Abstract
The death morphology commonly known as apoptosis results from a post-translational pathway driven largely by specific limited proteolysis. In the last decade the structural basis for apoptosis regulation has moved from nothing to ‘quite good’, and we now know the fundamental structures of examples from the initiator phase, the pre-mitochondrial regulator phase, the executioner phase, inhibitors and their antagonists, and even the structures of some substrates. The field is as well advanced as the best known of proteolytic pathways, the coagulation cascade. Fundamentally new mechanisms in protease regulation have been disclosed. Structural evidence suggests that caspases have an unusual catalytic mechanism, and that they are activated by apparently unrelated events, depending on which position in the apoptotic pathway they occupy. Some naturally occurring caspase inhibitors have adopted classic inhibition strategies, but other have revealed completely novel mechanisms. All of the structural and mechanistic information can, and is, being applied to drive therapeutic strategies to combat overactivation of apoptosis in degenerative disease, and underactivation in neoplasia. We present a comprehensive review of the caspases, their regulators and inhibitors from a structural and mechanistic point of view, and with an aim to consolidate the many threads that define the rapid growth of this field.
Keywords: apoptosis, caspase, inhibitor, inhibitor of apoptosis protein (IAP), protease, zymogen
Abbreviations: ALPS, autoimmune lymphoproliferative syndrome; APAF-1, apoptotic protease activating factor-1; ASC/PYCARD, apoptosis-associated speck-like protein containing a CARD/PYD- and CARD-containing molecule; BIR, baculoviral IAP repeat; CAD, caspase-activated DNase; CARD, caspase-recruitment domain; CARP, caspase-associated RING protein; CED, cell death-defective; CLARP, caspase-like apoptosis-regulatory protein; CRADD/RAIDD, caspase-2 and RipK1 domain-containing adaptor with death domain/Rip-associated protein with a death domain; CrmA, cytokine response modifier A; DD, death domain; DED, death effector domain; DFF, DNA fragmentation factor; DIABLO, direct IAP-binding protein with low pI; DIAP1, Drosophila inhibitor of apoptosis 1; DISC, death-inducing signalling complex; DRONC, Drosophila Nedd2-like caspase; FADD, Fas (TNFRSF6)-associated via death domain; FLICE, FADD-like ICE; FLIP, FLICE inhibitory protein; IAP, inhibitor of apoptosis protein; IBM, IAP binding motif; ICAD, inhibitor of CAD; ICE, interleukin-1β-converting enzyme; Ipaf/CLAN, ICE-protease-activating factor/CARD, LRR and NACHT-containing protein; LRR, leucine-rich repeat; NACHT, NTPase-domain named after NAIP, CIITA, HET-E and TP1; NALP1, NACHT, LRR and Pyrin domain containing 1; NBD, nucleotide-binding domain; NF-κB, nuclear factor-κB; NOD, nucleotide-binding and oligomerization domain-containing protein; PARP, poly(ADP-ribose) polymerase; PIDD, p53-induced protein with a death domain; RICK/CARDIAK, Rip-like interacting CLARP kinase/CARD-containing ICE-associated kinase; RING, really interesting new gene; Rip, receptor-interacting protein; serpin, serine protease inhibitor; Smac, second mitochondrial activator of caspases; TFPI, tissue factor pathway inhibitor; TLR, Toll-like receptor; TNF, tumour necrosis factor; TRADD, TNFRSF1A-associated via death domain; TRAF, TNF receptor-associated factor; TRAIL, TNF-related apoptosis-inducing ligand; XIAP/BIRC4, X-linked IAP/baculoviral IAP repeat-containing 4; P1, P2, …Pn and P1′, P2′, …Pm′ designate the side chains in substrates and inhibitors in the N- and C-terminal direction respectively from the P1–P1′ scissile peptide bond; S1, S2, …Sn and S1′, S2′, …Sm′ refer to the cognate pockets on the protease that accept these side chains [1]
SCOPE OF THE REVIEW AND A LITTLE HISTORY
“Life is pleasant. Death is peaceful. It's the transition that's troublesome.”
—Isaac Asimov
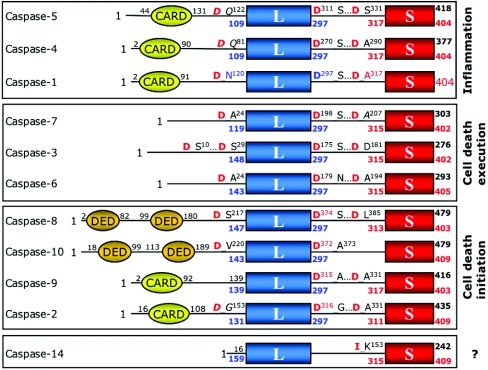
In 1992 two groups simultaneously reported the identity of the human protease responsible for activating the precursor of interleukin-1β, naming it ICE (interleukin-1β-converting enzyme) [2,3]. Several months later the product of the CED3 gene governing the commitment to apoptosis in Caenorhabditis elegans was demonstrated to show identity with ICE [4]. These publications initiated a successful search by many groups over the ensuing years for mammalian ICE homologues that should govern cell death. Today we call these proteases caspases [5], and they constitute a family of cysteine proteases (peptidases that employ a cysteine residue as the catalytic nucleophile) that share a stringent specificity for cleaving their substrates after aspartic acid residues in target proteins. In mammals, seven caspases are probably involved in apoptosis, three are probably involved in pro-inflammatory cytokine activation, and one is probably involved in keratinocyte differentiation (Figure 1).
Figure 1. Domain organization of human caspases.
Human caspases have been grouped according to their sequence similarities. Notice that sequence identity divides caspases-1 to -10 into three subfamilies, in accordance with the physiological distinction between inflammatory, initiator and effector caspases. In contrast with the widespread distribution of these family members, caspase-14 is found mainly in the epidermis, may be involved in keratinocyte differentiation [292–294], and is not activated in vivo at an Asp residue [295]. The positions of maturation cleavage sites are given, with the P1 aspartate residue highlighted in red (in italics in cases where the usage of the site has not been confirmed experimentally). Numberings correspond either to the Swiss-Prot entries (with exception of caspase-10, for which the sequence of the more commonly expressed isoform 10/a is given [296]) or to the caspase-1-based system used throughout this work (colour-coded).
The first structures of caspases were published in 1994 [6,7], displaying a novel protease fold family and suggesting an activation mechanism. Structural elucidations over the last few years of caspases, caspase inhibitors and caspase–inhibitor complexes have supported the early theories of caspase catalytic activity, but overturned many of the ideas of caspase activation and regulation. This review focuses on caspase structure/function studies over the last 10 years, and sets them in the context of current concepts of the biological role and regulation of cell death and pro-inflammatory cytokine activation. In a sense, this is a companion to the excellent general review published in the Biochemical Journal in 1997 by Gerry Cohen [8].
DEFINITIONS AND CLASSIFICATION
The caspases constitute family C14 of peptidase clan CD, as defined by Rawlings and Barrett [9], forming part of the extensive MEROPS peptidase database, to which readers are referred (http://merops.sanger.ac.uk). The Nomenclature Committee of the IUBMB places them in enzyme category 3.4.22, but to date has only officially classified caspase-1. Clan CD has other families that contain distant homologues in the bacterial, plant and animal kingdoms (see Supplemental Table 1 at http://www.BiochemJ.org/bj/384/bj3840201add.htm), but caspases are found strictly in metazoan animals, with the earliest identifiable members in nematodes. The term caspases, as well as recommendations for definitions of chain assemblies, was introduced in 1996 [5]. The literature is divided regarding the terminology of various structural elements, with some groups confusingly using the same designation for different loops or different designations for the same loop. For example, the 381-loop defined in this review is called loop 3 [10] or loop 4 [11] in caspase-3, and loop 3 [12] or loop 5 [13] in caspase-8. Therefore, throughout this review, in order to facilitate structural comparisons, we employ the caspase-1-based numbering convention for caspase catalytic domains, which is somewhat of a standard solution to this sort of problem throughout structural biology. Insertions in caspase sequences compared with caspase-1 are defined by a letter following the insertion point (see Figure 2). However, because some publications and Protein Data Base entries utilize individual caspase numbering, we have attempted to satisfy individual taste by also including these numberings. Thus the number in parentheses, when used, refers to the number specific for the particular caspase. In addition, we have included in supplementary data PDB files converted to the caspase-1 numbering system (http://www.BiochemJ.org/bj/384/bj3840201add.htm). Caspases are dimers of catalytic domains in the active state, so we use a prime designation to identify the same residue in symmetrically related units; such that the catalytic cysteine is Cys-285 in one domain, and Cys-285′ in the adjacent domain of the functional dimer, for example. Unless otherwise stated, all individual numberings refer to the human proteins, as deposited with the Swiss-Prot database (http://www.expasy.org/sprot/).
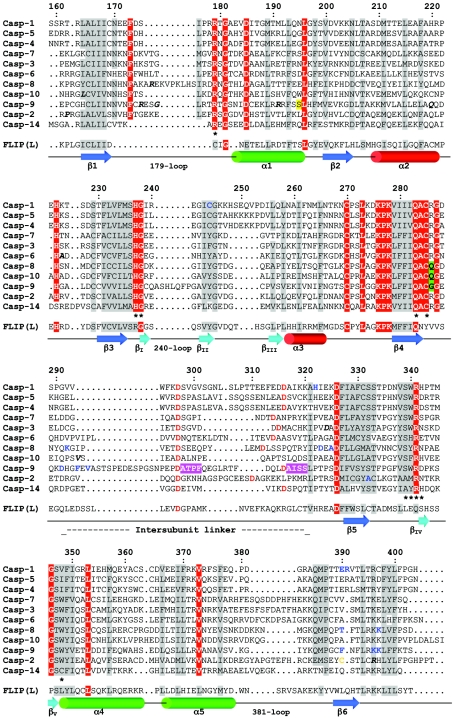
Figure 2. Structure-based sequence alignment of human caspase domains.
Strictly conserved residues are shown in white with a red background; other conserved residues have a grey background. Single-nucleotide polymorphisms are indicated in bold italics. Residues involved in substrate recognition and catalysis are marked (*). Residues representing autolytic cleavage sites in the intersubunit linker are in red, and the cysteine residue that forms a dimer interface disulphide in caspase-2 is in orange. Caspase-14 is not activated by autolysis [295], and the cleavage site detected in vivo is presumably from another cellular protease [294,295]. Residues that have been mutated are in bold blue letters. The Ser residue reportedly phosphorylated in caspase-9 is shadowed yellow. The secondary structure representations (arrow, β-strand; cylinder, α-helix) follow the CATCH classification (http://www.biochem.ucl.ac.uk/bsm/cath/) for 1QTN [13].
As an aid to following the sometimes cryptic names of apoptotic proteins, Table 1 may be helpful, in addition to the list of abbreviations.
Table 1. Explanation of some commonly used terms for proteins involved in death pathways.
| Name | Derivation | Property/function |
|---|---|---|
| Activation regulators | ||
| Caspase | Cysteine-dependent aspartic specific protease | Protease |
| FLIP | FLICE inhibitory protein | Inhibitor or activator homologue of caspase-8 |
| Apoptosome | Activation platform for caspase-9 | |
| APAF-1 | Apoptotic protease-activating factor-1 | Protein component of the apoptosome |
| DISC | Death-inducing signalling complex | Activation platform for caspase-8 (and -10) |
| CARD | Caspase-recruitment domain | Recruitment domain (caspase-9 and APAF-1 components of the apoptosome, among several other proteins) |
| DED | Death effector domain | Recruitment domain (caspases-8 and -10 and FLIP) |
| DD | Death domain | Recruitment domain (Fas and FADD components of the DISC) |
| Inhibition regulators | ||
| IAP | Inhibitor of apoptosis protein | Protein inhibitor of apoptosis |
| BIR | Baculoviral IAP repeat | Domain of an IAP |
| IBM | IAP-binding motif | Short stretch of amino acids, usually at the N-terminus of a protein, that interacts with BIR domains |
| Smac | Second mitochondrial activator of caspases | Protein antagonist of IAPs |
| DIABLO | Direct IAP-binding protein with low pI | Mouse orthologue of Smac |
CASPASES: PHYSIOLOGICAL ROLES
Cytokine activators
The pro-inflammatory cytokines interleukin-1β and interleukin-18 are stored in the cytosol of cells, where they await proteolytic activation and release. The release mechanism is debated and does not resemble known secretory mechanisms, but the proteolytic mechanism is due, at least in part, to caspase-1. This transcription/translation-independent event presumably acts as a rapid response to infection, and as such is an extremely beneficial part of innate immunity. The cytokine activator caspases could simply be defined as those showing the closest sequence similarity and intrinsic substrate specificity to caspase-1 (Figure 1). In humans, these equate to caspases-4 and -5, and in mice they equate to caspases-11 and (possibly)-12. Caspase-12 is a conundrum, since the important residue Arg-341 that accounts largely for the defining Asp-specificity of caspases is replaced by a Lys in mice, and in humans the caspase-12 gene appears not to be expressed as a protease in the majority of the population due to a premature stop codon [14]. Moreover, the expression pattern of mouse caspase-12 differs substantially from that of caspases-1 and -11, lessening the likelihood that caspase-12 participates in cytokine activation [15]. However, the immune response to endotoxins such as lipopolysaccharide is attenuated in individuals that express the full-length enzyme, thus increasing their risk of developing sepsis [16].
Genetic data link caspases-1 and -11 in cytokine activation in mice [17], and biochemical data link caspases-1 and -5 in humans [18]. A possible mechanism for the interesting dual requirement of caspases-1 and -5 (the human orthologue of mouse caspase-11) in cytokine processing is described later.
Apoptotic initiators
The apoptotic caspases are initiators or effectors, depending on their point of entry into the apoptotic pathway. The initiator caspases are the first to be activated in a particular death pathway [Figure 3 and Supplemental Figure 1 (http://www.BiochemJ.org/bj/384/bj3840201add.htm)], and they constitute the opening step in a minimal two-step cascade by activating the effector caspases. One characteristic of the initiators is the presence of homotypic CARD (caspase-recruitment domain) or DED (death effector domain) interaction domains at their N-termini (Figure 1). These modules direct procaspases to oligomeric activation assemblies in the cell. Perhaps the most important characteristic of the initiators is their inherent substrate specificity which, as explained by Thornberry and colleagues [19], allows them to recognize their targets, the effector caspases (Figure 1). In humans and mice we recognize two independent initiation pathways: the extrinsic pathway, defined by the activation of caspase-8 via transmembrane receptors of the TNF (tumour necrosis factor) type-I receptor family; and the intrinsic pathway, that responds to stress, genotoxic damage and some developmental cues by the activation of caspase-9 (Figure 3). Humans contain a close relative of caspase-8, namely caspase-10, which is absent from mice [20]. Once activated within their oligomeric assemblies, caspases-8–10 then directly activate the effector caspases-3 and -7.
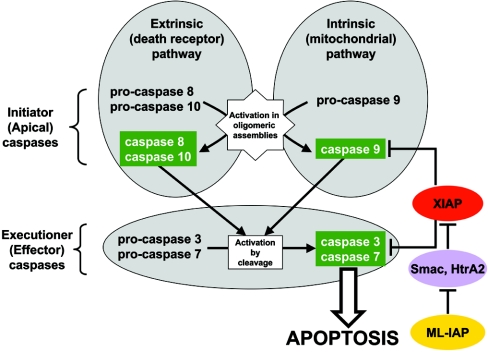
Figure 3. The framework of apoptosis.
Death is signalled by ligand-enforced clustering of receptors at the cell surface, which leads to the activation of initiator caspases-8 and -10 [297]. These caspases then directly cleave and activate the effector caspases-3 and -7 (and possibly caspase-6), which are predominantly responsible for the limited proteolysis that characterizes apoptotic morphology in a cell. On the other hand, genotoxic damage – transmitted by a mechanism thought to involve the release of cytochrome c from mitochondria – engages the same effector caspases [298]. The latter events progress through the initiator caspase-9 and its activator platform APAF-1 [55]. The common execution phase is regulated through direct caspase inhibition by XIAP, which can also regulate the active form of caspase-9. XIAP is under the influence of antagonist proteins such as Smac/DIABLO and HtrA2 that compete with caspases for IAPs [251]. Finally, these IAP antagonists may be sequestered by other IAPs, such as ML-IAP (melanoma IAP), acting as competitive sinks for the availability of the pro-apoptotic antagonists [299]. Although other modulators may regulate the apoptotic pathway in a cell-specific manner, this framework is considered to be common to most mammalian cells.
Apoptotic effectors
The apoptotic effectors are defined by the absence of recognizable homotypic recruitment domains (Figure 1). Together they are responsible for the majority of the limited proteolytic events that combine to create the morphological cellular changes known as apoptosis. Caspases-3 and -7 are closely related and have a virtually indistinguishable substrate and inhibitor specificity. The largest difference between these effectors is in their N-terminal peptides, regions thought to be involved in subcellular targeting [21,22], where they diverge completely. Caspase-6 is frequently considered to be an effector, based on a short N-terminal peptide, but its inherent substrate specificity differs substantially from that of caspases-3 and -7 (see below), suggesting a non-overlapping subset of natural substrates. Various data have implicated caspases-3, -6 and -7 in the execution phase; however, the data may not be truly representative, because caspase-3 is studied much more frequently than the other two, possibly because it has a role in development, as demonstrated by genetic ablation in certain mice strains.
Mouse knockouts: roles in development and in mature animals
Genetic ablation of caspases has been instructive, and has helped to place them in the hierarchical scheme portrayed in Figure 3. Animals ablated in caspases-1 or -11 are deficient in cytokine processing [17,23], but without any overt developmental or apoptotic phenotype. In contrast, the phenotypes of some apoptotic caspase knockouts are very gross, evidently anti-apoptotic, and vary from early embryonic lethality (caspase-8) to perinatal lethality (caspases-3 and -9) [24–26], to relatively mild (e.g. defects in the process of normal oocyte ablation for caspase-2) [27]. Currently, mice ablated in caspases-6 and -7 are without a known developmental phenotype [28,29], but care should be taken in equating lack of a developmental phenotype to a less important physiological role, because of the influence of strain background. For example, on a 129×1/SvJ genetic background, caspase-3 null mice are uniformly affected with a severe and perinatally lethal neurodevelopmental phenotype characterized by massive expansion of neuronal progenitor cells in the forebrain, consistent with failed apoptosis. In stark contrast, on a C57BL/6J background, caspase-3 null mice reach adulthood and have a minimal neuronal phenotype [30]. The origins and meaning of this marked strain dependence are unclear, but argue for caution in ascribing functions to caspases based simply on developmental phenotype.
Sometimes a role in development is not correlated with a pathological phenotype. For example, caspase-2-deficient mice have a phenotype characterized by excessive oocytes due to inhibition of cell deletion during development [27,31]. The nervous systems of caspase-2-deficient mice seem unaffected. However, experiments with explanted primary neurons suggest that caspase-2 programmes death induced by trophic factor withdrawal, but that in caspase-2 null mice this role is subsumed by caspase-9 [32]. Thus there exists a degree of compensation and redundancy, emphasized by other studies [29], that complicates the assessment of caspase physiological roles using mice genetics. The answers are simply more complex than many current interpretations, and one can conclude that the roles of individual caspases are strain-dependent, cell type-dependent, and even context-dependent.
Given the contradictions observed in some mouse caspase knockouts, it is useful to gain insight from lessons learned from humans with caspase deficiencies. Humans with the mutations L164(285)F and V293a(410)I in the caspase-10 coding sequence have been reported to exhibit an ALPS (autoimmune lymphoproliferative syndrome) caused by defective lymphocyte apoptosis [33]. The association of the L164F mutation with ALPS is supported by other data, but the predicted homozygosity frequency of the V293aI mutation is approx. 1 in 200 in the Danish population, thus arguing against a major contribution to the ALPS phenotype [34].
Humans with mutant caspase-8 [R175b(248)W], while also exhibiting defects in lymphocyte apoptosis, have pronounced defects in their ability to activate lymphocytes, with resulting immunodeficiency [35]. Significantly, this latter study suggested that caspase-8 deficiency is compatible with development in humans, although it is embryonic lethal in mice [24]. Taken together, these studies suggest that although there is major overlap and redundancy, caspases-8 and -10 probably have distinct functions in T-cell subsets.
Caspases in disease: degeneration and proliferation
Current data on the role of caspases in neurodegenerative diseases are confusing and even contradictory, as reviewed in [36,37]. Examination of post-mortem tissue has implicated caspases in multiple neurodegenerative diseases, but whether the activation of apoptosis represents a cause of degeneration, or merely disposal of already dysfunctional neurons, remains to be established. With respect to this, there is a distinction to be made between chronic neurodegenerative diseases (Alzheimer's, Huntington's, Parkinson's, for example) and acute ones, such as nerve crush injury and stroke. The difficulty of constructing animal models of human neurodegenerative disease makes the assessment of caspase participation and apoptosis difficult in chronic diseases. On a more definitive note, in the acute cases there is good evidence that neurons in the brain can be spared in specific caspase knockouts or by ablating caspase activity (reviewed in [37]). This bodes well for anti-caspase therapy as a means of sparing acutely damaged neurons, thereby giving them a chance to recover following injury.
On the flip side of inappropriate apoptosis leading to degenerative disease is the role of caspases in proliferative disease, especially cancer. An example is the silencing of the human caspase-8 gene that is frequently associated with neuroblastoma, giving support to the concept that apoptotic caspases are tumour suppressors [38]. In general, signals that drive cell proliferation also prime the apoptotic machinery [39]. This inherent propensity of rapidly cycling cells secures cell expansion to locations governed by specific cell–cell interactions. When cells exceed the local interaction environment, they die by a process known as ‘anoikis’, stemming from loss of cell–cell contacts [40]. Tumour cells must adapt by inactivating the apoptotic forces that would normally signal their demise, and it is now clear that a significant event in many (possibly all) neoplastic transformations leading to cancer is the inactivation of apoptosis at one of the several control points in the apoptotic pathways (reviewed in [41,42]). Therefore therapeutic measures designed to intervene to restore the apoptotic process have potential as specific anti-cancer agents. Indeed, many chemotherapeutic pharmaceuticals work by inducing apoptosis in tumour cells, and specific intervention to ablate known anti-caspase controls causes regression of tumours in vivo [43]. The trick, as usual, is to target this Achilles' heel of cancer cells in a way that does not cause apoptosis in non-tumour proliferating cell populations [39].
MECHANISMS OF CASPASES: CATALYSIS AND SPECIFICITY
Since the pioneering work on the structure of caspase-1 [6,7], several crystal structures of inflammatory, initiator and effector caspases have been reported (Table 2). In this section, we discuss the major characteristics of the caspase fold as a basis for understanding the subsequent sections.
Table 2. Structures of caspase catalytic domains deposited with the Protein Data Bank*.
*Released before August 15, 2004. Eight structures of caspase-1 (PDB entries 1RWK, 1RWM, 1RWN, 1RWO, 1RWP, 1RWV, 1RWX and 1RWW), as well as two structures of caspase-7 (1SHJ and 1SHL), in complex with different inhibitors have not yet been released. PDB entries are linked to the supplemental co-ordinate files (http://www.BiochemJ.org/bj/384/bj3840201add.htm) converted into the caspase-1 numbering system. Abbreviations: Ac, acetyl; BOP, 1-bromo-4-methoxybenzene; CHO, aldehyde; dcmk, dichloromethyl ketone; fmk, fluoromethyl ketone; IL, interleukin; PTF, [(methylsulphanyl)methyl]benzene; Z, benzyloxycarbonyl.
| Protein | Entry/Resolution | Comments | Ref. |
|---|---|---|---|
| Caspase-1 | 1ICE/2.60 Å | Bound to Ac-YVAD-CHO. First structure of a caspase. The bound aldehyde mimics the activation cleavage site in pro-IL-1β. | [7] |
| Caspase-1 | 1IBC/2.73 Å | Bound to Ac-WEHD-CHO. Highly potent inhibitor (Ki 56 pM) identified in combinatorial screening. | [74] |
| Caspase-1 | 1BMQ/2.50 Å | Bound to the irreversible inhibitor (3S)-N-methanesulphonyl-3-({1-[N-(2-naphthoyl)-L-Val]-L-Pro}amino)-4-oxobutanamide. First example of an inhibitor that lacks an acidic carboxylate at P1. | [300] |
| Caspase-1 | 1SC1/2.60 Å | Free C285A mutant. Loop bundle formed, but active site disrupted (‘closed’ conformation). | [301] |
| Caspase-1 | 1SC3/1.80 Å | C285A mutant in complex with malonate. The malonate moiety binds tightly in the S1 pocket, inducing the active (‘open’) conformation. | [301] |
| Caspase-1 | 1SC4/2.10 Å | C285A mutant, after removal of malonate (see 1SC3). Active site disrupted, 341-loop disordered. | [301] |
| Caspase-2 | 1PYO/1.65 Å | Bound to Ac-LDESD-CHO. Disulphide-linked dimer (C-390−C-390′). Subsite S5 occupied. | [75] |
| Caspase-3 | 1QX3/1.90 Å | Unliganded caspase. The Y338 side chain occludes the S2 pocket. | [67] |
| Caspase-3 | 1GFW/2.80 Å | Bound to the reversible, competitive non-peptide inhibitor 1-methyl-5-(2-phenoxymethylpyrrolidine-1-sulphonyl)-1h-indole-2,3-dione. Binds primarily to the S2, but not the S1 pocket. | [302] |
| Caspase-3 | 1CP3/2.30 Å | Bound to Ac-DVAD-fmk. Oxyanion hole occupied by ketone oxygen. | [10] |
| Caspase-3 | 1PAU/2.50 Å | Bound to Ac-DEVD-CHO. First structure of an effector caspase. | [76] |
| Caspase-3 | 1NME/1.60 Å | Bound to a non-peptide inhibitor identified using ‘extended tethering’, formed by a 3-(2-mercapto-acetylamino)-4-oxopentanoic acid moiety (occupies S1 pocket) and a 2-hydroxy-5-(2-mercapto-ethylsulphamoyl)-benzoic acid moiety (S4 pocket). | [303] |
| Caspase-3 | 1NMQ/2.40 Å | Bound to 3-(3-{2-[(5-methanesulphonylthiophene-2-carbonyl)amino]ethyldisulphanylmethyl}benzenesulphonylamino)-4-oxo-pentanoic acid. | [303] |
| Caspase-3 | 1NMS/1.70 Å | Bound to 5-[4-(1-carboxymethyl-2-oxopropylcarbamoyl)benzylsulphamoyl]-2-hydroxybenzoic acid. | [303] |
| Caspase-3 | 1RE1/2.50 Å | Bound to (3S)-3-{[(5-bromopyridin-3-yl)carbonyl]amino}-4-oxobutanoic acid. Caspase Y338 fills the S2 pocket as in 1QX3, and interacts with the pyridyl ring of the inhibitor. | [68] |
| Caspase-3 | 1RHJ/2.20 Å | Bound to 3-(2-{5-tert-butyl-3-[(4-methylfurazan-3-ylmethyl)amino]-2-oxo-2H-pyrazin-1-yl}butyrylamino)-5-(hexylmethylamino)-4-oxopentanoic acid. Alkylamine moiety not visible in S1′ pocket. | |
| Furazan moiety makes significant contribution to binding strength (S4 pocket). | [68] | ||
| Caspase-3 | 1RHK/2.50 Å | Bound to the phenylpropyl ketone derivative of Ac-DEVD-CHO. Binding to S1′ subsite increases inhibitor potency by 50-fold. | [68] |
| Caspase-3 | 1RHM/2.50 Å | Bound to 4-[5-(2-carboxy-1-formylethylcarbamoyl)pyridin-3-yl]benzoic acid. Caspase Y338 fills the S2 pocket and interacts with the pyridyl ring of the inhibitor. | |
| First description of a edge-to-face interaction between an inhibitor bromoanisole moiety and indole moiety of W-340. | [68] | ||
| Caspase-3 | 1RHQ/3.00 Å | Bound to Ac-BOP-PTF. Conversion of P1′ alkyl linker into a thioether increases binding affinity (also valid for 1RHR, 1RHU). | [68] |
| Caspase-3 | 1RHR/3.00 Å | Bound to (3S)-5-[(2-chloro-6-fluorobenzyl)sulphanyl]-3-{[N-({2-ethoxy-5-[(1E)-3-methoxy-3-oxoprop-1-enyl]phenyl}acetyl)-D-valyl]amino}-4-oxopentanoic acid. Benzene derivative partially disordered in S1′ pocket. | [68] |
| Caspase-3 | 1RHU/2.50 Å | Bound to (3S)-3-[({(2S)-5-[(N-acetyl-L-α-aspartyl)amino]-4-oxo-1,2,4,5,6,7-hexahydroazepino[3,2,1-hi]indol-2-yl}-carbonyl)amino]-5-(benzylsulphanyl)-4-oxopentanoic acid. | [68] |
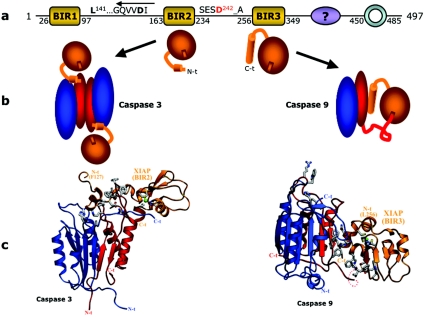
| Caspase-3 | 1I3O/2.70 Å | Complex with XIAP (BIR2 domain and linker to BIR1). Parallel binding mode, BIR domain defined. | [255] |
| Caspase-7 | 1F1J/2.35 Å | Bound to Ac-DEVD-CHO. First structure of active caspase-7. | [63] |
| Caspase-7 | 1I51/2.45 Å | Complex with XIAP (BIR2 domain and linker to BIR1). Parallel binding mode. BIR domain flexibly disordered | [133] |
| Caspase-7 | 1I4O/2.40 Å | Complex with XIAP (BIR2 domain and linker to BIR1). Parallel binding mode. BIR domain flexibly disordered. | [134] |
| Caspase-7 | 1KMC/2.90 Å | Complex with XIAP (BIR2 domain and linker to BIR1). Parallel binding mode. BIR domain flexibly disordered. | † |
| Caspase-7 | 1K86/2.60 Å | Free caspase. Active site region unoccupied. The catalytic machinery is not completely formed. | [11] |
| Procaspase-7 | 1GQF/2.90 Å | Free zymogen. Disordered substrate-binding loops, intersubunit linker partially inserted in central cavity. | [124] |
| Procaspase-7 | 1K88/2.70 Å | Free zymogen. Disordered substrate-binding loops, intersubunit linker partially inserted in central cavity. | [11] |
| Caspase-8 | 1QTN/1.20 Å | Bound to Ac-IETD-CHO. Highest-resolution structure of a caspase reported to date. | [13] |
| Caspase-8 | 1QDU/2.80 Å | Bound to Z-EVD-dcmk. Additional interaction of Arg-177 with the Glu P3 residue. Stresses importance of S3 subsite for substrate recognition. | [12] |
| Caspase-8 | 1F9E/2.90 Å | Bound to Z-DEVD-CHO. Suggests important role of the S3 subsite in determining caspase specificity. | [65] |
| Caspase-8 | 1I4E/3.00 Å | Complex with baculovirus suicide inhibitor p35. Novel mechanism of proteinase inhibition. | [240] |
| Caspase-9 | 1JXQ/2.80 Å | Complex with Glu-Val-dehydrohymethyl aspartate. One active site occupied by inhibitor, the other free and distorted as in procaspase-7. | [73] |
| Caspase-9 | 1NW9/2.40 Å | Complex with XIAP (BIR3). Inhibitor blocks dimerization region of the (pro)caspase. | [122] |
| Caspase-1 | 1M72/2.30 Å | First structure of a non-human caspase (from Spodoptera frugiperda). Bound to Ac-DEV-Ask-methyl ketone. | [304] |
† S. J. Riedl, W. Bode, G. S. Salvesen and P. Fuentes-Prior, unpublished work.
Structure of active caspases
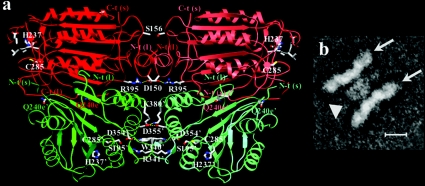
The fundamental catalytic domain seen in all caspase structures comprises a large (17–20 kDa) and a small (10–12 kDa) subunit. The obligate catalytic domain is composed of a large and a small subunit tightly packed into a compact ellipsoid of approximate dimensions 25 Å×50 Å×30 Å. A two-fold axis, that in most cases constitutes an exact or crystallographic axis, relates the pairs of catalytic domains (Figure 4a). Active caspases have been referred to as ‘homodimers of heterodimers’ [6,7], but, as we describe later, the initiator caspases do not require proteolytic processing to achieve the active form, and so this somewhat confusing terminology is replaced in this review by the concept of a catalytic domain that is often composed of two subunits or chains.
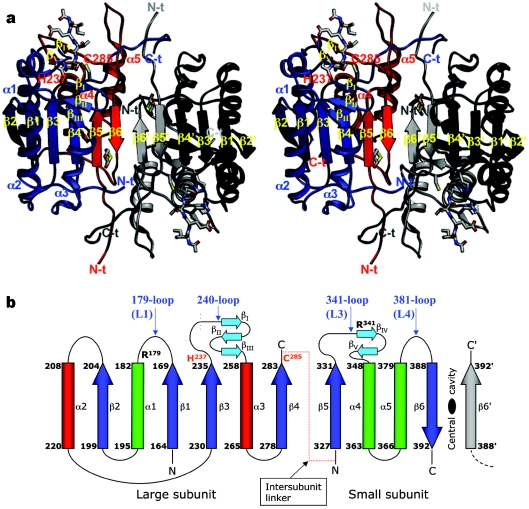
Figure 4. Structure of active caspases.
(a) The crystal structure of human caspase-8 exemplifies the fundamental caspase fold, and is shown bound to the tetrapeptide aldehyde inhibitor acetyl-Ile-Glu-Thr-Asp-CHO ([13]; PDB entry 1QTN), which represents the highest-resolution structure of a caspase reported to date. Note the three-layer structure of a twisted, 12-stranded β-sheet that is sandwiched by α-helices. Most of the interdomain contact area is built by the central small subunits, with additional interactions (the characteristic ‘loop bundle’) tying together the C- and N-termini of large and small subunits from neighbouring domains. The bound inhibitor is represented with a ball-and-stick model, as are dithiane diol molecules trapped in the cleft between the two monomers (termed the central cavity, for obvious reasons). (b) Simplified topological diagram of the caspase structure, following the CATCH definition of secondary structure elements for 1QTN. An additional N-terminal α-helix of variable length (α0; not shown) is present in caspases-1 [6,7], -2 [75] and -9 [73], and closes the ‘bottom’ of the α/β barrel. Also not depicted is an additional α-helix found solely in the long 179-loop of caspase-8. The positions of catalytic dyad residues His-237 and Cys-285 (red), along with those of the specificity-determining arginine residues (Arg-179 and Arg-341), are indicated. The location of loops that contain important functional elements is indicated in blue text using the numbering convention designated throughout this review, along with an alternative designation [145].
Each catalytic domain derives from a single procaspase molecule (see below), and is composed of a twisted, mostly parallel β-sheet sandwiched between two layers of α-helices (see Figure 4b for a schematic topology diagram). Two domains fit together by alignment of the C-terminal strands β6 in an antiparallel manner (Figure 4), thus generating a continuous, 12-stranded β-sheet. The caspase fold (or caspase–haemoglobinase fold, as defined in [44]) is conserved in the bacterial relative, gingipain R [45], and is predicted in a number of distantly related proteases (summarized in Supplemental Table 1; http://www.BiochemJ.org/bj/384/bj3840201add.htm). Significantly, similar open α/β structures are encountered in over 100 different domains, most of which catalyse enzymic reactions (see http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.d.html for the SCOP classification [46] of α/β proteins).
Recruitment domains
In addition to the catalytic domain, both inflammatory and initiator caspases carry at their N-termini one or two copies of CARD or DED modules (Figure 1), which are critical for their activation in vivo. Despite lack of obvious sequence similarity, CARDs, DEDs, the DDs (death domains) found for example in Fas, and the Pyrin/DAPIN (domain in apoptosis and interferon response) domains identified in other adaptor proteins such as NALP1 [NACHT (NTPase-domain named after NAIP, CIITA, HET-E and TP1), LRR (leucine-rich repeat) and Pyrin domain-containing 1] share a common fold. These globular domains are mainly composed of six antiparallel α-helices, with helices α1–α5 building an α-helical Greek key [47–49]. The structural similarity thus strongly suggests a common evolutionary origin for all members of this so-called DD or DEATH superfamily [50,51]. Comparative structural analysis of these recruitment domains has been reviewed previously [52,53].
Active-site architecture
The active sites of enzymes with open α/β structures are commonly found at ‘topological switch points’, i.e. at positions within or close to the C-terminal end of the central β-sheet, in which loops originating in two adjacent β-strands lead to the opposite helical layers. The location of these switch points can thus be derived from topology diagrams [54]. As first appreciated in the structure of caspase-1 [6] (see also Figure 4), this general rule is valid also for caspases, since the catalytic residue His-237 is located in the loop that connects β-strand β3 to the ‘front’ helix α3, while the neighbouring strand β1 is followed by the ‘back’ helix α1 (‘back’ and ‘front’ refer to the standard caspase orientation shown in Figure 4a). The β1–α1 loop harbours one of the residues that determine the characteristic P1 specificity, Arg-179. Although less obvious in active caspases, a similar consideration applies to the second residue of the catalytic dyad, Cys-285, which is found in the large, highly variable β4–β5 linker (Figure 2). This loop protrudes from strand β4 (terminating in another conserved substrate-binding residue, Gln-283) towards the front helical layer, while the one emanating from the adjacent strand β5 leads to the back helix α4 (Figure 4b). Finally, the β5–α4 loop contains the second arginine residue involved in substrate fixation, Arg-341 (see also below).
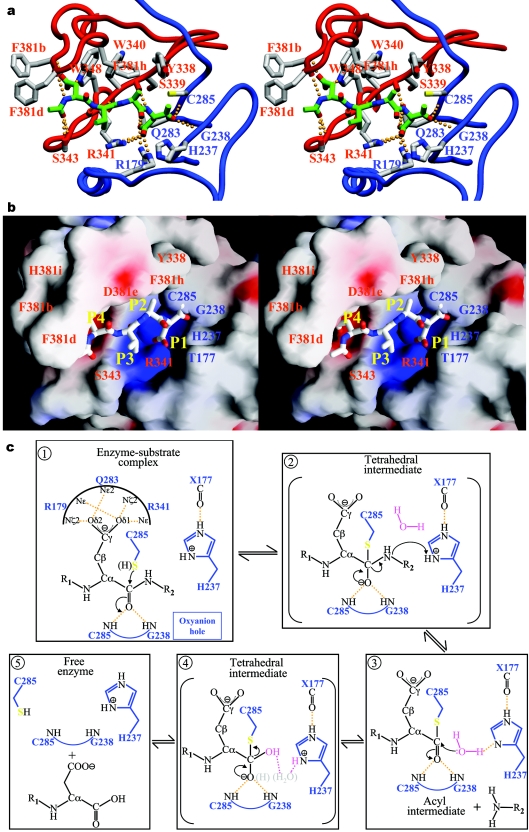
The identities of His-237 and Cys-285 as catalytic residues were confirmed in a mutagenesis study that accompanied the determination of the caspase-1 crystal structure [7]. In the ensuing years, Cys-285→Ala/Ser mutants of most caspases have been generated, and shown to be essentially inactive in vitro. Of note, the Cys-285 mutants of initiator caspases confer a dominant negative apoptotic phenotype when overexpressed in multiple cell lines (see for example [55,56]). Mutation of the topologically equivalent Cys residue in legumain [57] and separin [58,59] abrogates catalysis by these distant relatives of caspases. Finally, covalent linkage of the Sγ atom of Cys-285 to substrate-like tetrapeptide inhibitors in the crystal structures of human caspase-1 conclusively established the latter residue as the active-site nucleophile ([6,7]; see also Figure 5a).
Figure 5. Caspase catalytic mechanism.
Close-up of the active-site region in acetyl-Asp-Val-Ala-Asp-methyl ketone-inhibited caspase-3 (PDB code 1CP3; [10]) shown in standard orientation, i.e. with the active-site residues facing the viewer, and substrates running from left to right. The stereo plots display (a) a ribbon representation of the caspase (large subunit, blue; small subunit, red), and (b) the GRASP electrostatic surface potential of the caspase (contoured between −25 and +25 kBT/e) with stick inhibitor. Important residues are labelled in both panels. Hydrogen bonds were calculated using HBPLUS (http://www.biochem.ucl.ac.uk/bsm/hbplus/home.html) and are indicated with orange dotted lines in (a). Note that the inhibitor binds in an extended conformation, with backbone atoms of P3 and P1 residues hydrogen-bonded to strictly (Arg-341) and highly (Ser-339) conserved caspase residues. The guanidinium groups of Arg-179 and Arg-341 engage in strong salt bridges with the carboxylate of the P1 aspartate, which is further hydrogen-bonded to the side-chain carboxyamide of Gln-283. The combination of extended, β-sheet-like hydrogen bonding to the enzyme and of substrate recognition based mainly on interactions with the S1 and S4 pockets places caspases in a mechanistic sense closer to serine proteases, in particular those of the subtilisin clan. (c) Proposed substrate-hydrolysis mechanistic scheme. During the acylation step (1), the carbonyl oxygen of the non-covalently bound P1 residue is anchored through hydrogen bonds to the nitrogen atoms of Gly-238 and Cys-285 (the oxyanion hole). This increases the polarization of the C–O bond, and therefore facilitates nucleophilic attack of the sulphur atom of Cys-285 on the highly electrophilic carbonyl carbon. The result is a covalent enzyme–substrate adduct, the high-energy tetrahedral intermediate (2), as visualized in crystal structures of methyl ketone-inhibited caspases (see a). The imidazole moiety of His-237 acts as a general acid at this stage of catalysis by protonating the α-amino group of the leaving peptide product, thus avoiding re-formation of the peptide bond. Deacylation of the acyl-enzyme complex occurs then in a similar manner: the deprotonated His-237 side chain abstracts a proton from a water molecule, the hydrolytic water (3), which is thus activated to attack the thioester bond. Deacylation proceeds through a second tetrahedral intermediate (4), formed upon nucleophilic attack of the hydroxy group on the carbonyl carbon. (A putative, neutral gem-diol intermediate found in a recent quantum mechanics/molecular mechanics simulation of the hydrolysis of the acyl-enzyme complex in caspase-3 [79] is shown by grey atoms in parentheses. These authors also predicted that the catalytic histidine is activated by the hydroxy group of Ser-178, but this residue is not conserved in other caspases.) Rupture of the Sγ–C bond regenerates the enzyme in a non-covalent complex with the N-terminal peptide product (5). By analogy with serine proteases, it is conceivable that movements of the 341- and/or 381- substrate-binding loops are coupled to the latter reaction, thus allowing disruption of the main-chain–main-chain hydrogen bonds with the P1/P3 residues, and of the P1 carbonyl oxygen atom with the oxyanion hole. In other words, thioester hydrolysis and product release may be synchronized to ensure a high efficiency of catalysis.
Another critical element in the catalytic machinery of diverse proteolytic enzymes is the oxyanion hole, a pocket that hydrogen-bonds the carbonyl oxygen of the P1 residue of the substrate during catalysis. The concept of an oxyanion hole was propounded in trypsin-like serine proteases, where it comprises the main-chain nitrogen atoms of residues Gly-193 and Ser-195. Recent time-resolved crystallographic studies, molecular dynamics simulations and calculations based on a combined quantum mechanical/molecular mechanical approach underscore its critical role in catalysis, in particular the hydrogen bond donated by the Gly-193 amide group [60–62]. The crystal structures of methyl ketone-inhibited caspases reveal oxyanion holes that consist of the backbone nitrogen atoms of the strictly conserved Gly-238 and of the catalytic Cys-285, which are thus similar to those of trypsin-like enzymes (Figure 5a; [6,10,12]).
Substrate specificity and selectivity
The availability of crystal structures for members of all three caspase subfamilies bound to the same or highly similar substrate-analogue peptidyl inhibitors (Table 2) allows detailed comparisons of specificity determinants, seen most effectively in the analysis of interactions with the inhibitor acetyl-Asp-Glu-Val-Asp-CHO [63].
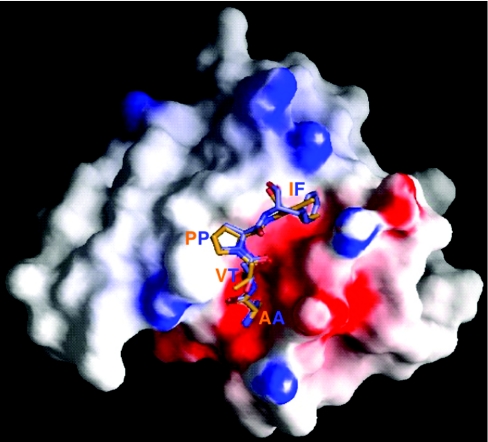
The S1 pocket
Caspases are among the proteases with the most stringent specificity for a P1 residue. Remarkably, their S1 pockets are almost identical, being constructed of the side chains of the strictly conserved residues Arg-179, Arg-341 and Gln-283 (Figure 5a). This deep, highly basic pocket is ideally shaped to accommodate an aspartate side chain, explaining the up to four orders of magnitude lower catalytic efficiency for cleavage of peptides with a glutamate residue at P1 [64]. Significantly, mouse caspase-12 contains Lys at the position equivalent to residue 341, lessening the likelihood that this is a catalytically efficient caspase.
The S2 pocket
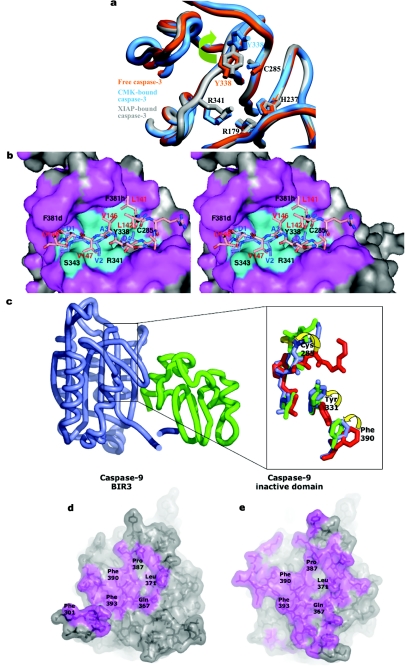
The S2 subsite can also contribute to substrate differentiation [63,65,66]. Most notably, the aromatic S2 pockets of caspases-3 and -7, formed by the side chains of Tyr-338, Trp-340 and Phe-381h, preferentially accommodate small aliphatic residues (Ala, Val). By contrast, subsite S2 is larger in inflammatory and initiator caspases due to the substitution Tyr-338→Val (Ala in caspase-2), and therefore tolerates well residues with bulkier side chains. The S2 pocket is the only site to show a considerable alteration during substrate binding. In caspase-3 with an unoccupied active site [67], the side chain of Tyr-338 occupies the pocket. During binding of tetrapeptide inhibitors, the Tyr-338 side chain rotates ∼90° around the Cα–Cβ bond to allow occupancy of the pocket. This degree of plasticity has been confirmed in a recent study of inhibitor binding to caspase-3 [68]. Interestingly, an analogous rotation of Tyr-99 in the completely unrelated serine protease Factor IXa occurs during substrate binding, opening the S4 pocket [69]. In the case of Factor IX, Tyr-99 prevents premature onset of blood coagulation [70]. Currently there is no function ascribed to the rotation of Tyr-338 in caspases-3 and -7.
The S3 pocket
Residue Arg-341 plays a dual role in the anchoring of substrates and peptidyl inhibitors. In addition to its role in the S1 pocket, and perhaps more importantly, it is engaged in main-chain–main-chain hydrogen bonds with the P3 residue. Secondly, its guanidinium group interacts with the carboxylate of a P3 glutamate [13,63], explaining the preference for this residue in small peptide substrates and inhibitors [19,71,72]. Adjacent basic residues such as Arg-177 in caspases-8 [12] and -9 [73] allow tighter binding of inhibitors containing a glutamic acid residue at position P3, although without direct salt bridge formation. Indeed, caspases-8 and -9 show 500-fold weaker Ki values for inhibition with benzyloxycarbonyl-Ala-Val-Asp-CHO compared with benzyloxycarbonyl-Glu-Val-Asp-CHO, whereas caspases-1 and -2 show an only 40-fold discrimination between the two aldehyde inhibitors [66].
The S4 pocket
While subsites S1 and S3 have a similar character in all caspases, the S4 pockets provide major specificity-conferring elements to the different subclasses. In the inflammatory caspase-1, and predictably in caspases-4 and -5, this subsite is an extended, shallow hydrophobic depression that best accommodates large aromatic side chains. In particular, the indole moiety of a P4 Trp residue can engage in multiple interactions with residues of the 341-loop [74]. These interactions explain why inflammatory caspases optimally process substrates after a Trp-Glu-Xaa-Asp sequence (Xaa denotes any amino acid residue). Based on this specificity, these proteases were classified as group I caspases [19,72]. In contrast with this one-to-one correspondence of group I and inflammatory caspases, there is a less clear-cut distinction between initiator and effector caspases based on their preferred cleavage sequences. The initial classification into group II (caspases-2, -3, and -7; preferentially cleaving after Asp-Glu-Xaa-Asp) and group III (caspases-6, -8, -9 and -10; specific for Leu/Ile/Val-Glu-Xaa-Asp) caspases was challenged by the observation that caspase-8 tolerates equally well both small hydrophobic (Ile) [13] and acidic (Asp) residues at P4 [65].
Apoptotic caspases possess a bulky Trp residue at position 348 (compared with the smaller Ile/Val found in caspases-1, -4 and -5; Figure 2), which reduces considerably the size of the S4 pocket. The indole moiety of this residue contributes to the preference for short branched aliphatic side chains by group III caspases through van der Waals contacts, but can also hydrogen-bond to the carboxylate of a P4 aspartate residue [10,65,75,76]. Additional van der Waals or polar interactions strengthen the recognition of chemically diverse groups at P4. Hydrophobic side chains can, for example, contact the aromatic side chain of Tyr-340, as observed in caspase-8 [13]. Alternatively, a P4 aspartate can interact with the carboxyamides of Asn-342 (as in caspases-2 [75], -3 [10,76] and -8 [65]) or Gln-381b (in caspase-7; [63]). It is also noteworthy that similar insertions of 10 residues at position 381 (compared with caspase-1; Figure 2) generate enlarged loops (‘flaps’) in caspases-3 and -7, which further narrow their S4 pockets. The presence of these flaps could partially explain the increased discrimination of Asp over Glu at P4 (≈100- or ≈275-fold for caspases-3 and -7 respectively) [64]. Interestingly, it has been suggested that phosphorylation of the flaps may down-regulate the activity of effector caspases [77].
The S5 pocket (caspase-2)
Caspase-2 is currently exceptional in its requirement for an occupied S5 subsite for efficient substrate cleavage. In contrast with caspases-3 and -7 [71], the presence of a P5 residue conferred a 35-fold increase in catalytic efficiency, which may, at least partly, reflect a better burial of the P4 aspartate [75]. The origin of this extended specificity has been unveiled in the recently solved, high-resolution structure of caspase-2 in complex with acetyl-Leu-Asp-Glu-Ser-Asp-CHO [75]. The P5 Leu side chain occupies a small groove formed by two residues of the 381-loop, Tyr-381a and Pro-381c, thus explaining the preference of caspase-2 for small hydrophobic residues at this position.
The S1′ pocket
The primed subsites have only recently been characterized crystallographically [68], and appear to be less restrictive than the S1–S4 pockets. A systematic study using fluorescent peptidyl substrates, however, indicated a clear degree of discrimination for small (Gly, Ala, Ser) residues at position P1′ in all caspases tested Large aromatic side chains (Phe/Tyr) were also well tolerated, but not polar residues or proline [64]. These observations verify and extend previous investigations on the P1′ substrate specificity of caspases-1 and -4 [71,78].
Proposed catalytic mechanism
Neither experimental investigations using time-resolved crystallography nor detailed theoretical calculations of substrate cleavage have been reported for caspases (for a recent quantum mechanics/molecular mechanics study of the deacylation reaction, see [79]). However, the similarity of chemical groups involved in catalysis and the crystal structures of active caspases bound to substrate-like inhibitors suggests a similar course of the catalytic reaction as in serine proteases and other families of cysteine proteases (summarized in Figure 5c). Alternatively, a role for the His-237 Nδ atom in stabilizing the developing charge on the leaving group has been proposed by Brady and co-workers [80]. Further, these authors postulated that a water molecule hydrogen-bonded to the Gly-238 amide, but not seen directly in crystal structures, donates a proton to the leaving amine group. This mechanism thus practically reverses the roles of the ‘classic’ oxyanion hole and His-237. The controversy is spurred on by a distinguishing characteristic of the caspases when compared with serine proteases and cysteine proteases of other clans: the side chain of the second component of the catalytic dyad, His-237, is located over 5 Å away from the Sγ atom of Cys-285. Clearly at this unusual Nδ1/Nε2–Sγ distance His-237 cannot accept the thiol proton of Cys-285. This implies that Cys-285 may not be prepolarized, but that the nucleophile may develop along the reaction co-ordinate [81], in accordance with the pH optima of all caspases lying in the narrow range 6.8–7.4 [82].
Is there substrate-induced activation?
The crystal structure of non-inhibited (unliganded) caspase-7 featured a Cys-285 displaced by approx. 3 Å from its active position, and an active-site cleft partially invaded by residues from the 341-loop. Therefore the authors suggested that substrate binding induces the final step(s) in active-site formation [11]. This is in contrast with the crystal structures of uninhibited caspases-3 and -9, which show the same arrangements of catalytic residues as seen in the inhibitor-bound form [67,73] (see also the discussion on caspase activation below). Whether the latent conformation observed in free caspase-7 is specific to this caspase is currently unknown. Stable intermediates between the zymogen and the enzyme with a fully developed active site are uncommon in other protease classes, and might not be highly populated in an effector caspase in solution.
Is there a third catalytic residue in caspases?
This issue refers to the possible participation of the carbonyl oxygen of residue 177 as an important element of the catalytic machinery, as first proposed by Wilson and colleagues for caspase-1 [7], and later by others [13,76]. This oxygen atom accepts a hydrogen bond from the Nε atom of His-237, and might either affect the basicity of the imidazole moiety and/or orientate it similarly to the carboxylate of Asp-102 in trypsin-like proteases and the side chains of catalytic Asn/Asp residues in clan CA cysteine peptidases. Regardless, it is noteworthy that investigations on the role of this third residue in the clan CA cysteine protease papain [83] revealed a comparatively minor role in catalysis. Caspase-1 hydrolyses tetrapeptide p-nitroanilide substrates with a low kcat of approx. 1 s−1, which is similar to values reported for mutants of the catalytic aspartate in papain [7]. The carboxylate of the P1 residue might also help to orientate His-237 during catalysis, as proposed by Wilson and co-workers [7]. Finally, a His/Cys catalytic dyad has been reported in several viral proteases, notwithstanding the lower Nδ1/Nε2–Sγ distances of ≈4 Å [84,85], although in this case also the involvement of a third residue is disputed [86]. Heuristic considerations may be brought in to either support or refute a role for the carbonyl group of residue 177 in catalysis. An attractive consideration in favour of this possibility is that a direct engagement in peptide bond cleavage might allow for a better connection to product release, for example through weakening of the interactions of the neighbouring Arg-179 with the side chain of the P1 residue. Rigorous quantum mechanics/molecular dynamics calculations, along with structural and mutagenesis studies, will be needed to settle the issues discussed above and finally characterize the caspase catalytic mechanism.
CASPASE SUBSTRATES IN VIVO
The common occurrence of sequences that match the preferred inherent substrate specificity of caspases in intracellular proteins would suggest a multitude of substrates in vivo – somewhere in the order of several hundred. Indeed, the list of proteins that are cleaved by caspases either in vivo or in vitro is ever growing (for detailed analysis of caspase substrates, see [77,87,88]). However, only a few of these proteins have been rigorously established as biologically relevant, bona fide death substrates, and many of them may represent just ‘innocent bystanders’ [89]. Readers are referred to the above reviews for specific details on the many putative natural caspase substrates, and we will cover some overall concepts here.
Enzyme regions located distant from the active site that participate in substrate binding (exosites) are essential determinants of substrate recognition and processing by other regulatory proteases, most notably those involved in blood coagulation [90] and matrix regulation [91]. It is therefore remarkable that similar exosites have not been identified so far on caspases, although they have been proposed to explain the discrepancies between cleavage preferences in vitro and substrate cleavage sites actually found [87,92,93]. In particular, a large number of caspase-3 substrates are processed at non-canonical sites [77,87,89]. These findings imply that an exosite(s) in caspases recognizes specific tertiary motifs in their substrates, in addition to and in some cases ‘overruling’ the specificity requirements derived from studies with short synthetic peptides. In support of this, recent investigations reveal that caspase recognition by some natural inhibitors might also rely on exosite interactions (see below).
On the other hand, it is conceivable that solvent-exposed, disordered loops that present non-optimal cleavage sequences might be processed by active caspases without the need of additional interactions. For instance, heat-denatured pro-interleukin-1β was cleaved by caspase-1 as efficiently as the folded precursor [78]. We finish our overview of caspase activity by briefly discussing three protein substrates whose cleavage has been repeatedly associated with the execution of programmed cell death [77]. We discuss in the next section further important substrates, the caspase zymogens themselves.
Bid cleavage and the cross-talk between extrinsic and intrinsic pathways
The pro-apoptotic Bcl-2 family member Bid is an early substrate of caspase-8, and the cleaved protein translocates rapidly to the mitochondria [94,95]. Processed Bid then engages two further pro-apoptotic Bcl-2 members, Bax and Bak, which are hypothesized to selectively permeabilize the mitochondrial membrane, amplifying the death signal transmitted through Fas/FasL at the mitochondrial level [96].
The solution structures of Bid showed that the caspase-8 cleavage site, Leu-Gln-Thr-Asp-60↓Gly, maps to an unusually long, highly flexible loop [97,98]. Therefore the Asp-60↓Gly scissile bond could be easily targeted by caspase-8, with the P4–P1′ residues conforming to the inherent substrate specificity against short peptidyl substrates in vitro (see above). Notably, in vitro-translated Bid is targeted at the same site by active caspase-2 [99]. What makes the active-site-only specificity arguable in this case is that caspase-8 does not cleave a second site found in the same loop (Ile-Glu-Ala-Asp-75↓Ser). This more C-terminal site is certainly accessible, as it is the target of the cytotoxic serine protease granzyme B, unleashing the mitochondrial pathway via a similar Bax/Bak-mediated mechanism [100,101]. Importantly, this second site is almost identical to the activation cleavage site in the intersubunit linker of procaspase-3 (Figure 2). Thus we are tempted to speculate that important exosite-mediated interactions preferentially guide the Asp-60↓Gly site of Bid into the active-site cleft of caspase-8, or conversely steer the caspase away from the Asp-75↓Ser site. Recombinant variants of Bid with mutated or swapped cleavage sites might help to clarify this point.
Finally, it is not clear how cleavage in the long flexible loop of Bid that lies between helices 2 and 3 would lead to a conformational change that should, by analogy with other Bcl-2 family members, remove helix 2. Gel filtration analysis reveals that, in vitro at least, helix 2 does not even dissociate from cleaved Bid, and so there is some uncertainty about the functional consequence of Bid cleavage [97].
Activation of DFF40/CAD – how to get rid of an inhibitor
Internucleosomal DNA fragmentation (visualized in the form of a DNA ladder) is a hallmark of cells undergoing apoptosis. The nuclease responsible for these double-strand cleavages, DFF40 (DNA fragmentation factor)/CAD (caspase-activated DNase), exists in a tight non-covalent complex with its endogenous chaperone/inhibitor, DFF45/ICAD (inhibitor of CAD). This complex is disrupted when caspase-3 cleaves DFF45 at positions Asp-Glu-Thr-Asp-117↓Ser [linker between the N-terminal (D1) and central (D2) domains] and Asp-Ala-Val-Asp-224↓Thr (located between D2 and the C-terminal domain, D3), allowing the free nuclease to dimerize into the catalytically competent form [102–106]. Both cleavage sites thus map to presumably highly flexible and exposed linkers, and may be well accessible to caspase-3 without the need for additional enzyme–substrate interactions.
Solution structures of DFF45 domain D1 bound to the homologous N-terminal domain of DFF40 [107,108] and of the isolated domain D3 [109] have been reported. However, just as in the case of Bid, it is difficult to understand how the severing of two poorly ordered loops that most probably do not contact DFF40 leads to dissociation of the resulting DFF45 fragments from the nuclease. Possibly cleavage serves to convert a first-order interaction into a second-order one, although the kinetics of such events remain to be clarified. Large, complementary interfaces between the N-terminal domains of the two moieties have been revealed in the solution structures [107,108], or inferred from multiple observations at the independent interfaces between the DFF40 catalytic domain and domains D2 and D3 of the inhibitor. For example, the recombinant fragments D1–D2 and D2–D3 are strong repressors of the nuclease activity, and show only 4–5-fold lower affinities for DFF40 than full-length DFF45 [110,111]. Furthermore, a truncated natural variant of the chaperone that lacks a folded C-terminal domain, DFF35, binds more strongly to DFF40 than does the full-length form [112]. Remarkably, most single point mutations of charged interface residues in the N-terminal domains do not disrupt complex formation [107].
More recently, the crystal structure of activated DFF40/CAD has been reported, featuring a Zn2+-mediated dimer [106]. These authors also reported that the central D2 domain of DFF45 is critical for the inhibitory activity, which is relieved after caspase-mediated cleavage due to the low affinity of the isolated domain for the enzyme, although the kinetics of dissociation have yet to be explained, as mentioned above. Thus the mechanism of inhibition by ICAD basically relies on enzyme monomerization. Curiously, a similar principle applies to the inhibition of capase-9 by its physiological inhibitor, XIAP [X-linked IAP (inhibitor of apoptosis protein)] (see below).
Cleavage of PARP – the influence of protein modifications
The abundant nuclear enzyme PARP [poly(ADP-ribose) polymerase] catalyses the attachment of poly(ADP-ribose) to several acceptor proteins, including itself, in response to DNA strand breaks. PARP cleavage by caspases-3 and -7 at the Asp-Glu-Val-Asp-213↓Gly site bisects a bipartite nuclear localization signal [113], and results in a form that cannot synthesize ADP-ribose polymers in response to damaged DNA [114]. PARP was one of the first identified examples of a substrate processed by an effector caspase much more efficiently than by either the inflammatory caspase-1 [115] or the initiator caspase-8 [116]. Notwithstanding the fact that the PARP cleavage site perfectly matches the substrate specificity of effector caspases, secondary interactions seem to modulate its processing in vivo. Indeed, PARP molecules modified with long, branched poly(ADP-ribose) chains possess a much higher affinity for caspase-7 than for the close homologue caspase-3 in vivo, thus reversing the observed catalytic efficiency of caspases against small synthetic substrates. The enhanced affinity results from specific interactions between the large subunit of caspase-7, but not caspase-3, and poly(ADP-ribose) [117]. Notably, the potential acceptor of (poly)ADP-ribose closest to Asp-213, i.e. Glu-406, is located almost 200 amino acid residues downstream of the caspase-3 and -7 cleavage site.
Non-canonical substrates
Although caspases are thought to be exquisitely selective for Asp in their S1 pocket, the transcription factor Max is cleaved by caspase-5 at a non-canonical Ile-Glu-Val-Glu-10↓Ser site following dephosphorylation of Ser-11 [118]. In this case, the negative effect of the protein modification might be explained by unfavourable contacts of the phosphoryl moiety attached to the P1′ residue with the enzyme S1′ pocket [64]. However, it is worth mentioning that caspase-5 does not cleave a peptide comprising Max residues 7–16, suggesting that the tertiary/quaternary structure of the properly folded transcription factor is responsible for the use of this atypical site. In particular, Max homodimerization and the presence of some residues C-terminal of the cleavage site (amino acids 13–21) appear to be important for proteolysis after Glu-10 [118].
The findings discussed above, among others, illustrate that exosite interactions, in addition to contacts with residues in the immediate surroundings of the P1–P1′ scissile bond, modulate caspase specificity against macromolecular substrates. Inspection of caspase structures (see Figure 4a) would suggest that the characteristic loop-bundle, but also residues from the more distant α2 and α3 helices of a neighbouring monomer, may provide additional substrate recognition surfaces.
MECHANISMS OF CASPASE-ZYMOGEN ACTIVATION: CONSERVATION AND VARIATION
Unregulated caspase activity would be lethal for a cell, and so to prevent this occurrence the cell stores caspases as latent precursors (zymogens, procaspases) that require an activating event. Recent advances suggest that the activation mechanisms of initiator and effector caspases are entirely distinct, but the device for stabilizing the latent zymogens is fundamentally conserved. Although the details of activation pathways are not yet fully worked out, it is now established that activation of initiator apoptotic caspases, and probably also inflammatory caspases, requires dimerization of inactive monomeric proforms, which occurs in vivo upon recruitment to large macromolecular assemblies called ‘activation platforms’ (Supplemental Figure 1; (http://www.BiochemJ.org/bj/384/bj3840201add.htm). In stark contrast with the initiators, the zymogens of the effector caspases-3 and -7 exist within the cytosol as inactive dimers [10,11,119–123]. They are activated by limited proteolysis within their β4–β5 intersubunit linker, carried out by an initiator caspase, and occasionally by other proteases under specific circumstances.
The crystal structures of the initiator caspase-9 [73], and the structures of zymogen caspase-7, active caspase-7 and inhibitor-bound caspase-7 [11,63,124], serve as models with which to rationalize the apparent dichotomy between the dimerization mechanism for initiator caspase activation and the cleavage mechanism for effector caspase activation. This is in addition to several investigations that have addressed activation biochemically, characterizing in particular the activation/maturation mechanism of inflammatory and initiator caspases [123,125–129]. Because of the more extensive structural dataset, we start our analysis with the activation process of effector caspases.
Activation of effector caspases: lessons from the structures of procaspase-7
There are two major mechanisms by which the zymogen of a proteolytic enzyme can be kept in a latent conformation, so that it can be converted rapidly into the active enzyme upon cleavage of a few, and usually a single, peptide bond. In the simplest conceivable situation, the catalytic machinery is preformed, but a prodomain sterically blocks substrate access, as observed for instance in several metalloproteases, subtilisin-like serine proteases, and clan CA cysteine proteases (reviewed in [130,131]). A second mechanism, typical of trypsin-like proteases, involves insertion of the N-terminal peptide liberated upon activation cleavage into the main body of the enzyme to stabilize its catalytically competent conformation (reviewed in [130]).
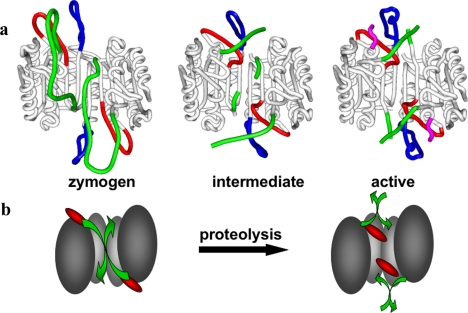
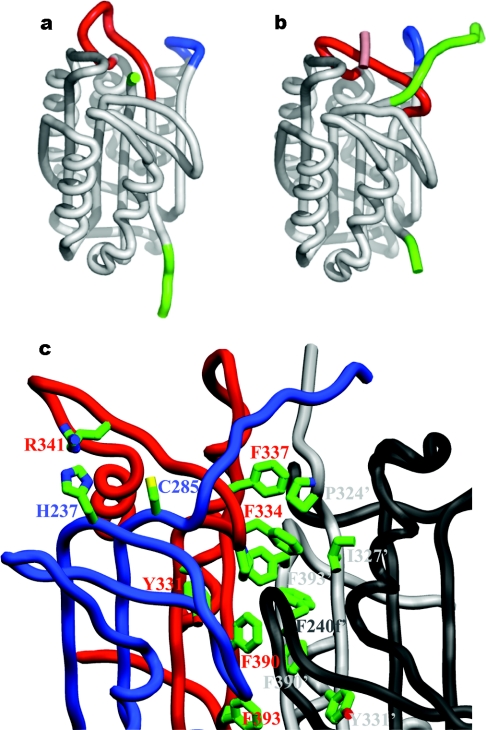
Procaspase-3 originates as a result of the association of two monomeric folding intermediates, and the monomeric domains are extremely tightly associated in the procaspase-3 dimer [132]. The same is probably true for its close homologue and co-effector caspase-7. The crystal structures of procaspase-7 [11,124] showed that the domain organization of the zymogen and the overall constitution of secondary structure elements are indistinguishable from those of the active enzyme [63,133,134]. Furthermore, the residues that form the catalytic dyad are displaced only slightly (Cys-285; ≤2.5 Å) or not at all (His-237) in the zymogen. The displacement of Cys-285 disrupts the oxyanion hole in particular. However, the most evident structural differences between active caspase-7 and the single-chain zymogen relate to the critical determinants of substrate specificity, the 341- and 381-loops, which protrude into the bulk solvent in procaspase-7 (Figure 6a). These loops are disordered in the zymogen, in striking contrast to their well-defined conformations in inhibitor-bound caspases. Disorder particularly affects the crucial residue Arg-341, which contacts substrate residues at positions P1 and P3, but also the neighbouring Trp-340, which shapes the S2 pocket in active caspases (see above and Figures 5a and 5b). Trp-348, located at the bottom of the S4 pocket, is only slightly displaced from the conformation observed in inhibitor-bound caspases, but most of the ‘flap’, including Gln-381b and the side chain of Phe-381h, is disordered. Altogether, the substrate-binding pockets of the two monomers are essentially disrupted in procaspase-7, and this explains its complete lack of catalytic activity. The activation mechanism of effector caspases thus appears to be more closely related to trypsin-like serine proteases.
Figure 6. Mechanisms of procaspase-7 activation.
(a) Ribbon plots showing the crystal structures of human procaspase-7 ([124]; PDB code 1GQF), and both free ([11]; 1K86) and inhibitor-bound ([63]; 1F1J) active caspase-7. Loops that display significant changes during activation are coloured red for the 341-loop, blue for the 381-loop, and green for the intersubunit linker. Notice the turn of almost 180° at the Val-323–Glu-324 peptide bond in the intersubunit linker in the left and middle structures compared with the right (inhibitor-bound) form, leading to the insertion of residues N-terminal of Val-323 into the central cavity of the zymogen. Residues Thr-288 to Arg-318 were only poorly or not defined at all by electron density, suggesting enhanced flexibility. However, the distances between the defined N- and C-termini in the zymogen (left) suggest that the spatially adjacent large and small subunits derive from the same domain. (b) Cartoon version of the activation, showing the critical loop transitions. In this context, the unliganded active form is omitted as an intermediate in the generation of a fully functional active site.
Superposition with structures of the active enzyme immediately reveals that the partial insertion of the intersubunit linkers into the central cavity accounts for this unexpected expulsion of the substrate-binding loops. This ‘linker in-conformation’ does not allow the characteristic 341-loop to adopt the conformation observed in active caspases (compare Figures 6a and 7a). In particular, there would be major clashes between residues Val-334–Tyr-337 from a β5–βIV ‘elbow loop’ and the inserted linkers from a neighbouring monomer. We note that the two independent crystal structures of procaspase-7 were determined using crystals grown from rather different solutions, and show differences in the extent of insertion of the linker peptide [11,124]. Thus the two linkers compete with each other and with the elbow loops for occupancy of the central cavity. The conformations revealed in the crystal structures represent snapshots of particularly stable conformations in the linker regions that are almost certainly in dynamic motion. Because of the limited size of the central cavity (Figure 7a), it cannot accommodate the two linkers simultaneously, and this implies that a procaspase-7 dimer is intrinsically asymmetrical. This is a major difference from the symmetrical arrangement of domains in active caspases (see above and compare Figures 4a and 6a). The consequences of this symmetry breaking for the mechanism of activation, in particular the frustrated state of the linkers in the uncleaved zymogen, have not been explored so far.
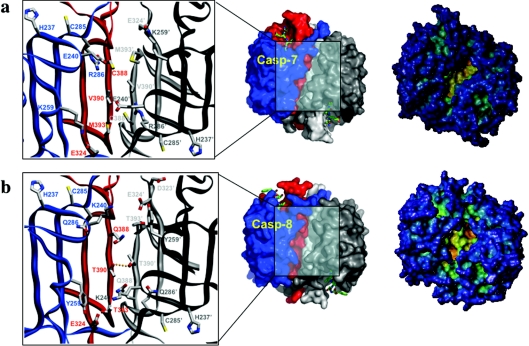
Figure 7. Details of interdomain regions of an effector and an initiator caspase.
Close-ups of the interdomain interfaces (left), surface (middle) and cavity depth (right) in caspase-7 (a) and caspase-8 (b). Caspase-7 is the acetyl-Asp-Glu-Val-Asp-CHO-inhibited form (PDB 1F1J; [63]) and caspase-8 is the acetyl-Ile-Glu-Thr-Asp-CHO-inhibited form (PDB 1QTN; [13]). Several interdomain residues are shown with all of their non-hydrogen atoms (colour-coded). Hydrogen bonds are denoted by orange dotted lines. The central cavity, formed at the dimer interface, is significantly larger in caspase-8, with the attendant possibility that the intersubunit linker that would be in the zymogen, and therefore not visible here, may partially occupy it without producing the steric clashes seen in the smaller caspase-7 central cavity.
While cleavage of the Asp-297↓Ser-298 sites in procaspase-7 promotes the release of the intersubunit linkers from the central cavity, contacts between the newly formed N- and C-termini of the small and large subunits from the neighbouring domains stabilize the active enzyme conformation by forming a bundle with each other and with the 381-loop [11]. The net free energy gain provided by loop-bundle formation seems to prevent the N-termini of the small subunit from sliding back into the central cavity, in line with a recent theoretical analysis of residue contribution to dimer stability [135]. Interestingly, the change from the zymogen to the active conformation in caspase-7 can be reversed by the binding of small molecules in the central cavity of the active conformation, where they act to inhibit the protease allosterically [135a]. The mechanism of such allosteric inhibitors supports the importance of translation of the 341-loop and organization of the loop bundle formed in the activation of procaspase-7, and implies that the central cavity of caspases is an alternative target to the active site for small molecule targeting and drug design.
A puzzling observation made with the crystal structure of cleaved, uninhibited caspase-7 is that the linker peptide remains partially inserted in the central cavity, thus impairing formation of fully competent active sites [11]. By contrast, the catalytic machinery is almost perfectly arranged in the corresponding caspase-3 structure, with the important exception of Tyr-338 [67] (see Supplemental Movie 1; http://www.BiochemJ.org/bj/384/bj3840201add.htm). Perhaps the high salt concentrations used to grow crystals of free caspase-7 had disrupted the interactions between the newly formed chain termini, favouring a pseudo-zymogenic conformation instead. Whatever the reason for this observation might be, it underscores the fundamental correlation between linker insertion and catalytic state in caspases.
We also note that it is possible to activate procaspase-7 efficiently in vitro by treatment with the serine protease cathepsin G, which processes the zymogen after Gln-295, two residues upstream of the physiological cleavage site [136]. This finding indicates that cleavage after a particular Asp residue is not needed for activation; this is a notable difference from the activation mechanism of trypsin-like serine proteases [130]. Finally, we recall that biochemical data place caspase-7 as the most downstream caspase in apoptosis, at least in some cell types. Caspase-3 appears to be necessary for removing the N-terminal peptide of procaspase-7 before an initiator caspase (or granzyme B) gains access to the β4–β5 cleavage site [137]. This observation may be explained by the finding that this highly charged N-terminal peptide sequesters the zymogen in a cytosolic location, which is inaccessible to the active initiator caspases-8, -9 and -10 [22]. The identity of the factor(s) that interacts with procaspase-7 has not been established.
Implications for the activation of procaspases-3 and -6
The high degree of sequence identity between the catalytic domains of caspases-3, -6 and -7 (Figure 2) strongly suggests that the major features of the inactive zymogen and the mechanism of activation described above for caspase-7 are also essentially valid for the two other caspases. More importantly, both of the residues that line the central cavity in procaspase-7 and those of the intersubunit linker that intrude into this cavity are conserved or conservatively replaced (e.g. Met-393 is strictly conserved, while Val-323 and Val-390 are conserved in caspase-3, and replaced by alanines in caspase-6). Thus it may be expected that the substrate-binding loops in procaspases-3 and -6 are also disordered, due to the partial insertion of the intersubunit linker into the central cavity. There might, however, be some differences in the degree of linker insertion, as caspase-6 possesses alanine residues at positions 323 and 334, and this would alleviate the steric clashes between the elbow loop and the linker peptide. Notwithstanding, a biochemical investigation confirmed the validity of the model for procaspase-6, including a change in the environment of Trp-348 upon proteolytic activation [121]. Also in support of this model, constitutively active, recombinant caspases-3 and -6 have been generated by exchanging the order of large and small subunits in the expression constructs [138,139]. In addition to these elegant experiments, mixtures of separately expressed subunits allowed to refold also form active enzymes spontaneously (e.g. see [74,76]).
Finally, we stress that cleavage of the intersubunit linker, as an essential element of the activation process, appears to be limited to effector caspases and eukaryotic haemoglobinases [140]. Gingipain R is the most closely related clan CD protease for which structural information exists [45], and an analysis of the activation process has been reported [141]. Cleavage of the intersubunit linker is not required for activation of gingipain R. In addition, the reported autocatalytic processing sites in human separase precede the cysteine protease domain [59,142,143].
Activation of initiator caspases: the induced proximity model revisited
Since the active form of caspases seen in all crystal structures contains large and small subunits comprising the catalytic domain, it was assumed that all caspases were activated by proteolytic cleavage within their linker region (reviewed in [53,144]). However, recent studies have revealed that cleavage is neither required nor sufficient for the activation of initiator caspases-8 and -9. Their activation mechanism relies on a previously unappreciated property of procaspases-8 and -9: that they reside in the latent state as monomers, and require dimerization to assume an active conformation. This property has now been demonstrated both for recombinant material [73,129] and for the natural endogenous zymogen [123]. This is in stark contrast to the zymogens of the effector caspases-3 and -7, which are already dimeric in their latent forms. The reason for this important difference, which at first glance is incongruous [145], is clarified below.
From an evolutionary point of view, cofactor-mediated oligomerization constitutes the primordial mechanism of procaspase activation, as exemplified by the C. elegans caspase CED3 (cell death-defective 3) and the APAF-1 (apoptotic protease activating factor-1) orthologue CED4 [146,147]. The nature of this activation mechanism has puzzled researchers, in particular following the realization that caspases are activated in an ordered cascade, with active initiator caspases processing the zymogens of effector enzymes [55,56,148,149]. The reason is clear: there is no proteolytic enzyme upstream of the initiator caspases. Thus it is apparent that their zymogens must possess a certain degree of latent activity, as suggested by the labelling of single-chain caspases-1 [150] and -9 [73] using relatively high concentrations of biotinylated methyl ketone inhibitors. Quantitatively, initiator caspases show low zymogenicity (i.e. ratio of activity of the cleaved compared with the uncleaved form) values of at most 10 (caspase-9) or 100 (caspase-8), in contrast with values of >10000 for caspase-3 [81,123,129,151,152]. This means that the initiator caspase zymogens have a much greater tendency to become activated than do the effector zymogens.
These features are shared by mechanistically unrelated serine proteases of the blood clotting and fibrinolytic cascades, which also proceed through the sequential activation of inactive proforms. In particular, the upstream factor of the latter cascade, tissue-type plasminogen activator, possesses a similar zymogeneicity to caspase-9 [153], and its physiological activity (plasminogen activation) depends upon co-localization to the fibrin clot. In a similar manner, the initiator of the coagulation cascade, Factor VIIa, only processes its substrate Factor X when bound to its cofactor, tissue factor [154]. What sets initiator procaspases apart from these upstream serine proteases is that the enforced oligomerization of natural (cofactor-induced) or chimaeric variants results in their activation [56,125,126,128,151,152,155,156]. In striking contrast to the effector caspases, activation of initiator caspases does not require processing of the intersubunit linker [123,125,126,129,151,157,158], and cleavage may simply provide stability to the active dimer. In summary, oligomerization of inflammatory and initiator procaspases suffices to trigger their processing, and this constitutes the central postulate of the induced proximity model [152].
Before discussing the accumulated structural and biochemical evidence in support of this mechanism, we recall that oligomerization events occur in vivo at multiprotein activating complexes (activation platforms), which engage the caspase zymogens by virtue of their N-terminal CARD or DED recruitment domains. The activating complex involved depends on the origin of the death stimulus: either extrinsic [the DISC (death-inducing signalling complex)] or intrinsic (the apoptosome) [Figure 3 and Supplemental Figure 1 (http://www.BiochemJ.org/bj/384/bj3840201add.htm)]. The crystal structure of the CARD of procaspase-9 in complex with the CARD of APAF-1 revealed that complex formation is mediated by an exquisite balancing of complementary charges on the surface of the two modules [159,160]. The structure of the corresponding DED·DED or DD·DD complexes that constitute the DISC are unknown to date, but those of isolated domains have been reported, along with extensive mutagenesis studies [47–49]. This is in addition to the structure of the heterodimer formed by the DD modules of the Drosophila kinase Pelle and its adaptor, Tube [161], which represents a valuable template for the related Fas·FADD [Fas (TNF receptor SF6)-associated via death domain] complex. Critical reassessment of the structural and mutagenesis evidence has allowed the recent development of working models for the DISC, which feature three-cornered aggregates centred on either a trimer of the Fas DD or FADD DD modules [162,163]. A similar arrangement has been proposed for the heterohexameric complex of procaspase-8 and FADD DED modules [164].
Although the DEDs/CARDs and the catalytic domains of procaspases are separated by relatively long linkers (Figure 1), it is apparent that the productive encounter of two zymogen monomers would be facilitated enormously by their prodomain-mediated recruitment to an adaptor platform. This would convert a bimolecular interaction with a relatively weak Kd into a much more productive unimolecular reaction. While this event and the concomitant dimerization appear understandable, a major question remains unanswered: why does a dimeric effector procaspase persist in a dormant state, while dimers of initiator procaspases, once formed, are either fully active or at least active enough to catalyse their own maturation?
Activation of procaspase-9: what a crystal structure tells us
The location of the intersubunit linker correlates with the activity of (pro)caspase-7 (linker inserted=substrate-binding site disordered; linker exposed=substrate-binding site formed). This observation immediately suggests that generation of a central cavity that is not accessible to this linker may also allow at least transient formation of stable catalytic machinery, independently of linker cleavage.
Indeed, the crystal structure of caspase-9 [73] revealed that the unique insertion 240-loops (Figure 2) from the two monomers intrude into the central cavity stabilized through important contacts with each other [Figure 8 and Supplemental Figure 2 (http://www.BiochemJ.org/bj/384/bj3840201add.htm)]. Hydrophobic contacts across the dimer interface prime the translocation of the all-important 341-loop to adopt its active, substrate-binding conformation, without the need for intersubunit cleavage. This hydrophobic interaction would be expected to stabilize the active conformation in uncleaved procaspase-9, in place of the loop bundle of the processed effector enzymes and independently of its formation. Indeed, uncleavable procaspase-9 either with Asp-297 [157,165] or with both processing sites (Asp-297 and Asp-316) mutated to alanine [157,158,166] still recruited and activated procaspase-3 within reconstituted apoptosomes in vitro. Further, uncleavable procaspase-9 develops activity against small synthetic substrates that is indistinguishable from that of the wild type [158].
Figure 8. Details of interdomain regions of caspase-9 reveal its activation mechanism.
The crystal structure of caspase-9 is unusual in that it contains one catalytic domain in the active conformation (a) and one in a zymogen-like conformation (b) ([73]; PDB code 1JXQ). Loops that display significant changes between the two forms are coloured red for the 341-loop, blue for the 381-loop and green for the intersubunit linker, as in Figure 6(a). (c) Ribbon plot showing a close-up of the central cavity in human caspase-9. The unique insertion 240-loops (see also Figure 2) from both caspase-9 monomers intrude into the central cavity stabilized through important contacts with each other. In addition, two aliphatic residues that line the central cavity in procaspase-7, Val-390 and Met-393, are replaced by the bulkier aromatic side chains of Phe-390 and Phe-393 in the initiator caspase, thus effectively ‘sealing’ its central cavity. The well-conserved Arg-286, whose guanidinium group is sandwiched between Tyr-331 and Val-334 in active caspases-3 and -7, is substituted in caspase-9 by a glycine, leaving additional empty space for the elbow loop. Most notably, a hydrophobic pocket formed by some of the very same residues that seal the central cavity, i.e. Phe-240f′ and Phe-393′, together with Pro-324′ accepts the phenyl moiety of the elbow loop residue Phe-334 from the neighbouring monomer, which is thus anchored into the central cavity. In this manner, residues from one monomer indirectly prime the 341-loop of a neighbouring procaspase-9 molecule to adopt its active, substrate-binding conformation, without the need for intersubunit cleavage.
Mutagenesis studies underscore the critical role of zymogen dimerization in activation. Point mutants of interface residues, i.e. Phe-390→Asp [122,123], Phe-390→Val [123] and the double mutant Lys-396→Asp/Lys-397→Asp [125], have been reported. Most convincingly, replacement of Phe-390 with a hydrophobic valine did not compromise induction of cell death by caspase-9, while mutant Phe-390→Asp failed to trigger caspase-9 activation and apoptosis, and blocked apoptosis induced by overexpression of Bax [123]. Furthermore, the double mutant with reversed charges at positions 396 and 397 failed to self-associate, was not activated by APAF-1, and failed to induce cell death when transfected in 293 or HeLa cells [125]. These observations are explained by the fact the electrostatic repulsion between equally charged groups at the domain interface would impair dimerization. It is less straightforward to rationalize the effects of mutations that aim at a disruption of the loop bundle, i.e. Asp-291→Ala [122] and (Phe-293a→Ala/Val-293c→Ala) [125], as they alter structures of the protein that differ significantly between the monomer, the uncleaved dimer and the fully processed dimer. The Asp-291 mutant was reported to have a low level of procaspase-3-converting activity [122], while the double mutant showed the same deficits as reversal-of-charge variants [125]. We notice that the important polar/van der Waals contacts made by Asp-291/Phe-293a in the cleaved caspase-9 dimer [73] are predictably formed in the dimer of uncleaved procaspase-9 molecules, and so these mutants might simply add to the relevance of dimerization for activation.
However, caution must be used in interpreting the consequence of mutations in a caspase dimer interface, since there is an unexpected tendency for substitutions to be compensated for by unanticipated interactions. Thus multiple-site mutants in the caspase-3 interface all produced a reasonably functional dimeric enzyme [167], probably explained by the inherently strong interface of this effector caspase compared with the initiator caspases. The mutations are simply not sufficient to radically disrupt dimerization.
Importantly, steric clashes at the dimer interface prohibit the two elbow loops from occupying the central cavity simultaneously, in line with the observation that only half of the caspase-9 monomers were labelled by Val-Ala-Asp-fluoromethyl ketone in solution [73]. The crystal structure of human caspase-9 confirmed this unanticipated finding, featuring a second domain in a zymogen-like conformation with disabled specificity determinants and catalytic apparatus [Figure 8 and Supplemental Movie 2 (http://www.BiochemJ.org/bj/384/bj3840201add.htm)]. The conformation of the inactive domain is almost identical to that of the zymogen form of caspase-7, as described above. This finding again suggests that there might be only two major, mutually exclusive conformations of the linker/substrate binding loops (Supplemental Movie 3; http://www.BiochemJ.org/bj/384/bj3840201add.htm).
A putative assembly of (pro)caspase-9 molecules in the apoptosome
The central component of the apoptosome is a protein known as APAF-1, which recruits procaspase-9 via its N-terminal CARD [55]. In its quiescent state, APAF-1 is a compact molecule with the ‘head’ (CARD) tucked between its ‘feet’ (two β-propellers formed by sets of WD40 repeats). Cytochrome c (which is conveniently about the same size as the CARD) displaces the head, allowing the compact structure to stretch out into a more linear molecule that polymerizes upon binding ATP [165]. Electron cryomicroscopy studies show the apoptosome to be a seven-spoked wheel, with a central hub that contains the caspase-9 recruitment domain, provided by the CARD of APAF-1.
Interestingly, procaspase-9 induces the stacking of APAF-1 rings, and the density assigned to full-length procaspase-9 in the electron micrographs of apoptosome complexes [165] resembles the tetrameric arrangement of caspase-9 domains in the crystal structure of CARD-less caspase-9 [73] (Figure 9). The centre of the interface between the two active domains (to the top in Figure 9a) is occupied by the antiparallel arranged C-termini of the CARD–(large subunit) linkers, including helices α0, and is stabilized by strong salt bridges between the side chains of residues conserved in caspase-9 from different species, i.e. Asp-150 and Arg-395. Contacts between the ‘down’ inactive monomers are mostly mediated by the exposed 341-loops, which lean against the neighbouring helix α4, forming important van der Waals interactions (most notably, edge-to-edge contacts between the two Trp-348 indole rings), as well as polar bonds (e.g. hydrogen bonds between Asp-354 and the indole nitrogen atom of Trp-340). Altogether, the protected surface at this dimer–dimer interface (≈1100 Å2, or 6% of the area of a dimer) is not particularly large, but the presence of conserved Trp and Arg residues points to a biologically relevant role [168], which is also suggested by the observation that the charge-reversal variant, Asp-150→Lys, behaves like the interface mutants discussed above [125]. Additionally, phosphorylation of the interface residue Ser-195 (Figure 9a) by the Ser/Thr protein kinase Akt/protein kinase B also inhibits procaspase-9 processing [169]. We also observe that phosphorylation of Thr-125 in the CARD–(large subunit) linker inhibits procaspase-9 processing both in vivo and in vitro [170]. Mutation Thr-125→Glu (which mimics the phosphorylated threonine side chain), but not Thr-125→Ala, reduced activation of procaspase-9 by a constitutively active APAF-1 variant in vitro. These findings point to important electrostatic interactions of the procaspase-9 linkers within the apoptosome. Conceivably, either the linkers of two procaspase molecules symmetrically approach each other or they are sandwiched between the globular CARD and catalytic domains in the apoptosome.
Figure 9. Caspase-9 packing and the apoptosome.
(a) Tetrameric arrangement observed in the crystal structure of caspase-9 ([73]; PDB code 1JXQ), colour-coded to show the active (red tones) and inactive (green tones) domain of each standard caspase dimer. (b) Average map of the APAF-1·procaspase-9 complex [165]. The arrowed region of central density represents bound procaspase-9 molecules sandwiched between two APAF-1 rings. The scale bar corresponds to 100 Å. Reprinted from Molecular Cell, Vol. 9, D. Acehan, X. Jiang, D. G. Morgan, J. E. Heuser, X. Wang and C. W. Akey, Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation, pp. 423–432, Copyright (2002), with permission from Elsevier.
We are therefore tempted to speculate that the domain arrangement in the caspase-9 crystals [73] mimics a functional apoptosome assembly, in which the restricted orientations of the CARD and catalytic domains might help docking of incoming procaspase-3/7 substrate molecules. Regardless, the combined structural and biochemical evidence suggests that the half-of-sites reactivity of caspase-9 is a biologically relevant feature. A flip-flop mechanism in vivo is conceivable, where the ‘upper’ and ‘lower’ monomers (Figure 9a) change synchronously from the active into the inactive state.
A balancing act at the cell membrane: DISC formation and activation of procaspases-8 and -10
The crystal structures of procaspases -8 and -10 have not yet been reported. However, it is possible to extend the model of activation discussed above to the initiators of the extrinsic apoptotic pathway, thanks to a large body of evidence accumulated from multiple biochemical and biophysical studies. As for procaspase-9, current evidence indicates that procaspase-8 molecules exist as inactive monomers in solution [123,129], and their enhanced oligomerization close to the cell membrane allows transient stabilization of the active conformation in vivo [148,149,151,155,171]. Accordingly, the dimerization interface mutant Thr-390→Asp [123] failed to induce apoptosis upon Fas ligation when transfected in different cell lines, whereas the conservative mutation Thr-390→Ser even enhanced caspase activity generated via the extrinsic pathway [123]. Similarly, the reversal-of-charge mutants Lys-292→Asp, Asp-323→Lys and Lys-397→Asp showed a severely decreased potential for homodimerization in vitro and were not processed upon enforced dimerization of N-terminal FV domains, and the full-length forms containing these mutations failed to induce apoptosis upon Fas engagement [126]. The results obtained with the latter mutants, however, have to be viewed with some caution, as these side chains do not point directly into the dimerization interface, and in addition the Asp-323 carboxylate hydrogen-bonds the main-chain N atom of Leu-268. Finally, and also underscoring the similarity of activation mechanisms with procaspase-9, cleavage of the intersubunit linker is not required for activation, and neither is processing of the intersubunit linker followed by activation. For example, the double mutant (Asp-297→Ala/Asp-312→Ala) did not abrogate cell death in transfection experiments with intrinsically dimeric variants [151,155]. Furthermore, when these mutations were incorporated in a prodomain-less form, it still displayed 1% of activity of mature caspase-8 [123,129].
How would the loop conformations in the procaspase-8 dimer compare with those of the corresponding caspase-9 zymogen? We observe that the higher zymogenicity of procaspase-8 indicates that the insertion of the elbow loop into the central cavity is favoured in the uncleaved procaspase-8. In other words, the equilibrium is displaced towards a ‘linker in, substrate-binding loops out’ state. The absence of the 240-loop found in caspase-9 and the presence of Thr residues at positions 390 and 393 [Figure 2 and Supplemental Figure 2 (http://www.BiochemJ.org/bj/384/bj3840201add.htm)] appear to be mainly responsible for this feature. It is remarkable that the central cavity of caspase-8 is deeper than in other caspases [13] (see also Figure 7b), and this increase in size of the intermonomer space might alleviate steric clashes between linker and elbow loops. Furthermore, the side chain of Gln-286 does not intrude into the central cavity, but hydrogen-bonds the main-chain N and O atoms of Gly-241 and Ile-242 respectively [12]. The overall result is similar to the (more radical) replacement of the highly conserved Arg-286 by glycine in caspase-9: liberation of space in the central cavity to allow transient insertion of the elbow loop, without previous proteolytic cleavage of the linker peptide.
The non-enzymic procaspase-8 homologue, c-FLIPL [FLIP is FLICE (FADD-like ICE) inhibitory protein] (Figure 2), was recently identified as an activator of the extrinsic pathway, lending additional support to the induced proximity model of cofactor-induced activation, but also adding a new level of complexity to the activation/maturation process in vivo. Initial studies had already indicated that c-FLIPL overexpression results in spontaneous cell death [172], and c-FLIPL knockout mice showed a phenotype that was strikingly similar to those of FADD(−/−) and procaspase-8(−/−) animals [173]. Nevertheless, c-FLIPL was long considered a bona fide inhibitor of DISC formation. Recent results have corrected this view and indicate that c-FLIPL is recruited preferentially to the DISC, and its subsequent heterodimerization with a procaspase-8 molecule induces active-site formation in the latter, without severing of the intersubunit linker [127,174]. Indeed, at least in vitro, c-FLIPL has a lower barrier for heterodimerization with caspase-8, leading to full activation with no apparent differences in inherent substrate specificity, than homodimerization of caspase-8 itself [175]. c-FLIPL residues Leu-390/Gln-391 (Figure 2), perhaps along with the Tyr-285-Val-Val triplet, would occlude the central cavity in a c-FLIPL·procaspase-8 heterodimer and disfavour insertion of the linker peptides into the central cavity to a greater extent than in homodimeric procaspase-8. Therefore we predict that the elbow loop of a procaspase-8 molecule in a heterodimer with c-FLIPL intrudes preferentially into the central cavity and generates a form with a higher intrinsic activity than a procaspase-8 homodimer [174], which effectively ignites the extrinsic apoptotic pathway. We discuss below further consequences of differential recruitment of procaspase-8 and c-FLIPL to the DISC.
In the absence of structural information, the conformation of monomeric procaspase-8 in the cytosol remains speculative. Association with a chaperone protein appears as an attractive possibility, but has not been reported to date, and no cognate IAP for caspase-8 is known, in contrast with the control of caspase-9 by XIAP (see below). On a more positive note, proteins termed CARPs [caspase-associated RING (‘really interesting new gene’) proteins] are suspected of regulating caspase-8 and -10 levels through ubiquitin-mediated proteasomal degradation, although no mechanism has been reported [176]. Alternatively, it is conceivable that either the intersubunit linker and/or the N-terminal DEDs block the dimerization interface in the small subunit, thus avoiding unregulated activation in vivo. In this regard, it is noteworthy that an N-terminally extended variant of procaspase-8 possesses enhanced dimerization potential in solution, and autoactivates spontaneously [126]. Finally, a natural truncated form lacking the catalytic domain is recruited more efficiently to the DISC, and acts as a potent inhibitor of apoptosis even at relatively low concentrations compared with full-length variants [177], also suggesting that full-length procaspase-8 exist in a somehow ‘closed’ conformation with less accessible DEDs.
Beyond activation: processing of initiator caspases to their mature forms
As stated above, from a mechanistic point of view proteolysis of inflammatory and initiator procaspases constitutes an epiphenomenon and is not needed for generation of an active enzyme. However, within their physiological activation complexes, but also at supraphysiological concentrations achieved in overexpression experiments, the intersubunit and/or the prodomain–(large subunit) linkers are cleaved rapidly after specific aspartate residues (Figure 1). While at least the former cleavage confers additional stability to the caspase dimer through formation of the loop bundles [11,129], it is noteworthy that cleavage of the β4–β5 intersubunit linker of procaspase-9 at the Asp-297↓Ala site represents a means for inactivation via binding to XIAP [158] (see below).
Two questions arise in connection with the mechanism of proteolytic maturation of initiator/inflammatory caspases. First, which active procaspase is responsible for these cleavages in vivo? In other words, are these cleavages performed by the same, transiently activated molecule or its non-covalently bound ‘partner’ (intradimer mechanism), or by an adjacent dimer (interdimer mechanism)? Furthermore, these cleavages have to proceed in a sequential manner [first intersubunit, then prodomain–(large subunit) linker], if release of inactive forms is to be avoided. This poses a second challenge: is this sequential processing regulated by the accessibility of cleavage sites (sequential accessibility model), or because the intermediate form with a cleaved β4–β5 linker specifically recognizes the more N-terminal site (sequential activity model)? A model that answers these questions has to account also for the apparent paradox that mature initiator caspases are not able to process their proforms in vivo [178].
These important open questions have recently been addressed for the maturation of procaspase-8. The intradimer mechanism is favoured in one study [128] that further suggests an intramolecular processing as the initial maturation step (i.e. an activated procaspase-8 molecule cleaves its own β4–β5 linker). By contrast, another study presented evidence in support of an interdimer, sequential accessibility mechanism of caspase-8 maturation [126]. Inspection of a procaspase-8 model developed based on the caspase-7 zymogen indicates that major structural rearrangements would be needed for an intersubunit linker to bind productively to its own active-site cleft, and binding to the more distant active site of the second domain can be practically excluded. Furthermore, the proposed interdimer mechanism seems to better capture the actual activation events in vivo, because this analysis included full-length, DED-containing procaspase-8 variants and their processing by the purified DISC. The finding that the Ala-325→Val mutant failed to be processed, although it possessed enzymic activity upon dimerization [126], might be related to a disruption of the ‘pre-loop bundle’, which impairs the conformation of the intersubunit linker.
The interdimer mechanism of caspase-8 maturation offers, among other advantages, an elegant explanation for the apparent mismatch between the number of binding sites in a single Fas· FADD complex (composed of three molecules each) and the dimeric nature of active (pro)caspases. Indeed, the third, lone procaspase-8 molecule bound to the Fas–FADD heterotrimer would be in the position to dimerize with a second ‘unmatched’ monomer from a neighbouring DISC (Supplemental Figure 1a; http://www.BiochemJ.org/bj/384/bj3840201add.htm). These interactions might be further strengthened by aggregation of the membrane-proximal Fas DD and FADD DD modules [163], and perhaps also by contacts between the more C-terminal DEDs of neighbouring procaspase-8 molecules. Support for this model comes from the observation that clustering of death receptor complexes enhances apoptosis [171,179]. Furthermore, this model immediately suggests interesting possibilities for regulating the cell response to extracellular stimuli [126]. For example, it allows for an alternative onset of either apoptosis (by a high level of receptor oligomerization, with concomitant generation of mature caspase-8) or a proliferative pathway(s), if only a small proportion of Fas molecules is engaged, and procaspase-8 activation is not followed by the massive release of the mature (p20)2(p10)2 heterotetramer. Finally, we notice that the presence of up to five different active caspase-8 isoforms, which differ only in the length of the DED1–DED2 linker, along with 14 c-FLIP variants, would suggest an impressive richness of possible DISC arrangements.
Engagement of the catalytically inactive c-FLIPL adds a level of complexity to both the initiation and course of the death receptor pathways. The c-FLIPL·procaspase-8 complex preferentially processes the ‘local substrate’ Rip (receptor-interacting protein), which may then stimulate cell survival pathways via activation of NF-κB (nuclear factor-κB) [127]. This proposal is in line with the early observation that overexpressed, intrinsically dimeric forms of procaspase-8 in which the Asp-210/Asp-216 maturation sites were mutated to alanine did not induce cell death [155]. However, it is at odds with the finding that caspase-8-mediated cleavage of Rip promotes apoptosis by enhancing the recruitment of FADD to TRADD (TNF receptor-associated death domain protein), with concomitant caspase-8 activation [180]. The substrates of a c-FLIPL-enriched DISC and the pathways that are stimulated preferentially by active, membrane-restricted caspase-8 remain to be elucidated.
Implications for other inflammatory and initiator caspases
We postulate that mechanisms similar to the ones discussed above for the activation/maturation of procaspases-8 and -9 will also capture the general features of these processes in all inflammatory and initiator procaspases – with a few differences, as we will briefly discuss below.
Activation of inflammatory procaspases: the inflammasome
Somewhat ironically, the mechanism of activation of the first member of the caspase family to be identified and characterized crystallographically, caspase-1, has been less intensively studied. That oligomerization is needed for activation, as implied from the crystal structures [6,7] and initial mutagenesis studies [7], was rigorously demonstrated soon afterwards [181]. Furthermore, and as observed in other long-prodomain caspases, overexpression of procaspase-1 or refolding from denaturing conditions results in spontaneous autoactivation, which requires the presence of the N-terminal CARD [182,183].
The seminal work of Yamin and colleagues [150] showed that procaspase-1 has extremely low but detectable intrinsic activity (also detected by others; [184]), while forms cleaved in the β4–β5 and the CARD–(large subunit) linker possess moderate and full activity respectively. Thus the same principles that govern activation of initiator caspases-8 and -9 seem to apply to the inflammatory caspases. Caspase-1 possesses an additional residue in strand β6 (Arg-391) (also conserved in caspases-4 and -5; Figure 2), which ‘bulges out’ and reduces the size of the central cavity. The guanidinium group of Arg-391 is part of an extended hydrogen-bond/salt-bridge network across the central cavity, which includes two further arginine residues, Arg-240 and Arg-286, the carboxylates of Glu-324 and Asp-336, along with the carboxyamide of Asn-259 (Supplemental Figure 2; http://www.BiochemJ.org/bj/384/bj3840201add.htm). Thus polar/charged side chains occlude the central cavity in (pro)caspase-1 (and presumably also in caspases-4 and -5), in stark contrast to the aromatic/hydrophobic nature of topologically equivalent residues in (pro)caspase-9. Conceivably, this network can be formed upon dimerization of procaspase-1 molecules, thus displacing the intersubunit linker from the central cavity, but this event would be more dependent on salt concentration and pH than the hydrophobic ‘priming pockets’ of caspases-8 and -9.
The presence of an N-terminal CARD implies that adaptor proteins structurally related to APAF-1 recruit procaspase-1 and promote its activation by the induced proximity model. The Ser/Thr kinase Rip2/RICK/CARDIAK [Rip-like interacting CLARP (caspase-like apoptosis-regulatory protein) kinase/CARD-containing ICE-associated kinase], containing a C-terminal CARD [185,186], was the first adaptor reported to bind specifically to the CARD of procaspase-1 [187]. The direct involvement of Rip2 for caspase-1 activation, however, is questionable. As shown recently, the kinase plays a critical role in both innate and adaptive immunoresponses, including T-cell differentiation, via a pathway that involves activation of NF-κB downstream of several TLRs (Toll-like receptors) and NODs (nucleotide-binding and oligomerization domain-containing proteins) [188,189]. A second putative adaptor, the APAF-1-related protein CARD2/Ipaf/CLAN (ICE-protease-activating factor/CARD, LRR and NACHT-containing protein), associates specifically with procaspase-1 via its N-terminal CARD, and induces autocatalytic processing of the inflammatory caspase [190]. Ipaf contains in addition NBD (nucleotide-binding domain) and LRR motifs and oligomerizes in a CARD-dependent manner, suggesting the formation of apoptosome-like structures, and appears to be critical for transmitting caspase-1 activation induced by macrophage infection by Salmonella typhimurium, but not lipopolysaccharide [191].
The actual activation/maturation process in vivo may be more complicated, however. In a recent investigation, procaspases-1 and -5 were co-activated preferentially within a large macromolecular complex organized around two adaptor proteins, ASC/PYCARD (apoptosis-associated speck-like protein containing a CARD/PYD- and CARD-containing molecule) and NALP1, and termed the inflammasome [18]. ASC is required for efficient caspase-1 activation following either S. typhimurium infection or lipopolysaccharide, indicating that it and Ipaf have overlapping, but non-identical, roles in the host's inflammatory response to pathogens [191]. In a mechanism that is conceptually similar to the extrinsic pathway, delivery of lipopolysaccharide promotes adaptor heterodimerization through association of their N-terminal Pyrin domains. This allows their C-terminal CARDs to interact specifically with the homologous domains of procaspases-1 and -5 respectively. Activation of these caspase zymogens then occurs via the induced proximity mechanism, and the activated procaspases are able to process themselves at the intersubunit linker. This occurs presumably via interdimer cleavage, as overexpression of procaspase-1 results in its autoactivation, in a strict CARD-dependent manner [182,183]. In vivo, the Pyrin domain of ASC seems to be responsible for the oligomerization of procaspase-1 molecules [192]. In addition, maturation at each CARD large subunit site depends upon the presence of the complementary procaspase [18]. In support of this model, the mouse orthologue of human caspase-4/5, caspase-11, is indispensable for the generation of active caspase-1 in mice, through physical interactions with procaspase-1 [17]. Additionally, the Cys-285→Ser variant of procaspase-1 is converted into the ‘mature’ form when co-expressed with procaspase-4 in COS cells [193]. Whether the adaptor molecules discussed above represent complementary or concurrent pathways in vivo depending on cell type or apoptotic/inflammatory stimulus remains to be clarified.
Interestingly, active (cleaved) caspases-1 and -5 are detected in the extracellular medium, but the externalization mechanism remains uncharacterized. It is also currently unknown how the inflammatory signal is transmitted to trigger assembly of the inflammasome [18]. These authors suggested that the LRR domain of NALP1 may recognize pathogen-associated molecular patterns and/or endogenous ‘alarm signals’, perhaps analogous to cytochrome c binding by WD40 propellers in the apoptosome [165]. Circumstantial support for this hypothesis comes from the existence of a large family of NALP1-related intracellular proteins (about 25 members in humans; recently reviewed in [194]), which may account for the differential recognition of specific pathogen-specific markers and/or second messengers of their presence. Thus it might be more than just a structural coincidence that the same motif used by TLRs and plant disease-resistance proteins for the recognition of pathogen-associated molecular patterns in the extracellular environment is also present in NALP1 and related adaptors. Recent work suggests indeed that LRRs of another family member, NOD2/CARD15, recognize bacterial muramyl dipeptide [195].
These pathways are perhaps best appreciated in the light of the interplay between bacterial factors and host cell death/inflammatory responses. For instance, bifurcating mechanisms downstream of TLR2 engagement by bacterial lipoproteins either stimulate caspase-8 activation and apoptosis, a pro-inflammatory response through caspase-1 activation and processing of interleukin-1β, or promote NF-κB activation and cell survival (see e.g. [196,197]). Similarly, lipopolysaccharide promotes FADD-dependent cell death in endothelial and monocytic cells upon binding to TLR4 [196,198], but also triggers a caspase activity in dendritic cells that is critical for their survival and maturation [199]. Finally, several bacterial pathogens modulate the apoptotic differentiation programme in human neutrophils, caspase-1 being one of the genes that is up-regulated following phagocytosis, while caspase-8 is repressed [200]. Understanding the interplay of inflammatory and apoptotic responses elicited to resolve pathogen infections, and the mechanisms used by the pathogens to evade and/or subvert these pathways, is likely to contribute important insights into the infection process and its clearance, with obvious potential clinical applications. These interesting topics are, however, outside the realm of this review.
Activation of procaspase-2: viewing the primordial caspase?
Although the absence of severe abnormalities in caspase-2(−/−) mice pointed to a less important role in cell death [201], this CARD-containing caspase has been suggested to act upstream of the mitochondria within the intrinsic apoptotic pathway [99,202,203]. Caspase-2 appears to be necessary in particular for the onset of apoptosis upon several insults, including UV irradiation and treatment with the DNA-damaging agents etoposide and cisplatin [202,203]. The identity of the direct mitochondrial target of caspase-2 is disputed, as release of cytochrome c from the intermembrane space, among other pro-apoptotic factors, does not seem to require Bid cleavage [99]. The recent identification of histone H1.2 as a cytochrome c-releasing factor following DNA double-strand breaks [204] suggests that not only caspase-2, but perhaps also some nuclear substrate(s), might be involved in membrane permeabilization.
The adaptors involved in procaspase-2 activation are also the subject of some controversy. Zymogen recruitment to the CARD-containing protein, CRADD/RAIDD (caspase-2 and RipK1 domain-containing adaptor with death domain/Rip-associated protein with a death domain), couples to the TNF receptor 1 signalling complex via two further adaptor proteins, Rip and TRADD [205–207]. Complex formation with CRADD/RAIDD is probably mediated by similar electrostatic interactions of the N-terminal CARDs of procaspase-2 and CRADD/RAIDD [208], as demonstrated for the procaspase-9·APAF-1 complex [159]. However, and since caspase-2 appears to be irrelevant for receptor-mediated apoptosis [201,202], an independent pathway that directly couples genotoxic stress to procaspase-2 activation may be expected. Such a pathway may require the assembly of a large adaptor platform [209], and recent evidence suggests the adaptor PIDD (p53-induced protein with a death domain) can collaborate with RAIDD in activating caspase-2 in such a complex [210]. Since PIDD was originally identified as a p53-regulated gene [211], it is likely that the complex assembled by PIDD and caspase-2 would be an initiator of apoptosis following DNA damage. In contrast, suppression of caspase-2 using RNA interference was found to inhibit the extrinsic pathway triggered by TRAIL (TNF-related apoptosis-inducing ligand), via a pathway that suggested a role for caspase-2 in the cleavage of Bid [212]. Additionally, a role for processed, but catalytically inactive, caspase-2 in the release of cytochrome c from mitochondria spices this mix of reported functionalities for this fascinating caspase [213].
Both the endogenous zymogen and overexpressed forms of caspase-2 are found constitutively in the cell nucleus (see e.g. [214–216]). Nuclear localization signals that flank the CARD are responsible for this unique feature of procaspase-2, in particular a conserved Pro-132–Asp-142 motif that mediates interactions with the importin α/β heterodimer [217,218].
In contrast with all other caspases, the crystal structure of caspase-2 revealed an intrinsic, covalent dimer, in which the opposing central residues of strands β6, Cys-390 and Cys-390′, form an interdomain disulphide bond {[75] and Supplemental Figure 2 (http://www.BiochemJ.org/bj/384/bj3840201add.htm)}. This covalent linkage appears to be rather stable, as reducing agents were used throughout protein purification and crystallization, and furthermore is not disrupted after separation of the large and small subunits in reverse-phase chromatography. However, cytosolic procaspase-2 is found as a monomer in the cytosol of transfected cells [209]. It is tempting to speculate that caspase-2 represents the evolutionary link between an original, intrinsically dimeric caspase that required oligomerization for activation and current members of the family. Along these lines, CARDs might be more closely related to the primordial DD from which all other subfamilies evolved [162], also indicated by their presence in C. elegans CED3 and CED4.
NATURAL INHIBITORS OF CASPASES
It is now widely appreciated that endogenous inhibitors strictly regulate the activity of mature caspases in vivo. Not surprisingly in view of their critical role in the immune response that eventually clears virally infected cells, several viral inhibitors also target their host caspases. For instance, several γ-herpesviruses and the tumorigenic molluscipoxvirus block the extrinsic pathway at the level of zymogen recruitment to the DISC with the help of decoy molecules (v-FLIPs). These inhibitors are essentially composed of one or two copies of domains similar to DDs of endogenous proteins; their preferential interaction with the adaptor protein FADD or with procaspase-8 interrupts assembly of a functional DISC [219–221]. Similar strategies are employed by the endogenous inhibitor c-FLIPS, which competes with caspase-8 recruitment to the DISC [172], as well as the CARD-containing inhibitor of caspase-1, ICEBERG [222]. We start this section with a brief discussion of two other viral, active-site-directed inhibitors: the serpin CrmA (cytokine response modifier A), from cowpox virus, and the baculovirus inhibitor p35. Finally, we discuss recent advances in our understanding of the mechanisms of caspase inhibition by their major cellular inhibitors, the IAPs.
Suicide inhibitors
Most natural inhibitors of proteolytic enzymes directly block substrate access to the active site of the cognate protease, usually employing substrate-like sequences as specific ‘baits’. As revealed in several crystal structures, these stable enzyme–inhibitor complexes are formed without gross structural distortions of either moiety (reviewed in [223]). By contrast, for some biologically important natural inhibitors (collectively termed suicide inhibitors or mechanism-based inactivators), cleavage of the bait is followed by major structural rearrangements of the inhibitor and/or the cognate protease. The two viral inhibitors discussed below, although otherwise completely unrelated in both sequence and structure, are mechanistically related members of this class. We briefly discuss the main features of caspase inhibition by CrmA and p35; these are reviewed in more detail elsewhere [224].
A viral serpin, CrmA
Serpins form a widespread superfamily of large inhibitors (350–500 amino acid residues) which employ a unique suicide mechanism for inactivation of their target proteases (for a recent review, see [225]). The inhibitor initially forms a non-covalent Michaelis complex with the protease through interactions with an exposed loop [226]. In the well understood case of serine protease inhibition, attack of the active-site Ser-195 on the P1–P1′ scissile bond and concomitant formation of a covalent ester linkage between the Ser-195 Oγ atom and the backbone carbonyl of the P1 residue proceed as in ‘normal’ catalysis. The subsequent cleavage of the peptide bond, however, is followed by rapid insertion of the N-terminal part of the loop into the main body of the serpin, with synchronous displacement of the covalently bound protease to the opposite pole of the inhibitor. The crystal structure of the antitrypsin–trypsin complex showed that serine protease inhibition results from clashes between the loops that surround the active site of the enzyme and the main body of the serpin. These loops are thereby disordered, and catalytic residues His-57 and Ser-195 are separated by approx. 6 Å, with the serpin acting almost as a protease ‘denaturant’ [227].
Although most known serpins are specific for serine proteases, cross-class inhibitors are also known. A serpin from cowpox virus, CrmA [228], inhibits not only the cytotoxic serine protease granzyme B [229], but also caspases-1 and -8 (with Ki values of 0.01 and 0.95 nM respectively; [230]). The serpin also inhibits caspase-9 in vitro, yet fails to block caspase-9-mediated cell death [231]. Two independently solved crystal structures of cleaved CrmA [232,233] showed that the viral serpin possesses the common structural features found in all members of the superfamily studied to date. CrmA, however, constitutes a ‘minimal’ serpin, because it lacks completely an α-helix, helix D, which in mammalian members of the superfamily contributes to allosteric activity regulation [225]. In addition, other helices are also shortened compared with mammalian serpins. The capability of other serpins to target caspases extends, at least in vitro, to some endogenous serpins as well. For instance, the physiological inhibitor of granzyme B, PI-9 (protease inhibitor-9), could be transformed into a potent caspase inhibitor that is able to block Fas-mediated apoptosis by simply mutating its P1 residue from glutamate to aspartate [234].
Experimental proof for a similar mechanism of serpin-mediated ‘inhibition by compression’ of caspases is lacking as yet. Gettins [225] discussed the possibility in detail, and concluded that cysteine proteases fulfil the major prerequisites for inhibition according to the standard suicide mechanism. For example, the active site is accessible, the thioester linkage is stable, and the protease may be pulled out to the ‘bottom’ of the serpin upon cleavage of the P1–P1′ scissile bond. Nevertheless, further studies are certainly needed to establish the general validity of the mechanism. An attractive hypothesis is that translocation of the covalently bound caspase forces expulsion of its substrate binding loops, as observed in caspase zymogens (see above).
A bowstring mechanism of inhibition: p35 and p49
The baculovirus protein p35 is the only known natural inhibitor that targets most caspases, but does not inhibit granzyme B [235–238]. Crystal structures of p35, both free [239] and in complex with human caspase-8 [240], together with a recent investigation by site-directed mutagenesis, have revealed a previously unobserved mechanism of inhibition referred to as the ‘bowstring model’ [241]. Engagement of a caspase-recognition sequence (Asp-Gln-Met-Asp-87↓Gly) within a protruding loop is followed by the formation of a thioester linkage between the catalytic Cys-285 and the carbonyl of the P1 residue [240]. Cleavage of the scissile peptide bond, however, leads to the repositioning of the p35 N-terminus close to the active site of the bound caspase. This prevents access of the hydrolytic water molecule to the catalytic residues, consequently avoiding deacylation of the thioester bond. This mechanism of inhibition presents some analogy with that of serpins, in that (1) an intermediate of peptide hydrolysis remains kinetically trapped in a complex with a disabled enzyme, and (2) there is some restructuring of the inhibitor after cleavage of the ‘bait’ loop. However, the structure of p35-bound caspase-8 is essentially unaffected, compared with the extensive unfolding of serpin-inhibited trypsin-like proteases [225,227].
A baculoviral homologue of p35, known as p49, inhibits a p35-insensitive initiator caspase in insects, Sf-caspase-X, as well as human caspase-9 in vitro [242,243]. Interestingly, p49 represents the first example of a caspase inhibitor that appears to interact with regions distant from the active site (exosites) to modulate caspase specificity. Indeed, swapping of the caspase targeting sequences (residues at positions P4–P1) did not change the specificity of chimaeric p35/p49 variants [243]. These authors concluded that other structural determinants of p49 are responsible for its restricted specificity for initiator caspases, in particular an inserted domain located C-terminally to the P1 site (residues 226–346). Interestingly, in contrast with the widespread distribution of serpins, no homologues of p35/p49 have been identified to date in animals.
XIAP: a non-standard design for a very specific caspase inhibitor
Studies on the regulation of insect cell death induced by baculovirus that lacked a functional p35 gene led to the discovery of another potent inhibitor of apoptosis, IAP. The protein is characterized by a small (∼70–80 residues), highly conserved Zn2+-binding module termed the BIR (baculoviral IAP repeat) [244]. Endogenous homologues of IAP were identified shortly afterwards in animals [245–248], with eight members characterized to date in humans (reviewed in [249–251]). The current evidence indicates that endogenous IAPs are one of the most important control points, if not the control point, for apoptosis execution, as they not only effectively counteract low amounts of adventitiously activated caspases, but, perhaps most importantly, so establish a threshold above which extensive caspase generation can rapidly occur.
Both caspase-dependent and -independent anti-apoptotic activities of IAPs have been reported. A review of caspase-independent anti-apoptotic mechanisms can be found in [251]; however, the pathways are poorly understood as yet. Consequently, we shall concentrate in this review on the paradigm of human IAPs, XIAP, for which available crystal structures showing its constituent domains bound to different caspases and IAP regulators allow an understanding of its direct effect on apoptosis (see Figure 10a for a schematic representation of XIAP domain organization). These structural investigations, in turn, followed biochemical studies that mapped the caspase-binding sites in XIAP. Thus a fragment comprising the BIR2 domain and the linker to the N-terminal BIR1 module targets the effector caspases-3 and -7 [252,253]. On the other hand, the BIR3–RING tandem specifically inhibits caspase-9 [253], an activity that depends almost exclusively on the BIR3 domain [254]. The completely unrelated mechanisms of caspase inhibition employed by the two XIAP subunits will be discussed below.
Figure 10. Mechanisms of caspase inhibition by XIAP.
(a) Schematic diagram of the domain organization of human XIAP. The sequence critical for the inhibition of effector caspases by XIAP is given [254,256]; the arrow above the sequence indicates that it binds in a ‘reverse mode’ to effector caspases (see text and Figure 11b). A caspase cleavage site in XIAP [253,263] is indicated; other cleavages are also detected [166]. (b) Cartoon depicting the mode of binding of BIR2 to caspase-3 and of BIR3 to caspase-9 (caspase large subunits in blue, and small subunits in red) bound to the respective BIR (orange). The shallow groove on each BIR domain symbolizes the Smac pocket into which the four N-terminal residues constituting an IAP-binding motif fit. Although important for caspase-9 inhibition [122], the significance of this groove in BIR2 is less well understood in terms of caspase-3 or -7 inhibition [255]. (c) Ribbon diagram of the inhibitory interactions, with the same colour coding. The N- and C-termini of the subunits of the complex are labelled, and important side chains in the interactions are shown, but for simplicity are not labelled.
XIAP inhibition of effector caspases
Crystal structures of caspases-3 and -7 bound to the minimum caspase-inhibiting region of XIAP have been reported [133,134,255] (see Figures 10 and 11 for a representation of the caspase-3·XIAP complex). The results of these structural investigations are rather unexpected on three counts. First, the region of the inhibitor that makes the most important contacts with the enzyme corresponds to the linker peptide and not to the globular BIR domain, in agreement with a previous mutagenesis study [256]. Secondly, this peptide docks in a reverse orientation into the caspase active-site cleft in relation to observed binding mode of productively bound substrates and inhibitors ([223]; compare Figures 5a, 5b and 11). Thirdly, it is particularly remarkable that no carbonyl carbon of XIAP approaches the catalytic Cys-285 Sγ atom closer than 5.8 Å, and the oxyanion pocket of the enzyme remains unoccupied.
Figure 11. Details of caspase inhibition by XIAP.
(a) The catalytic cleft of caspase-3 in three different forms, i.e. inhibitor [CMK (chloromethyl ketone)]-bound (blue; PDB code 1CP3; [10]), active with free catalytic site (red; PDB 1QX3; [67]) or bound to BIR2 (grey; PDB 1I30; [255]), is highly conserved. There are no substantial alterations in main-chain or side-chain conformations with the docking of BIR2 to the cleft, and it is only when the tetrapeptide CMK inhibitor binds that Tyr-338 must rotate to accept the P2 side chain (green arrow). Consequently, the linker region of BIR2 appears perfectly adapted to the unliganded active form of caspase-7. (b) Superposition of the XIAP inhibitory region (pink sticks) and acetyl-DVAD-CMK (blue sticks) on the active-site cleft of caspase-3 from PDB file 1I30 [255]. The orientation is as for Figure 5 and shows the small 4 Å footprint made by acetyl-DVAD-CMK side-chains (cyan) compared with that made by XIAP side chains (magenta). Note the manifold interactions of the two residues in the BIR1–BIR2 linker that are critical for activity, i.e. Leu-141 and Asp-148 [256]. The former engages in van der Waals contacts with Tyr-338 and Phe-381h, while Asp-148 occupies the S4 pocket of the caspase, hydrogen-bonding in particular residues of the 341 substrate-binding loop (Arg-341, Ser-343 and Trp-348). In addition, the carboxylate of Asp-148 is clamped to the C-terminal residue of XIAP (BIR2), Arg-233 (not shown). (c) Crystal structure of BIR3 (green)-bound caspase-9 (blue) taken from PDB 1NW9 [122]. The BIR domain takes the position of the dimeric partner domain of caspase-9, essentially monomerizing the caspase, thus reversing the process of zymogen activation. The blow-up depicts a conformational relay upon BIR2 binding (green residues) that forces Phe-390 from the active conformation (red) to the inactive one (blue). A rotation of Phe-390 requires a compensatory rotation in Tyr-331 and a concomitant expulsion of the catalytic Cys-285 into a non-catalytic location. (d, e) The 4 Å footprint of caspase-9 residues (shaded magenta) in contact with either its dimeric partner caspase-9 (d) or BIR3 (e) at the dimer interface. Note that although the contacts are more extensive in the caspase-9 dimer, most of the crucial contacts are the same. An exception is the segment Ala-298–Phe-301, which forms the IBM interacting with the Smac pocket on BIR3. Rendered with PyMol.
XIAP exploits the peculiarities of the caspase-3/7 structure by engaging in important van der Waals interactions with residues of the inserted 381-loop (the ‘flap’). Most notably, the aromatic side chains of Phe-381b and Phe-381d contact Ile-149 and Ile-153 of the inhibitor, whereas Phe-381h interacts with the side chain of the critical residue Leu-141, as well as with Val-146. These manifold interactions suggest that the 381-loop of these caspases may have evolved to ensure inhibition by IAPs, rather than as a means to increase their substrate selectivity. In addition, they explain why XIAP targets active effector caspases, but does not bind their inactive zymogens [256,257], which possess disordered substrate-binding loops (see above). The carboxylate of the second XIAP residue that is critical for inhibition, Asp-148 [256,258], occupies the S4 pocket of the enzymes, although the reverse conformation of the linker peptide precludes an exact mimicking of substrate-like inhibitors (compare Figures 5a, 5b and 11).
It is also noteworthy that the globular BIR domain is only defined by electron density in the caspase-3·XIAP (BIR2) structure [255], while it is disordered in the independently determined complexes with caspase-7 ([133,134]; S. J. Riedl, W. Bode, G. S. Salvesen and P. Fuentes-Prior, unpublished work). The solvent-protected areas between caspase-3 and the BIR domain (≈550 Å2) seem too small to support formation of a stable interaction. Perhaps more importantly, the BIR2/caspase-3 interface lacks conserved ‘hot spot’ residues (in particular Trp, but also the aromatic side chains of Phe/Tyr, as well as Met and Arg) which define protein–protein interactions [168].
Although these observations suggest a minor role of the BIR2 domain in caspase inhibition, studies with truncated variants cannot easily reproduce the actual situation in vivo, as pointed out by others [258]. Moreover, the crystal structure of the caspase-3·XIAP (BIR2) complex showed that the N-terminal residues of the caspase small subunit occupy a surface pocket of a symmetry-related inhibitor molecule [255], engaging in interactions that closely resemble those seen in the caspase-9·XIAP (BIR3) complex (see below). Together, these findings suggest that the interaction between the BIR1–BIR2 linker and the active-site cleft of caspases-3 and -7 do not tell the whole story, and that additional strengthening interactions not seen directly in the crystal structures may occur in solution.
XIAP inhibition of caspase-9
The crystal structure of caspase-9·XIAP (BIR3) complex nicely explained the inhibitory mechanism by showing that the globular BIR domain binds to a caspase-9 monomer, in a manner that blocks the surface patch corresponding to the intermonomer area in the active caspase dimer [122] (see Figure 11). The most important contacts in this interface include residues of the unique 240-loop, together with the pair of phenyl rings that occupy the central cavity in the caspase-9 dimer [see also Figure 8 and Supplemental Figure 2 (http://www.BiochemJ.org/bj/384/bj3840201add.htm)]. Moreover, the C-terminal α5 helices of the two moieties align in an antiparallel manner, allowing the side chains of Leu-370 and Leu-371 (caspase-9) and Leu-344 (XIAP) to interdigitate, while the neighbouring inhibitor side chains of Asn-340 and His-343 engage in hydrogen bonds with main-chain atoms of the caspase. Because of these multiple interactions, XIAP (BIR3) can effectively counteract procaspase-9 dimerization and concomitant activation (see above).
Perhaps the most striking interaction is seen between the N-terminal tetrapeptide Ala-298(316)-Thr-Pro-Phe in the small subunit of caspase-9 and a groove on the surface of BIR3, as predicted [158] [Figure 12 and Supplemental Movie 4 (http://www.BiochemJ.org/bj/384/bj3840201add.htm)]. This groove of the XIAP moiety, frequently called the Smac (second mitochondrial activator of caspases) pocket, is structurally equivalent to the area on BIR2 that engages the N-terminal tetrapeptide in the small subunit of caspase-3 [255], hinting at a conservation of mechanism. However, the caspase-9–BIR3 interaction is much more direct, as it occurs within the molecule that will be inhibited. This interaction is vital for the inhibitory function of XIAP [158], and a site utilized by IAP antagonists to derepress inhibition (see below).
Figure 12. IAP domains bound to Smac/DIABLO and caspase neoepitopes.
The GRASP electrostatic surface potential of XIAP BIR3 (contoured between −15 and +15 kBT/e) with stick representation of the neoepitope generated following cleavage of caspase-9 at Asp-297 (blue; PDB 1NW9; [122]) superimposed on the N-terminal tetrapeptide of mature Smac/DIABLO (orange; PDB 1G73; [278]). Residues are identified by the corresponding colour. Notice the highly negative end of the Smac pocket, which helps to counter the energetic penalty of burying the positive charge on the N-terminal amine of IBM peptides.
Mutation of most of the XIAP side chains that contact caspase-9 (Trp-310→Ala, Glu-314→Ser, His-343→Ala [254]; Trp-323→Ala, Pro-325→Gly, Gly-326→Glu and Leu-344→Ala [122]) confirm their critical role for caspase-9 inhibition. In addition, investigations with full-length XIAP molecules that incorporate some of these BIR3 mutations together with Asp-148→Ala corroborated the results of the structural investigations in a physiological frame [258,259].
Interestingly, the substrate-binding loops of XIAP (BIR3)-bound caspase-9 are solvent-exposed in a conformation almost identical to that in the inactive caspase-9 zymogen-like monomer [73]. In particular, the conformation of the 341-loop reproduces that in inactive caspase-9 monomers [73], and is stabilized by similar contacts with a neighbouring caspase molecule (Figure 11), while the 381-loop is also exposed and mobile, as judged from the high-temperature values. These findings indirectly confirm the monomeric nature of zymogen caspase-9 in vivo (see above), and explain how the relatively large hydrophobic patches that form the intermonomer region are protected from bulk solvent, thus avoiding unwanted procaspase-9 dimerization and activation. The mode of binding of XIAP (BIR3) to active, dimeric caspase-9 [158] remains unresolved. Either it is different or, more likely, BIR3 binds first to the free N-terminus of the small subunit, perhaps aiding monomerization. In essence, BIR3 reverses the zymogen→active-enzyme transition of caspase-9.
On the basis of conservation of mechanism, one might expect that the Drosophila equivalent of caspase-9, DRONC (Drosophila Nedd2-like caspase), should be inhibited by its cognate IAP, DIAP1, in a similar manner to caspase-9 and XIAP [260]. However, DRONC does not utilize its cleaved intersubunit linker to bind DIAP1, but rather a segment N-terminal to the caspase catalytic domain that nevertheless binds to the equivalent Smac pocket of DIAP1, although in the reverse orientation [261]. This unexpected finding suggests that the Smac pockets of BIR domains can do more than simply bind specific N-termini of proteins.
Together we stand…: XIAP retains activated caspase-3 bound to the apoptosome
It is apparent that a single XIAP molecule can simultaneously bind and inhibit caspase-3 and -9 moieties [259], and that XIAP remains bound to activated caspases-3 and -9 within the apoptosome [158,166]. The enhanced affinity of caspase-3-bound XIAP for caspase-9 [259] suggests a tight packaging of activated caspase-3, caspase-9 and XIAP within the apoptosome. Considering that the larger caspase-9–BIR3 interface is not accessible to XIAP in dimeric (APAF-1-bound) forms of the initiator caspase (compare Figures 9a and 10c), sequestering activated caspase-3 through interactions with the BIR1–BIR2 linker would contribute to stabilize the apoptosome in vivo. In this context, the Smac pocket of a BIR2 domain would be free to accept the N-terminal tetrapeptide from a neighbouring caspase-3 small subunit, as observed in the crystal structure of the caspase-3· XIAP (BIR2) complex (see above), and also supported by modelling exercises. These interactions might explain the preserved inhibitory activity of XIAP against caspase-9 cleaved at Asp-316 [262]. We also note that XIAP interacts with oligomerized APAF-1 [166], but the domain(s) responsible for this interaction are unknown to date. A predicted α-helical domain located between the BIR3 and the RING domain (Figure 10a) might be involved in adaptor binding.
In a mechanistic sense, this situation resembles regulation of the blood coagulation cascade via a highly specific multidomain inhibitor, TFPI (tissue factor pathway inhibitor) [154]. Similar to interactions of XIAP with both initiator and effector caspases, one TFPI domain blocks the upstream Factor VIIa (equivalent to caspase-9), while a second domain targets the activated Factor Xa (which corresponds to caspase-3/7). Importantly, in both cascades, the generation of small amounts of the activated downstream protease is required for formation of the final complex.
The large surface areas protected by XIAP at the interfaces with caspase-3/-7 and caspase-9 would suggest that their synergistic binding effectively blocks apoptosome-mediated apoptosis. Indeed, current evidence suggests that XIAP has to be removed from caspase complexes, including its proteolytic degradation, for apoptosis to progress ([262,263] and see below).
IAPs clearly have functions in addition to caspase inhibition, because they have been found in organisms such as yeast, which neither contain caspases nor undergo apoptosis [264]. Moreover, although most of the mechanistic studies on IAPs relate to their binding and direct inhibition of caspase catalytic activity, it is likely that some of the IAPs function to down-regulate caspases not by inhibiting them, but by acting as E3 ligases for their rapid removal via the proteasomal route [265] (see also below). Irrespective of the actual mechanism for terminating caspase activity, the IAPs set the scene for the final phase of caspase control – derepression of IAP-inhibited complexes as a method of caspase activation.
…divided we fall: how Smac/DIABLO and Omi/HtrA2 relieve IAP inhibition
IAP-interacting proteins were first identified in Drosophila by genetic screens [266]. The four known activators of apoptosis in flies that employ IAP binding [Rpr, Grim, Hid (heat involution defective) and Skl] encode partially redundant functions and share an N-terminal amino acid motif referred to as the IBM (IAP-binding motif) [267]. More details of IAP-binding proteins can be found in recent reviews [42,251,268,269] in keeping with the scope of this review, we focus here mainly on structural considerations. Proteins liberated from the mitochondrial intermembrane space of mammals after signalling through pro-apoptotic Bcl-2 proteins play roles in derepressing caspase inhibition, at least in vitro. These are the homodimeric Smac proteins [also known in mice as DIABLO (direct IAP-binding protein with low pI)] [270–272], the mammalian multimeric serine protease Omi/HtrA2 [273–276], and the thioredoxin peroxidase Jafrac2 from Drosophila [267]. Common to these otherwise unrelated proteins is the presence of similar N-terminal tetrapeptide sequences that are able to antagonize the inhibitory action of XIAP [277–279]. These tetrapeptide sequences are similar to the neoepitopes generated in the small subunit of caspase-9 following its proteolysis (Figures 2 and 12), and in part explain the derepression of inhibition.
Support for the derepression model comes from the crystal structure of the Smac·XIAP (BIR3) complex [278] and the solution structure of the BIR3 domain bound to the Smac N-terminus [277]. These structural investigations revealed that the N-terminal knob occupies the Smac pocket in an extended conformation that is almost identical to that assumed by the N-terminal peptide of the caspase-9 small subunit [Figure 12 and Supplemental Movie 4 (http://www.BiochemJ.org/bj/384/bj3840201add.htm)]. Ala-1 of Smac establishes an intricate network of hydrogen bonds with several side chains of the inhibitor, in particular Glu-314 and Gln-319. More importantly, a second interface between two elongated Smac helices and XIAP (BIR3) buries an additional ≈2000 Å2 surface area [278]. Notably, an alternatively spliced form of Smac that lacks the N-terminal BIR3-binding knob, the cytosolic Smac, as well as a recombinant form lacking the first 75 residues still promote apoptosis in vivo to the same extent as mitochondrial Smac [280]. The discovery of the second contact interface would help to explain these findings.
Importantly, Smac displaces the BIR2 domain from its complex with caspase-7 in vitro [133]. These authors suggested that Smac binding to the BIR2 domain may affect the conformation of the linker peptide such that it can no longer bind and inhibit caspase-7. An alternative explanation is that the N-terminus of the caspase-7 small subunit may form a stabilizing interaction with the BIR2 Smac groove, much as seen in the caspase-3·BIR2 structure, which can be destabilized by Smac (see Supplemental Movie 4; http://www.BiochemJ.org/bj/384/bj3840201add.htm). However, it is not known whether Smac displaces full-length XIAP from similar complexes in vivo. Most biochemical and studies of Smac binding have been performed on individual BIR domains of XIAP, demonstrating relatively weak Kds in the range of 500 nM. However, a recent investigation demonstrated that Smac bound with subnanomolar affinity to the connected BIR2 and BIR3 domains [281]. This observation demonstrates the importance of two-site interactions in the competition of caspases with Smac, and underscores the likelihood that only oligomeric proteins containing IBMs derepress caspase activity efficiently in vivo.
The hexameric mitochondrial chaperone/degradative protease HtrA2 contains IBMs at its N-termini, and is likely to recapitulate the same derepression properties as Smac. In addition to its physical competition of XIAP binding to caspases, HtrA2 was recently shown to degrade most IAPs in vitro, an activity that also depends on its N-terminal peptide [282,283]. Although these authors fail to detect degradation products in vivo (which are presumably targeted to further degradation by the proteasome), Yang and co-workers [282] demonstrated that Omi/HtrA2 inactivation through RNA interference abolished c-IAP1 cleavage in cells and desensitized cells to apoptosis induced by TRAIL. The protease cleaves, for instance, c-IAP1 within the BIR1–BIR2 linker, in line with the previous finding of a ≈53 kDa degradation product [284]. The degradation of IAPs, either by HtrA2 and/or by activated caspases observed in Fas-induced apoptosis [253,263], may contribute to overcoming the hurdle posed by these inhibitors. In addition, caspase-3 cleavage after Asp-316 would weaken the interaction of the APAF-1-bound caspase-9 with XIAP, thus facilitating the displacement of the inhibitor via Smac and/or HtrA2.
E3 ubiquitin ligase activity of RING domains
While the BIR domains of XIAP appear to be exclusively responsible for its direct caspase inhibitory activity, E3 ubiquitin ligase activity has been demonstrated for the RING domains found in several members of the family. The latter seems to represent an additional level in the regulation of apoptosis. Two alternative, but not mutually exclusive, scenarios have been proposed: (1) auto-ubiquitylation of the IAP leads to its subsequent degradation by the proteasome [285], or (2) the RING domain modifies IAP-bound molecules and signals their degradation. IAP-mediated ubiquitylation of caspases-3 and -7 [139,265], Smac/DIABLO and other IBM-containing proteins [286–289], and TRAF2 (TNF receptor-associated factor 2) [290] has been reported. These ubiquitylation processes may either potentiate or counteract the anti-apoptotic potential of the IAPs. At least in the case of XIAP, however, the major effect of ubiquitylation in vivo appears to be anti-apoptotic [291].
CONCLUSIONS AND PERSPECTIVES
Comprehension of the structural basis for regulating apoptosis is quite advanced, probably more so than for any field that describes a signalling pathway from beginning to end. We now know the fundamental structures of examples from the initiator phase, the pre-mitochondrial regulator phase (not covered in this review), the effector (or execution) phase, inhibitors and their antagonists, and even the structures of some substrates. The field is about as advanced as the best known of proteolytic pathways, the coagulation cascade. New paradigms for the initiation of proteolytic pathways have been revealed, along with unexpected ways of utilizing interactions for proteolytic inhibition. However, from the point of view of understanding mechanism, there remain several important gaps that only structural biology can solve. In this context we see the following challenges for the future: (1) structure determination of the zymogens of initiator caspases; (2) high-resolution images of activator platforms such as the apoptosome, DISC and inflammasome; (3) identification of the conformational requirements that define a natural protein caspase–substrate recognition site; and (4) elucidation of three-dimensional structures for all components of an entire apoptotic signalling pathway, perhaps the primitive CED9/CED4/CED3 paradigm of C. elegans.
Scientists involved in high-throughput crystallography initiatives promise that within 5 years structural determination will be as simple as sequencing DNA – place your order and wait a couple of weeks for the results. Although this may be a bit optimistic for complex assemblies, it bodes well for an exciting next few years as the current hypotheses for how a cell decides its fate are proven, or otherwise.
Online data
Supplementary movies
Supplementary PDB Structures
Acknowledgments
We thank past and present members of our laboratories for insightful discussions. Some of the work described here was supported by NIH grants HL51399, NS37878, AG15402 and CA69381.
References
- 1.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 2.Thornberry N. A., Bull H. G., Calaycay J. R., Chapman K. T., Howard A. D., Kostura M. J., Miller D. K., Molineaux S. M., Weidner J. R., Aunins J. A novel heterodimeric cysteine protease is required for interleukin-1β processing in monocytes. Nature (London) 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 3.Cerretti D. P., Kozlosky C. J., Mosley B., Nelson N., Van Ness K., Greenstreet T. A., March C. J., Kronheim S. R., Druck T., Cannizzaro L. A., et al. Molecular cloning of the interleukin-1β converting enzyme. Science. 1992;256:97–100. doi: 10.1126/science.1373520. [DOI] [PubMed] [Google Scholar]
- 4.Yuan J., Shaham S., Ledoux S., Ellis H. M., Horvitz H. M. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1β-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 5.Alnemri E. S., Livingston D. J., Nicholson D. W., Salvesen G., Thornberry N. A., Wong W. W., Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 6.Walker N. P., Talanian R. V., Brady K. D., Dang L. C., Bump N. J., Ferenz C. R., Franklin S., Ghayur T., Hackett M. C., Hammill L. D. Crystal structure of the cysteine protease interleukin-1β-converting enzyme: A (p20/p10)2 homodimer. Cell. 1994;78:343–352. doi: 10.1016/0092-8674(94)90303-4. [DOI] [PubMed] [Google Scholar]
- 7.Wilson K. P., Black J. A., Thomson J. A., Kim E. E., Griffith J. P., Navia M. A., Murcko M. A., Chambers S. P., Aldape R. A., Raybuck S. A. Structure and mechanism of interleukin-1β converting enzyme. Nature (London) 1994;370:270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 8.Cohen G. M. Caspases: The executioners of apoptosis. Biochem. J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawlings N. D., Barrett A. J. Evolutionary families of peptidases. Biochem. J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mittl P. R., Di Marco S., Krebs J. F., Bai X., Karanewsky D. S., Priestle J. P., Tomaselli K. J., Grütter M. G. Structure of recombinant human CPP32 in complex with the tetrapeptide Acetyl-Asp-Val-Ala-Asp fluoromethyl ketone. J. Biol. Chem. 1997;272:6539–6547. doi: 10.1074/jbc.272.10.6539. [DOI] [PubMed] [Google Scholar]
- 11.Chai J., Wu Q., Shiozaki E., Srinivasula S. M., Alnemri E. S., Shi Y. Crystal structure of a procaspase-7 zymogen: Mechanisms of activation and substrate binding. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 12.Blanchard H., Kodandapani L., Mittl P. R., Marco S. D., Krebs J. F., Wu J. C., Tomaselli K. J., Grütter M. G. The three-dimensional structure of caspase-8: An initiator enzyme in apoptosis. Struct. Fold. Des. 1999;7:1125–1133. doi: 10.1016/s0969-2126(99)80179-8. [DOI] [PubMed] [Google Scholar]
- 13.Watt W., Koeplinger K. A., Mildner A. M., Heinrikson R. L., Tomasselli A. G., Watenpaugh K. D. The atomic-resolution structure of human caspase-8, a key activator of apoptosis. Struct. Fold. Des. 1999;7:1135–1143. doi: 10.1016/s0969-2126(99)80180-4. [DOI] [PubMed] [Google Scholar]
- 14.Fischer H., Koenig U., Eckhart L., Tschachler E. Human caspase 12 has acquired deleterious mutations. Biochem. Biophys. Res. Commun. 2002;293:722–726. doi: 10.1016/S0006-291X(02)00289-9. [DOI] [PubMed] [Google Scholar]
- 15.Kalai M., Lamkanfi M., Denecker G., Boogmans M., Lippens S., Meeus A., Declercq W., Vandenabeele P. Regulation of the expression and processing of caspase-12. J. Cell Biol. 2003;162:457–467. doi: 10.1083/jcb.200303157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saleh M., Vaillancourt J. P., Graham R. K., Huyck M., Srinivasula S. M., Alnemri E. S., Steinberg M. H., Nolan V., Baldwin C. T., Hotchkiss R. S., et al. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature (London) 2004;429:75–79. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- 17.Wang S., Miura M., Jung Y. K., Zhu H., Li E., Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92:501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 18.Martinon F., Burns K., Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 19.Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 20.Reed J. C., Doctor K., Rojas A., Zapata J. M., Stehlik C., Fiorentino L., Damiano J., Roth W., Matsuzawa S., Newman R., et al. Comparative analysis of apoptosis and inflammation genes of mice and humans. Genome Res. 2003;13:1376–1388. doi: 10.1101/gr.1053803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meergans T., Hildebrandt A. K., Horak D., Haenisch C., Wendel A. The short prodomain influences caspase-3 activation in HeLa cells. Biochem. J. 2000;349:135–140. doi: 10.1042/0264-6021:3490135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denault J.-B., Salvesen G. S. Human caspase-7 activity and regulation by its N-terminal peptide. J. Biol. Chem. 2003;278:34042–34050. doi: 10.1074/jbc.M305110200. [DOI] [PubMed] [Google Scholar]
- 23.Kuida K., Lippke J. A., Ku G., Harding M. W., Livingston D. J., Su M. S., Flavell R. A. Altered cytokine export and apoptosis in mice deficient in interleukin-1β converting enzyme. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 24.Varfolomeev E. E., Schuchmann M., Luria V., Chiannilkulchai N., Beckmann J. S., Mett I. L., Rebrikov D., Brodianski V. M., Kemper O. C., Kollet O., et al. Targeted disruption of the mouse caspase 8 gene ablates cell death induction by the TNF receptors, Fas/APO1, and DR3 and is lethal prenatally. Immunity. 1998;9:267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- 25.Kuida K., Haydar T. F., Kuan C. Y., Gu Y., Taya C., Karasuyama H., Su M. S., Rakic P., Flavell R. A. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- 26.Kuida K., Zheng T. S., Na S., Kuan C.-y., Yang D., Karasuyama H., Rakic P., Flavell R. A. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature (London) 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 27.Morita Y., Maravei D. V., Bergeron L., Wang S., Perez G. I., Tsutsumi O., Taketani Y., Asano M., Horai R., Korsmeyer S. J., et al. Caspase-2 deficiency prevents programmed germ cell death resulting from cytokine insufficiency but not meiotic defects caused by loss of ataxia telangiectasia-mutated (Atm) gene function. Cell Death Differ. 2001;8:614–620. doi: 10.1038/sj.cdd.4400845. [DOI] [PubMed] [Google Scholar]
- 28.Zheng T. S., Hunot S., Kuida K., Flavell R. A. Caspase knockouts: Matters of life and death. Cell Death Differ. 1999;6:1043–1053. doi: 10.1038/sj.cdd.4400593. [DOI] [PubMed] [Google Scholar]
- 29.Zheng T. S., Hunot S., Kuida K., Momoi T., Srinivasan A., Nicholson D. W., Lazebnik Y., Flavell R. A. Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nat. Med. 2000;6:1241–1247. doi: 10.1038/81343. [DOI] [PubMed] [Google Scholar]
- 30.Leonard J. R., Klocke B. J., D'sa C., Flavell R. A., Roth K. A. Strain-dependent neurodevelopmental abnormalities in caspase-3-deficient mice. J. Neuropathol. Exp. Neurol. 2002;61:673–677. doi: 10.1093/jnen/61.8.673. [DOI] [PubMed] [Google Scholar]
- 31.Wang L., Miura M., Bergeron L., Zhu H., Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 32.Troy C. M., Rabacchi S. A., Hohl J. B., Angelastro J. M., Greene L. A., Shelanski M. L. Death in the balance: alternative participation of the caspase-2 and -9 pathways in neuronal death induced by nerve growth factor deprivation. J. Neurosci. 2001;21:5007–5016. doi: 10.1523/JNEUROSCI.21-14-05007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Zheng L., Lobito A., Chan F. K., Dale J., Sneller M., Yao X., Puck J. M., Straus S. E., Lenardo M. J. Inherited human caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 34.Gronbaek K., Dalby T., Zeuthen J., Ralfkiaer E., Guidberg P. The V410I (G1228A) variant of the caspase-10 gene is a common polymorphism of the Danish population. Blood. 2000;95:2184–2185. [PubMed] [Google Scholar]
- 35.Chun H. J., Zheng L., Ahmad M., Wang J., Speirs C. K., Siegel R. M., Dale J. K., Puck J., Davis J., Hall C. G., et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature (London) 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- 36.Troy C. M., Salvesen G. S. Caspases on the brain. J. Neurosci. Res. 2002;69:145–150. doi: 10.1002/jnr.10294. [DOI] [PubMed] [Google Scholar]
- 37.Yuan J., Yankner B. A. Apoptosis in the nervous system. Nature (London) 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- 38.Teitz T., Wei T., Valentine M. B., Vanin E. F., Grenet J., Valentine V. A., Behm F. G., Look A. T., Lahti J. M., Kidd V. J. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat. Med. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- 39.Green D. R., Evan G. I. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 40.Frisch S. M., Screaton R. A. Anoikis mechanisms. Curr. Opin. Cell Biol. 2001;13:555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 41.Kaufmann S. H., Vaux D. L. Alterations in the apoptotic machinery and their potential role in anticancer drug resistance. Oncogene. 2003;22:7414–7430. doi: 10.1038/sj.onc.1206945. [DOI] [PubMed] [Google Scholar]
- 42.Salvesen G. S., Abrams J. M. Caspase activation – stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 2004;23:2774–2784. doi: 10.1038/sj.onc.1207522. [DOI] [PubMed] [Google Scholar]
- 43.Fulda S., Wick W., Weller M., Debatin K. M. Smac agonists sensitize for Apo2L/TRAIL- or anticancer drug-induced apoptosis and induce regression of malignant glioma in vivo. Nat. Med. 2002;8:808–815. doi: 10.1038/nm735. [DOI] [PubMed] [Google Scholar]
- 44.Aravind L., Koonin E. V. Classification of the caspase-hemoglobinase fold: Detection of new families and implications for the origin of the eukaryotic separins. Proteins. 2002;46:355–367. doi: 10.1002/prot.10060. [DOI] [PubMed] [Google Scholar]
- 45.Eichinger A., Beisel H. G., Jacob U., Huber R., Medrano F. J., Banbula A., Potempa J., Travis J., Bode W. Crystal structure of gingipain R: An Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 1999;18:5453–5462. doi: 10.1093/emboj/18.20.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murzin A. G., Brenner S. E., Hubbard T., Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 47.Huang B., Eberstadt M., Olejniczak E. T., Meadows R. P., Fesik S. W. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature (London) 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- 48.Eberstadt M., Huang B., Chen Z., Meadows R. P., Ng S. C., Zheng L., Lenardo M. J., Fesik S. W. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature (London) 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- 49.Jeong E. J., Bang S., Lee T. H., Park Y. I., Sim W. S., Kim K. S. The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD. J. Biol. Chem. 1999;274:16337–16342. doi: 10.1074/jbc.274.23.16337. [DOI] [PubMed] [Google Scholar]
- 50.Hofmann K. The modular nature of apoptotic signaling proteins. Cell. Mol. Life Sci. 1999;55:1113–1128. doi: 10.1007/s000180050361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fairbrother W. J., Gordon N. C., Humke E. W., O'Rourke K. M., Starovasnik M. A., Yin J. P., Dixit V. M. The PYRIN domain: A member of the death domain-fold superfamily. Protein Sci. 2001;10:1911–1918. doi: 10.1110/ps.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fesik S. W. Insights into programmed cell death through structural biology. Cell. 2000;103:273–282. doi: 10.1016/s0092-8674(00)00119-7. [DOI] [PubMed] [Google Scholar]
- 53.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 54.Branden C.-I., Tooze J. New York: Garland Publishing Inc.; 1998. Introduction to Protein Structure. [Google Scholar]
- 55.Li P., Nijhawan D., Budihardjo I., Srinivasula S. M., Ahmad M., Alnemri E. S., Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 56.Srinivasula S. M., Ahmad M., Fernandes-Alnemri T., Alnemri E. S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 57.Chen J. M., Rawlings N. D., Stevens R. A., Barrett A. J. Identification of the active site of legumain links it to caspases, clostripain and gingipains in a new clan of cysteine endopeptidases. FEBS Lett. 1998;441:361–365. doi: 10.1016/s0014-5793(98)01574-9. [DOI] [PubMed] [Google Scholar]
- 58.Uhlmann F., Wernic D., Poupart M. A., Koonin E. V., Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 59.Chestukhin A., Pfeffer C., Milligan S., DeCaprio J. A., Pellman D. Processing, localization, and requirement of human separase for normal anaphase progression. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4574–4579. doi: 10.1073/pnas.0730733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilmouth R. C., Edman K., Neutze R., Wright P. A., Clifton I. J., Schneider T. R., Schofield C. J., Hajdu J. X-ray snapshots of serine protease catalysis reveal a tetrahedral intermediate. Nat. Struct. Biol. 2001;8:689–694. doi: 10.1038/90401. [DOI] [PubMed] [Google Scholar]
- 61.Topf M., Varnai P., Schofield C. J., Richards W. G. Molecular dynamics simulations of the acyl-enzyme and the tetrahedral intermediate in the deacylation step of serine proteases. Proteins. 2002;47:357–369. doi: 10.1002/prot.10097. [DOI] [PubMed] [Google Scholar]
- 62.Topf M., Varnai P., Richards W. G. Ab initio QM/MM dynamics simulation of the tetrahedral intermediate of serine proteases: Insights into the active site hydrogen-bonding network. J. Am. Chem. Soc. 2002;124:14780–14788. doi: 10.1021/ja026219q. [DOI] [PubMed] [Google Scholar]
- 63.Wei Y., Fox T., Chambers S. P., Sintchak J., Coll J. T., Golec J. M., Swenson L., Wilson K. P., Charifson P. S. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem. Biol. 2000;7:423–432. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 64.Stennicke H. R., Renatus M., Meldal M., Salvesen G. S. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem. J. 2000;350:563–568. [PMC free article] [PubMed] [Google Scholar]
- 65.Blanchard H., Donepudi M., Tschopp M., Kodandapani L., Wu J. C., Grütter M. G. Caspase-8 specificity probed at subsite S4: Crystal structure of the caspase-8-Z-DEVD-cho complex. J. Mol. Biol. 2000;302:9–16. doi: 10.1006/jmbi.2000.4041. [DOI] [PubMed] [Google Scholar]
- 66.Chereau D., Kodandapani L., Tomaselli K. J., Spada A. P., Wu J. C. Structural and functional analysis of caspase active sites. Biochemistry. 2003;42:4151–4160. doi: 10.1021/bi020593l. [DOI] [PubMed] [Google Scholar]
- 67.Ni C.-Z., Li C. Y., Wu J. C., Spada A. P., Ely K. R. Conformational restriction in the active site of unliganded human caspase-3. J. Mol. Recog. 2003;16:121–124. doi: 10.1002/jmr.615. [DOI] [PubMed] [Google Scholar]
- 68.Becker J. W., Rotonda J., Soisson S. M., Aspiotis R., Bayly C., Francoeur S., Gallant M., Garcia-Calvo M., Giroux A., Grimm E., et al. Reducing the peptidyl features of caspase-3 inhibitors: A structural analysis. J. Med. Chem. 2004;47:2466–2474. doi: 10.1021/jm0305523. [DOI] [PubMed] [Google Scholar]
- 69.Hopfner K. P., Lang A., Karcher A., Sichler K., Kopetzki E., Brandstetter H., Huber R., Bode W., Engh R. A. Coagulation factor IXa: The relaxed conformation of Tyr99 blocks substrate binding. Struct. Fold. Des. 1999;7:989–996. doi: 10.1016/s0969-2126(99)80125-7. [DOI] [PubMed] [Google Scholar]
- 70.Sichler K., Kopetzki E., Huber R., Bode W., Hopfner K.-P., Brandstetter H. Physiological fIXa activation involves a cooperative conformational rearrangement of the 99-loop. J. Biol. Chem. 2003;278:4121–4126. doi: 10.1074/jbc.M210722200. [DOI] [PubMed] [Google Scholar]
- 71.Talanian R. V., Quinlan C., Trautz S., Hackett M. C., Mankovich J. A., Banach D., Ghayur T., Brady K. D., Wong W. W. Substrate specificities of caspase family proteases. J. Biol. Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 72.Garcia-Calvo M., Peterson E. P., Leiting B., Ruel R., Nicholson D. W., Thornberry N. A. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J. Biol. Chem. 1998;273:32608–32613. doi: 10.1074/jbc.273.49.32608. [DOI] [PubMed] [Google Scholar]
- 73.Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., Salvesen G. S. Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rano T. A., Timkey T., Peterson E. P., Rotonda J., Nicholson D. W., Becker J. W., Chapman K. T., Thornberry N. A. A combinatorial approach for determining protease specificities: Application to interleukin-1β converting enzyme (ICE) Chem. Biol. 1997;4:149–155. doi: 10.1016/s1074-5521(97)90258-1. [DOI] [PubMed] [Google Scholar]
- 75.Schweizer P., Briand C., Grütter M. G. Crystal structure of caspase-2, apical initiator of the intrinsic apoptotic pathway. J. Biol. Chem. 2003;278:42441–42447. doi: 10.1074/jbc.M304895200. [DOI] [PubMed] [Google Scholar]
- 76.Rotonda J., Nicholson D. W., Fazil K. M., Gallant M., Gareau Y., Labelle M., Peterson E. P., Rasper D. M., Ruel R., Vaillancourt J. P., et al. The three-dimensional structure of apopain/CPP32, a key mediator of apoptosis. Nat. Struct. Biol. 1996;3:619–625. doi: 10.1038/nsb0796-619. [DOI] [PubMed] [Google Scholar]
- 77.Earnshaw W. C., Martins L. M., Kaufmann S. H. Mammalian caspases: Structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 78.Sleath P. R., Hendrickson R. C., Kronheim S. R., March C. J., Black R. A. Substrate specificity of the protease that processes human interleukin-1β. J. Biol. Chem. 1990;265:14526–14528. [PubMed] [Google Scholar]
- 79.Sulpizi M., Laio A., VandeVondele J., Cattaneo A., Rothlisberger U., Carloni P. Reaction mechanism of caspases: Insights from QM/MM Car-Parrinello simulations. Proteins. 2003;52:212–224. doi: 10.1002/prot.10275. [DOI] [PubMed] [Google Scholar]
- 80.Brady K. D., Giegel D. A., Grinnell C., Lunney E., Talanian R. V., Wong W., Walker N. A catalytic mechanism for caspase-1 and for bimodal inhibition of caspase-1 by activated aspartic ketones. Bioorg. Med. Chem. 1999;7:621–631. doi: 10.1016/s0968-0896(99)00009-7. [DOI] [PubMed] [Google Scholar]
- 81.Stennicke H. R., Salvesen G. S. Catalytic properties of the caspases. Cell Death Differ. 1999;6:1054–1059. doi: 10.1038/sj.cdd.4400599. [DOI] [PubMed] [Google Scholar]
- 82.Stennicke H. R., Salvesen G. S. Biochemical characteristics of caspases-3, -6, -7, and -8. J. Biol. Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 83.Vernet T., Tessier D. C., Chatellier J., Plouffe C., Lee T. S., Thomas D. Y., Storer A. C., Menard R. Structural and functional roles of asparagine 175 in the cysteine protease papain. J. Biol. Chem. 1995;270:16645–16652. doi: 10.1074/jbc.270.28.16645. [DOI] [PubMed] [Google Scholar]
- 84.Bergmann E. M., Mosimann S. C., Chernaia M. M., Malcolm B. A., James M. N. The refined crystal structure of the 3C gene product from hepatitis A virus: Specific proteinase activity and RNA recognition. J. Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anand K., Palm G. J., Mesters J. R., Siddell S. G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Phan J., Zdanov A., Evdokimov A. G., Tropea J. E., Peters H. K., III, Kapust R. B., Li M., Wlodawer A., Waugh D. S. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 2002;277:50564–50572. doi: 10.1074/jbc.M207224200. [DOI] [PubMed] [Google Scholar]
- 87.Nicholson D. W. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 88.Fischer U., Jänicke R. U., Schulze-Osthoff K. Many cuts to ruin: A comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Denault J.-B., Salvesen G. S. Caspases: Keys in the ignition of cell death. Chem. Rev. 2002;102:4489–4500. doi: 10.1021/cr010183n. [DOI] [PubMed] [Google Scholar]
- 90.Bode W., Brandstetter H., Mather T., Stubbs M. T. Comparative analysis of haemostatic proteinases: Structural aspects of thrombin, factor Xa, factor IXa and protein C. Thromb. Haemostasis. 1997;78:501–511. [PubMed] [Google Scholar]
- 91.Lopez-Otin C., Overall C. M. Protease degradomics: A new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 92.Thornberry N. A., Lazebnik Y. Caspases: Enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 93.Stennicke H. R., Salvesen G. S. Caspases – controlling intracellular signals by protease zymogen activation. Biochim. Biophys. Acta. 2000;1477:299–306. doi: 10.1016/s0167-4838(99)00281-2. [DOI] [PubMed] [Google Scholar]
- 94.Li H., Zhu H., Xu C. J., Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 95.Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 96.Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. Proapoptotic Bax and Bak: A requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chou J. J., Li H., Salvesen G. S., Yuan J., Wagner G. Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell. 1999;96:615–624. doi: 10.1016/s0092-8674(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 98.McDonnell J. M., Fushman D., Milliman C. L., Korsmeyer S. J., Cowburn D. Solution structure of the proapoptotic molecule BID: A structural basis for apoptotic agonists and antagonists. Cell. 1999;96:625–634. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 99.Guo Y., Srinivasula S. M., Druilhe A., Fernandes-Alnemri T., Alnemri E. S. Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J. Biol. Chem. 2002;277:13430–13437. doi: 10.1074/jbc.M108029200. [DOI] [PubMed] [Google Scholar]
- 100.Barry M., Heibein J. A., Pinkoski M. J., Lee S. F., Moyer R. W., Green D. R., Bleackley R. C. Granzyme B short-circuits the need for caspase 8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly cleaving Bid. Mol. Cell. Biol. 2000;20:3781–3794. doi: 10.1128/mcb.20.11.3781-3794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heibein J. A., Goping I. S., Barry M., Pinkoski M. J., Shore G. C., Green D. R., Bleackley R. C. Granzyme B-mediated cytochrome c release is regulated by the Bcl-2 family members Bid and Bax. J. Exp. Med. 2000;192:1391–1401. doi: 10.1084/jem.192.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu X., Zou H., Slaughter C., Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 103.Enari M., Sakahira H., Yokoyama H., Okawa K., Iwamatsu A., Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature (London) 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 104.Sakahira H., Enari M., Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature (London) 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 105.Liu X., Zou H., Widlak P., Garrard W., Wang X. Activation of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease). Oligomerization and direct interaction with histone H1. J. Biol. Chem. 1999;274:13836–13840. doi: 10.1074/jbc.274.20.13836. [DOI] [PubMed] [Google Scholar]
- 106.Woo E. J., Kim Y. G., Kim M. S., Han W. D., Shin S., Robinson H., Park S. Y., Oh B. H. Structural mechanism for inactivation and activation of CAD/DFF40 in the apoptotic pathway. Mol. Cell. 2004;14:531–539. doi: 10.1016/s1097-2765(04)00258-8. [DOI] [PubMed] [Google Scholar]
- 107.Otomo T., Sakahira H., Uegaki K., Nagata S., Yamazaki T. Structure of the heterodimeric complex between CAD domains of CAD and ICAD. Nat. Struct. Biol. 2000;7:658–662. doi: 10.1038/77957. [DOI] [PubMed] [Google Scholar]
- 108.Zhou P., Lugovskoy A. A., McCarty J. S., Li P., Wagner G. Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6051–6055. doi: 10.1073/pnas.111145098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fukushima K., Kikuchi J., Koshiba S., Kigawa T., Kuroda Y., Yokoyama S. Solution structure of the DFF-C domain of DFF45/ICAD. A structural basis for the regulation of apoptotic DNA fragmentation. J. Mol. Biol. 2002;321:317–327. doi: 10.1016/s0022-2836(02)00588-0. [DOI] [PubMed] [Google Scholar]
- 110.McCarty J. S., Toh S. Y., Li P. Multiple domains of DFF45 bind synergistically to DFF40: Roles of caspase cleavage and sequestration of activator domain of DFF40. Biochem. Biophys. Res. Commun. 1999;264:181–185. doi: 10.1006/bbrc.1999.1498. [DOI] [PubMed] [Google Scholar]
- 111.McCarty J. S., Toh S. Y., Li P. Study of DFF45 in its role of chaperone and inhibitor: Two independent inhibitory domains of DFF40 nuclease activity. Biochem. Biophys. Res. Commun. 1999;264:176–180. doi: 10.1006/bbrc.1999.1497. [DOI] [PubMed] [Google Scholar]
- 112.Gu J., Dong R. P., Zhang C., McLaughlin D. F., Wu M. X., Schlossman S. F. Functional interaction of DFF35 and DFF45 with caspase-activated DNA fragmentation nuclease DFF40. J. Biol. Chem. 1999;274:20759–20762. doi: 10.1074/jbc.274.30.20759. [DOI] [PubMed] [Google Scholar]
- 113.Schreiber V., Molinete M., Boeuf H., de Murcia G., Menissier-de Murcia J. The human poly(ADP-ribose) polymerase nuclear localization signal is a bipartite element functionally separate from DNA binding and catalytic activity. EMBO J. 1992;11:3263–3269. doi: 10.1002/j.1460-2075.1992.tb05404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lazebnik Y. A., Kaufmann S. H., Desnoyers S., Poirier G. G., Earnshaw W. C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature (London) 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 115.Gu Y., Sarnecki C., Aldape R. A., Livingston D. J., Su M. S. Cleavage of poly(ADP-ribose) polymerase by interleukin-1β converting enzyme and its homologs TX and Nedd-2. J. Biol. Chem. 1995;270:18715–18718. doi: 10.1074/jbc.270.32.18715. [DOI] [PubMed] [Google Scholar]
- 116.Nicholson D. W., Ali A., Thornberry N. A., Vaillancourt J. P., Ding C. K., Gallant M., Gareau Y., Griffin P. R., Labelle M., Lazebnik Y. A. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature (London) 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 117.Germain M., Affar E. B., D'Amours D., Dixit V. M., Salvesen G. S., Poirier G. G. Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis. Evidence for involvement of caspase-7. J. Biol. Chem. 1999;274:28379–28384. doi: 10.1074/jbc.274.40.28379. [DOI] [PubMed] [Google Scholar]
- 118.Krippner-Heidenreich A., Talanian R. V., Sekul R., Kraft R., Thole H., Ottleben H., Luscher B. Targeting of the transcription factor Max during apoptosis: Phosphorylation-regulated cleavage by caspase-5 at an unusual glutamic acid residue in position P1. Biochem. J. 2001;358:705–715. doi: 10.1042/0264-6021:3580705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Talanian R. V., Dang L. C., Ferenz C. R., Hackett M. C., Mankovich J. A., Welch J. P., Wong W. W., Brady K. D. Stability and oligomeric equilibria of refolded interleukin-1β converting enzyme. J. Biol. Chem. 1996;271:21853–21858. doi: 10.1074/jbc.271.36.21853. [DOI] [PubMed] [Google Scholar]
- 120.Pop C., Chen Y. R., Smith B., Bose K., Bobay B., Tripathy A., Franzen S., Clark A. C. Removal of the pro-domain does not affect the conformation of the procaspase-3 dimer. Biochemistry. 2001;40:14224–14235. doi: 10.1021/bi011037e. [DOI] [PubMed] [Google Scholar]
- 121.Kang B. H., Ko E., Kwon O. K., Choi K. Y. The structure of procaspase 6 is similar to that of active mature caspase 6. Biochem. J. 2002;364:629–634. doi: 10.1042/BJ20011787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shiozaki E. N., Chai J., Rigotti D. J., Riedl S. J., Li P., Srinivasula S. M., Alnemri E. S., Fairman R., Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 123.Boatright K. M., Renatus M., Scott F. L., Sperandio S., Shin H., Pederson I., Ricci J.-E., Edris W. A., Sutherlin D. P., Green D. R., et al. A unified model for apical caspase activation. Mol. Cell. 2003;11:1–20. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 124.Riedl S. J., Fuentes-Prior P., Renatus M., Kairies N., Krapp S., Huber R., Salvesen G. S., Bode W. Structural basis for the activation of human procaspase-7. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14790–14795. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chang D. W., Ditsworth D., Liu H. T., Srinivasula S. M., Alnemri E. S., Yang X. L. Oligomerization is a general mechanism for the activation of apoptosis initiator and inflammatory procaspases. J. Biol. Chem. 2003;278:16466–16469. doi: 10.1074/jbc.C300089200. [DOI] [PubMed] [Google Scholar]
- 126.Chang D. W., Xing Z., Capacio V. L., Peter M. E., Yang X. L. Interdimer processing mechanism of procaspase-8 activation. EMBO J. 2003;22:4132–4142. doi: 10.1093/emboj/cdg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Micheau O., Thome M., Schneider P., Holler N., Tschopp J., Nicholson D. W., Briand C., Grütter M. G. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J. Biol. Chem. 2002;277:45162–45171. doi: 10.1074/jbc.M206882200. [DOI] [PubMed] [Google Scholar]
- 128.Chen M., Orozco A., Spencer D. M., Wang J. Activation of initiator caspases through a stable dimeric intermediate. J. Biol. Chem. 2002;277:50761–50767. doi: 10.1074/jbc.M210356200. [DOI] [PubMed] [Google Scholar]
- 129.Donepudi M., MacSweeney A., Briand C., Grütter M. G. Insights into the regulatory mechanism for caspase-8 activation. Mol. Cell. 2003;11:543–549. doi: 10.1016/s1097-2765(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 130.Khan A. R., James M. N. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 1998;7:815–836. doi: 10.1002/pro.5560070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Turk B., Turk D., Turk V. Lysosomal cysteine proteases: More than scavengers. Biochim. Biophys. Acta. 2000;1477:98–111. doi: 10.1016/s0167-4838(99)00263-0. [DOI] [PubMed] [Google Scholar]
- 132.Bose K., Clark A. C. Dimeric procaspase-3 unfolds via a four-state equilibrium process. Biochemistry. 2001;40:14236–14242. doi: 10.1021/bi0110387. [DOI] [PubMed] [Google Scholar]
- 133.Chai J., Shiozaki E., Srinivasula S. M., Wu Q., Dataa P., Alnemri E. S., Shi Y. Structural basis of caspase-7 inhibition by XIAP. Cell. 2001;104:769–780. doi: 10.1016/s0092-8674(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 134.Huang Y., Park Y. C., Rich R. L., Segal D., Myszka D. G., Wu H. Structural basis of caspase inhibition by XIAP: Differential roles of the linker versus the BIR domain. Cell. 2001;104:781–790. [PubMed] [Google Scholar]
- 135.Piana S., Sulpizi M., Rothlisberger U. Structure-based thermodynamic analysis of caspases reveals key residues for dimerization and activity. Biochemistry. 2003;42:8720–8728. doi: 10.1021/bi034032l. [DOI] [PubMed] [Google Scholar]
- 135a.Hardy J. A., Lam J., Nguyen J. T., O'Brien T., Wells J. A. Discovery of an allosteric site in the caspases. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhou Q., Salvesen G. S. Activation of pro-caspase-7 by serine proteases includes a non-canonical specificity. Biochem. J. 1997;324:361–364. doi: 10.1042/bj3240361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang X., Stennicke H. R., Wang B., Green D. R., Jänicke R. U., Srinivasan A., Seth P., Salvesen G. S., Froelich C. J. Granzyme B mimics apical caspases. Description of a unified pathway for trans-activation of executioner caspase-3 and -7. J. Biol. Chem. 1998;273:34278–34283. doi: 10.1074/jbc.273.51.34278. [DOI] [PubMed] [Google Scholar]
- 138.Srinivasula S. M., Ahmad M., MacFarlane M., Luo Z., Huang Z., Fernandes-Alnemri T., Alnemri E. S. Generation of constitutively active recombinant caspases-3 and -6 by rearrangement of their subunits. J. Biol. Chem. 1998;273:10107–10111. doi: 10.1074/jbc.273.17.10107. [DOI] [PubMed] [Google Scholar]
- 139.Suzuki Y., Nakabayashi Y., Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen J. M., Fortunato M., Barrett A. J. Activation of human prolegumain by cleavage at a C-terminal asparagine residue. Biochem. J. 2000;352:327–334. [PMC free article] [PubMed] [Google Scholar]
- 141.Mikolajczyk J., Boatright K. M., Stennicke H. R., Nazif T., Potempa J., Bogyo M., Salvesen G. S. Sequential autolytic processing activates the zymogen of Arg-gingipain. J. Biol. Chem. 2003;278:10458–10464. doi: 10.1074/jbc.M210564200. [DOI] [PubMed] [Google Scholar]
- 142.Hornig N. C., Knowles P. P., McDonald N. Q., Uhlmann F. The dual mechanism of separase regulation by securin. Curr. Biol. 2002;12:973–982. doi: 10.1016/s0960-9822(02)00847-3. [DOI] [PubMed] [Google Scholar]
- 143.Waizenegger I., Gimenez-Abian J., Wernic D., Peters J. Regulation of human separase by securin binding and autocleavage. Curr. Biol. 2002;12:1368–1378. doi: 10.1016/s0960-9822(02)01073-4. [DOI] [PubMed] [Google Scholar]
- 144.Salvesen G. S., Dixit V. M. Caspases: Intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 145.Shi Y. Caspase activation: Revisiting the induced proximity model. Cell. 2004;117:855–858. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 146.Chinnaiyan A. M., Chaudhary D., O'Rourke K., Koonin E. V., Dixit V. M. Role of CED-4 in the activation of CED-3. Nature (London) 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- 147.Yang X., Chang H. Y., Baltimore D. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 148.Muzio M., Chinnaiyan A. M., Kischkel F. C., O'Rourke K., Shevchenko A., Ni J., Scaffidi C., Bretz J. D., Zhang M., Gentz R., et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 149.Boldin M. P., Goncharov T. M., Goltsev Y. V., Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 150.Yamin T. T., Ayala J. M., Miller D. K. Activation of the native 45-kDa precursor form of interleukin-1-converting enzyme. J. Biol. Chem. 1996;271:13273–13282. doi: 10.1074/jbc.271.22.13273. [DOI] [PubMed] [Google Scholar]
- 151.Muzio M., Stockwell B. R., Stennicke H. R., Salvesen G. S., Dixit V. M. An induced proximity model for caspase-8 activation. J. Biol. Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 152.Salvesen G. S., Dixit V. M. Caspase activation: The induced-proximity model. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10964–10967. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Renatus M., Engh R. A., Stubbs M. T., Huber R., Fischer S., Kohnert U., Bode W. Lysine 156 promotes the anomalous proenzyme activity of tPA: X-ray crystal structure of single-chain human tPA. EMBO J. 1997;16:4797–4805. doi: 10.1093/emboj/16.16.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Davie E. W., Fujikawa K., Kisiel W. The coagulation cascade: Initiation, maintenance and regulation. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 155.Martin D. A., Siegel R. M., Zheng L., Lenardo M. J. Membrane oligomerization and cleavage activates the caspase-8 (FLICE/MACHα1) death signal. J. Biol. Chem. 1998;273:4345–4349. doi: 10.1074/jbc.273.8.4345. [DOI] [PubMed] [Google Scholar]
- 156.Yang X., Chang H. Y., Baltimore D. Autoproteolytic activation of pro-caspases by oligomerization. Mol. Cell. 1998;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 157.Stennicke H. R., Deveraux Q. L., Humke E. W., Reed J. C., Dixit V. M., Salvesen G. S. Caspase-9 can be activated without proteolytic processing. J. Biol. Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 158.Srinivasula S. M., Hegde R., Saleh A., Datta P., Shiozaki E., Chai J., Lee R. A., Robbins P. D., Fernandes-Alnemri T., Shi Y., et al. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature (London) 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 159.Qin H., Srinivasula S. M., Wu G., Fernandes-Alnemri T., Alnemri E. S., Shi Y. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature (London) 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 160.Zhou P., Chou J., Olea R. S., Yuan J., Wagner G. Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: A structural basis for specific adaptor/caspase interaction. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11265–11270. doi: 10.1073/pnas.96.20.11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xiao T., Towb P., Wasserman S. A., Sprang S. R. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell. 1999;99:545–555. doi: 10.1016/s0092-8674(00)81542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Weber C. H., Vincenz C. The death domain superfamily: A tale of two interfaces? Trends Biochem. Sci. 2001;26:475–481. doi: 10.1016/s0968-0004(01)01905-3. [DOI] [PubMed] [Google Scholar]
- 163.Weber C. H., Vincenz C. A docking model of key components of the DISC complex: Death domain superfamily interactions redefined. FEBS Lett. 2001;492:171–176. doi: 10.1016/s0014-5793(01)02162-7. [DOI] [PubMed] [Google Scholar]
- 164.Kaufmann M., Bozic D., Briand C., Bodmer J. L., Zerbe O., Kohl A., Tschopp J., Grütter M. G. Identification of a basic surface area of the FADD death effector domain critical for apoptotic signaling. FEBS Lett. 2002;527:250–254. doi: 10.1016/s0014-5793(02)03146-0. [DOI] [PubMed] [Google Scholar]
- 165.Acehan D., Jiang X., Morgan D. G., Heuser J. E., Wang X., Akey C. W. Three-dimensional structure of the apoptosome: Implications for assembly, procaspase-9 binding, and activation. Mol. Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- 166.Bratton S. B., Walker G., Srinivasula S. M., Sun X. M., Butterworth M., Alnemri E. S., Cohen G. M. Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J. 2001;20:998–1009. doi: 10.1093/emboj/20.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pop C., Feeney B., Tripathy A., Clark A. C. Mutations in the procaspase-3 dimer interface affect the activity of the zymogen. Biochemistry. 2003;42:12311–12320. doi: 10.1021/bi034999p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ma B., Elkayam T., Wolfson H., Nussinov R. Protein-protein interactions: Structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cardone M. H., Roy N., Stennicke H. R., Salvesen G. S., Franke T. F., Stanbridge E., Frisch S., Reed J. C. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 170.Allan L. A., Morrice N., Brady S., Magee G., Pathak S., Clarke P. R. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat. Cell Biol. 2003;5:647–654. doi: 10.1038/ncb1005. [DOI] [PubMed] [Google Scholar]
- 171.Kischkel F. C., Hellbardt S., Behrmann I., Germer M., Pawlita M., Krammer P. H., Peter M. E. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Irmler M., Thome M., Hahne M., Schneider P., Hofmann K., Steiner V., Bodmer J. L., Schroter M., Burns K., Mattmann C., et al. Inhibition of death receptor signals by cellular FLIP. Nature (London) 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 173.Yeh W. C., Itie A., Elia A. J., Ng M., Shu H. B., Wakeham A., Mirtsos C., Suzuki N., Bonnard M., Goeddel D. V., et al. Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity. 2000;12:633–642. doi: 10.1016/s1074-7613(00)80214-9. [DOI] [PubMed] [Google Scholar]
- 174.Chang D. W., Xing Z., Pan Y., Algeciras-Schimnich A., Barnhart B. C., Yaish-Ohad S., Peter M. E., Yang X. c-FLIPL is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21:3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Boatright K. M., Deis C., Denault J.-B., Sutherlin D. P., Salvesen G. S. Activation of caspases 8 and 10 by FLIPL. Biochem. J. 2004;382:651–657. doi: 10.1042/BJ20040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.McDonald E. R., III, El-Deiry W. S. Suppression of caspase-8- and -10-associated RING proteins results in sensitization to death ligands and inhibition of tumor cell growth. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6170–6175. doi: 10.1073/pnas.0307459101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Himeji D., Horiuchi T., Tsukamoto H., Hayashi K., Watanabe T., Harada M. Characterization of caspase-8L: A novel isoform of caspase-8 that behaves as an inhibitor of the caspase cascade. Blood. 2002;99:4070–4078. doi: 10.1182/blood.v99.11.4070. [DOI] [PubMed] [Google Scholar]
- 178.Medema J. P., Scaffidi C., Kischkel F. C., Shevchenko A., Mann M., Krammer P. H., Peter M. E. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC) EMBO J. 1997;16:2794–2804. doi: 10.1093/emboj/16.10.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Siegel R. M., Frederiksen J. K., Zacharias D. A., Chan F. K., Johnson M., Lynch D., Tsien R. Y., Lenardo M. J. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 180.Lin Y., Devin A., Rodriguez Y., Liu Z. G. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13:2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gu Y., Wu J., Faucheu C., Lalanne J. L., Diu A., Livingston D. J., Su M. S. Interleukin-1β converting enzyme requires oligomerization for activity of processed forms in vivo. EMBO J. 1995;14:1923–1931. doi: 10.1002/j.1460-2075.1995.tb07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Van Criekinge W., Beyaert R., Van de Craen M., Vandenabeele P., Schotte P., De Valck D., Fiers W. Functional characterization of the prodomain of interleukin-1β-converting enzyme. J. Biol. Chem. 1996;271:27245–27248. doi: 10.1074/jbc.271.44.27245. [DOI] [PubMed] [Google Scholar]
- 183.Ramage P., Cheneval D., Chvei M., Graff P., Hemmig R., Heng R., Kocher H. P., Mackenzie A., Memmert K., Revesz L., et al. Expression, refolding, and autocatalytic proteolytic processing of the interleukin-1β-converting enzyme precursor. J. Biol. Chem. 1995;270:9378–9383. doi: 10.1074/jbc.270.16.9378. [DOI] [PubMed] [Google Scholar]
- 184.Ghayur T., Hugunin M., Talanian R. V., Ratnofsky S., Quinlan C., Emoto Y., Pandey P., Datta R., Huang Y., Kharbanda S., et al. Proteolytic activation of protein kinase C delta by an ICE/CED 3-like protease induces characteristics of apoptosis. J. Exp. Med. 1996;184:2399–2404. doi: 10.1084/jem.184.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.McCarthy J. V., Ni J., Dixit V. M. RIP2 is a novel NF-κB-activating and cell death-inducing kinase. J. Biol. Chem. 1998;273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 186.Inohara N., del Peso L., Koseki T., Chen S., Nunez G. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J. Biol. Chem. 1998;273:12296–12300. doi: 10.1074/jbc.273.20.12296. [DOI] [PubMed] [Google Scholar]
- 187.Thome M., Hofmann K., Burns K., Martinon F., Bodmer J. L., Mattmann C., Tschopp J. Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr. Biol. 1998;8:885–888. doi: 10.1016/s0960-9822(07)00352-1. [DOI] [PubMed] [Google Scholar]
- 188.Kobayashi K., Inohara N., Hernandez L. D., Galan J. E., Nunez G., Janeway C. A., Medzhitov R., Flavell R. A. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature (London) 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 189.Chin A. I., Dempsey P. W., Bruhn K., Miller J. F., Xu Y., Cheng G. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature (London) 2002;416:190–194. doi: 10.1038/416190a. [DOI] [PubMed] [Google Scholar]
- 190.Poyet J. L., Srinivasula S. M., Tnani M., Razmara M., Fernandes-Alnemri T., Alnemri E. S. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 191.Mariathasan S., Newton K., Monack D. M., Vucic D., French D. M., Lee W. P., Roose-Girma M., Erickson S., Dixit V. M. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature (London) 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 192.Srinivasula S. M., Poyet J. L., Razmara M., Datta P., Zhang Z., Alnemri E. S. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J. Biol. Chem. 2002;277:21119–21122. doi: 10.1074/jbc.C200179200. [DOI] [PubMed] [Google Scholar]
- 193.Faucheu C., Diu A., Chan A. W., Blanchet A. M., Miossec C., Herve F., Collard-Dutilleul V., Gu Y., Aldape R. A., Lippke J. A., et al. A novel human protease similar to the interleukin-1β converting enzyme induces apoptosis in transfected cells. EMBO J. 1995;14:1914–1922. doi: 10.1002/j.1460-2075.1995.tb07183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Inohara N., Nunez G. NODs: Intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 195.Tanabe T., Chamaillard M., Ogura Y., Shu L., Qiu S., Masumoto J., Ghosh P., Moran A., Predergast M. M., Tromp G., et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Aliprantis A. O., Yang R. B., Mark M. R., Suggett S., Devaux B., Radolf J. D., Klimpel G. R., Godowski P., Zychlinsky A. Cell activation and apoptosis by bacterial lipoproteins through Toll-like receptor-2. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 197.Aliprantis A. O., Yang R. B., Weiss D. S., Godowski P., Zychlinsky A. The apoptotic signaling pathway activated by Toll-like receptor-2. EMBO J. 2000;19:3325–3336. doi: 10.1093/emboj/19.13.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Choi K. B., Wong F., Harlan J. M., Chaudhary P. M., Hood L., Karsan A. Lipopolysaccharide mediates endothelial apoptosis by a FADD-dependent pathway. J. Biol. Chem. 1998;273:20185–20188. doi: 10.1074/jbc.273.32.20185. [DOI] [PubMed] [Google Scholar]
- 199.Franchi L., Condo I., Tomassini B., Nicolo C., Testi R. A caspase-like activity is triggered by LPS and is required for survival of dendritic cells. Blood. 2003;102:2910–2915. doi: 10.1182/blood-2003-03-0967. [DOI] [PubMed] [Google Scholar]
- 200.Kobayashi S. D., Braughton K. R., Whotney A. R., Voyich J. M., Schwan T. G., Musser J. M., DeLeo F. R. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10948–10953. doi: 10.1073/pnas.1833375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bergeron L., Perez G. I., Macdonald G., Shi L., Sun Y., Jurisicova A., Varmuza S., Latham K. E., Flaws J. A., Salter J. C., et al. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev. 1998;12:1304–1314. doi: 10.1101/gad.12.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lassus P., Opitz-Araya X., Lazebnik Y. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science. 2002;297:1352–1354. doi: 10.1126/science.1074721. [DOI] [PubMed] [Google Scholar]
- 203.Robertson J. D., Enoksson M., Suomela M., Zhivotovsky B., Orrenius S. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J. Biol. Chem. 2002;277:29803–29809. doi: 10.1074/jbc.M204185200. [DOI] [PubMed] [Google Scholar]
- 204.Konishi A., Shimizu S., Hirota J., Takao T., Fan Y., Matsuoka Y., Zhang L., Yoneda Y., Fujii Y., Skoultchi A. I., et al. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell. 2003;114:673–688. doi: 10.1016/s0092-8674(03)00719-0. [DOI] [PubMed] [Google Scholar]
- 205.Duan H., Dixit V. M. RAIDD is a new ‘death’ adaptor molecule. Nature (London) 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- 206.Ahmad M., Srinivasula S. M., Wang L., Talanian R. V., Litwack G., Fernandes-Alnemri T., Alnemri E. S. CRADD, a novel human apoptotic adaptor molecule for caspase-2, and FasL/tumor necrosis factor receptor-interacting protein RIP. Cancer Res. 1997;57:615–619. [PubMed] [Google Scholar]
- 207.Shearwin-Whyatt L. M., Harvey N. L., Kumar S. Subcellular localization and CARD-dependent oligomerization of the death adaptor RAIDD. Cell Death Differ. 2000;7:155–165. doi: 10.1038/sj.cdd.4400632. [DOI] [PubMed] [Google Scholar]
- 208.Chou J. J., Matsuo H., Duan H., Wagner G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell. 1998;94:171–180. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- 209.Read S. H., Baliga B. C., Ekert P. G., Vaux D. L., Kumar S. A novel Apaf-1-independent putative caspase-2 activation complex. J. Cell Biol. 2002;159:739–745. doi: 10.1083/jcb.200209004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tinel A., Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- 211.Lin Y., Ma W., Benchimol S. PIDD, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat. Genet. 2000;26:122–127. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- 212.Wagner K. W., Engels I. H., Deveraux Q. L. Caspase-2 can function upstream of Bid cleavage in the TRAIL-apoptosis pathway. J. Biol. Chem. 2004;279:35047–35052. doi: 10.1074/jbc.M400708200. [DOI] [PubMed] [Google Scholar]
- 213.Robertson J. D., Gogvadze V., Kropotov A., Vakifahmetoglu H., Zhivotovsky B., Orrenius S. Processed caspase-2 can induce mitochondria-mediated apoptosis independently of its enzymatic activity. EMBO Rep. 2004;5:643–648. doi: 10.1038/sj.embor.7400153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhivotovsky B., Samali A., Gahm A., Orrenius S. Caspases: Their intracellular localization and translocation during apoptosis. Cell Death Differ. 1999;6:644–651. doi: 10.1038/sj.cdd.4400536. [DOI] [PubMed] [Google Scholar]
- 215.Shikama Y., U M., Miyashita T., Yamada M. Comprehensive studies on subcellular localizations and cell death-inducing activities of eight GFP-tagged apoptosis-related caspases. Exp. Cell Res. 2001;264:315–325. doi: 10.1006/excr.2000.5153. [DOI] [PubMed] [Google Scholar]
- 216.O'Reilly L. A., Ekert P., Harvey N., Marsden V., Cullen L., Vaux D. L., Hacker G., Magnusson C., Pakusch M., Cecconi F., et al. Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9. Cell Death Differ. 2002;9:832–841. doi: 10.1038/sj.cdd.4401033. [DOI] [PubMed] [Google Scholar]
- 217.Paroni G., Henderson C., Schneider C., Brancolini C. Caspase-2 can trigger cytochrome c release and apoptosis from the nucleus. J. Biol. Chem. 2002;277:15147–15161. doi: 10.1074/jbc.M112338200. [DOI] [PubMed] [Google Scholar]
- 218.Baliga B. C., Colussi P. A., Read S. H., Dias M. M., Jans D. A., Kumar S. Role of prodomain in importin-mediated nuclear localization and activation of caspase-2. J. Biol. Chem. 2003;278:4899–4905. doi: 10.1074/jbc.M211512200. [DOI] [PubMed] [Google Scholar]
- 219.Thome M., Schneider P., Hofmann K., Fickenscher H., Meinl E., Neipel F., Mattmann C., Burns K., Bodmer J. L., Schroter M., et al. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature (London) 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 220.Bertin J., Armstrong R. C., Ottilie S., Martin D. A., Wang Y., Banks S., Wang G. H., Senkevich T. G., Alnemri E. S., Moss B., et al. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hu S., Vincenz C., Buller M., Dixit V. M. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J. Biol. Chem. 1997;272:9621–9624. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 222.Humke E. W., Shriver S. K., Starovasnik M. A., Fairbrother W. J., Dixit V. M. ICEBERG: A novel inhibitor of interleukin-1β generation. Cell. 2000;103:99–111. doi: 10.1016/s0092-8674(00)00108-2. [DOI] [PubMed] [Google Scholar]
- 223.Bode W., Huber R. Structural basis of the endoproteinase-protein inhibitor interaction. Biochim. Biophys. Acta. 2000;1477:241–252. doi: 10.1016/s0167-4838(99)00276-9. [DOI] [PubMed] [Google Scholar]
- 224.Stennicke H. R., Ryan C. A., Salvesen G. S. Reprieval from execution: The molecular basis of caspase inhibition. Trends Biochem. Sci. 2002;27:94–101. doi: 10.1016/s0968-0004(01)02045-x. [DOI] [PubMed] [Google Scholar]
- 225.Gettins P. G. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 226.Ye S., Cech A. L., Belmares R., Bergstrom R. C., Tong Y., Corey D. R., Kanost M. R., Goldsmith E. J. The structure of a Michaelis serpin-protease complex. Nat. Struct. Biol. 2001;8:979–983. doi: 10.1038/nsb1101-979. [DOI] [PubMed] [Google Scholar]
- 227.Huntington J. A., Read R. J., Carrell R. W. Structure of a serpin-protease complex shows inhibition by deformation. Nature (London) 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 228.Ray C. A., Black R. A., Kronheim S. R., Greenstreet T. A., Sleath P. R., Salvesen G. S., Pickup D. J. Viral inhibition of inflammation: Cowpox virus encodes an inhibitor of the interleukin-1β converting enzyme. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 229.Quan L. T., Caputo A., Bleackley R. C., Pickup D. J., Salvesen G. S. Granzyme B is inhibited by the cowpox virus serpin cytokine response modifier A. J. Biol. Chem. 1995;270:10377–10379. doi: 10.1074/jbc.270.18.10377. [DOI] [PubMed] [Google Scholar]
- 230.Zhou Q., Snipas S., Orth K., Muzio M., Dixit V. M., Salvesen G. S. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J. Biol. Chem. 1997;272:7797–7800. doi: 10.1074/jbc.272.12.7797. [DOI] [PubMed] [Google Scholar]
- 231.Ryan C. A., Stennicke H. R., Nava V. E., Burch J. B., Hardwick J. M., Salvesen G. S. Inhibitor specificity of recombinant and endogenous caspase-9. Biochem. J. 2002;366:595–601. doi: 10.1042/BJ20020863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Renatus M., Zhou Q., Stennicke H. R., Snipas S. J., Turk D., Bankston L. A., Liddington R. C., Salvesen G. S. Crystal structure of the apoptotic suppressor Crma in its cleaved form. Struct. Fold. Des. 2000;8:789–797. doi: 10.1016/s0969-2126(00)00165-9. [DOI] [PubMed] [Google Scholar]
- 233.Simonovic M., Gettins P. G. W., Volz K. Crystal structure of viral serpin Crma provides insights into its mechanism of cysteine proteinase inhibition. Protein Sci. 2000;9:1423–1427. doi: 10.1110/ps.9.8.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Bird C. H., Sutton V. R., Sun J., Hirst C. E., Novak A., Kumar S., Trapani J. A., Bird P. I. Selective regulation of apoptosis: The cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol. Cell. Biol. 1998;18:6387–6398. doi: 10.1128/mcb.18.11.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Clem R. J., Fechheimer M., Miller L. K. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 236.Bump N. J., Hackett M., Hugunin M., Seshagiri S., Brady K., Chen P., Ferenz C., Franklin S., Ghayur T., Li P., et al. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 237.Xue D., Horvitz H. R. Inhibition of the Caenorhabditis elegans cell-death protease CED-3 by a CED-3 cleavage site in baculovirus p35 protein. Nature (London) 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 238.Zhou Q., Krebs J. F., Snipas S. J., Price A., Alnemri E. S., Tomaselli K. J., Salvesen G. S. Interaction of the baculovirus anti-apoptotic protein p35 with caspases: Specificity, kinetics, and characterization of the caspase/p35 complex. Biochemistry. 1998;37:10757–10765. doi: 10.1021/bi980893w. [DOI] [PubMed] [Google Scholar]
- 239.Fisher A. J., Cruz W., Zoog S. J., Schneider C. L., Friesen P. D. Crystal structure of baculovirus p35: Role of a novel reactive site loop in apoptotic caspase inhibition. EMBO J. 1999;18:2031–2039. doi: 10.1093/emboj/18.8.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Xu G., Cirilli M., Huang Y., Rich R. L., Myszka D. G., Wu H. Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex. Nature (London) 2001;410:494–497. doi: 10.1038/35068604. [DOI] [PubMed] [Google Scholar]
- 241.Xu G., Rich R. L., Steegborn C., Min T., Huang Y., Myszka D. G., Wu H. Mutational analyses of the p35-caspase interaction. A bowstring kinetic model of caspase inhibition by p35. J. Biol. Chem. 2003;278:5455–5461. doi: 10.1074/jbc.M211607200. [DOI] [PubMed] [Google Scholar]
- 242.Jabbour A. M., Ekert P. G., Coulson E. J., Knight M. J., Ashley D. M., Hawkins C. J. The p35 relative, p49, inhibits mammalian and Drosophila caspases including DRONC and protects against apoptosis. Cell Death Differ. 2002;9:1311–1320. doi: 10.1038/sj.cdd.4401135. [DOI] [PubMed] [Google Scholar]
- 243.Zoog S. J., Schiller J. J., Wetter J. A., Chejanovsky N., Friesen P. D. Baculovirus apoptotic suppressor p49 is a substrate inhibitor of initiator caspases resistant to p35 in vivo. EMBO J. 2002;21:5130–5140. doi: 10.1093/emboj/cdf520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Crook N. E., Clem R. J., Miller L. K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Roy N., Mahadevan M. S., McLean M., Shutler G., Yaraghi Z., Farahani R., Baird S., Besner-Johnston A., Lefebvre C., Kang X., et al. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 246.Duckett C. S., Nava V. E., Gedrich R. W., Clem R. J., Van Dongen J. L., Gilfillan M. C., Shiels H., Hardwick J. M., Thompson C. B. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- 247.Uren A. G., Pakusch M., Hawkins C. J., Puls K. L., Vaux D. L. Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4974–4978. doi: 10.1073/pnas.93.10.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Rothe M., Pan M. G., Henzel W. J., Ayres T. M., Goeddel D. V. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 249.Deveraux Q. L., Reed J. C. IAP family proteins – suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 250.Fesik S. W., Shi Y. Structural biology. Controlling the caspases. Science. 2001;294:1477–1478. doi: 10.1126/science.1062236. [DOI] [PubMed] [Google Scholar]
- 251.Salvesen G. S., Duckett C. S. IAP proteins: Blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 252.Takahashi R., Deveraux Q., Tamm I., Welsh K., Assa-Munt N., Salvesen G. S., Reed J. C. A single BIR domain of XIAP sufficient for inhibiting caspases. J. Biol. Chem. 1998;273:7787–7790. doi: 10.1074/jbc.273.14.7787. [DOI] [PubMed] [Google Scholar]
- 253.Deveraux Q. L., Leo E., Stennicke H. R., Welsh K., Salvesen G. S., Reed J. C. Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J. 1999;18:5242–5251. doi: 10.1093/emboj/18.19.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sun C., Cai M., Meadows R. P., Xu N., Gunasekera A. H., Herrmann J., Wu J. C., Fesik S. W. NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP. J. Biol. Chem. 2000;275:33777–33781. doi: 10.1074/jbc.M006226200. [DOI] [PubMed] [Google Scholar]
- 255.Riedl S. J., Renatus M., Schwarzenbacher R., Zhou Q., Sun C., Fesik S. W., Liddington R. C., Salvesen G. S. Structural basis for the inhibition of caspase-3 by XIAP. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- 256.Sun C., Cai M., Gunasekera A. H., Meadows R. P., Wang H., Chen J., Zhang H., Wu W., Xu N., Ng S. C., et al. NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature (London) 1999;401:818–822. doi: 10.1038/44617. [DOI] [PubMed] [Google Scholar]
- 257.Deveraux Q. L., Takahashi R., Salvesen G. S., Reed J. C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature (London) 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 258.Silke J., Hawkins C. J., Ekert P. G., Chew J., Day C. L., Pakusch M., Verhagen A. M., Vaux D. L. The anti-apoptotic activity of XIAP is retained upon mutation of both the caspase 3- and caspase 9-interacting sites. J. Cell Biol. 2002;157:115–124. doi: 10.1083/jcb.200108085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Bratton S. B., Lewis J., Butterworth M., Duckett C. S., Cohen G. M. XIAP inhibition of caspase-3 preserves its association with the Apaf-1 apoptosome and prevents CD95- and Bax-induced apoptosis. Cell Death Differ. 2002;9:881–892. doi: 10.1038/sj.cdd.4401069. [DOI] [PubMed] [Google Scholar]
- 260.Wu J. W., Cocina A. E., Chai J., Hay B. A., Shi Y. Structural analysis of a functional DIAP1 fragment bound to Grim and Hid peptides. Mol. Cell. 2001;8:95–104. doi: 10.1016/s1097-2765(01)00282-9. [DOI] [PubMed] [Google Scholar]
- 261.Chai J., Yan N., Huh J. R., Wu J. W., Li W., Hay B. A., Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent DRONC ubiquitination. Nat. Struct. Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- 262.Zou H., Yang R. M., Hao J. S., Wang J., Sun C. H., Fesik S. W., Wu J. C., Tomaselli K. J., Armstrong R. C. Regulation of the Apaf-1/caspase-9 apoptosome by caspase-3 and XIAP. J. Biol. Chem. 2003;278:8091–8098. doi: 10.1074/jbc.M204783200. [DOI] [PubMed] [Google Scholar]
- 263.Johnson D. E., Gastman B. R., Wieckowski E., Wang G. Q., Amoscato A., Delach S. M., Rabinowich H. Inhibitor of apoptosis protein hILP undergoes caspase-mediated cleavage during T lymphocyte apoptosis. Cancer Res. 2000;60:1818–1823. [PubMed] [Google Scholar]
- 264.Uren A. G., Coulson E. J., Vaux D. L. Conservation of baculovirus inhibitor of apoptosis repeat proteins (BIRPs) in viruses, nematodes, vertebrates and yeasts. Trends Biochem. Sci. 1998;23:159–162. doi: 10.1016/s0968-0004(98)01198-0. [DOI] [PubMed] [Google Scholar]
- 265.Huang H., Joazeiro C. A., Bonfoco E., Kamada S., Leverson J. D., Hunter T. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 2000;275:26661–26664. doi: 10.1074/jbc.C000199200. [DOI] [PubMed] [Google Scholar]
- 266.White K., Grether M. E., Abrams J. M., Young L., Farrell K., Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 267.Tenev T., Zachariou A., Wilson R., Paul A., Meier P. Jafrac2 is an IAP antagonist that promotes cell death by liberating Dronc from DIAP1. EMBO J. 2002;21:5118–5129. doi: 10.1093/emboj/cdf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Abrams J. M. Competition and compensation: Coupled to death in development and cancer. Cell. 2002;110:403–406. doi: 10.1016/s0092-8674(02)00904-2. [DOI] [PubMed] [Google Scholar]
- 269.Liston P., Fong W. G., Korneluk R. G. The inhibitors of apoptosis: There is more to life than Bcl2. Oncogene. 2003;22:8568–8580. doi: 10.1038/sj.onc.1207101. [DOI] [PubMed] [Google Scholar]
- 270.Du C., Fang M., Li Y., Li L., Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 271.Verhagen A. M., Ekert P. G., Pakusch M., Silke J., Connolly L. M., Reid G. E., Moritz R. L., Simpson R. J., Vaux D. L. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 272.Deng Y., Lin Y., Wu X. TRAIL-induced apoptosis requires Bax-dependent mitochondrial release of Smac/DIABLO. Genes Dev. 2002;16:33–45. doi: 10.1101/gad.949602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Verhagen A. M., Silke J., Ekert P. G., Pakusch M., Kaufmann H., Connolly L. M., Day C. L., Tikoo A., Burke R., Wrobel C., et al. HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J. Biol. Chem. 2002;277:445–454. doi: 10.1074/jbc.M109891200. [DOI] [PubMed] [Google Scholar]
- 274.Hegde R., Srinivasula S. M., Zhang Z., Wassell R., Mukattash R., Cilenti L., DuBois G., Lazebnik Y., Zervos A. S., Fernandes-Alnemri T., et al. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J. Biol. Chem. 2002;277:432–438. doi: 10.1074/jbc.M109721200. [DOI] [PubMed] [Google Scholar]
- 275.Martins L. M., Iaccarino I., Tenev T., Gschmeissner S., Totty N. F., Lemoine N. R., Savopoulos J., Gray C. W., Creasy C. L., Dingwall C., et al. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a Reaper-like motif. J. Biol. Chem. 2002;277:439–444. doi: 10.1074/jbc.M109784200. [DOI] [PubMed] [Google Scholar]
- 276.Suzuki Y., Imai Y., Nakayama H., Takahashi K., Takio K., Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- 277.Liu Z., Sun C., Olejniczak E. T., Meadows R. P., Betz S. F., Oost T., Herrmann J., Wu J. C., Fesik S. W. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature (London) 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- 278.Wu G., Chai J., Suber T. L., Wu J. W., Du C., Wang X., Shi Y. Structural basis of IAP recognition by Smac/DIABLO. Nature (London) 2000;408:1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- 279.Chai J., Du C., Wu J. W., Kyin S., Wang X., Shi Y. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature (London) 2000;406:855–862. doi: 10.1038/35022514. [DOI] [PubMed] [Google Scholar]
- 280.Roberts D. L., Merrison W., MacFarlane M., Cohen G. M. The inhibitor of apoptosis protein-binding domain of Smac is not essential for its proapoptotic activity. J. Cell Biol. 2001;153:221–228. doi: 10.1083/jcb.153.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Huang Y., Rich R. L., Myszka D. G., Wu H. Requirement of both the second and third BIR domains for the relief of X-linked inhibitor of apoptosis protein (XIAP)-mediated caspase inhibition by Smac. J. Biol. Chem. 2003;278:49517–49522. doi: 10.1074/jbc.M310061200. [DOI] [PubMed] [Google Scholar]
- 282.Yang Q. H., Church-Hajduk R., Ren J., Newton M. L., Du C. Omi/HtrA2 catalytic cleavage of inhibitor of apoptosis (IAP) irreversibly inactivates IAPs and facilitates caspase activity in apoptosis. Genes Dev. 2003;17:1487–1496. doi: 10.1101/gad.1097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Srinivasula S. M., Gupta S., Datta P., Zhang Z. J., Hegde R., Cheong N. E., Fernandes-Alnemri T., Alnemri E. S. Inhibitor of apoptosis proteins are substrates for the mitochondrial serine protease Omi/HtrA2. J. Biol. Chem. 2003;278:31469–31472. doi: 10.1074/jbc.C300240200. [DOI] [PubMed] [Google Scholar]
- 284.Shu H. B., Takeuchi M., Goeddel D. V. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13973–13978. doi: 10.1073/pnas.93.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Yang Y., Fang S., Jensen J. P., Weissman A. M., Ashwell J. D. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 286.MacFarlane M., Merrison W., Bratton S. B., Cohen G. M. Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro. J. Biol. Chem. 2002;277:36611–36616. doi: 10.1074/jbc.M200317200. [DOI] [PubMed] [Google Scholar]
- 287.Ditzel M., Wilson R., Tenev T., Zachariou A., Paul A., Deas E., Meier P. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 288.Hu S., Yang X. Cellular inhibitor of apoptosis 1 and 2 are ubiquitin ligases for the apoptosis inducer Smac/DIABLO. J. Biol. Chem. 2003;278:10055–10060. doi: 10.1074/jbc.M207197200. [DOI] [PubMed] [Google Scholar]
- 289.Olson M. R., Holley C. L., Yoo S. J., Huh J. R., Hay B. A., Kornbluth S. Reaper is regulated by IAP-mediated ubiquitination. J. Biol. Chem. 2003;278:4028–4034. doi: 10.1074/jbc.M209734200. [DOI] [PubMed] [Google Scholar]
- 290.Li X., Yang Y., Ashwell J. D. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature (London) 2002;416:345–347. doi: 10.1038/416345a. [DOI] [PubMed] [Google Scholar]
- 291.Shin H., Okada K., Wilkinson J. C., Solomon K. M., Duckett C. S., Reed J. C., Salvesen G. S. Identification of ubiquitination sites on the X-linked inhibitor of apoptosis protein. Biochem. J. 2003;373:965–971. doi: 10.1042/BJ20030583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Lippens S., Kockx M., Knaapen M., Mortier L., Polakowska R., Verheyen A., Garmyn M., Zwijsen A., Formstecher P., Huylebroeck D., et al. Epidermal differentiation does not involve the pro-apoptotic executioner caspases, but is associated with caspase-14 induction and processing. Cell Death Differ. 2000;7:1218–1224. doi: 10.1038/sj.cdd.4400785. [DOI] [PubMed] [Google Scholar]
- 293.Eckhart L., Ban J., Fischer H., Tschachler E. Caspase-14: Analysis of gene structure and mRNA expression during keratinocyte differentiation. Biochem. Biophys. Res. Commun. 2000;277:655–659. doi: 10.1006/bbrc.2000.3698. [DOI] [PubMed] [Google Scholar]
- 294.Chien A. J., Presland R. B., Kuechle M. K. Processing of native caspase-14 occurs at an atypical cleavage site in normal epidermal differentiation. Biochem. Biophys. Res. Commun. 2002;296:911–917. doi: 10.1016/s0006-291x(02)02015-6. [DOI] [PubMed] [Google Scholar]
- 295.Mikolajczyk J., Scott F. L., Krajewski S., Sutherlin D. P., Salvesen G. S. Activation and substrate specificity of caspase-14. Biochemistry. 2004;43:10560–10569. doi: 10.1021/bi0498048. [DOI] [PubMed] [Google Scholar]
- 296.Kischkel F. C., Lawrence D. A., Tinel A., LeBlanc H., Virmani A., Schow P., Gazdar A., Blenis J., Arnott D., Ashkenazi A. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J. Biol. Chem. 2001;276:46639–46646. doi: 10.1074/jbc.M105102200. [DOI] [PubMed] [Google Scholar]
- 297.Ashkenazi A., Dixit V. M. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 298.Green D. R., Reed J. C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 299.Vucic D., Deshayes K., Ackerly H., Pisabarro M. T., Kadkhodayan S., Fairbrother W. J., Dixit V. M. SMAC negatively regulates the anti-apoptotic activity of melanoma inhibitor of apoptosis (ML-IAP) J. Biol. Chem. 2002;277:12275–12279. doi: 10.1074/jbc.M112045200. [DOI] [PubMed] [Google Scholar]
- 300.Okamoto Y., Anan H., Nakai E., Morihira K., Yonetoku Y., Kurihara H., Sakashita H., Terai Y., Takeuchi M., Shibanuma T., et al. Peptide based interleukin-1β converting enzyme (ICE) inhibitors: synthesis, structure activity relationships and crystallographic study of the ICE-inhibitor complex. Chem. Pharm. Bull. 1999;47:11–21. doi: 10.1248/cpb.47.11. [DOI] [PubMed] [Google Scholar]
- 301.Romanowski M. J., Scheer J. M., O'Brien T., McDowell R. S. Crystal structures of a ligand-free and malonate-bound human caspase-1: implications for the mechanism of substrate binding. Structure. 2004;12:1361–1371. doi: 10.1016/j.str.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 302.Lee D., Long S. A., Adams J. L., Chan G., Vaidya K. S., Francis T. A., Kikly K., Winkler J. D., Sung C. M., Debouck C., et al. Potent and selective nonpeptide inhibitors of caspases 3 and 7 inhibit apoptosis and maintain cell functionality. J. Biol. Chem. 2000;275:16007–16014. doi: 10.1074/jbc.275.21.16007. [DOI] [PubMed] [Google Scholar]
- 303.Erlanson D. A., Lam J. W., Wiesmann C., Luong T. N., Simmons R. L., DeLano W. L., Choong I. C., Burdett M. T., Flanagan W. M., Lee D., et al. In situ assembly of enzyme inhibitors using extended tethering. Nat. Biotechnol. 2003;21:308–314. doi: 10.1038/nbt786. [DOI] [PubMed] [Google Scholar]
- 304.Forsyth C. M., Lemongello D., LaCount D. J., Friesen P. D., Fisher A. J. Crystal structure of an invertebrate caspase. J. Biol. Chem. 2004;279:7001–7008. doi: 10.1074/jbc.M312472200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.