Abstract
The metalloisomerase glyoxalase I (GlxI) catalyses the conversion of methylglyoxal-glutathione hemithioacetal and related derivatives into the corresponding thioesters. In contrast with the previously characterized GlxI enzymes of Homo sapiens, Pseudomonas putida and Saccharomyces cerevisiae, we recently determined that Escherichia coli GlxI surprisingly did not display Zn2+-activation, but instead exhibited maximal activity with Ni2+. To investigate whether non-Zn2+ activation defines a distinct, previously undocumented class of GlxI enzymes, or whether the E. coli GlxI is an exception to the previously established Zn2+-activated GlxI, we have cloned and characterized the bacterial GlxI from Yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitidis. The metal-activation profiles for these additional GlxIs firmly establish the existence of a non-Zn2+-dependent grouping within the general category of GlxI enzymes. This second, established class of metal activation was formerly unidentified for this metalloenzyme. Amino acid sequence comparisons indicate a more extended peptide chain in the Zn2+-dependent forms of GlxI (H. sapiens, P. putida and S. cerevisiae), compared with the GlxI enzymes of E. coli, Y. pestis, P. aeruginosa and N. meningitidis. The longer sequence is due in part to the presence of additional regions situated fairly close to the metal ligands in the Zn2+-dependent forms of the lyase. With respect to sequence alignments, these inserts may potentially contribute to defining the metal specificity of GlxI at a structural level.
Keywords: glyoxalase, lyase, metal activation, metalloenzyme, nickel, zinc
Abbreviations: DTT, dithithreitol; GlxI, glyoxalase I; MG, methylglyoxal; VOC, vicinal oxygen chelate
INTRODUCTION
The glyoxalase system has been recognized for over 90 years as a critical enzymic detoxification route that is widely distributed across major taxonomic divisions [1–9]. This two-enzyme system accepts α-oxoaldehydes (α-ketoaldehydes) as substrates for conversion into corresponding α-hydroxyacids [10]. GlxI [glyoxalase I; S-D-lactoylglutathione methylglyoxal lyase (isomerizing), EC 4.4.1.5] is a metalloisomerase that converts the non-enzymically formed MG (methylglyoxal)–GSH hemithioacetal adduct into the corresponding thioester (Scheme 1). This product is subsequently hydrolysed by glyoxalase II (S-2-hydroxyacylglutathione hydrolase, EC 3.1.2.6) generating D-lactate and regenerating GSH [11].
Scheme 1. Reactions of the glyoxalase system.
GlxI is considered to be a member of the βαβββ and VOC (vicinal oxygen chelate) superfamilies [12–16]. The former superfamily is distinguished by proteins that possess tandem arrangements of βαβββ that form a cleft, which, in the case of GlxI, comprises the metal-binding active site. The VOC superfamily refers to a grouping of enzymes defined by the mechanistic feature of co-ordination by vicinal substrate oxygen atoms to the catalytic metal centre. The defining attributes of these two superfamilies as they manifest in GlxI imply the importance of metal-ion binding for the function of this lyase.
The GlxI enzyme has been isolated from several sources, mostly of eukaryotic origin. Characterization of GlxI enzymes, particularly those from Homo sapiens and Saccharomyces cerevisiae, has revealed selectivity for Zn2+ activation [17,18]. In fact, the classification of GlxI as a Zn2+-dependent lyase has persisted. However, initial enzymological investigation of E. coli GlxI revealed that this metalloisomerase was not activated by Zn2+ ions, but instead was optimally activated in the presence of Ni2+ ions [19].
The question arises as to whether Ni2+ (or rather non-Zn2+) activation as observed in the E. coli enzyme is mirrored by GlxI enzymes from other bacterial sources, potentially indicating a second, previously unrecognized class of this metalloenzyme. A putative division of metal selectivity for GlxI enzymes on the basis of biological origin (eukaryotic compared with bacterial) could provide an interesting structure–function target for developing antibacterial agents, if this difference is proven and is biochemically exploitable, given the continuing interest in the glyoxalase system from a medicinal perspective [20–27]. However, the Pseudomonas putida (prokaryotic) GlxI has been shown to be a Zn2+ enzyme, implying that a division in metal activation along evolutionary lines may not be quite so clearly defined [28]. To investigate this possibility, we have cloned and overexpressed several putative bacterial GlxI-encoding sequences from the micro-organisms Yersinia pestis, P. aeruginosa and Neisseria meningitidis [29].
The results of our biochemical characterization of these purified enzymes indicate that the Y. pestis, P. aeruginosa and N. meningitidis putative GlxI sequences do indeed code for MG–GSH-isomerizing GlxI enzymes. Of seminal importance, our results show that these prokaryotic GlxI enzymes are maximally active in the presence of Ni2+ and do not exhibit activity in the presence of Zn2+. This assembly of metal activation data establishes the existence of a previously undetected class of GlxI enzymes, specifically a class of non-Zn2+-dependent metalloisomerases compared with a Zn2+-dependent class. A biochemical study of GlxI in this respect exemplifies the limits of assigning enzymic characteristics from bioinformatic methods. Although amino acid sequence comparison was sufficient to assign putative GlxI functions to these proteins, metal activation could not be inferred except by investigation of the isolated enzyme. The characterization of these bacterial enzymes therefore represents a critical contribution to the identification of the scope of differential metal activation in a metalloenzyme class formerly considered to be exclusively Zn2+-dependent.
EXPERIMENTAL
Materials
Nickel (II) chloride hexahydrate (99.9999% pure) was from Aldrich (Oakville, Ontario, Canada), cobalt chloride hexahydrate (assay, 100.4%; specification, 98–102%), cadmium chloride (assay, 99.4%), manganous chloride tetrahydrate (assay, 98.8%) and zinc chloride (assay, 99.3%) were obtained from J. T. Baker (Toronto, Ontario, Canada), and magnesium chloride hexahydrate (assay, 99.2%) and calcium chloride dihydrate (assay, 75.5%) was obtained from BDH (Toronto, Ontario, Canada).
Chelex 100 resin was obtained from Bio-Rad (Mississauga, Ontario, Canada). Q-Sepharose Fast Flow anion exchange resin was obtained from Amersham Biosciences (Uppsala, Sweden). For the isoelectric focusing stage of protein purification, a Bio-Rad Rotofor Isoelectric Focusing Unit was employed. This apparatus was used with Bio-Rad Rotolytes (pH 3.9–5.6), with a Mes/Gly-Gly ratio of 1:1. In purifying N. meningitidis GlxI, 200 mM Mopso [3-(N-morpholino)-2-hydroxypropanesulphonic acid; Sigma-Aldrich, St. Louis, MO, U.S.A.] and 200 mM β-alanine solutions (Sigma-Aldrich) were mixed at a 7:3 ratio for use as the isoelectric focusing solution as suggested by Bio-Rad [30]. Water used in all experiments was purified using a Milli-Q RG Ultrapure water system (18 MΩ-cm; Waters Associates, Milford, MA, U.S.A.).
Methods
Protein samples were concentrated using an Amicon Ultrafiltration cell with a YM10 or PM10 membrane, or by using a Centricon 10 (Amicon, Beverley, MA, U.S.A.). Protein quantification was performed using the method of Bradford, with BSA as the standard as described previously [31].
Bacterial strains and sources of template DNA
E. coli strains BL21(λDE3) and DH5α were used as GlxI overexpression and plasmid propagation hosts respectively. E. coli strain MG1655 was used as an overexpression host for the variant E. coli GlxI enzymes. Genomic template DNA used for cloning of the Y. pestis GlxI gene was from the strain CO92. A sample of this DNA was obtained from Dr K. Isherwood at the Biomedical Sciences Department of Chemical and Biological Defence (CBD) establishment at Porton Down (Wiltshire, U.K.). Genomic DNA was isolated from wild-type P. aeruginosa strain PA01. This strain was obtained as a gift from Dr J. Lam of the University of Guelph (Ontario, Canada). Genomic DNA from a N. meningitidis clinical isolate was obtained from Dr D.A.A. Ala'Aldeen, from the Division of Microbiology at the University of Nottingham (Nottingham, U.K.) [32]. Although identification of a GlxI from N. meningitidis has been reported, detailed biochemical characterization of the enzyme was not carried out [32].
Oligonucleotides and DNA sequencing
DNA primers for isolating GlxI-encoding genes of Y. pestis, P. aeruginosa and N. meningitidis were obtained from MOBIX (McMaster University, Hamilton, Ontario, Canada) or Sigma Genosys (Oakville, Ontario, Canada). Sequencing of the plasmid constructs for overexpression of Y. pestis, P. aeruginosa and N. meningitidis GlxI-encoding genes was performed by MOBIX.
DNA manipulation
All manipulation of DNA and plasmid isolation was performed according to the protocols of Sambrook et al. [33].
Cloning of Y. pestis, P. aeruginosa and N. meningitidis GlxI-encoding genes
The putative GlxI DNA sequences used to design oligonucleotide primers were those determined from previous database searches [29]. The Y. pestis GlxI gene was amplified from the aforementioned template DNA by PCR, using the following primer pair: (+) 5′-CCAGAATTCCATATGCGCTTACTCCATACCATG-3′, (−) 5′-CCAAAGCTTGGATCCTCAGTTTCCGAGGCAGTCACC-3′.
The P. aeruginosa GlxI gene was amplified from the aforementioned template DNA by PCR, using the following primer pair: (+) 5′-CCAGAATTCCATATGCGCATTCTCCATACCATG-3′, (−) 5′-CCAAAGCTTGGATCCTCAGGAAGACTTCTGGATCAG-3′.
The N. meningitidis GlxI gene was amplified from the aforementioned template DNA by PCR, using the following primer pair: (+) 5′-CCACTGCAGCATATGCGCTTACTCCATACTATGC-3′, (−) 5′-CCAGGATCCTCAGGCAGTTTGATAGGCAACC-3′.
All three amplification products and the pET22b expression vector (ampR, T7 lac promoter, f1origin of replication; Novagen, Madison, WI, U.S.A.) were subjected to restriction endonuclease digestion with NdeI and BamHI. The digestion products were subsequently ligated to generate the overexpression constructs designated pYPG1, pPAG1 and pNMG1 respectively. These plasmids denote constructs carrying the cloned Y. pestis, P. aeruginosa and N. meningitidis GlxI-encoding genes respectively. All three plasmid constructs were transformed into E. coli BL21(λDE3) cells, yielding the final overexpression systems for these GlxI enzymes.
Growth and induction of GlxI-overexpressing cells
The protocol for induction of Y. pestis [using cell line BL21-(λDE3)/pYPG1] and P. aeruginosa [BL21(λDE3)/pPAG1] GlxI was similar to that used for induction of the E. coli GlxI-overexpression system [19], except that no NiCl2 was added to the growth medium. Also, cells were induced with IPTG (isopropyl β-D-thiogalactoside) for only 4 h.
Growth and induction of BL21(λDE3)/pNMG1 for overexpression of N. meningitidis GlxI was also similar to the protocol outlined in [19], including the aforementioned modifications for BL21(λDE3)/pYPG1 and BL21(λDE3)/pPAG1. TB (terrific broth) was employed as the growth medium for BL21(λDE3)/pNMG1 as opposed to LB (Luria–Bertani) medium, and again the medium was not supplemented with NiCl2. Cells were harvested and stored as indicated in [19].
Purification of Y. pestis, P. aeruginosa and N. meningitidis GlxI
Purification of GlxI from BL21(λDE3)/pYPG1, BL21(λDE3)/pPAG1 and BL21(λDE3)/pNMG1 cells was performed using the protocol optimized for E. coli GlxI [19], with the following modifications indicated in this section. For Y. pestis GlxI purification, the Q-Sepharose Fast Flow (Amersham Biosciences) elution buffers [no-salt: 20 mM Tris/HCl, 30% (v/v) glycerol, pH 7.0; high-salt: no-salt+1 M KCl] and the storage buffer (50 mM Chelex-treated Mops, pH 7.0) contained DTT (dithiothreitol) at a final concentration of 1 mM. This reducing agent was employed since initial attempts at purifying Y. pestis yielded GlxI as a tetramer (confirmed by electrospray ionization MS and native PAGE), probably attributable to an intermolecular disulphide bond between dimers as a consequence of a second cysteine residue in the amino acid sequence at position 132 (Figure 1). This residue is likely to be an exposed cysteine, based on comparison with the E. coli GlxI X-ray crystal structure determined previously [34]. DTT concentrations were decreased by dilution of the GlxI sample with storage buffer (50 mM Chelex-treated Mops, pH 7) not containing the thiol reducing agent, such that DTT concentrations during enzymic assays did not inhibit the isomerization reaction (i.e. GSH concentration was at least 120-fold in molar excess of DTT during assay). Initial chromatographic purification of N. meningitidis GlxI was performed on a Q-Sepharose Fast Flow column with the no-salt and high-salt elution buffers. The GlxI-containing fractions were pooled and dialysed against 20 mM Tris, 10% (v/v) glycerol, pH 7.5, and then applied to a Mono-Q (Amersham Biosciences) column. The elution buffers used with this column were identical with those used in the initial chromatographic step, except that the glycerol content was 10% (v/v). The GlxI-containing fractions were pooled and dialysed into 10% (v/v) glycerol before being subjected to the isoelectric focusing step, with the modifications as indicated above. Purified apo-GlxI was concentrated and stored in 50 mM Chelex-treated Mops, pH 7.0, in acid-treated plasticware at 4 °C as described in [19].
Figure 1. Amino acid sequence alignment for H. sapiens, E. coli, Y. pestis, P. aeruginosa, N. meningitidis and P. putida GlxI enzymes.
Conserved metal ligands are indicated in bold lettering with the position marked by an asterisk. The additional regions of the H. sapiens and P. putida GlxI enzymes are indicated in bold with underlining. The configuration of sequences in this alignment was obtained from a previous comparative study [29].
Molecular-mass determination
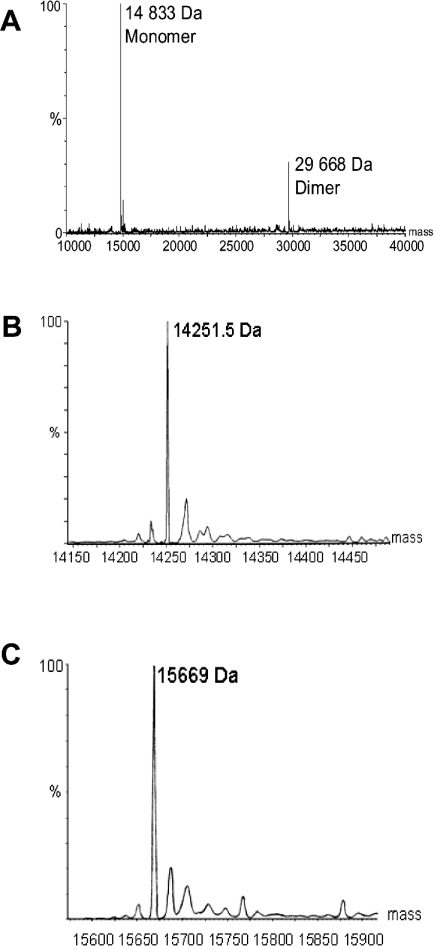
Electrospray ionization MS was used to confirm the molecular mass of the overproduced bacterial GlxI enzymes in crude lysate and in purified samples. Analysis was performed on protein samples using a Micromass Q-TOF Ultima Global (Manchester, U.K.) mass spectrometer at the Waterloo Chemical Analysis Facility, University of Waterloo. Samples were introduced using eluants consisting of 1:1 water/acetonitrile plus 0.2% methanoic (formic) acid. Electrospray ionization was performed in positive ion mode. The peaks of higher molecular mass following the major GlxI peaks in the obtained spectra were attributed to sodium adducts of the purified enzymes (see Figure 2).
Figure 2. Electrospray ionization mass spectra of (A) Y. pestis (reconstructed spectrum) (B) P. aeruginosa GlxI and (C) N. meningitidis GlxI enzymes.
The expected molecular masses of the monomeric enzymes are 14834 Da, 14251 Da and 15669 Da respectively.
SDS/PAGE was employed to screen for the presence of GlxI enzyme during purification. Separation (using precast homogeneous 20% gels) and visualization (Coomassie Brilliant Blue staining) of protein samples by SDS/PAGE was performed using a PhastSystem™ (Amersham Biosciences).
Gel-filtration chromatography was employed to assess whether the Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes were dimeric in native form, as is the case with the E. coli enzyme. The Y. pestis and P. aeruginosa GlxI samples were purified for this analysis using Q-Sepharose Fast Flow and Mono-Q columns. The N. meningitidis GlxI analysed was prepared as an apoenzyme, as described above. A Superdex 75 10/30 column was employed for molecular-mass determination, using 20 mM Tris/HCl, pH 7, and 100 mM KCl as the eluent. The column was calibrated using commercially obtained protein standards.
GlxI enzymic assay
The assays and assay conditions used for quantification of GlxI activity were identical with those described in [19]. Initial rate data was fitted by non-linear regression analysis using the software GraFit version 3.01 (Erithacus Software).
Metal titration of apo-GlxI
Metal-titration experiments were conducted as an initial assessment of metal activation for the GlxI enzymes under study. The standard GlxI activity assay was used to survey activation when enzyme was pre-incubated with differing concentrations of metal chloride. A sample containing a particular GlxI enzyme was diluted in 50 mM Mops and metal chloride was added to an approximate range of 0–7 molar equivalents of metal to dimeric enzyme (see Figure 4). Assays were performed using a MG–GSH concentration of 0.5 mM, with 0.7–2.5 μg of protein per assay (see Figure 4). The purified Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes were confirmed to be active as homodimers by gel-filtration chromatography (results not shown), as observed with E. coli GlxI (gel-filtration chromatography, X-ray diffraction methods) [19,34]. Activity assays were performed in triplicate at a substrate concentration of 0.5 mM, with samples assayed in order of increasing metal concentration. For the apoenzyme control assays, cuvettes were prepared by soaking in concentrated nitric acid for 10 min, followed by soaking in 1 mM EDTA for 10 min, followed by liberal rinsing with Chelex-treated Milli-Q water before assaying the first replicate. This pre-treatment of the cuvettes was administered to minimize recorded enzyme activity for the apo-control due to the tendency for metal abstraction from the cuvette itself [19,35]. Before the second and third replicates of the apo-control sample, cuvettes were soaked in 1 mM EDTA, followed by liberal rinsing with Chelex-treated Milli-Q water. No further cuvette treatment was employed for the remaining samples assayed during the titration.
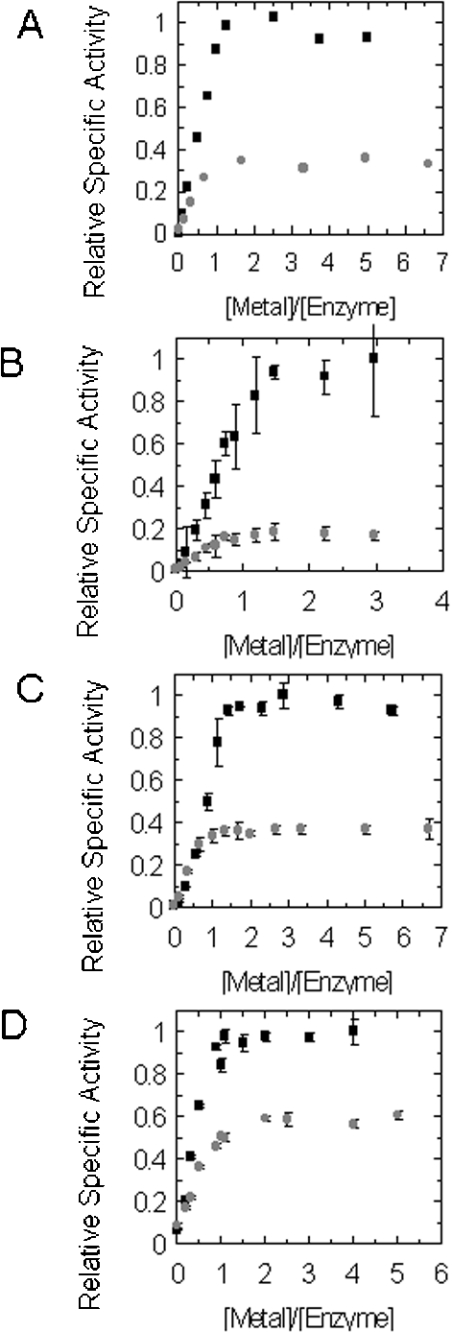
Figure 4. Titration of GlxI apoenzymes with various NiCl2 (black squares) and CoCl2 (grey circles) ratios: (A) E. coli [19], (B) Y. pestis, (C) P. aeruginosa and (D) N. meningitidis GlxI enzymes.
Data points and error bars in (B), (C) and (D) represent the specific activity±S.D. for triplicate readings. Relative specific activity values were calculated as the proportion of activity relative to the highest specific activity values obtained during the NiCl2 titration.
Metal-activation studies
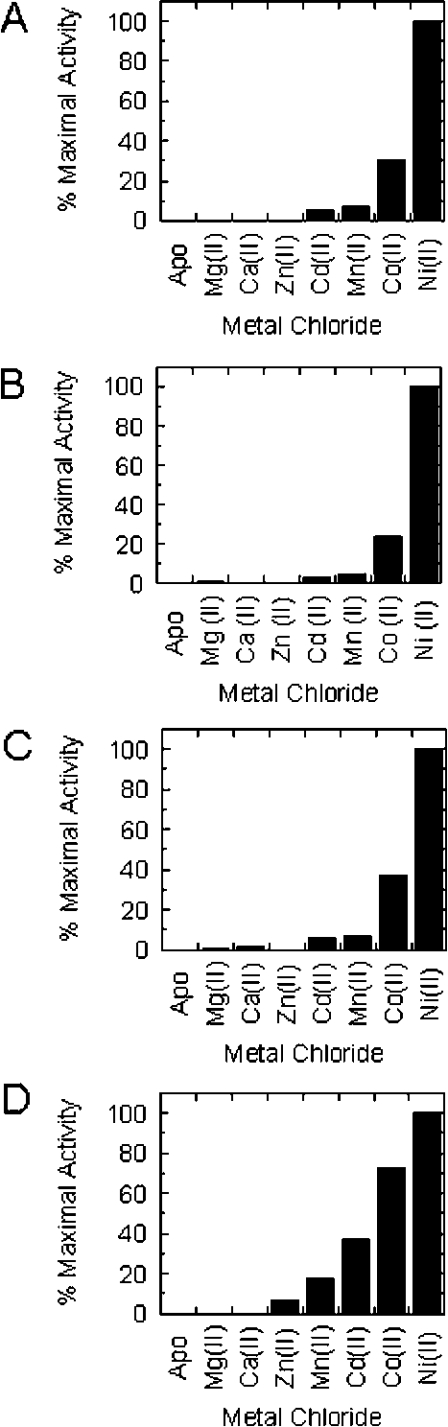
Profiles for metal activation for each of the purified bacterial GlxI enzymes with various bivalent metal chlorides were obtained using the methods described previously [19,35]. Samples of a particular apo-GlxI were diluted in 50 mM Chelex-treated Mops, pH 7, and pre-incubated with either 10 molar equivalents of metal chloride or activated with a 0.5 mM final concentration of metal chloride in the enzyme dilution. Enzyme activity was assayed at a 0.5 mM substrate concentration in triplicate. Amounts of GlxI used per assay ranged from 0.2 to 0.9 μg. Activation was assessed using the following metal chlorides: Mg2+, Ca2+, Zn2+, Cd2+, Mn2+, Co2+ and Ni2+. It must be noted that assays for Mn2+ activation of Y. pestis GlxI were performed in Mes buffer. Also, the maximal activity values for Ni2+-, Co2+- and Zn2+-reconstituted Y. pestis GlxI were obtained from metal-titration data (see Figure 3).
Figure 3. Profiles of apoenzyme reactivation with various bivalent ions for (A) E. coli [19] (B) Y. pestis (C) P. aeruginosa and (D) N. meningitidis GlxI enzymes.
To obtain the relative maximal activity values for these graphs, baseline apo-control assays were subtracted from all activities so that relative maximal activity was calculated using the specific activity−apo-specific activity values.
Determination of kinetic parameters
The Michaelis–Menten constants (Km) and maximal enzyme velocities (Vmax) for Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes were determined experimentally by measurement of initial reaction rate [19]. Rate data was collected at substrate concentration ranges between 0.025 and 1 mM. Stock enzyme was diluted with 50 mM Chelex-treated Mops pH 7.0 and pre-incubated with NiCl2 or CoCl2 at a final concentration of 0.5 mM. Enzyme activity at each concentration was measured in triplicate. The entire set of triplicates was measured twice, and the values from the duplicate experiments were averaged to obtain the final parameter values.
RESULTS
Overexpression and purification of Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes
Robust expression of GlxI enzymes was observed with induction of all three overexpressing clones [BL21(λDE3)/pYPG1, BL21-(λDE3)/pPAG1 and BL21(λDE3)/pNMG1]. Sequencing of all three plasmids confirmed that these GlxI-encoding genes were isolated in entirety with correct amplification of the template gene (results not shown). Metal activation for all three GlxI enzymes was determined on a preliminary basis using the crude lysate from small-scale induced cultures (25 or 50 ml). The lysates were assayed for GlxI activity in the presence of NiCl2 and ZnCl2 at final concentrations of 0.5 mM metal chloride. The results of this screening indicated that Y. pestis, P. aeruginosa and N. meningitidis GlxI sequences do indeed encode for GlxI enzymes capable of isomerizing a MG–GSH hemithioacetal substrate. Furthermore, the MG–GSH hemithioacetal isomerizing activity associated with all three lysates was enhanced in the presence of NiCl2. This result also indicates that some of the Y. pestis, P. aeruginosa and N. meningitidis GlxI enzyme was produced in the apo form. Enhancement of enzyme activity was not observed during the assay of lysate in the presence of ZnCl2. Observed relative increases in GlxI activity were based on comparison with lysate activity with no metal chlorides added to the substrate.
The protocol employed for purification of Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes was similar to that optimized for isolation of E. coli GlxI. The GlxI purification protocol as applied to Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes, yielded substantial quantities of each protein as purified isolates characterized by electrospray ionization MS.
The molecular mass of the Y. pestis, P. aeruginosa and N. meningitidis GlxI monomers were confirmed by electrospray ionization MS to be 14833 Da, 14251 Da and 15669 Da respectively, in agreement with their predicted values (14834 Da, 14251 Da and 15669 Da respectively; Figure 2). Although each of these enzymes was overproduced in an E. coli host strain, the electrospray ionization mass spectra did not indicate the presence of any E. coli GlxI (predicted mass, 14919 Da) in any of the purified enzyme preparations. Furthermore, endogenous levels of E. coli GlxI are extremely low [19,35].
Relative metal activation of Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes
The results of these experiments are summarized in Figure 3. The propensity toward Ni2+-activation of all three GlxI enzymes is readily apparent from these profiles. The series of activating metals (Ni2+>Co2+>Mn2+>Cd2+) for each of these newly characterized bacterial GlxI enzymes is similar to that documented for the E. coli lyase. Comparison of these four bacterial GlxI enzymes together reveals some slight differences in relative activation conferred by the aforementioned metal ions.
The activation profiles for Y. pestis and P. aeruginosa GlxI enzymes show a similar trend towards activation with Ni2+ over Co2+, Mn2+ and Cd2+, as observed with E. coli GlxI (see Figure 3). Notable features of N. meningitidis GlxI are the higher levels of activation, specifically with Co2+, but also with Mn2+ and Cd2+ relative to the maximal activity obtained by reconstitution with Ni2+ (Figure 3). Although it appears in this data set that the activity level in the presence of Zn2+ is elevated, no increase in activity relative to the apo-control was observed during titration of N. meningitidis GlxI with this metal (results not shown).
The second defining attribute of these characterized bacterial GlxI enzymes, which is of critical importance, is the non-activation of these apoenzymes in the presence of Zn2+. This differing metal-dependence observed with E. coli GlxI in contrast with previously characterized GlxI enzymes (H. sapiens, P. putida and S. cerevisiae) is also exhibited by the Y. pestis, P. aeruginosa and N. meningitidis enzymes investigated. Titrations with ZnCl2 were also performed on Y. pestis, P. aeruginosa and N. meningitidis apo-GlxI, confirming that this bivalent ion is ineffectual as a co-factor, as no increase in activity was observed with increasing amounts of Zn2+ (results not shown).
Ni2+ and Co2+ titration of Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes
These titrations in comparison with similar reconstitutions of E. coli apo-GlxI reveal the overall similarity in selectivity of Ni2+ over Co2+ for activation (in roughly similar proportions) (Figure 4). The higher relative Co2+ reconstitution of N. meningitidis GlxI observed in the previous activation profile is supported by the ratio of Ni2+ to Co2+ activation observed in the titration experiments. Furthermore, titrations for Y. pestis, P. aeruginosa and N. meningitidis GlxI in comparison with the E. coli enzyme reveal that all four enzymes are strong recruiters of metal ions, based on the low stoichiometric equivalents of Ni2+ or Co2+ relative to amount of enzyme, that confer maximal GlxI activation (Figure 4).
Kinetic parameters of Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes
Initial comparison of the kinetic data obtained for the Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes reveals overall similarities in metal selectivity to E. coli GlxI (Table 1). Greater activation with Ni2+ over Co2+ is characteristic of all four enzymes. In contrast, the selectivity between Ni2+ and Co2+ activation is narrowed for the N. meningitidis enzyme. The deviations in measured kinetic parameters (Km and Vmax) of Y. pestis, P. aeruginosa and N. meningitidis GlxI are slight, not exceeding an order of magnitude in comparison with the E. coli lyase. The catalytic efficiency values (kcat/Km) obtained for the Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes indicate another similar attribute to that of E. coli GlxI, namely catalytic function at a close to diffusion-controlled rate (Table 1).
Table 1. Kinetic parameters for E. coli, Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes with MG–GSH substrate.
E. coli GlxI kinetic data with Ni2+ was obtained from [19] and with Co2+ was obtained from [35]. Results are means±S.D.
| GlxI source | Metal chloride | Km (μM) | Vmax (μmol/min per mg of protein) | kcat (s−1) | kcat/Km (M−1·s−1) |
|---|---|---|---|---|---|
| E. coli | Ni2+ | 27.0±0.4 | 676±17 | 338 | 1.2×107 |
| Co2+ | 12±2 | 213±33 | 106 | 8.8×106 | |
| Y. pestis | Ni2+ | 56.0±0.6 | 618±48 | 306 | 5.5×106 |
| Co2+ | 29±5 | 140±6 | 69 | 2.4×106 | |
| P. aeruginosa | Ni2+ | 32±2 | 571±28 | 271 | 8.5×106 |
| Co2+ | 16±3 | 180±7 | 86 | 5.4×106 | |
| N. meningitidis | Ni2+ | 45±5 | 390±5 | 204 | 4.5×106 |
| Co2+ | 28.0±0.5 | 279±24 | 146 | 5.2×106 |
DISCUSSION
We have previously investigated the E. coli GlxI, including enzymological aspects and three-dimensional structure, and our results revealed a novel metal-activation profile in contrast with previously studied GlxI enzymes [19,35–38]. However, further biochemical study of other prokaryotic GlxI enzymes was imperative to categorize the metal activation of E. coli GlxI, either as an exception to the previously established Zn2+-selectivity of this protein or as a member of a novel, but previously unrecognized, non-Zn2+-activated class. The characterization of three bacterial GlxI enzymes in addition to the E. coli enzyme has now established that non-Zn2+-activation of this lyase occurs within the prokaryotic domain of life. The results of this biochemical investigation indicate the existence of two classes of GlxI.
A structural basis for differential metal selectivity of GlxI enzymes might be implied from amino acid sequence analysis. Comparing the four metal ligands of E. coli (His5, Glu56, His74 and Glu122) and H. sapiens GlxI (Gln34, Glu100, His127 and Glu173), all positions are conserved with the exception of the His5/Gln34 variation. The presence of a histidine residue at a position analogous to Gln34 of the human enzyme can be observed in a number of bacterial GlxI sequences including those examined in the present study. This ligand variation does not represent an unambiguous correlation with metal activation however, since P. putida GlxI has a histidine residue at the ‘Gln34’ position and is Zn2+-dependent (Figure 1). Furthermore, enzymological studies of the E. coli H5Q (His5→Gln) GlxI variant has shown that the ‘humanized’ active site promotes a minor shift towards Zn2+ activation, yet confers a severely compromised affinity for all activating metal ions [35]. So in spite of the influence of this ligand on GlxI metal selectivity, it is clearly not a dominant contributor to promoting shifts in metal activation while maintaining the catalytic capacity of the enzyme.
The initial study of E. coli GlxI revealed marked sequence conservation to H. sapiens GlxI (36% identity) [19]. However, the H. sapiens and P. putida GlxI amino acid sequences are significantly longer than those for E. coli, Y. pestis, P. aeruginosa and N. meningitidis GlxI enzymes (Figure 1). Alignment of these bacterial GlxI sequences makes apparent the additional regions of the H. sapiens and P. putida lyases that are systematically absent in the non-Zn2+-dependent counterparts investigated here. The P. putida GlxI primary sequence more closely resembles the H. sapiens enzyme in this respect, in contrast with other bacterial GlxI sequences. The striking architectural similarity between the E. coli and H. sapiens GlxI active sites, however, does not provide any evidence by visual inspection of any obvious secondary or tertiary dissimilarities that would influence metal selectivity on a differential basis [39,34].
These findings emphasize that caution must be exercised in attempts at inferring enzymic attributes, in this case metal selectivity, from primary structure data. The consequences of assuming that zinc activation is a widespread feature of GlxI enzymes can be potentially deleterious in attempts for identifying this enzyme in other biological sources. Specifically, attempts at identifying GlxI activity in cell/tissue lysates using metal reconstitution would be best performed using a variety of bivalent metals during screening, which would take into account the potential for novel metal activation in newly identified forms of this metalloisomerase. This caveat would also extend to more systematic assessments of metal utilization of other metalloenzymes whose properties may or may not resemble characterized counterparts from which its putative function was deduced.
In conclusion, our characterization of several bacterial GlxI enzymes has now firmly established the existence of two classes of this lyase that differ in metal activation. This difference in metal selectivity cannot yet be associated with any specific structural feature of GlxI enzymes, but this class division is a real occurrence amongst these lyases, only recently discovered after 91 years of investigation of the glyoxalase system. Further investigation of GlxI enzymes is essential from a structure–function perspective, to elucidate the fundamental structural parameters that have given rise to these two metal activation classes. Intense current interest is focused on the additional inserts found in the Zn2+-activated GlxI enzymes as potential mediators of metal specificity.
Acknowledgments
We thank Dr K. Isherwood for providing us with Y. pestis CO92 DNA. We also extend thanks to Dr J. Lam for the gift of P. aeruginosa strain PA01 and to Dr D. A. A. Ala'Aldeen for N. meningitidis genomic DNA. Thanks are extended to Ms A. So and Dr R. W. Smith for electrospray ionization MS analysis of protein samples. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (J. F. H.). N. S. was supported by a postgraduate scholarship from NSERC.
References
- 1.Neuberg C. The destruction of lactic aldehyde and methylglyoxal by animal organisms. Biochem. Z. 1931;49:502–506. [Google Scholar]
- 2.Dakin H. D., Dudley H. W. An enzyme concerned with the formation of hydroxy acids from ketonic aldehydes. J. Biol. Chem. 1913;14:155–157. [Google Scholar]
- 3.Thornalley P. J. Glyoxalase I – structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 2003;31:1343–1348. doi: 10.1042/bst0311343. [DOI] [PubMed] [Google Scholar]
- 4.Becker K., Rahlfs S., Nickel C., Schirmer R. H. Glutathione – functions and metabolism in the malarial parasite Plasmodium falciparum. Biol. Chem. 2003;384:551–566. doi: 10.1515/BC.2003.063. [DOI] [PubMed] [Google Scholar]
- 5.Rose I. A., Nowick J. S. Methylglyoxal synthetase, enol-pyruvaldehyde, glutathione and the glyoxalase system. J. Am. Chem. Soc. 2002;124:13047–13052. doi: 10.1021/ja027065h. [DOI] [PubMed] [Google Scholar]
- 6.Creighton D. J., Hamilton D. S. Brief history of glyoxalase I and what we have learned about metal ion-dependent, enzyme-catalyzed isomerizations. Arch. Biochem. Biophys. 2001;387:1–10. doi: 10.1006/abbi.2000.2253. [DOI] [PubMed] [Google Scholar]
- 7.Kalapos M. P. On the promine/retine theory of cell division: now and then. Biochim. Biophys. Acta. 1999;1426:1–16. doi: 10.1016/s0304-4165(98)00141-x. [DOI] [PubMed] [Google Scholar]
- 8.Thornalley P. J. Glutathione-dependent detoxification of α-oxoaldehydes by the glyoxalase system: involvement in disease mechanisms and antiproliferative activity of glyoxalase I inhibitors. Chem. Biol. Interact. 1998;111–112:137–151. doi: 10.1016/s0009-2797(97)00157-9. [DOI] [PubMed] [Google Scholar]
- 9.Inoue Y., Kimura A. Methylglyoxal and regulation of its metabolism in microorganisms. Adv. Microb. Physiol. 1995;37:177–227. doi: 10.1016/s0065-2911(08)60146-0. [DOI] [PubMed] [Google Scholar]
- 10.Racker E. The mechanism of action of glyoxalase. J. Biol. Chem. 1951;190:685–696. [PubMed] [Google Scholar]
- 11.Mannervik B., Ridderstrom M. Catalytic and molecular properties of glyoxalase I. Biochem. Soc. Trans. 1993;21:515–517. doi: 10.1042/bst0210515. [DOI] [PubMed] [Google Scholar]
- 12.Bergdoll M., Eltis L. D., Cameron A. D., Dumas P., Bolin J. T. All in the family: structural and evolutionary relationships among three modular proteins with diverse functions and variable assembly. Protein Sci. 1998;7:1661–1670. doi: 10.1002/pro.5560070801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armstrong R. N. Mechanistic diversity in a metalloenzyme superfamily. Biochemistry. 2000;39:13625–13632. doi: 10.1021/bi001814v. [DOI] [PubMed] [Google Scholar]
- 14.Pakhomova S., Rife C. L., Armstrong R. N., Newcomer M. E. Structure of fosfomycin resistance protein FosA from transposon Tn2921. Protein Sci. 2004;13:1260–1265. doi: 10.1110/ps.03585004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rife C. L., Pharris R. E., Newcomer M. E., Armstrong R. N. Crystal structure of a genomically encoded fosfomycin resistance protein (FosA) at 1.19 Å resolution by MAD phasing off the L-III edge of Tl+ J. Am. Chem. Soc. 2002;124:11001–11003. doi: 10.1021/ja026879v. [DOI] [PubMed] [Google Scholar]
- 16.Martin T. W., Dauter Z., Devedjiev Y., Sheffield P., Jelen F., He M., Sherman D. H., Otlewski J., Derewenda Z. S., Derewenda U. Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein. Structure. 2002;10:933–942. doi: 10.1016/s0969-2126(02)00778-5. [DOI] [PubMed] [Google Scholar]
- 17.Ridderstrom M., Mannervik B. Optimized heterologous expression of the human zinc enzyme glyoxalase I. Biochem. J. 1996;314:463–467. doi: 10.1042/bj3140463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aronsson A. C., Marmstal E., Mannervik B. Glyoxalase I, a zinc metalloenzyme of mammals and yeast. Biochem. Biophys. Res. Commun. 1978;81:1235–1240. doi: 10.1016/0006-291x(78)91268-8. [DOI] [PubMed] [Google Scholar]
- 19.Clugston S. L., Barnard J. F., Kinach R., Miedema D., Ruman R., Daub E., Honek J. F. Overproduction and characterization of a dimeric non-zinc glyoxalase I from Escherichia coli: evidence for optimal activation by nickel ions. Biochemistry. 1998;37:8754–8763. doi: 10.1021/bi972791w. [DOI] [PubMed] [Google Scholar]
- 20.Chen F., Wollmer M. A., Hoerndli F., Munch G., Kuhla B., Rogaev E. I., Tsolaki M., Papassotiropoulos A., Gotz J. Role for glyoxalase I in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7687–7692. doi: 10.1073/pnas.0402338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vander Jagt D. L., Hunsaker L. A. Methylglyoxal metabolism and diabetic complications: roles of aldose reductase, glyoxalase-I, betaine aldehyde dehydrogenase and 2-oxoaldehyde dehydrogenase. Chem. Biol. Interact. 2003;143–144:341–351. doi: 10.1016/s0009-2797(02)00212-0. [DOI] [PubMed] [Google Scholar]
- 22.Beard K. M., Shangari N., Wu B., O'Brien P. J. Metabolism, not autoxidation, plays a role in α-oxoaldehyde- and reducing sugar-induced erythrocyte GSH depletion: relevance for diabetes mellitus. Mol. Cell. Biochem. 2003;252:331–338. doi: 10.1023/a:1025544309616. [DOI] [PubMed] [Google Scholar]
- 23.Iozef R., Rahlfs S., Chang T., Schirmer H., Becker K. Glyoxalase I of the malarial parasite Plasmodium falciparum: evidence for subunit fusion. FEBS Lett. 2003;554:284–288. doi: 10.1016/s0014-5793(03)01146-3. [DOI] [PubMed] [Google Scholar]
- 24.Creighton D. J., Zheng Z. B., Holewinski R., Hamilton D. S., Eiseman J. L. Glyoxalase I inhibitors in cancer chemotherapy. Biochem. Soc. Trans. 2003;31:1378–1382. doi: 10.1042/bst0311378. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson G. P., Totemeyer S., MacLean M. J., Booth I. R. Methylglyoxal production in bacteria: suicide or survival? Arch. Microbiol. 1998;170:209–218. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 26.Thornalley P. J., Strath M., Wilson R. J. Antimalarial activity in vitro of the glyoxalase I inhibitor diester, S-p-bromobenzylglutathione diethyl ester. Biochem. Pharmacol. 1994;47:418–420. doi: 10.1016/0006-2952(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 27.Barnard J. F., Vander Jagt D. L., Honek J. F. Small molecule probes of glyoxalase I and glyoxalase II. Biochim. Biophys. Acta. 1994;1208:127–135. doi: 10.1016/0167-4838(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 28.Saint-Jean A. P., Phillips K. R., Creighton D. J., Stone M. J. Active monomeric and dimeric forms of Pseudomonas putida glyoxalase I: evidence for 3D domain swapping. Biochemistry. 1998;37:10345–10353. doi: 10.1021/bi980868q. [DOI] [PubMed] [Google Scholar]
- 29.Clugston S. L., Honek J. F. Identification of sequences encoding the detoxification metalloisomerase glyoxalase I in microbial genomes from several pathogenic organisms. J. Mol. Evol. 2000;50:491–495. doi: 10.1007/s002390010052. [DOI] [PubMed] [Google Scholar]
- 30.Bier M., Ostrem J., Marquez R. B. A new buffering system and its use in electrophoresis and isoelectric focusing. Electrophoresis. 1993;14:1011–1018. doi: 10.1002/elps.11501401161. [DOI] [PubMed] [Google Scholar]
- 31.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 32.Kizil G., Wilks K., Wells D., Ala'Aldeen D. A. A. Detection and characterisation of the genes encoding glyoxalase I and II from Neisseria meningitidis. J. Med. Microbiol. 2000;49:669–673. doi: 10.1099/0022-1317-49-7-669. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J., Fritsch E. F., Maniatis T. Plainsview: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 34.He M. M., Clugston S. L., Honek J. F., Matthews B. W. Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation. Biochemistry. 2000;39:8719–8727. doi: 10.1021/bi000856g. [DOI] [PubMed] [Google Scholar]
- 35.Clugston S. L., Yajima R., Honek J. F. Investigation of metal binding and activation of Escherichia coli glyoxalase I: kinetic, thermodynamic and mutagenesis studies. Biochem. J. 2004;377:309–316. doi: 10.1042/BJ20030271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidson G., Clugston S. L., Honek J. F., Maroney M. J. XAS investigation of the nickel active site structure in Escherichia coli glyoxalase I. Inorg. Chem. 2000;39:2962–2963. doi: 10.1021/ic0001208. [DOI] [PubMed] [Google Scholar]
- 37.Stokvis E., Clugston S. L., Honek J. F., Heck A. J. Characterization of glyoxalase I (E. coli)-inhibitor interactions by electrospray time-of-flight mass spectrometry and enzyme kinetic analysis. J. Protein. Chem. 2000;19:389–397. doi: 10.1023/a:1026439531005. [DOI] [PubMed] [Google Scholar]
- 38.Davidson G., Clugston S. L., Honek J. F., Maroney M. J. An XAS investigation of product and inhibitor complexes of Ni-containing GlxI from Escherichia coli: mechanistic implications. Biochemistry. 2001;40:4569–4582. doi: 10.1021/bi0018537. [DOI] [PubMed] [Google Scholar]
- 39.Cameron A. D., Olin B., Ridderstrom M., Mannervik B., Jones T. A. Crystal structure of human glyoxalase I – evidence for gene duplication and 3D domain swapping. EMBO J. 1997;16:3386–3395. doi: 10.1093/emboj/16.12.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]