Abstract
The effect of the length of ROCK (Rho-associated kinase) on its oligomerization state has been investigated by analysing full-length protein and four truncated constructs using light-scattering and analytical ultracentrifugation methods. Changes in size correlate with the kinetic properties of the kinase. Sedimentation velocity, sedimentation equilibrium and light-scattering data analyses revealed that protein constructs of size Ser6–Arg415 and larger exist predominantly as dimers, while smaller constructs are predominantly monomeric. The amino acid segments comprising residues 379–415 and 47–78 are shown to be necessary to maintain the dimeric ROCK structure. kcat values ranged from 0.7 to 2.1 s−1 and from 1.0 to 5.9 s−1 using ROCK peptide (KKRNRTLSV) and the 20000 Da subunit of myosin light chain respectively as substrate, indicating that the effect of the ROCK oligomerization state on the kcat is minor. Values of ATP Km for monomeric constructs were increased by 50–80-fold relative to the dimeric constructs, and Ki comparisons using the specific competitive ROCK inhibitor Y-27632 also showed increases of at least 120-fold, demonstrating significant perturbations in the ATP binding site. The corresponding Km values for the ROCK peptide and myosin light chain substrates increased in the range 1.4–16-fold, demonstrating that substrate binding is less sensitive to the ROCK oligomerization state. These results show that the oligomerization state of ROCK may influence both its kinase activity and its interactions with inhibitors, and suggest that the dimeric structure is essential for normal in vivo function.
Keywords: analytical ultracentrifugation, dimer, light scattering, oligomerization, protein interactions, Rho-associated kinase (ROCK)
Abbreviations: DFDNB, 1,5-difluoro-2,4-dinitrobenzene; DMPK, myotonic dystrophy kinase; GTP[S], guanosine 5-[γ-thio]triphosphate; MLC, myosin light chain; MRCK, myotonic dystrophy kinase-related Cdc42 binding kinase; PH, pleckstrin homology; RI, refractive index; ROCK, Rho-associated kinase; SEC, size-exclusion chromatography
INTRODUCTION
RhoA is a small GTPase exhibiting both GDP/GTP binding and GTPase activities, and its activation plays a key role in Ca2+-independent smooth muscle contraction. Two important downstream targets of activated RhoA are the two isoforms of ROCK (Rho-associated kinase), known as ROCK II and I or Rho-kinase α and β respectively. These kinases regulate muscle MLC (myosin light chain) proteins both by direct phosphorylation [1,2] and indirectly by phosphorylation of the myosin binding subunit of myosin phosphatase. This inhibits the phosphatase activity, leading to increased levels of phosphorylated MLCs, followed by subsequent muscle contraction [3]. The Rho/ROCK pathway is also involved in non-muscle myosin regulation, and has been implicated in stress fibre and focal adhesion formation [4,5], neurite retraction [6,7] and tumour cell invasion [8,9] in non-muscle cells. Given the importance of ROCK in regulating such key regulatory processes involved in cytoskeleton rearrangement and cell adhesion, considerable effort has been expended on delineating the details of its mechanism of action.
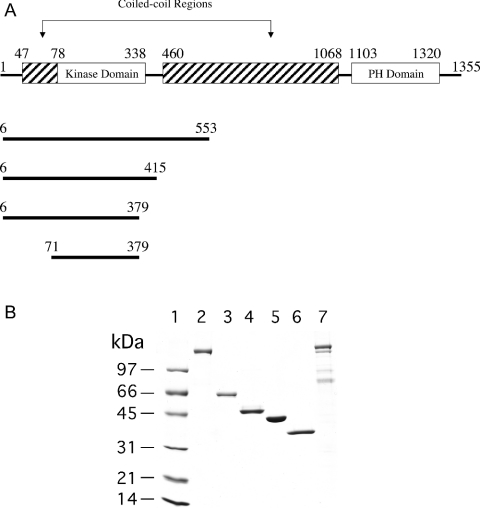
ROCK I and II have 64% sequence identity overall, and kinase domains that are 90% identical. ROCK is composed of N-terminal catalytic, coiled-coil and C-terminal PH (pleckstrin homology) domains. These domain features are shared by other closely related kinases, including DMPK (myotonic dystrophy kinase), MRCK (myotonic dystrophy kinase-related Cdc42 binding kinase) and citron kinase. For illustrative purposes, the predicted functional domains of full-length ROCK I are shown in Figure 1. This schematic representation is similar to that presented by Nakagawa et al. [10], except that an additional short coiled-coil domain segment N-terminal to the catalytic domain has been identified and added. This segment, comprising residues 47–78, was found using ‘Coils’, a program that compares sequences with a database of known parallel two-stranded coiled-coils and derives a similarity score [11]. This segment and the corresponding ROCK II Arg63–Asp93 segment are unique to both ROCK isoforms, and are absent from DMPK and MRCK.
Figure 1. (A) Schematic representation of the predicted functional domains of ROCK I, and (B) SDS/PAGE of purified full-length ROCK I and II and the four truncated ROCK I constructs.
(A) The limits of the four ROCK truncations are shown below the full-length sequence. Both full-length ROCK I (1354 amino acids) and ROCK II (1388 amino acids) and the four ROCK I constructs were cloned and expressed in insect cells. (B) Standard protein markers (lane 1) in the order of increasing molecular mass are: lysozyme (14.4 kDa), soybean trypsin inhibitor (21.5 kDa), carbonic anhydrase (31 kDa), ovalbumin (45 kDa), BSA (66.2 kDa), and phosphorylase b (97.4 kDa). The identities of the sample proteins are: lane 2, full-length ROCK II; lane 3, S6–L553; lane 4, S6–R415; lane 5, S6–E379; lane 6, M71–E379; lane 7, full–length ROCK I.
It has been demonstrated previously that the C-terminus of ROCK negatively regulates its kinase activity [12]. This regulatory domain consists of the Rho binding and PH domains. Binding of GTP-bound Rho is believed to disrupt the negative regulatory interaction between the catalytic domain and the autoinhibitory C-terminal region, resulting in activation of the enzyme. The domain structure of ROCK has more recently been probed with antibodies specific to the various different regions [13]. Results from immunoprecipitation experiments using various tagged ROCK constructs spanning different segments of the protein sequence have indicated that ROCK activity is regulated by its own aggregation state, which in turn is affected by its domain structure. These experiments, along with gel filtration and cross-linking studies, suggest that ROCK forms multimeric structures by virtue of interactions between the coiled-coil and Rho binding domains. The crystal structure of RhoA bound to the RhoA binding domain of ROCK has recently been solved by two groups [14,15], and both structures show this ROCK segment forming a parallel coiled-coil dimer, providing strong evidence that the full-length enzyme is also a dimer.
Oligomeric interactions between the coiled-coil and RhoA binding domains, together with the autoinhibitory C-terminal region, are thought to play a crucial role in regulating the catalytic activity of the enzyme. This is analogous to the oligomeric interactions and autoinhibitory two distal coiled-coil domains (CC2 and CC3) proposed to regulate the catalytic activity of the two isoforms of MRCK [16]. The coiled-coil region of the closely related DMPK has also been shown to be required for its oligomerization, which correlates with enhanced catalytic activity. DMPK also possesses an autoinhibitory domain at the C-terminus [17]. Recently, Zhang and Epstein [18] extensively characterized full-length dimeric DMPK and three truncation mutants using analytical ultracentrifugation and sucrose density gradient sedimentation methods. On eliminating the coiled-coil region, the full-length enzyme DMPK became monomeric.
Given the significance of the self-associative interactions of ROCK, we have undertaken the characterization of ROCK oligomerization using a combination of classical light scattering and analytical ultracentrifugation. The properties of four ROCK truncation mutants of successively decreasing length were analysed and compared with those of the full-length species in order to investigate the role of the domains in oligomerization (see Figure 1A for a schematic diagram of the truncations). An SDS/PAGE gel of all proteins used in this study is shown in Figure 1(B).
Kinetic data were collected for each construct in order to correlate oligomerization with function. These studies have characterized the transition from dimer to monomer as a function of the primary protein sequence length, with the amino acid regions comprising residues 47–78 and 379–415 being shown to be critical for the maintenance of the ROCK dimer. Although the monomers are shown to retain enzymic activity, kinetic data indicate that the dimeric structure is required to maintain high affinity towards both ATP and the ATP site inhibitor Y-27632. The effect on affinity is much less pronounced for binding of the small ROCK peptide, and is almost entirely absent when using the large protein substrate MLC.
MATERIALS AND METHODS
Cloning and expression
ROCK I (NM 005406) and ROCK II (NM 004850) were isolated from human leucocytes and a mix of liver, brain, bone marrow cDNA libraries respectively. The cDNAs encoding the various ROCK proteins used in this study were cloned into a baculoviral transfer vector, pBEV10, and expressed in insect cells as described previously [19]. In construction, the proteins were engineered to include a hexa-histidine (His6) tag at the N-terminus to facilitate purification.
Protein purification
Both full-length ROCK I and ROCK II proteins and the four truncated ROCK I constructs S6–L553 (Ser6–Leu553), S6–R415 (Ser6–Arg415), S6–E379 (Ser6–Glu379) and M71–E379 (Met71–Glu379) were isolated from insect cells and metal-affinity-purified as described previously [20]. After thrombin cleavage of the His tag, the protein was diluted 10-fold with 20 mM Hepes, pH 7.4, and loaded on to a MonoQ HR 5/5 ion-exchange column (Amersham Biosciences). The protein was eluted with a 100–400 mM NaCl gradient over 40 column volumes, and the ROCK fraction was pooled and concentrated to 2 ml. This was then loaded on to a HiLoad 16/60 Superdex 200 column (Amersham Biosciences) equilibrated with 20 mM Hepes, 200 mM NaCl, 2 mM β-mercaptoethanol, pH 7.4. The main peak was pooled and typically appeared to be ∼99% pure, as judged by SDS/PAGE analysis (Figure 1B). Due to low yields, purification of full-length ROCK I was limited to a two-step purification of Talon affinity chromatography followed by gel filtration. The final product was <50% pure, but this was sufficient for determination of Km and Vmax values. Since Vmax is proportional to kcat, it is possible to assess the effect of RhoA on the ROCK I catalytic constant.
The 20000 Da subunit of MLC was purified from chicken gizzard smooth muscle according to the procedure of Hathaway and Haeberle [21]. The final preparation used for kinetic studies was >90% pure, as judged by SDS/PAGE analysis, with the remainder being composed of the 17000 Da subunit.
Classical Raleigh light-scattering data collection
A model 1090 HPLC system (Hewlett Packard, Wilmington, DE, U.S.A.) was connected to a model 2410 Differential Refractometer (Waters Corp., Milford, MA, U.S.A.) equipped with a PD2020 static light-scattering system (Precision Detectors, Bellingham, MA, U.S.A.). Light-scattering data were collected at an angle of 90°. A TSK-GEL super SW 2000 column (4.6 mm×30 cm; Tosohaas, Montgomeryville, PA, U.S.A.) was used with a mobile phase of 20 mM Hepes, 200 mM NaCl and 5 mM β-mercaptoethanol, pH 7.5, delivered at a flow rate of 0.3 ml/min. The system was calibrated using transferrin (77000 Da; Calbiochem, San Diego, CA, U.S.A.) as a standard. An injection volume of 25 μl was used for each run, using a protein sample concentration of 5 mg/ml. The data were collected and analysed using PrecisionAcquire32 and Discovery32 software from Precision Detectors.
Sedimentation equilibrium and sedimentation velocity data acquisition and analysis
Sedimentation velocity and sedimentation equilibrium measurements were performed in a Beckman Coulter Optima XL-I instrument using an An Ti 60 rotor. The sedimentation velocity experiments were performed at 42000 rev./min for the truncated constructs and 15000 rev./min for the full-length protein using charcoal-filled Epon double-sector cells. Concentrations of the four ROCK truncations and of the full-length enzyme were 0.5 and 0.25 mg/ml respectively. Sedimentation velocity data were analysed using the SVEDBERG (version 6.39) [22] and DCDT+ (version 1.14) [23] programs to obtain the weight average sedimentation coefficients and distribution of sedimenting species respectively.
Sedimentation equilibrium data were collected in three sample cells with six-channel centerpieces. Data were acquired every 0.001 cm with 30 replicates in a step scan mode at speeds of 12000, 14000 and 16000 rev./min for all four of the truncated ROCK I constructs, and 8000, 12000 and 14000 rev./min for full-length ROCK II. Data sets for three concentrations were collected for each enzyme. Concentrations used were 0.50, 0.25, and 0.125 mg/ml for the ROCK I constructs. ROCK II concentrations used were 0.25, 0.125 and 0.063 mg/ml. Buffer conditions for all samples were 20 mM Hepes, 200 mM NaCl, and 2 mM β-mercaptoethanol, pH 7.4, and all experiments were performed at 20 °C. Data were analysed with the appropriate mathematical models using the Optima XL-A data analysis software (BeckmanCoulter, Palo Alto, CA, U.S.A.) running under the Origin (version 5.0) platform (Microcal, Northampton, MA, U.S.A.).
Steady-state kinetics
A coupled enzyme assay, as described previously [24], was used to quantify the ADP generated in the kinase reaction with ROCK peptide (KKRNRTLSV; American Peptide Company, Sunnyvale, CA, U.S.A.). The ROCK concentration was 100 nM in the assay. Y-27632 (Biomol, Plymouth Meeting, PA, U.S.A.) was dissolved in DMSO and incubated for 10 min at 30 °C with enzyme prior to initiation of the reaction by ATP addition. For Ki determinations, ATP and ROCK peptide concentrations were at the previously determined Kms for each enzyme construct. Kinetic data for full-length ROCK I and II and for assays using MLC as a substrate were acquired using [γ-33P]ATP. Samples were assayed in 50 μl of 100 mM Hepes, 10 mM MgCl2, 2 mM dithiothreitol and 0.15% (w/v) CHAPS, pH 7.4. Full-length ROCK assays were done with and without 5 μM GTP[S] (guanosine 5-[γ-thio]triphosphate)–RhoA. RhoA (Cytoskeleton, Denver, CO, U.S.A.) was activated with GTP[S] (Sigma Chemical Company, St Louis, MO, U.S.A.) to generate the GTP[S]–RhoA complex as described previously [25]. For determination of peptide or protein Km values, the ATP concentration was 1 mM (2 μCi of [γ-33P]ATP per reaction). Reactions were initiated by addition of ATP, and after 15 min reactions were quenched with 175 μl of 10% (v/v) trichloroacetic acid for protein substrate, or with 5% (v/v) phosphoric acid for peptide substrate. Trichloroacetic acid-quenched reactions were filtered through Whatman GF/F 96-well filter plates, and phosphoric acid-quenched reactions were filtered through Whatman p81 filter 96-well filter plates. Plates were washed four times each with their corresponding stop solution, then with methanol. Then 200 μl of scintillant was added to all wells and the plates were counted for radioactivity in a Packard 96-well plate γ-radiation counter. Kinetic constants were determined by non-linear least-squares analysis using Prism (GraphPad Software, San Diego, CA, U.S.A.).
Cross-linking studies
Cross-linking studies were conducted using the reagent DFDNB (1,5-difluoro-2,4-dinitrobenzene; Pierce, Rockford, IL, U.S.A.). The protein concentration used was 1 mg/ml. Reactions were initiated by the addition of either 0.1 mM or 5 mM DFDNB (final concentration), and after the designated time interval the reaction was stopped by the addition of SDS sample buffer. The degree of cross-linking was visualized by SDS/PAGE analysis.
Sequence alignments
Alignments were done using Clustal W, a publicly available multiple sequence alignment program (http://clustalw.genome.ad.jp).
RESULTS
ROCK characterization by SEC (size-exclusion chromatography) with classical light scattering and RI (refractive index) detection
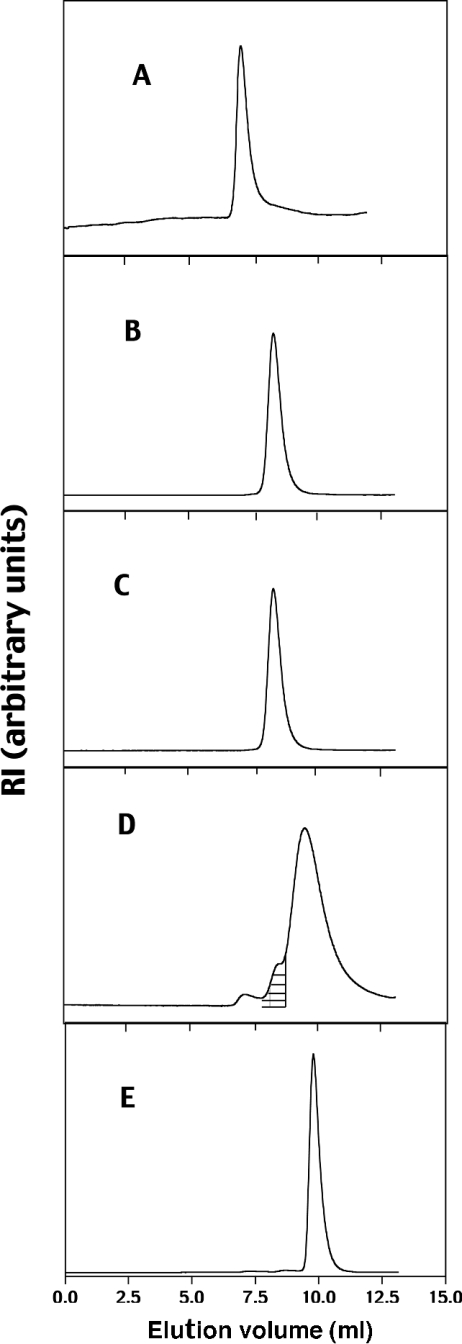
The solution-phase size distributions of all four constructs were examined by SEC coupled with RI and static light-scattering detection. Both RI and light-scattering data were collected in triplicate for each ROCK construct, and representative elution profiles for the RI signals are shown in Figure 2. All of the constructs, with the exception of the S6–E379 construct, gave symmetrical peaks with close to Gaussian distributions (Figures 2A, 2B, 2C and 2E). The RI signal for S6–E379 shows a shoulder on the left-hand side of the main peak (hatched portion of Figure 2D), indicating protein heterogeneity in the sample. The smallest extreme left shoulder is indicative of high-order molecular mass aggregates.
Figure 2. SEC chromatograms of full-length and truncated ROCK constructs.
(A) Full-length ROCK II, (B) S6–L553, (C) S6–R415, (D) S6–E379, (E) M71–E379. The shaded portion in (D) represents the area integrated to give a value of 90000 Da for the dimeric portion of S6–E379.
The experimental molecular mass values determined from the RI and light-scattering data are listed in Table 1, along with the corresponding calculated theoretical molecular masses. Full-length ROCK II and the S6–L553, S6–R415, S6–E379 and M71–E379 constructs gave apparent molecular masses ranging from 426000 to 36800 Da. Dividing the experimental values by the theoretical molecular masses gave ratios ranging from 1.0 to 2.6. These data suggest that both full-length ROCK II (426000 Da) and S6–L553 (164066 Da) are predominantly dimeric, with some higher-order oligomers present. S6–R415 behaves almost entirely as a dimer, and S6–E379 appears to be predominantly monomeric, with some dimer present. The apparent molecular mass of the S6–E379 monomer band is significantly greater than the theoretical monomer weight due to the close overlap of the dimer band with the monomer band. The presence of dimer is confirmed when only the left shoulder on the main band is integrated to give a value of 90000 Da, very close to the expected dimer value of 87000 Da. The value of unity for M71–E379 indicates that it is monomeric.
Table 1. Apparent molecular masses from light scattering analyses of full-length ROCK II and the four ROCK I constructs.
The theoretical molecular mass of each construct was calculated using the ProtParam Tool available on the ExPASy Molecular Biology Server (www.expasy.org).
| Molecular mass (Da) | |||
|---|---|---|---|
| Construct | Theoretical | Apparent | Ratio (apparent/theoretical) |
| Full-length ROCK II | 161308 | 426000±21300 | 2.6 |
| S6–L553 | 64210 | 164066±5400 | 2.6 |
| S6–R415 | 47840 | 100467±153 | 2.1 |
| S6–E379 | 43705 | 54490±220 | 1.2 |
| M71–E379 | 35995 | 36800±630 | 1.0 |
ROCK characterization by analytical ultracentrifugation
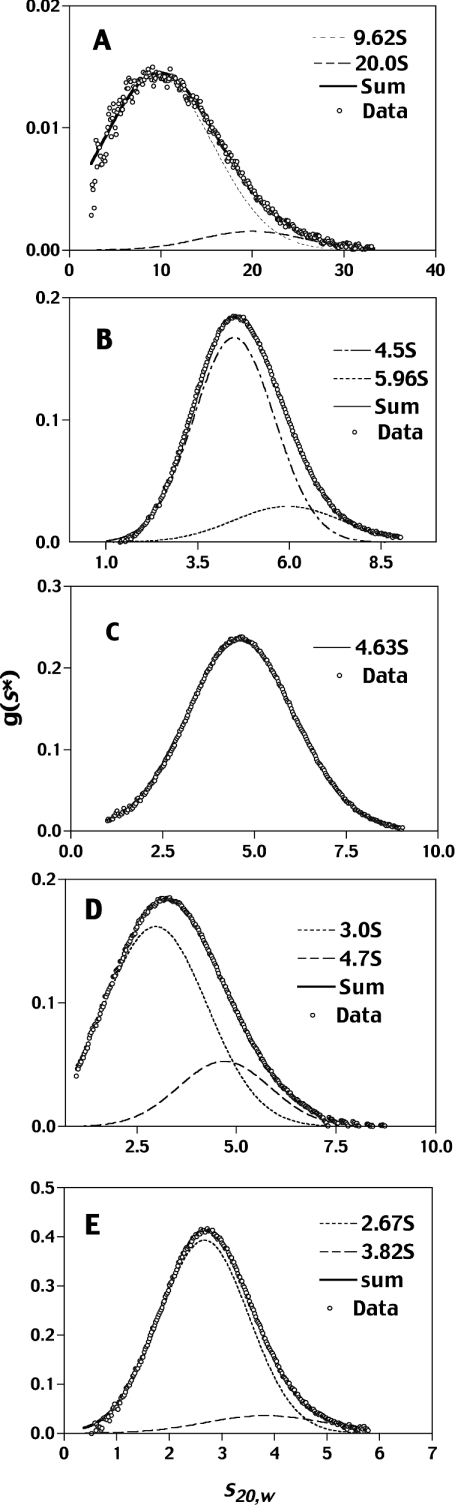
Both sedimentation velocity and sedimentation equilibrium experiments were performed with all five enzyme forms, and the data are presented in Tables 2 and 3. A rigorous interpretation of the sedimentation velocity data is not possible, since there was insufficient material to perform a detailed analysis of the dependence of the sedimentation coefficient on concentration, a necessary prerequisite for revealing evidence of a self-associating system. However, rough estimates of the degree of homogeneity and the relative abundance of protein species in solution are possible with data obtained at a single concentration [22]. Figure 3(C) shows that construct S6–R415 gave a single symmetrical peak that was best fit to a model representing a single species, consistent with the data from both light-scattering and sedimentation equilibrium experiments. All other samples showed asymmetrical and broadened peaks that were best fit using a model representing two independently sedimenting species (Figures 3A, 3B, 3D and 3E).
Table 2. Summary of parameters obtained from analyses of the sedimentation velocity data.
| Construct | Amount (%) | s20,w (S) |
|---|---|---|
| Full-length ROCK II | ||
| Species 1 | 92 | 9.66±0.01 |
| Species 2 | 8 | 20.3±0.5 |
| S6–L553 | ||
| Species 1 | 81 | 4.49±0.04 |
| Species 2 | 19 | 5.96±1.1 |
| S6–R415 | ||
| Species 1 | 100 | 4.63±0.003 |
| S6–E379 | ||
| Species 1 | 78 | 2.97±0.01 |
| Species 2 | 22 | 4.7±0.1 |
| M71–E379 | ||
| Species 1 | 89 | 2.67±0.04 |
| Species 2 | 11 | 3.66±1.9 |
Table 3. Summary of parameters obtained from analyses of the sedimentation equilibrium data.
| Molecular mass (Da) | ||||
|---|---|---|---|---|
| Construct | Apparent | Ratio(apparent/theoretical) | Model used for data fitting | Kd (μM) |
| Full-length ROCK II | 318658±20828 | 2.0 | Single species | – |
| S6–L553 | 132093±6713 | 2.1 | Single species | – |
| S6–R415 | 91257±4230 | 1.9 | Monomer/dimer | 0.4 |
| S6–E379 | 57618±4230 | 1.3 | Monomer/dimer | 17.6 |
| M71–E379 | 38394±460 | 1.1 | Monomer/dimer | 357.0 |
Figure 3. Sedimentation velocity experiments with five ROCK constructs.
Plots of the apparent sedimentation coefficient g(s*) against the sedimentation coefficient s20,w corrected to the viscosimetric properties of water at 20 °C are shown. They were fitted using the DCDT+ program to models representing one or two sedimenting species. The circles represent the experimental data. The solid lines represent the best fit and are the sum of the contributions to the sedimenting boundary from individual species (dashed and dotted lines). (A) Full-length ROCK II, (B) S6–L553, (C) S6–R415, (D) S6–E379, (E) M71–E379.
The sedimentation coefficients are summarized in Table 2. As expected, they increase as both construct sequence length and the degree of oligomerization increase, with s20,w values ranging from 2.67 and 2.97 for the S6–E379 and M71–E379 constructs respectively to 20.3 for full-length ROCK II. Although the S6–L553 is considerably larger than the S6–R415 construct (by over 16000 Da), they share very similar s20,w values. The sedimentation coefficient depends on molecular shape and degree of hydration as well as molecular mass; hence the similar values suggest that S6–R415 may be more compact and/or less hydrated than S6–L553.
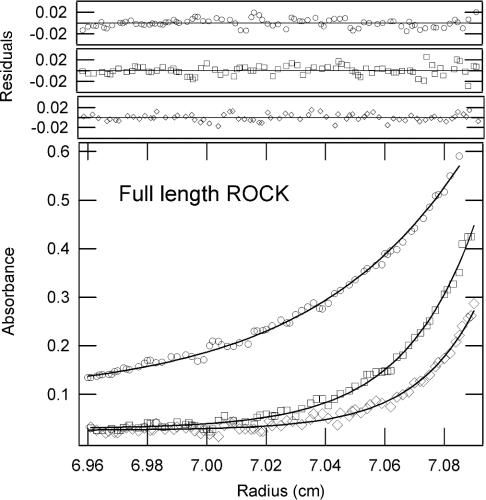
Sedimentation equilibrium experiments were performed to both confirm and extend the molecular mass determinations and inferred association states determined from light-scattering analysis. These data permit a reliable determination of the molecular mass of species at true thermodynamic equilibrium. The data analysis also provides direct information on the association state by virtue of the model used to provide the best fit, and in cases where there is a reversible self-association it is also possible to evaluate the dissociation constant. The sedimentation equilibrium data obtained from three different concentrations and three rotor speeds were analysed as described in the Materials and methods section. Typical sedimentation equilibrium profiles at three different speeds using full-length ROCK II are shown in Figure 4. The apparent molecular masses determined from the equilibrium experiments and the ratios calculated by dividing the apparent molecular mass by the theoretical molecular mass are shown in Table 3. The values in Table 3 are in close agreement with the values determined by light scattering (Table 1). The data for both full-length ROCK II and S6–L553 fit best to a model describing a single species, making them predominantly dimeric. It is probable that the higher ratios seen for the light-scattering data for these two enzymes are due to small amounts of higher oligomers, which can have a large effect on the light-scattering measurements but are probably sedimented out of the optical data collection zone prior to sedimentation equilibrium data collection. The best fits for constructs S6–R415, S6–E379 and M71–E379 were obtained with a model representing a monomer/dimer system, with calculated Kd values ranging from 0.43 to 357 μM. The S6–R415 construct is a tightly associated dimeric complex, as demonstrated by its low Kd of 0.43 μM, consistent with the homogeneous bands seen in both the size exclusion separation profile in the light-scattering experiments and in the sedimentation velocity data. The S6–E379 enzyme resolves itself into two components in the SEC fractionation data (see the shaded shoulder in Figure 2D). This heterogeneity is reflected in both the sedimentation velocity and sedimentation equilibrium data, and suggests the presence of both monomer and dimers. The apparent molecular mass of M71–E379 is approx. 10% higher than that of the theoretical monomer, suggesting the presence of a small amount of dimer.
Figure 4. Sedimentation equilibrium experiments with full-length ROCK II.
The data are shown as absorbance against radial position at 20 °C. The raw data (○, 8000 rev./min; □, 10000 rev./min; ◇, 12000 rev./min) and the fit (lines) are shown. The distribution of the residuals for each of these fits is shown in the upper panels.
Steady-state kinetics
The steady-state kinetic parameters measured using both ROCK peptide and MLC as substrates for full-length ROCK I and ROCK II and the four truncated ROCK I constructs are presented in Table 4. The MLC Km values for full-length ROCK I and II ranged from 6.0 to 8.0 and from 2.2 to 5.0 μM in the absence and presence respectively of GTP[S]–RhoA, and both kcat values increased 1.5–2-fold in the presence of GTP[S]–RhoA. These Kms compare favourably with the reported MLC Kms of 12.6 and 2.6 μM [2] and 7.0 and 4.5 μM [26] measured in the absence and presence respectively of GTP[S]–GST–RhoA; a 2-fold stimulation of kcat was also observed by these workers. These latter studies were performed using ROCK II purified from natural sources, suggesting that both of our recombinant full-length enzymes behave similarly to endogenous ROCK. For the other four ROCK I constructs, MLC Km and kcat values did not show significant changes, ranging from 11 to 18 μM and from 1.0 to 5.9 s−1 respectively.
Table 4. Kinetic parameters for full-length ROCK I and II and the four ROCK I constructs.
FL, full-length; RP, ROCK peptide; ND, not determined.
| Km (μM) | kcat (s−1) or Vmax (μmol/min)‡ | |||||
|---|---|---|---|---|---|---|
| Construct | ATP* ([RP]=2 mM) | RP* ([ATP]=1 mM) | MLC† ([ATP]=1 mM) | ATP* ([RP]=2 mM) | MLC† ([ATP]=1 mM) | Y-27632 Ki (μM) |
| FL ROCK I (−Rho) | 100±21 | 458±46 | 6.0±1.4 | 9.0±1‡ | 9.0*±1.3‡ | ND |
| FL ROCK I (+Rho) | 140±32 | 325±52 | 2.2±0.6 | 7.0±0.8‡ | 14±1.8‡ | ND |
| FL ROCK II (−Rho) | 82±25 | 174±35 | 8.0±1.0 | 1.8±0.15 | 2.6±0.3 | 0.6±0.15 |
| FL ROCK II (+Rho) | 100±17 | 146±20 | 5.0±1.0 | 2.1±0.2 | 3.9±0.4 | 0.8±0.1 |
| S6–L553 | 13±2.0 | 25±8.0 | 11±2.7 | 1.3±0.2 | 4.1±0.7 | 0.25±0.04 |
| S6–R415 | 8±0.6 | 92±14 | 12±1.2 | 1.7±0.1 | 5.9±0.5 | 1.40±0.07 |
| S6–E379 | 695±160 | 141±32 | 7.1±1.6 | 0.7±0.08 | 1.0±0.1 | >30 |
| M71–E379 | 613±67 | 405±52 | 18±3.5 | 1.1±0.1 | 1.9±0.4 | >30 |
* Measured with the coupled-kinase assay.
† Measured with the [γ-33P]ATP assay.
A much greater effect was seen on the Km for ATP when using the ROCK peptide as a substrate. Both S6–L553 and S6–R415 constructs were shown to have similar ATP Kms, of 13 and 8 μM respectively, whereas those of S6–E379 and M71–E379 were much greater (approx. 600 μM). A similar, although much less pronounced, trend was seen with the ROCK peptide Km, ranging from 25 to 405 μM for the S6–L553 through to the M71–E379 construct. The kcat values ranged from 0.7 to 1.7 s−1, demonstrating that the four truncated constructs have similar catalytic-centre activities. Using S6 peptide as a substrate, Trauger et al. [27] reported kcat and ATP Km values of 1.5 s−1 and 21 μM respectively for the ROCK II construct M1–L543 (Met1–Leu543), comparable with the values found in the present study for ROCK I S6–L553. Ki values measured in the presence of ROCK peptide were also evaluated using the specific ROCK inhibitor Y-27632 [28]. The Ki value increased progessively as the protein was truncated, with S6–L553 giving the tightest binding at 0.25 μM. This value is close to the Ki of 0.1 μM reported by Trauger et al. [27] using the analogous ROCK II M1–L543 construct. Y-27632 was at least a 120-fold less potent inhibitor for both S6–E379 and M71–E379, and 5.6-fold less potent for S6–R415.
ROCK cross-linking
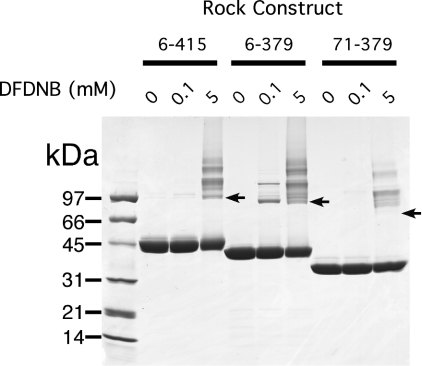
The small size of DFDNB makes it suitable for cross-linking across the short distances expected between dimers [29]. An SDS/PAGE profile illustrating the effects of the cross-linking reagent DFDNB on ROCK constructs S6–R415, S6–E379 and M71–E379 is shown in Figure 5. The results are consistent with the trends from the sedimentation equilibrium data in Table 3. The M71–E379 construct showed no dimer even at a concentration of 5 mM DFDNB; only non-specific higher-order aggregates were seen, consistent with its essentially complete monomeric nature. The S6–E379 construct showed cross-linked dimer at 0.1 mM DFDNB, demonstrating the presence of dimer. The relatively low concentration of DFDNB needed to cross-link the dimer is consistent with the high Kd of 17.6 μM calculated for the monomer/dimer equilibrium. The S6–R415 construct showed no cross-linked dimer at 0.1 mM DFDNB, and this was only seen at the higher concentration of 5 mM. This reflects the tighter binding between subunits in the S6–R415 dimer relative to the S6–E379 dimer.
Figure 5. SDS/PAGE of cross-linking of ROCK with DFDNB.
Standard protein markers were as in Figure 1(b). The three indicated constructs were incubated with either 0.1 or 0.5 mM DFDNB for 15 min, then the reaction was quenched with SDS sample buffer. Arrows indicate where the dimers of each construct are expected to appear.
DISCUSSION
In the present study, full-length ROCK was shown unequivocally to be dimeric in vitro, with some higher-order oligomers present. Previous SEC experiments by Chen et al. [13] suggested a tetrameric structure; however, more error is likely using the empirical SEC method compared with the combination of light-scattering and analytical ultracentrifugation methodologies, which are derived from theoretical well-established principles, and are not complicated by protein–matrix interactions. In the presence of GTP[S]–RhoA, a 40–60% decrease in MLC Km and a 1.5–2-fold increase in kcat was observed for both full-length species, consistent with reported literature results for ROCK II. To our knowledge, our results are the first to directly compare the kinetics of full-length ROCK I and ROCK II, which we have demonstrated to be very similar. This suggests that the different isoforms exist so that they may be localized to different intracellular targets, as proposed by Nakagawa et al. [10]. There are specific sequence differences between ROCK I and ROCK II that have been implicated in differential regulation. For example, caspase-3 has been shown to cleave ROCK I at a caspase-3 recognition sequence that is absent from ROCK II, providing a tentative mechanism for ROCK I activation during apoptotic membrane blebbing [30].
ROCK maintains its dimeric structure even after complete removal of both the PH and coiled-coil domains (S6–R415 construct). This is in agreement with the results of Chen et al. [13], where immunoprecipitation of cell extracts transfected with ROCK II constructs corresponding to residues 1–432 indicated dimerization. Removal of another 36 C-terminal residues had a much more dramatic effect on the association state, yielding a predominantly monomeric species with about 20% dimer. Although there is no specific information predicted via sequence analysis to describe this segment, it is clear that these residues contribute significantly to maintaining the dimeric structure of ROCK. The presence of 20% dimeric species is most probably due to interactions between residues 47–78, as this segment has high similarity to the coiled-coil domain. Removal of another 65 N-terminal residues resulted in an entirely monomeric kinase, consistent with this hypothesis. M71–E379 required the monomer/dimer self-association model to best fit the sedimentation equilibrium data, implying the presence of dimer. However, the calculated Kd for this association is high (357 μM), suggesting that this interaction is weak. The weakness of this interaction is verified by the inability of the reagent DFDNB to cross-link this construct to a detectable dimer even at 5 mM concentration. The relatively high Kd values of S6–E379 and M71–E379 also suggest that they will essentially be completely monomeric under the conditions of the enzyme assay at 100 nM.
The association state of ROCK has little effect on turnover number, which was similar for all truncated proteins. However, dramatic increases in both ATP Km and the Ki for Y-27632 were found when comparing the monomeric and dimeric constructs. The complementary loss of Y-27632 inhibitory potency is not surprising, since this inhibitor is known to bind at the ATP site of ROCK. The effect of the dimer-to-monomer transition on the Km values of ROCK peptide and MLC was much less pronounced. The higher potency conferred by the more physiologically relevant substrate correlates with a greatly decreased sensitivity to the oligomerization state of ROCK. Using MLC as substrate, the ATP Km for M71–E379 was 314 μM and the Ki for Y-27632 was >30 μM (results not shown), suggesting that MLC does not induce dimerization of M71–E379. It is possible that more numerous and/or tighter contacts with a protein substrate are responsible for this decreased sensitivity.
Taken together, these results indicate that residues 379–415 of ROCK play a crucial role both in modulating the accessibility of the ATP binding site (and to a much lesser extent the substrate binding site) and in maintaining the dimeric structure. This segment may be in close proximity to the ATP active site, and may also interact directly at the dimer interface between two ROCK monomers. The identity between ROCK I and ROCK II is >80% in this segment, making it probable that the two isoforms behave similarly. This segment, which fails to register as part of any known protein domain, comprises part of the separation between the large coiled-coil and the kinase domains. We speculate that this conserved segment could be part of a hinge region that helps to modulate interactions between the C-terminal segments of both isoforms and their kinase domains.
It was possible to make cross-linked dimers of both S6–E379 and S6–R415 with DFDNB. These dimers were purified by gel filtration and assayed, but no activity could be detected (results not shown). Hence we cannot differentiate whether the kinetic changes are due to the deletions themselves, to the change in oligomerization, or to both factors simultaneously. Nevertheless, the significance of residues 379–415 in maintaining the dimeric structure and their absence contributing to alterations in Km is clear.
It is interesting that Zhang and Epstein [18] found that truncating the closely related DMPK to residues 1–432 resulted in a monomeric enzyme. This DMPK construct has over 75% identity with the S6–R415 ROCK construct, yet we have convincingly demonstrated the latter to be a dimer. Apparently, removal of the coiled-coil domain of DMPK is sufficient to induce monomer formation, while ROCK retains its dimeric structure after a similar deletion of its coiled-coil domain. This is a remarkable difference in oligomerization state between two closely related proteins. Zhang and Epstein [18] also observed 3- and 10-fold decreases in peptide substrate Km and kcat values respectively for monomeric DMPK compared with the dimer. For ROCK, the effect on Km (a 1.5–16-fold increase) was opposite to that seen for DMPK, and the decrease in kcat was less significant. Evaluation of the ATP Km for the monomeric DMPK would be informative, since we have shown that this Km is affected far more by the quaternary state of ROCK than is the peptide substrate Km. By extending these studies to other homologues, such as MRCK [16], citron kinase [31] and LET-502 kinase from Caenorhabditis elegans [32], one could assess if truncation of native kinase dimers of this family leads to active monomers with altered kinetic parameters. Given the differences in monomer properties, it is reasonable to speculate that the dimeric structure of full-length ROCK is a critical prerequisite for normal in vivo interactions with endogenous molecular partners such as RhoA, ATP and myosin.
Dimers such as those found with ROCK have evolved to optimize interactions with other molecular components. Our results are consistent with the conclusions of others that changes in the quaternary structure of an enzyme can have a significant impact on its functional properties. Cox et al. [33] disrupted the dimer interface of creatine kinase via site-directed mutagenesis and discovered that the resultant monomeric enzyme showed significant activity, with the kcat being 60% of that of the native full-length dimer. However, this was accompanied by a 10-fold reduction in substrate affinity, results similar to those reported here. Owen et al. [34] reported two truncated monomeric species of hsp90, one whose catalytic efficiency was >100 times greater than that of the dimeric full-length species, while the other failed to hydrolyse ATP, demonstrating that the association state can have a wide range of effects on enzyme kinetic properties.
In conclusion, we have shown that amino acids 379–415, and to a lesser extent residues 47–78, are intimately involved in maintaining the dimeric structure of ROCK. The fact that the monomers have similar turnover numbers as the dimers shows that dimerization is not essential for ROCK activity, and may be necessary to enhance its affinity towards ATP. Hence the dimerization motif for this kinase may be essential for normal interactions with ATP, and may be important for its in vivo regulation of cytoskeletal dynamics. The correlation between ROCK oligomerization state and ligand affinity is also an important consideration in inhibitor design.
Acknowledgments
We thank Dr Dan Frantz, Catherine Couture, Jim Jernee and Judy Lippke for help cloning the ROCK constructs, Dr Marc Jacobs for useful discussions about construct design, and Dr John Thomson, Dr Scott Raybuck and Dr Steve Chambers for critical reading of the manuscript.
References
- 1.Kureishi Y., Kobayashi S., Amano M., Kimura K., Kanaide H., Nakano T., Kaibuchi K., Ito M. Rho-associated kinase directly induces smooth muscle contraction through myosin light chain phosphorylation. J. Biol. Chem. 1997;272:12257–12260. doi: 10.1074/jbc.272.19.12257. [DOI] [PubMed] [Google Scholar]
- 2.Amano M., Ito M., Kimura K., Fukata Y., Chihara K., Nakano T., Matsuura Y., Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J. Biol. Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 3.Totsukawa G., Yamakita Y., Yamashiro S., Hartshorne D. J., Sasaki Y., Matsumura F. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J. Cell Biol. 2000;150:797–806. doi: 10.1083/jcb.150.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishizaki T., Naito M., Fujisawa K., Maekawa M., Watanabe N., Saito Y., Narumiya S. p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett. 1997;404:118–124. doi: 10.1016/s0014-5793(97)00107-5. [DOI] [PubMed] [Google Scholar]
- 5.Kawano Y., Fukata Y., Oshiro N., Amano M., Nakamura T., Ito M., Matsumura F., Inagaki M., Kaibuchi K. Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J. Cell Biol. 1999;147:1023–1038. doi: 10.1083/jcb.147.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amano M., Chihara K., Nakamura N., Fukata Y., Yano T., Shibata M., Ikebe M., Kaibuchi K. Myosin II activation promotes neurite retraction during the action of Rho and Rho-kinase. Genes Cells. 1998;3:177–188. doi: 10.1046/j.1365-2443.1998.00181.x. [DOI] [PubMed] [Google Scholar]
- 7.Hirose M., Ishizaki T., Watanabe N., Uehata M., Kranenburg O., Moolenaar W. H., Matsumura F., Maekawa M., Bito H., Narumiya S. Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells. J. Cell Biol. 1998;141:1625–1636. doi: 10.1083/jcb.141.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshioka K., Matsumura F., Akedo H., Itoh K. Small GTP-binding protein Rho stimulates the actomyosin system, leading to invasion of tumor cells. J. Biol. Chem. 1998;273:5146–5154. doi: 10.1074/jbc.273.9.5146. [DOI] [PubMed] [Google Scholar]
- 9.Sahai E., Marshall C. J. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa O., Fujisawa K., Ishizaki T., Saito Y., Nakao K., Narumiya S. ROCK-I and ROCK-II, two isoforms of Rho-associated coiled-coil forming protein serine/threonine kinase in mice. FEBS Lett. 1996;392:189–193. doi: 10.1016/0014-5793(96)00811-3. [DOI] [PubMed] [Google Scholar]
- 11.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 12.Amano M., Chihara K., Nakamura N., Kaneko T., Matsuura Y., Kaibuchi K. The COOH terminus of Rho-kinase negatively regulates rho-kinase activity. J. Biol. Chem. 1999;274:32418–32424. doi: 10.1074/jbc.274.45.32418. [DOI] [PubMed] [Google Scholar]
- 13.Chen X. Q., Tan I., Ng C. H., Hall C., Lim L., Leung T. Characterization of RhoA-binding kinase ROKalpha: implication of the pleckstrin homology domain in ROKalpha function using region-specific antibodies. J. Biol. Chem. 2002;277:12680–12688. doi: 10.1074/jbc.M109839200. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu T., Ihara K., Maesaki R., Amano M., Kaibuchi K., Hakoshima T. Parallel coiled-coil association of the RhoA-binding domain in Rho-kinase. J. Biol. Chem. 2003;278:46046–46051. doi: 10.1074/jbc.M306458200. [DOI] [PubMed] [Google Scholar]
- 15.Dvorsky R., Blumenstein L., Vetter I. R., Ahmadian M. R. Structural insights into the interaction of ROCKI with the switch regions of RhoA. J. Biol. Chem. 2004;279:7098–7104. doi: 10.1074/jbc.M311911200. [DOI] [PubMed] [Google Scholar]
- 16.Tan I., Seow K. T., Lim L., Leung T. Intermolecular and intramolecular interactions regulate catalytic activity of myotonic dystrophy kinase-related Cdc42-binding kinase alpha. Mol. Cell. Biol. 2001;21:2767–2778. doi: 10.1128/MCB.21.8.2767-2778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bush E. W., Helmke S. M., Birnbaum R. A., Perryman M. B. Myotonic dystrophy protein kinase domains mediate localization, oligomerization, novel catalytic activity, and autoinhibition. Biochemistry. 2000;39:8480–8490. doi: 10.1021/bi992142f. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R., Epstein H. F. Homodimerization through coiled-coil regions enhances activity of the myotonic dystrophy protein kinase. FEBS Lett. 2003;546:281–287. doi: 10.1016/s0014-5793(03)00601-x. [DOI] [PubMed] [Google Scholar]
- 19.Chambers S. P. High-throughput protein expression for the post-genomic era. Drug Discov. Today. 2002;7:759–765. doi: 10.1016/s1359-6446(02)02361-9. [DOI] [PubMed] [Google Scholar]
- 20.ter Haar E., Coll J. T., Austen D. A., Hsiao H. M., Swenson L., Jain J. Structure of GSK3beta reveals a primed phosphorylation mechanism. Nat. Struct. Biol. 2001;8:593–596. doi: 10.1038/89624. [DOI] [PubMed] [Google Scholar]
- 21.Hathaway D. R., Haeberle J. R. Selective purification of the 20,000-Da light chains of smooth muscle myosin. Anal. Biochem. 1983;135:37–43. doi: 10.1016/0003-2697(83)90726-1. [DOI] [PubMed] [Google Scholar]
- 22.Philo J. S. An improved function for fitting sedimentation velocity data for low-molecular-weight solutes. Biophys. J. 1997;72:435–444. doi: 10.1016/S0006-3495(97)78684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philo J. S. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal. Biochem. 2000;279:151–163. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 24.Fox T., Coll J. T., Xie X., Ford P. J., Germann U. A., Porter M. D., Pazhanisamy S., Fleming M. A., Galullo V., Su M. S., Wilson K. P. A single amino acid substitution makes ERK2 susceptible to pyridinyl imidazole inhibitors of p38 MAP kinase. Protein Sci. 1998;7:2249–2255. doi: 10.1002/pro.5560071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond S. M., Jenco J. M., Nakashima S., Cadwallader K., Gu Q., Cook S., Nozawa Y., Prestwich G. D., Frohman M. A., Morris A. J. Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-alpha. J. Biol. Chem. 1997;272:3860–3868. doi: 10.1074/jbc.272.6.3860. [DOI] [PubMed] [Google Scholar]
- 26.Feng J., Ito M., Kureishi Y., Ichikawa K., Amano M., Isaka N., Okawa K., Iwamatsu A., Kaibuchi K., Hartshorne D. J., Nakano T. Rho-associated kinase of chicken gizzard smooth muscle. J. Biol. Chem. 1999;274:3744–3752. doi: 10.1074/jbc.274.6.3744. [DOI] [PubMed] [Google Scholar]
- 27.Trauger J. W., Lin F. F., Turner M. S., Stephens J., LoGrasso P. V. Kinetic mechanism for human Rho-Kinase II (ROCK-II) Biochemistry. 2002;41:8948–8953. doi: 10.1021/bi0258243. [DOI] [PubMed] [Google Scholar]
- 28.Uehata M., Ishizaki T., Satoh H., Ono T., Kawahara T., Morishita T., Tamakawa H., Yamagami K., Inui J., Maekawa M., Narumiya S. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature (London) 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 29.Kornblatt J. A., Lake D. F. Cross-linking of cytochrome oxidase subunits with difluorodinitrobenzene. Can. J. Biochem. 1980;58:219–224. doi: 10.1139/o80-030. [DOI] [PubMed] [Google Scholar]
- 30.Sebbagh M., Renvoize C., Hamelin J., Riche N., Bertoglio J., Breard J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat. Cell Biol. 2001;3:346–352. doi: 10.1038/35070019. [DOI] [PubMed] [Google Scholar]
- 31.Yamashiro S., Totsukawa G., Yamakita Y., Sasaki Y., Madaule P., Ishizaki T., Narumiya S., Matsumura F. Citron kinase, a Rho-dependent kinase, induces di-phosphorylation of regulatory light chain of myosin II. Mol. Biol. Cell. 2003;14:1745–1756. doi: 10.1091/mbc.E02-07-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spencer A. G., Orita S., Malone C. J., Han M. A RHO GTPase-mediated pathway is required during P cell migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13132–13137. doi: 10.1073/pnas.241504098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox J. M., Davis C. A., Chan C., Jourden M. J., Jorjorian A. D., Brym M. J., Snider M. J., Borders C. L., Jr, Edmiston P. L. Generation of an active monomer of rabbit muscle creatine kinase by site-directed mutagenesis: the effect of quaternary structure on catalysis and stability. Biochemistry. 2003;42:1863–1871. doi: 10.1021/bi027083b. [DOI] [PubMed] [Google Scholar]
- 34.Owen B. A., Sullivan W. P., Felts S. J., Toft D. O. Regulation of heat shock protein 90 ATPase activity by sequences in the carboxyl terminus. J. Biol. Chem. 2002;277:7086–7091. doi: 10.1074/jbc.M111450200. [DOI] [PubMed] [Google Scholar]