Abstract
The yeast SNARE (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor) protein Ykt6 was shown to mediate palmitoylation of the fusion factor Vac8 in a reaction essential for the fusion of vacuoles. Here I present evidence that hYkt6 (human Ykt6) has self-palmitoylating activity. Incubation of recombinant hYkt6 with [3H]Pal-CoA ([3H]palmitoyl-CoA) leads to covalent attachment of palmitate to C-terminal cysteine residues. The N-terminal domain of human Ykt6 contains a Pal-CoA binding site and is required for the reaction.
Keywords: longin, palmitoylation, palmitoyl-CoA, soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE) protein, vesicular trafficking, Ykt6
Abbreviations: (v-)SNARE, (vesicular) soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor; Br-Pal, 2-bromopalmitate; DHHC-CRD proteins, polytopic membrane proteins with the sequence DHHC and a cysteine-rich domain; DTT, dithiothreitol; F42E, Phe42→Glu mutation; hYkt6, human Ykt6; MEGA-8, octanoyl N-methylglucamide; NSF, N-ethylmaleimide-sensitive factor; Pal-CoA, palmitoyl-CoA
INTRODUCTION
Palmitoylation is the attachment of fatty acids to cysteine residues of membrane proteins. The fatty acids can mediate protein–lipid interactions, thereby targeting the modified protein to membranes or membrane subdomains. In contrast with other hydrophobic modifications, i.e. myristoylation or isoprenylation, palmitoylation is often a dynamic event, with cycles of acylation and deacylation [1]. The enzymology of protein palmitoylation is poorly understood. Several enzymic activities have been characterized, but none of them was purified to homogeneity [2–4]. It has even been questioned whether enzymes were required for palmitoylation. Several proteins can be palmitoylated in vitro by simply incubating them with Pal-CoA (palmitoyl-CoA), which serves as an acyl donor in the palmitoylation reaction [5–7]. This non-enzymic palmitoylation often occurs on the same cysteine residues that are targets for palmitoylation in vivo. Recently, progress in this field was obtained in yeast with a genetic screen. The family of DHHC-CRD proteins (polytopic membrane proteins with the sequence DHHC and a cysteine-rich domain) has been presented as potential acyltransferases for proteins with a C-terminal CAAX box [8,9].
Pal-CoA is furthermore of interest because it has a stimulatory effect on vesicular transport [10,11]. This effect was attributed to protein palmitoylation, but only recently did myself and others identify the fusion factor Vac8 as the first substrate of palmitoylation required for a vesicular trafficking reaction, namely the fusion of yeast vacuoles [12]. Subsequent studies showed that a palmitoylating activity is present on vacuoles [13], and this was subsequently identified as Ykt6 [14].
Ykt6 belongs to the family of SNAREs (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptors), which are essential for vesicular trafficking in yeast and higher organisms. Fusion of vesicles with the membrane is driven by complex formation of cognate SNARE proteins [15,16]. Ykt6, the most conserved SNARE protein, is unique because it is present in the cytosol as well as on membranes. It is required for multiple transport steps, e.g. trafficking to and within the Golgi, to the yeast vacuole and for vacuole fusion. Ykt6 has a wide tissue distribution, and appears to have a specialized function in neuronal cells [17–20].
Ykt6 consists of the C-terminal SNARE motif and a regulatory N-terminal domain, which classifies Ykt6 as a member of the longin group of v (vesicular)-SNAREs [21]. The crystal structure of the N-terminal domain of Ykt6 shows the unusual hydrophobic surface that has been proposed to fold back and interact with the SNARE motif, thereby preventing its participation in SNARE complex formation [22].
In contrast with most other SNARE proteins, Ykt6 does not contain a transmembrane region, but a C-terminal CAAX box which is predicted to be farnesylated [23]. However, farnesylation alone is not sufficient to confer membrane binding of intrinsically hydrophilic proteins. In the case of the Ras protein, which is best characterized in this regard, additional sequences located in the vicinity of the CAAX-box are required for stable membrane anchorage. These are either basic amino acids or one or more cysteine residues, which are reversibly palmitoylated. Farnesylation at the CAAX box occurs co-translationally and is followed by proteolysis of AAX, carboxymethylation and palmitoylation [24,25].
Stimulated by the observation that Ykt6 also contains a conserved cysteine residue in the vicinity of the CAAX box and by the described palmitoylating activity of the protein, I have analysed whether hYkt6 (human Ykt6) might catalyse its own palmitoylation.
METHODS
Molecular biology
The hYkt6 gene was obtained from Dr James McNew (Department of Biochemistry and Cell Biology, Rice University, Houston, TX, U.S.A.). It was amplified by PCR using an N-terminal primer equipped with a SacI site and a C-terminal primer equipped with a PstI site. The N-terminal domain of hYkt6 was created with a C-terminal primer in which a stop codon was inserted after Thr140. The cysteine mutants were constructed with PCR using C-terminal primers to convert Cys194, Cys195, or both, into serine. The PCR products were cloned into the plasmid pQE30, resulting in proteins with the amino acids MRGSHHHHHHGSACE upstream of the start codon of hYkt6. The C-terminal domain of hYkt6 was created with an N-terminal primer equipped with a BamHI site and a C-terminal primer equipped with an XhoI site. The PCR product was cloned into plasmid pGEX-6P-1. Expression of the SNARE domain started at Ala136. The F42E (Phe42→Glu) mutant of hYkt6 was constructed with the QuikChange® Site-Directed Mutagenesis Kit (Stratagene).
Purification of hYkt6
PQE plasmids were transformed into M15(pREP4) cells (Qiagen) and pGEX-plasmids into BL-21 cells. Purification of hYkt6 on Ni-NTA (Ni2+-nitrilotriacetate)–agarose or glutathione–Sepharose was performed as described in [7], with the following buffers: sonification buffer (50 mM Tris/HCl/300 mM NaCl/1 mM PMSF), wash buffer (50 mM Tris/HCl/300 mM NaCl/20 mM imidazole) and elution buffer (20 mM Tris/HCl/300 mM NaCl, containing 250 mM imidazole or 10 mM glutathione). Proteins were desalted using PD-10 columns (Pharmacia) with 20 mM Tris/HCl (pH 8.4)/120 mM NaCl or 20 mM Pipes/KOH (pH 6.8)/120 mM KCl as eluent.
Affinity chromatography
A 100 μl portion of immobilized Cibacron Blue F3GA (Pierce) or Pal-CoA–agarose (Sigma) were equilibrated with 20 mM Pipes/KOH (pH 6.8)/120 mM KCl. The beads were then incubated with 40 μg of hYkt6 for 1 h at 4 °C. Beads with bound protein were pelleted (3 min, 500 g) and the supernatant (containing unbound proteins) was removed. Beads with bound proteins were then resuspended in 20 mM Pipes/KOH (pH 6.8)/120 mM KCl, again pelleted, and the supernatant was removed (wash fractions). Washing of the beads was repeated eight times. The pelleted beads were then incubated with different CoA compounds, as indicated, for 15 min at 4 °C. Beads were yet again pelleted, and the supernatant (eluate) was removed. Elution with CoA compounds was repeated once. Comparable aliquots of the input material, of the unbound proteins, of several wash fractions and of the eluates were analysed by SDS/PAGE and silver staining.
Palmitoylation assay
[3H]Pal-CoA, prepared as described in [6], was dried in a Savant SpeedVac vacuum concentrator and dissolved in 20 mM Pipes (pH 6.8)/120 mM KCl/0.1% Triton to yield a final sp. radioactivity of 100000 c.p.m./μl. The standard palmitoylation reaction mixture contained hYkt6 (20 μl, 8 μg, 3.2 μM), 10 μl of [3H]Pal-CoA (10 pmol, 100 nM), 1 mM DTT (dithiothreitol) in a final volume of 100 μl of Tris/NaCl buffer [20 mM Tris/HCl (pH 8.5)/120 mM NaCl] and was incubated for 60 min at 30 °C. Deviations from these standard conditions are indicated in the Figure legends. A 1 ml portion of chloroform/methanol (1:1, v/v) was added to the samples, and precipitated proteins were pelleted (15 min, 14000 g). Pellets were washed with 1 ml of methanol, air-dried, then resuspended in 20 μl of non-reducing SDS/PAGE sample buffer and boiled for 5 min. Samples were subjected to SDS/PAGE, and the gels were then stained with Coomassie Brilliant Blue and subjected to fluorography [7].
RESULTS AND DISCUSSION
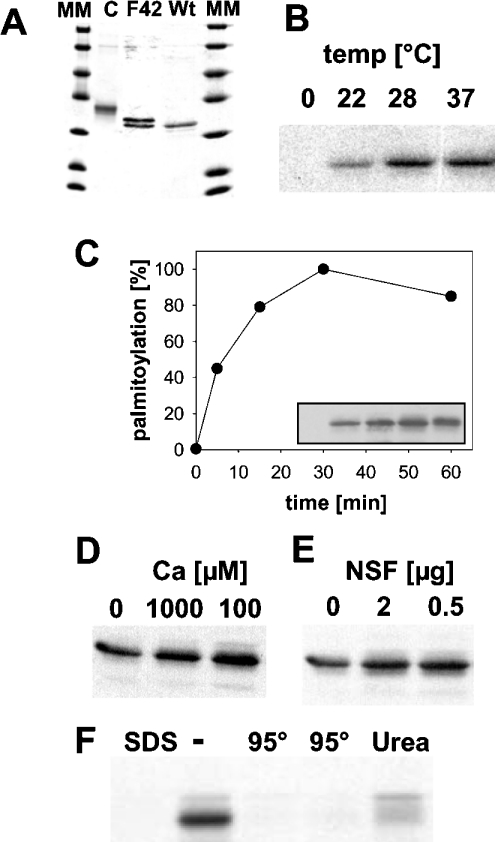
His6-tagged hYkt6 purified from Escherichia coli (Figure 1A) was incubated with [3H]Pal-CoA. Samples were then extracted with chloroform/methanol and were subjected to SDS/PAGE and fluorography. Palmitoylation of hYkt6 does not occur on ice, but requires physiological temperatures (Figure 1B). Acylation proceeds rapidly, with 50% of total hYkt6 acylation being completed after 10 min of labelling (Figure 1C). Several compounds were tested to see whether they could stimulate the palmitoylation reaction. Neither calcium (Figure 1D) nor NSF (N-ethylmaleimide-sensitive factor) (Figure 1E), regulators of many SNARE-dependent fusion events, had a significant effect on palmitoylation of hYkt6. Heating of hYkt6, as well as inclusion of SDS and urea in the reaction buffer, inhibits palmitoylation, which is evidence for the specificity of the reaction (Figure 1F). Liquid-scintillation counting of the radioactivty in excised bands revealed that approx. 1% of the Ykt6 molecules were acylated under optimal conditions. This might indicate that more efficent palmitoylation requires cofactors, e.g. lipids or other proteins, which would stimulate the reaction in vivo. Substoichiometric acylation of their substrate proteins were also reported for members of the DHHC-CRD family [8,9] and for other in vitro palmitoylation reactions [2–4].
Figure 1. hYkt6 is palmitoylated in vitro.
(A) Coomassie Blue-stained gel with recombinant His6-hYkt6 wild-type (Wt), His6-[F42E]Ykt6 (F42) and GST–hYkt6–C-terminal domain (C) purified from E. coli. MM, molecular-mass (14.4, 18.4, 25, 35, 45, 66 and 116 kDa) markers. (B) hYkt6 (32 μM) was incubated with [3H]Pal-CoA (100 nM) for 60 min at the indicated temperatures. Samples were then extracted with chloroform/methanol and subjected to SDS/PAGE and fluorography. (C) hYkt6 was incubated with [3H]Pal-CoA at 30 °C, aliquots were removed immediately or 5, 15, 30 and 60 min later. The absorbances of the bands in the resulting fluorogram (inset) were quantified. Relative palmitoylation is plotted against the period of incubation. (D) CaCl2 at the indicated concentration was added to the palmitoylation reaction. (E) Purified NSF (0, 2 or 0.5 μg) was added to the palmitoylation reaction. (F) SDS (0.1%) or urea (7 M) was added to the palmitoylation reaction. Two lanes, marked with 95° contained Ykt6 boiled for 2 (left lane) or 5 min (right lane) prior to starting the palmitoylation reaction.
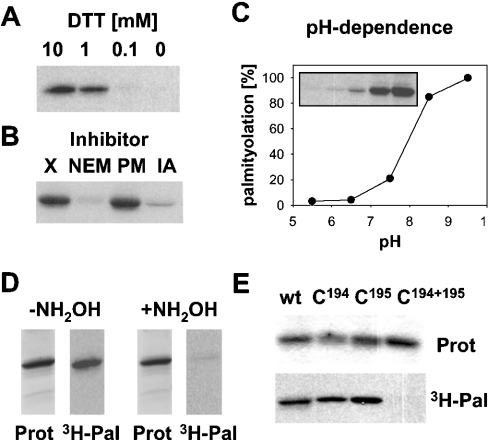
The next set of experiments was performed to identify the palmitoylation site of hYkt6. Palmitoylation of proteins occurs on cysteine residues, which must be present in their reduced form to act as an acceptor for the fatty acid. Omission of DTT from the reaction buffer inhibited palmitoylation of hYkt6, indicating that free cysteine residues are required (Figure 2A). Likewise, adding N-ethylmaleimide and iodoacetamide, which covalently react with free cysteine residues, completely blocked palmitoylation of hYkt6 (Figure 2B). Palmitoylation of hYkt6 has a pH optimum at a slightly basic pH (Figure 2C). Acyl-chain transfer likely occurs through nucleophilic attack of the fatty acid binding site in the protein on the carbonyl of the C–S bond in the Pal-CoA molecule [27]. To act as good nucleophile, the fatty acid acceptor should be present in a deprotonated form. The pH optimum of the palmitoylation reaction corresponds to the pKa of cysteine (pH 8.5), implicating this amino acid as a fatty acid acceptor. Furthermore, the fatty acid bond in hYkt6 can be cleaved with hydroxylamine at neutral pH, indicating a thioester-type fatty acid linkage (Figure 2D). The hydroxylamine-sensitivity of the fatty acid bond, in combination with its resistance against extraction with chloroform/methanol, is also definitive proof for the covalent nature of the fatty acid attachment. Finally, the cysteine residue of the CAAX box (Cys195) and the upstream cysteine residue, Cys194, were replaced by serine residues and the hYkt6 mutants were analysed for palmitoylation. Replacement of Cys194 decreased acylation to 55%, whereas replacement of Cys195 had only a minor effect, with a 13% decrease in [3H]palmitate incorporation. Replacement of both cysteine residues simultaneously completely blocked palmitoylation (Figure 2E). Thus both cysteine residues in the C-terminus of hYkt6 are substrates for palmitoylation in vitro, but Cys194 is apparently the preferred site. Inside eukaryotic cells, Cys195 of the CAAX box is farnesylated and is therefore blocked for palmitoylation. However, it is not known whether prenylation occurs quantitatively or whether a subpopulation of hYkt6 molecules exist with a free cysteine residue at position 195 which would then be a potential substrate for palmitoylation. These experiments also show that farnesylation is not an absolute requirement for palmitoylation. Similar results have been reported for a mutant form of the Ras protein, which is palmitoylated without prior farnesylation [28].
Figure 2. Palmitoylation of hYkt6 occurs on Cys194 and Cys195.
(A) DTT at the indicated concentration was added to the palmitoylation reaction. (B) N-Ethylmaleimide (NEM, 5 mM), PMSF (PM, 1 mM) and iodoacetamide (IA, 1 mM) were added to the palmitoylation reaction. X, no additions. (C) Palmitoylation reactions took place in 20 mM Pipes/120 mM KCl, pH 5.5 or 6.5, or in 20 mM Tris/120 mM KCl at pH 7.5, 8.5 or 9.5. The resulting fluorogram (inset) was quantified. Relative palmitoylation was plotted against the pH of the reaction mixture. (D) Two identical palmitoylation reactions were subjected to SDS/PAGE and Coomassie Blue staining (Prot). One part of the gel was treated with 1 M hydroxylamine (+NH2OH), the other part with 1 M Tris (−NH2OH). Gels were then processed for fluorography (3H-Pal). (E) Wild-type Ykt6 and the cysteine→serine mutants (C194, C195 and C194+195) were incubated with [3H]Pal-CoA. Samples were then subjected to SDS/PAGE, Coomassie Blue staining (Prot) and fluorography (3H-Pal). Fluorograms from three experiments were quantified to determine acylation. The Cys194→Ser mutant showed 55%, and the Cys195→Ser mutant 87%, palmitoylation relative to the wild-type (wt) protein.
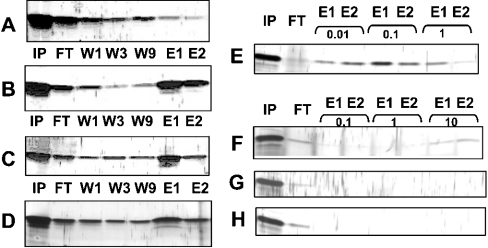
The following experiments were performed to analyse whether acylation of hYkt6 occurs by a spontaneous (chemical) or an enzymic (autocatalytic) mechanism. Assuming an enzymic mechanism, hYkt6 should contain a binding pocket for Pal-CoA at a site distinct from its palmitoylation site. Myself and others have recently demonstrated, with a filter-binding assay, that the N-terminal domain of yeast Ykt6 binds Pal-CoA [14]. hYkt6 protein shows chromatographic properties that are consistent with this observation. The N-terminal domain of hYkt6 binds efficiently to immobilized Cibacron Blue, an affinity medium for enzymes requiring adenine-containing cofactors. The protein can be eluted from the matrix with Pal-CoA (Figure 3B), but not with CoA (Figure 3A). Full-length hYkt6 shows the same chromatographic properties, indicating that the binding site is accessible in the complete protein (Figure 3C). The N-terminal domain also binds to Pal-CoA–agarose (Figure 3D). I then analysed which CoA compounds were able to elute the N-terminal domain from the Cibacron Blue matrix. Pal-CoA at a concentration of 10 μM did compete with hYkt6 for the binding site on Cibacron Blue (Figure 3E). In contrast, only traces of hYkt6 were eluted with 10 mM octanoyl-CoA (Figure 3F), but not with acetyl-CoA (Figure 3G) or CoA alone (Figure 3H). Thus only Pal-CoA binds to the N-terminal domain of hYkt6.
Figure 3. The N-terminal domain of hYkt6 contains a Pal-CoA binding site.
The N-terminal domain of hYkt6 or the full-length protein was added to immobilized Cibacron Blue or to Pal-CoA–agarose and an aliquot was immediately removed (input, IP). Samples were incubated for 1 h. Beads with bound proteins were then pelleted and the supernatant was removed (flowthrough, FT). Beads were resuspended in buffer, again pelleted, and the supernatant was removed (wash 1, W1). Washing of the beads was repeated eight times. The pelleted beads were then incubated with various CoA compounds for 15 min. Beads were pelleted and the supernatant was removed (eluate 1, E1). Elution was repeated once (eluate 2, E2). Comparable aliquots of input, flowthrough, wash 1, wash 3, wash 9 and from the eluates were subjected to SDS/PAGE and silver staining. The analysed protein, the material of the beads and the eluents were as follows: (A) binding of the N-terminal domain to Cibacron Blue and elution with CoA (1 mM); (B) binding of the N-terminal domain to Cibacron Blue and elution with Pal-CoA (1 mM); (C) Binding of the full-length protein to Cibacron Blue and elution with Pal-CoA (1 mM); (D) Binding of the N-terminal domain to Pal-CoA–agarose and elution with Pal-CoA (10 mM). (E–H) Binding of the N-terminal domain to Cibacron Blue, elution with Pal-CoA (E), octanoyl-CoA (F), acetyl-CoA (G) or CoA (H) at the concentrations in mM as indicated above the lanes.
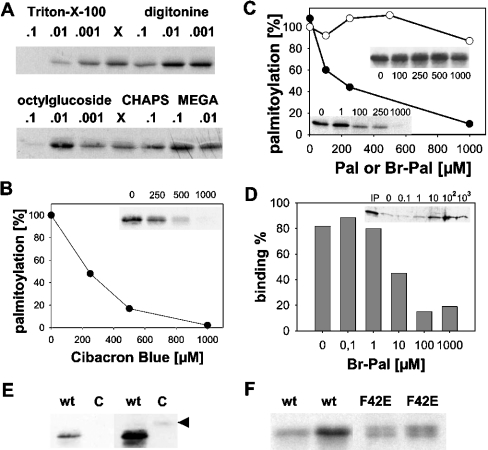
Next, the effect of various detergents on the palmitoylation reaction was tested. Addition of Triton X-100 and octyl glucoside was found to inhibit palmitoylation of hYkt6. Other detergents, such as digitonine, MEGA-8 (octanoyl N-methylglucamide) and CHAPS had no effect on, or did slightly stimulate, palmitoylation (Figure 4A). Whether a certain detergent inhibited the palmitoylation reaction was not dependent on whether it was present above or below its critical micellar concentration. Detergents with an inhibitory effect on the palmitoylation reaction contained a hydrophobic structure that resembles a fatty acid – an alkyl group in the case of octyl glucoside or a polyoxyethylene in the case of Triton. Thus inhibition of palmitoylation might be due to binding of those detergents to hydrophobic pockets on hYkt6, e.g. the Pal-CoA binding site.
Figure 4. Acylation of hYkt6 requires it N-terminal domain.
(A) Triton X-100, digitonine, octyl glucoside, CHAPS and MEGA-8 were added to the palmitoylation reaction at the indicated concentrations (%, v/v). X, no additions. (B) The palmitoylation reaction was incubated with soluble Cibacron Blue at a final concentration of 0, 250, 500 or 1000 μM. (C) The palmitoylation reaction was incubated with Br-Pal (● and lower left inset) at a final concentration of 0, 1, 100, 250 or 1000 μM. Palmitate (○ and upper right inset) added at a final concentration of 0, 100, 250, 500 or 1000 μM served as control. (D) hYkt6 (4 μg) was incubated with immobilized Cibacron Blue (30 μl) in the presence of 0, 0.1, 1, 10, 100 and 1000 μM Br-Pal. Cibacron Blue with bound hYkt6 was pelleted, the supernatant was precipitated with trichloracetic acid and analysed by SDS/PAGE and Coomassie Blue staining. IP, input material. (E) An 8 μg portion of GST–hYkt6–C-terminal domain (C) or full-length hYkt (wt) were incubated with [3H]Pal-CoA prior to SDS/PAGE and fluorography. A short (left) and a long (right) exposure of the same gel is shown. Arrowhead, GST–hYkt6–C-terminal domain. (F) Portions (2 and 4 μg) of wild-type hYkt6 (wt) and 8 and 16 μg of [F42E]hYkt6 (F42E) were incubated with [3H]Pal-CoA. Note that purified [F42E]hYkt6 consists of two bands (see Figure 1a) which are both acylated.
Blocking the Pal-CoA binding site more specifically with soluble Cibacron Blue completely blocked palmitoylation of hYkt6, with 50% inhibition at a concentration of 250 μM (Figure 4B). Likewise, adding Br-Pal (2-bromopalmitate), an established inhibitor of protein palmitoylation, also blocked acylation of Ykt6 at the same concentration. In contrast, palmitate had only a marginal effect on the palmitoylation reaction (Figure 4C). Inside cells, it is feasible that Br-Pal can be converted into a CoA derivative and subsequently transferred to proteins [13,29]. However, under the experimental conditions I used, activation of Br-Pal, and thus competition with [3H]Pal-CoA in a chemical acylation reaction, can be excluded. It is more likely that Br-Pal, but not palmitate, binds to the Pal-CoA binding site of hYkt6. Indeed, Br-Pal inhibited binding of hYkt6 to immobilized Cibacron Blue (Figure 4D). Furthermore, replacing the N-terminal domain of hYkt6 with glutathione S-transferase almost completely abolished palmitoylation (Figure 4E). Thus palmitoylation of hYkt6 is dependent on its N-terminal domain, which provides further evidence for an enzymic mechanism for the reaction.
My observation with purified hYkt6 is likely to be of physiological relevance. Recent results from Rothman and co-workers [30] showed that hYkt6 is palmitoylated at Cys194 inside cells. Palmitoylation confers stable membrane binding to hYkt6, by which vesicular transport through the Golgi is stimulated. The F42E mutant of hYkt6, representing the open conformation of hYkt, is the preferred substrate for palmitoylation in vivo. In my assay, [F42E]hYkt6 is also palmitoylated, but with slightly lower efficiency compared with the wild-type protein (Figure 4F). Thus enhanced palmitoylation of [F42E]hYkt6 in vivo is not an intrinsic property of the protein, but is rather dependent on other factors such as interacting proteins. Considering the participation of hYkt6 in many vesicular trafficking reactions it is important to identify components which regulate its autoacylation.
Acknowledgments
I am grateful to Professor Michael F.G. Schmidt (Institut fur Immunologie und Molekularbiologie, FB Veterinarmedizin, Freie Universitat Berlin, Berlin, Germany) for encouragement, to Dr James McNew (Rice University), Dr Jim Rothman and Dr Thomas Söllner (both at the Memorial Sloan-Kettering Cancer Center, New York, NY, U.S.A.) for providing the hYkt6 clone and to Dr Christian Ungermann [Biochemie-Zentrum Heidelberg (BZH), University of Heidelberg, Heidelberg, Germany] for providing comments on the manuscript before its submission. The excellent technical help of Ms Ellen Lyhs and Ms Ingrid Poese is acknowledged. The project was supported by the Deutsche Forschungsgemeinschaft (DFG) through grant Ve 141/5-1.
References
- 1.Bijlmakers M. J., Marsh M. The on-off story of protein palmitoylation. Trends Cell Biol. 2003;13:32–42. doi: 10.1016/s0962-8924(02)00008-9. [DOI] [PubMed] [Google Scholar]
- 2.Berger M., Schmidt M. F. Cell-free fatty acid acylation of Semliki Forest viral polypeptides with microsomal membranes from eukaryotic cells. J. Biol. Chem. 1984;259:7245–7252. [PubMed] [Google Scholar]
- 3.Berthiaume L., Resh M. D. Biochemical characterization of a palmitoyl acyltransferase activity that palmitoylates myristoylated proteins. J. Biol. Chem. 1995;270:22399–22405. doi: 10.1074/jbc.270.38.22399. [DOI] [PubMed] [Google Scholar]
- 4.Veit M., Sachs K., Heckelmann M., Maretzki D., Hofmann K. P., Schmidt M. F. Palmitoylation of rhodopsin with S-protein acyltransferase: enzyme catalyzed reaction versus autocatalytic acylation. Biochim. Biophys. Acta. 1998;1394:90–98. doi: 10.1016/s0005-2760(98)00097-6. [DOI] [PubMed] [Google Scholar]
- 5.Bano M. C., Jackson C. S., Magee A. I. Pseudo-enzymatic S-acylation of a myristoylated yes protein tyrosine kinase peptide in vitro may reflect non-enzymatic S-acylation in vivo. Biochem. J. 1998;330:723–731. doi: 10.1042/bj3300723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan J. A., Gilman A. G. Autoacylation of G protein α subunits. J. Biol. Chem. 1996;271:23594–23600. doi: 10.1074/jbc.271.38.23594. [DOI] [PubMed] [Google Scholar]
- 7.Veit M. Palmitoylation of the 25-kDa synaptosomal protein (SNAP-25) in vitro occurs in the absence of an enzyme, but is stimulated by binding to syntaxin. Biochem. J. 2000;345:145–151. [PMC free article] [PubMed] [Google Scholar]
- 8.Linder M. E., Deschenes R. J. Model organisms lead the way to protein palmitoyltransferases. J. Cell Sci. 2004;117:521–526. doi: 10.1242/jcs.00989. [DOI] [PubMed] [Google Scholar]
- 9.Roth A. F., Feng Y., Chen L., Davis N. G. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfanner N., Orci L., Glick B. S., Amherdt M., Arden S. R., Malhotra V., Rothman J. E. Fatty acyl-coenzyme A is required for budding of transport vesicles from Golgi cisternae. Cell. 1989;59:95–102. doi: 10.1016/0092-8674(89)90872-6. [DOI] [PubMed] [Google Scholar]
- 11.Pfanner N., Glick B. S., Arden S. R., Rothman J. E. Fatty acylation promotes fusion of transport vesicles with Golgi cisternae. J. Cell Biol. 1990;110:955–961. doi: 10.1083/jcb.110.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veit M., Laage R., Dietrich L., Wang L., Ungermann C. Vac8p release from the SNARE complex and its palmitoylation are coupled and essential for vacuole fusion. EMBO J. 2001;20:3145–3155. doi: 10.1093/emboj/20.12.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veit M., Dietrich L. E., Ungermann C. Biochemical characterization of the vacuolar palmitoyl acyltransferase. FEBS Lett. 2003;540:101–105. doi: 10.1016/s0014-5793(03)00232-1. [DOI] [PubMed] [Google Scholar]
- 14.Dietrich L. E., Gurezka R., Veit M., Ungermann C. The SNARE Ykt6 mediates protein palmitoylation during an early stage of homotypic vacuole fusion. EMBO J. 2004;23:45–53. doi: 10.1038/sj.emboj.7600015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jahn R., Sudhof T. C. Membrane fusion and exocytosis. Annu. Rev. Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- 16.Rothman J. E., Wieland F. T. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 17.Dilcher M., Kohler B., von Mollard G. F. Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6. J. Biol. Chem. 2001;276:34537–34544. doi: 10.1074/jbc.M101551200. [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa H., Zinsser S., Rhee Y., Vik-Mo E. O., Davanger S., Hay J. C. Mammalian ykt6 is a neuronal SNARE targeted to a specialized compartment by its profilin-like amino terminal domain. Mol. Biol. Cell. 2003;14:698–720. doi: 10.1091/mbc.E02-09-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNew J. A., Sogaard M., Lampen N. M., Machida S., Ye R. R., Lacomis L., Tempst P., Rothman J. E., Sollner T. H. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum–Golgi transport. J. Biol. Chem. 1997;272:17776–17783. doi: 10.1074/jbc.272.28.17776. [DOI] [PubMed] [Google Scholar]
- 20.Zhang T., Hong W. Ykt6 forms a SNARE complex with syntaxin 5, GS28, and Bet1 and participates in a late stage in endoplasmic reticulum–Golgi transport. J. Biol. Chem. 2001;276:27480–27487. doi: 10.1074/jbc.M102786200. [DOI] [PubMed] [Google Scholar]
- 21.Filippini F., Rossi V., Galli T., Budillon A., D'Urso M., D'Esposito M. Longins: a new evolutionary conserved VAMP family sharing a novel SNARE domain. Trends Biochem. Sci. 2001;26:407–409. doi: 10.1016/s0968-0004(01)01861-8. [DOI] [PubMed] [Google Scholar]
- 22.Tochio H., Tsui M. M., Banfield D. K., Zhang M. An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science. 2001;293:698–702. doi: 10.1126/science.1062950. [DOI] [PubMed] [Google Scholar]
- 23.McNew J. A., Sogaard M., Lampen N. M., Machida S., Ye R. R., Lacomis L., Tempst P., Rothman J. E., Sollner T. H. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum–Golgi transport. J. Biol. Chem. 1997;272:17776–17783. doi: 10.1074/jbc.272.28.17776. [DOI] [PubMed] [Google Scholar]
- 24.Berthiaume L. G. Insider information: how palmitoylation of Ras makes it a signaling double agent. Sci. STKE 2002. 2002:E41. doi: 10.1126/stke.2002.152.pe41. [DOI] [PubMed] [Google Scholar]
- 25.Magee T., Marshall C. New insights into the interaction of Ras with the plasma membrane. Cell. 1999;98:9–12. doi: 10.1016/S0092-8674(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 26.Reference deleted [Google Scholar]
- 27.Bizzozero O. A., Bixler H. A., Pastuszyn A. Structural determinants influencing the reaction of cysteine-containing peptides with palmitoyl-coenzyme A and other thioesters. Biochim. Biophys. Acta. 2001;1545:278–288. doi: 10.1016/s0167-4838(00)00291-0. [DOI] [PubMed] [Google Scholar]
- 28.Booden M. A., Baker T. L., Solski P. A., Der C. J., Punke S. G., Buss J. E. A non-farnesylated Ha-Ras protein can be palmitoylated and trigger potent differentiation and transformation. J. Biol. Chem. 1999;274:1423–1431. doi: 10.1074/jbc.274.3.1423. [DOI] [PubMed] [Google Scholar]
- 29.Webb Y., Hermida-Matsumoto L., Resh M. D. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J. Biol. Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 30.Fukasawa M., Varlamov O., Eng W. S., Sollner T. H., Rothman J. E. Localization and activity of the SNARE Ykt6 determined by its regulatory domain and palmitoylation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4815–4820. doi: 10.1073/pnas.0401183101. [DOI] [PMC free article] [PubMed] [Google Scholar]