes, possibly owing to its faster accumulation of locally adaptive and/or incompatible substitutions. In contrast, the asymmetric introgression of the W/neo-W suggests either asymmetric mating success (e.g., an excess of hybrid crosses involving Mcard females), mitonuclear or W/neo-W linked incompatibilities which limit introduction of Mtris W/neo-W and/or mtDNA into Mcard genomic backgrounds, or some form of selection that favors Mcard W/neo-W and/or mitochondrial haplotypes in Mtris. Further work is of course needed to determine the specific behaviors and/or loci that underlie these patterns. By revisiting the Myzomela honeyeater system in Makira and testing predictions of the allopatric model of speciation with genomic data, we build on the work started by Mayr and Diamond [4], providing genomic insights into the “moment of truth” for speciation.
Materials and methods
Ethics statement
Sampling methodology was approved by the Institutional Animal Care and Use Committees at the University of Miami (Protocol 12–100) and University of Kansas (Protocol AUS 174–01).
Samples and sequencing
Between 2008 and 2015 we sampled 40 allopatric Myzomela cardinalis from two island groups adjacent to the region of sympatry on Makira (i.e., Ugi and Three Sisters) and 20 allopatric M. tristrami from high elevation regions on the island of Makira (Fig 1). We also sampled 82 birds from the low elevation region of sympatry on Makira. Of these, 40 were identified as M. cardinalis, 30 were identified as M. tristrami, and 12 were identified as hybrids, based on a phenotype of mostly black plumage with patches of red feathers. In addition, we used a sample of Myzomela pulchella collected on New Ireland, Papua New Guinea and held at the University of Kansas Biodiversity Institute and Natural History Museum (see S2 Table for details on all samples).
We collected whole blood using brachial venipuncture from birds captured in mist nets at flowering trees. We added blood to lysis buffer [90] and stored it at room temperature until arrival to the lab, where it was subsequently stored at -80°C. We extracted DNA using a Qiagen DNeasy kit with an RNase step.
Extracted genomic DNA was sequenced at Novogene (Sacramento, CA). Following quality and concentration assessment using Agarose Gel Electrophoresis and Qubit 2.0, genomic DNA was randomly fragmented and fragments were end polished, A-tailed, and ligated with Illumina adapters. Further PCR amplification preceded library construction and purification with the AMPure XP system. Finally, size distribution of libraries was checked using Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). Libraries were then pooled and sequenced by synthesis using the Illumina platform to generate 150 bp paired end reads. The M. pulchella sample was sequenced at the Oklahoma Medical Research Foundation. Libraries were constructed using the Swift 2S Turbo DNA Library Kit prior to sequencing by synthesis of 150 bp paired end reads using the Illumina Novaseq machine.
Reference genome assembly
We sequenced a M. tristrami female at the University of Delaware DNA sequencing & Genotyping Cener. HiFi libraries were prepared with SMRTbell prep kit, followed by Blue Pippin size selection (15-20Kbp) before sequencing on a PacBio Sequel IIe. We generated a de novo assembly using hifiasm v0.13-r308 with default parameters using the resulting long reads [91,92]. We used GeMoMa (v1.8) and the annotation from zebra finch genome bTaeGut1.4.pri to infer a rough annotation of genes in the Myzomela tristrami genome. We then used these rough annotations, comparing contigs against both zebra finch and the chicken genome bGalGal1.mat.broiler.GRCg7b to infer synteny relationships, remove duplicate haplotigs, and, finally, scaffold contigs into chromosomes in Myzomela tristrami. The resulting assembly uses the zebra finch numbering system for chromosomes 1–29; chromosome 30–40 were named in descending order of size. Final chromosomes and contigs were aligned with those of related species—helmeted honeyeater (Lichenostomus melanops cassidix), and blue-faced honeyeater (Entomyzon cyanotis)—using Mauve (version 2015-02-25), and visualized using FastANI (v1.33) [62,70,93,94]. We generated repetitive DNA libraries using the RepeatModeler v2 pipeline [95]. RepeatModeler employs a combination of de novo and homology-based characterization of different classes of repeats. The repeat library was annotated and combined with Repbase, and manually curated repeat libraries from other studies [96–99]. We then used RepeatMasker (v4.1.0) to identify and mask repetitive regions of the genome [100].
Alignment and variant calling
We used Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to process raw reads. Trim Galore first removes low quality reads from the 3’ end and then trims adapter sequences using the program Cutadapt [101] before running FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to check adapter content after trimming. We aligned trimmed reads to the Myzomela tristrami reference genome using Burrows-Wheeler-Aligner (bwa-mem, v0.7.17; [102], mapping 26,893,270,155 reads and yielding a mean coverage of 16.95x. After alignment, we sorted the resulting mapped reads by coordinate using samtools, v1.7 [103]. We continued with processing following the Genome Analysis Toolkit best practices workflow (GATK 4.2.6.1 [104]). First, we used AddOrReplaceReadGroups (Picard v.2.12.0, http://broadinstitute.github.io/picard) to denote flow cell and lane of each read. We used MarkDuplicates (Picard v12.2.0) to identify duplicate reads resulting from PCR amplification. Next, we used FixMateInformation (Picard v12.2.0) to verify and correct information between mate-pairs. At this point we assessed coverage using qualimap (v2.2.1; [105]) and confirmed sex of individuals based on coverage of Z scaffolds (diploid in males, haploid in females). In addition, we extracted mean coverage of sex-linked scaffolds and chromosomes 1–10 for allopatric Mtris individual to verify that mapping and raw read depth supported assignment of chr5 derived sequence in the Mtris reference assembly as Z- or W-linked (Fig 2).
We then called variants using the GATK pipeline, starting with HaplotypeCaller, on the diploid (default) setting. Due to hemizygosity of female sex chromosomes, we first ran HaplotypeCaller on autosomes and pseudo-autosomal regions across both sexes. We then separated males and females, running HaplotypeCaller on males for Z sequence only and on females for both Z and W sequence. We then used CombineGVCFs to facilitate joint genotyping for each region of the genome. Finally, using GenotypeGVCFs we generated an all-sites VCF file that contained both variant and invariant sites for downstream filtering and analysis.
Filtering
We used VariantFiltration and GATK recommendations for hard filtering in non-model organisms to flag any sites that had QD < 2.0, SOR > 3.0, FS > 60.0, MQ < 40.0, MQRankSum < 12.5, or ReadPosRankSum < -80.0. We also flagged any sites overlapping known repetitive regions in the reference genome. We then used bcftools [103] to recode any flagged low-quality sites or sites in repetitive regions as missing. We used vcftools [106] to remove indels and assess depth of coverage prior to further filtering. We used bcftools to recode any autosomal genotypes as missing if they had less than 10x or more than 34x (twice the mean before filtering for depth) coverage averaged across all samples. For sex chromosome genotypes we adjusted our depth of coverage filters to accommodate the reduced coverage for hemizygous female samples, recoding as missing any genotypes less than 6x or more than 24x averaged across all samples. In addition, we further filtered sex chromosomes to remove any spurious heterozygous sites on the female Z and W sequence (excluding the new pseudo-autosomal region), recoding any such sites as missing [107]. For mitochondrial genotypes we imposed only a minimum depth filter of 10x, and also masked any regions showing heteroplasmy, resulting in 14,122 remaining sites.
For individual-level assessment of genomic variation and admixture we did further filtering, imposing a minimum minor allele frequency of 0.05 in vcftools and pruning for linkage disequilibrium (LD) in plink [108]. Our pruning procedure calculated LD between each pair of single nucleotide polymorphisms (SNPs) in 50 SNP windows, removing one SNP of each pair with an r2 > 0.5. The window was then shifted 5 SNPs forward before repeating the procedure.
Population genomic analyses
To infer demographic history and effective population sizes for M. cardinalis, M. tristrami and the outgroup M. pulchella, we used individuals sampled in allopatry to construct a pairwise sequential Markovian coalescent (PSMC) model [71]. We first generated a consensus sequence in fastq format for each individual using samtools mpileup and bcftools call [103], followed by limiting to autosomes and using the vcf2fq in the vcfutil.pl of bcftools. We then ran PSMC using default settings (https://github.com/lh3/psmc), and plotted output in the R v4.1.1 [109] package ggplot2 [110], using generation length of 2.37, 2.25, and 2.51 years for M. pulchella, M. cardinalis, and M. tristrami respectively from [111], and a per generation mutation rate of 4.6*10−9 from [112].
For all remaining analyses we separated results for autosomes, neo-PAR, Z/neo-Z, W/neo-W, and the mitochondrial genome. For windowed estimates we further parsed ancestral sex chromosome regions from neo-sex chromosome regions. We used the program pixy [113] and an allsites VCF filtered for quality and depth to calculate nucleotide diversity (π), absolute divergence (dxy) and pairwise genetic differentiation (FST) across 50kb windows for each population of phenotypically cardinalis and tristrami individuals. Using a quality and depth filtered VCF containing only variant sites, we used vcftools [106] to calculate Tajima’s D in 50kb windows. For all windowed analyses we limited our calculation of average estimates to 50kb windows containing a minimum of 10,000 genotyped sites, with the exception of estimates for mitochondrial sequence (which is a single window). When calculating ratios of nucleotide diversity of sex chromosomes to autosomes (S6 Table), we restricted our autosomal windows to chromosomes 1–10. Chromosomes 1–10 are similar in size to the Z/neo-Z sex chromosome, allowing more direct comparison of nucleotide diversity, as recombination rate and nucleotide diversity is expected to be elevated in smaller michrochromosomes [114]. All other estimates involving autosomes include all autosomes.
To quantify the degree of admixture across the genome we used the quality and depth filtered dataset to conduct ABBA-BABA tests in Dsuite [73,74]. We calculated D statistics for autosomes, neo-PAR, Z, neo-Z, and W/neo-W and used a block-jackknife procedure to determine if the D statistic was significantly different from zero. To quantify the proportion of introgression for each region we calculated f4 admixture ratio, which compares the observed excess of ABBA sites to the expected value if admixture was complete, by substituting a subset of P3 individuals for P2 individuals. The value of f4 therefore provides an estimate of the proportion of admixture assuming unidirectional introgression from P3 into P2 [74,115]. The assumption of unidirectional introgression may be inaccurate, so we also calculated the fdM statistic, which is agnostic to the direction of gene flow. Instead, it uses the frequency of the derived allele to determine whether P2 or P3 is the donor population. The fdM statistic is symmetric about zero, with positive values indicating gene flow between P2 and P3 and negative values indicating gene flow between P1 and P3. Because we have allopatric populations for both parental species, we can construct and analyze two topologies to compare amount (fdM) and direction (f4) of introgression between sympatric Mcard and Mtris. The Mcard P3 topology places allopatric and sympatric Mtris as P1 and P2 respectively and sympatric Mcard as P3, therefore estimating gene flow from Mcard into Mtris for f4. The Mtris P3 topology places allopatric and sympatric Mcard as P1 and P2 respectively, and sympatric Mtris as P3, giving an estimate of gene flow from Mtris into Mcard using the f4 admixture proportion. In addition to calculating D, f4 and fdM for all autosomes combined, we also calculated these for each chromosome individually to further visualize patterns of introgression across the genome. We calculated the fdM statistic in 100 SNP non-overlapping windows and averaged across windows in which the fdM statistic ≥ 0, indicating potential gene flow between P2 and P3, sympatric Mcard and sympatric Mtris.
To understand genomic variation at an individual level and assess degree of admixture we conducted a principal components analysis (PCA) using the package SNPRelate [116] in R to investigate genomic variation of autosomes, Z/neo-Z, and W/neo-W among individuals sampled on Makira, Ugi and Three Sisters. For characterization of individuals as F1 or advanced generation backcross in the triangle plot we first identified 2,449 autosomal and 37,613 Z/neo-Z SNPs fixed between species (FST = 1) using allopatric individuals in vcftools. Using only the fixed SNPs we then calculated interspecific heterozygosity and hybrid index on the autosomes for all sympatric individuals and on the Z/neo-Z using only sympatric males in the package introgress in R [117]. Finally, we used ADMIXTURE [76] which estimates a maximum likelihood proportion of ancestry per individual and uses cross-validation error to determine the optimal number of populations or groups (K) present in sample of individuals. We ran replicate analyses using values of K from 1 to 7, and performed fivefold cross-validation to estimate error associated with each value of K.
We generated a mitochondrial haplotype network using variant sites from the full mitochondrial genome filtered for quality, minimum depth, and heteroplasmy. We used vcf2phylip [118] to convert the VCF to a phylip file for importing into the program PopART [119], where we constructed a TCS network [120] to visualize haplotype sharing and mutations separating all individuals in our dataset (including phenotypic hybrids and the outgroup M. pulchella).
To assess the number of private alleles for each species and sampling location we used a custom perl script (https://github.com/ehshogren/MyzomelaPopulationGenomics) which identified monomorphic or biallelic sites that were found only in the species under consideration and present in at least five individuals, reporting which location(s) the individuals carrying that private allele were sampled from. To identify the number of fixed differences and shared polymorphisms between species/sampling locations we used another custom perl script which considered two groups of phenotypically Mcard or Mtris individuals (from different species and/or sampling locations) and required a minimum of 5 individuals in each group with a genotype at the site in question.
Supporting information
Summary of Myzomela tristrami reference genome statistics, including both the raw and final scaffolded assembly.
(PDF)
Number of whole genome sequences by species, sampling site, and sex.
(PDF)
Number of single nucleotide polymorphisms per genomic region for different filtering of datasets used in analyses.
(PDF)
Nucleotide diversity (π) averaged across 50 kb windows, standard error in parentheses for each sampled population of phenotypic parental Myzomela cardinalis and M. tristrami.
(PDF)
Ratio of nucleotide diversity for sex chromosome regions to large autosomes (chr1–10), and for the new pseudo-autosomal region (neoPAR) to a comparatively sized autosome (chr 14). Nucleotide diversity averaged across 50kb windows.
(PDF)
Differentiation (FST), averaged across 50kb windows for each region of the genome, standard error in parentheses for pairwise comparisons of phenotypic parental populations of Myzomela cardinalis and M. tristrami. Allopatry and sympatry abbreviated as Allo. and Sym. respectively.
(PDF)
Divergence (dxy) averaged across 50kb windows for each region of the genome, standard error in parentheses for pairwise comparisons of phenotypic parental populations of Myzomela cardinalis and M. tristrami. Allopatry and sympatry abbreviated as Allo. and Sym. respectively.
(PDF)
Summary of expectations for genomic compartments with respect to recombination, ploidy, and introgression (relative to autosomes).
(PDF)
Contingency tables of number of individuals with matching or mismatched phenotype and W/neo-W or mitochondrial genotype. Number of individuals differs between W/neo-W and mitochondria because W/neo-W includes only females while mitochondria includes both females and males. Fisher’s exact test significant for W/neo-W (p = 0.026) and for mitochondria (p = 0.005).
(PDF)
Number of captured and molecularly sexed males and females for each sampled population, used to calculate a sex ratio.
(PDF)
Number of alleles per genomic region private to each population of the respective focal species, using SNPs filtered for depth and quality only.
(PDF)
Number of fixed and shared alleles in each region of the genome for population comparisons, using SNPs filtered for depth and quality only. Allopatry and sympatry abbreviated as Allo. and Sym. respectively.
(PDF)
Comparative structure of neo-sex chromosomes across Lichenostomus melanops cassidix (no neo-sex chromosome), Myzomela tristrami (neo-sex chromosome), and Entomyzon cyanotis (neo-sex chromosome, described in Burley and Orzechowski et al. 2023 [62]).
(PDF)
Admixture ratio (f4 statistic) for each autosome and sex chromosome regions. Color of the point indicates for which topology the statistic was calculated, and shape of the point indicates whether the D statistic for that chromosome was significantly different from zero, using the block-jackknife procedure.
(PDF)
Cross-validation error for ADMIXTURE of K = 1–7 for autosomes (A), Z/neo-Z (B), and W/neo-W (C). ADMIXTURE plot for autosomal sequence with K = 2 (D). Phenotypic hybrids are outlined in yellow, with phenotypic cardinalis to the left and phenotypic tristrami to the right of phenotypic hybrids.
(PDF)
Metric for quantifying admixture (fdM) plotted against length of chromosome or chromosomal region (shape indicates genomic compartment). We calculated and averaged fdM across 100 SNP non-overlapping windows using both the cardinalis P3 and the tristrami P3 topology (color of shapes indicates topology). We included only windows where D ≥ 0 indicating no introgression or sharing of alleles between sympatric populations (P2 and P3).
(PDF)
Principal component 1 (PC1) of autosomal PCA plotted against PC1 of Z/neo-Z region (A) and W/neo-W region (B). Symbol color represents phenotypic species assignment while symbol shape indicates sampling locality. Plot for Z/neo-Z is separated by sex to distinguish homogametic males and heterogametic females. All individuals are female in (B).
(PDF)
Metric for quantifying admixture (fdM) plotted against nucleotide diversity of chromosome or chromosomal region for the Mtris allopatric population (shape indicates genomic compartment). We calculated and averaged fdM across 100 SNP non-overlapping windows using the cardinalis P3 topology. We included only windows where D ≥ 0 indicating no introgression or gene flow between sympatric Mtris (P2) and sympatric Mcard (P3). When considering only autosomes there is a significant correlation between fdM and nucleotide diversity (Pearson’s correlation coefficient = 0.41, p = 0.007).
(PDF)
Mitochondrial haplotype TCS network show that Mcard haplotypes are much less diverse than Mtris haplotypes but are shared with hybrids and phenotypic Mtris individuals in sympatry. Mutations between nodes shown in parentheses, and number of individuals sharing haplotype denoted by size of circles (see legend).
(PDF)
Acknowledgments
We thank the Solomon Islands Ministries of Environment and Education for granting research permits to work in the Solomon Islands. We also thank John and Joyce Murray, J. Tauni, J. Waihuru, K. Rupen, H. Ha’aina, J. Pepare, L. Taka, P. Teo, J. Suafuria, G. Wabeasi, and J. Waihuru for invaluable assistance in data collection.
Data Availability
Genomic data are available on Sequence Read Archive under BioProject PRJNA1095846. Reference genome files are available on dryad at doi:10.5061/dryad.612jm64c9. Analysis pipeline and scripts are available at https://github.com/ehshogren/MyzomelaPopulationGenomics.
Funding Statement
Fieldwork was funded by the Aresty Chair in Tropical Ecology and an NSF CAREER award (IOS 1137624/0643606) to JACU, and awards from the Society of Systematic Biologists, Kushlan Graduate Research Support Fund, and Jay M. Savage Graduate Research Support Fund to JMS. Additional support was provided by NSF-PRFB to EHS (2010748), and NSF-DEB 2112474 to JACU and DCP. The NSF-PRFB 2010748 funded salary for EHS and the NSF-DEB 2112474 funded salary for CAM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mayr E. Systematics and the origin of species. New York, NY: Columbia University Press; 1942. [Google Scholar]
- 2.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 3.Price TD. Speciation in birds. Greenwood Village, CO, USA: Roberts and Company; 2008. [Google Scholar]
- 4.Mayr E, Diamond J. The birds of northern Melanesia: speciation, ecology, and biogeography. New York, NY: Oxford University Press; 2001. [Google Scholar]
- 5.Payseur BA, Rieseberg LH. A genomic perspective on hybridization and speciation. Molecular Ecology. 2016;25: 2337–2360. doi: 10.1111/mec.13557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C-I. The genic view of the process of speciation. Journal of Evolutionary Biology. 2001;14: 851–865. doi: 10.1046/j.1420-9101.2001.00335.x [DOI] [Google Scholar]
- 7.Barton NH, Hewitt GM. Analysis of hybrid zones. Annual review of ecology and systematics Vol 16. 1985; 113–148. doi: 10.1146/annurev.es.16.110185.000553 [DOI] [Google Scholar]
- 8.Payseur BA. Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Molecular Ecology Resources. 2010;10: 806–820. doi: 10.1111/j.1755-0998.2010.02883.x [DOI] [PubMed] [Google Scholar]
- 9.Rand DM, Harrison RG. Ecological genetics of a mosaic hybrid zone: mitochondrial, nuclear, and reproductive differentiation of crickets by soil type. Evolution. 1989;43: 432–449. doi: 10.1111/j.1558-5646.1989.tb04238.x [DOI] [PubMed] [Google Scholar]
- 10.Rohwer S, Wood C. Three hybrid zones between Hermit and Townsend’s Warblers in Washington and Oregon. The Auk. 1998;115: 284–310. doi: 10.2307/4089188 [DOI] [Google Scholar]
- 11.Marques DA, Lucek K, Meier JI, Mwaiko S, Wagner CE, Excoffier L, et al. Genomics of rapid incipient speciation in sympatric Threespine Stickleback. PLoS Genetics. 2016;February: 1–34. doi: 10.1371/journal.pgen.1005887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semenov GA, Linck E, Enbody ED, Harris RB, Khaydarov DR, Alström P, et al. Asymmetric introgression reveals the genetic architecture of a plumage trait. Nature Communications. 2021;12: 1–9. doi: 10.1038/s41467-021-21340-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campagna L, Mo Z, Siepel A, Uy JAC. Selective sweeps on different pigmentation genes mediate convergent evolution of island melanism in two incipient bird species. PLOS Genetics. 2022;18: e1010474. doi: 10.1371/journal.pgen.1010474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravinet M, Faria R, Butlin RK, Galindo J, Bierne N, Rafajlović M, et al. Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. Journal of Evolutionary Biology. 2017;30: 1450–1477. doi: 10.1111/jeb.13047 [DOI] [PubMed] [Google Scholar]
- 15.Moran BM, Payne C, Langdon Q, Powell DL, Brandvain Y, Schumer M, et al. The genomic consequences of hybridization. 2021; 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell DL, Moran BM, Kim BY, Banerjee SM, Aguillon SM, Fascinetto-Zago P, et al. Two new hybrid populations expand the swordtail hybridization model system. Evolution. 2021;75: 2524–2539. doi: 10.1111/evo.14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldassarre DT, White TA, Karubian J, Webster MS. Genomic and morphological analysis of a semipermeable avian hybrid zone suggests an asymmetrical introgression of a sexual signal. Evolution. 2014;68: 2644–2657. doi: 10.1111/evo.12457 [DOI] [PubMed] [Google Scholar]
- 18.Pardo-Diaz C, Salazar C, Baxter SW, Merot C, Figueiredo-Ready W, Joron M, et al. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genetics. 2012;8. doi: 10.1371/journal.pgen.1002752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: massive introgression by local genes. Evolution. 2008;62: 1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x [DOI] [PubMed] [Google Scholar]
- 20.Stein AC, Uy JAC. Unidirectional introgression of a sexually selected trait across an avian hybrid zone: a role for female choice? Evolution. 2006;60: 1476–1485. doi: 10.1111/j.0014-3820.2006.tb01226.x [DOI] [PubMed] [Google Scholar]
- 21.Yang W, Feiner N, Laakkonen H, Sacchi R, Zuffi MAL, Scali S, et al. Spatial variation in gene flow across a hybrid zone reveals causes of reproductive isolation and asymmetric introgression in wall lizards. Evolution. 2020;74: 1289–1300. doi: 10.1111/evo.14001 [DOI] [PubMed] [Google Scholar]
- 22.Thompson KA, Peichel CL, Rennison DJ, McGee MD, Albert AYK, Vines TH, et al. Analysis of ancestry heterozygosity suggests that hybrid incompatibilities in threespine stickleback are environment-dependent. PLoS Biology. 2021;20: e3001469. doi: 10.1371/journal.pbio.3001469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnegard ME, McGee MD, Matthews B, Marchinko KB, Conte GL, Kabir S, et al. Genetics of ecological divergence during speciation. Nature. 2014;511: 307–311. doi: 10.1038/nature13301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobzhansky T. Genetics and the origin of species. Columbia University Press; 1937. [Google Scholar]
- 25.Muller HJ. Isolating mechanisms, evolution, and temperature. Biological Symposia. 1942;6: 71–125. [Google Scholar]
- 26.Bateson W. Heredity and variation in modern lights. In: Seward AC, editor. Darwin and Modern Science. Cambridge: Cambridge University Press; 1909. pp. 85–101. [Google Scholar]
- 27.Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. The American Naturalist. 1987;130: 113–146. [Google Scholar]
- 28.Kirkpatrick M, Hall DW. Male-Biased mutation, sex linkage, and the rate of adaptive evolution. Evolution. 2004;58: 437–440. doi: 10.1111/j.0014-3820.2004.tb01659.x [DOI] [PubMed] [Google Scholar]
- 29.Mank JE, Axelsson E, Ellegren H. Fast-X on the Z: rapid evolution of sex-linked genes in birds. 2007; 618–624. doi: 10.1101/gr.6031907.618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meisel RP, Connallon T. The faster-X effect: integrating theory and data. Trends in Genetics. 2013;29: 537–544. doi: 10.1016/j.tig.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the faster-X effect: empirical results and their interpretation. Evolution. 2010;64: 663–674. doi: 10.1111/j.1558-5646.2009.00853.x [DOI] [PubMed] [Google Scholar]
- 32.Dean R, Harrison PW, Wright AE, Zimmer F, Mank JE. Positive selection underlies faster-Z evolution of gene expression in birds. Molecular Biology and Evolution. 2015;32: 2646–2656. doi: 10.1093/molbev/msv138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bechsgaard J, Schou MF, Vanthournout B, Hendrickx F, Knudsen B, Settepani V, et al. Evidence for faster X chromosome evolution in spiders. Molecular Biology and Evolution. 2019;36: 1281–1293. doi: 10.1093/molbev/msz074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coyne JA, Orr HA. Two rules of speciation. In: Otte D, Endler J, editors. Speciation and Its Consequences. Sunderland, MA: Sinauer Associates; 1989. pp. 180–207. [Google Scholar]
- 35.Irwin DE. Sex chromosomes and speciation in birds and other ZW systems. Molecular Ecology. 2018;27: 3831–3851. doi: 10.1111/mec.14537 [DOI] [PubMed] [Google Scholar]
- 36.Sæther SA, Sætre GP, Borge T, Wiley C, Svedin N, Andersson G, et al. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science. 2007;318: 95–97. doi: 10.1126/science.1141506 [DOI] [PubMed] [Google Scholar]
- 37.Prowell DP. Sex linkage and speciation in Lepidoptera. Endless forms: species and speciation. Oxford University Press; 1998. pp. 309–319. [Google Scholar]
- 38.Uy JAC, Irwin DE, Webster MS. Behavioral isolation and incipient speciation in birds. Annual Review of Ecology and Systematics. 2018;49: 1–24. [Google Scholar]
- 39.Haldane JBS. Sex ratio and unisexual sterility in hybrid animals. Journal of Genetics. 1922;12: 101–109. [Google Scholar]
- 40.Coyne JA. Genetics and speciation. Nature. 1992;355: 511–515. doi: 10.1038/355511a0 [DOI] [PubMed] [Google Scholar]
- 41.Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends in Genetics. 2008;24: 336–343. doi: 10.1016/j.tig.2008.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muirhead CA, Presgraves DC. Hybrid incompatibilities, local adaptation, and the genomic distribution of natural introgression between species. American Naturalist. 2016;187: 249–261. doi: 10.1086/684583 [DOI] [PubMed] [Google Scholar]
- 43.Fraïsse C, Sachdeva H. The rates of introgression and barriers to genetic exchange between hybridizing species: Sex chromosomes vs autosomes. Genetics. 2021;217. doi: 10.1093/genetics/iyaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiong T, Tarikere S, Rosser N, Li X, Yago M, Mallet J. A polygenic explanation for Haldane’s rule in butterflies. Proceedings of the National Academy of Sciences. 2023;120: e2300959120. doi: 10.1073/pnas.2300959120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigeman H, Ponnikas S, Chauhan P, Dierickx E, Brooke MDL, Hansson B. Repeated sex chromosome evolution in vertebrates supported by expanded avian sex chromosomes. Proceedings of the Royal Society B: Biological Sciences. 2019;286: 20192051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei KHC, Bachtrog D. Ancestral male recombination in Drosophila albomicans produced geographically restricted neo-Y chromosome haplotypes varying in age and onset of decay. PLoS Genetics. 2019;15: 1–28. doi: 10.1371/journal.pgen.1008502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beaudry FEG, Rifkin JL, Peake AL, Kim D, Jarvis-Cross M, Barrett SCH, et al. Effects of the neo-X chromosome on genomic signatures of hybridization in Rumex hastatulus. Molecular Ecology. 2022;31: 3708–3721. doi: 10.1111/mec.16496 [DOI] [PubMed] [Google Scholar]
- 48.Huang Z, De O. Furo I, Liu J, Peona V, Gomes AJB, Cen W, et al. Recurrent chromosome reshuffling and the evolution of neo-sex chromosomes in parrots. Nature Communications. 2022;13: 1–11. doi: 10.1038/s41467-022-28585-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461: 1079–1083. doi: 10.1038/nature08441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bracewell RR, Bentz BJ, Sullivan BT, Good JM. Rapid neo-sex chromosome evolution and incipient speciation in a major forest pest. Nature Communications. 2017;8. doi: 10.1038/s41467-017-01761-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Filatov DA. The two “rules of speciation” in species with young sex chromosomes. Molecular Ecology. 2018;27: 3799–3810. doi: 10.1111/mec.14721 [DOI] [PubMed] [Google Scholar]
- 52.Wang S, Nalley MJ, Chatla K, Aldaimalani R, MacPherson A, Wei K, et al. Neo-sex chromosome evolution shapes sex-dependent asymmetrical introgression barrier. Proceedings of the National Academy of Sciences. 2022;119: e2119382119. doi: 10.1073/pnas.2119382119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nanda I, Schlegelmilch K, Haaf T, Schartl M, Schmid M. Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenet Genome Res. 2008;122: 150–156. doi: 10.1159/000163092 [DOI] [PubMed] [Google Scholar]
- 54.Ellegren H. Evolutionary stasis: the stable chromosomes of birds. Trends in Ecology & Evolution. 2010;25: 283–291. doi: 10.1016/j.tree.2009.12.004 [DOI] [PubMed] [Google Scholar]
- 55.Pala I, Naurin S, Stervander M, Hasselquist D, Bensch S, Hansson B. Evidence of a neo-sex chromosome in birds. Heredity. 2011;108: 264–272. doi: 10.1038/hdy.2011.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gunski RJ, Cañedo AD, Garnero ADV, Ledesma MA, Coria N, Montalti D, et al. Multiple sex chromosome system in penguins (Pygoscelis, Spheniscidae). Comp Cytogenet. 2017;11: 541–552. doi: 10.3897/CompCytogen.v11i3.13795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sigeman H, Zhang H, Ali S, Bengt A. A novel neo-sex chromosome in Sylvietta brachyura (Macrosphenidae) adds to the extraordinary avian sex chromosome diversity among Sylvioidea songbirds. 2022; 1–9. doi: 10.1111/jeb.14096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gan HM, Falk S, Morales HE, Austin CM, Sunnucks P, Pavlova A. Genomic evidence of neo-sex chromosomes in the eastern yellow robin. GigaScience. 2019;8: giz111. doi: 10.1093/gigascience/giz111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dierickx EG, Wa Sin SY, Van Veelen PJH, De Brooke LM, Liu Y, Edwards SV, et al. Genetic diversity, demographic history and neo-sex chromosomes in the Critically Endangered Raso lark. Proceedings of the Royal Society B: Biological Sciences. 2020;287. doi: 10.1098/rspb.2019.2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kretschmer R, Gunski RJ, Garnero A del V, de Freitas TRO, Toma GA, Cioffi M de B, et al. Chromosomal analysis in Crotophaga ani (Aves, Cuculiformes) reveals extensive genomic reorganization and an unusual Z-Autosome Robertsonian Translocation. Cells. 2020;10: 4. doi: 10.3390/cells10010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leroy T, Anselmetti Y, Tilak M-K, Bérard S, Csukonyi L, Gabrielli M, et al. A bird’s white-eye view on avian sex chromosome evolution. Peer Community Journal. 2021;1. doi: 10.24072/pcjournal.70 [DOI] [Google Scholar]
- 62.Burley JT, Orzechowski SCM, Sin SYW, Edwards SV. Whole-genome phylogeography of the blue-faced honeyeater (Entomyzon cyanotis) and discovery and characterization of a neo-Z chromosome. Molecular Ecology. 2023;32: 1248–1270. doi: 10.1111/mec.16604 [DOI] [PubMed] [Google Scholar]
- 63.Kvistad L, Falk S, Austin L. Widespread genomic signatures of reproductive isolation and sex-specific selection in the Eastern Yellow Robin, Eopsaltria australis. G3 Genes|Genomes|Genetics. 2022;12: jkac145. doi: 10.1093/g3journal/jkac145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sardell JM. Evolutionary consequences of recent secondary contact between Myzomela honeyeaters. Open Access Dissertations. Miami, FL; 2016. [Google Scholar]
- 65.Sardell JM, Uy JAC. Hybridization following recent secondary contact results in asymmetric genotypic and phenotypic introgression between island species of Myzomela honeyeaters. Evolution. 2016;70: 257–269. doi: 10.1111/evo.12864 [DOI] [PubMed] [Google Scholar]
- 66.Marki PZ, Jønsson KA, Irestedt M, Nguyen JMT, Rahbek C, Fjeldså J. Supermatrix phylogeny and biogeography of the Australasian Meliphagides radiation (Aves: Passeriformes). Molecular Phylogenetics and Evolution. 2017;107: 516–529. doi: 10.1016/j.ympev.2016.12.021 [DOI] [PubMed] [Google Scholar]
- 67.Diamond JM. Dispersal, Mimicry, and Geographic Variations in Northern Melanesian Birds. 2002;56: 1–22. [Google Scholar]
- 68.Mayr E. Birds of the Southwest Pacific. New York, NY: Macmillan; 1945. [Google Scholar]
- 69.Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol Biol Evol. 2021;38: 4647–4654. doi: 10.1093/molbev/msab199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robledo-Ruiz DA, Gan HM, Kaur P, Dudchenko O, Weisz D, Khan R, et al. Chromosome-length genome assembly and linkage map of a critically endangered Australian bird: the helmeted honeyeater. GigaScience. 2022;11: giac025. doi: 10.1093/gigascience/giac025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475: 493–496. doi: 10.1038/nature10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pool JE, Nielsen R. Population size changes reshape genomic patterns of diversity. Evolution. 2007;61: 3001–3006. doi: 10.1111/j.1558-5646.2007.00238.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Malinsky M, Matschiner M, Svardal H. Dsuite—Fast D-statistics and related admixture evidence from VCF files. Molecular Ecology Resources. 2021;21: 584–595. doi: 10.1111/1755-0998.13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, et al. Ancient admixture in human history. Genetics. 2012;192: 1065–1093. doi: 10.1534/genetics.112.145037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malinsky M, Challis RJ, Tyers AM, Schiffels S, Terai Y, Ngatunga BP, et al. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science. 2015;350: 1493–1498. doi: 10.1126/science.aac9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alexander DH, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics. 2011;12: 246. doi: 10.1186/1471-2105-12-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quilodrán CS, Tsoupas A, Currat M. The spatial signature of introgression after a biological invasion with hybridization. Frontiers in Ecology and Evolution. 2020;8. Available from: https://www.frontiersin.org/articles/10.3389/fevo.2020.569620. [Google Scholar]
- 78.Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin’s corollary to Haldane’s rule. Genetics. 2007;176: 1059–1088. doi: 10.1534/genetics.106.065979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tobler M, Barts N, Greenway R. Mitochondria and the origin of species: bridging genetic and ecological perspectives on speciation processes. Integrative and Comparative Biology. 2019;59: 900–911. doi: 10.1093/icb/icz025 [DOI] [PubMed] [Google Scholar]
- 80.Presgraves DC, Meiklejohn CD. Hybrid sterility, genetic conflict and complex speciation: lessons from the Drosophila simulans clade species. Frontiers in Genetics. 2021;12: 1–18. doi: 10.3389/fgene.2021.669045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Price TD, Bouvier MM. The evolution of F1 postzygotic incompatibilities in birds. Evolution. 2002;56: 2083–2089. doi: 10.1111/j.0014-3820.2002.tb00133.x [DOI] [PubMed] [Google Scholar]
- 82.Hill GE. Mitonuclear coevolution as the genesis of speciation and the mitochondrial DNA barcode gap. Ecology and Evolution. 2016;6: 5831–5842. doi: 10.1002/ece3.2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meiklejohn CD, Holmbeck MA, Siddiq MA, Abt DN, Rand DM, Montooth KL. An incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLOS Genetics. 2013;9: e1003238. doi: 10.1371/journal.pgen.1003238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Toews DPL, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Molecular Ecology. 2012;21: 3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x [DOI] [PubMed] [Google Scholar]
- 85.Dagilis AJ, Matute DR. The fitness of an introgressing haplotype changes over the course of divergence and depends on its size and genomic location. PLOS Biology. 2023;21: e3002185. doi: 10.1371/journal.pbio.3002185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Toews DPL, Mandic M, Richards JG, Irwin DE. Migration, mitochondria, and the yellow-rumped warbler. Evolution. 2014;68: 241–255. doi: 10.1111/evo.12260 [DOI] [PubMed] [Google Scholar]
- 87.Hulsey CD, Bell KL, García-de-León FJ, Nice CC, Meyer A. Do relaxed selection and habitat temperature facilitate biased mitogenomic introgression in a narrowly endemic fish? Ecology and Evolution. 2016;6: 3684–3698. doi: 10.1002/ece3.2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meiklejohn CD, Landeen EL, Gordon KE, Rzatkiewicz T, Kingan SB, Geneva AJ, et al. Gene flow mediates the role of sex chromosome meiotic drive during complex speciation. eLife. 2018;7: 1–31. doi: 10.7554/eLife.35468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou Q, Bachtrog D. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science. 2012;337: 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Longmire JL, Maltbie M, Baker RJ. Use of “lysis buffer” in DNA isolation and its implication for museum collections. Occasional Papers—Museum of Texas Tech University. 1997;163. [Google Scholar]
- 91.Cheng H, Concepcion GT, Feng X, Zhang H, Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature Methods. 2021;18: 170–175. doi: 10.1038/s41592-020-01056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheng H, Jarvis ED, Fedrigo O, Koepfli KP, Urban L, Gemmell NJ, et al. Haplotype-resolved assembly of diploid genomes without parental data. Nature Biotechnology. 2022;40: 1332–1335. doi: 10.1038/s41587-022-01261-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14: 1394–1403. doi: 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9: 5114. doi: 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proceedings of the National Academy of Sciences of the United States of America. 2020;117: 9451–9457. doi: 10.1073/pnas.1921046117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suh A, Smeds L, Ellegren H. Abundant recent activity of retrovirus-like retrotransposons within and among flycatcher species implies a rich source of structural variation in songbird genomes. Molecular Ecology. 2018;27: 99–111. doi: 10.1111/mec.14439 [DOI] [PubMed] [Google Scholar]
- 97.Boman J, Frankl-Vilches C, dos Santos MDS, de Oliveira EHC, Gahr M, Suh A. The genome of Blue-capped Cordon-bleu uncovers hidden diversity of LTR retrotransposons in Zebra Finch. Genes. 2019;10. doi: 10.3390/genes10040301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weissensteiner MH, Bunikis I, Catalán A, Francoijs KJ, Knief U, Heim W, et al. Discovery and population genomics of structural variation in a songbird genus. Nature Communications. 2020;11: 1–11. doi: 10.1038/s41467-020-17195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peona V, Palacios-Gimenez OM, Blommaert J, Liu J, Haryoko T, Jønsson KA, et al. The avian W chromosome is a refugium for endogenous retroviruses with likely effects on female-biased mutational load and genetic incompatibilities. Philosophical Transactions of the Royal Society B: Biological Sciences. 2021;376. doi: 10.1098/rstb.2020.0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Smit A, Hubley R, Green P. RepeatMasker Open-4.0. 2013. Available from: <http://www.repeatmasker.org> [Google Scholar]
- 101.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17: 10–12. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 102.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25: 1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25: 2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Van der Auwera G, O’Connor B. Genomics in the cloud: using Docker, GATK, and WDL in Terra. 1st ed. O’Reilly Media; 2020. [Google Scholar]
- 105.Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32: 292–294. doi: 10.1093/bioinformatics/btv566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, Depristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27: 2156–2158. doi: 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schield DR, Scordato ESC, Smith CCR, Carter JK, Cherkaoui SI, Gombobaatar S, et al. Sex-linked genetic diversity and differentiation in a globally distributed avian species complex. Molecular Ecology. 2021; 2313–2332. doi: 10.1111/mec.15885 [DOI] [PubMed] [Google Scholar]
- 108.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81: 559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.R Core Team. R: a language and environment for statistical computing. 2018. Available from: https://www.r-project.org/ [Google Scholar]
- 110.Wickham H. ggplot2: Elegant graphics for data analysis. New York, NY: Springer-Verlag; 2016. Available from: https://ggplot2.tidyverse.org [Google Scholar]
- 111.Bird JP, Martin R, Akçakaya HR, Gilroy J, Burfield IJ, Garnett ST, et al. Generation lengths of the world’s birds and their implications for extinction risk. Conservation Biology. 2020;34: 1252–1261. doi: 10.1111/cobi.13486 [DOI] [PubMed] [Google Scholar]
- 112.Smeds L, Qvarnström A, Ellegren H. Direct estimate of the rate of germline mutation in a bird. 2016; 1211–1218. doi: 10.1101/gr.204669.116.Freely [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Korunes KL, Samuk K. pixy: Unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Molecular Ecology Resources. 2021; 1–10. doi: 10.1111/1755-0998.13326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Srikulnath K, Ahmad SF, Singchat W, Panthum T. Why do some vertebrates have microchromosomes? Cells. 2021;10: 2182. doi: 10.3390/cells10092182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martin SH, Davey JW, Jiggins CD. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Molecular Biology and Evolution. 2015;32: 244–257. doi: 10.1093/molbev/msu269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28: 3326–3328. doi: 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gompert Z, Alex Buerkle C. introgress: a software package for mapping components of isolation in hybrids. Molecular Ecology Resources. 2010;10: 378–384. doi: 10.1111/j.1755-0998.2009.02733.x [DOI] [PubMed] [Google Scholar]
- 118.Ortiz EM. vcf2phylip v2.0: convert a VCF matrix into several matrix formats for phylogenetic analysis. 2019. doi: 10.5281/zenodo.2540861 [DOI] [Google Scholar]
- 119.Leigh JW, Bryant D. POPART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution. 2015;6: 1110–1116. doi: 10.1111/2041-210X.12410 [DOI] [Google Scholar]
- 120.Clement M, Posada D, Crandall KA. TCS: A computer program to estimate gene genealogies. Molecular Ecology. 2000;9: 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of Myzomela tristrami reference genome statistics, including both the raw and final scaffolded assembly.
(PDF)
Number of whole genome sequences by species, sampling site, and sex.
(PDF)
Number of single nucleotide polymorphisms per genomic region for different filtering of datasets used in analyses.
(PDF)
Nucleotide diversity (π) averaged across 50 kb windows, standard error in parentheses for each sampled population of phenotypic parental Myzomela cardinalis and M. tristrami.
(PDF)
Ratio of nucleotide diversity for sex chromosome regions to large autosomes (chr1–10), and for the new pseudo-autosomal region (neoPAR) to a comparatively sized autosome (chr 14). Nucleotide diversity averaged across 50kb windows.
(PDF)
Differentiation (FST), averaged across 50kb windows for each region of the genome, standard error in parentheses for pairwise comparisons of phenotypic parental populations of Myzomela cardinalis and M. tristrami. Allopatry and sympatry abbreviated as Allo. and Sym. respectively.
(PDF)
Divergence (dxy) averaged across 50kb windows for each region of the genome, standard error in parentheses for pairwise comparisons of phenotypic parental populations of Myzomela cardinalis and M. tristrami. Allopatry and sympatry abbreviated as Allo. and Sym. respectively.
(PDF)
Summary of expectations for genomic compartments with respect to recombination, ploidy, and introgression (relative to autosomes).
(PDF)
Contingency tables of number of individuals with matching or mismatched phenotype and W/neo-W or mitochondrial genotype. Number of individuals differs between W/neo-W and mitochondria because W/neo-W includes only females while mitochondria includes both females and males. Fisher’s exact test significant for W/neo-W (p = 0.026) and for mitochondria (p = 0.005).
(PDF)
Number of captured and molecularly sexed males and females for each sampled population, used to calculate a sex ratio.
(PDF)
Number of alleles per genomic region private to each population of the respective focal species, using SNPs filtered for depth and quality only.
(PDF)
Number of fixed and shared alleles in each region of the genome for population comparisons, using SNPs filtered for depth and quality only. Allopatry and sympatry abbreviated as Allo. and Sym. respectively.
(PDF)
Comparative structure of neo-sex chromosomes across Lichenostomus melanops cassidix (no neo-sex chromosome), Myzomela tristrami (neo-sex chromosome), and Entomyzon cyanotis (neo-sex chromosome, described in Burley and Orzechowski et al. 2023 [62]).
(PDF)
Admixture ratio (f4 statistic) for each autosome and sex chromosome regions. Color of the point indicates for which topology the statistic was calculated, and shape of the point indicates whether the D statistic for that chromosome was significantly different from zero, using the block-jackknife procedure.
(PDF)
Cross-validation error for ADMIXTURE of K = 1–7 for autosomes (A), Z/neo-Z (B), and W/neo-W (C). ADMIXTURE plot for autosomal sequence with K = 2 (D). Phenotypic hybrids are outlined in yellow, with phenotypic cardinalis to the left and phenotypic tristrami to the right of phenotypic hybrids.
(PDF)
Metric for quantifying admixture (fdM) plotted against length of chromosome or chromosomal region (shape indicates genomic compartment). We calculated and averaged fdM across 100 SNP non-overlapping windows using both the cardinalis P3 and the tristrami P3 topology (color of shapes indicates topology). We included only windows where D ≥ 0 indicating no introgression or sharing of alleles between sympatric populations (P2 and P3).
(PDF)
Principal component 1 (PC1) of autosomal PCA plotted against PC1 of Z/neo-Z region (A) and W/neo-W region (B). Symbol color represents phenotypic species assignment while symbol shape indicates sampling locality. Plot for Z/neo-Z is separated by sex to distinguish homogametic males and heterogametic females. All individuals are female in (B).
(PDF)
Metric for quantifying admixture (fdM) plotted against nucleotide diversity of chromosome or chromosomal region for the Mtris allopatric population (shape indicates genomic compartment). We calculated and averaged fdM across 100 SNP non-overlapping windows using the cardinalis P3 topology. We included only windows where D ≥ 0 indicating no introgression or gene flow between sympatric Mtris (P2) and sympatric Mcard (P3). When considering only autosomes there is a significant correlation between fdM and nucleotide diversity (Pearson’s correlation coefficient = 0.41, p = 0.007).
(PDF)
Mitochondrial haplotype TCS network show that Mcard haplotypes are much less diverse than Mtris haplotypes but are shared with hybrids and phenotypic Mtris individuals in sympatry. Mutations between nodes shown in parentheses, and number of individuals sharing haplotype denoted by size of circles (see legend).
(PDF)
Data Availability Statement
Genomic data are available on Sequence Read Archive under BioProject PRJNA1095846. Reference genome files are available on dryad at doi:10.5061/dryad.612jm64c9. Analysis pipeline and scripts are available at https://github.com/ehshogren/MyzomelaPopulationGenomics.