Abstract
Fertilization begins with interaction between the sperm and the egg. The surface of the vertebrate oocyte is covered with the egg envelope, which is composed of ZP (zona pellucida) glycoproteins. We have identified two glycoproteins, ZP1/gp97 and ZPC/gp42, as the major components of the chicken egg envelope. In the present study, another 42 kDa protein, designated ZPD, has been found as a new major component of the chicken egg envelope. ZPD was specifically released from the egg envelope by ultrasonication treatment without urea. ZPD cDNA was cloned using a chicken granulosa cell cDNA pool. The deduced amino acid sequence showed that preproprotein of ZPD is composed of 418 amino acid residues with four potential N-glycosylation sites and includes a ZP domain, common in vertebrate ZP glycoproteins, and a transmembrane domain. ZPD belongs phylogenetically to a distinct group from known ZP glycoprotein subfamilies, ZPA, ZPB, and ZPC. In two-dimensional gel electrophoresis ZPD proteins were identified to be several isoforms with different pI values between 5 and 7. ZP1, ZPC and the newly identified ZPD were confirmed to be the major components of chicken egg envelope by MS of proteolytic digests of whole egg envelope. The in vitro incubation of chicken sperm with calcium ionophore A23187 induced sperm activation, resulting in the fragmentation and release of a 41 kDa PNA (peanut agglutinin)-positive glycoprotein and the decrease or loss of sperm PNA-stainability. The incubation with ZPD and dimeric ZP1, but not ZPC and monomeric ZP1, also induced the decrease or loss of sperm PNA-stainability, suggesting the in vitro sperm activation by these ZP components. Collectively, ZPD might bind loosely to egg envelope matrix and play a key role in the sperm activation on avian sperm–egg interaction.
Keywords: acrosome reaction, fertilization, glycoprotein, mass spectrometry, sperm, zona pellucida
Abbreviations: CBB, Coomassie Brilliant Blue; MALDI, matrix-assisted laser-desorption ionization; PNA, peanut agglutinin; poly(A)+, polyadenylated; 5′-RACE, rapid amplification of 5′ cDNA ends; TOF, time-of-flight; ZP, zona pellucida
INTRODUCTION
Fertilization is composed of multiple sequential steps to transfer the genome of one generation to the next generation [1]. The first step is the interaction of the sperm with the egg surface. The oocytes in vertebrates are covered by the envelope, which forms a hard extracellular matrix structure and is called the ZP (zona pellucida) in mammals [2]. The binding between the sperm and the egg envelope initiates exocytosis of the acrosomal contents, including proteases and glycosidases to lyse the egg envelope, from the sperm head, and activates sperm signal pathways [3]. This change in the sperm is called the acrosome reaction. During and after the acrosome reaction, the activated sperm detaches from the egg envelope, penetrates the envelope, and binds to and fuses with the oocyte plasma membrane. The acrosome reaction is therefore essential for subsequent fertilization processes to inject the sperm nucleus into the egg cytosol. The initiation mechanism of acrosome reaction is thus inevitable to describe the whole story of vertebrate fertilization.
The egg envelope in vertebrates has been reported to be composed of ZP glycoproteins [1,2,4]. The ZP glycoproteins have a common ZP domain, which is about 260 residues long and contains eight or ten strictly conserved cysteine residues [5]. This domain has additional conservation of hydrophobicity, polarity, and turn-forming tendency, suggesting a common three-dimensional structure. Although the function of this domain is largely unclear, a recent paper has demonstrated its importance in polymerization of ZP glycoproteins [6]. To date, many ZP glycoproteins have been isolated and cloned, and form a large ZP glycoprotein family. Mammalian ZP glycoproteins have been classified by amino acid sequence comparison into three subfamilies, ZPA, ZPB and ZPC [7]. The composition of the ZP glycoproteins expressed in one species shows obvious variations among vertebrates, especially non-mammalian ones. For example, while three kinds of ZP glycoproteins (ZPA/ZP2, ZPB/ZP1 and ZPC/ZP3) are expressed in mice [2], eight and five genes encoding ZP glycoproteins have been found in Oryzias latipes [8] and Xenopus laevis [9] respectively. The biological significance of these variations is not understood. For example, links to species-specific morphology of the egg envelope and interaction between the egg and the sperm are still speculative. Study on non-mammalian vertebrates such as fish, amphibians and birds is considered to point out the evolutionally conserved parts and modified parts in vertebrate fertilization, providing deeper insights on this complex phenomenon.
We have reported previously that the chicken egg envelope includes two glycoproteins, gp97 and gp42 (designated after their apparent molecular masses on SDS/PAGE) [4,10]. gp42 has been cloned in our recent study and, based on peptide sequence homology, is regarded as a chicken counterpart of mammalian ZPC [10]. gp97 has been cloned by another group and has been termed ZP1 [11]. In the present study, we have cloned a new chicken ZP glycoprotein, identified it as a component of the egg envelope, and suggested the involvement of this protein in sperm activation on sperm–egg interaction.
EXPERIMENTAL
cDNA cloning of a new ZP glycoprotein
Total RNA was prepared from chicken pre-ovulatory ovarian follicles as described previously [10]. Purification of poly(A)+ (polyadenylated) RNA and preparation of a cDNA pool with an adaptor-combined oligo(dT) primer were described in our previous paper [12]. PCR was performed to amplify DNA including a ZP domain sequence from the cDNA pool under the following conditions. The forward degenerate primer, 5′-GA(C/T)CCCAACATCAAGCTGGT-3′, was designed on the basis of the conserved nucleotide sequences of ZP domains among human ZPA (GenBank® accession no. M90366), mouse ZPA (GenBank® accession no. M34148), pig ZPA (GenBank® accession no. D45064) and frog ZPA/gp69 (GenBank® accession no. AF038151). The reverse adaptor primer was 5′-CAGAATTCAGCTGCAGGATCC-3′. Amplification was carried out with recombinant Taq polymerase (Takara Biomedicals, Otsu, Japan) by 30 cycles of denaturation at 94 °C for 0.5 min, annealing at 55 °C for 0.5 min and extension at 72 °C for 1 min. Reaction products were separated on a 1.5% agarose gel and stained with ethidium bromide to visualize DNA bands under UV light. Amplified cDNA fragments were isolated from the gel by use of the QIAEXII gel extraction kit (Qiagen, Hilden, Germany), subcloned into pGEM-T Easy vector (Promega, Madison, WI, U.S.A.) according to the manufacturer's instructions and sequenced using the ABI PRISM 310 DNA sequencer (Applied Biosystems, Foster City, CA, U.S.A.). 5′-RACE (rapid amplification of 5′ cDNA ends) was performed using reverse transcriptase (Superscript II, Invitrogen, Carlsbad, CA, U.S.A.) and the ovarian follicle total RNA. The following reverse primers recognizing our new cDNA were used: 5′-GATGCGGTCTTGTACAGCCT-3′, for first-strand cDNA synthesis, and 5′-CATGCTGACGTTGAAGTGTCC-3′, for the subsequent PCR. Amplified DNA was isolated, subcloned and sequenced as described above.
The cDNA sequence was subjected to a BLAST search (National Center for Biotechnology Information, Bethesda, MD, U.S.A.). The signal peptide region of a translated product was predicted by the PROSITE database search (at http://www.expasy.org/tools/scanprosite/).
Collection and solubilization of the chicken egg envelope
The egg envelope was isolated from the largest pre-ovulatory mature follicles of laying White Leghorn hens as described previously [10]. Briefly, the granulosa cell layer composed of the perivitelline layer (egg envelope), the monolayer of granulosa cells and the basal lamina (basement membrane) was mechanically separated from the oocyte with forceps. The granulosa cells and the basement membrane were removed from the egg envelope by shaking it gently in distilled water with forceps. After checking for isolation under a stereoscopic microscope, the egg envelope was stored at −20 °C until use.
The isolated egg envelope was suspended in 500 μl of cold PBS and subjected to sonication (output, 3; duty cycle, 30%) using Sonifier 250 (Branson Ultrasonics Corporation, Danbury, CT, U.S.A.). The egg envelope suspension was centrifuged at 13000 g for 20 min to separate the supernatant (fraction X) and the precipitate. After being washed with cold PBS twice, the precipitate was sonicated in 7 ml of the solubilization buffer [50 mM sodium acetate buffer, pH 5.0, containing 0.2 M NaCl, 50 mM glycine and 8 M urea (ultrapure grade; ICN Biomedicals, Irvine, CA, U.S.A.)] and kept on ice for 60 min for further solubilization. This solution was then centrifuged at 10000 g for 15 min to remove insoluble materials, and the supernatant was filtered through a 0.45-μm-pore-size cellulose acetate filter. The filtrate (fraction Y) was immediately used for gel filtration. To solubilize all envelope proteins for electrophoretic analysis, the egg envelope was sonicated in the presence of 8 M urea and, after centrifugation, the supernatant was subjected to electrophoresis.
Gel electrophoresis, immunoblotting and lectin blotting
Mouse polyclonal anti-(chicken ZPD) antibody was raised against glutathione S-transferase fused with full-length ZPD protein, which was prepared using pGEM 4T-3 vector (Amersham Biosciences, Piscataway, NJ, U.S.A.) and our ZPD cDNA. Mouse monoclonal antibody against chicken ZP1 [4] and mouse polyclonal antibody against chicken ZPC [10] were described previously.
SDS/PAGE was performed according to the method of Laemmli [13]. Under reducing conditions, protein samples were boiled for 3 min in SDS/PAGE sample buffer including 2-mercaptoethanol. Under non-reducing conditions, samples were boiled in the same way, but in the absence of 2-mercaptoethanol. To detect protein bands, gels were stained with CBB (Coomassie Brilliant Blue) R-250. For immunoblotting, proteins were electroblotted on to a PVDF membrane (Immobilon; Millipore, Bedford, MA, U.S.A.). After blocking with 2.5% gelatin, the membrane was incubated with primary antibody and then with horseradish-peroxidase-labelled anti-(mouse IgG) antibody (Cappel, Costa Mesa, CA, U.S.A.). The signal was detected by the ECL® (enhanced chemiluminescence) Western Blotting Detection Reagents (Amersham Biosciences). For lectin blotting analysis, electroblotting and blocking were performed as described above. The membrane was incubated with PNA [peanut (Arachis hypogaea) agglutinin] (Seikagaku Cooperation, Tokyo, Japan), with rabbit anti-PNA IgG (ICN Biomedicals), and finally with peroxidase-labelled anti-(rabbit IgG) antibody. Detection was performed using the ECL® reagents. Two-dimensional gel electrophoresis was performed according to the method of O’Farrell [14] using the Bio-Rad two-dimensional electrophoresis apparatus (Hercules, CA, U.S.A.). Separated proteins were visualized using the silver staining kit (Wako Pure Chemical Industries, Osaka, Japan).
Gel filtration
Fraction Y was applied on a column of Sephacryl S-400 SF (Amersham Biosciences) (gel bed: 2.6 cm×88 cm) equilibrated previously with the elution buffer: 50 mM sodium acetate buffer, pH 5.0, containing 6 M urea, 0.2 M NaCl and 50 mM glycine. The column was eluted at a rate of 18 ml/h, and 5.9-ml fractions were collected. The absorbance at 280 nm of each fraction was measured by a spectrophotometer U-2001 (Hitachi, Tokyo, Japan). Peak fractions were analysed by SDS/PAGE followed by CBB staining. Pooled fractions were dialysed extensively against distilled water [15], freeze-dried and dissolved in PBS before use.
Amino acid sequencing analysis
Fraction X and its tryptic digest were subjected to SDS/PAGE, and the proteins in the gel were electroblotted on to a PVDF membrane. The bands of ZPD and its fragment on the membrane stained with CBB were excised and applied to a peptide sequencer, Procise HT (Applied Biosystems).
Protease digestion of egg envelope
The egg envelope (1.04 mg wet mass) was solubilized with 15 μl of 0.1% SDS, 5 mM 2-mercaptoethanol and 50 mM Tris/HCl (pH 8.0), and heated at 95 °C for 20 min for protein denaturation. Then 4.7 units of trypsin (Trypsin Gold, MS Grade; Promega) or 0.1 μg of endoproteinase Lys-C (Sigma-Aldrich, St. Louis, MO, U.S.A.) in 5 μl of 50 mM Tris/HCl (pH 8.0) was added to the solubilized and denatured egg envelope, and the mixture was incubated at 37 °C for 2–18 h.
MS and data analysis
The proteolytic digests (0.5 μl) were spotted on to the MALDI (matrix-assisted laser-desorption ionization) sample target plate with a matrix consisting of a saturated solution of α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) prepared in 50% acetonitrile/0.1% trifluoroacetic acid. MS and MS/MS (tandem MS) analyses were performed in the positive ion reflector mode by using a MALDI–TOF/TOF (TOF is time-of-flight) mass spectrometer (4700 Proteomics Analyzer; Applied Biosystems). Peptide mass spectra were obtained in the mass range 850–4000 Da. Mass accuracy of the instrument was externally calibrated to the 4700 Proteomics Analyzer Calibration Mixture (Applied Biosystems). Amino acid sequences of the major MS peaks were deduced from the MS/MS data using the DeNovo Explorer™ software (Applied Biosystems). The obtained peptide sequences were assigned to known proteins by using a BLAST search, while theoretical masses of the peptides were calculated with the PeptideMass software (http://us.expasy.org/tools/peptide-mass.html).
Chicken sperm preparation and sperm activation by calcium ionophore
Semen was collected from White Leghorn cocks after ejaculation induced by lumbar massage. The sperm concentration was calculated by using a haemocytometer (Nitirin, Tokyo, Japan) under a phase-contrast microscope (IMT-2; Olympus, Tokyo, Japan).
Semen (200 μl) including 9.7×108 spermatozoa was centrifuged at 250 g for 10 min, and the supernatant was collected as seminal plasma. The precipitate (sperm) was washed twice with cold PBS, centrifugation at 250 g for 10 min, and finally resuspended in 500 μl of cold PBS. Half of this sperm suspension was centrifuged again at 250 g for 10 min, and the precipitate (4.9×108 spermatozoa) was used as the intact sperm. The other half of the sperm suspension was also centrifuged at 250 g for 10 min, and the precipitated sperm was suspended in cold PBS containing 20 μM calcium ionophore (A23187; Wako Pure Chemical Industries). This sperm suspension containing A23187 was incubated at 39 °C for 30 min to induce sperm activation leading to acrosome reaction [16] and centrifuged at 13000 g for 5 min. The precipitate was used as the A23187-treated sperm.
Sperm activation by ZP components and PNA staining
Sperm washed with PBS was prepared as described above. The sperm suspension (1×106 spermatozoa in 10 μl of PBS) was mixed with an equal volume of each ZP preparation and incubated at 39 °C for 30 min. As the positive and negative controls, the sperm suspensions were incubated with 10 μl of PBS containing 40 μM A23187 and PBS respectively. Sperm solution (10 μl) was then smeared on to a poly(L-lysine)-coated glass slide and kept for 10 min at room temperature (25 °C). The spermatozoa on slides were washed with PBS and incubated with PBS in the presence or absence of 10 μg/ml PNA for 60 min at room temperature. After washing with PBS, the spermatozoa on slides were fixed with 2% (w/v) paraformaldehyde in PBS for 15 min at room temperature and then washed with PBS. The fixed spermatozoa on slides were incubated with 3 μg/ml anti-PNA rabbit IgG antibody overnight at 4 °C, washed with PBS and finally incubated with 2 μg/ml Alexa Fluor® 488 goat anti-rabbit IgG (Molecular Probes, Eugene, OR, U.S.A.) overnight at 4 °C in darkness. After washing with PBS, the spermatozoa on slides were examined under a fluorescence microscope (BX-60; Olympus). The number of spermatozoa whose heads were labelled with Alexa Fluor® 488 (=PNA-positive) was counted for each field. The total number of spermatozoa (43–185 per field) was also counted under phase contrast.
CD measurement
Each protein sample was dissolved in PBS at a concentration of 280 and 171 μg/ml for fractions I and II respectively, and transferred to a quartz cell with a 1-mm path length. The CD spectra were recorded on a Jasco J-720 spectropolarimeter with Jasco J-700 Spectra Manager version 1.35 software (Tokyo, Japan). The CD spectrum of each sample was measured three times in accumulation mode at room temperature in the wavelength range 200–240 nm, corrected by subtraction of the background solvent spectrum obtained under similar experimental conditions and smoothed for clarity of display. All data were converted to give molar ellipticity values ([θ], deg·cm2·dmol−1) based upon the concentrations of the samples.
Protein determination
Protein concentration was estimated by using the BCA (bicinchoninic acid) protein assay kit (Pierce, Rockford, IL, U.S.A.), except that densitometric analysis of the CBB-stained polyacrylamide gel by an ATTO Lane and Spot Analyzer 6.0 (ATTO corporation, Tokyo, Japan) was used for the ZPD preparation because of its slight turbidity. BSA was used as the standard.
RESULTS
Chicken ZPD is a new member of the ZP glycoprotein family
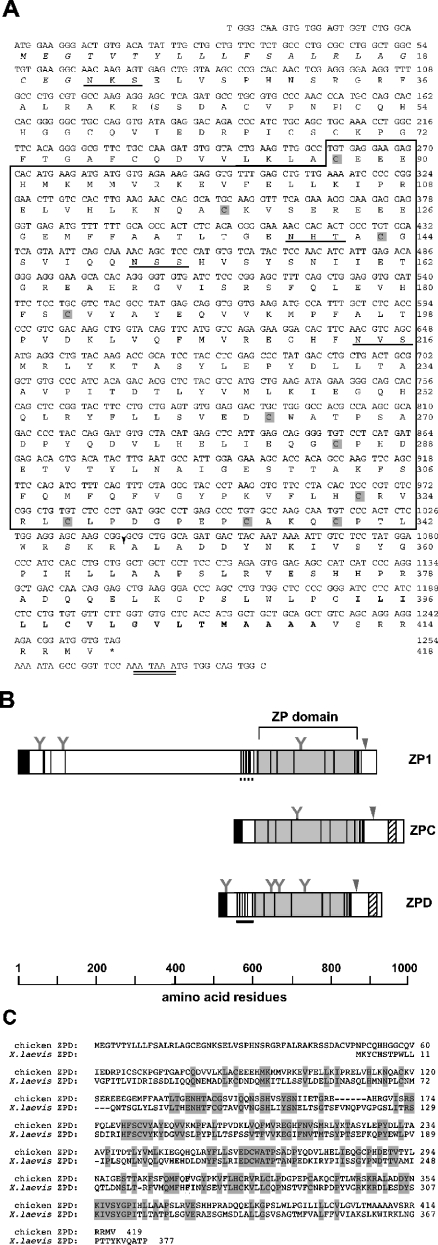
In search of cDNA encoding a new ZP glycoprotein by degenerate PCR (see the Experimental section for detailed methods), we found a DNA fragment (0.8 kb) including a nucleotide sequence of the ZP domain and a poly(A)+ tail (results not shown). Extension of the 5′-end of the cDNA was achieved by 5′-RACE. The fulllength cDNA thus obtained (1.4 kb) had an open reading frame encoding 418 amino acids (Figure 1A). The nucleotide sequence flanking the first ATG was in accordance with the Kozak consensus sequence for translation initiation [17], and a consensus polyadenylation signal [18] was present in the 3′ region of the cDNA (Figure 1A). The region of N-terminal 21 amino acid residues is hydrophobic and is likely to be a signal peptide according to the prediction by the PROSITE database search. The calculated molecular mass of the full-length pre-pro-form of this protein is 47069 Da. The translated sequence includes four potential N-glycosylation sites and is rich in serine and threonine residues (a total of 47 residues), implying O-glycosylation of the protein. The domain structures of this protein and other chicken ZP glycoproteins are depicted in Figure 1(B). In the middle of the new protein, the ZP domain with ten conserved cysteine residues is identified. Between the ZP domain and the signal sequence, there is a cysteine-rich region similar to the EGF (epidermal growth factor)-like domain. The hydrophobic transmembrane region is predicted to be in a C-terminal region. Upstream of the transmembrane region, there is a cleavage site recognized by furin protease, which works for processing of secretory proteins [consensus cleavage site, RX(K/R)R] [19]. Therefore it appears that newly synthesized ZPD is cleaved by furin and that its C-terminally truncated form is secreted into an extracellular space. The molecular masses of its pro- and mature forms generated by the removal of the putative signal peptide and the C-terminal transmembrane domain are calculated to be 44843 and 37091 Da respectively. In addition to the presence of the ZP domain, the structural properties described here are commonly found in vertebrate ZP glycoproteins, which suggests that our clone is an authentic member of the ZP glycoprotein family.
Figure 1. Structure and characterization of chicken ZPD.
(A) Nucleotide and deduced amino acid sequences of ZPD. The ZP domain is boxed, and conserved cysteine residues in this domain are shaded. The putative signal sequence and the putative transmembrane region are shown in italics and in bold respectively. Potential N-glycosylation sites and a putative polyadenylation signal sequence are underlined and double underlined, respectively. A putative furin cleavage site is indicated by the arrowhead. The sequence corresponding to the N-terminal sequence of a ZPD-tryptic peptide is indicated within parentheses. These sequence data are available from GenBank® under accession no. AB114441. (B) Schematic representation of chicken ZP1 [11], ZPC [10] and ZPD. Light grey squares represent ZP domains. Cysteine residues are shown as vertical lines. N-terminal signal peptide regions and C-terminal transmembrane regions are indicated by dark grey squares and hatched squares respectively. Potential furin cleavage sites are indicated by the arrowheads. N-glycosylation sites are also indicated as ‘Y’ marks. The trefoil [27] and EGF (epidermal growth factor)-like domains are shown by horizontal dashed and solid lines respectively. (C) Alignment of amino acid sequences of chicken ZPD and Xenopus ZPD [20]. Conserved residues are boxed and shaded. Gaps inserted for maximum matching are represented by dashes.
In spite of common features of the domain structure of our protein, searches through the database indicated that the peptide sequence outside the ZP domain was not significantly similar to that of ZP glycoproteins belonging to the ZPA, ZPB and ZPC subfamilies. Therefore we have tentatively designated this protein ZPD. Chicken ZPD has only limited similarity to a recently reported Xenopus laevis egg envelope protein, ZPD [20], which is shown in Figure 1(C) (identities, 37%; positives, 56%).
Chicken ZPD is a component of the egg envelope
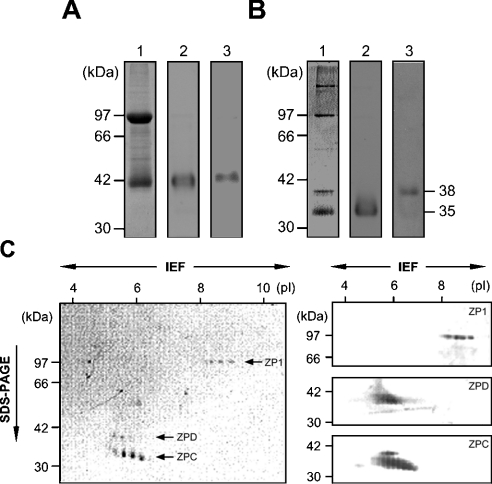
We then determined whether ZPD protein was a component of the egg envelope by electrophoretic methods. SDS/PAGE of total envelope proteins under reducing conditions followed by CBB staining (left-hand panel of Figure 2A) showed two major bands, ZP1/gp97 and ZPC/gp42, which is consistent with our previous report [4]. No additional ZPD-like band was identified under these conditions. Under non-reducing conditions, however, a total of four major protein bands were detected (left-hand panel of Figure 2B). To identify these bands, immunoblotting with antibodies against ZP1, ZPC and ZPD was performed. The upper two bands under non-reducing conditions (97 kDa and 180 kDa) were stained with anti-ZP1 antibody (results not shown) [4]. Because ZP1 forms a homodimer covalently linked with disulphide bonds [4], the two bands that cross-reacted with anti-ZP1 antibody were judged to be ZP1 dimer and ZP1 monomer. Under non-reducing conditions, ZPC migrated much faster (compare the middle panels of Figures 2A and 2B), so that the ZPC band was observed as a 35-kDa band at the bottom of the gel (left-hand panel of Figure 2B). Immunoblotting with anti-ZPD antibody revealed that the cross-reactive materials of 42 kDa and 38 kDa exist under reducing and non-reducing conditions respectively, and that the protein that was cross-reactive with anti-ZPD antibody co-migrated with ZPC under reducing conditions (right-hand panels of Figures 2A and 2B). These two bands were separated under non-reducing conditions mainly due to the drastic shift of the ZPC band described above. These data indicate that ZPD is included in the egg envelope as its main constituent. The N-terminal amino acid sequence for the ZPD band could not be determined even when a larger amount of protein was subjected to analysis. Therefore ZPD was digested with trypsin, resulting in the production of a 37 kDa fragment. The N-terminal amino acid sequence was determined to be SSDAXVPNP, which corresponds to the sequence Ser43–Pro51 in the deduced amino acid sequence of ZPD (see Figure 1A).
Figure 2. Electrophoresis of egg envelope proteins.
SDS/PAGE under reducing (A) and non-reducing (B) conditions. Proteins separated on 10% gels were stained with CBB (lane 1) or subjected to immunostaining with anti-ZPC antibody (lane 2) and anti-ZPD antibody (lane 3). The migration positions of the protein markers are shown in the far left-hand panels. (C) Two-dimensional gel electrophoresis. Proteins were first separated by isoelectric focusing (IEF; pH 3–10), then subjected to SDS/PAGE (10% gel, non-reducing conditions), and finally detected by silver staining (left-hand panel) and immunoblotting (right-hand panels). Theoretical pI values are indicated on top of the gel. The migration positions of the protein mass markers are shown on the left of the panels. The righthand panels are the blotted membranes stained with anti-ZP1 (top), anti-ZPD (middle) and anti-ZPC (bottom) antibodies respectively.
To test the possibility that other unidentified proteins with identical or similar molecular masses still existed in the egg envelope, two-dimensional electrophoresis was performed. Each of the three bands (97 kDa, 38 kDa and 35 kDa under non-reducing conditions) was separated further into 4–6 protein spots by isoelectric focusing (left-hand panel of Figure 2C). These spots were stained specifically with each corresponding antibody (right-hand panel of Figure 2C), indicating that all these proteins with similar molecular masses and different pIs are isoforms of each ZP glycoprotein. ZP1 was detected in the region between pI 8 and pI 9, whereas ZPC and ZPD were between pI 5 and pI 7. These estimated pIs of the three ZP glycoproteins roughly agreed with the theoretical pI values for mature proteins of ZP1 (7.93), ZPC (6.38) and ZPD (6.40). Multiple spots of ZP1, ZPC and ZPD appeared in isoelectric focusing, probably due to post-translational modifications such as heterogeneous sialylation of these proteins. In two-dimensional electrophoresis, neither silver staining nor immunoblotting could detect a ZP1 dimer, due to low solubility of ZP1 dimer or polymer in the isoelectric focusing sample buffer (results not shown).
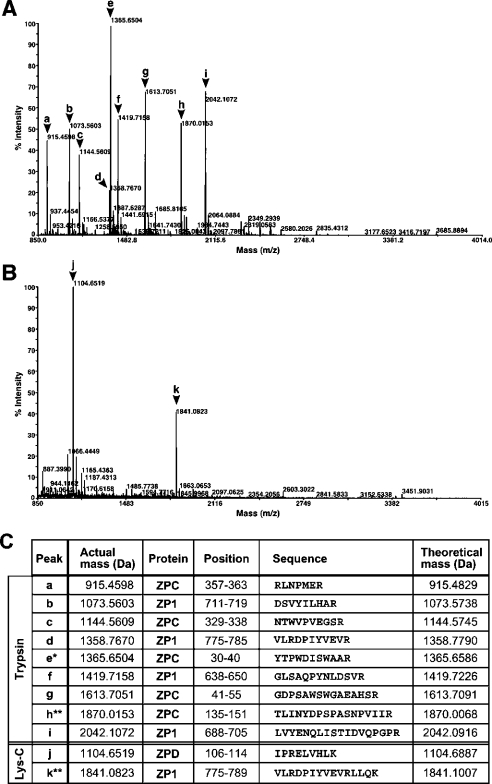
To confirm that the major constituents of chicken egg envelope are ZP1, ZPC and the newly identified ZPD, whole egg envelope was degraded by tryptic or Lys-C limited hydrolysis, and resultant peptide fragments were subjected to MS analyses. Figures 3(A) and 3(B) show typical mass spectra of the tryptic and Lys-C digests. Nine major MS peaks were detected in the tryptic digest (2 h digestion), while only two major ones were seen in the Lys-C digest (18 h digestion). Almost the same mass spectra with lower mass intensities were obtained by the proteolytic hydrolysis for longer or shorter periods. As shown in Figure 3(C), all these MS peaks except two were identified to be peptide fragments from ZP1, ZPC or ZPD by MS/MS sequencing, while the mass data of the two peak exceptions (h and k) agreed well with theoretical masses of ZP1 and ZPC fragments. Thus all tryptic and Lys-C peptides could be assigned to ZP1, ZPC or ZPD.
Figure 3. Identification of peptides derived from tryptic and Lys-C digests of egg envelope constituents by MS.
The tryptic digest (2 h incubation) (A) and the Lys-C digest (18 h incubation) (B) were analysed by using a MALDI–TOF/TOF mass spectrometer. The major peaks (arrowheads a to k) were subjected to further MS/MS analysis to obtain sequence data. The mass and sequence data for the major 11 peptides are summarized in (C). *, N-terminal end fragment of ZPC mature protein. **, no sequence data could be obtained for these two peptides by MS/MS analysis.
ZPD and dimeric ZP1 are key components of sperm–egg envelope interaction
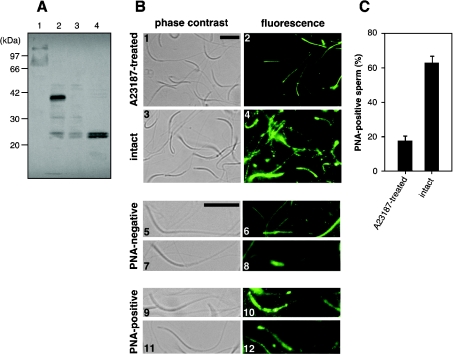
Because all major ZP glycoproteins in chicken egg envelope were identified, we started to examine which of the envelope glycoproteins was involved in sperm–egg envelope interaction. For this purpose, we constructed a quantitative assay for sperm activation utilizing the reactivity of PNA to the sperm [21], which shows high affinity to the disaccharide structure, Gal(β1-3)-GalNAc- [22]. First, biochemical changes in PNA-positive sperm glycoprotein(s) were examined by the lectin blotting analysis before and after activation with the calcium ionophore, A23187, which was reported to be a chemical inducer of the acrosome reaction [16]. By PNA-staining, a 41 kDa band was detected from the lysates of the intact sperm, but became undetectable after incubation with A23187 for 30 min (lanes 2 and 3 of Figure 4A). This 41 kDa band was not detected from the seminal plasma of untreated sperm samples (lane 1 of Figure 4A), excluding the possibility that the 41 kDa glycoprotein was a contaminated seminal plasma protein. Furthermore, two PNA-positive bands of approx. 24 kDa were detected in the supernatant of the A23187-treated sperm (lane 4 of Figure 4A), indicating that the 41 kDa glycoprotein was degraded and released from sperm during sperm activation. Our data suggested that the 41 kDa glycoprotein recognized by PNA could be used as a marker to discriminate the intact sperm from the sperm activated by interaction with egg envelope.
Figure 4. Establishment of a sperm activation assay.
(A) Lectin blotting. Proteins from seminal plasma (lane 1), intact sperm (lane 2), sperm incubated in the presence of A23187 (lane 3) and the supernatant of the A23187-treated sperm (lane 4) were subjected to SDS/PAGE (12% polyacrylamide gel), electroblotted on to a PVDF membrane and stained with PNA. The migration positions of the protein markers are shown on the left of the panel. (B) Typical observation by fluorescence microscopy. Detailed assay conditions are described in the Experimental section. Sperm samples treated with A23187 (panels 1 and 2) and intact sperm samples (panels 3 and 4) were visualized by phase-contrast (panels 1 and 3) and staining with PNA and immunofluorescence (panels 2 and 4). Typical staining patterns of PNA-negative (panels 5, 6, 7 and 8) and -positive (panels 9, 10, 11 and 12) sperm were shown. Scale bar, 12 μm. The ratio of the number of PNA-positive cells to total cell number is summarized by solid bars in (C). Results are means±S.D. for five fields. Another independent experiment was performed with similar results.
Next, a histochemical method was adopted for in situ detection of the binding of PNA to the sperm. Namely, spermatozoa were fixed on glass slides and treated sequentially with PNA, rabbit anti-PNA antibody and Alexa Fluor® 488 goat anti-rabbit IgG. PNA-bound spermatozoa were visualized under a fluorescence microscope. Figure 4(B) shows typical microscopic views in control experiments. Fluorescence was strongly detected from the head of intact sperm, especially the tip portion (panels 9, 10, 11 and 12 of Figure 4B). Such PNA-stainability was diminished from the activated sperm that had been treated with A23187. These suggest the preferential binding of PNA to the intact sperm through the 41 kDa glycoprotein described above. The PNA-stainability of sperm was lost by A23187 treatment, probably due to the induction of morphological and/or biochemical changes in the sperm so as to degrade and release the 41 kDa glycoprotein (Figure 4A). The quantitative data of these control experiments are summarized in Figure 4(C). The number of PNA-positive sperm obviously decreased in sperm samples treated with A23187, as compared with PBS-treated sperm samples (Figure 4C). Taken together, it was concluded that sperm activation by the calcium ionophore A23187 resulted in release of a PNA-positive glycoprotein from the sperm surface.
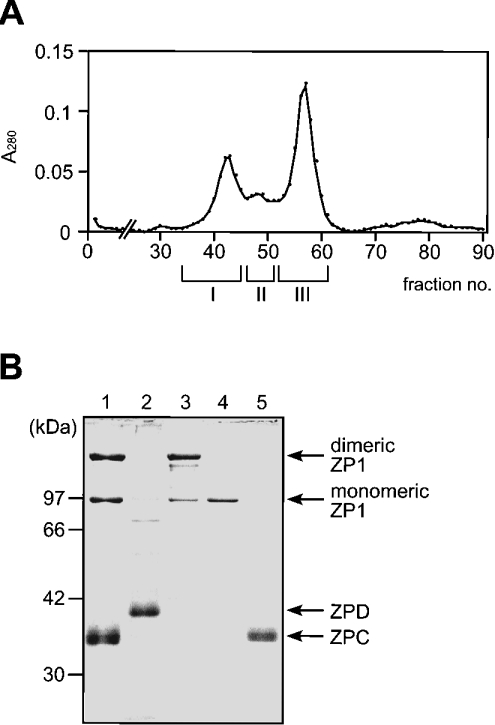
In order to examine the biological activity of egg envelope components on the sperm–egg envelope interaction, the ZP glycoproteins were isolated from the mature egg envelope (Figure 5). As mentioned below in the Discussion section, we found that, distinct from other ZP glycoproteins, ZPD was released quantitatively from the egg envelope by sonication in the absence of urea. ZPD prepared by this method (fraction X) was almost homogeneous as judged by SDS/PAGE followed by CBB staining (lane 2 of Figure 5B). After the removal of ZPD from the egg envelope, ZP1 and ZPC were solubilized successfully by sonication at pH 5.0 in the presence of 8 M urea (under non-reducing conditions), and could be separated from each other by gel filtration (Figure 5A). As shown in Figure 5(B) (lanes 3–5), SDS/PAGE under non-reducing conditions indicated that fractions I, II, and III contained dimeric and monomeric forms of ZP1, monomeric ZP1, and ZPC respectively. However, fraction I, as well as fraction II, showed a single band of monomeric ZP1 by SDS/PAGE under reducing conditions (results not shown), indicating the presence of intermolecular disulphide bond(s) in the dimeric form. The identification of all ZP glycoproteins was confirmed by immunoblotting with their specific antibodies (results not shown). Then, these four fractions (I, II, III and X) were used as dimeric/monomeric ZP1, monomeric ZP1, ZPC and ZPD in the following experiments. The yields of fraction I (dimeric/monomeric ZP1), fraction II (monomeric ZP1), fraction III (ZPC) and fraction X (ZPD) were 1.70, 0.92, 1.67, and 0.88 mg respectively from 89.3 mg (wet mass) of the egg envelope.
Figure 5. Preparation of ZP glycoproteins from the egg envelope.
Fraction X was obtained as the supernatant fraction of the egg envelope sonicated in the absence of urea. The precipitated material was then sonicated in the acetate buffer containing 8 M urea and centrifuged at 10000 g for 15 min. The supernatant (fraction Y) was subjected to gel filtration. (A) Gel filtration. Fractions were pooled as indicated by the horizontal bars. Chromatographic conditions are described in the Experimental section. (B) SDS/PAGE of the fractions thus obtained. Proteins were separated on a 10% gel under non-reducing conditions and stained with CBB. The migration positions of the protein markers are shown on the left of the gel. A typical PAGE pattern of egg envelope and the protein bands identified by immunoblotting with antibodies against each ZP glycoprotein are indicated on the right. Lane 1, fraction Y; lane 2, fraction X; lane 3, fraction I; lane 4, fraction II; lane 5, fraction III.
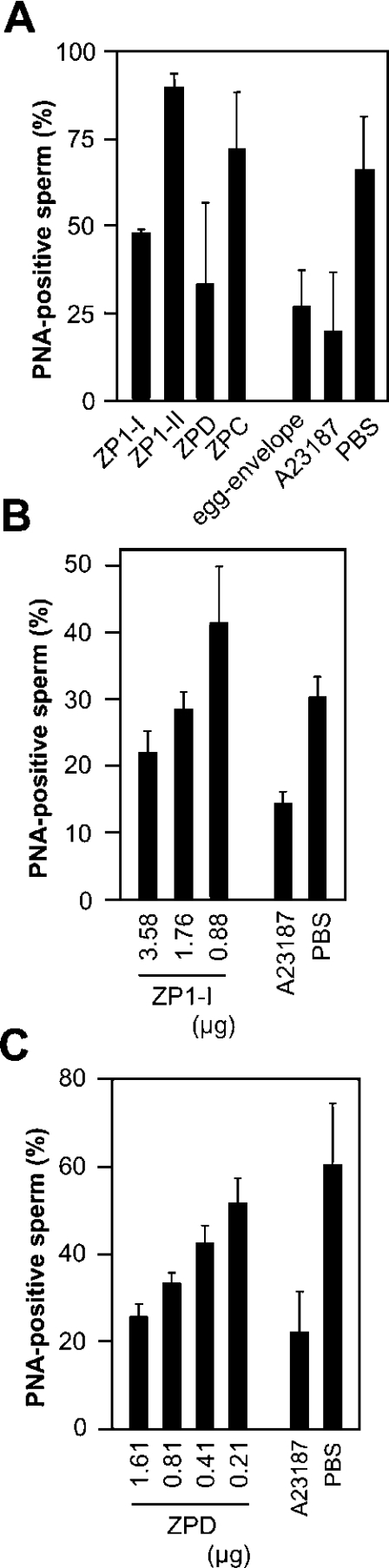
The reactivity of the purified ZP glycoproteins with the sperm was finally analysed by the PNA-staining assay. Incubation of sperm with dimeric ZP1 and ZPD, but not by monomeric ZP1 or ZPC, induced the loss of PNA-stainability from sperm. Fraction I contained the dimeric ZP1 as the major constituent and the monomeric ZP1 as the minor one. Interestingly, no induction was observed for monomeric ZP1 (fraction II), suggesting that dimeric ZP1 or the mixture of dimeric and monomeric ZP1s was the active components in fraction I. Together with dose-dependent induction by dimeric/monomeric ZP1 and ZPD (Figures 6B and 6C), the result suggests that this induction by dimeric ZP1 and ZPD is direct. Because small amounts of dimeric/monomeric ZP1 (3.6 μg) and ZPD (1.6 μg) triggered the sperm activation as crude egg envelope (400 μg wet mass) and A23187 did, dimeric ZP1 and ZPD are considered to be true inducers of sperm activation on sperm–egg envelope interaction.
Figure 6. Sperm activation assay for the ZP glycoprotein preparations.
(A) Sperm activation induced by the four ZP protein preparations. Dimeric/monomeric ZP1 (ZP1-I, 3.58 μg), monomeric ZP1 (ZP1-II, 2.19 μg), ZPC (1.56 μg), ZPD (1.61 μg) and the egg envelope suspended by ultrasonication (400 μg by wet mass) were used for every measurement. (B and C) Dose-dependent induction by dimeric/monomeric ZP1 and ZPD. The amounts of these proteins used for the assay are indicated. The calcium ionophore A23187 and PBS were used instead of the ZP proteins as positive and negative controls respectively. Results are means±S.D. for three (A and B) or five (C) fields. Another independent experiment was performed with similar results.
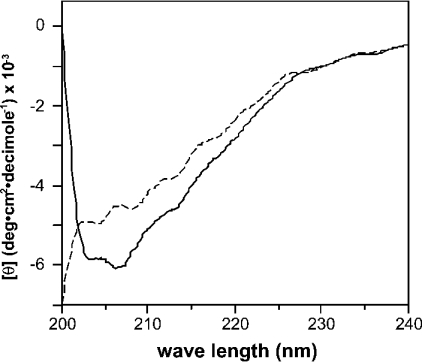
Figure 6 suggests that ZP1 induces the sperm activation when it refolds after removal of urea by dialysis. To analyse the structural feature of ZP1, we measured the far-UV CD spectra of the isolated dimeric and monomeric ZP1s (Figure 7). Because the far-UV CD spectra provide information about the polypeptide backbone and its conformation, CD analysis can be a sensitive assay of changes in protein conformation [23]. The CD spectrum of ZP1 in fraction I showed a negative peak around 205 nm (solid line in Figure 7). No such peak or other apparent peak was observed in the spectrum of ZP1 in fraction II (broken line in Figure 7), suggesting that monomeric ZP1 is rich in unordered structure. Thus the dimeric or polymeric ZP1 was estimated to have ordered secondary structure more than the monomeric one. Although, due to the limitation of our purified materials, NMR or other high-resolution analysis for three-dimensional protein structure was not applicable, the difference in CD spectra suggests that conformational change is accompanied by ZP1 dimerization or polymerization, in addition to covalent disulphide-bond formation between monomers [4], which may contribute to the biological activity of dimeric and polymeric ZP1.
Figure 7. CD spectra of ZP1 in fractions I (solid line) and II (broken line).
The dimeric/monomeric ZP1 in fraction I and the monomeric one in fraction II were dissolved in PBS, and subjected to CD spectrum measurement. Detailed conditions for CD analysis are described in the Experimental section.
DISCUSSION
In the present study, we identified a new egg envelope glycoprotein, ZPD. ZP1 (monomer and dimer) and ZPC have been known to be included in the envelope [4,10,11], but ZPD has not been found until this study. Usually SDS/PAGE of ZP glycoproteins has been performed under reducing conditions. As shown in Figure 2(A), ZPD co-migrates with ZPC on SDS/PAGE under these conditions and is not identified as a separate band. Furthermore, in the present study, the N-terminal amino acid sequence of ZPD could not be determined, probably due to α-amino group blocking, indicating that ZPD hidden behind the ZPC band had escaped the detection by the amino acid sequencing analysis for ZPC in our previous study [10]. ZPC and ZPD are separated under non-reducing conditions (Figure 2B). This difference in electrophoretic patterns is caused mainly by a drastic increase in mobility of ZPC in the absence of a reducing reagent. Although we do not know exactly why this mobility shift occurs, ZPC may have a compact structure supported by intramolecular disulphide bonds. Actually, the CD spectrum of ZPC purified in the present study showed a strong negative peak at 210 nm, suggesting its β-sheet-rich structure (results not shown). Considering together with the implied conformational change of ZP1 accompanied by intermolecular disulphide bond formation (Figure 7), some ZP glycoproteins might utilize disulphide bonds to maintain particular high-order structures.
Although ZPD is a component of the hard egg envelope, ZPD is easily released and separated from the egg envelope matrix by the ultrasonication treatment in the absence of urea (Figure 5B). This unexpected property of ZPD rendered it difficult to evaluate this protein as a main component of the egg envelope. In our initial experiments, some amounts of ZPD were lost during an unnecessarily harsh wash of the collected envelope, so that the yield of ZPD varied from preparation to preparation. We then found that this protein was quantitatively and specifically released under the conditions described here (gentle wash and sonication without urea). According to this procedure, the yield of isolated ZPD has been constant and comparable with that of other ZP proteins in every preparation. Therefore we now believe that ZPD is a major component of the mature egg envelope. Because no other distinct bands and spots were detected by SDS/PAGE (Figures 2A and 2B) and in two-dimensional electrophoresis of total egg envelope proteins (Figure 2C), it is very unlikely that the yet unidentified protein exists as a major component of the mature envelope. This conclusion is supported by the fact that all of the proteolytic peptides from whole egg envelope were assigned to ZP1, ZPC or ZPD (Figure 3). However, the possibility that egg envelope contains such unidentified protein(s) as those with resistance against proteolytic digestion could not completely be ruled out. In fact, the ZPD-derived peptide was only one of the 11 identified peptides, suggesting that proteolytic digestibility of ZP proteins largely differ.
Although many mammalian ZP glycoproteins, which are classified into the ZPA, ZPB and ZPC subfamilies, have been cloned, no ZPD homologue has been found in mammals. ZPD is not unique in birds, because ZPD homologue has been found in Xenopus laevis (Figure 1C) [20]. Therefore ZPD is considered to be distributed in some non-mammalian vertebrates and to form a new subfamily. The finding of ZPD is a new example of diversity of egg envelope components among vertebrates. A recent review has suggested the unexpectedly complex variations in genes encoding vertebrate ZP glycoproteins, especially in non-mammalian ones [9]. We do not know how the variations including ZPD are linked to the evolution of vertebrates and phylogenetic distance among current vertebrate species. To delineate the generation, modification and deletion of genes encoding ZP glycoproteins during the course of vertebrate evolution, a more intensive survey of ZP glycoproteins in many kinds of vertebrates is essential. Recent rapid progress in genomics will help the comprehensive identification of ZP protein genes. The discovery of ZPD stimulates us further to examine in which tissue ZPD is produced. Mammalian ZPA, ZPB and ZPC are produced in the oocyte [2], whereas in birds and fish, some ZP glycoproteins, such as chicken ZP1, are produced in the liver and are transported via the bloodstream to the follicle [11]. While Xenopus laevis ZPD was reported to be produced in the oocyte [20], mRNA of chicken ZPD was expressed in the granulosa cells at the latter stages of folliculogenesis (Y. Kohno, N. Aoki, K. Kitajima and T. Matsuda, unpublished work). Therefore the production of ZPD is unlikely to be the same story between frogs and birds.
The papers on the initial sperm–egg interaction in vertebrates have suggested that there is no paradigm regarding the ZP glycoprotein that interacts with and activates the sperm. In mice, ZPC is an absolute player for sperm activation [2]. Pig ZPC has activity only when it forms hetero-oligomers with ZPB [24]. In Xenopus laevis, gp69/64, homologous with mammalian ZPA, works as a sperm receptor [25]. We have here provided the evidence that ZPD as well as dimeric or polymeric ZP1 stimulates sperm activation (Figure 6), although mechanisms of such stimulation by two different components remain to be investigated. It is unclear that these different interactions may contribute to species specificity. For our activation assay using the PNA-positive glycoprotein as an indicator, ZP proteins were prepared here from the egg envelope by classical biochemical methods (Figure 5). Another possible approach for preparation was the use of recombinant ZP proteins since ZP1, ZPC and ZPD were cloned. However, sugar chains of ZP glycoproteins have been indicated to be important for the sperm–egg interaction in mammals [1,2] and the frog [25]. The involvement of envelope N-glycans in sperm–egg interaction has been reported also in the chicken, although the carrier of N-glycans was not identified [26]. In the present study, ZPD and ZP1, which had four and three potential N-glycosylation sites in their sequences (Figure 1B for N-glycosylation), were shown to stimulate sperm activation. Glycosylation of chicken ZP proteins is indicated also by molecular heterogeneity revealed by isoelectric focusing (Figure 2C). Therefore it is doubtful that the results obtained by use of chicken ZP proteins produced in bacteria would be conclusive. Interestingly, whereas N-glycans are suggested to be involved in the chicken [26], O-glycans have been reported to be involved in the sperm–egg interaction in mice [2] and the frog [25]. Therefore diversity in sperm receptor ZP glycoproteins appears quite complex, including structural differences in sugar chains and their carrier polypeptides. This complexity may be related to the current unconvincing knowledge on the sperm proteins that interact with the egg envelope. In mammals, more than ten sperm proteins have been published as the candidate. However, the biological importance of these proteins is still controversial [1,3]. Microgram-order amounts of the isolated ZP glycoproteins will directly be utilized for further molecular explanation on the sperm–egg interaction, such as the identification of receptors for dimeric ZP1 and ZPD on the sperm surface.
The chicken egg envelope has been visualized as a well-organized porous matrix structure under a scanning electron microscope [4]. In spite of the progress in determining the primary structures of ZP glycoproteins, their association process to finally form the ultrastructure of extracellular matrix remains elusive. As described above, ZPD is released from the envelope without urea and other ZP glycoproteins are not. ZPD is considered to be loosely associated with a strong matrix structure composed of dimeric and monomeric ZP1 and ZPC. To confirm this and to delineate the egg envelope generation process, we first need to study how ZP glycoproteins and their domains bind each other and form a large stable complex. Our ZP proteins and clones will be invaluable to reconstitute the association process in vitro.
Acknowledgments
This work was supported in part by the Grant-in-Aid for The 21st Century COE Program (14COEA12) for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (to T. M., D. N. and K. K.). We thank Dr T. Yamagata and Mr T. Mano (Department of Animal Genetics) for their kind help on chicken semen collection, and Dr N. Tanphaichitr (Ottawa Health Research Institute, Ottawa, Ontario, Canada) for helpful suggestions on the sperm activation assay.
References
- 1.Primakoff P., Myles D. G. Penetration, adhesion, and fusion in mammalian sperm–egg interaction. Science. 2002;296:2183–2185. doi: 10.1126/science.1072029. [DOI] [PubMed] [Google Scholar]
- 2.Wassarman P. M. Zona pellucida glycoproteins. Annu. Rev. Biochem. 1988;57:415–442. doi: 10.1146/annurev.bi.57.070188.002215. [DOI] [PubMed] [Google Scholar]
- 3.Howes L., Jones R. Interaction between zona pellucida glycoproteins and sperm proacrosin/acrosin during fertilization. J. Reprod. Immunol. 2002;53:181–192. doi: 10.1016/s0165-0378(01)00101-2. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi Y., Cho R., Iwata Y., Nishimura K., Kato T., Aoki N., Kitajima K., Matsuda T. Morphological and biochemical changes of isolated chicken egg-envelope during sperm penetration: degradation of the 97-kilodalton glycoprotein is involved in sperm-driven hole formation on the egg-envelope. Biol. Reprod. 2001;64:822–830. doi: 10.1095/biolreprod64.3.822. [DOI] [PubMed] [Google Scholar]
- 5.Bork P., Sander C. A large domain common to sperm receptors (Zp2 and Zp3) and TGF-β type III receptor. FEBS Lett. 1992;300:237–240. doi: 10.1016/0014-5793(92)80853-9. [DOI] [PubMed] [Google Scholar]
- 6.Jovine L., Qi H., Williams Z., Litscher E., Wassarman P. M. The ZP domain is a conserved module for polymerization of extracellular proteins. Nat. Cell Biol. 2002;4:457–461. doi: 10.1038/ncb802. [DOI] [PubMed] [Google Scholar]
- 7.Harris J. D., Hibler D. W., Fontenot G. K., Hsu K. T., Yurewicz E. C., Sacco A. G. Cloning and characterization of zona pellucida genes and cDNAs from a variety of mammalian species: the ZPA, ZPB and ZPC gene families. DNA Seq. 1994;4:361–393. doi: 10.3109/10425179409010186. [DOI] [PubMed] [Google Scholar]
- 8.Kanamori A., Naruse K., Mitani H., Shima A., Hori H. Genomic organization of ZP domain containing egg envelope genes in medaka (Oryzias latipes) Gene. 2003;305:35–45. doi: 10.1016/s0378-1119(02)01211-8. [DOI] [PubMed] [Google Scholar]
- 9.Spargo S. C., Hope R. M. Evolution and nomenclature of the zona pellucida gene family. Biol. Reprod. 2003;68:358–362. doi: 10.1095/biolreprod.102.008086. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi Y., Nishimura K., Aoki N., Adachi T., Sato C., Kitajima K., Matsuda T. A 42-kDa glycoprotein from chicken egg-envelope, an avian homolog of the ZPC family glycoproteins in mammalian zona pellucida: its first identification, cDNA cloning and granulosa cell-specific expression. Eur. J. Biochem. 1999;260:736–742. doi: 10.1046/j.1432-1327.1999.00203.x. [DOI] [PubMed] [Google Scholar]
- 11.Bausek N., Waclawek M., Schneider W. J., Wohlrab F. The major chicken egg envelope protein ZP1 is different from ZPB and is synthesized in the liver. J. Biol. Chem. 2000;275:28866–28872. doi: 10.1074/jbc.275.37.28866. [DOI] [PubMed] [Google Scholar]
- 12.Usui Y., Nakase M., Hotta H., Urisu A., Aoki N., Kitajima K., Matsuda T. A 33-kDa allergen from rice (Oryza sativa L. japonica): cDNA cloning, expression, and identification as a novel glyoxalase I. J. Biol. Chem. 2001;276:11376–11381. doi: 10.1074/jbc.M010337200. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 15.Bleil J. D., Wassarman P. M. Mammalian sperm–egg interaction: identification of a glycoprotein in mouse egg zonae pellucidae possessing receptor activity for sperm. Cell. 1980;20:873–882. doi: 10.1016/0092-8674(80)90334-7. [DOI] [PubMed] [Google Scholar]
- 16.Bleil J. D., Wassarman P. M. Sperm–egg interactions in the mouse: sequence of events and induction of the acrosome reaction by a zona pellucida glycoprotein. Dev. Biol. 1983;95:317–324. doi: 10.1016/0012-1606(83)90032-5. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 18.Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 19.Williams Z., Wassarman P. M. Secretion of mouse ZP3, the sperm receptor, requires cleavage of its polypeptide at a consensus furin cleavage-site. Biochemistry. 2001;40:929–937. doi: 10.1021/bi002275x. [DOI] [PubMed] [Google Scholar]
- 20.Lindsay L. L., Yang J. C., Hedrick J. L. Identification and characterization of a unique Xenopus laevis egg envelope component, ZPD. Dev. Growth Differ. 2002;44:205–212. doi: 10.1046/j.1440-169x.2002.00635.x. [DOI] [PubMed] [Google Scholar]
- 21.Robertson L., Wishart G. J. Detection of the acrosome reaction of chicken sperm. J. Reprod. Fertil. Abs. Ser. 1996;17:41. [Google Scholar]
- 22.Lotan R., Skutelsky E., Danon D., Sharon N. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea) J. Biol. Chem. 1975;250:8518–8523. [PubMed] [Google Scholar]
- 23.Adler A. J., Greenfield N. J., Fasman G. D. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 1973;27:675–735. doi: 10.1016/s0076-6879(73)27030-1. [DOI] [PubMed] [Google Scholar]
- 24.Yurewicz E. C., Sacco A. G., Gupta S. K., Xu N., Gage D. A. Hetero-oligomerization-dependent binding of pig oocyte zona pellucida glycoproteins ZPB and ZPC to boar sperm membrane vesicles. J. Biol. Chem. 1998;273:7488–7494. doi: 10.1074/jbc.273.13.7488. [DOI] [PubMed] [Google Scholar]
- 25.Tian J., Gong H., Lennarz W. J. Xenopus laevis sperm receptor gp68/64 glycoprotein is a homolog of the mammalian sperm receptor ZP2. Proc. Natl. Acad. Sci. U.S.A. 1999;96:829–834. doi: 10.1073/pnas.96.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson L., Wishart G. J., Horrocks A. J. Identification of perivitelline N-linked glycans as mediators of sperm–egg interaction in chickens. J. Reprod. Fertil. 2000;120:397–403. [PubMed] [Google Scholar]
- 27.Thim L. Trefoil peptides: from structure to function. Cell. Mol. Life Sci. 1997;53:888–903. doi: 10.1007/s000180050108. [DOI] [PMC free article] [PubMed] [Google Scholar]