Abstract
Interferons are cytokines that play a complex role in the resistance of mammalian hosts to pathogens. IFNγ (interferon-γ) is secreted by activated T-cells and natural killer cells. IFNγ is involved in a wide range of physiological processes, including antiviral activity, immune response, cell proliferation and apoptosis, as well as the stimulation and repression of a variety of genes. IFNγ activity is modulated by the binding of its C-terminal domain to HS (heparan sulphate), a glycosaminoglycan found in the extracellular matrix and at the cell surface. In the present study, we analysed the interaction of isolated heparin-derived oligosaccharides with the C-terminal peptide of IFNγ by NMR, in aqueous solution. We observed marked changes in the chemical shifts of both peptide and oligosaccharide compared with the free state. Our results provide evidence of a binding through electrostatic interactions between the charged side chains of the protein and the sulphate groups of heparin that does not induce specific conformation of the C-terminal part of IFNγ. Our data also indicate that an oligosaccharide size of at least eight residues displays the most efficient binding.
Keywords: heparin-derived oligosaccharide, interferon-γ (IFNγ), NMR spectroscopy, protein–carbohydrate interaction
Abbreviations: 1D, one-dimensional; dp, degree of polymerization; DSS, sodium 2,2-dimethyl-2-silapentane-5-sulphonate; HBS, Hepes-buffered saline; ΔHexA, 4-deoxy-α-L-threo-hex-4-enepyranosyluronic acid; HS, heparan sulphate; IdoA, L-iduronic acid; IFNγ, interferon-γ; C-IFNγ, C-terminal domain of IFNγ; IFNγR, IFNγ receptor; nOe, nuclear Overhauser effect; RU, resonance units; NS, 2-N-sulphate; 2S, 2-O-sulphate; 6S, 6-O-sulphate
INTRODUCTION
IFNγ (interferon-γ) is a highly pleiotropic protein secreted mainly by activated T-lymphocytes and natural killer cells. It plays a central role in modulating most phases of the inflammatory and immune response, and displays antiproliferative and antiviral activities [1]. IFNγ mediates its activities through binding to a specific transmembrane receptor (IFNγR) expressed at the surface of almost all cells [2,3]. This cytokine is a homodimer consisting of two 143-amino-acid polypeptides. Both the X-ray structure and the NMR solution structure showed that the monomeric subunits interact in an antiparallel orientation and consist of six helices, followed by an unfolded sequence at the C-terminal side (amino acids 124–143). This part of the molecule extends away from the protein core, and is believed to be highly flexible and/or to adopt multiple conformations [4,5]. Owing to proteolytic cleavages, human IFNγ is often found as a mixture of C-terminal-truncated molecules [5–7]. The conditions and role of the cytokine processing remain unclear. However, the C-terminal tail was shown to be critical for biological activity. The C-terminal domain (C-IFNγ) that appears to be the most important is a conserved basic stretch from residue 128 to 131, called the D1 domain, since its removal inactivates the cytokine [6,8]. Downstream of the D1 domain is another cluster of basic amino acids, called D2, cleavage of which increases the biological activity of the protein. The C-IFNγ also confers to the protein a high affinity for HS (heparan sulphate), a glycosaminoglycan found in extracellular matrix and at the cell surface [9]. Our previous studies showed that, in vivo, the binding of IFNγ to HS is involved in the plasma clearance of the cytokine, but also limits the extent of the C-terminus degradation, which thus in turn enhances the cytokine activity [10]. HS (or the chemically related glycosaminoglycan, heparin) is a highly sulphated, linear polysaccharide with a basic structure composed of a repeat disaccharide unit [4IdoAα/GlcAβ1-4GlcNα1-(IdoA is L-iduronic acid)]. This widely distributed polysaccharide has a unique molecular design in which highly sulphated domains (called S or heparin-like domains) alternate with more homogeneous and poorly sulphated regions [11]. Using a footprinting strategy, in which HS was depolymerized in the presence of IFNγ, we isolated an HS fragment composed of a poorly sulphated internal domain, flanked by two small highly sulphated oligosaccharides. Our previous study showed that these two sulphated heparin-like sequences directly interacted with an IFNγ dimer [12]. In the present study, we have prepared and characterized heparin-derived oligosaccharides and analysed their interaction with the C-terminal peptide of the cytokine by NMR spectroscopy.
EXPERIMENTAL
Heparin immobilization and binding assay
Heparin (9 kDa; a gift from Dr M. Petitou, Sanofi-Synthelabo, Paris, France) at 1 mM in PBS was reacted for 24 h at room temperature (22 °C) with 10 mM biotin–LC-hydrazine (Pierce). The mixture was extensively dialysed against water to remove unreacted biotin and was freeze-dried. Two flow cells of an F1 sensorchip were activated with 50 μl of 0.2 M EDC [N-ethyl-N′-(diethylaminopropyl)-carbodi-imide] (Biacore) and 0.05 M NHS [N-hydroxysuccinimide] (Biacore), after which 50 μl of streptavidin (Sigma-Aldrich) at 0.2 mg/ml in 10 mM sodium acetate buffer, pH 4.2, was injected. The remaining activated groups were blocked with 50 μl of 1 M ethanolamine, pH 8.5. Typically, this procedure yielded a coupling of 2000–2500 RU (resonance units) of streptavidin, on which approx. 100 RU of biotinylated heparin were immobilized. The other surface was left untreated and served as negative control surface. Before use, the chip was submitted to several injections of HBS (Hepes-buffered saline) containing 1.25 M NaCl, followed by continuous flow washing with HBS-EP buffer (10 mM Hepes, 0.15 M NaCl, 3 mM EDTA and 0.05% P20 detergent, pH 7.4). Binding experiments were carried out at 25 °C and at a constant flow rate of 50 μl/min. IFNγ was injected over the negative-control surface or the heparin surface for 5 min. The heparin-sensor surface was regenerated with a 250 μl pulse of 1.25 M NaCl.
The binding of heparin to the C-IFNγ was analysed using a filter binding assay [13]. Briefly, the C-IFNγ (0–5 μg) and biotinylated heparin (0.5 μg/ml) were co-incubated for 45 min at room temperature in 500 μl of PBS. For the competition assay, unlabelled heparin (4 μg/ml) was added to the mixture. The C-IFNγ plus any bound biotinylated heparin was then trapped on a nitrocellulose membrane (Bio-Rad), by drawing the incubation mixture through the membrane with a vacuum-assisted dot-blot apparatus. The membrane was washed twice with 200 μl of PBS and blocked with 5% (w/v) dry milk in PBS/0.05% (v/v) Tween 20. The peptide-bound biotinylated heparin was revealed by incubating the membrane with 0.5 μg/ml extravidin peroxidase (Sigma-Aldrich), followed by ECL® (enhanced chemiluminescence) detection (Amersham Biosciences).
C-IFNγ
The human C-IFNγ (residues 123–143), AAKTGKRKRSQMLFRGRRASQ, was synthesized by Neosystem (Strasbourg, France).
Preparation of heparin oligosaccharides
Porcine mucosal heparin (Sigma-Aldrich) (10 g) was digested with heparinase I (Grampian Enzymes, Orkney, U.K.) (8 m-units/ml) in 150 ml of 0.1 mg/ml BSA, 2 mM CaCl2, 50 mM NaCl and 5 mM Tris buffer, pH 7.5, for 54 h at 25 °C. The enzymic reaction was stopped by heating the digest at 100 °C for 5 min. Digestion products were then size-separated using a Bio-Gel P-10 column (Bio-Rad) (4.4 cm×150 cm), equilibrated with 0.25 M NaCl and run at 1 ml/min. Eluted material, detected by absorbance at 232 nm, consisted of a graded series of size-uniform oligosaccharides resolved from disaccharide [dp (degree of polymerization) 2] to octadecasaccharide (dp18). To ensure homogeneity, only the top fractions of each peak were pooled, and each isolated fraction was re-chromatographed on a gel-filtration column to eliminate possible contamination further. Samples were dialysed against distilled water and quantified. Tetrasaccharides (dp4) and octasaccharides (dp8) were purified further by strong-anion-exchange HPLC, on a 9 mm×250 mm preparative ProPac PA1 column (Dionex). After equilibration in the mobile phase (distilled water adjusted to pH 3.5 with HCl) at 1 ml/min, samples (7.5 mg) were injected and eluted with a gradient of NaCl (0–1.4 M over 30 min, then 1.4–1.8 M over 60 min) in the same mobile phase. The eluate was monitored on-line for UV absorbance at 232 nm, and the most anionic species (eluted at 1.23 and 1.49 M NaCl for dp4 and dp8 respectively) were selected for further analysis.
NMR spectroscopy
A C-IFNγ peptide sample was prepared at 2.5 mM in a 20 mM phosphate buffer, pH 5, and 10% 2H2O (Euriso-top, Saint-Aubin, France). Oligosaccharide samples were prepared at a concentration of 6, 3 and 1.4 mM for the octa-, tetra- and di-saccharide respectively in the same buffer as for the peptide. All samples were degassed under argon before recording. The oligosaccharide–peptide complexes were prepared by direct addition of several aliquots of a concentrated solution of unlabelled sugar (typically 15 mM) to the NMR peptide sample.
NMR spectra were recorded at 27 °C, on Varian Inova 600 and 800 MHz spectrometers equipped with a triple-resonance (1H, 13C, 15N) probe including shielded z-gradients. To compare the free and sugar-bound C-IFNγ, the backbone chemical shifts of the sugar–peptide complex were assigned under the same conditions as the free C-IFNγ. Peptide and the peptide–sugar complex assignments were performed with COSY, TOCSY and NOESY experiments. All data were processed, using the FELIX 2000 software (Accelrys). Proton chemical shifts were reported with respect to the H2O signal relative to DSS (sodium 2,2-dimethyl-2-silapentane-5-sulphonate) at 27 °C.
RESULTS
The heparin-binding site of IFNγ is located in the C-IFNγ
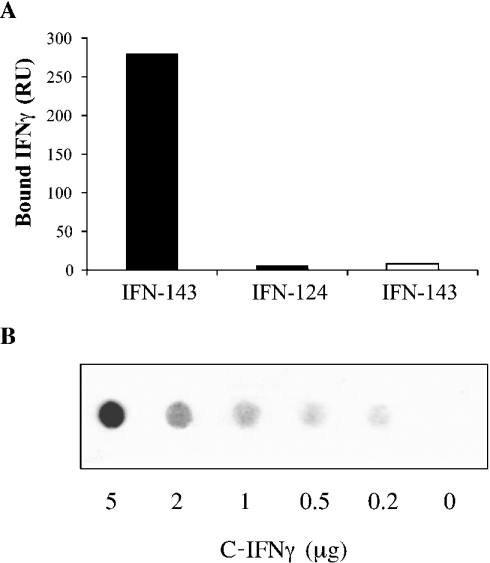
IFNγ comprises four basic clusters of amino acids (KLFKNFK, KKKR, KTGKRKR and RGRR) being putatively HS binding domains. Two of them (KTGKRKR and RGRR, termed D1 and D2 respectively) are clustered in the C-IFNγ. In order to analyse IFNγ binding to heparin, we used surface plasmon resonance techniques. Injection of full-length IFNγ (IFN-143) over a Biacore sensorchip containing 100 RU of immobilized heparin produced, at equilibrium, a binding response of 280 RU, while no binding was observed upon injection of the cytokine over a control surface. Injection of C-terminal-deleted (last 19 amino acids) IFNγ (IFN-124) produced virtually no response (Figure 1A), indicating that the remaining basic clusters of the cytokine (KLFKNFK and KKKR, residues 55–61 and 86–89 respectively) do not permit binding to heparin, and thus that the C-IFNγ (AAKTGKRKRSQMLFRGRRASQ) contains the heparin-binding activity. To demonstrate further direct interaction between the C-IFNγ and heparin, the peptide was incubated with biotinylated heparin. The reaction mixture was then drawn through a nitrocellulose membrane. Free heparin was washed away, while the C-IFNγ-bound heparin remained trapped on the membrane and was detected by exposing the blots to peroxidase-conjugated extravidin, followed by ECL® detection (Figure 1B). The absence of signal for the negative control confirmed that free biotinylated heparin was not retained by the nitrocellulose membrane. In contrast, binding of heparin to the C-IFNγ was clearly shown by spots, the density of which increased with the C-IFNγ concentration. The C-IFNγ–biotinylated heparin interaction could be inhibited with an excess of non-biotinylated heparin (results not shown). The C-IFNγ was then used in the following NMR experiments.
Figure 1. Surface-plasmon-resonance-based binding of IFNγ to immobilized heparin and filter analysis of C-IFNγ–heparin interaction.
(A) IFNγ (IFN-143) or C-terminal-domain-deleted IFNγ (IFN-124), both at 0.5 μg/ml, were injected over a Biacore F1 sensorchip containing streptavidin alone (open bar) or streptavidin plus 100 RU of heparin (closed bars) for 5 min. The surface plasmon resonance signals (in RU) were recorded at the end of the injection phase. (B) Biotin-labelled heparin (0.5 μg/ml) was co-incubated with a range of C-IFNγ concentrations, and bound material was recovered by filtration through a nitrocellulose membrane. Retained biotinylated heparin was revealed by chemiluminescent assay.
NMR characterization of the oligosaccharides
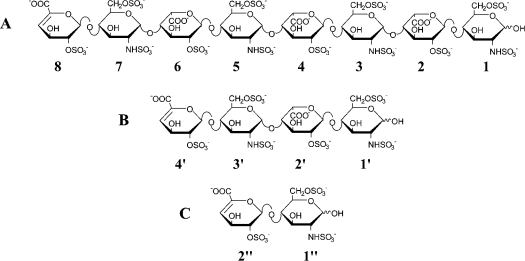
Heparin-derived oligosaccharides (dp8) corresponding to the IFNγ-binding domain in HS [12] were obtained by enzymic depolymerization, and purified by molecular sieving and ion-exchange chromatography. Chemical shifts of the octasaccharide (dp8) were assigned by two-dimensional TOCSY and COSY analyses, as reported for the sulphated oligosaccharides isolated previously from heparin [14] and HS [15]. The NMR data obtained in the present study for dp8 are summarized in Table S1 (available at http://www.BiochemJ.org/bj/384/bj3840093add.htm). The internal uronic acid residues of dp8 were unambiguously identified on the basis of the chemical shifts of anomeric proton signals and coupling constants 3JH1,H2. Anomeric proton signals observed around δ 5.2–5.0 and coupling constants 3JH1,H2 of approx. 3 Hz clearly indicated [16] the presence of three αIdoA residues in dp8. The chemical shifts of H1 and H2 of IdoA residues were shifted downfield by approx. 0.2 and 0.6 p.p.m. respectively when compared with those of non-sulphated IdoA residues [15], supporting the 2-sulphation of the three IdoA residues of dp8. Regarding the sulphation of GlcN residues, large upfield shifts for chemical shifts of H2 and downfield shifts for chemical shifts of H6 and H6′ indicate the N- and 6-sulphation of the four GlcN residues in dp8. Based upon these NMR data, the following structure was proposed for dp8: ΔHexA (2S)-[GlcN(NS,6S)-IdoA(2S)]3-GlcN(NS,6S) (where ΔHexA is 4-deoxy-α-L-threo-hex-4-enepyranosyluronic acid, 2S is 2-O-sulphate, NS is 2-N-sulphate and 6S is 6-O-sulphate) (Figure 2A). Chemical shift assignment of each residue was performed using NOESY experiments by identifying nOe (nuclear Overhauser effect) correlations through the glycosidic linkages.
Figure 2. Chemical structures of heparin-derived oligosaccharides.
Structures of oligosaccharides: dp8 (A), dp4 (B) and dp2 (C). Residues are labelled 1, 2, 3, 4, 5, 6, 7 and 8 for glucosamine, iduronate, glucosamine, iduronate, glucosamine, iduronate, glucosamine and uronate, as discussed in the text. Note that ring 8 uronate has an un-natural double bond resulting from heparinase digestion.
1H NMR characterization of the C-IFNγ–dp8 complex
To gain more structural insights into the interaction, the C-IFNγ–dp8 complex was analysed by NMR. The assignment of the C-IFNγ (Table S2; see http://www.BiochemJ.org/bj/384/bj3840093add.htm) was performed using the usual approach described by Wüthrich [17]. HN and Hα proton chemical shifts were in the random-coil range, indicating that the peptide was not structured in the absence of dp8 [18]. This was corroborated further by the absence in the NOESY experiment of any medium- or long-range inter-residue correlations, although intra-residue correlations were observed.
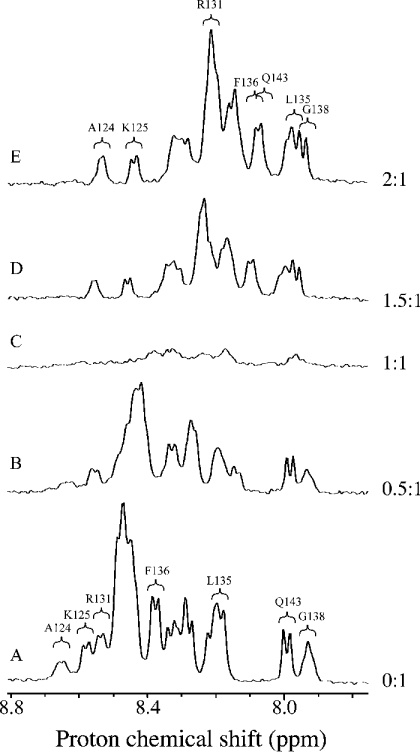
Addition of dp8 to the C-IFNγ resulted for the peptide in large changes in the 1D (one-dimensional)-spectra amidic part as illustrated in Figure 3. Such a result clearly indicated that the oligosaccharide formed a complex with the C-IFNγ. Moreover, a transitory formation of a gel phase was observed, characterized macroscopically by a large increase in sample viscosity, the maximum occurring at a C-IFNγ/dp8 ratio of approx. 1:1. Spectroscopically, below this value, we observed variations of the peptide resonances (Figure 3B) concomitant with a decrease of the signal resonances (see Figure 3C), without appearance of the oligosaccharide signals. In a second phase, the gel disappeared upon addition of the oligosaccharide. At this stage, dp8 signals were observed, whereas peptide signals re-increased without significant variations of the chemical shifts (see Figures 3D and 3E). At some point, no further variation of the peptide signal was observed upon oligosaccharide additions, indicating C-IFNγ–dp8 complex formation. This was obtained for a stoichiometry of 2:1. Interestingly, we re-observed the initial 1D-spectrum of the peptide after addition of 500 mM NaCl, indicating a complete dissociation of the complex under these conditions.
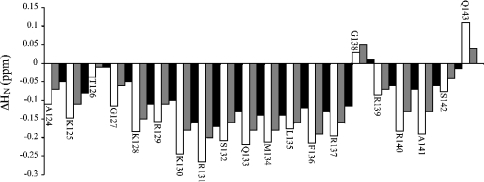
Figure 3. NMR spectra of the C-IFNγ in the free state and in complex.
1H NMR spectra (800 MHz) of the C-IFNγ (A) and mixtures of the C-IFNγ with heparin octasaccharide (dp8) at the molar ratios ([dp8]/[C-IFNγ]) of 0.5 (B), 1 (C), 1.5 (D) and 2 (E). Only the HN region of the spectra is shown.
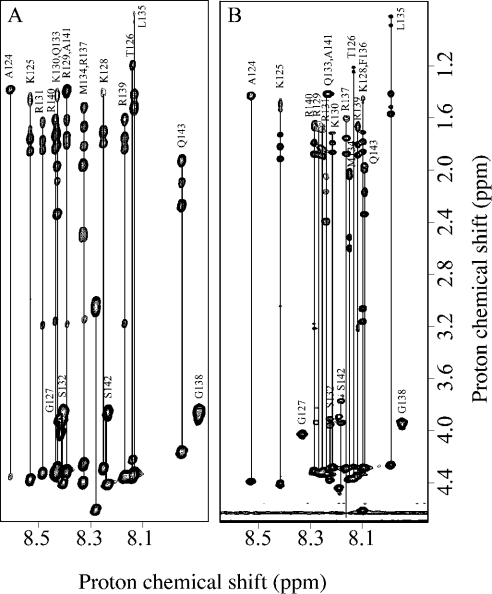
The assignment of the peptide in complex was made following the proton amide changes during the titration, and was confirmed directly using the same assignment protocol as for the peptide alone, as illustrated in Figure 4. Assignments of the complexed form are reported in Table S2 (see http://www.BiochemJ.org/bj/384/bj3840093add.htm). As for the peptide alone, we did not observe any nOe correlations indicative of a structuration of the peptide. Similarly, the Hα chemical shifts did not experience any systematic shielding or unshielding upon binding. All of the HN protons were significantly shielded downfield apart from residues Thr126, Gly138 and Gln143, which is the last residue of the sequence (Figure 5, open bars). For the octasaccharide within the complex, the assignment was performed as for free dp8 and reported in Table S1 (see http://www.BiochemJ.org/bj/384/bj3840093add.htm). Variations of the dp8 1H chemical shifts during complex formation are represented in Figure 6(A). Major downfield variations were observed for IdoA(2S) H5 protons and upfield for GlcN(NS,6S) H3- and ΔHexA(2S)-8 H4-protons. All these protons, except H4-8, are in the proximity of glycosidic linkages. We did not observe any nOe correlations between dp8 and the C-IFNγ, indicating that the average distance between a dp8 proton and a C-IFNγ proton is likely to be greater than 5 Å (1 Å=0.1 nm).
Figure 4. Two-dimensional TOCSY NMR spectra of the C-IFNγ in the free state and in complex.
Amide-aliphatic part of the 800 MHz TOCSY spectra of the C-IFNγ (A) and in complex with dp8 at the molar ratio of 2:1 dp8/C-IFNγ (B).
Figure 5. Chemical-shift variations on complex formation for the C-IFNγ.
Variations of the 1H chemical shifts of the C-IFNγ on complex formation with the three oligosaccharides: dp8 (open bars), dp4 (grey bars) and dp2 (closed bars). Final molar ratios were of 1:2 C-IFNγ/oligosaccharide. The difference in the chemical shifts observed in the TOCSY spectra is displayed as a function of the sequence number.
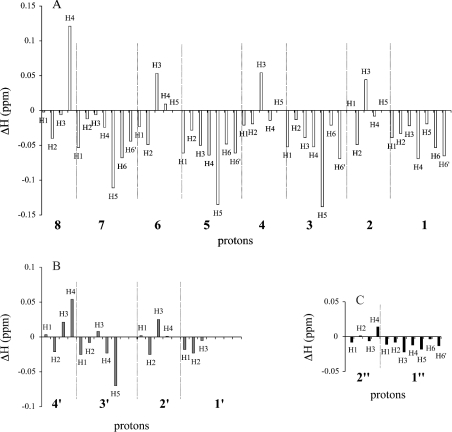
Figure 6. Chemical-shift variations on complex formation for oligosaccharides.
Variations of the 1H chemical shifts of oligosaccharides on complex formation with C-INFγ–dp8 (open bars, A), dp4 (grey bars, B) and dp2 (closed bars, C). Final molar ratios of C-IFNγ/oligosaccharide were 1:2. The difference of the chemical shifts observed in the TOCSY spectra is displayed.
Effect of the oligosaccharide size upon binding
To investigate further the general features of this interaction, we studied the impact of the oligosaccharide size. Two other oligosaccharides were prepared, tetrasaccharide (dp4) and disaccharide (dp2), as described in the Experimental section. The following structures were proposed for dp4 and dp2 respectively: ΔHexA(2S)-GlcN(NS,6S)-IdoA(2S)-GlcN(NS,6S) and ΔHexA(2S)-GlcN(NS,6S) (Figures 2B and 2C). Additions of dp4 and dp2 to the C-IFNγ resulted, for the peptide, in similar changes in the NMR spectra as for dp8, in particular in the HN region.
Interestingly, if the general features were similar for the three oligosaccharides dp2, dp4 and dp8, we observed some striking differences between them. In the case of di- and tetra-saccharide, a molar ratio of 1:1 for the complex was observed in both cases. Moreover, the formation of the gel was observed in presence of dp4, but not with dp2. Variations of 1H chemical shifts of peptide upon complex formation showed that the peptide was less affected by the presence of dp4 and dp2 than dp8 (Figure 6). Regarding oligosaccharide chemical shift variations, protons of dp4 were less affected than those of dp8, and dp2 proton chemical shifts remained quite unchanged.
DISCUSSION
IFNγ comprises a number of basic amino acid clusters that could all function as HS/heparin-binding sequences [19]. We reported previously that the C-IFNγ, which contains two of these clusters, was involved in the interaction [20]. In the present study, we have shown that IFNγ with its C-terminal domain eliminated was no longer able to bind to heparin. This indicated that the binding activity was contained entirely within the C-IFNγ, which thus was used as a model to analyse the binding of IFNγ to heparin. Similarly, as we have already described that the binding of IFNγ to HS involved heparin-like oligosaccharides [12,21], heparin was used in the present study to prepare oligosaccharides that could mimic the binding sequence. This involved enzymic depolymerization of heparin and size-separation of the generated fragments, from disaccharide (dp2) to octadecasaccharide (dp18). Fractions corresponding to octa- (dp8) and tetra- (dp4) saccharides were resolved further by strong-anion-exchange HPLC. Since we did not know the importance of the number and position of sulphate groups within these oligosaccharides for the binding process, we selected the most anionic species, i.e. the molecules that contain the highest number of sulphate groups, and characterized their sequences by NMR spectroscopy. NMR spectral analyses of the oligosaccharides prepared by enzymic depolymerization of heparin showed that they consisted of three sulphated disaccharide repeating units [i.e. 4IdoA(2S)1-4GlcN(NS,6S)α1-].
As expected from the high sensitivity to proteolysis of the C-IFNγ, which indicated a large flexibility of this part in the context of the full-length protein [4,5], the NMR spectral analyses showed that the peptide alone was unstructured. Heparin-binding domains in proteins are often helical structures, and it has been demonstrated for different natural or synthetic peptides, such as poly(L-lysine), that the interaction with heparin promoted an α-helix conformation in the polypeptide [22–25]. Although we clearly observed by NMR spectroscopy the formation of a complex between the oligosaccharide and the peptide, we demonstrated that the formation of the C-IFNγ–dp8 complex did not induce any secondary structure of the peptide. Our results also corroborated the implication of charged side chain groups in the binding. The complex formation was sensitive to the salt concentration, in agreement with a binding driven by ionic interactions between sulphate groups and the positively charged groups of lysine and arginine side chains. The absence of any nOe correlation between protons of dp8 and the C-IFNγ also suggested that the distance between the protons was greater than 5 Å, which supports such a model of interaction further. The absence of structuration consequently indicated that all the positively charged side chains can be implicated in the binding. This also strongly suggests that, in the full-length IFNγ the C-terminus has not the propensity to adopt any particular secondary-structural motif and keeps a level of flexibility sufficient to adapt different binding domains in oligosaccharides. Such a property has been also observed for the transduction domain of the HIV-1 Tat protein. The binding of heparin to this domain only induced a decrease in the mobility without any significant structural change [26].
Interestingly, we observed the formation of a gel between the C-IFNγ and dp4 or dp8. This implicated the formation of an oligomeric spatial structure and thus the presence of at least two interaction sites by peptide and oligosaccharide dp4 units. Variations observed for the peptide and oligosaccharides could be interpreted as follows. As recognition takes place between dp8 and the C-IFNγ, the different binding sites of the oligosaccharides interact with the binding sites of different peptides, leading to the formation of a supramolecular structure. This explains the absence of dp4 or dp8 signals as well as the decrease of peptide signals, both being implicated in a large structure whose signals are not observable by NMR. At a determined concentration, all the binding sites of the peptides are occupied. As the concentration of sugar increases, a competition for the binding sites of the peptide takes place between the oligosaccharides, leading to the release of species composed of only one peptide interacting with one or few oligosaccharides. In this case, the apparent mass of the peptide–oligosaccharide complex decreases strongly without significant changes in the chemical environment of the peptide atoms, allowing the observation of oligosaccharide signals and an increase in peptide signals. This point will require further investigation.
The question we wanted to address is the consequences of the binding on the solution structure of both partners. Our results clearly indicate that the complex does not adopt a unique rigid conformation. However, with regard to dp8, we observed major variations for all the H5 protons of IdoA(2S) and H3 protons of GlcN(NS,6S) which are at the proximity of glycosidic linkages. The IdoA residue exists in an equilibrium of three conformations, namely 1C4, 4C1 and 2S0 [27]. The conformer population depends on substitution at C-2 and on the structure of neighbouring units. We tried to investigate this point with the help of coupling constants and nOe measurements in order to estimate the influence of conformations on chemical-shift modifications. Unfortunately, the overlapping of IdoA anomeric signals makes any accurate measurement impossible. However, such a study was possible for the ΔHexA residue. The 3JH1,H2 and 3JH2,H3 coupling constants are both inferior to 4 Hz in free dp8, indicating a 1H2 conformation of the cycle. No relevant change was observed for dp8 in complex with the peptide. Thus the conformation of the ΔHexA residue was not influenced by the presence of the C-IFNγ. For IdoA, it was shown that when this residue is inserted between two N,6-disulphated D-glucosamine residues, the skew-boat form 2S0 has an important contribution (40%) to the conformation of the sulphated IdoA residues [27]. The 3JH2,H3 coupling constants of dp8 IdoA seem to be in agreement with this feature. Moreover, a thorough comparison of dp8 signals in the free state and in complex with the C-IFNγ did not show significant differences in vicinal interproton coupling constants (3JH,H; results not shown). Thus the presence of the C-IFNγ does not stabilize the IdoA(2S) residue into any specific conformation. Consequently, the observed modifications of chemical shifts can be assigned to changes at the glycosidic linkages. Such chemical-shift variations have been observed in a tetrasaccharide–antithrombin complex and interpreted as resulting from conformational changes at the glycosidic linkages [28]. It could then be speculated that the C-IFNγ–dp8 binding induces a reciprocal fitting of the two partner conformations, leading to a restriction of the accessible conformations for oligosaccharide glycosidic linkages. Interestingly, it must be noted that the peptide chemical-shift variations in the complex are greater for dp8 than for dp4, and similarly greater for dp4 than for dp2. Regarding the modifications in oligosaccharides, we have clearly observed that proton chemical shifts are disturbed more by the presence of the C-IFNγ in the case of dp8 than for the smaller oligosaccharides (dp4 and dp2). These differences of behaviour between dp8 on one hand, and dp4 and dp2 on the other could be due to different chemical environments. As pointed out above, binding is very probably related to charge–charge interactions located far away from the peptide and oligosaccharide backbones. We then suggest that the larger the heparin fragment is, the more a modification of torsion angles is necessary for an optimal interaction. Thus dp8 seems to correspond to the optimal length proposed in a previous study, in which it was shown that two sulphated hexa- to octa-saccharides interact simultaneously with the two C-termini of an IFNγ dimer [12]. So, even if smaller oligosaccharides bind, the interaction is weaker. The fact that smaller oligosaccharides were not observed to interact with the C-IFNγ could probably be explained by the different ranges of the concentrations in experiments. Indeed, competition experiments between HS and heparin of different molecular masses for binding to IFNγ were performed using concentrations in the micromolar range [12], whereas NMR solutions were in the millimolar range.
The basic residues in the C-IFNγ are clustered in two domains, D1 and D2. Binding of IFNγ to heparin limits the extent of the C-terminal degradation of the cytokine, and in particular protects the D1 domain from proteolysis [10]. Based on the importance of this cluster for biological activity [6], it has been proposed that specific association between D1 and heparin or HS oligosaccharides was functionally relevant. The D2 basic cluster did not appear to be required for this biological activity [6], but its deletion induced a strong increase of activity, suggesting a possible regulatory role. Our NMR results suggest that both of these basic clusters were involved in oligosaccharide binding, as the resonances of the residues of both domains D1 and D2 were affected. For D1, this binding is in agreement with the biochemical observation. In contrast, the implication of the D2 domain was unexpected and is difficult to rationalize in view of the biological results. However, as discussed above, the C-IFNγ exhibits at least two HS/heparin-binding sites, these being presumably the D1 and D2 segments. Taking into account the different contents in positively charged side chains of D1 (+5 charges) and D2 (+3 charges), it could be proposed that the respective affinity of these domains for HS is different. Then, in the dimeric form, if the distance between the two C-terminal parts does not match two binding motifs in HS precisely, the competition between D1 and D2 will lead, under physiological conditions, to a preferential binding of the D1 domain.
In conclusion, our results favour a model in which the C-IFNγ binds to HS by electrostatic interactions between lysine or arginine charged side chains and HS sulphate groups, and reinforce the view that at least an octasaccharide represents the minimum length for efficient binding.
Online data
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique, the Commissariat à l'Energie Atomique (CEA) and the University Joseph Fourier. We thank J.P. Andrieu (Laboratoire d'Enzymologie Moleculaire, Institut de Biologie Structurale) for amino acid analyses and R. Vives for critical reading of the manuscript. C.V. was recipient of a CEA fellowship.
References
- 1.Boehm U., Klamnp T., Groot M., Howard J. C. Cellular responses to interferon-γ. Annu. Rev. Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 2.Farrar M. A., Shreiber R. D. The molecular cell biology of interferon-γ and its receptor. Annu. Rev. Immunol. 1993;11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 3.Van Loon A. P., Ozmen L., Fountoulakis M., Kania M., Haiker M., Garotta G. High-affinity receptor for interferon-γ (IFN-γ), a ubiquitous protein occurring in different molecular forms on human cells: blood monocytes and eleven different cell lines have the same IFN-γ receptor protein. J. Leukocyte Biol. 1991;49:462–473. doi: 10.1002/jlb.49.5.462. [DOI] [PubMed] [Google Scholar]
- 4.Ealick S. E., Cook W. J., Vijay-Kumar S., Carson M., Nagabhushan T. L., Trotta P. P., Bugg C. E. Three-dimensional structure of recombinant human interferon-γ. Science. 1991;252:698–702. doi: 10.1126/science.1902591. [DOI] [PubMed] [Google Scholar]
- 5.Grzesiek S., Döbeli H., Gentz R., Garotta G., Labhardt A. M., Bax A. 1H, 13C, and 15N NMR backbone assignments and secondary structure of human interferon-γ. Biochemistry. 1992;31:8180–8190. doi: 10.1021/bi00150a009. [DOI] [PubMed] [Google Scholar]
- 6.Döbeli H., Gentz R., Jucker W., Garotta G., Hartmann D. W., Hochuli E. Role of the C-terminal sequence on the biological activity of human immune interferon (IFN) J. Biotechnol. 1988;7:199–216. [Google Scholar]
- 7.Curling E. M., Hayter P. M., Baines A. J., Bull A. T., Gull K., Strange P. G., Jenkins N. Recombinant human interferon-γ: differences in glycosylation and proteolytic processing lead to heterogeneity in batch culture. Biochem. J. 1990;272:333–337. doi: 10.1042/bj2720333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadir R., Forest E., Lortat-Jacob H. The heparan sulfate binding sequence of interferon-γ increased the on rate of the interferon-γ–interferon-γ receptor complex formation. J. Biol. Chem. 1998;273:10919–10925. doi: 10.1074/jbc.273.18.10919. [DOI] [PubMed] [Google Scholar]
- 9.Lortat-Jacob H., Kleinman H. K., Grimaud J. A. High affinity binding of interferon-γ to a basement membrane complex (Matrigel) J. Clin. Invest. 1991;87:878–883. doi: 10.1172/JCI115093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lortat-Jacob H., Baltzer F., Grimaud J. A. Heparin decreases the blood clearance of interferon-γ and increases its activity by limiting the processing of its carboxyl-terminal sequence. J. Biol. Chem. 1996;271:16139–16143. doi: 10.1074/jbc.271.27.16139. [DOI] [PubMed] [Google Scholar]
- 11.Lindahl U., Kusche-Gullberg M., Kjellén L. Regulated diversity of heparan sulfate. J. Biol. Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 12.Lortat-Jacob H., Turnbull J. E., Grimaud J. A. Molecular organization of the interferon-γ binding domain in heparan sulphate. Biochem. J. 1995;310:497–505. doi: 10.1042/bj3100497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadir R., Baleux F., Grosdidier A., Imberty A., Lortat-Jacob H. Characterization of the stromal cell-derived factor-1α/heparin complex. J. Biol. Chem. 2001;276:8288–8296. doi: 10.1074/jbc.M008110200. [DOI] [PubMed] [Google Scholar]
- 14.Yamada S., Yoshida K., Sugiura M., Sugahara K., Khoo K. H., Morris H. R., Dell A. Structural studies on the bacterial lyase-resistant tetrasaccharides derived from the antithrombin III-binding site of porcine intestinal heparin. J. Biol. Chem. 1993;268:4780–4787. [PubMed] [Google Scholar]
- 15.Sugahara K., Tohno-oka R., Yamada S., Khoo K. H., Morris H. R., Dell A. Structural studies on the oligosaccharides isolated from bovine kidney heparan sulphate and characterization of bacterial heparitinases used as substrates. Glycobiology. 1994;4:535–544. doi: 10.1093/glycob/4.4.535. [DOI] [PubMed] [Google Scholar]
- 16.Yamada S., Murakami T., Tsuda H., Yoshida K., Sugahara K. Isolation of the porcine heparin tetrasaccharides with glucuronate 2-O-sulfate: heparinase cleaves glucuronate 2-O-sulfate-containing disaccharides in highly sulfated blocks in heparin. J. Biol. Chem. 1995;270:8696–8705. [PubMed] [Google Scholar]
- 17.Wüthrich K. New York: John Wiley & Sons; 1986. NMR of proteins and nucleic acids. [Google Scholar]
- 18.Wishart D. S., Sykes B. D., Richards F. M. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 19.Cardin A. D., Weintraub H. J. Molecular modeling of protein–glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 20.Lortat-Jacob H., Grimaud J. A. Interferon-γ binds to heparan sulfate by a cluster of amino acids located in the C-terminal part of the molecule. FEBS Lett. 1991;280:152–154. doi: 10.1016/0014-5793(91)80225-r. [DOI] [PubMed] [Google Scholar]
- 21.Lortat-Jacob H., Grimaud J. A. Binding of interferon-γ to heparan sulfate is restricted to the heparin-like domains and involves carboxylic – but not N-sulfated – groups. Biochim. Biophys. Acta. 1992;1117:126–130. doi: 10.1016/0304-4165(92)90069-7. [DOI] [PubMed] [Google Scholar]
- 22.Ferran D. S., Sobel M., Harris R. B. Design and synthesis of a helix heparin-binding peptide. Biochemistry. 1992;31:5010–5016. doi: 10.1021/bi00136a014. [DOI] [PubMed] [Google Scholar]
- 23.Taylor G. J., Yorke S. C., Harding D. R. Glycosaminoglycan specificity of a heparin-binding peptide. Pept. Res. 1995;8:286–293. [PubMed] [Google Scholar]
- 24.Mulloy B., Crane D. T., Drake A. F., Davies D. B. The interaction between heparin and polylysine: a circular dichroism and molecular modelling study. Braz. J. Med. Biol. Res. 1996;29:721–729. [PubMed] [Google Scholar]
- 25.Pimenta D. C., Nantes I. L., de Souza E. S., Le Bonniec B., Ito A. S., Tersariol I. L., Oliveira V., Juliano M. A., Juliano L. Interaction of heparin with internally quenched fluorogenic peptides derived from heparin-binding consensus sequences, kallistatin and anti-thrombin III. Biochem. J. 2002;366:435–446. doi: 10.1042/BJ20020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hakansson S., Caffrey M. Structural and dynamic properties of the HIV-1 tat transduction domain in the free and heparin-bound states. Biochemistry. 2003;42:8999–9006. doi: 10.1021/bi020715+. [DOI] [PubMed] [Google Scholar]
- 27.Ferro D. R., Provasoli A., Ragazzi M., Torri G., Casu B., Gatti G., Jacquinet J.-C., Sinaÿ P., Petitou M., Choay J. Evidence for conformational equilibrium of the sulphated L-iduronate residue in heparin and in synthetic heparin mono- and oligosaccharides: NMR and force-field studies. J. Am. Chem. Soc. 1986;108:6773–6778. [Google Scholar]
- 28.Hricovini M., Guerrini M., Bisio A. Structure of heparin-derived tetrasaccharide complexed to the plasma protein antithrombin derived from NOEs, J-couplings and chemical shifts. Eur. J. Biochem. 1999;261:789–801. doi: 10.1046/j.1432-1327.1999.00335.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.