Abstract
The DD4 mRNA of the penaeid prawn Penaeus japonicus was shown previously to be expressed in the epidermis adjacent to the exoskeleton specifically during the post-moult period, when calcification of the exoskeleton took place. The encoded protein possessed a Ca2+-binding site, suggesting its involvement in the calcification of the exoskeleton. In the present study, an additional ORF (open reading frame) of 289 amino acids was identified at the 5′ end of the previous ORF. The newly identified part of the encoded protein included a region of approx. 120 amino acids that was highly rich in glutamate residues, and contained one or more Ca2+-binding sites. In an immunohistochemical study, signals were detected within calcified regions in the endocuticular layer of the exoskeleton. Bacterially expressed partial segments of the protein induced CaCO3 crystallization in vitro. Finally, a reverse transcription-PCR study showed that the expression was limited to an early part of the post-moult period, preceding significant calcification of the exoskeleton. These observations argue for the possibility that the encoded protein, renamed crustocalcin (CCN), promotes formation of CaCO3 crystals in the exoskeleton by inducing nucleation.
Keywords: biomineralization, calcification, calcium-binding protein, calcium carbonate, nucleation, Penaeus japonicus (penaeid prawn)
Abbreviations: ACC, amorphous calcium carbonate; ALP, alkaline phosphatase; BSP, bone sialoprotein; CCN, crustocalcin; EBSD, electron back-scattered diffraction; EDS, energy dispersive X-ray spectrometer; EMP, electron microprobe; HA, hydroxyapatite; lacZα, β-galactosidase α-fragment; MBP, maltose-binding protein; ORF, open reading frame; poly(A)+, polyadenylated; 5′ RACE, 5′ rapid amplification of cDNA ends; R–R, Rebers–Riddiford; RT, reverse transcription; TBS, Tris-buffered saline; TBSN, TBS with normal goat serum; UTR, untranslated region
INTRODUCTION
Deposition of calcium-bearing crystals in hard tissues is widely observed in the animal kingdom [1]. The calcified hard tissues contain the organic matrix comprising proteins, lipids and polysaccharides, some of which are implicated in aspects of the calcification process, such as promotion of nucleation, selection of crystal polymorph (i.e. calcite or aragonite in CaCO3) and control of crystal growth [2–5].
Biochemical analyses of the organic matrices in vertebrate and invertebrate calcified hard tissues demonstrated the presence of acidic proteins and glycoproteins which are implicated in nucleation of calcium-bearing crystals. For example, a mammalian glycoprotein named bone sialoprotein (BSP) is expressed specifically in calcified hard tissues, and has been shown to induce nucleation of HA (hydroxyapatite) crystals in vitro [6–8]. This protein contains two E-rich (glutamate-rich) regions, and in vitro studies with the porcine BSP demonstrated that both of these regions had the nucleation activity [9,10]. A polyglutamate sequence is found in one of the E-rich sequences, and the homopolymeric sequence confers the HA nucleation activity [10]. An immunohistochemical study has revealed the presence of BSP at the mineralization front in bone [11]. Similarly, acidic proteins named phosphophoryn and starmaker, highly rich in aspartate and phosphorylated serine residues, are implicated in the formation of HA and CaCO3 crystals during the dentinogenesis and otolith formation in vertebrates respectively [12,13].
In invertebrate hard tissues, involvement of acidic macromolecules has also been suggested in the nucleation of CaCO3 crystals [3,14]. Amino acid composition analyses of the organic matrix in the exoskeleton of invertebrates such as molluscs and corals revealed high contents of aspartate [15–19]. During the past decade, the amino acid sequence was determined in a number of proteins associated with calcified hard tissues in invertebrates, and some of them in fact had regions of high aspartate content [20,21].
In the present paper, we report the characterization of a Ca2+-binding protein, named crustocalcin (CCN), in the penaeid prawn Penaeus japonicus. The cDNA encoding CCN was originally identified as DD4, together with two other cDNAs, in a search for genes expressed in the epidermal cells adjacent to the exoskeleton specifically during the post-moult period of the moult cycle, when calcification of the exoskeleton takes place [22–24]. The DD4/CCN protein was found to contain a Ca2+-binding region [24]. These observations suggested the possibility that the DD4/CCN protein was involved in the calcification process. The present study aimed to address this possibility by investigating its distribution in the exoskeleton, in vitro activity in the formation of CaCO3 crystals and the temporal change in the level of its cognate mRNA during the post-moult stage. Furthermore, re-examination of the cDNA found an additional ORF (open reading frame) that included a coding sequence for a highly E-rich region. Results of these analyses suggest strongly that the protein promotes formation of CaCO3 crystals in the crustacean exoskeleton by facilitating nucleation.
EXPERIMENTAL
Prawns and moulting stages
Live adult prawns were purchased from Washizu fishery market, Shizuoka, Japan, and kept in 500-litre tanks under running sea water until sampled. The prawns were grouped into the following three moult periods based on the cuticular morphology of the mandibular expodite [25]: post-moult, intermoult, and pre-moult periods.
5′ RACE (5′ rapid amplification of cDNA ends) and nucleotide sequence analysis of the DD4/CCN cDNA
Total RNA was extracted from tail fans (uropods) of prawns at an early post-moult stage (within 2 h after moulting), as described previously [22]. 5′ RACE was also carried out according to the method of Watanabe et al. [22] with the following modifications: two antisense primers, 5′-GCTGAGAAATCTCTGCAGTAGG-3′ and 5′-CACAGGCGCAGCAGGAGTCT-3′, were used in the RT (reverse transcription) and PCR respectively. The PCR program consisted of the initial denaturation at 95 °C for 5 min; 3 cycles of PCR at 94 °C for 1 min, 50 °C for 1 min and 72 °C for 3 min; 37 cycles of PCR at 94 °C for 1 min, 55 °C for 1 min and 72 °C for 3 min; and the final extension at 72 °C for 15 min. The PCR products were subcloned into the pGEM-T Easy vector (Promega) and sequenced on both strands.
cDNA containing the entire ORF was isolated by reverse-transcribing T12-primed total RNA with Power Script Reverse Transcriptase (Clontech), and subjecting the resulting cDNA to PCR using the following primers: 5′ UTR1 (5′-GAGTGCGCCGGCGCCGTGTCT-3′) and 3′ UTR1 (5′-CGGGATCCTGGCTCATGGCCTGG-3′). The latter was designed based on the 3′ UTR (untranslated region) sequence of DD4 [24]. The PCR consisted of the initial denaturation at 95 °C for 5 min; 30 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 3 min; and the final extension at 72 °C for 15 min. The PCR product was sequenced as described above.
Northern blot analysis
Preparation of poly(A)+ (polyadenylated) RNA and Northern blot analysis were performed as described previously [24], except for the following modifications: only prawns within 4 h of moulting were used as the RNA source. A 10 cm agarose gel was used for better resolution than in the previous analysis [24], in which a 6 cm gel was used.
Antibody generation and Western blot analysis
Synthesis of an antigen peptide and production of an antibody against CCN were carried out by Sawady Technology (Qiagen). Briefly, a polypeptide consisting of the C-terminal 20 amino acids of CCN with an additional cysteine residue (CKEIQSPPLLPPLMKPSVLLN) was synthesized and coupled to BSA. A rabbit was injected with the BSA-coupled antigen peptide, and the antiserum was affinity-purified using an antigen-coupled agarose gel column.
For Western blot analysis of exoskeletal proteins, the exoskeleton of the tail fan was decalcified in 10% ethanoic (acetic) acid at 4 °C for 2 h for decalcification, ground in liquid nitrogen, boiled in a sample buffer [25 mM Tris/HCl, pH 6.5, 5% (v/v) glycerol, 2% (w/v) SDS, 0.01% (w/v) Bromophenol Blue, 1% (w/v) 2-mercaptoethanol] for 10 min, and centrifuged at 10000 g for 10 min. Proteins in the supernatant were run on an SDS/12% polyacrylamide gel and blotted on to a PVDF membrane (Schleicher & Schuell). The blot was incubated in a blocking solution [TBS (Tris-buffered saline) with 1% (w/v) dried skimmed milk] at room temperature (approx. 25 °C) for 30 min, and then reacted at room temperature for 1 h with the antibody diluted 1:5000 in the same solution. After washing, the membrane was incubated at room temperature for 1 h with an ALP (alkaline phosphatase)-conjugated anti-rabbit IgG (Boehringer Mannheim) that was diluted 1:10000 in the blocking solution. Immunoreactive protein bands were visualized with 188 μg/ml BCIP (5-bromo-4-chloroindol-3-yl phosphate) and 376 μg/ml NBT (Nitro Blue Tetrazolium) in 0.1 M Tris/HCl, pH 9.5, 0.1 M NaCl and 50 mM MgCl2. In a control experiment, the primary antibody was pre-incubated with 50 μg/ml of a fusion protein consisting of MBP (maltose-binding protein) and containing the C-terminal 30 amino acids of CCN including the antigen sequence [26].
Light-microscopic immunohistochemistry and detection of calcified regions in the exoskeleton
Tail fans of intermoult prawns were fixed with 4% (w/v) formaldehyde in 0.1 M phosphate buffer (pH 7.2) at room temperature for 2 h with gentle shaking, dehydrated through graded ethanol series, and embedded using a JB-4 embedding kit (Polysciences, Warrington, PA, U.S.A.). Tissue sections were cut at 3 μm and mounted on slides coated with Biobond® (British Biocell International, Cardiff, U.K.). The specimens were blocked in TBSN [TBS with 2% (v/v) normal goat serum (Dako)] for 30 min, and subsequently reacted at 4 °C for 16 h with the anti-CCN antibody diluted 1:5000 in TBSN. They were then rinsed three times with double-distilled water and reacted for 1 h with an ALP-conjugated anti-rabbit IgG diluted 1:5000 in TBSN. After washing three times with double-distilled water, the ALP activity was detected as described above. The specificity of the antibody was examined by incubating the diluted primary antibody with the above fusion protein (100 μg/ml) for 12 h at 4 °C before used in the immunohistochemical analysis.
In order to detect calcified regions in the exoskeleton, tissue sections were subjected to EMP (electron microprobe) analysis using a wavelength dispersive X-ray EMP (JXA-8900R, JEOL) according to Kotake et al. [27]. The accelerating voltage and beam current were 15 kV and 12 nA respectively.
Protein expression in Escherichia coli and 45Ca2+-overlay analysis
To characterize Ca2+-binding of the newly identified part of CCN, overlapping partial proteins were expressed in E. coli. Four cDNA fragments encoding Fragment 1 (Fr1), 2a, 2b and 3 (see the Results section) were amplified using PCR with the following primer pairs; F1f (5′-CGGGATCCAGAACCATCTTCGACTA-3′) and F1r (5′-CGGGATCCTATCCCAGGACCTGCGTTC-3′) for Fr1, F2f (5′-CGGGATCCACCCAGTCATCAGGTGC-3′) and F2r (5′-CGGGATCCTACCCTTCTCCTTCGGCTT3′) for Fr2a and 2b, F3f (5′-CGGGATCCAAGTCAGACGCGGAAGC-3′) and F3r (5′-CGGGATCCTACGCACTATCGGCAGAGT-3′) for Fr3. PCRs were performed as follows: denaturation at 94 °C for 5 min, 30 cycles of 94 °C for 0.5 min, 55 °C for 0.5 min and 72 °C for 1.5 min, followed by extension at 72 °C for 7 min. The amplification products were digested with BamHI, and subcloned into pMAL c2 (New England Biolabs) except for Fr1 that was subcloned into pET5a (Promega). The constructs were introduced into E. coli cells, and the expression of the proteins was induced as described previously [24]. 45Ca2+-overlay analyses were performed according to the method of Maruyama et al. [28].
In vitro nucleation assay
Cells expressing three fusion proteins [MBP–Fr2a, –Fr3 and –lacZα (β-galactosidase α-fragment)] were harvested and sonicated, and the supernatants of the cell lysates were incubated with amylose resin beads (New England Biolabs) at 4 °C for 16 h. The beads were collected by centrifugation at 1000 g for 1 min, washed with column buffer (20 mM Tris/HCl, pH 7.2, 200 mM NaCl and 1 mM EDTA) and incubated in a calcifying solution (5 mM CaCl2 and 5 mM NaHCO3 at pH 8.0) to allow CaCO3 crystal formation at 4 °C for 3 days. The crystals formed on the beads were examined using a scanning electron microscope (S-4500, Hitachi) with an EDS (energy dispersive X-ray spectrometer) (Super Quantum; Kevex, Valencia, CA, U.S.A.) and an EBSD (electron back-scattered diffraction) detector (Phase ID; ThermoNoran, Madison, WI, U.S.A.). A total of 100 beads were randomly selected, and the numbers of CaCO3 crystals on them were counted. Statistical analysis for the numbers of the crystals was carried out using Tukey's method. The polymorph in the crystals was determined by the EBSD technique which can obtain crystallographic information (crystalline phase, crystal orientation, etc.) by analysing the diffraction patterns [29].
To quantify the amount of the fusion proteins bound to the beads, 10 μl of the coated beads was incubated in the column buffer containing 10 mM maltose, and the eluates were subjected to SDS/PAGE. Protein bands were stained with Sypro-Red (Sigma), and the fluorescence was measured using a computerized densitometer scanner (FLA 2000, Fujifilm), using the fluorescence level of ovalbumin in the LMW Marker Kit (Amersham Biosciences) as a standard.
RT-PCR for detecting the expression of CCN
Total RNA was prepared from staged post-moult prawns and cDNA was prepared as described previously [22]. The following primer pairs were used to detect the expression of CCN and PjL26 [30] respectively: 5′-CCTGTTGAGCCTATTGCATA-3′ (forward) and 5′-TAATTCAATAACACCGATG-3′ (reverse), and 5′-AAGATGGTAACGGAGTCCCG-3′ (forward) and 5′-ACCTTTGATGGGTGGATGCC-3′ (reverse). The PCRs were carried out using the following program: denaturation at 95 °C for 5 min; 26 cycles of PCR reaction at 94 °C for 1 min, 55 °C for 1 min and 72 °C for 1 min; and extension at 72 °C for 7 min. The PCR products were run on 2% agarose gels and stained with ethidium bromide. Fluorescence was measured as described above, and the fluorescence level due to the CCN expression was normalized relative to that of PjL26 expression.
RESULTS
5′ RACE and revision of the DD4 ORF sequence
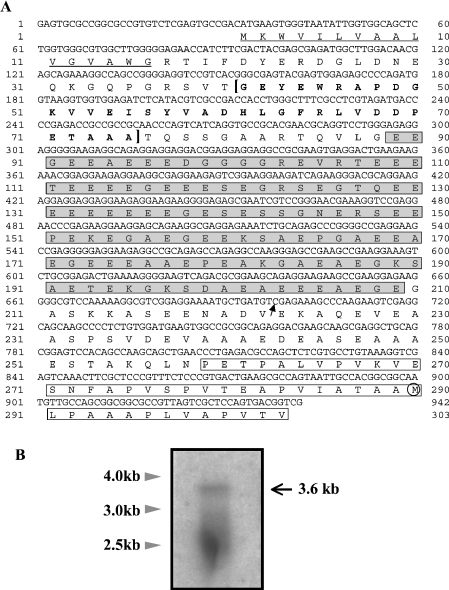
5′ RACE was performed to search for an ORF sequence which may be present on the 5′ side of the previously reported DD4 ORF [24]. A cDNA clone with an insert of 942 bp (excluding the primers) was identified, and a 3′ part (245 bp) of the sequence was found to be identical with a 5′ part of the DD4 sequence, except for nucleotide substitution at nine positions (results not shown). As the 5′ RACE product sequence was merged with the DD4 sequence, a cDNA sequence of 3198 bp was obtained [excluding poly(A)+] (GenBank® accession number AB049148). In the merged cDNA sequence, an ORF for 831 amino acids was found, which consisted of the previously reported DD4 ORF (542 amino acids) and additional 289 amino acids (derived from the 5′ RACE product) on the N-terminal side. This ORF was open at the 5′ end of the cDNA sequence, as an in-frame stop codon was not found upstream from the putative initiator codon (Figure 1A).
Figure 1. A 5′ sequence of the DD4 cDNA and Northern blot analysis.
(A) Nucleotide sequence and conceptually translated amino acid sequence of a 5′ RACE product. The sequence of 245 bp on the 3′ side [from Cys698 (arrow) to Gly942] overlaps with the previously reported DD4 sequence. The potential signal peptide (Met1-Gly16) is underlined, and the R–R-like sequence (Gly41-Ala75) in bold letters is bracketed. The E-rich region (Glu89-Gly209) is in shaded boxes, and a part of the P-rich region is in open boxes. The Met290 residue that was presumed to be the N-terminal residue in the previous report is circled. (B) Northern blot analysis. Positions of molecular mass standards are indicated on the left.
Northern blot analysis was performed to determine the size of the DD4 mRNA, and a band was detected at approx. 3.6 kb, as well as a smeary signal at 2.3–2.9 kb (Figure 1B). Assuming that the size of the full-length DD4 mRNA was 3.6 kb, this observation suggested that the above cDNA sequence of approx. 3.2 kb still lacked 200 bp or more at the 5′ end, given that the size of a poly(A)+ tail is usually 200 bases or less. Thus we carried out another round of 5′ RACE, using cDNA from two individuals as the template. In analysing cloned PCR products, three sequences were identified, all of which had the same 5′ end as the sequence shown in Figure 1(A) (results not shown). In one of the products, a deletion of 1 bp was found upstream from the Met1 codon. Since insertions or deletions which cause frameshifts are rare in ORFs, this observation supported the possibility that the Met1 codon in Figure 1(A) was the initiator codon.
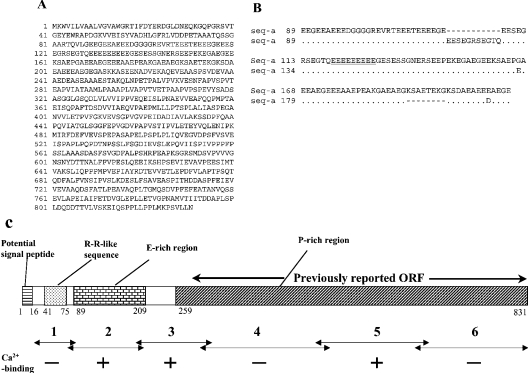
To confirm that the merged ORF is not a chimaeric sequence consisting of sequences derived from different splicing products or related genes, RT-PCR was performed using primers derived from the putative 5′ and the 3′ UTRs, and the PCR products were sequenced. Two types of ORF sequences, named seq-a and b, were thereby identified (GenBank® accession numbers AB114444 and AB114445). Translated seq-a (831 amino acids; Figure 2A) was highly similar to the above sequence, with amino acid substitution at 15 positions (results not shown). Assuming that these minor differences are due to intra-specific polymorphism, this observation indicated that the merged ORF was derived from a single transcript. The translated seq-b (834 amino acids) was less similar, as an insertion of 11 amino acids and a deletion of eight amino acids were observed in the E-rich region (Figure 2B, and see below). Outside of this region, amino acid substitution was observed at 14 positions, but no insertions or deletions (results not shown). A computer-aided analysis [31] predicted that the encoded protein contained a signal peptide of 16 amino acids. Excluding the putative signal sequence, the predicted molecular mass of the protein was 85731 (seq-a) or 86175 (seq-b), and the predicted pI was 3.67 (seq-a) or 3.66 (seq-b). Analyses of the translated sequence found no consensus sequence for N-linked glycosylation, but 27 potential target sequences for protein kinases in seq-a (cAMP- and cGMP-dependent protein kinase, protein kinase C, tyrosine kinase and casein kinase II).
Figure 2. Primary structure of the encoded protein.
(A) Translated amino acid sequence of seq-a. Positions of amino acid residues are indicated on the left. (B) Alignment of the conceptual amino acid sequences of seq-a and b in the E-rich region. A dot and a hyphen indicate an identical amino acid and a gap respectively. The nine consecutive E residues are underlined. (C) A schematic drawing of the primary structure of CCN. Numbers below the boxes indicate positions of amino acid residues (in seq-a) at the boundaries of regions. Ca2+-binding (see text and Figure 4) was detected in the three fragments marked with +, but not in those marked with − (see Figure 5 and [24]).
The encoded protein was renamed CCN, due to the Ca2+ binding (see below). CCN contains the following three regions (Figure 2C): (i) the Gly41-Ala75 sequence (also see Figure 1A), which exhibited similarity to the Rebers–Riddiford (R–R) consensus sequence (results not shown), commonly found in arthropod cuticular proteins and involved in chitin binding [32,33]. It was therefore named the R–R-like sequence. (ii) A region of about 120 amino acids (Glu89-Glu209 in seq-a and Glu89-Glu212 in seq-b), which is highly rich in glutamate residues (48% in seq-a), and was therefore named the E-rich region. This region is also rich in glycine, alanine and serine. (iii) A large region in the C-terminal part of the protein (Pro259-Asn831 in seq-a and Pro262-Asn834 in seq-b), which was named the P-rich region, due to the abundance of proline (14% in seq-a), as well as valine, alanine and serine. In contrast with the E-rich region, it is highly hydrophobic. Though aspartate (5%) and glutamate (9%) are not prominent in this region, the predicted pI is low (3.54), since basic residues make up a very small fraction (3%) of this region.
Western blot analysis and immunohistochemistry
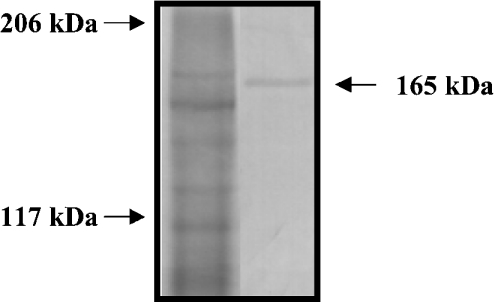
In a Western blot analysis using an affinity-purified polyclonal antibody, an immunoreactive band was detected at about 165 kDa (Figure 3; see the Discussion for discrepancy with the predicted molecular mass). This antibody was used in an immunohistochemical study of tissue sections containing the epidermis and exoskeleton.
Figure 3. Western blot analysis of CCN.
An extract of an intermoult exoskeleton was subjected to SDS/PAGE, and proteins were detected with Coomassie Brilliant Blue (left-hand lane). Positions of molecular-mass standards are indicated on the left. In a Western blot (right-hand lane), an immunoreactive band is observed at 165 kDa (arrow).
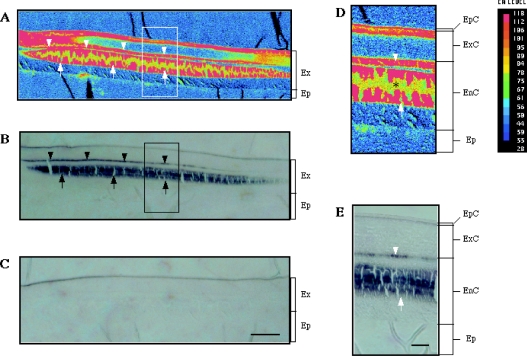
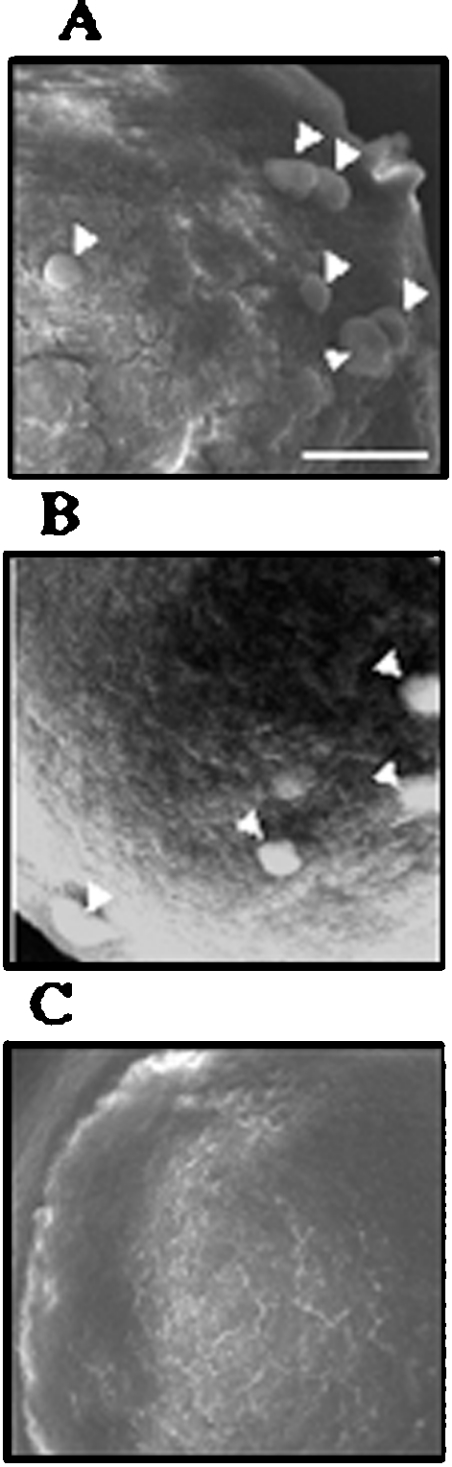
Tissue samples from a medial part of the tail fan were fixed, sectioned without decalcification and subjected to EMP and immunohistochemical analyses, in order to find whether CCN was distributed in calcified regions of the exoskeleton. The intermoult crustacean exoskeleton consists of three cuticular layers: the epicuticle, exocuticle and endocuticle (from the apical to basal order) [34]. The EMP analysis showed that the Ca2+ content in the exocuticle and endocuticle were not uniform in the tail fan of P. japonicus, and heavy calcification in these two layers was seen in discrete regions (Figure 4A). Immunoreactive signals were seen in two areas in the exoskeleton (Figure 4B). The thin stained area (arrowheads in Figure 4B) was on the endocuticular side of the exo/endo boundary, and the thick area (arrows) was located in the medial part of the endocuticle (Figure 4E). Both of these areas were within the heavily calcified parts of the endocuticle (compare Figures 4D and 4E). The immunostaining observed in the epicuticle was unlikely to be due to CCN, since the staining was not eliminated by pre-absorption of the antibody with a fusion protein containing the antigen sequence (Figure 4C).
Figure 4. Calcification and CCN distribution in the integument.
(A) An X-ray intensity map for Ca content in a section of the integument which comprises the exoskeleton (Ex) and epidermis (Ep). The red and blue indicate the highest and lowest Ca contents respectively. (B) An immunostaining for CCN in an adjacent section. The two immunoreactive regions (see text) are indicated by arrowheads and arrows. (C) A negative control. Scale bar, 100 μm. (D) A magnified image of an enclosed area in (A). The exoskeleton consists of three layers, epicuticle (EpC), exocuticle (ExC) and endocuticle (EnC). The low signal levels in yellow/green parts of calcified regions (*) is due to loss of the specimen during the EMP analysis. (E) A magnified image of an enclosed area in (B). Scale bar, 25 μm.
45Ca2+-overlay analysis
Ca2+-binding in the newly identified part of the protein was studied (see [24], and Figure 2C for Ca2+-binding in the previously identified part of the protein). Three overlapping fragments covering amino acid positions Arg17–Ala321 (the putative signal peptide was excluded) in seq-a and one fragment corresponding to the E-rich region in seq-b were expressed in E. coli. Fr1, amino acid position Arg17–Gly88 in seq-a, was successfully expressed when its coding sequence was cloned in the pET5 vector (Figure 5A). That was not the case with Fr2a (Thr76–Gly210 in seq-a), 2b (Thr76–Gly213 in seq-b) and 3 (Lys197–Ala321 in seq-a), so these fragments were expressed as fusion proteins to MBP. We also attempted to express the full-length ORF (excluding the potential signal sequence) using the pET5 and pMAL vectors, but expression of the encoded protein could not be observed.
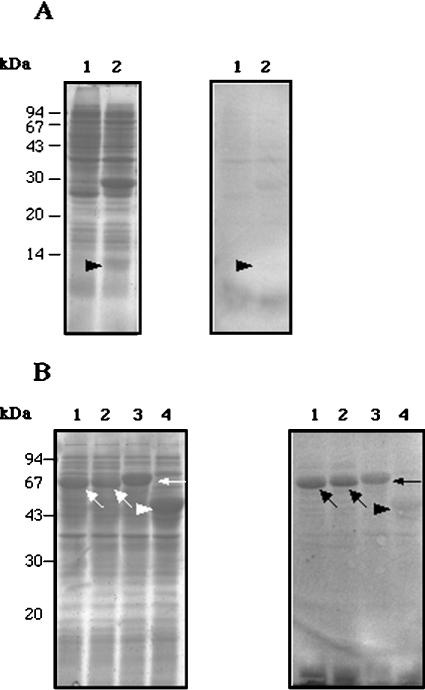
Figure 5. Ca2+ binding to bacterially expressed CCN fragments.
(A) Left-hand gel: SDS/PAGE of extracts of the following E. coli cells: lane 1, extracts of E. coli without an expression construct (a negative control); lane 2, extracts of E. coli expressing Fr1. Right-hand gel: 45Ca2+ overlay of a Western blot. Significant binding of Ca2+ to Fr1 is not detected (arrowhead). (B) Left-hand gel: SDS/PAGE of extracts of E. coli expressing the following proteins: lane 1, MBP–Fr2a; lane 2, MBP–Fr2b; lane 3, MBP–Fr3, lane 4, MBP–lacZα (negative control). The three fragment fusion proteins and MBP–lacZα are indicated with arrows and an arrowhead respectively. Right-hand gel: 45Ca2+ overlay of a Western blot. Binding of Ca2+ to the three fragment fusion proteins is detected in lanes 1, 2 and 3 (arrows), whereas no significant Ca2+-binding to MBP–lacZα is observed (arrowhead, lane 4).
As shown in Figure 5(B), Ca2+-binding to MBP–Fr2a, –Fr2b and –Fr3, but not to MBP–lacZα, was observed, indicating that the three fragments bound Ca2+. Binding to the Fr1 band was not detected.
In vitro nucleation assay
The roles of two Ca2+-binding fragments (Fr2a and Fr3) in CaCO3 crystallization were studied using an in vitro nucleation assay. Three fusion proteins, MBP–Fr2a, –Fr3 and –lacZα (a negative control), were bound to amylose beads and incubated in the calcifying solution to allow CaCO3 crystals to form on the bead surface. Formation of crystals (2–10 μm in diameter) was observed on the beads coated with the three proteins (Figure 6). EDS analysis showed that the majority of the crystals were CaCO3, while NaCl and NaHCO3 crystals were rarely found (results not shown). The number of CaCO3 crystals on 100 beads were summed, and normalized with the amount of the polypeptides bound to the beads. As shown in Table 1, MBP–Fr2a and –Fr3 induced formation of more CaCO3 crystals than the control polypeptide (P<0.001). The number of CaCO3 crystals formed on MBP–Fr3-coated beads was significantly larger than on MBP–Fr2a beads (P<0.001). No prominent difference could be noted between the sizes of crystals formed on the experimental and control beads, arguing against the possibility that CCN, as bound to solid substrate, is involved in the regulation of crystal growth. Thus the result of this analysis suggested that Fr2a and Fr3 promoted formation of CaCO3 crystals by inducing nucleation.
Figure 6. Scanning electron micrographs of CaCO3 crystals (arrowheads) formed on the amylose beads to which MBP–Fr2a (A), MBP–Fr3 (B) and MBP–lacZα (C) were bound.
Scale bar, 10 μm.
Table 1. CaCO3 crystal nucleation activities of CCN fragments.
Results are means±S.D.
| Fusion protein | Number of crystals/100 beads (a) | Amount of fusion protein (mg) bound to 10 ml of beads (b) | Relative values of b (c) | (a)/(c) |
|---|---|---|---|---|
| MBP–Fr2a | 113.7±12.01 | 3.071±0.066 | 0.53 | 214.47±22.67 |
| MBP–Fr3 | 209.3±32.02 | 3.168±0.079 | 0.54 | 387.65±59.29 |
| MBP–LacZα | 31.3±7.02 | 5.835±0.083 | 1.00 | 31.33±7.02 |
The polymorph was determined on subsets (ten crystals in each of the three experiments) of observed CaCO3 crystals formed on beads coated with three polypeptides. All of the 90 examined crystals were calcite (results not shown).
RT-PCR
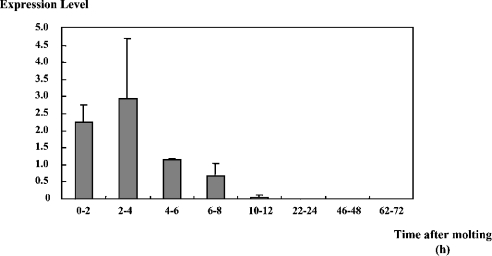
In a previous report [24], the expression of DD4/CCN was detectable during the post-moult period, but not in the rest of the moult cycle. In this study, changes in the level of the CCN mRNA during the post-moult period were studied, using semi-quantitative RT-PCR. As shown in Figure 7, expression of CCN was detected within 2 h of moulting. The expression level was high from 2 to 4 h after moulting, decreased thereafter, and became undetectable by 24 h after moulting. Since the post-moult stage lasts at least 4 days [25], this result indicates that the main expression of CCN was limited to the initial part of the post-moult stage.
Figure 7. Changes in the level of the CCN expression during the post-moult period.
The y-axis represents the level of fluorescence due to the expression of CCN normalized by that of PjL-26 (a constitutively expressed gene [30]) on a relative scale. Results are means±S.D. (n=3). The difference between the 2–4 h and 4–6 h samples was statistically significant (P<0.001).
DISCUSSION
CCN: a putative promoter of calcification in the crustacean exoskeleton
The results of the present study argue strongly for a positive regulatory role of CCN in the calcification of the crustacean exoskeleton. The results of the immunohistochemical study (Figure 4) and the 45Ca2+-overlay analysis (Figure 5) suggest involvement of CCN in the calcification of the endocuticle. The results of the nucleation assay (Figure 6 and Table 1) indicate that CCN promotes formation of CaCO3 crystals in vitro. The detection of CCN expression before significant calcification of the exoskeleton (Figure 7) is consistent with the possibility that CCN promotes, rather than represses, the calcification of the exoskeleton. Based on these observations, we propose that CCN promotes formation of CaCO3 crystals in the endocuticle by facilitating nucleation. The present paper is the first report of a positive function of a crustacean exoskeletal protein in calcification.
In invertebrate hard tissues mineralized with CaCO3, proteins with regions that are rich in acidic residues such as aspartate [20,21] or phosphoserine [35,36] have been found, but no protein with an E-rich region was identified previously. CCN is similar to the vertebrate BSP in possessing an E-rich segment and also containing a polyglutamate sequence (see the Introduction), although lack of sequence similarity outside of the E-rich sequences argues against a common ancestry of the two proteins. The results of the in vitro nucleation assay of Fr2a, which spanned the E-rich region, suggest that the E-rich sequence can nucleate CaCO3 crystals (Table 1). The analysis on Fr3 suggests the presence of another nucleation site outside of the E-rich region, since Fr3 contained only 12 residues from that region.
It is yet to be determined which parts of these fragments carry the nucleation activity. No known Ca2+-binding motifs such as E-F hand could be found in Fr2 and Fr3, and so it remains unknown what parts of those fragments interacts with Ca2+. The abundance of negatively charged residues in Fr2a and Fr2b suggests that the Ca2+ binding may be simply due to electrostatic interaction, rather than presence of a Ca2+-binding motif. On the other hand, the density of negative charges in Fr3 was about the same as that in Fr6, which did not bind Ca2+ [26]. Thus the Ca2+ binding to this fragment and Fr5 (Figure 2C), whose negative charge density is even lower, is likely to be due to formation of specific Ca2+-binding structures.
In the in vitro analysis, all of the examined CaCO3 crystals that were formed on the bead surface were calcite (see the Results section), and an X-ray diffraction analysis has detected calcite, but not aragonite, in the exoskeleton in the tail fan of P. japonicus (H. Endo, T. Watanabe and T. Kogure, unpublished work). These observations may suggest that CCN induces calcite nucleation in vivo as well as in vitro. However, ACC (amorphous calcium carbonate) is known to commonly occur in crustacean hard tissues, such as the exoskeleton and gastrolith [37], and the possibility is open that CCN promotes formation of ACC rather than crystalline CaCO3 in vivo.
Results of the sequence analyses and the Western blot analysis on CCN suggest that CCN is a component of the insoluble fraction of the organic matrix, rather than an intra-crystalline protein. First, CCN contains a variant of the R–R consensus sequence, suggesting binding of the protein to chitin fibres in the exoskeleton. Secondly, the P-rich region, which occupies about two thirds of the protein, is highly hydrophobic, and so it is likely that CCN interacts with other hydrophobic macromolecules of the organic matrix. Finally, in the Western blot analysis, an immunoreactive band was detected in skeletal samples that had been treated with ethanoic acid for decalcification, indicating that at least some of the CCN molecules remained insoluble after the treatment. We presume that CCN interacts with other constituents of the insoluble matrix through the R–R-like sequence and the P-rich region. The highly hydrophilic E-rich region and the hinge between the E- and P-rich regions are likely to be exposed to the aqueous milieu and provide nucleation sites for CaCO3 crystals.
In the present study, strong calcification was detected in both the exocuticle and endocuticle in the tail fan exoskeleton. Immunostaining for CCN, however, was limited to the latter, suggesting that different molecular mechanisms may be operating in the regulation of calcification of the two layers. Roer and collaborators [38–40] have proposed the possibility that in the exoskeleton of the blue crab Callinectes sapidas, certain proteins act as inhibitors of calcification in the pre-moult exoskeleton, and are altered to lose the inhibitory activity by modification (e.g. deglycosylation) soon after the ecdysis. Such a regulatory mechanism may operate in the exocuticle, which is synthesized during the pre-moult period and remain uncalcified until the post-moult period [2]. In contrast, the endocuticle, built and calcified following ecdysis, may not require such negative regulation, and its calcification is likely to be induced by a macromolecule(s), such as CCN, that is synthesized during the post-moult period.
The size of the protein and mRNA
In the Northern blot analyses in the previous (Figure 2 in [24]) and present studies, bands of different sizes were observed, although the same probe was used. In Figure 1(B) of the present study, a distinct band of approx. 3.6 kb was detected. This band was not observed in the previous Northern blot analysis [24] in which strong hybridization was seen at approx. 3.0 kb, as well as a weaker diffusive signal at 3.0–4.0 kb. This apparent inconsistency is likely to be due to the difference in the stage of the prawn samples. In the previous analysis, RNA was prepared from a mixture of post-moult prawns of a wide time window (6–30 h after moulting), whereas in the present study, we used prawns at an early stage (0–4 h after moulting) when CCN expression was at its highest level (Figure 7). We presume that the 3.6 kb band in the present study corresponds to the full-length CCN mRNA, and the smeary signal of lower molecular masses represents degradation products. In the previous study [24], presumably, the majority of the detected mRNA was degraded, since RNA was prepared from individuals which had passed the major expression period.
The predicted molecular mass of CCN based on the translated sequence (approx. 86 kDa) is about half of the apparent molecular mass in the Western analysis (165 kDa). Although the discrepancy might be explained in part by post-translational modifications such as phosphorylation, we presume that it was caused mostly by aberrance in the electrophoretic mobility due to abundance of certain amino acid residues. Overestimation of the molecular mass based on SDS/PAGE analysis has been observed in a number of proteins including highly negatively charged proteins (e.g. [35,41]) and calphotin [42,43], a Drosophila protein which exhibits similarity to the P-rich region of CCN in the amino acid composition [24]. Although our attempts to bacterially express the entire ORF of CCN were not successful, analyses of expressed fragments of CCN are consistent with the presumption. The apparent molecular masses of Fr3 and a partial segment of Fr2a (separated from MBP by protease digestion) on SDS/PAGE were 2.5 and 3.5 times higher respectively than the values determined by MS (H. Endo and T. Watanabe, unpublished work).
Acknowledgments
We are grateful to Dr Michiko Kono and Dr Kiyoshi Furukawa for acquisition and culture of prawns, to Dr Yoshio Takei and Dr Susumu Hyodo for the use of the computerized densitometer scanner, and to Ms Aya Kotake and Dr Katsumi Tsukamoto for the use of the X-ray EMP analyser. This work was in part supported by a Grant-in-Aid (12NP0201) from the Ministry of Education, Science, Culture and Sports, Japan.
References
- 1.Lowenstam H. A. Minerals formed by organisms. Science. 1981;211:1126–1131. doi: 10.1126/science.7008198. [DOI] [PubMed] [Google Scholar]
- 2.Simkiss K., Wilbur K. M. San Diego: Academic Press; 1989. Biomineralization: Cell Biology and Mineral Deposition. [Google Scholar]
- 3.Addadi L., Weiner S. Biomineralization – Chemical and Biological Perspectives. New York: VCH Press; 1989. Stereochemical and structural relations between macromolecules and crystals in biomineralization; pp. 133–156. [Google Scholar]
- 4.Belcher A. M., Wu X. H., Christensen R. J., Hansma P. K., Stucky G. D., Morse D. E. Control of crystal phase switching and orientation by soluble mollusk-shell proteins. Nature (London) 1996;381:56–58. [Google Scholar]
- 5.Falini G., Albeck S., Weiner S., Addadi L. Control of aragonite or calcite polymorphism by mollusk-shell macromolecules. Science. 1996;271:67–69. [Google Scholar]
- 6.Hunter G. K., Goldberg H. A. Nucleation of hydroxyapatite by bone sialoprotein. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8562–8565. doi: 10.1073/pnas.90.18.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter G. K., Goldberg H. A. Modulation of crystal formation by bone phosphoproteins: role of glutamic acid-rich sequences in the nucleation of hydroxyapatite by bone sialoprotein. Biochem. J. 1994;302:175–179. doi: 10.1042/bj3020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter G. K., Hauschka P. V., Poole A. R., Rosenberg L. C., Goldberg H. A. Nucleation and inhibition of hydroxyapatite formation by mineralized tissue protein. Biochem. J. 1996;317:59–64. doi: 10.1042/bj3170059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldberg H. A., Warner K. J., Stilman M. J., Hunter G. K. Determination of the hydroxyapatite-nucleation region of bone sialoprotein. Connect. Tissue Res. 1996;35:439–446. doi: 10.3109/03008209609029216. [DOI] [PubMed] [Google Scholar]
- 10.Harris N. L., Rattray K. R., Tye C. E., Underhill T. M., Somerman M. J., D'Errico J. A., Chambers A. F., Hunter G. K., Goldberg H. A. Functional analysis of bone sialoprotein: identification of the hydroxyapatite-nucleating and cell-binding domains by recombinant peptide expression and site-directed mutagenesis. Bone. 2000;27:795–802. doi: 10.1016/s8756-3282(00)00392-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen J., McKee M. D., Nanci A., Sodek J. Bone sialoprotein mRNA expression and ultrastructural localization in fetal porcine calvarial bone: comparisons with osteopontin. Histochem. J. 1994;26:67–78. [PubMed] [Google Scholar]
- 12.Butler W. T. Dentin matrix proteins. Eur. J. Oral Sci. 1998;106:204–210. doi: 10.1111/j.1600-0722.1998.tb02177.x. [DOI] [PubMed] [Google Scholar]
- 13.Söllner C., Burghammer M., Busch-Nentwich E., Berger J., Schwarz H., Riekel C., Nicolson T. Control of crystal size and lattice formation by starmaker in otolith biomineralization. Science. 2003;302:282–286. doi: 10.1126/science.1088443. [DOI] [PubMed] [Google Scholar]
- 14.Wheeler A. P., Sikes C. S. Biomineralization – Chemical and Biological Perspectives. New York: VCH Press; 1989. Matrix–crystal interactions in CaCO3 biomineralization; pp. 95–132. [Google Scholar]
- 15.Weiner S., Hood L. Soluble-protein of organic matrix of mollusk shells: potential template for shell formation. Science. 1975;190:987–988. doi: 10.1126/science.1188379. [DOI] [PubMed] [Google Scholar]
- 16.Mitterer R. M. Amino acid composition and metal binding capability of the skeletal protein of corals. Bull. Mar. Sci. 1978;28:173–180. [Google Scholar]
- 17.Young S. D. Organic material from scleractinian coral skeletons: I. Variation in composition between several species. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1971;40:113–120. [Google Scholar]
- 18.Cuif J. P., Dauphin Y. Occurrence of mineralization disturbances in nacreous layers of cultivated pearls produced by Pinctada margaritifera var cumingi from French Polynesia. Comparison with reported shell alterations. Aquat. Living Resour. 1996;9:187–193. [Google Scholar]
- 19.Watanabe T., Fukuda I., China K., Isa Y. Molecular analyses of protein components of the organic matrix of the exoskeleton of two scleractinian coral species. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003;136:767–774. doi: 10.1016/s1096-4959(03)00177-5. [DOI] [PubMed] [Google Scholar]
- 20.Sarashina I., Endo K. Primary structure of a soluble matrix protein of scallop shell: implications for calcium carbonate biomineralization. Am. Mineral. 1998;83:1510–1515. [Google Scholar]
- 21.Inoue H., Ozaki N., Nagasawa H. Purification and structural determination of a phosphorylated peptide with anti-calcification and chitin-binding activities in the exoskeleton of the crayfish, Procambarus clarkii. Biosci. Biotechnol. Biochem. 2001;65:1840–1848. doi: 10.1271/bbb.65.1840. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe T., Persson P., Endo H., Kono M. Molecular analysis of two genes, DD9A and B, which are expressed during the postmoult stage in the decapod crustacean Penaeus japonicus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2000;125:127–136. doi: 10.1016/s0305-0491(99)00153-4. [DOI] [PubMed] [Google Scholar]
- 23.Ikeya T., Persson P., Kono M., Watanabe T. The DD5 gene of the decapod crustacean Penaeus japonicus encodes a putative exoskeletal protein with a novel tandem repeat structure. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001;128:379–388. doi: 10.1016/s1096-4959(00)00335-3. [DOI] [PubMed] [Google Scholar]
- 24.Endo H., Persson P., Watanabe T. Molecular cloning of the crustacean DD4 cDNA encoding a Ca2+-binding protein. Biochem. Biophys. Res. Commun. 2000;276:286–291. doi: 10.1006/bbrc.2000.3446. [DOI] [PubMed] [Google Scholar]
- 25.Hong D. I. University of Tokyo: 1977. Studies on the sex maturation and oviposition in Penaeus japonicus. Ph.D. Thesis. [Google Scholar]
- 26.Endo H. University of Tokyo; 2004. Molecular analysis of calcification in the crustacean exoskeleton. Ph.D. Thesis. [Google Scholar]
- 27.Kotake A., Arai T., Ozawa T., Nojima S., Miller M. J., Tsukamoto K. Variation in migratory history of Japanese eels, Anguilla japonica, collected in coastal waters of the Amakusa Islands, Japan, inferred from otolith Sr/Ca ratios. Mar. Biol. 2003;142:849–854. [Google Scholar]
- 28.Maruyama K., Mikawa T., Ebashi S. Detection of the calcium-binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J. Biochem. (Tokyo) 1984;95:511–519. doi: 10.1093/oxfordjournals.jbchem.a134633. [DOI] [PubMed] [Google Scholar]
- 29.Kogure T. A program to assist Kikuchi pattern analyses. J. Cryst. Soc. Jpn. 2003;45:391–395. [Google Scholar]
- 30.Watanabe T. Isolation of a cDNA encoding a homologue of ribosomal protein L26 in the decapod crustacean Penaeus japonicus. Mol. Mar. Biol. Biotechnol. 1998;7:259–262. [PubMed] [Google Scholar]
- 31.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Rebers J. E., Riddiford L. M. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J. Mol. Biol. 1988;203:411–423. doi: 10.1016/0022-2836(88)90009-5. [DOI] [PubMed] [Google Scholar]
- 33.Rebers J. E., Willis J. H. A conserved domain in arthropod cuticular proteins binds chitin. Insect Biochem. Mol. Biol. 2001;31:1083–1093. doi: 10.1016/s0965-1748(01)00056-x. [DOI] [PubMed] [Google Scholar]
- 34.Roer R., Dillaman R. The structure and calcification of crustacean cuticle. Am. Zool. 1984;24:893–909. [Google Scholar]
- 35.Testeniere O., Hecker A., Gurun S. L., Quennedey B., Graf F., Luquet G. Characterization and spatiotemporal expression of orchestin, a gene encoding an ecdysone-inducible protein from a crustacean organic matrix. Biochem. J. 2002;361:327–335. doi: 10.1042/0264-6021:3610327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hecker A., Testeniere O., Marin F., Luquet G. Phosphorylation of serine residues is fundamental for the calcium-binding ability of Orchestin, a soluble matrix protein from crustacean calcium storage structures. FEBS Lett. 2003;535:49–54. doi: 10.1016/s0014-5793(02)03856-5. [DOI] [PubMed] [Google Scholar]
- 37.Addadi L., Raz S., Weiner S. Taking advantage of disorder: amorphous calcium carbonate and its roles in biomineralization. Adv. Mater. 2003;15:959–970. [Google Scholar]
- 38.Coblentz F. E., Shafer T. H., Roer R. D. Cuticular proteins from the blue crab alter in vitro calcium carbonate mineralization. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1998;121:349–360. [Google Scholar]
- 39.Roer R. D., Halbrook K. E., Shafer T. H. Glycosidase activity in the post-ecdysial cuticle of the blue crab, Callinectes sapidus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001;128:683–690. doi: 10.1016/s1096-4959(00)00363-8. [DOI] [PubMed] [Google Scholar]
- 40.Pierce D. C., Butler K. D., Roer R. D. Effects of exogenous N-acetylhexosaminidase on the structure and mineralization of the post-ecdysial exoskeleton of the blue crab, Callinectes sapidus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001;128:691–700. doi: 10.1016/s1096-4959(00)00362-6. [DOI] [PubMed] [Google Scholar]
- 41.Dunker A. K., Ruckert R. R. Observations on molecular weight determinations on polyacrylamide gel. J. Biol. Chem. 1969;244:5074–5080. [PubMed] [Google Scholar]
- 42.Martin J. H., Benzer S., Rudnicka M., Miller C. A. Calphotin: a Drosophila photoreceptor cell calcium-binding protein. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1531–1535. doi: 10.1073/pnas.90.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballinger D. G., Xue N., Harshman K. D. A Drosophila photoreceptor cell-specific protein, calphotin, binds calcium and contains a leucine zipper. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1536–1540. doi: 10.1073/pnas.90.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]