Abstract
During liver fibrogenesis, quiescent HSC (hepatic stellate cells) become active, a transformation that is associated with enhanced cell proliferation and overproduction of ECM (extracellular matrix). Inhibition of cell proliferation and induction of apoptosis are potential strategies to block the activation of HSC for the prevention and treatment of liver fibrosis. Levels of PPARγ (peroxisome proliferator-activated receptor γ) are dramatically diminished in parallel with HSC activation. Stimulation of PPARγ by its agonists inhibits HSC activation in vitro and in vivo. We demonstrated recently that curcumin, the yellow pigment in curry, inhibited HSC activation in vitro, reducing cell proliferation, inducing apoptosis and inhibiting ECM gene expression. Further studies indicated that curcumin induced the gene expression of PPARγ and stimulated its activity in activated HSC in vitro, which was required for curcumin to inhibit HSC proliferation. The aims of the present study were to evaluate the roles of PPARγ activation in the induction of apoptosis and suppression of ECM gene expression by curcumin in activated HSC, and to elucidate the underlying mechanisms. Our results demonstrated that blocking PPARγ activation abrogated the effects of curcumin on the induction of apoptosis and inhibition of the expression of ECM genes in activated HSC in vitro. Further experiments demonstrated that curcumin suppressed the gene expression of TGF-β (transforming growth factor-β) receptors and interrupted the TGF-β signalling pathway in activated HSC, which was mediated by PPARγ activation. Taken together, our results demonstrate that curcumin stimulated PPARγ activity in activated HSC in vitro, which was required for curcumin to reduce cell proliferation, induce apoptosis and suppress ECM gene expression. These results provide novel insight into the mechanisms responsible for the inhibition of HSC activation by curcumin. The characteristics of curcumin, which has no adverse health effects, make it a potential candidate for prevention and treatment of hepatic fibrosis.
Keywords: apoptosis, extracellular matrix, hepatic fibrosis, hepatic stellate cell, peroxisome proliferator-activated receptor γ (PPARγ), phytochemical
Abbreviations: α-SMA, α-smooth muscle actin; ECM, extracellular matrix; HSC, hepatic stellate cells; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; FBS, fetal bovine serum; NF-κB, nuclear factor-κB; PPARγ, peroxisome proliferator-activated receptor γ; PPRE, peroxisome proliferator response element; TGF-β, transforming growth factor-β; Tβ-RI and Tβ-RII, type I and II TGF-β receptors respectively
INTRODUCTION
Hepatic fibrogenesis occurs as a wound-healing process after many forms of chronic hepatic injury, including viral infection, drug-induced hepatitis and sustained alcohol abuse [1]. The latter accounts for approx. 50% of deaths due to cirrhosis in Western societies [2]. During the process of fibrosis, production of the ECM (extracellular matrix) exceeds its degradation in the liver. Without effective treatment at an early stage, reversible hepatic fibrosis progresses to irreversible cirrhosis [1]. HSC (hepatic stellate cells), previously called fat- or vitamin A-storing cells, or Ito cells, are the primary source of excessive production of ECM during hepatic fibrogenesis [1]. HSC normally reside in the space of Disse in a quiescent, non-proliferative state. During hepatic injury, HSC become active and undergo profound phenotypic changes, including enhanced cell proliferation, de novo expression of α-SMA (α-smooth muscle actin) and excessive production of ECM. Activation of HSC is associated with the sequential expression of several key cytokines and their surface receptors, including TGF-β (transforming growth factor-β) and its type I and II receptors (Tβ-RI and Tβ-RII respectively) [3,4]. TGF-β is the most potent inducer of HSC activation and ECM production [5–8]. Blocking TGF-β signalling results in a marked decrease in ECM production in activated HSC in vitro and in vivo [5–8]. Inhibition of HSC proliferation and induction of HSC apoptosis have been proposed as strategies for the elimination of activated HSC for the prevention and treatment of hepatic fibrosis [9].
In response to a variety of endogenous and exogenous ligands, the nuclear transcription factor PPARγ (peroxisome proliferator-activated receptor γ) forms heterodimers with the retinoid X receptor and binds to PPREs (peroxisome proliferator response elements) in gene promoters to regulate the transcription of genes [10]. PPARγ activation has effects on diverse physiological and pathophysiological events, including stimulation of adipocyte differentiation, activation of insulin, regulation of lipid metabolism, inhibition of cell proliferation and induction of apoptosis [10–12]. Recent studies have started a new page for evaluating the effects of PPARγ on HSC activation and hepatic fibrogenesis. PPARγ is highly expressed in quiescent HSC in the normal liver [13–15]. However, the level of PPARγ and its activity are dramatically reduced during HSC activation in vitro and in vivo [13–15]. Stimulation of PPARγ activity by its agonists inhibits HSC proliferation and α1(I) collagen expression in vitro and in vivo [14,16]. Furthermore, the adenoviral vector-mediated expression of PPARγ itself is sufficient to reverse the morphology of activated HSC to the quiescent phenotype [17].
Curcumin, the yellow pigment of turmeric in curry derived from the rhizome of the plant Curcuma longa, is a potent antioxidant [18]. Besides its role as a dietary spice, turmeric has been used for centuries as an anti-inflammatory remedy in Chinese medicine. In addition, it inhibits lipid peroxidation [19], nitric oxide synthase activity [20], production of reactive oxygen species [21], protein kinase C activity [22], and NF-κB (nuclear factor-κB) activity [23]. Curcumin has received attention as a promising dietary supplement for cancer prevention [18] and liver protection [24]. We observed recently that curcumin inhibited the cell growth of activated HSC in vitro by reducing cell proliferation and inducing apoptosis [25]. In addition, curcumin dramatically increased the level of PPARγ and induced its transcriptional activity in cultured HSC, without the need to introduce exogenous PPARγ or its agonists [25]. Previous experiments have demonstrated that activation of PPARγ mediates the inhibition of HSC cell proliferation by curcumin [25].
The aims of the present study were to evaluate the role of PPARγ activation in the induction of apoptosis and the suppression of ECM gene expression by curcumin in activated HSC, and to elucidate the underlying mechanisms. We demonstrated that activation of PPARγ by curcumin was a necessary step, and contributed to the induction of HSC apoptosis by stimulating caspase 3 activity, increasing the abundance of pro-apoptotic Bax and reducing the level of anti-apoptotic Bcl-2 in activated HSC in vitro. In addition, activation of PPARγ was required for curcumin to inhibit ECM gene expression. Activation of PPARγ mediated the blockade of the TGF-β signalling pathway by curcumin, by suppressing TGF-β receptors. These results provided novel insight into the roles and mechanisms of the inhibition of HSC activation by curcumin.
MATERIALS AND METHODS
Isolation and culture of HSC
HSC were isolated from male Sprague–Dawley rats (200–250 g) as described previously [26]. Passaged HSC were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) FBS (fetal bovine serum). Unless indicated otherwise, activated HSC were used at passages 4–8. Curcumin (purity >94%) was purchased from Sigma (St. Louis, MO, U.S.A.). PD 68235, a specific PPARγ antagonist, was kindly provided by Pfizer (Ann Arbor, MI, U.S.A.) [27].
Western blotting analyses
Pre-confluent HSC were pretreated with or without PD 68235 (10 or 20 μM) for 30 min prior to the addition of curcumin at the indicated concentrations for 24 h. Cells were lysed in ice-cold RIPA lysis buffer. Cell lysates were collected after centrifugation at 9000 g for 15 min at 4 °C and stored at −80 °C. The protein concentration was determined using a Micro BCA™ Protein Assay Reagent Kit following the protocol provided by the manufacturer (Pierce, Rockford, IL, U.S.A.). SDS/PAGE (10% resolving gel) was used to separate proteins (25 μg/well). Separated proteins were detected using primary antibodies and horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.). Protein bands were visualized by utilizing chemiluminescence reagent (Kirkegaard & Perry Laboratories, Gaithersburg, MD, U.S.A.).
Caspase 3 activity assays
Caspase 3 activity was measured using a caspase 3 activity assay kit (Promega), as we described recently [25]. In brief, semi-confluent HSC were treated as indicated. Cell lysates were incubated with the substrate DEVD-p-nitroanilide at 37 °C for 60 min. The results of the reaction were read by a spectrophotometer at 405 nm. The level of caspase enzymic activity in the cell lysates is directly proportional to the colour reaction. A standard calibration curve of p-nitroanilide was established by a series of dilutions of p-nitroanilide solution provided with the kit. Each treatment was performed at least three times.
Flow cytometric analyses of apoptotic HSC
These experiments were carried out as described previously [25]. Briefly, semi-confluent HSC were treated as indicated. Cells were harvested by brief trypsin/EDTA treatment and washed several times with cold PBS. HSC (≥1×106 cells/sample) were suspended in 2 ml of FACS buffer [1% FA buffer (Difco), 0.1% sodium azide and 1% FBS]. Cells were fixed with ethanol, and then labelled with propidium iodide (Sigma). Cells that were positively labelled with propidium iodide were detected by a Coulter® EPICS® XL-MCL flow cytometer and analysed using its System II™ software.
RNA isolation and real-time PCR
Total RNA was isolated using TRI-Reagent (Sigma), following the protocol provided by the manufacturer. Real-time PCR was carried out as described [25]. DNase I-treated total RNA (1 μg) was used for synthesis of the first strand of cDNA. Reverse transcription conditions were as follows: 42 °C for 15 min, 95 °C for 5 min and 5 °C for 5 min (one cycle). Real-time PCR was carried out in 25 μl of reaction solution [2.5 μl of 10×buffer, 5 mM of each dNTP, 10 mM MgCl2, 200 nM primers and 0.75 unit of platinum® Taq polymerase; all from Invitrogen] plus 1 μl of SYBR Green (1:2000; BioWhittaker, Richland, ME, U.S.A.). No genomic DNA contamination or pseudogenes were detected by PCR in the absence of the reverse transcription step in the total RNA used. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as an invariant control. The reactions started at 95 °C for 7 min, followed by 40 cycles of 95 °C for 20 s, 54 °C for 30 s and 72 °C for 30 s. Melting peaks of PCR products were determined by heat denaturation from 60 to 95 °C at 0.2 °C/s. Fold changes in the mRNA levels of target genes relative to the endogenous GAPDH control were calculated as suggested by Schmittgen et al. [27a]. Primers used in real-time PCR were as followsa: α1(I) collagen: forward, 5′-CCTCAAGGGCTCCAACGAG-3′; reverse, 5′-TCAATCACTGTCTTGCCCCA-3′; PPARγ: forward, 5′-ATTCTGGCCCACCAACTTCGG-3′; reverse, 5′-TGGAAGCCTGATGCTTTATCCCCA-3′; Bcl-2: forward, 5′-ATGGGGTGAACTGGGGGATTG-3′; reverse, 5′-TTCCGAATTTGTTTGGGGCAGGTC-3′; Bax: forward, 5′-GGGTGGTTGCCCTTTTCTACT-3′; reverse, 5′-CCCGGAGGAAGTCCAGTGTC-3′; cyclin D1: forward, 5′-GCACAACGCACTTTCTTTCTTTCCA-3′; reverse, 5′-CGCAGGCTTGACTCCAGAAG-3′; Tβ-RI: forward, 5′-ATCCATGAAGACTATCAGTTGCCT-3′; reverse, 5′-CATTTTGATGCCTTCCTGTTGGCT-3′; Tβ-RII: forward, 5′-TGTGCTCCTGTAACACAGAG-3′; reverse, 5′-GATCTTGACAGCCACGGTCT-3′; GAPDH: forward, 5′-GGCAAATTCAACGGCACAGT-3′; reverse, 5′-AGATGGTGATGGGCTTCCC-3′.
Plasmids and transient transfection assays
The PPARγ reporter plasmid pPPRE-TK-Luc contains three copies of the PPRE from the acyl-CoA oxidase gene linked to the herpes virus thymine kinase promoter (−105/+51) and a luciferase vector, and was a gift from Dr Kevin J. McCarthy (Louisiana State University Health Sciences Center in Shreveport). The plasmid pCMV-cyclinD1, containing cyclin D1 cDNA, was a gift from Dr Richard G. Pestell (Department of Oncology, Lombardi Cancer Center, Georgetown University, Washington, DC, U.S.A.). The PPARγ expression plasmid pPPARγcDNA was a gift from Dr Reed Graves (Department of Medicine, University of Chicago, Chicago, IL, U.S.A.). The plasmid p3TP-Lux is a TGF-β-inducible luciferase reporter, containing the plasminogen activator inhibitor-1 gene promoter, and was kindly provided by Dr Joan Massague (Memorial Sloan-Kettering Cancer Center, New York NY, U.S.A.). The luciferase reporter plasmid Tβ-RI (pES1.0), a gift from Dr Michael Centrella (Yale University, New Haven, CT, U.S.A.), contains 965 bp of the gene promoter of Tβ-RI [28]. The luciferase reporter plasmid pTβ-RII was generously provided by Dr Seong-Jin Kim (Laboratory of Chemoprevention, NCI, NIH, Bethesda, MD, U.S.A.), and contains 1670 bp of the gene promoter of Tβ-RII [29].
Semi-confluent cells in six-well plastic plates were transiently transfected using the LipofectAMINE® reagent (Life Technologies, Grand Island, NY, U.S.A.). A total of 3.0–4.5 μg of DNA was added to each well. Luciferase assays were performed as described previously [26]. Transfection efficiency was determined by co-transfection of a β-galactosidase reporter, pSV-β-gal (0.5 μg/well) (Promega). β-Galactosidase activities were measured using a chemiluminescence assay kit (Tropix, Bedford, MA, U.S.A.), according to the manufacturer's instructions. Each treatment was performed in triplicate. Results from three independent experiments were combined.
TGF-β1 immunoassays
These assays were performed as described previously [5]. To evaluate the effects of curcumin on the release and/or activation of latent TGF-β1, passaged HSC were treated with curcumin at the indicated concentrations for 24 h. Conditioned media were collected and centrifuged at ∼5600 g for 10 min at 4 °C. The supernatants were analysed for the total amount of TGF-β1 and the active form of TGF-β1 using the TGF-β1 Emax ImmunoAssay System (Promega), following the protocol provided by the manufacturer. This system was designed for the sensitive and specific detection of biologically active TGF-β1. The antibodies in it recognize only the active form of TGF-β1. To determine the total amount of TGF-β1 in the media, samples were pretreated with 1 M HCl for 15 min, followed by neutralization with 1 M NaOH. This procedure converted all latent TGF-β1 into the active form.
Statistical analysis
Differences between means were evaluated using an unpaired two-sided Student's t test, with P<0.05 considered significant. Where appropriate, comparisons of multiple treatment conditions with controls were analysed by ANOVA with Dunnett's test for post hoc analysis.
RESULTS
Activation of PPARγ mediates the suppression by curcumin of cyclin D1 expression, which, in turn, facilitates the activation of PPARγ
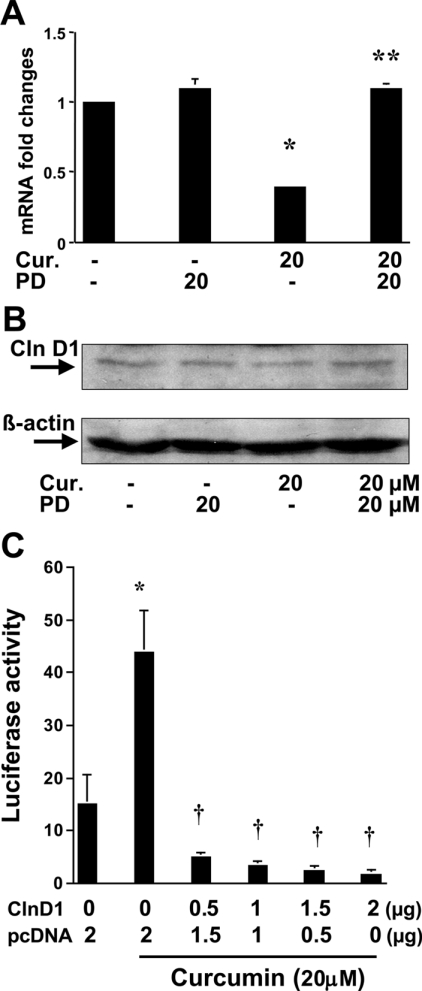
We demonstrated previously that expression of cyclin D1 was significantly inhibited by curcumin in activated HSC [25]. In addition, curcumin induced the gene expression of PPARγ and stimulated its activity in activated HSC [25]. The latter was required in order for curcumin to inhibit HSC proliferation. It was hypothesized that activation of PPARγ might mediate the suppression of cyclin D1 expression by curcumin. To test this hypothesis, preconfluent HSC were pretreated with or without the PPARγ antagonist PD 68235 (20 μM) for 30 min prior to the addition of curcumin at 20 μM for an additional 24 h. Total RNA or whole-cell protein extracts were prepared for real-time PCR (Figure 1A) and Western blotting analyses (Figure 1B) respectively. The results indicated that the expression of cyclin D1 was significantly inhibited at both the transcriptional and translational levels by curcumin (Figures 1A and 1B). Pretreatment of cells with the PPARγ antagonist PD 68235 abrogated this inhibitory effect, suggesting that the activation of PPARγ might be required for curcumin to inhibit the gene expression of cyclin D1 in passaged HSC.
Figure 1. Activation of PPARγ mediates curcumin suppression of cyclin D1 expression in passaged HSC, which, in turn, facilitates the activation of PPARγ.
Passaged HSC were pretreated with or without the PPARγ antagonist PD 68235 (20 μM) for 30 min prior to the addition of curcumin (Cur.) at 20 μM for an additional 24 h. Total RNA and protein extracts were prepared for real-time PCR (A) (n=3) and Western blotting analyses (B) (n=3) respectively. Significance: *P<0.05 compared with cells without curcumin; **P<0.05 compared with cells with curcumin. (C) To assess the effects of cyclin D1 (ClnD1) on PPARγ activity, HSC in 6-well culture plates were co-transfected with a total of 4.5 μg of plasmid DNA per well, including 2 μg of pPPRE-TK-Luc, 0.5 μg of pSV-β gal, pCMV-cyclinD1 at the indicated doses and an empty vector. The amount of DNA in pCMV-cyclinD1 plus the empty vector was 2 μg. After recovery, cells were treated with or without curcumin at 20 μM for 36 h. Luciferase activities were expressed as relative units after β-galactosidase normalization (n=6). Significance: *P<0.05 compared with cells without curcumin; †P<0.05 compared with cells transfected with no pCMV-cyclinD1, with curcumin treatment (second column).
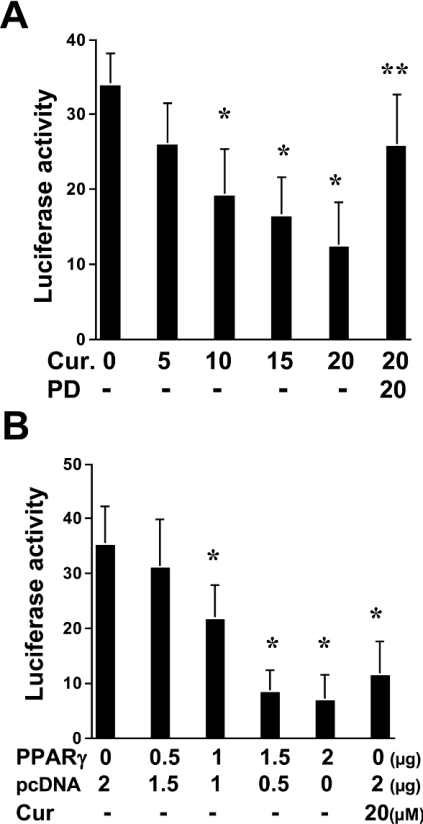
Other previous studies have indicated that the expression of cyclin D1 inhibits ligand-induced PPARγ activation [30]. We therefore further hypothesized that, in addition to the induction of PPARγ gene expression, inhibition of cyclin D1 expression by curcumin might facilitate the activation of the receptor. To study this hypothesis, preconfluent HSC in 6-well culture plates were co-transfected with a total of 4.5 μg of plasmid DNA per well, including 2 μg of the PPARγ luciferase reporter plasmid pPPRE-TK-Luc, 0.5 μg of pSV-β-gal, pCMV-cyclinD1 at the indicated doses and an empty vector. The amount of DNA in pCMV-cyclinD1 plus the empty vector was equal to 2 μg. After transfection, cells were treated with or without curcumin at 20 μM for 36 h. As shown in Figure 1(C), curcumin, as expected, dramatically increased PPARγ activity, demonstrated by an increase in luciferase activity. Alongside the increase in the amount of pCMV-cyclinD1, luciferase activity induced by curcumin was gradually diminished (Figure 1C), suggesting that high-level expression of cyclin D1 in activated HSC might inhibit endogenous PPARγ activity. Taken together, these results indicated that activation of PPARγ by curcumin might be required for the suppression of cyclin D1 gene expression, which might, in turn, facilitate PPARγ activation in activated HSC, since high expression of cyclin D1 inhibits expression of the receptor and its activity.
Blocking the activation of PPARγ abrogates the pro-apoptotic effect of curcumin on activated HSC
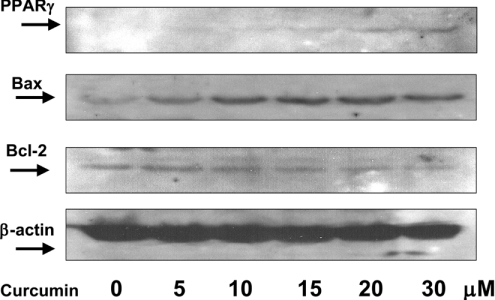
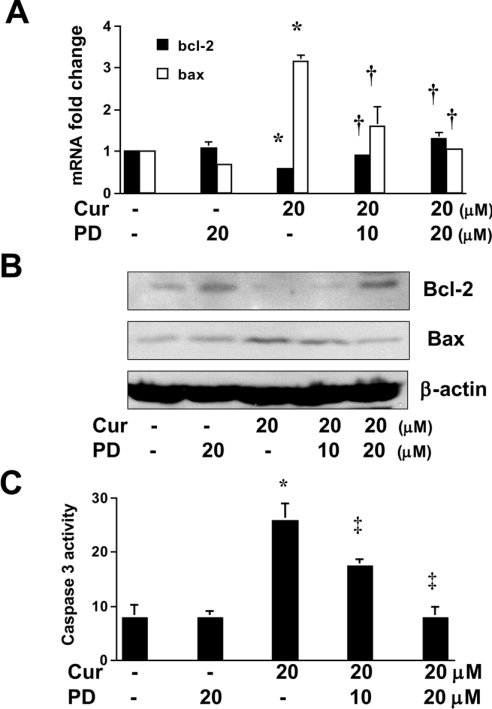
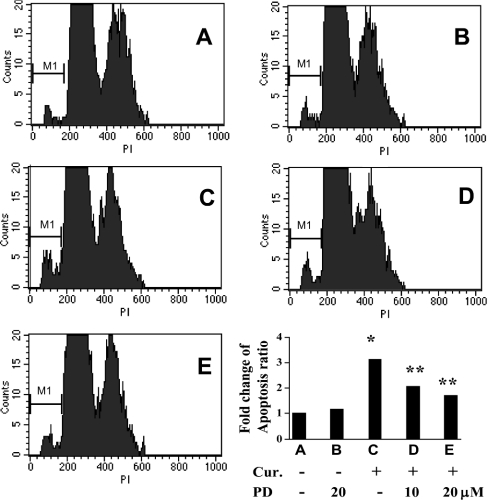
We demonstrated previously that curcumin induced the apoptosis of activated HSC in vitro [25]. As shown in Figure 2, curcumin caused a dose-dependent increase in the abundance of PPARγ, and had opposite effects on the abundance of a pair of apoptosis-related proteins, i.e. an increase in the abundance of pro-apoptotic Bax and a reduction in the level of anti-apoptotic Bcl-2. It was hypothesized that the induction of apoptosis of activated HSC by curcumin might be mediated by activation of PPARγ. To test the hypothesis, passaged HSC were pretreated with or without the specific PPARγ antagonist PD 68235 (10 or 20 μM) for 30 min prior to the addition of curcumin (20 μM) for an additional 24 h. Total RNA or whole-cell extracts were prepared for real-time PCR, Western blotting analyses or caspase 3 activity assays. As expected, curcumin increased the gene expression of pro-apoptotic Bax and suppressed the expression of anti-apoptotic Bcl-2 in activated HSC, as demonstrated by real-time PCR (Figure 3A) and Western blotting analyses (Figure 3B). Pretreatment of cells with PD 68235 eliminated the effects of curcumin on the expression of these genes. PD 68235 (20 μM) itself had little effect, if any. Additional experiments demonstrated that blocking PPARγ activation by PD 68235 attenuated the ability of curcumin to increase caspase 3 activity (Figure 3C). Further flow cytometric assays confirmed the necessity of PPARγ activation for the induction of apoptosis of activated HSC by curcumin (Figure 4). Compared with the control (Figure 4A), curcumin induced the apoptosis of passaged HSC by 3.2-fold (Figure 4C). Blocking PPARγ activation by pretreatment of cells with PD 68235 caused a dose-dependent reduction in the rate of apoptosis (Figures 4D and 4E). PD 68235 itself had no apparent effect on the cell cycle or apoptosis (Figure 4B). TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling) staining of these cells gave similar results (not shown). Taken together, these results demonstrated that blocking the activation of PPARγ abrogated the effect of curcumin on the induction of HSC apoptosis in vitro, suggesting the necessity of PPARγ activation for this action.
Figure 2. Curcumin dose-dependently increases the abundance of PPARγ and pro-apoptotic Bax and decreases that of anti-apoptotic Bcl-2 in passaged HSC.
Passaged HSC were treated with curcumin at indicated concentrations for 24 h. Whole-cell protein extracts were prepared for Western blotting analyses (n=3). β-Actin was an internal control for equal loading.
Figure 3. Blocking PPARγ activation abrogates the pro-apoptotic effects of curcumin on activated HSC in vitro.
Passaged HSC were pretreated or not with the specific PPARγ antagonist PD 68235 (10 or 20 μM) for 30 min prior to the addition of curcumin (Cur.; 20 μM) for an additional 24 h. Total RNA or whole-cell protein extracts were prepared. (A) Real-time PCR. GAPDH was used as an invariant control for calculating fold changes in mRNA levels (n=3). Values are expressed as means±S.D. Significance: *P<0.05 compared with cells with no treatment; †P<0.05 compared with cells treated only with curcumin. (B) Western blotting analyses. β-Actin was an internal control for equal loading (n=3). (C) Caspase 3 activity assays. Values are expressed as means±S.D. (n=3 independent experiments). Significance: *P<0.05 compared with cells with no treatment; ‡P<0.05 compared with cells treated only with curcumin.
Figure 4. PPARγ activation by curcumin is required for induction of HSC apoptosis.
Passaged HSC were pretreated or not with the specific PPARγ antagonist PD 68235 for 30 min prior to the addition of curcumin (Cur.; 20 μM) for an additional 24 h. HSC (≥1×106 cells/sample) were fixed with ethanol and then labelled with propidium iodide (PI) for flow cytometric analyses (n=3). Results representative of three independent experiments are shown. Values are expressed as means±S.D. Significance: *P<0.05 compared with cells with no treatment; **P<0.05 compared with cells treated only with curcumin.
PPARγ activation mediates the inhibitory effect of curcumin on the expression of ECM genes
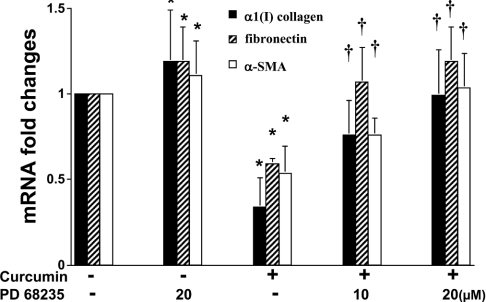
We demonstrated previously that curcumin significantly suppressed the expression of ECM genes, including those encoding α1(I) collagen and fibronectin in passaged HSC [25]. The underlying mechanisms are largely to be defined. Other previous reports indicated that activation of PPARγ by its agonists apparently inhibited α1(I) collagen gene expression in activated HSC in vitro and in vivo [16]. These results prompted us to propose that activation of PPARγ by curcumin might be involved in its inhibitory effect on ECM gene expression in activated HSC. Accordingly, passaged HSC were pretreated with or without PD 68235 (10 or 20 μM) for 30 min prior to the addition of curcumin (20 μM) for an additional 24 h. Real-time PCR indicated that, as expected, curcumin significantly reduced the steady-state mRNA levels of α1(I) collagen, fibronectin and α-SMA, a marker of activated HSC (Figure 5). Pretreatment of cells with the PPARγ antagonist PD 68235 abrogated, in a dose-dependent manner, the inhibitory effect of curcumin on the expression of these genes, suggesting that PPARγ activation might mediate the inhibitory effect of curcumin on the expression of ECM genes in activated HSC.
Figure 5. PPARγ activation mediates the inhibitory effects of curcumin on the gene expression of α1(I) collagen, fibronectin and α-SMA.
Passaged HSC were pretreated or not with the specific PPARγ antagonist PD 68235 (10 or 20 μM) for 30 min prior to the addition of curcumin (20 μM) for an additional 24 h. Total RNA was prepared for real-time PCR (n=3). Fold changes in mRNA levels were calculated by using GAPDH as an invariant control. Values are expressed as means±S.D. Significance: *P<0.05 compared with cells with no treatment; †P<0.05 compared with cells treated only with curcumin.
Activation of PPARγ by curcumin blocks TGF-β1 signalling in activated HSC
TGF-β is the most fibrogenic factor in stimulating ECM production in HSC [31]. It is, therefore, hypothesized that curcumin might inhibit the expression of ECM genes in activated HSC by blocking TGF-β signalling. To test this hypothesis, passaged HSC were transiently transfected with the TGF-β-inducible luciferase reporter plasmid p3TP-Lux, containing the plasminogen activator inhibitor-1 gene promoter. Cells were then treated with curcumin at the indicated concentrations. Curcumin reduced luciferase activity in these cells in a dose-dependent manner, suggesting that the phytochemical might block TGF-β signalling (Figure 6A). To determine further whether PPARγ is involved in blockade of TGF-β signalling, transfected cells were pretreated with the PPARγ antagonist PD 68235 (20 μM) prior to the addition of curcumin. This abrogated the inhibitory effect of curcumin on the TGF-β signalling pathway (Figure 6A). To verify the inhibitory role of PPARγ in blocking the TGF-β signal pathway, HSC were co-transfected with p3TP-Lux and pPPARγcDNA, containing PPARγ cDNA in a CMV (cytomegalovirus)-driven expression vector. Pilot experiments showed that forced expression of PPARγ cDNA resulted in an increase in PPARγ transcriptional activity in HSC co-transfected with pPPRE-TK-Luc and pPPARγ cDNA (results not shown). The increase in the amount of the PPARγ expression plasmid led to an apparent reduction in luciferase activity in these cells (Figure 6B). This indicated that the increase in the expression of PPARγ interrupted TGF-β signalling, which mimicked the role of curcumin in blocking TGF-β signalling in activated HSC. Taken together, our results demonstrated that activation of PPARγ by curcumin blocked TGF-β signalling in activated HSC, which might lead to inhibition of ECM gene expression. PPARγ agonists are presumed to exist in media containing 10% FBS [17,32].
Figure 6. Activation of PPARγ by curcumin blocks TGF-β signalling in activated HSC.
(A) Passaged HSC were transiently transfected with the TGF-β-inducible luciferase reporter plasmid p3TP-Lux. Cells were then pretreated with or without PD 68235 (20 μM) prior to the addition of curcumin (Cur.) at the indicated concentrations for an additional 36 h. Luciferase activities are expressed in relative units after β-galactosidase normalization (n=6). Significance: *P<0.05 compared with cells without curcumin; **P<0.05 compared with cells treated with 20 μM curcumin. (B) HSC in 6-well culture plates were co-transfected with a total of 4.5 μg of plasmid DNA per well, including 2 μg of p3TP-Lux, 0.5 μg of pSV-β gal, pPPARγcDNA at the indicated doses and an empty vector. The amount of DNA in pPPARγcDNA plus the empty vector was 2 μg. Cells were then treated with or without 20 μM curcumin for 36 h. Luciferase activities are expressed in relative units after β-galactosidase normalization (n=6). Significance: *P<0.05 compared with cells transfected with no pPPARγ cDNA and not treated with curcumin.
Curcumin has no effect on the release and/or activation of latent TGF-β1 in activated HSC
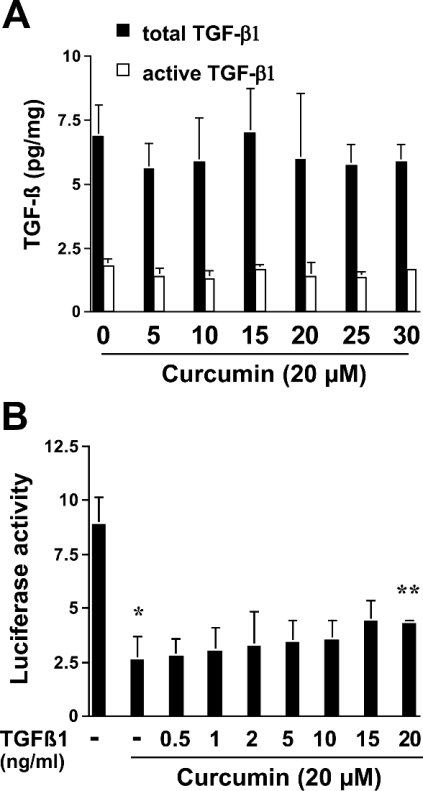
TGF-β1 is synthesized and secreted in a latent, biologically inactive form, which must be activated before being able to bind to TGF-β receptors [33]. TGF-β signalling is initiated by binding of active TGF-β1 to Tβ-RII, which leads to the phosphorylation and activation of Tβ-RI [34]. The latter phosphorylates Smad 2/3 proteins, which subsequently form a complex with Smad 4 and migrate into the nucleus to regulate the expression of target genes, including that encoding α1(I) collagen [35,36]. To elucidate the mechanisms underlying curcumin blockade of the TGF-β signalling pathway, it was initially hypothesized that curcumin might inhibit the release and/or activation of latent TGF-β1 in activated HSC. TGF-β1 immunoassays of passaged HSC-conditioned media demonstrated that curcumin at the indicated concentrations caused no significant change in the amount of total TGF-β1 or active TGF-β1 (Figure 7A). Additional assays of HSC transfected with the TGF-β-inducible plasmid p3TP-Lux demonstrated that exogenous active TGF-β1 did not significantly eliminate the inhibitory effect of curcumin on TGF-β signalling, except at very high doses (Figure 7B). Taken together, these results did not support our initial hypothesis, and indicated that curcumin has no effect on the release and/or activation of latent TGF-β1 in activated HSC.
Figure 7. Curcumin has no effect on the release and/or activation of latent TGF-β1 in activated HSC.
(A) Passaged HSC were treated with curcumin at the indicated concentrations for 24 h. Conditioned media were collected and analysed for the amounts of total TGF-β1 and the active form of TGF-β1 by the TGF-β1 Emax ImmunoAssay System (ELISA). Values are expressed as means±S.D. (n=6). (B) Passaged HSC were transfected with the TGF-β-inducible plasmid p3TP-Lux. Cells were then pretreated with 20 μM curcumin for 24 h prior to the addition of exogenous active TGF-β1 for an additional 24 h. Luciferase activities are expressed as relative units after β-galactosidase normalization (n=6). Significance: *P<0.05 compared with cells with no treatment; **P<0.05 compared with cells treated with curcumin but without exogenous TGF-β1.
Activation of PPARγ by curcumin results in inhibition of TGF-β receptor gene expression
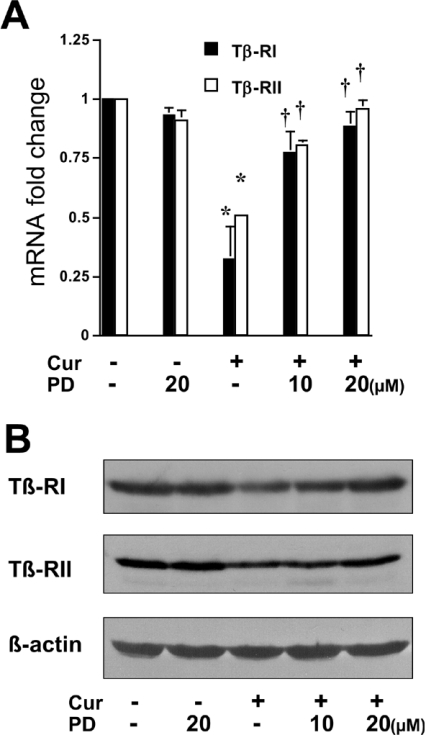
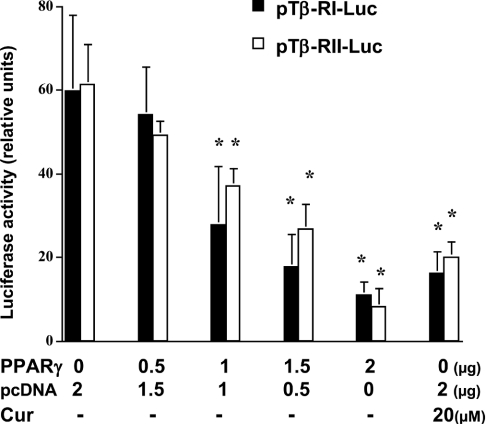
To explore the mechanisms by which activation of PPARγ by curcumin blocks TGF-β signalling in activated HSC, it was then hypothesized that curcumin might suppress the gene expression of TGF-β receptors in activated HSC, which might be mediated by activation of PPARγ. To test these hypotheses, passaged HSC were treated with or without 20 μM curcumin for 24 h. Curcumin significantly reduced the levels of Tβ-RI and Tβ-RII mRNA and proteins in activated HSC, as demonstrated by real-time PCR and Western blotting analyses respectively (Figure 8). Pretreatment of cells with the PPARγ antagonist eliminated these inhibitory effects. To verify the inhibitory effect of PPARγ activation on the expression of the Tβ-RI and Tβ-RII genes, passaged HSC were co-transfected with the pPPARγcDNA and the TGF-β receptor luciferase reporter pTβ-RI or pTβ-RII. The last two plasmids contain a portion of the promoter of the Tβ-RI or Tβ-RII genes respectively (see the Materials and methods section). Luciferase assays demonstrated that forced expression of exogenous PPARγ cDNA mimicked the role of curcumin in reducing luciferase activity in a dose-dependent manner (Figure 9), suggesting an inhibitory effect of PPARγ on the expression of Tβ-RI or Tβ-RII genes in activated HSC in vitro. Taken together, these results indicated that curcumin might inhibit the expression of TGF-β receptors in activated HSC, and that activation of PPARγ by curcumin might be required for this action. These results supported our hypothesis that curcumin blocks the TGF-β signalling pathway by inhibiting the gene expression of TGF-β receptors mediated by activation of PPARγ.
Figure 8. Curcumin inhibits the expression of TGF-β receptor genes in passaged HSC, which requires the activation of PPARγ.
Passaged HSC were pretreated with or without the specific PPARγ antagonist PD 68235 (10 or 20 μM) for 30 min prior to the addition of curcumin (Cur.; 20 μM) for an additional 24 h. Total RNA and whole-cell protein extracts were prepared. (A) Real-time PCR of the TGF-β receptors Tβ-RI and Tβ-RII. GAPDH was used as an invariant control for calculating fold changes in mRNA levels (n=3). Values are expressed as means±S.D. Significance: *P<0.05 compared with cells with no treatment; †P<0.05 compared with cells treated only with curcumin. (B) Western blotting analyses of Tβ-RI and Tβ-RII. β-Actin was an internal control for equal loading (n=3).
Figure 9. Forced expression of PPARγ mimics the inhibitory effect of curcumin on the expression of TGF-β receptor genes in passaged HSC.
Passaged HSC were co-transfected with the pPPARγcDNA and a TGF-β receptor luciferase reporter plasmid (pTβ-RI or pTβ-RII). HSC in 6-well culture plates were transfected with a total of 4.5 μg of plasmid DNA per well, including 2 μg of the TGF-β receptor luciferase reporter pTβ-RI or pTβ-RII, 0.5 μg of pSV-β gal, pPPARγcDNA at the indicated doses and an empty vector. The amount of DNA in pPPARγcDNA plus the empty vector was 2 μg. Cells were then treated with or without 20 μM curcumin (Cur) for 36 h. Luciferase activities are expressed as relative units after β-galactosidase normalization (n=6). Significance: *P<0.05 compared with cells not transfected with pPPARγ cDNA and not treated with curcumin.
DISCUSSION
Endogenous PPARγ is highly expressed in quiescent HSC and is functionally active [13–15]. However, the abundance of PPARγ protein is dramatically reduced in activated HSC in vitro and in vivo [13–15]. Activation of PPARγ transcriptional activity by its specific agonists inhibits HSC activation in vivo and in vitro [13,15]. PPARγ has been implicated as a repressor for maintaining HSC in the quiescent state. The adenoviral vector-mediated expression of PPARγ itself is sufficient to reverse the morphology of activated HSC to the quiescent phenotype [17]. We recently reported that curcumin inhibited HSC activation in vitro by inducing cell cycle arrest and apoptosis, and suppressing ECM gene expression [25]. Further studies indicated that curcumin increased PPARγ activity by inducing PPARγ gene expression in activated HSC in vitro. In addition, we demonstrated the requirement for receptor activation in the inhibition of cell proliferation of activated HSC in vitro [25], which partially explained the inhibitory effect of curcumin on the cell growth of HSC. In the present study, we further explored the mechanisms by which curcumin inhibits HSC activation in vitro. It was found that PPARγ activation was required not only for inhibiting cell proliferation, but also for inducing apoptosis and suppressing ECM gene expression.
The D-group cyclin proteins play critical roles in the progression of cells through the G1 phase of the cell cycle. Our previous study demonstrated that curcumin significantly inhibited the expression of cyclin D1 in activated HSC [25], which might make an important contribution to the induction of cell cycle arrest and inhibition of cell proliferation. The inhibitory effect of curcumin on cyclin D1 has been observed in many other cell types [37–39]. Our present experiments demonstrated that activation of PPARγ mediated the inhibition by curcumin of cyclin D1 gene expression in activated HSC. This result is supported by other reports that the PPARγ agonist prostaglandin J2 inhibited the expression of cyclin D1 [40]. On the other hand, our additional experiments suggested that high expression of cyclin D1 in activated HSC might inhibit endogenous PPARγ activity. Co-transfection assays demonstrated that that forced expression of cyclin D1 ameliorated, in a dose-dependent manner, the stimulatory effect of curcumin on PPARγ transcriptional activity in passaged HSC (Figure 1). This was consistent with a recent observation that cyclin D1 repressed PPARγ gene expression and its transcriptional activity in vitro and in vivo [30]. Our results collectively indicate that activation of PPARγ by curcumin might be required for the suppression of cyclin D1 gene expression in activated HSC. In addition, inhibition of cyclin D1 might, in turn, bring relief to PPARγ activity from suppression and facilitate PPARγ activation in activated HSC, since high expression of cyclin D1 inhibits PPARγ activity.
Activation of PPARγ has been demonstrated to exert an inhibitory effect on cell growth in most cell types studied. For example, PPARγ activation is associated with growth inhibition and increased levels of markers of cellular differentiation in cultured colon cancer cells [41]. This nuclear receptor often functions as a tumour suppressor [42]. Studies have shown that activation of PPARγ is associated with the expression of Bcl-2 and NF-κB in human colon cancer [43]. Ligand activation of PPARγ resulted in apoptosis, accompanied by reduced levels of NF-κB and Bcl-2. Overexpression of Bcl-2 significantly protected the cells from apoptosis, suggesting that a PPARγ/Bcl-2 feedback loop may control the life/death continuum [43]. We demonstrated previously that curcumin inhibited NF-κB activity and reduced the abundance of anti-apoptotic Bcl-2 in activated HSC [25]. We demonstrate further here that inhibition of Bcl-2 by curcumin required PPARγ activation. However, how Bcl-2 is negatively regulated by PPARγ activation remains largely unknown. A recent study reported an interesting observation that the human bcl-2 gene contains a functional PPRE in the 3′ untranslated region [44]. Additional studies are necessary to elucidate the mechanisms by which activation of PPARγ mediates the inhibitory effects of curcumin on the expression of Bcl-2 in activated HSC.
Activation of TGF-β signalling has been implicated as a key event in up-regulating ECM gene expression and stimulating HSC activation in vitro and in vivo [3,45,46]. Accumulating evidence indicates that activation of PPARγ disrupts the TGF-β signalling pathway and inhibits its pro-fibrotic effect [47,48]. However, the underlying mechanisms remain largely elusive. Our present study demonstrated that curcumin apparently inhibits the activity of TGF-β signalling in passaged HSC. Further experiments suggested that the inhibitory effect might be PPARγ-activation-dependent. To elucidate the underlying mechanisms, we investigated the effects of curcumin on the ligand TGF-β1 and its receptors in passaged HSC. TGF-β1 is synthesized and secreted in a latent, biologically inactive form, which must be activated before being able to bind to TGF-β receptors [33]. TGF-β1 immunoassays (Figure 7A) did not show a significant effect of curcumin on the amounts of total or active TGF-β1 in the conditioned media. Treatment of cells with exogenous active TGF-β1 did not eliminate the inhibitory effect of curcumin on TGF-β signalling (Figure 7B). These results did not support our initial assumption that curcumin might reduce the release and/or activation of latent TGF-β1 in activated HSC. Additional experiments demonstrated that curcumin suppressed the expression of the two TGF-β receptors Tβ-RI and Tβ-RII. Pretreatment of cells with the PPARγ antagonist eliminated the inhibitory effect. Forced expression of exogenous PPARγ mimicked the effect of curcumin, inhibiting the promoter activity of the TGF-β receptors in passaged HSC. These results collectively indicated that activation of PPARγ by curcumin interrupts the TGF-β signalling pathway in activated HSC possibly by inhibiting the gene expression of TGF-β receptors. PPARγ has been shown previously to be capable of negatively regulating gene transcription [49–51]. PPARγ interfered with Smad 3, an inter-mediator in the TGF-β signalling pathway, and inhibited TGF-β-induced gene expression of connective tissue growth factor, resulting in inhibition of α1(I) collagen gene expression [52]. Other studies have observed that activation of TGF-β signalling shows bi-phasic effects on regulating PPARγ gene expression, i.e. early stimulation and late repression [53]. The complex relationship between PPARγ and the TGF-β signalling pathway is far from understood. Additional experiments are necessary to elucidate the mechanisms by which PPARγ activation regulates the expression of TGF-β receptors as well as possible additional interactions between the signalling pathways of PPARγ and TGF-β in the regulation of ECM gene expression.
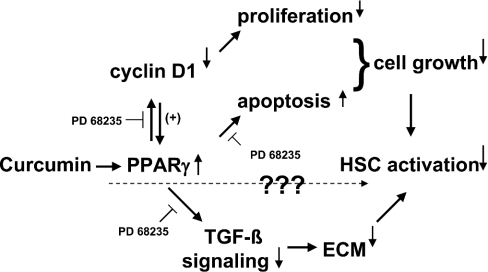
Based on our previous and present observations, a model has been proposed to explain the inhibitory effects of curcumin on HSC activation, including cell growth and ECM production (Figure 10). Curcumin induces the gene expression of PPARγ and its activation in activated HSC, which is required for inhibition of cell growth via the induction of cell cycle arrest and apoptosis. PPARγ activation mediates curcumin inhibition of cyclin D1 expression in activated HSC, which, in turn, facilitates the expression and activation of PPARγ. In addition, curcumin suppresses, in a PPARγ-activation-dependent manner, the expression of ECM genes, including those encoding α1(I) collagen and fibronectin, possibly by interrupting the TGF-β signalling pathway, including inhibition of the expression of TGF-β receptors. Taken together, activation of PPARγ is required for curcumin to inhibit HSC activation, including inhibiting cell proliferation, inducing cell apoptosis and suppressing ECM expression. It should be emphasized, however, that our results and this model do not exclude the possibility that other mechanisms are involved in the inhibition of HSC activation by curcumin.
Figure 10. Possible mechanisms underlying the inhibition of HSC activation by curcumin.
See the text for details.
Acknowledgments
This work was supported by grant DK 47995 from NIH/NIDDK to A. C., and starting funds from the Department of Pathology, LSUHSC-S. Thanks are due to Dr Qian-Jing Zhang, Department of Cellular Biology & Anatomy, LSUHSC-S) for his technical support in FACS.
References
- 1.Friedman S. L. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J. Biol. Chem. 2000;275:2247–2250. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- 2.Schuppan D., Atkinson J., Ruehl M., Riecken E. O. Alcohol and liver fibrosis – pathobiochemistry and treatment. Z. Gastroenterol. 1995;33:546–550. [PubMed] [Google Scholar]
- 3.Dooley S., Delvoux B., Lahme B., Mangasser-Stephan K., Gressner A. M. Modulation of transforming growth factor beta response and signaling during transdifferentiation of rat hepatic stellate cells to myofibroblasts. Hepatology. 2000;31:1094–1106. doi: 10.1053/he.2000.6126. [DOI] [PubMed] [Google Scholar]
- 4.Friedman S. L., Yamasaki G., Wong L. Modulation of transforming growth factor beta receptors of rat lipocytes during the hepatic wound healing response. Enhanced binding and reduced gene expression accompany cellular activation in culture and in vivo. J. Biol. Chem. 1994;269:10551–10558. [PubMed] [Google Scholar]
- 5.Chen A. Acetaldehyde stimulates the activation of latent transforming growth factor-beta1 and induces expression of the type II receptor of the cytokine in rat cultured hepatic stellate cells. Biochem. J. 2002;368:683–693. doi: 10.1042/BJ20020949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura T., Sakata R., Ueno T., Sata M., Ueno H. Inhibition of transforming growth factor beta prevents progression of liver fibrosis and enhances hepatocyte regeneration in dimethylnitrosamine-treated rats. Hepatology. 2000;32:247–255. doi: 10.1053/jhep.2000.9109. [DOI] [PubMed] [Google Scholar]
- 7.Qi Z., Atsuchi N., Ooshima A., Takeshita A., Ueno H. Blockade of type beta transforming growth factor signaling prevents liver fibrosis and dysfunction in the rat. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2345–2349. doi: 10.1073/pnas.96.5.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ueno H., Sakamoto T., Nakamura T., Qi Z., Astuchi N., Takeshita A., Shimizu K., Ohashi H. A soluble transforming growth factor beta receptor expressed in muscle prevents liver fibrogenesis and dysfunction in rats. Hum. Gene Ther. 2000;11:33–42. doi: 10.1089/10430340050016139. [DOI] [PubMed] [Google Scholar]
- 9.Iredale J. P., Benyon R. C., Pickering J., McCullen M., Northrop M., Pawley S., Hovell C., Arthur M. J. Mechanisms of spontaneous resolution of rat liver fibrosis. Hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J. Clin. Invest. 1998;102:538–549. doi: 10.1172/JCI1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houseknecht K. L., Cole B. M., Steele P. J. Peroxisome proliferator-activated receptor gamma (PPARgamma) and its ligands: a review. Domest. Anim. Endocrinol. 2002;22:1–23. doi: 10.1016/s0739-7240(01)00117-5. [DOI] [PubMed] [Google Scholar]
- 11.Takashima T., Fujiwara Y., Higuchi K., Arakawa T., Yano Y., Hasuma T., Otani S. PPAR-gamma ligands inhibit growth of human esophageal adenocarcinoma cells through induction of apoptosis, cell cycle arrest and reduction of ornithine decarboxylase activity. Int. J. Oncol. 2001;19:465–471. [PubMed] [Google Scholar]
- 12.Tontonoz P., Singer S., Forman B. M., Sarraf P., Fletcher J. A., Fletcher C. D., Brun R. P., Mueller E., Altiok S., Oppenheim H., et al. Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor gamma and the retinoid X receptor. Proc. Natl. Acad. Sci. U.S.A. 1997;94:237–241. doi: 10.1073/pnas.94.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galli A., Crabb D., Price D., Ceni E., Salzano R., Surrenti C., Casini A. Peroxisome proliferator-activated receptor gamma transcriptional regulation is involved in platelet-derived growth factor-induced proliferation of human hepatic stellate cells. Hepatology. 2000;31:101–108. doi: 10.1002/hep.510310117. [DOI] [PubMed] [Google Scholar]
- 14.Miyahara T., Schrum L., Rippe R., Xiong S., Yee H. F., Jr, Motomura K., Anania F. A., Willson T. M., Tsukamoto H. Peroxisome proliferator-activated receptors and hepatic stellate cell activation. J. Biol. Chem. 2000;275:35715–35722. doi: 10.1074/jbc.M006577200. [DOI] [PubMed] [Google Scholar]
- 15.Marra F., Efsen E., Romanelli R. G., Caligiuri A., Pastacaldi S., Batignani G., Bonacchi A., Caporale R., Laffi G., Pinzani M., Gentilini P. Ligands of peroxisome proliferator-activated receptor gamma modulate profibrogenic and proinflammatory actions in hepatic stellate cells. Gastroenterology. 2000;119:466–478. doi: 10.1053/gast.2000.9365. [DOI] [PubMed] [Google Scholar]
- 16.Galli A., Crabb D. W., Ceni E., Salzano R., Mello T., Svegliati-Baroni G., Ridolfi F., Trozzi L., Surrenti C., Casini A. Antidiabetic thiazolidinediones inhibit collagen synthesis and hepatic stellate cell activation in vivo and in vitro. Gastroenterology. 2002;122:1924–1940. doi: 10.1053/gast.2002.33666. [DOI] [PubMed] [Google Scholar]
- 17.Hazra S., Xiong S., Wang J., Rippe R., Chatterjee V. K., Tsukamoto H. PPARgamma induces a phenotypic switch from activated to quiescent hepatic stellate cells. J. Biol. Chem. 2003;279:11392–11401. doi: 10.1074/jbc.M310284200. [DOI] [PubMed] [Google Scholar]
- 18.Ruby A. J., Kuttan G., Babu K. D., Rajasekharan K. N., Kuttan R. Anti-tumour and antioxidant activity of natural curcuminoids. Cancer Lett. 1995;94:79–83. doi: 10.1016/0304-3835(95)03827-j. [DOI] [PubMed] [Google Scholar]
- 19.Sreejayan R. M. N. Curcuminoids as potent inhibitors of lipid peroxidation. J. Pharm. Pharmacol. 1994;46:1013–1016. doi: 10.1111/j.2042-7158.1994.tb03258.x. [DOI] [PubMed] [Google Scholar]
- 20.Brouet I., Ohshima H. Curcumin, an anti-tumour promoter and anti-inflammatory agent, inhibits induction of nitric oxide synthase in activated macrophages. Biochem. Biophys. Res. Commun. 1995;206:533–540. doi: 10.1006/bbrc.1995.1076. [DOI] [PubMed] [Google Scholar]
- 21.Joe B., Lokesh B. R. Role of capsaicin, curcumin and dietary n-3 fatty acids in lowering the generation of reactive oxygen species in rat peritoneal macrophages. Biochim. Biophys. Acta. 1994;1224:255–263. doi: 10.1016/0167-4889(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 22.Liu J. Y., Lin S. J., Lin J. K. Inhibitory effects of curcumin on protein kinase C activity induced by 12-O-tetradecanoyl-phorbol-13-acetate in NIH 3T3 cells. Carcinogenesis. 1993;14:857–861. doi: 10.1093/carcin/14.5.857. [DOI] [PubMed] [Google Scholar]
- 23.Singh S., Aggarwal B. B. Activation of transcription factor NF-kappa B is suppressed by curcumin (diferuloylmethane) J. Biol. Chem. 1995;270:24995–25000. doi: 10.1074/jbc.270.42.24995. [DOI] [PubMed] [Google Scholar]
- 24.Chuang S. E., Cheng A. L., Lin J. K., Kuo M. L. Inhibition by curcumin of diethylnitrosamine-induced hepatic hyperplasia, inflammation, cellular gene products and cell-cycle-related proteins in rats. Food Chem. Toxicol. 2000;38:991–995. doi: 10.1016/s0278-6915(00)00101-0. [DOI] [PubMed] [Google Scholar]
- 25.Xu J., Fu Y., Chen A. Activation of peroxisome proliferator-activated receptor-gamma contributes to the inhibitory effects of curcumin on rat hepatic stellate cell growth. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;285:G20–G30. doi: 10.1152/ajpgi.00474.2002. [DOI] [PubMed] [Google Scholar]
- 26.Chen A., Beno D. W. A., Davis B. H. Suppression of stellate cell type I collagen gene expression involves AP-2 transmodulation of nuclear factor-1-dependent gene transcription. J. Biol. Chem. 1996;271:25994–25998. doi: 10.1074/jbc.271.42.25994. [DOI] [PubMed] [Google Scholar]
- 27.Camp H. S., Chaudhry A., Leff T. A novel potent antagonist of peroxisome proliferator-activated receptor gamma blocks adipocyte differentiation but does not revert the phenotype of terminally differentiated adipocytes. Endocrinology. 2001;142:3207–3213. doi: 10.1210/endo.142.7.8254. [DOI] [PubMed] [Google Scholar]
- 27a.Schmittgen T. D., Zakrajsek B. A., Mills A. G., Gorn V., Singer M. J., Reed M. W. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 28.Ji C., Casinghino S., McCarthy T. L., Centrella M. Cloning, characterization, and expression of the transforming growth factor-beta type I receptor promoter in fetal rat bone cells. J. Cell. Biochem. 1996;63:478–490. doi: 10.1002/(SICI)1097-4644(19961215)63:4%3C478::AID-JCB9%3E3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 29.Bae H. W., Geiser A. G., Kim D. H., Chung M. T., Burmester J. K., Sporn M. B., Roberts A. B., Kim S. J. Characterization of the promoter region of the human transforming growth factor-beta type II receptor gene. J. Biol. Chem. 1995;270:29460–29468. doi: 10.1074/jbc.270.49.29460. [DOI] [PubMed] [Google Scholar]
- 30.Wang C., Pattabiraman N., Zhou J. N., Fu M., Sakamaki T., Albanese C., Li Z., Wu K., Hulit J., Neumeister P., et al. Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Mol. Cell. Biol. 2003;23:6159–6173. doi: 10.1128/MCB.23.17.6159-6173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Bleser P. J., Niki T., Rogiers V., Geerts A. Transforming growth factor-beta gene expression in normal and fibrotic rat liver. J. Hepatol. 1997;26:886–893. doi: 10.1016/s0168-8278(97)80257-7. [DOI] [PubMed] [Google Scholar]
- 32.McIntyre T. M., Pontsler A. V., Silva A. R., St Hilaire A., Xu Y., Hinshaw J. C., Zimmerman G. A., Hama K., Aoki J., Arai H., Prestwich G. D. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc. Natl. Acad. Sci. U.S.A. 2003;100:131–136. doi: 10.1073/pnas.0135855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis P. A., Rifkin D. B. Cellular activation of latent transforming growth factor beta requires binding to the cation-independent mannose 6-phosphate/insulin-like growth factor type II receptor. Proc. Natl. Acad. Sci. U.S.A. 1991;88:580–584. doi: 10.1073/pnas.88.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massague J., Chen Y. G. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 35.Ignotz R. A., Massague J. Transforming growth factor-beta stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J. Biol. Chem. 1986;261:4337–4345. [PubMed] [Google Scholar]
- 36.Czaja M. J., Weiner F. R., Flanders K. C., Giambrone M. A., Wind R., Biempica L., Zern M. A. In vitro and in vivo association of transforming growth factor-beta 1 with hepatic fibrosis. J. Cell Biol. 1989;108:2477–2482. doi: 10.1083/jcb.108.6.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moragoda L., Jaszewski R., Majumdar A. P. Curcumin induced modulation of cell cycle and apoptosis in gastric and colon cancer cells. Anticancer Res. 2001;21:873–878. [PubMed] [Google Scholar]
- 38.Mukhopadhyay A., Banerjee S., Stafford L. J., Xia C., Liu M., Aggarwal B. B. Curcumin-induced suppression of cell proliferation correlates with down-regulation of cyclin D1 expression and CDK4-mediated retinoblastoma protein phosphorylation. Oncogene. 2002;21:8852–8861. doi: 10.1038/sj.onc.1206048. [DOI] [PubMed] [Google Scholar]
- 39.Shishodia S., Potdar P., Gairola C. G., Aggarwal B. B. Curcumin (diferuloylmethane) down-regulates cigarette smoke-induced NF-kappaB activation through inhibition of IkappaBalpha kinase in human lung epithelial cells: correlation with suppression of COX-2, MMP-9 and cyclin D1. Carcinogenesis. 2003;24:1269–1279. doi: 10.1093/carcin/bgg078. [DOI] [PubMed] [Google Scholar]
- 40.Wang C., Fu M., D'Amico M., Albanese C., Zhou J. N., Brownlee M., Lisanti M. P., Chatterjee V. K., Lazar M. A., Pestell R. G. Inhibition of cellular proliferation through IkappaB kinase-independent and peroxisome proliferator-activated receptor gamma-dependent repression of cyclin D1. Mol. Cell. Biol. 2001;21:3057–3070. doi: 10.1128/MCB.21.9.3057-3070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarraf P., Mueller E., Jones D., King F. J., DeAngelo D. J., Partridge J. B., Holden S. A., Chen L. B., Singer S., Fletcher C., Spiegelman B. M. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nat. Med. 1998;4:1046–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- 42.Sporn M. B., Suh N., Mangelsdorf D. J. Prospects for prevention and treatment of cancer with selective PPARgamma modulators (SPARMs) Trends Mol. Med. 2001;7:395–400. doi: 10.1016/s1471-4914(01)02100-1. [DOI] [PubMed] [Google Scholar]
- 43.Chen G. G., Lee J. F., Wang S. H., Chan U. P., Ip P. C., Lau W. Y. Apoptosis induced by activation of peroxisome-proliferator activated receptor-gamma is associated with Bcl-2 and NF-kappaB in human colon cancer. Life Sci. 2002;70:2631–2646. doi: 10.1016/s0024-3205(02)01510-2. [DOI] [PubMed] [Google Scholar]
- 44.Butts B. D., Tran N. L., Briehl M. M. Identification of a functional peroxisome proliferator activated receptor response element in the 3′ untranslated region of the human bcl-2 gene. Int. J. Oncol. 2004;24:1305–1310. [PubMed] [Google Scholar]
- 45.Paradis V., Dargere D., Bonvoust F., Vidaud M., Segarini P., Bedossa P. Effects and regulation of connective tissue growth factor on hepatic stellate cells. Lab. Invest. 2002;82:767–774. doi: 10.1097/01.lab.0000017365.18894.d3. [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Trevijano E. R., Iraburu M. J., Fontana L., Dominguez-Rosales J. A., Auster A., Covarrubias-Pinedo A., Rojkind M. Transforming growth factor beta1 induces the expression of alpha1(I) procollagen mRNA by a hydrogen peroxide-C/EBPbeta-dependent mechanism in rat hepatic stellate cells. Hepatology. 1999;29:960–970. doi: 10.1002/hep.510290346. [DOI] [PubMed] [Google Scholar]
- 47.Isshiki K., Haneda M., Koya D., Maeda S., Sugimoto T., Kikkawa R. Thiazolidinedione compounds ameliorate glomerular dysfunction independent of their insulin-sensitizing action in diabetic rats. Diabetes. 2000;49:1022–1032. doi: 10.2337/diabetes.49.6.1022. [DOI] [PubMed] [Google Scholar]
- 48.Ghosh A. K., Bhattacharyya S., Lakos G., Chen S. J., Mori Y., Varga J. Disruption of transforming growth factor beta signaling and profibrotic responses in normal skin fibroblasts by peroxisome proliferator-activated receptor gamma. Arthritis Rheum. 2004;50:1305–1318. doi: 10.1002/art.20104. [DOI] [PubMed] [Google Scholar]
- 49.Subbaramaiah K., Lin D. T., Hart J. C., Dannenberg A. J. Peroxisome proliferator-activated receptor gamma ligands suppress the transcriptional activation of cyclooxygenase-2. Evidence for involvement of activator protein-1 and CREB-binding protein/p300. J. Biol. Chem. 2001;276:12440–12448. doi: 10.1074/jbc.M007237200. [DOI] [PubMed] [Google Scholar]
- 50.De Vos P., Lefebvre A. M., Miller S. G., Guerre-Millo M., Wong K., Saladin R., Hamann L. G., Staels B., Briggs M. R., Auwerx J. Thiazolidinediones repress ob gene expression in rodents via activation of peroxisome proliferator-activated receptor gamma. J. Clin. Invest. 1996;98:1004–1009. doi: 10.1172/JCI118860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schinner S., Dellas C., Schroder M., Heinlein C. A., Chang C., Fischer J., Knepel W. Repression of glucagon gene transcription by peroxisome proliferator-activated receptor gamma through inhibition of Pax6 transcriptional activity. J. Biol. Chem. 2002;277:1941–1948. doi: 10.1074/jbc.M109718200. [DOI] [PubMed] [Google Scholar]
- 52.Fu M., Zhang J., Zhu X., Myles D. E., Willson T. M., Liu X., Chen Y. E. Peroxisome proliferator-activated receptor gamma inhibits transforming growth factor beta-induced connective tissue growth factor expression in human aortic smooth muscle cells by interfering with Smad3. J. Biol. Chem. 2001;276:45888–45894. doi: 10.1074/jbc.M105490200. [DOI] [PubMed] [Google Scholar]
- 53.Fu M., Zhang J., Lin Y., Zhu X., Zhao L., Ahmad M., Ehrengruber M. U., Chen Y. E. Early stimulation and late inhibition of peroxisome proliferator-activated receptor gamma (PPAR gamma) gene expression by transforming growth factor beta in human aortic smooth muscle cells: role of early growth-response factor-1 (Egr-1), activator protein 1 (AP1) and Smads. Biochem. J. 2003;370:1019–1025. doi: 10.1042/BJ20021503. [DOI] [PMC free article] [PubMed] [Google Scholar]