Abstract
Spermidine/spermine-N1-acetyltransferase (SSAT1) is a short-lived polyamine catabolic enzyme inducible by polyamines and polyamine analogues. Induction of SSAT1 plays an important role in polyamine homoeostasis, since the N1-acetylated polyamines can be excreted or oxidized by acetylpolyamine oxidase. We have purified a recombinant human acetyltransferase (SSAT2) that shares 45% identity and 61% homology with human SSAT1, but is only distally related to other known members of the GNAT (GCN5-related N-acetyltransferase) family. Like SSAT1, SSAT2 is widely expressed, but did not turn over rapidly, and levels were unaffected by treatments with polyamine analogues. Despite similarity in sequence to SSAT1, polyamines were found to be poor substrates of purified SSAT2, having Km values in the low millimolar range and kcat values of <0.01 s−1. The kcat/Km values for spermine and spermidine for SSAT2 were <0.0003% those of SSAT1. Expression of SSAT2 in NIH-3T3 cells was not detrimental to growth, and did not reduce polyamine content or increase acetylpolyamines. These results indicate that SSAT2 is not a polyamine catabolic enzyme, and that polyamines are unlikely to be its natural intracellular substrates. A promising candidate for the physiological substrate of SSAT2 is thialysine [S-(2-aminoethyl)-L-cysteine], which is acetylated predominantly at the ε-amino group with Km and kcat values of 290 μM and 5.2 s−1. Thialysine is a naturally occurring modified amino acid that can undergo metabolism to form cyclic ketimine derivatives found in the brain and as urinary metabolites, which can undergo further reaction to form antioxidants. SSAT2 should be renamed ‘thialysine Nε-acetyltransferase’, and may regulate this pathway.
Keywords: acetylation, acetyl-CoA, polyamine, protein turnover, S-(2-aminoethyl)-L-cysteine (thialysine)
Abbreviations: AECK, aminoethyl-L-cysteine ketimine; AECK-DD, decarboxylated dimer of AECK; BE-3-3-3, N1,N11-bis(ethyl)norspermine; BE-3-4-3, N1,N12-bis(ethyl)spermine; BLAST, Basic Local Alignment Search Tool; DTNB, 5,5′-dithiobis-(2-nitrobenzoate); ESI, electrospray ionization; EST, expressed sequence tag; GNAT, GCN5-related N-acetyltransferase; IPTG, isopropyl β-D-thiogalactoside; MALDI–TOF-MS, matrix-assisted laser-desorption ionization–time-of-flight mass spectrometry; OPA, o-phthaldialdehyde; RT-PCR, reverse-transcriptase-PCR; SSAT, spermidine/spermine-N1-acetyltransferase; TMA, 1,4-thiomorpholine-3.5-dicarboxylic acid
INTRODUCTION
Polyamines are essential regulators of mammalian cell growth and differentiation [1–3]. Intracellular polyamine concentrations are adjusted to cellular requirements by the concerted action of biosynthetic and catabolic enzymes in addition to transport mechanisms. Spermidine/spermine N1-acetyltransferase (SSAT1) is a catabolic enzyme that plays a critical role in polyamine homoeostasis [4,5]. It contributes to the down-regulation of intracellular polyamine pools by catalysing the specific transfer of an acetyl group from acetyl-CoA to an aminopropyl group of spermine or spermidine. The N1-acetyl derivatives formed may be excreted from the cell or are subject to further catabolism by acetylpolyamine oxidase, forming spermidine or putrescine respectively. Acetylation of the aminobutyl moiety of spermidine to form N8-acetylspermidine also occurs physiologically, although the specific enzyme responsible for this reaction is not well defined. In general, acetylpolyamines do not persist in normal cells, although elevated levels of acetylpolyamines have been described in cancer cells [3] and may be associated with the progression of chemically induced skin tumours in transgenic mice that overexpress SSAT1 in the epidermis from a keratin-6 promoter [6].
SSAT1 is a short-lived enzyme whose activity is very low under basal cellular conditions. However, the levels of SSAT1 enzyme can be increased very readily in response to a number of physiological and pharmacological stimuli. Polyamine analogues such as BE-3-4-3 [N1,N12-bis(ethyl)spermine] and BE-3-3-3 [N1,N11-bis(ethyl)norspermine], currently undergoing clinical evaluation as antitumour agents [7,8], are potent inducers of cellular SSAT1 activity. A component of the elevated activity produced by these agents involves inhibition of SSAT1 degradation by the 26 S proteasome, and we have previously described amino acid residues in SSAT1 that play an important role in conferring this rapid turnover [9,10].
Studies by site-directed mutagenesis have provided some information identifying regions in the SSAT1 protein that are important for such properties as catalytic activity, substrate binding and stability of the enzyme [9–14]. Little additional information of key residues can be obtained by comparing SSAT1 sequences from different species, since these proteins are virtually identical. We have therefore attempted to identify other more distally related acetyltransferases that would facilitate the process of assigning properties to specific regions of the protein based on primary amino acid sequence similarity. To this end, we identified a related sequence in Schizosaccharomyces pombe using BLAST (basic local alignment search tool) homology searches as well as a human homologue that we originally termed SSAT2 [15] without reference to its functional properties. A recent paper also described SSAT2, and suggested that it may be compartmentalized in the cell, since it did not appear to influence polyamine levels in transiently transfected HEK-293 cells [16].
In the present study, we have studied the properties of human SSAT2 in detail using purified recombinant protein, studies of the turnover of the protein by the ubiquitin/proteasome system in reticulocyte lysates, and studies of cells stably transfected with a construct expressing SSAT2. The S. pombe SSAT2 was also studied using purified recombinant protein. Our findings show that, in contrast with SSAT1, polyamines are poor substrates for SSAT2 and that SSAT2, although widely expressed, is unlikely to influence cellular polyamine content. A probable physiological substrate is thialysine [S-(2-aminoethyl)-L-cysteine], which is very efficiently acetylated on the ε-amino group to form S-(2-acetylaminoethyl)-L-cysteine. SSAT2 does, however, provide a useful structural comparison with SSAT1, since it does not turn over rapidly or respond to polyamine analogues, and binds polyamine substrates very weakly.
MATERIALS AND METHODS
Materials
All reagents were purchased from Fisher Scientific (Pittsburgh, PA, U.S.A.) or Sigma Chemical Co. (St Louis, MO, U.S.A.), unless stated otherwise. Oligodeoxynucleotides were synthesized in the Macromolecular Core Facility, Hershey Medical Center, PA, U.S.A. Restriction enzymes were from New England Biolabs, Inc. (Beverly, MA, U.S.A.) and Roche Molecular Biochemicals (Indianapolis, IN, U.S.A.). L-[35S]Methionine and L-[35S]cysteine/[35S]methionine (both translation grade; 1000 Ci/mmol were obtained from DuPont NEN, Boston, MA, U.S.A.). [1-14C]acetyl-CoA (50 Ci/mol) was purchased from ICN Biochemicals (Costa Mesa, CA, U.S.A.). TNT T7-coupled rabbit reticulocyte lysate translation system and RNasin were from Promega (Madison, WI, U.S.A.). Anti-hSSAT2 antibody was derived from rabbits immunized with purified human SSAT2 protein at Cocalico Biologicals (Reamstown, PA, U.S.A.). Plasmid purification columns were from Qiagen Inc. (Valencia, CA, U.S.A.). BE-3-4-3 and BE-3-3-3 were kindly provided by Dr Raymond J. Bergeron (University of Florida, Gainesville, FL, U.S.A.). Multiple-tissue cDNA (MTC™) Panels and AdvanTaq Plus™ DNA polymerase were purchased from Clontech (Palo Alto, CA, U.S.A.). Transfection reagents: LIPOFECTAMINE™ Plus was from Invitrogen Life Technologies (Carlsbad, CA, U.S.A.). G418 sulphate was from Cellgro (Herndon, VA, U.S.A.). Reagents for Western blotting included PVDF membrane (Millipore Corp, Bedford, MA, U.S.A.) and the Phototop™-Horse Radish Peroxidase chemiluminescent detection system from Cell Signaling Technology Inc. (Beverly, MA, U.S.A.). Protein concentration was determined by the Bradford method using BSA (fraction V) as a standard [17].
Cloning of human SSAT2 and plasmid construction
All of the listed constructs were made by PCR. Forward (5′-GGGGATCCATATGGCTTCCGTGCGGATC-3′) and reverse (5′-AGTACTATCGATTCACTTTCCTGCCAACTTTCT-3′) oligodeoxynucleotides with unique Nde1 and Cla1 restriction sites were designed to flank the human SSAT2 cDNA sequence. PCR products of the anticipated size (534 bp) were amplified from human fetal liver and HeLa cell cDNA libraries (Stratagene, La Jolla, CA, U.S.A.). The product from the HeLa cell library was purified and digested with Nde1 and Cla1 for cloning into the same restriction sites of pT7 plasmids with or without an encoded N-terminal His6 epitope tag. The sequence of the pT7-hSSAT2 clones was verified in the Macromolecular Core Facility at Hershey Medical Center.
A clone for the S. pombe SSAT2 (described under the accession number AAB39945 in the GenBank® database as the ats1 gene product) was kindly provided by Dr Martin Lutzelberger and Dr Norbert F. Kaufer, Technical University, Braunschweig, Germany. The ats1 cDNA in the pUC plasmid was digested with BamH1 and cloned into the same sites of the pBluescript cloning vector (Stratagene, La Jolla, CA, U.S.A.) for in vitro translation reactions, and into the pQE30 bacterial expression plasmid (Qiagen Inc.).
Expression of SSAT1 and SSAT2 mRNA in human tissues
Human Multiple Tissue cDNA (MTC™) Panels I and II are sets of normalized, first-strand cDNA generated by RT-PCR (reverse transcriptase-PCR) using poly(A)+ (polyadenylated) RNA from different human tissues (Clontech). These cDNAs were used as templates for PCR to examine the expression profile of SSAT2 and SSAT1 in 16 different tissues. Forward PCR primers with the sequence 5′-ATTCTTCCAGCGCCCGGGAAGCTACTG-3′ for SSAT2 and 5′-GTGCCGAAAGAGCACTGGACTCCGGAA-3′ for SSAT1 were designed to anneal to the complementary cDNA strand that encodes amino acids 59–67 within each coding region. Reverse primers with sequence 5′-CTCAAGACAGAGATCCTCCTAGGGATG-3′ for SSAT2 and 5′-GGAGGTTGTCATCTACAGCAGCACTCC-3′ for SSAT1 were designed to anneal to a region within the respective 3′-UTR sequences. PCR products of the predicted size (369 bp) were generated for both SSAT2 and SSAT1 using the ‘hot-start’ PCR conditions recommended by Clontech. Control reactions were run in parallel and contained template and primers to amplify a 1 kb fragment of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase. PCR products were resolved on a 2% agarose/ethidium bromide gel.
Expression and purification of recombinant proteins in Escherichia coli
The pT7-hSSAT2 plasmid with an encoded N-terminal His6 epitope was expressed in BL21(DE3) lysogen E. coli (Novagen, Madison, WI, U.S.A). Bacterial cultures were grown in Luria–Bertani medium supplemented with 50 μg/ml ampicillin to a D600 of ≈0.5, and grown for an additional 3 h following induction with 1 mM IPTG (isopropyl β-D-thiogalactoside). Bacterial cell pellets were resuspended in 25 mM Tris/HCl, pH 8.0, lysed by sonication and centrifuged at 60000 gav for 60 min. The supernatant was applied to a TALON™ metal-affinity resin from Clontech, and the His6-tagged SSAT2 protein was eluted using an optimized imidazole gradient. Fractions that appeared as a single band on a denaturing 8% Tricine gel were pooled and dialysed extensively against 25 mM Tris/HCl, pH 8.0, containing 1 mM dithiothreitol and concentrated using an Amicon Centricon-10 microconcentrator (Millipore). Both the human SSAT1 and S. pombe SSAT proteins were expressed from the pQE30 plasmid in XL1-Blue E. coli, and purified as described previously [14].
Assay of acetyltransferase activity
Purified SSAT2 protein was assayed for polyamine acetyltransferase activity at 30 °C using the method of binding to cellulose phosphate discs to separate the 14C-acetylated product from the [1-14C]acetyl-CoA substrate [18]. A standard assay mixture contained 50 mM Tris/HCl, pH 8.0, 3 mM polyamine substrate (unless stated otherwise) and 16 μM [1-14C]acetyl-CoA in a total volume of 100 μl. This assay relies on the ability of the positively charged acetylated product to bind to cellulose phosphate, and cannot be used for potential substrates that form neutral or acidic products.
A spectrophotometric assay that relies on measuring the generation from acetyl-CoA of free CoA that reacts with DTNB [5,5′-dithiobis-(2-nitrobenzoate)] was used to determine the ability of thialysine to act as an acetyl acceptor [19]. Unless stated otherwise, the standard assay mixture contained 100 mM Tris/HCl, pH 8.0, 1 mM acetyl-CoA and 10 mM thialysine that was linear for up to 20 min when incubated at 30 °C using 100 ng of purified human or 50 ng of S. pombe SSAT2. Reactions were terminated by addition of 150 μl of ethanol, after which was added 0.5 ml of a 0.2 mM solution of DTNB dissolved in 100 mM Tris/HCl, pH 8.0. Samples were centrifuged at 16000 gav for 5 min, and the supernatant was used to determine the absorbance at 412 nm measured against a blank that lacked thialysine. The stoichiometric amount of 5-thio-2-nitrobenzoic acid generated from DTNB by reduction with CoASH released by the enzyme reaction was determined using a molar absorption coefficient of 14140 M−1·cm−1 [19]. Purified SSAT2 from both human and S. pombe were assayed for their ability to use thialysine as a substrate.
HPLC analysis
Polyamines were determined by reversed-phase HPLC using post-column derivatization with OPA (o-phthaldialdehyde) and fluorescence detection, as described previously [20]. Alternatively, the HPLC column was linked directly to a Canberra Packard Flow-one β-radiochemical detector for identification of [14C]acetyl-polyamine products of the acetyltransferase assays using Flo-Scint II scintillation cocktail (Packard Bioscience, Meridan, CT, U.S.A.) [21].
The following HPLC method was used to analyse thialysine and its α- and ε-acetyl-derivatives. Standards or aliquots of deproteinized enzyme assays were derivatized with equal volumes of OPA reagent [40 mM OPA/200 mM potassium phosphate buffer (pH 9.4)/0.14 mM 2-mercaptoethanol/10% methanol] and subjected to a Spherisorb ODS II (5 μM, 125 mm×3 mm) reversed-phase HPLC. The mobile phase consisted of solvent A (10 mM potassium dihydrogen phosphate, pH 5.9) and solvent B (acetonitrile/methanol/water; 4:3:3, by vol.). Separation was performed at a flow rate of 0.9 ml·min−1 under application of the following gradient (percentages of solvent B are shown): 0 min, 20%; 4 min, 27%; 11.5 min, 27%; 28 min, 100%; 31 min, 100%; 40 min, 20%; 58 min, 20%. OPA derivatives were detected with a fluorescence spectrophotometer (excitation wavelength 338 nm, emission wavelength 425 nm, SFM 25, Kontron, Neufahrn, Germany) and by a flow-through radiodetector (LB 506; Berthold, Pforzheim, Germany). The retention times of α- and ε-N-acetyl-thialysine were 9.52 min and 6.77 min respectively.
Mass spectrometry
Samples for analysis by MALDI–TOF-MS (matrix-assisted laserdesorption ionization–time-of-flight mass spectrometry) were mixed 1:1 with matrix solution containing 10 mg/ml dihydrobenozoic acid (Sigma) in 30% (v/v) acetonitrile/0.1% trifluoroacetic acid. After mixing each sample with the matrix, 0.8 μl of the mixture was spotted on to a stainless-steel MALDI target plate and allowed to air-dry, with control spots containing matrix alone spotted in contiguous spots. MALDI–TOF spectra in the range of m/z 10–1000 were acquired on an Applied Biosystems 4700 Proteomics Analyzer in positive-ion reflectron mode, using a laser power setting of 2900.
ESI (electrospray ionization) mass spectra were obtained in the positive-ion mode using a Quattro II mass spectrometer (Micromass, Beverly, MA, U.S.A.). Aliquots (20 μl) of each solution were mixed with 20 μl of HPLC-grade acetonitrile (Fisher Scientific), and 10 μl aliquots were introduced to the mass spectrometer by flow injection with 100 μl/min flow of acetonitrile/water (50:50, v/v). Full-scan spectra were obtained over the range of m/z 50–500 at 2 s/scan. Product ion MS/MS spectra were obtained using identical ionization conditions, except that 1.8×10−3 mbar of argon was used as the collision gas, with a collision cell potential of −20 V.
Construction of pLNCX-FLAG-SSAT2 and expression of SSAT2 in NIH3T3 cells
The human SSAT2 cDNA with an N-terminal FLAG epitope (encoding MDYKDDDDK) was cloned into the HindIII–ClaI restriction sites of the retroviral pLNCX mammalian expression vector (Clontech) using the following strategy: the pT7-His SSAT2 plasmid was digested with NdeI and BamHI to remove 15 bp of N-terminal sequence, and the desired large fragment was gel-purified. The N-terminal FLAG epitope was generated after annealing forward (5′-TAAAGCTTACCATGGATTACAAGGATGACGACGATAAGATGGCTTCCGTGCG-3′) and reverse (5′-GATCCGCACGGAAGCCATCTTATCGTCGTCATCCTTGTAATCCATGGTAAGCTT-3′) primers that encoded HindIII and BamHI restriction sites at the 5′ and 3′ ends respectively.
The annealed primers were ligated to the BamHI overhang of the NdeI–BamHI vector fragment to generate a linear template for PCR. The desired HindIII–ClaI FLAG-SSAT2 PCR product was generated using the above-mentioned forward primer and a reverse primer (5′-AGTACTATCGATTCACTTTCCTGCCAACTTTCT-3′) that encoded a ClaI restriction site downstream of the stop codon for SSAT2. The PCR product was digested with HindIII and ClaI, and ligated into the same sites of a mammalian expression vector to generate the construct pLNCX-FLAG-SSAT2.
NIH3T3 cells were seeded at 2×105 cells/60 mm dish and grown overnight in Dulbecco's modified Eagle's medium supplemented with 2 mM L-glutamine, 10% fetal-calf serum and 100 units/ml penicillin/100 μg/ml streptomycin. Cells were transfected with 2 μg of pLNCX-FLAG-SSAT2 plasmid for 5 h at 37 °C in a humidified atmosphere of 5% CO2 using the LIPOFECTAMINE Plus™ reagent, as described by the manufacturer (Invitrogen). The medium was changed 5 h after the start of transfection, and the cells were left to grow for an additional 48 h before harvesting. Cells were re-plated after this time at 5×104, 10×104 and 20×104 cells/100 mm dish in the presence of 0.5 mg/ml G418 selection medium, and maintained until resistant colonies were established. Individual G418-resistant colonies were selected by trypsin treatment using cloning cylinders, after which they were transferred to 24-well plates and allowed to expand.
Degradation assay
The degradation of [35S]cysteine/[35S]methionine-labelled SSAT2 and [35S]methionine-labelled SSAT1, generated as non-histidine-tagged proteins from pT7 and p9.3 plasmids respectively in a coupled transcription/translation TNT assay (Promega), was studied using a rabbit reticulocyte-based degradation assay, as described previously [9,10].
RESULTS
Identification of a human cDNA encoding a novel acetyltransferase (SSAT2)
We originally identified a novel human acetyltransferase (accession no. AAL83905) as SSAT2 [15], on the basis of sequence similarity to human SSAT1, without reference to the functional properties of this protein. SSAT2 was discovered using the human SSAT1 protein sequence to conduct a tBLASTn search of several eukaryotic EST (expressed sequence tag) databases. Several overlapping human sequences were identified and spliced together to form one contiguous sequence. The entire contiguous human sequence that comprised six exons with well-defined exon–intron boundaries (accession number AC008049) was found to be present in the high-throughput genomic sequence (HTGS) database, giving this gene a chromosomal location of 17p13.1, whereas human SSAT1 is localized to Xp22.1 [22]. Similar results have been published recently by Chen et al. [16].
Primary amino acid sequence comparison of acetyltransferases that have homology with human SSAT1
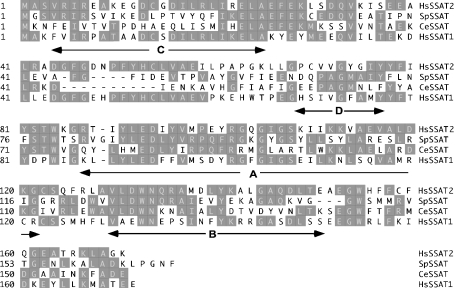
The human SSAT2 cDNA encodes a 170-amino-acid protein that is shown in Figure 1 in alignment with similar sequences from S. pombe and C. elegans for comparison with human SSAT1 (‘HsSSAT1’ in the Figure). The human SSAT2 protein (‘Hs-SSAT2’ in the Figure) contains the highly conserved acetyl-CoA-binding domain identified previously in SSAT1 as RGXGIGS [13], part of the universally conserved motif A in the GNAT superfamily [23], which includes Arg101 known to be essential for catalytic activity [12,13]. All of the enzymes shown have highly conserved residues found in human SSAT1 that forms part of motif D and motif B respectively in the GNAT superfamily. There is only weak similarity to motif C present in GNAT family members. Of the enzymes shown, human SSAT2 is closest in sequence similarity to human SSAT1 showing 46% identity and 61% homology with human SSAT1. The identity/homology values for S. pombe SSAT2 of 29%/43% and for C. elegans SSAT2 of 26%/45% are compared with human SSAT1. S. pombe SSAT2 and C. elegans SSAT2 are 38%/49% and 34%/49% identical/similar respectively when compared with the human SSAT2 sequence, making them closer in sequence to SSAT2 than to SSAT1. Notably, both the human SSAT2 and S. pombe SSAT2 sequences lack the two acidic residues present in the previously described C-terminal MATEE sequence motif of SSAT1 that was shown to be important in the rapid degradation of SSAT1 in vitro [9].
Figure 1. Alignment of N-acetyltransferase homologues.
Protein sequences were aligned using the ClustalW program. Human SSAT1(hsSSAT1) is compared with human SSAT2 (hsSSAT2), the S. pombe ats1 gene product designated SpSSAT (accession no. U82218) and the C. elegans protein designated CeSSAT (accession no. NP_505978). Arrows indicate motifs discussed further in the text. Conserved amino acids are shaded.
SSAT2 expression in human tissues
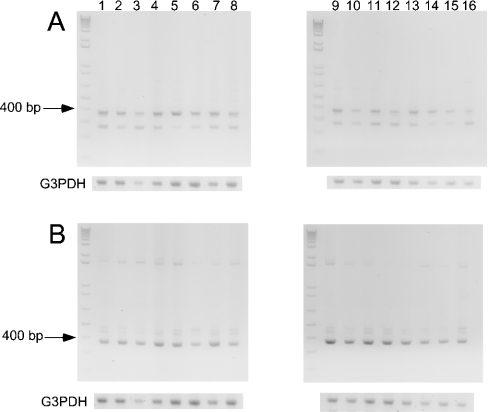
The expression profile of SSAT2 and SSAT1 mRNA was examined in a wide selection of human tissues using commercially available cDNA (generated by RT-PCR in the form of MTC™ panels) as a template for the PCR reaction. Both SSAT1 and SSAT2 were found to be widely expressed, as judged by a band corresponding to the predicted 369 bp product that was amplified in all 16 tissues examined (Figures 2A and 2B). The expression of SSAT2 mRNA was comparable with that of SSAT1 in all tissues tested. In the case of SSAT2, a smaller PCR product was also detected (Figure 2A) that may represent the amplification of an alternatively spliced form of the SSAT2 gene.
Figure 2. Comparison of the tissue expression pattern of human SSAT2 and SSAT1 mRNA in multiple human tissues.
MTC™ Panels I and II containing cDNAs derived by RT-PCR from the listed human tissue sources were used as templates for PCR to screen for expression of transcripts corresponding to SSAT2 (A) and SSAT1 (B). Primers were designed to amplify 369 bp products as described in the Materials and methods section. Lanes 1–16 are identical for each panel, and correspond to the following tissues: lane 1, heart; lane 2 brain; lane 3, placenta; lane 4, lung; lane 5, liver; lane 6, skeletal muscle; lane 7, kidney; lane 8, pancreas; lane 9, spleen; lane 10, thymus; lane 11, prostate; lane 12, testis; lane 13, ovary; lane 14, small intestine; lane 15, colon; and lane 16, peripheral blood leukocyte. G3PDH, glyceraldehyde-3-phosphate control reactions.
Kinetic properties of human SSAT2 and S. pombe SSAT2 purified from E. coli
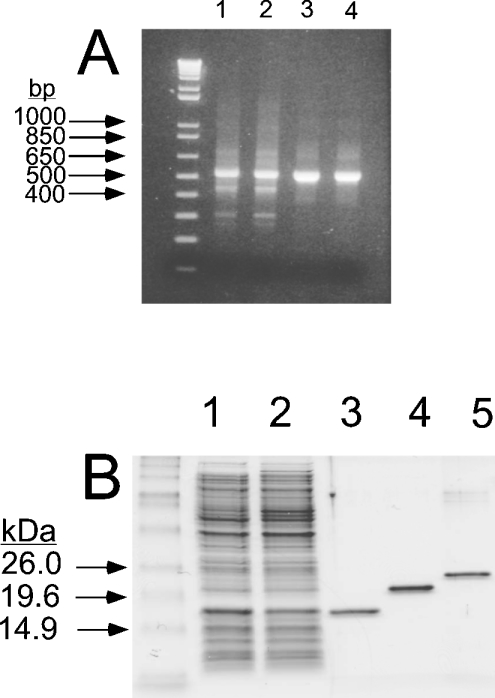
The human SSAT1 and the human and S. pombe SSAT2 proteins, all with (His)6 tags, were expressed in bacteria from the pT7 or pQE30 constructs described in the Materials and methods section and a gel showing purity and mobility of the purified proteins is shown in Figure 3B (lanes 3–5). (The predicted mass of each protein with epitope is 21.2 kDa, 20.1 kDa and 20.5 kDa for human SSAT1, human SSAT2 and S. pombe SSAT2 respectively). Although SSAT1 and SSAT2 are similar in size, there was a difference observed in their mobility when resolved on an SDS/15%-PAGE gel. In particular, the human SSAT2 protein showed a faster-than-predicted mobility under denaturing conditions. In order to check that the purified protein was full length, the molecular mass of the human SSAT2 protein was confirmed to be 20.1 kDa by MALDI–TOF-MS (results not shown).
Figure 3. Cloning of human SSAT2 and expression of human and S. pombe SSAT2 in E. coli.
The hsSSAT2 cDNA was cloned by PCR as described in the Materials and methods section, and the reaction products were resolved by 2% agarose gel electrophoresis. (A) shows PCR products (in duplicate) amplified from a human fetal liver library (lanes 1 and 2) and a HeLa cell library (lanes 3 and 4). (B) shows purified histidine-tagged proteins resolved on an SDS/15% polyacrylamide gel. Lanes 1 and 2 are, respectively, the crude soluble E. coli extract before and after induction by IPTG. Lane 3 shows purified His6-tagged human SSAT2 expressed from the pT7-7 plasmid in the BL21(DE3) E. coli strain, Lanes 4 and 5 are, respectively, purified His6-tagged human SSAT1 and His6-tagged S. pombe SSAT2, both expressed from the pQE30 plasmid in XL1-Blue cells.
It should also be noted that the human SSAT2 and S. pombe SSAT2 proteins ran with similar characteristics with respect to human SSAT1 in SDS/PAGE when expressed as 35S-labelled proteins from the pBluescript plasmid (results not shown). This suggests that the altered mobilities observed using SDS/PAGE are due to the unique composition of each protein.
The purified enzymes shown in Figure 3 were examined for their substrate specificity using the polyamines listed in Table 1 as putative acetyl acceptors. The polyamines were found to be poor acetyl acceptors in the reactions brought about by SSAT2 from human or S. pombe sources. The Km values were in the millimolar range (1.5–15 mM) and the kcat values were very low (<0.025 s−1). These results contrast strikingly with the known properties of SSAT1, which has Km values for spermidine and spermine in the 4–60 μM range [18] and kcat values of 5–8 s−1. For example, with spermine and spermidine as substrates for human SSAT2, the kcat/Km was <0.6 M−1·s−1, whereas with human SSAT1, the kcat/Km values are 150000 M−1·s−1 for spermidine and 1351350 M−1·s−1 for spermine [14]. Of the polyamines tested, the best diamine substrate for human SSAT2 was actually 1,3-diaminopropane, with a kcat/Km value of 8.7 M−1· s−1. This is a poor substrate for SSAT1, with a kcat/Km of 3.1 M−1·s−1 [18].
Table 1. Kinetic analysis of purified recombinant human and S. pombe SSAT2.
Apparent Km and kcat values were determined for the polyamine substrates using the filter binding assay described in the Materials and methods section at a fixed concentration of 16 μM [14C]acetyl-CoA, whereas the polyamine substrate concentration was varied from 0.05 mM up to 20 mM. Kinetic parameters shown using thialysine as an acetyl acceptor were determined in duplicate experiments using the spectrophotometric assay described in the Materials and methods section over a concentration range of 0.05–2 mM using 100 ng of human SSAT2 in the assay and from 0.05–8 mM using 50 ng of S. pombe SSAT2 in the presence of 1 mM acetyl-CoA. (Values agreed within a 5% error).
| Source | Substrate | Km (mM) | kcat (s−1) | kcat/Km (M−1·s−1) |
|---|---|---|---|---|
| Human SSAT2 | Putrescine* | 8.2 | 0.0023 | 0.28 |
| Spermidine† | 13.4 | 0.0072 | 0.54 | |
| Spermine† | 4.8 | 0.0020 | 0.42 | |
| 1,3-Diaminopropane* | 1.6 | 0.0137 | 8.7 | |
| Norspermidine* | 6.0 | 0.0168 | 2.8 | |
| Thialysine | 0.29 | 5.25 | 18100 | |
| S. pombe SSAT2 | Putrescine* | 4.3 | 0.0117 | 2.7 |
| Spermidine† | 4.6 | 0.0243 | 5.3 | |
| Spermine† | 1.8 | 0.0167 | 9.3 | |
| Thialysine | 12.7 | 46 | 3600 | |
| Human SSAT1 | Spermidine‡ | 0.058 | 8.7 | 150000 |
| Spermine‡ | 0.0037 | 5 | 1351350 | |
| Thialysine | No reaction |
Each value shown was determined in single* experiments (variation up to 28%).
Each value shown was determined in duplicate† experiments (variation up to 28%).
‡ Values shown for human SSAT1 are published data using the filter binding assay [14].
The predominant product of the action of SSAT2 on spermidine was found to be N1-acetylspermidine after separation of the products by HPLC. This is consistent with the preference for 1,3-diaminopropane over putrescine (1,4-diaminobutane) as a substrate and also with the finding that N8-acetylspermidine, but not N1-acetylspermidine, formed a product with a predicted retention time corresponding to the diacetylspermidine derivative (results not shown).
Many other compounds were tested as putative substrates for SSAT2, but were not active, including L-canavanine and taurine (results not shown). It was found that thialysine was a much better substrate than polyamines. Thus, with human SSAT2, the kcat/Km was 18100 M−1·s−1, and for S. pombe SSAT2 it was 3600 M−1·s−1. Thialysine was also shown to inhibit the acetylation of spermine with an ED50 value of 7.4 μM in an assay containing purified human SSAT2 at a fixed spermine concentration of 3 mM, confirming a preference for thialysine over spermine for the enzyme. Human SSAT2 did acetylate L-lysine, but less efficiently than thialysine (≈20% of the specific activity using 500 ng of purified enzyme, 1 mM acetyl-CoA with 10 mM L-lysine substituted for thialysine). Although the S. pombe SSAT2 had a relatively poor affinity for thialysine with a Km of 12.7 mM compared with 0.29 mM for human SSAT2, it did have a high turnover number of approx. 46 s−1 compared with the hSSAT2 enzyme for this substrate. There was no reaction of SSAT1 with thialysine, even in assays containing 1 μg of the hSSAT1, 10 mM thialysine and 1 mM acetyl-CoA. These results show a clear difference in substrate specificity for the respective purified enzymes.
Identification of Nε-acetylthialysine as the product formed using thialysine as a substrate
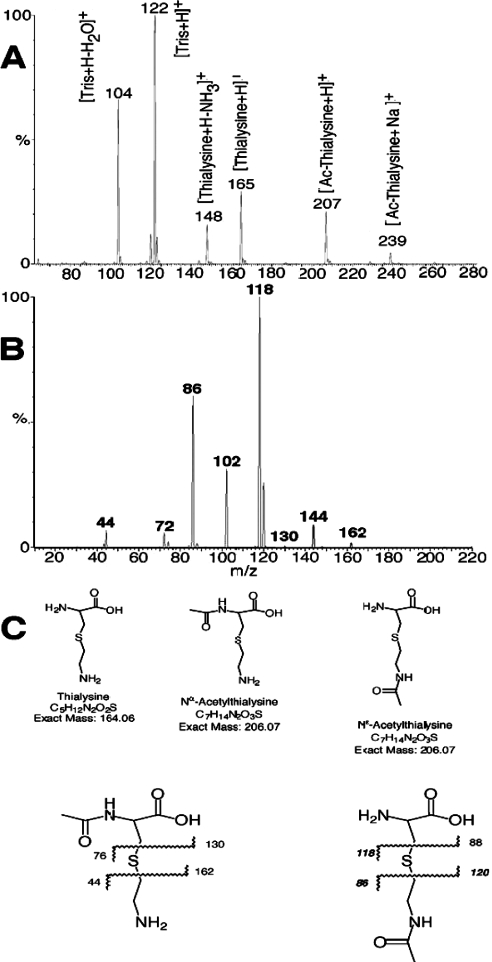
Since thialysine has two primary amine groups that can be acetylated, two approaches (MS and HPLC) were used to determine the product of the reaction. MALDI–TOF spectra of thialysine mixed with dihydrobenzoic acid showed the expected (M+H)+ peak of thialysine at 165. After reaction with acetyl-CoA and SSAT2, the resulting mass spectra showed peaks at both m/z 165 and 207, consistent with acetylation of thialysine. Spectra from control incubations in which thialysine was not included (SSAT2 and acetyl-CoA only) did not show peaks at either m/z 165 or 207, indicating that the m/z 207 peak observed was specific to acetylation of thialysine. A much smaller peak of m/z 207 was also observed in control incubations lacking enzyme (results not shown). Since non-enzymic acetylation of primary amines by acetyl-CoA occurs quite readily, this probably represents such non-enzymic acetylation of the thialysine occurring during the 1 h incubation with 1 mM acetyl-CoA.
MS/MS fragmentation of the acetylated thialysine at m/z 207 was performed using matrix-free ESI on a Quattro II triple-quadrupole mass spectrometer to determine whether the SSAT2-induced acetylation of thialysine was occurring on the α- or on the ε-amine moiety. As seen in Figure 4(A), the full-scan MS spectrum obtained on this instrument showed the same thialysine and acetylated thialysine peaks (m/z 165 and 207) as observed with the MALDI instrument. Typical collision-induced fragmentation spectra produced from the acetylated thialysine peak at m/z 207 are shown in Figure 4(B). The major fragment ions present in the MS/MS daughter spectrum (daughters of m/z 207) are at m/z 118 and 86. Corresponding fragmentation points are shown in Figure 4(C). These are consistent with the Nε-acetylthialysine, but not with the Nα-acetyl isomer. Minor fragments at m/z 130 and m/z 44 suggest a small amount of the latter isomer, but the majority appears to be acetylated on the side chain.
Figure 4. MS analysis of the reaction products formed using thialysine as a substrate for human SSAT2.
Reactions containing 1 μg of purified hsSSAT2 and 25 mM Tris/HCl, pH 8.0, 10 mM thialysine and 1 mM acetyl-CoA in a total volume of 50 μl were incubated at 30 °C for 1 h and analysed using MS to determine the nature of the N-acetylthialysine product formed as described in the Materials and methods section. Parallel control reactions included reactions containing inactivated protein and all other components, as well as reactions that contained active enzyme with acetyl-CoA but without thialysine. (A) shows a full-scan electrospray spectrum of product formed with the human SSAT2 enzyme. (B) shows the MS/MS spectrum of daughter ions generated from collision-induced dissociation of m/z 207 ([M+H]+ of acetylthialysine). (C) shows the potential fragmentation products from Nα-acetylthialysine (left) and Nε-acetylthialysine (right).
Further confirmation that the product of the reaction was Nε-acetylthialysine was obtained by separation of the products of the enzyme reaction using reversed-phase HPLC in collaboration with Dr Kai Lüersen at the Bernhard Nocht Institute for Tropical Medicine in Hamburg, Germany. Authentic standards of Nα-acetylthialysine and Nε-acetylthialysine were derivatized with OPA prior to injection, and the separated products were well resolved. The OPA-derivatized products from reactions containing either human SSAT2 or the S. pombe enzyme incubated in standard assays containing thialysine and [14C]acetyl-CoA were detected using a radiochemical detector connected downstream of the fluorescence detector. The resulting retention time of the labelled product from both the human SSAT2 and S. pombe enzymes were consistent with acetylation of the ε-amino group of thialysine (results not shown).
Overexpression of SSAT2 in NIH-3T3 cells
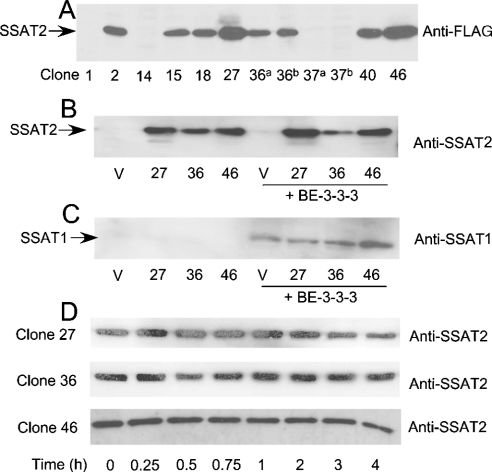
In order to study the effects of increased expression of SSAT2 on cell growth and polyamine metabolism, NIH-3T3 cells were stably transfected with a mammalian expression plasmid that encoded the human SSAT2 enzyme with an N-terminal FLAG epitope tag under the control of the strong CMV IE promoter. A number of G418-resistant clones were isolated, including empty vector controls. Cell extracts were prepared from ten clones, and SSAT2 expression was initially confirmed in seven of these cell lines using an anti-FLAG M5 antibody (Figure 5A). In contrast with the difficulty observed in establishing stable cell lines that overexpressed human SSAT1 [24], the human SSAT2 protein was readily expressed to high levels in NIH-3T3 cells. Three cell lines that were selected for further study included clones 27, 36 and 46, to reflect different levels of SSAT2 expression.
Figure 5. Western blots of cell extracts from NIH-3T3 cells stably expressing human SSAT2 and probed with anti-FLAG, anti-SSAT2 or anti-SSAT1 antibodies.
Cells were grown as indicated in the legend to Table 2. Cell extracts were prepared by three freeze–thaw cycles followed by centrifugation at 16000 gav for 20 min to remove insoluble cellular components. Extracts were resolved by SDS/PAGE (15% gels) and transferred on to a PVDF membrane for immunoblotting as described in the Materials and methods section. (A) shows extracts from a selection of G418-resistant clones numbered in order of isolation and probed with an anti-FLAG antibody. The total amount of protein loaded into each lane of a large gel was 25 μg (lanes with a superscript a) or 50 μg (lanes with a superscript b) as indicated. (B) shows the SSAT2 protein detected using an anti-SSAT2 antibody in 10 μg of total protein from clones 27, 36 and 46 either untreated or treated with 10 μM BE-3-3-3 for 48 h, as indicated. Control extracts (V) were from cells transfected with an empty vector. (C) shows the same extracts resolved in (B) that were probed with an anti-SSAT1 antibody. (D) illustrates the levels of SSAT2 protein (probed with an anti-SSAT2 antibody) in clones 27, 36 and 46 after treatment with 0.2 mM cycloheximide for up to 4 h.
A polyclonal antibody was raised in rabbits using the purified human SSAT2 enzyme as immunogen. This antibody was used to examine expression of SSAT2 in the three selected clones, as well as a control cell line that harboured the empty vector. As expected from the results with the anti-FLAG M5 antibody, ectopically expressed SSAT2 was detected in all three cell lines using the anti-SSAT2 antibody, but no signal was detected in extracts from empty vector controls (Figure 5B). This suggests that endogenous expression of SSAT2 is below detection under these conditions, but it cannot be discounted that the antibody may not react with the mouse SSAT2 protein, even though it is 89% identical with its human counterpart (alignment not shown). SSAT2 expression was unaffected by treatment with the polyamine analogue BE-3-3-3 (Figure 5B), in contrast with endogenous SSAT1, whose level of expression is greatly increased in the same NIH-3T3 cell lines under the same conditions (Figure 5C). SSAT1 was identified with an antiserum raised against human SSAT1 [25]. It appears, from Figures 5(B) and 5(C), that there is no cross-reactivity between the rabbit antibodies to human SSAT1 and SSAT2, with the caveat that the human SSAT2 antibody may or may not react with the mouse protein.
Overexpression of human SSAT2 in NIH-3T3 cells was not detrimental to cell growth. Clones 27, 36 and 46 divided with generation times of 20.8, 19.5 and 18.4 h respectively compared with 19.3 h for empty vector control cells. The same cell lines were grown for 48 h in the absence or presence of 10 μM BE-3-3-3, and extracts were assayed for acetyltransferase activity with spermidine as a substrate, and for polyamine content (Table 2). There was no difference in spermidine acetyltransferase activity in untreated control and clone 27, 36 and 46 cells, and this activity was very low in all cases (Table 2). Treatment with BE-3-3-3 caused an increase in spermidine acetyltransferase activity due to induction of endogenous SSAT1 (Figure 5C) that ranged from 185-fold for control cells to 105-, 161- and 210-fold for clones 27, 36 and 46 respectively.
Table 2. Polyamine content and spermidine acetyltransferase activity of control NIH-3T3 cells and stable clones that overexpress SSAT2.
The stable cell lines listed in the table were derived from NIH-3T3 cells by clonal selection as described in the Materials and methods section. Control (G418-resistant cells transfected with empty vector) and the three individual SSAT2 clones indicated were plated at a density of 2×105 cells/60 mm dish and incubated for 6 h to allow the cells to adhere to the plates. Cells were then treated with or without 10 μM BE-3-3-3 and harvested 48 h later for polyamine and enzyme assays, as described in the Materials and methods section. Polyamine content was determined by HPLC as described previously and values shown are the means±S.D. for five replicates. Spermidine acetyltransferase activity was determined using the [14C]acetyl-CoA filter binding assay described previously at a fixed spermidine concentration of 10 mM, and values shown are the means±S.D. for three replicates.
| Polyamine content (nmol/mg of protein) | |||||
|---|---|---|---|---|---|
| Cells | Treatment | Putrescine | Spermidine | Spermine | Acetyltransferase activity* (pmol/min per mg) |
| Control | None | 4.7±0.5 | 22.9±1.9 | 8.0±1.4 | 7.7±4.1 |
| Clone 27 | None | 4.0±0.5 | 25.6±2.6 | 9.2±1.3 | 20.3±0.8 |
| Clone 36 | None | 6.4±0.7 | 34.2±2.6 | 15.0±1.2 | 8.0±0.2 |
| Clone 46 | None | 9.3±0.9 | 34.3±2.7 | 10.9±0.4 | 9.9±1.2 |
| Control | BE-3-3-3 | 6.6±0.9 | 9.7±3.3 | <0.5 | 1429±55 |
| Clone 27 | BE-3-3-3 | 7.8±2.1 | 12.9±2.3 | 1.5±0.4 | 2129±233 |
| Clone 36 | BE-3-3-3 | 10.9±1.4 | 17.9±2.0 | 3.5±0.6 | 1290±166 |
| Clone 46 | BE-3-3-3 | 9.7±1.1 | 9.0±1.1 | <0.5 | 2075±242 |
* Measured with 10 mM spermidine as substrate.
As expected from the known ability of SSAT1 to act as a polyamine catabolic enzyme whose function is associated with decreasing spermidine and spermine levels, all of the cells responded to treatment with BE-3-3-3, with a decrease in spermine and spermidine pools. In contrast, overexpression of human SSAT2 increased, rather than decreased, the total polyamine content of clones 36 and 46, with little change in the polyamine profile of clone 27 when compared with empty vector controls (Table 2). This is not consistent with SSAT2 having a role in metabolizing polyamines, but is in keeping with the high Km values and low kcat values determined for polyamines using the purified enzyme (Table 1) and the absence of a detectable increase in the ability of extracts from clones 27, 36 and 46 to acetylate spermidine in vitro.
Turnover of human SSAT2 protein
Previous work showed that human SSAT1 is degraded by the ubiquitin/proteasome pathway in reticulocyte lysates with a half-life of approx. 30 min under standard assay conditions [9,10]. A similar half-life was measured in mammalian cells [24]. To test whether the differences in amino acid sequence between the human SSAT1 and SSAT2 enzymes had any effect on the turnover of the protein, their stability was examined both in reticulocyte lysates and in the stable cell lines 27, 36 and 46.
In order to examine the stability of human SSAT2 in a cellular context, the NIH-3T3 clones that overexpress human SSAT2 were grown for 48 h, before treatment with cycloheximide to inhibit further protein synthesis. Cells were harvested at times up to 4 h afterwards, and extracts were resolved by SDS/PAGE for analysis of remaining SSAT2 protein by Western blotting. The results shown in Figure 5(D) indicate that SSAT2 was relatively stable over the 4 h time period examined, in contrast with the published antigenic half-life of 30 min after cycloheximide treatment observed for SSAT1 in stably transfected CHO cells [24].
When studied in reticulocyte lysates described previously [9,10], 35S-labelled SSAT2 was found to be considerably more stable than SSAT1, and showed little accumulation of high-molecular-mass conjugates with ubiquitin in the presence of ubiquitin aldehyde and the proteasome inhibitor MG132 (results not shown).
DISCUSSION
Comparison of the GNAT family indicates that human SSAT2 is more closely related to SSAT1 than to any other member, but our results show clearly that, despite this similarity, SSAT2 is not likely to be involved in polyamine metabolism. Although acetylation of polyamines can be demonstrated in vitro with large amounts of the recombinant purified SSAT2 protein, the very low activity towards these substrates argues against any physiological role in regulating polyamine content. The results of expression of SSAT2 in NIH-3T3 cells are consistent with this, since there was no decrease in polyamine content and no acetylpolyamines detected. In contrast, expression of comparable levels of SSAT1 (assuming that the immunochemical detection is equally sensitive in both cases) in these cells by induction of the endogenous activity by BE-3-3-3 dramatically reduced the polyamine content.
An alternative explanation for the lack of effect of SSAT2 on polyamine content was suggested by Chen et al. [16], who hypothesized that compartmentation of the expressed SSAT2 might account for a lack of acetylation of cellular polyamines. Although we cannot absolutely rule out such compartmentation, a far more likely explanation is the very low activity of SSAT2 towards spermidine and spermine as substrates. Our experiments differ from theirs in that we assayed the kinetics of SSAT2 in detail using purified recombinant protein, whereas they only detected activity by measuring acetylation in lysates from HEK-293 cells transiently transfected with plasmids for SSAT1 and SSAT2 using an assay with the potential substrate at a single concentration of 3 mM. The inadequacy of this assay is demonstrated by the fact that they found that putrescine, which is not a substrate for purified SSAT1, had a relative activity of 9% that of spermidine. Extracts from SSAT2-transfected cells in the experiments of Chen et al. [16] had 25% of the activity with spermidine as a substrate that was seen with extracts from SSAT1-transfected cells, and 55% of the activity with spermine as a substrate, but these results were not corrected for the amount of protein expressed. Our results indicate that, on the basis of kcat/Km values, the SSAT2 activity is actually much less than 0.01% of the activity of SSAT1.
Previous studies using site-directed mutagenesis identified several residues in SSAT1 that are implicated in polyamine substrate binding to the enzyme active site. A notable difference between the aligned sequences is the absence in the human and S. pombe SSAT2 proteins of the C-terminal MATEE motif present in SSAT1, which was shown to be important for polyamine acetyltransferase activity and in mediating the stabilizing effects of polyamine analogues [9,11]. Another key region of importance with regard to polyamine binding in SSAT1 occurs between Glu152–Leu156. While Glu152 is conserved between human SSAT1 and human SSAT2, specific changes occur at residues 155 and 156, and neither of these residues are conserved in the S. pombe SSAT2. Mutation of Arg155 to alanine in SSAT1 was shown previously to decrease SSAT activity and increase the apparent Km for spermidine 10-fold [12], whereas a poorly active mutant form of SSAT1, L156F-SSAT, was isolated from Chinese hamster ovary cells selected for resistance to the cellular effects of the polyamine analogue BE-3-3-3 [14]. Since the corresponding residue to Leu156 in SSAT1 is Phe156 in human SSAT2, this may in part account for the poor affinity of polyamines for the SSAT2 protein.
SSAT1 has been shown to undergo rapid degradation by the ubiquitin/26 S proteasome pathway, and results from mutational analysis have also implicated the C-terminal MATEE sequence in conferring this lability [9,10]. The human SSAT2 protein appears to be more stable than human SSAT1, and also lacks the corresponding lysine residue at position 87 that may be a preferred site in SSAT1 for ubiquitination [10]. Taken together, these differences may impart some insight into residues that confer specific properties to the respective proteins. It is interesting that the E. coli spermidine acetyltransferase [26] and the spermidine/spermine acetyltransferase BltD from Bacillus subtilis [27] bear little resemblance to the primary amino acid sequence of human SSAT1. This may indicate the evolution of two distinct structural classes of polyamine acetyltransferases, such as occurs with bacterial and eukaryotic ornithine decarboxylases [28].
Splice variants of SSAT2 may exist as suggested by (a) Figure 2, (b) by the presence of additional sequences in the human EST clone (ID 503372) and (c) a cDNA clone isolated from a human testis library that lacked sequence corresponding to exon 4 (results not shown). Our results show clearly that polyamines are not the normal substrate for SSAT2, but the widespread expression of the mRNA for this enzyme and the presence of a similar enzyme in S. pombe and C. elegans suggest that it must play some physiological role. Since SSAT2 has a preferential activity towards thialysine, it would be more appropriately described as thialysine Nε-acetyltransferase. The accompanying paper by Abo-Dalo et al. [29] reaches the same conclusion based on studies of the equivalent enzyme from C. elegans.
A bacterial thialysine Nε-acetyltransferase, with a weak activity towards lysine, was previously identified from Aerobacter aerogenes [30]. This enzyme is inducible by thialysine, and allows growth with this lysine antagonist as the sole nitrogen source. Other enzymes have been isolated from yeast that acetylate lysine at the Nε-position and have activity towards thialysine. These enzymes are preferentially induced by lysine, and are involved in lysine catabolism [19,31].
Metabolites formed from thialysine have been characterized and identified in mammalian tissues including brain [32,33]. The formation of thialysine can occur via the reaction of serine and cysteamine or pantetheine catalysed by cystathionine-β-synthase [34]. Further metabolism of thialysine by L-amino-acid oxidase [33,35,36] or a mammalian glutamine transaminase [33,36] leads to the cyclic ketimine, AECK [S-(aminoethyl)-L-cysteine ketimine] [32]. AECK can be reduced to form TMA (1,4-thiomorpholine-3,5-dicarboxylic acid) by a reductase isolated from bovine brain [37], or it can dimerize spontaneously to form a tricyclic product that loses a carboxy group to form AECK-decarboxylated dimer (AECK-DD) [38,39]. AECK, TMA and AECK-DD have been found in bovine brain, and AECK-DD has been found in human urine and plasma and in a variety of vegetables and human cells [39]. The functions of AECK, TMA and AECK-DD are not known, although the former two compounds have been postulated to serve neurochemical roles, whereas the latter has strong antioxidant properties and could play a role in prevention of damage from hydroxyl radicals, peroxynitrite and other oxidative products. The Nε-acetylation of thialysine would prevent its conversion into AECK, and hence TMA and AECK-DD: the thialysine Nε-acetyltransferase may therefore control this pathway.
Acknowledgments
This work was supported by grants GM26290 and CA18138 from the National Institutes of Health, Bethesda, MD, U.S.A. We are grateful to Dr Kai Lüersen for helpful discussions and exchanging information, as well as for conducting the HPLC analysis to distinguish the N-acetylthialysine isomer formed in enzyme reactions using the human and S. pombe enzymes. C. S. C. wishes to thank Dr Vincent Chau for the discovery of the human and mouse SSAT2 nucleotide sequences, the construction of bacterial expression systems for SSAT2, the purification of bacterially expressed proteins as described herein, and for supplying purified SSAT2 protein for the present study.
References
- 1.Pegg A. E. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem. J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen S. S. New York, NY: Oxford University Press; 1998. A Guide to the Polyamines. [Google Scholar]
- 3.Wallace H. M., Fraser A. V., Hughes A. A perspective of polyamine metabolism. Biochem. J. 2003;376:1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casero R. A., Jr, Pegg A. E. Spermidine/spermine N1-acetyltransferase: the turning point in polyamine metabolism. FASEB J. 1993;7:653–661. [PubMed] [Google Scholar]
- 5.Seiler N. Functions of polyamine acetylation. Can. J. Physiol. Pharmacol. 1987;65:2024–2035. doi: 10.1139/y87-317. [DOI] [PubMed] [Google Scholar]
- 6.Coleman C. S., Pegg A. E., Megosh L. C., Guo Y., Sawicki J. A., O'Brien T. G. Targeted expression of spermidine/spermine N1-acetyltransferase increases susceptibility to chemically induced skin carcinogenesis. Carcinogenesis. 2002;23:359–364. doi: 10.1093/carcin/23.2.359. [DOI] [PubMed] [Google Scholar]
- 7.Casero R. A., Jr, Woster P. M. Terminally alkylated polyamine analogues as chemotherapeutic agents. J. Med. Chem. 2001;44:1–26. doi: 10.1021/jm000084m. [DOI] [PubMed] [Google Scholar]
- 8.Hahm H. A., Ettinger D. S., Bowling K., Hoker B., Chen T. L., Zabelina Y., Casero R. A., Jr Phase I study of N1,N11-diethylnorspermine in patients with non-small cell lung cancer. Clin. Cancer Res. 2002;8:684–690. [PubMed] [Google Scholar]
- 9.Coleman C. S., Pegg A. E. Proteasomal degradation of spermidine/spermine N1-acetyltransferase requires the carboxyl-terminal glutamic acid residues. J. Biol. Chem. 1997;272:12164–12169. doi: 10.1074/jbc.272.18.12164. [DOI] [PubMed] [Google Scholar]
- 10.Coleman C. S., Pegg A. E. Polyamine analogues inhibit the ubiquitination of spermidine/spermine N1-acetyltransferase and prevent its targeting to the proteasome for degradation. Biochem. J. 2001;358:137–145. doi: 10.1042/0264-6021:3580137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman C. S., Huang H., Pegg A. E. Role of the carboxyl terminal MATEE sequence of spermidine/spermine N1-acetyltransferase in the activity and stabilization by the polyamine analog N1,N12-bis(ethyl)spermine. Biochemistry. 1995;34:13423–13430. doi: 10.1021/bi00041a020. [DOI] [PubMed] [Google Scholar]
- 12.Coleman C. S., Huang H., Pegg A. E. Structure and critical residues at the active site of spermidine/spermine N1-acetyltransferase. Biochem. J. 1996;316:697–701. doi: 10.1042/bj3160697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu L., Berkey K. A., Casero R. A., Jr RGFGIGS is an amino acid sequence required for acetyl coenzyme A binding and activity of human spermidine/spermine N1-acetyltransferase. J. Biol. Chem. 1996;271:18920–18924. doi: 10.1074/jbc.271.31.18920. [DOI] [PubMed] [Google Scholar]
- 14.McCloskey D. E., Pegg A. E. Properties of the spermidine/spermine N1-acetyltransferase mutant L156F that decreases cellular sensitivity to the polyamine analogue N1,N11-bis(ethyl)norspermine. J. Biol. Chem. 2003;278:13881–12887. doi: 10.1074/jbc.M205689200. [DOI] [PubMed] [Google Scholar]
- 15.Coleman C. S., Chau V., Pegg A. E. Identification of a novel polyamine acetylase; FASEB Proceedings A169; 2001. [Google Scholar]
- 16.Chen Y., Vujcic S., Liang P., Diegelman P., Kramer D. L., Porter C. W. Genomic identification and biochemical characterization of a second spermidine/spermine N1-acetyltransferase. Biochem. J. 2003;373:661–667. doi: 10.1042/BJ20030734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Della Ragione F., Pegg A. E. Studies of the specificity and kinetics of rat liver spermidine/spermine N1-acetyltransferase. Biochem. J. 1983;213:701–706. doi: 10.1042/bj2130701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bode R., Thurau A.-M., Schmidt H. Characterization of acetyl-CoA:L-lysine N6-acetyltransferase, which catalyzes the first step of carbon catabolism from lysine in Saccharomyces cerevisiae. Arch. Microbiol. 1993;160:397–400. doi: 10.1007/BF00252227. [DOI] [PubMed] [Google Scholar]
- 20.Pegg A. E., Wechter R., Poulin R., Woster P. M., Coward J. K. Effect of S-adenosyl-1,12-diamino-3-thio-9-azadodecane, a multisubstrate inhibitor of spermine synthase, on polyamine metabolism in mammalian cells. Biochemistry. 1989;28:8446–8453. doi: 10.1021/bi00447a026. [DOI] [PubMed] [Google Scholar]
- 21.Coleman C. S., Hu G., Pegg A. E. Putrescine biosynthesis in mammalian tissues. Biochem. J. 2004;379:849–855. doi: 10.1042/BJ20040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao L., Celano P., Mank A. R., Griffin C., Jabs E. W., Hawkins A. L., Casero R. A., Jr Structure of the human spermidine/spermine N1-acetyltransferase gene. Biochem. Biophys. Res. Commun. 1992;187:1493–1502. doi: 10.1016/0006-291x(92)90471-v. [DOI] [PubMed] [Google Scholar]
- 23.Neuwald A. F., Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 24.McCloskey D. E., Coleman C. S., Pegg A. E. Properties and regulation of human spermidine/spermine N1-acetyltransferase stably expressed in Chinese hamster ovary cells. J. Biol. Chem. 1999;274:6175–6182. doi: 10.1074/jbc.274.10.6175. [DOI] [PubMed] [Google Scholar]
- 25.Casero R. A., Jr, Gabrielson E. W., Pegg A. E. Immunohistochemical staining of human spermidine/spermine N1-acetyltransferase superinduced in response to treatment with antitumor polyamine analogues. Cancer Res. 1994;54:3955–3958. [PubMed] [Google Scholar]
- 26.Fukuchi J., Kashiwagi K., Takio K., Igarashi K. Properties and structure of spermidine acetyltransferase in Escherichia coli. J. Biol. Chem. 1994;269:22581–22585. [PubMed] [Google Scholar]
- 27.Woolridge D. P., Martinez J. D., Stringer D. E., Gerner E. W. Characterization of a novel spermidine/spermine acetyltransferase, BltD, from Bacillus subtilis. Biochem. J. 1999;340:753–758. [PMC free article] [PubMed] [Google Scholar]
- 28.Grishin N. V., Phillips M. A., Goldsmith E. J. Modeling of the spatial structure of eukaryotic ornithine decarboxylase. Protein Sci. 1995;4:1291–1304. doi: 10.1002/pro.5560040705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abo-Dalo B., Ndjonka D., Pinnen F., Liebau E., Luersen K. A novel member of the GCN5-related N-acetyltransferase superfamily from Caenorhabditis elegans preferentially catalyses the N-acetylation of thialysine [S-(2-aminoethyl)-L-cysteine] Biochem. J. 2004;383:129–137. doi: 10.1042/BJ20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka H., Soda K. Enzymatic N-acetylation of lysine analogs. J. Biol. Chem. 1974;249:5285–5289. [PubMed] [Google Scholar]
- 31.Schmidt H., Bode R., Birnbaum D. Lysine degradation in Candida maltosa: occurrence of a novel enzyme, acetyl-CoA:L-lysine N-acetyltransferase. Arch. Microbiol. 1988;150:215–218. [Google Scholar]
- 32.Cavallini D., Ricci G., Dupre S., Pecci L., Costa M., Matarese R. M., Pensa B., Antonucci A., Solinas S. P., Fontana M. Sulfur-containing cyclic ketimines and imino acids. Eur. J. Biochem. 1991;202:217–223. doi: 10.1111/j.1432-1033.1991.tb16365.x. [DOI] [PubMed] [Google Scholar]
- 33.Cooper A. J. L. The role of glutamine transaminase K (GTK) in sulfur and α-keto acid metabolism in the brain, and in the possible bioactivation of neurotoxicants. Neurochem. Int. 2004;44:557–577. doi: 10.1016/j.neuint.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Pitari G., Maurizi G., Flati V., Ursini C. L., Spera L., Dupre S., Cavallini D. Enzymatic synthesis of S-aminoethyl-L-cysteine from pantetheine. Biochim. Biophys. Acta. 1992;1116:27–33. doi: 10.1016/0304-4165(92)90124-d. [DOI] [PubMed] [Google Scholar]
- 35.Cavallini D., Ricci G., Federici G., Costa M., Pensa B., Matarese R. M., Achilli M. The oxidation of sulfur-containing amino acids by L-amino acid oxidases. Adv. Exp. Med. Biol. 1982;148:359–374. doi: 10.1007/978-1-4615-9281-5_28. [DOI] [PubMed] [Google Scholar]
- 36.Ricci G., Nardini M., Federici G., Cavallini D. The transamination of L-cystathionine, L-cysteine and related compounds by a bovine kidney transaminase. Eur. J. Biochem. 1986;157:57–63. doi: 10.1111/j.1432-1033.1986.tb09637.x. [DOI] [PubMed] [Google Scholar]
- 37.Nardini M., Ricci G., Vesci L., Pecci L., Cavallini D. Bovine brain ketimine reductase. Biochim. Biophys. Acta. 1988;957:286–292. doi: 10.1016/0167-4838(88)90285-3. [DOI] [PubMed] [Google Scholar]
- 38.Matarese R. M., Macone A., Crescentini G., Dupre S., Cavallini D. Detection of decarboxylated dimer of aminoethylcysteine ketimine in bovine cerebellum. Neurochem. Int. 1998;32:365–368. doi: 10.1016/s0197-0186(97)00094-6. [DOI] [PubMed] [Google Scholar]
- 39.Nardini M., Macone A., Matarese R. M. Determination of aminoethylcysteine ketimine decarboxylated dimer in human plasma and cultured cells by high-performance liquid chromatography with electrochemical detection. J. Chromatogr. B Biomed. Sci. Appl. 2003;795:319–327. doi: 10.1016/s1570-0232(03)00597-x. [DOI] [PubMed] [Google Scholar]