Abstract
NALP1 (also called DEFCAP, NAC, CARD7) has been shown to play a central role in the activation of inflammatory caspases and processing of pro-IL1β (pro-interleukin-1β). Previous studies showed that NALP1 is highly expressed in peripheral blood mononuclear cells. In the present study, we report that expression of NALP1 is absent from CD34+ haematopoietic blast cells, and its levels are upregulated upon differentiation of CD34+ cells into granulocytes and to a lesser extent into monocytes. In peripheral blood cells, the highest levels of NALP1 were observed in CD3+ (T-lymphocytes), CD15+ (granulocytes) and CD14+ (monocytes) cell populations. Notably, the expression of NALP1 was significantly increased in the bone marrow blast cell population of some patients with acute leukaemia, but not among tissue samples from thyroid and renal cancer. A search for consensus sites within the NALP1 promoter revealed a sequence for CREB (cAMP-response-element-binding protein) that was required for transcriptional activity. Moreover, treatment of TF1 myeloid leukaemia cells with protein kinase C and protein kinase A activators induced CREB phosphorylation and upregulated the mRNA and protein levels of NALP1. Conversely, ectopic expression of a dominant negative form of CREB in TF1 cells blocked the transcriptional activity of the NALP1 promoter and significantly reduced the expression of NALP1. Thus NALP1 is transcriptionally regulated by CREB in myeloid cells, a mechanism that may contribute to modulate the response of these cells to pro-inflammatory stimuli.
Keywords: blast cell, cAMP-response-element-binding protein (CREB), inflammation, leukaemia, NALP1, tumour cell
Abbreviations: ATF1, activating transcription factor 1; CARD, caspase-recruitment domain; CRE, cAMP-response element; CREB, cAMP-response-element-binding protein; A-CREB, inhibitor of CREB DNA-binding activity; db-cAMP, dibutyryl-cAMP; EMSA, electrophoretic mobility-shift assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; G-CSF, granulocyte colony-stimulating factor; IL, interleukin; LRR, leucine-rich repeat; M-CSF, monocyte/macrophage colony-stimulating factor; NALP1L and NALP1S, long and short forms of NALP1 respectively; NALP1pt, promoter region of NALP1; NF-κB, nuclear factor κB; PKA, protein kinase A; PKC, protein kinase C; RACE, rapid amplification of cDNA ends; RT-PCR, reverse transcriptase PCR
INTRODUCTION
NALP1 (also called DEFCAP, NAC and CARD7) belongs to a newly identified family of cytoplasmic proteins that have been implicated in cell responses to apoptotic and inflammatory stimuli [1–3]. Two major isoforms have been identified for NALP1 which differ in a sequence of 132 bp that encodes an LRR (leucine-rich repeat) near the 3′ end of the coding region [2]. NALP1 has been shown to interact with caspases 5, -1, -2, -9 and Apaf1 [2–4], thus participating in the cytochrome c-mediated activation of apoptotic caspases, as well as in the activation of inflammatory caspases.
NALP1 differs from the other NALP proteins in that it contains a CARD (caspase-recruitment domain). CARD-containing proteins, including Apaf1, Ced-4, Nod1 and Nod2, are involved in the regulation of apoptotic and/or inflammatory responses. All these proteins have been grouped into the so-called CATERPILLER family, containing CARD, pyrin, nucleotide-binding and LRR domains [5]. Nod proteins have been shown to recognize bacterial components, including LPS (lipopolysaccharides) and/or bacterial muramyl dipeptide [6,7], and this interaction leads to the activation of NF-κB (nuclear factor κB), a transcription factor that plays a central role in innate immunity and cell survival [8,9]. Interestingly, mutations in the coding region of Nod2 have been associated with susceptibility to Crohn's disease and Blau syndrome, two chronic inflammatory disorders [10–12]. Furthermore, patients with hereditary fever syndromes and chronic inflammatory diseases, such as Muckle–Wells syndrome and familial cold urticaria, who carry mutations in the coding sequence of NALP3 have been identified recently [13]. Of note, these mutations are clustered in a highly conserved domain that shares significant similarity with NALP1.
Very little is known about the transcriptional regulation of NALPs and other genes of the CATERPILLER family. Of note, induction of Apaf1 mRNA and protein expression has been shown to be dependent on the transcriptional activity of p53 [14]. More recently, it has been described that Nod2 expression is enhanced by pro-inflammatory cytokines and bacterial components via NF-κB, a mechanism that may contribute to the amplification of the innate immune response [15]. Furthermore, much remains to be discovered to further elucidate the expression and function of these genes within the haematopoietic compartment. A detailed expression pattern in the different haematopoietic compartments has only been established for Nod2. This gene is expressed upon differentiation of CD34+ cells into granulocyte or monocyte/macrophages, and the highest levels were detected in mature myelomonocytes [15]. Previous studies showed that NALP1 is highly expressed in peripheral blood mononuclear cells [2,3]. In the present study we report that NALP1 is absent from CD34+ blast cells, but readily detected in T-lymphocytes, monocytes and granulocytes. Of note, the levels of NALP1 increased in bone marrow samples from some patients with acute leukaemia, but not in samples of solid tumours. Moreover, we have demonstrated that induction of NALP1 by a PKC (protein kinase C) activator or cAMP analogues is mediated through CREB (cAMP-response-element-binding protein), a transcription factor involved in inflammatory responses.
EXPERIMENTAL
Cells
Human leukaemia cell lines TF1 and K562 were maintained in RPMI 1640 medium (Seromed Biochrom KG, Berlin, Germany) supplemented with 10% fetal calf serum (Flow Laboratories, Irvine, CA, U.S.A.) and 5 ng/ml of recombinant human IL-3 (interleukin-3) (Immunex, Seattle, WA, U.S.A.).
Mobilized peripheral blood progenitors were obtained from normal donors undergoing mobilization for allogeneic progenitor cell transplantation, as described previously [15]. Purified CD34+ cells (more than 95%) were seeded into 24-well culture plates at 5×104 cells/ml in Iscove's modified Dulbecco's medium (Invitrogen, Carlsbad, CA, U.S.A.), containing 20% fetal calf serum and recombinant human SCF (stem cell factor), IL-3 and IL-6 (Immunex) at a final concentration of 100 ng/ml. Granulocyte and monocyte/macrophage cell populations were generated as described previously [15].
Mononuclear cells obtained from bone marrow of patients with acute leukaemia and snap-frozen tissue from patients with different solid tumours were analysed for the expression of NALP1 by real-time PCR.
All patients and normal donors gave their written informed consent according to Guidelines from the Committee for the Protection of Human Subjects at the Clinica Universitaria de Navarra.
TF1 cells were treated with PMA, db-cAMP (dibutyryl-cAMP) or staurosporine (all from Sigma, St. Louis, MO, U.S.A.) for the various time intervals indicated.
Gene reporter assays
A genomic PCR fragment of 297 bp from the promoter region of NALP1 (NALP1pt), was cloned into KpnI and XhoI sites of the pGL2-basic luciferase reporter vector (Promega, Madison, WI, U.S.A.). The authenticity of the construct was confirmed by sequencing. TF1 cells were co-transfected with 1 μg of pGL2-NALP1pt and 50 ng of pRSV-β-gal in triplicate by nucleofection following the manufacturer's instructions (Amaxa, Cologne, Germany). When indicated, cells were co-transfected with pGL2-NALP1pt and 1 μg of a vector containing A-CREB, a potent and selective inhibitor of CREB DNA-binding activity [16]. Post-transfection (24 h), cells were incubated with 50 ng/ml PMA for 6 h, and then cell extracts were prepared and analysed for the relative luciferase activity by a dual-light reporter gene assay system (Applied Biosystems, Foster City, CA, U.S.A.). Results were normalized for transfection efficiency with values obtained with pRSV-β-gal. Site-directed mutagenesis of the CRE (cAMP-response element) site in the pGL2-NALP1pt vector was carried out by using the QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) with the following primers: 5′-CCTCTGGGGAATGTTTCCTTC-3′ and 5′-GAAGGAAACATTCCCCAGAGG-3′. The NALP1pt DNA insert was sequenced to verify the mutation.
RT (reverse transcriptase)-PCR and RACE (rapid amplification of cDNA ends) analysis
Total RNA was prepared using TRIzol® reagent (Invitrogen). To assess mRNA expression, a semiquantitative RT-PCR method was used as described previously [17]. The generated cDNA was amplified by using primers for human NALP1 (5′-AAGTGACTGCTCCATTCGGAA-3′, and 5′-CTCCGAGAACAGCTGGTCTTCT-3′, located in different exons), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) [17]. After 20 (GAPDH) or 30 (NALP1) amplification cycles, the expected PCR products (205 bp for the long isoform and 73 bp for the short isoform of NALP1) were size fractionated on a 4–20% gradient polyacrylamide gel and stained with ethidium bromide.
Expression data were quantified by real-time PCR in a 7000 Sequence Detection System (Applied Biosystems). The ratio of the abundance of NALP1 transcripts to that of GAPDH transcripts was calculated as 2n, where n is the CT (threshold cycle) value of GAPDH minus the CT value of NALP1, and normalized by the value of the sample with the lowest expression level of NALP1. Specificity of the desired PCR products was determined with melting curve analysis.
For analysis of sequences at the 5′ end of NALP1 mRNA, a 5′-RACE kit was used according to the manufacturer's instructions (Roche, Mannheim, Germany), with the following NALP1 primers: 5′-GCTTGGTAGAGGAGTGAGGCA-3′ and 5′-CCACTCGTCTTCTCTGGCTGA-3′.
Western blot analysis and antibody generation
Cell extracts (60 μg of protein) were separated on an 8% polyacrylamide gel, and transferred on to nitrocellulose as described previously [17]. Blots were blocked with 3% BSA and incubated with the corresponding primary antibodies followed by incubation with goat anti-rabbit or anti-mouse antibodies conjugated to alkaline phosphatase. Bound antibody was detected by a chemiluminescence system (Applied Biosystems). A polyclonal anti-NALP1 serum was generated by repeated immunization of rabbits with a keyhole-limpet haemocyanin-conjugated synthetic peptide (amino acids 17–35 of NALP1). Rabbit polyclonal antibodies against CREB and phospho-CREB were purchased from Upstate (Charlottesville, VA, U.S.A.), and mouse anti-β-tubulin antibody was obtained from Sigma.
EMSAs (electrophoretic mobility-shift assays)
TF1 cells were lysed and nuclear fractions were resuspended in 20 mM Hepes, pH 7.9, 420 mM NaCl, 1 mM EDTA, 1 mM EGTA and 20% glycerol. Nuclear extracts (5 μg of total protein) were incubated with a 32P-labelled double-stranded DNA probe from the promoter region of the NALP1 gene (5′-TGGGTGACGTTT-3′). Samples were run on a 5% non-denaturing polyacrylamide gel in 200 mM Tris/borate and 2 mM EDTA. Gels were dried and visualized by autoradiography. Supershifts were performed using antibodies specifically against CREB, ATF1 (activating transcription factor 1) (both from Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) and phospho-CREB (Upstate).
RESULTS
NALP1 is expressed in mature haematopoietic cells
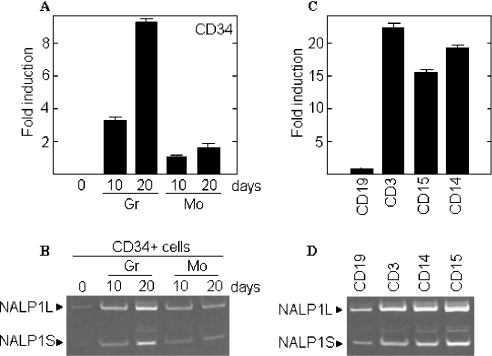
NALP1 has been shown to be expressed in peripheral blood leucocytes [18]. We aimed to extend these data by analysing the mRNA expression of NALP1 in cells derived from in vitro-differentiated CD34+ progenitors and in peripheral blood populations. We obtained the CD34+ cell fraction from the peripheral blood of normal donors, and the selected population was cultured with either G-CSF (granulocyte colony-stimulating factor) or M-CSF (monocyte/macrophage colony-stimulating factor) to induce granulocyte or monocyte/macrophage maturation, as previously described [19]. CD34+ progenitors expressed no detectable levels of NALP1 mRNA, as assessed by real-time and semiquantitative PCR (Figures 1A and 1B). However, by day 10 of culture the expression of NALP1 was readily detectable, with the highest levels observed in the granulocyte lineage (3-fold higher than in monocytic cells). At day 20 of culture the expression of NALP1 increased and the mRNA levels in granulocyte cells were about 5-fold higher than in monocytes (Figure 1A).
Figure 1. Analysis of NALP1 mRNA in CD34+-derived cells and peripheral blood subpopulations.
Total RNA was purified from CD34+ cells cultured in the presence of M-CSF (Mo) or G-CSF (Gr) (A and B) and from immune-purified peripheral blood cell populations (C and D), and subjected to real-time PCR (A and C) and semiquantitative RT-PCR analysis (B and D). PCR products were subjected to electrophoresis on a gradient polyacrylamide gel and stained with ethidium bromide. The histograms show the means±S.D. for triplicate analyses.
Then we analysed the expression of NALP1 mRNA in peripheral blood populations of B-cells (91.4% of CD19+ cells), T-cells (99.0% of CD3+ cells), granulocytes (84.0% of CD15+ cells) and monocytes (99% of CD14+ cells) (Figures 1C and 1D). The mRNA analysis showed that B-lymphocytes express low levels of NALP1, whereas granulocytes, monocytes and T-lymphocytes express 16- to 23-fold as much NALP1 mRNA as the B-cells by real-time PCR (Figure 1C). Semiquantitative RT-PCR analyses demonstrated that the two major isoforms of NALP1 (NALP1L and NALP1S, long and short forms respectively) were present in both differentiated CD34+ cells and peripheral blood populations. However, NALP1L consistently gave the highest signal on gels (Figures 1B and 1D).
Differential expression of NALP1 in tumour cells
NALP1 has been shown to be highly expressed in the chronic myeloid leukaemia cell line K562 [2]. Additionally, we have demonstrated that NALP1 is absent in normal progenitor blast cells (CD34+) and its levels increased along the myelomonocytic differentiation. With this in mind, we studied the expression of NALP1 in mononuclear cells from the bone marrow of 40 patients with acute myeloid leukaemia (Figure 2A) or acute lymphocytic leukaemia (Figure 2B). All selected patients had more than 80% blast cells in the bone marrow. Samples from 9 patients showed increased mRNA levels of NALP1, and differences among samples were as high as 60-fold. Interestingly, some of the samples preferentially expressed the short isoform, although the functional meaning of this pattern is presently unknown. In view of these data, we studied the expression of NALP1 in solid tumours. Figure 2(C) shows that the mRNA levels of NALP1 were homogeneous in all samples from thyroid and renal cancer as determined by semiquantitative RT-PCR. Furthermore, in all cases the predominant isoform was NALP1L. These data suggest that the differences in mRNA levels and isoforms of NALP1 are the result of genetic and/or signalling alterations occurring preferentially in haematologic malignancies such as acute leukaemia.
Figure 2. Expression of NALP1 in tumour cells.
Total RNA was obtained from the bone marrow of patients with acute myeloid (A) or lymphocytic (B) leukaemia and from frozen tissue sections of different solid tumours (C), and analysed for the expression of NALP1 by real-time PCR (upper panels in A and B) and semiquantitative RT-PCR (lower panels in A and B). PCR products were subjected to electrophoresis on a gradient polyacrylamide gel and stained with ethidium bromide. GAPDH mRNA was used as an amplification control. The histograms show the means±S.D. for triplicate analyses.
Promoter region of NALP1 contains a CRE consensus sequence
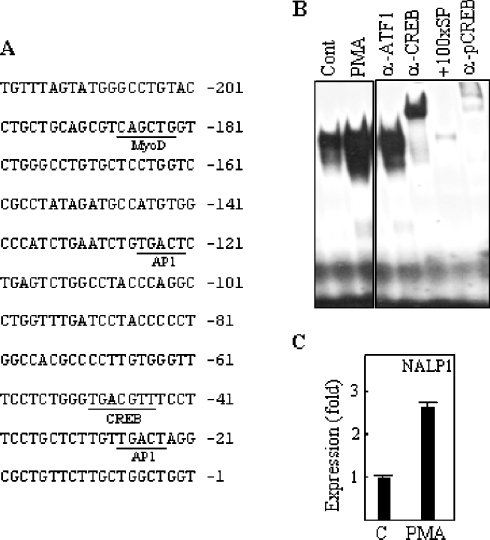
In order to study the transcriptional regulation of NALP1, we first located the promoter region. A 5′-RACE analysis revealed several transcription start sites in K562 and TF1 myeloid leukaemia cell lines. Thus we decided to focus on the upstream sequence from the most 5′ transcript identified in these cell lines, which coincides with the promoter region described for NALP1 by the Transcript Sequence Retrieval (TRASER) database (D. Relman, Department of Microbiology and Immunology, Stanford University, Stanford, CA, U.S.A.) (Figure 3A). We found a putative CRE sequence 47 bases upstream from the transcription start site (TGACGTT) that is not identical to the CRE consensus sequence (TGACGTC). However, it is described that CREB does not distinguish between the canonical CRE and other sequences, even with more significant differences, such as the Ces2/E2A-HLF-binding element (TTACGTA). Furthermore, the same CRE sequence found in the NALP1 promoter is present in the Bcl-2 5′ region, and this site demonstrates functional activity [20]. Thus we analysed the capacity of the CRE site to bind to CREB protein. Nuclear extracts from TF1 cells were incubated with a radiolabelled CRE probe from the NALP1 promoter, and then subjected to EMSA. As shown in Figure 3(B), a single protein–DNA complex was detected, which was supershifted in the presence of specific antibodies against CREB and active phospho-CREB, but not ATF1, a transcription factor that shares high sequence similarity with CREB, indicating that CREB specifically binds to the sequence found in the NALP1 promoter.
Figure 3. Binding of CREB to the NALP1 promoter is increased by PMA.
(A) Partial sequence of the NALP1 promoter. Putative transcription factor-binding sites are underlined. (B) Control (Cont; untreated) or PMA-treated TF1 cells were analysed for the formation of CREB–DNA complexes by EMSA using a radiolabelled probe from the NALP1 promoter. Nuclear extracts were pre-incubated with antibodies (α-) specific for CREB, phospho-CREB (pCREB) or ATF1, or with 100-fold molar excess of unlabelled CREB-specific probe (+100xSP). (C) Cells were treated with 10 ng/ml PMA for 24 h and then total RNA was extracted and analysed for the expression of NALP1 by real-time PCR. The histogram shows the means±S.D. for triplicate analyses.
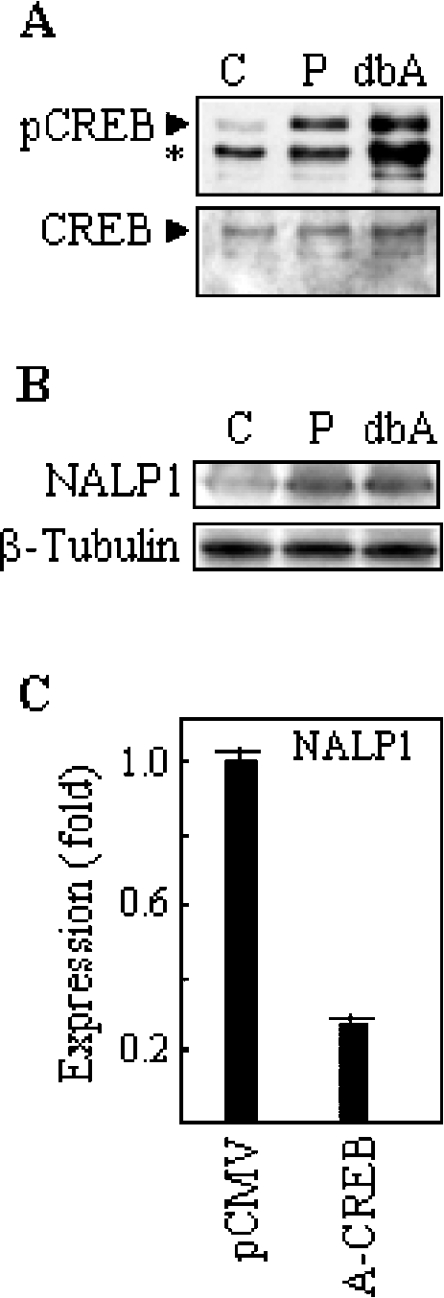
It has been shown that growth factors induce CREB activation in TF1 cells through PKC [21]. Thus we studied the effects of stimulation with PMA, a PKC activator, on the expression of NALP1 and CREB phosphorylation. PMA induced the binding of CREB to the NALP1 promoter, as shown in Figure 3(B). Furthermore, after 24 h of stimulation with PMA, the levels of NALP1 mRNA in TF1 cells were increased about 2.5-fold, as determined by real-time PCR (Figure 3C). Consistently, this treatment resulted in the rapid phosphorylation of CREB without a significant change in the protein levels (Figure 4A). In order to extend the expression data to other CREB-activating stimuli, we treated TF1 cells with db-cAMP, a cAMP analogue shown to induce CREB phosphorylation in a number of cell systems [22]. As expected, db-cAMP upregulated phospho-CREB in TF1 (Figure 4A). After 24 h of incubation, the NALP1 protein levels were determined by Western blotting with an antiserum raised against a synthetic NALP1 peptide. Specificity analyses revealed that pre-immune sera failed to produce any signal and an NALP1-specific neutralizing peptide abrogated the antibody reaction (results not shown). As shown in Figure 4(B), treatment with db-cAMP induced NALP1 protein, and the level of expression was similar to that reached with PMA. Conversely, treatment with the PKC inhibitor staurosporine reduced both phosphorylation of CREB and expression of NALP1 (results not shown).
Figure 4. Regulation of NALP1 levels in response to CREB modulators.
(A) TF1 cells were incubated with 50 ng/ml PMA (P) and 1 mM db-cAMP (dbA) and analysed for the presence of phospho-CREB by Western blotting. The asterisk denotes a cross-reactive protein recognized by the anti-pCREB antibody. The levels of CREB protein were also determined. Lane C, untreated control cells. (B) Cells were cultured in the presence of 10 ng/ml PMA (P), and 1 mM db-cAMP (dbA), and then analysed for the expression of NALP1 protein by Western blotting with an antiserum against an NALP1 peptide. The levels of β-tubulin were also determined to verify equal loading. (C) TF1 cell line was transiently transfected with a control vector (pCMV) or A-CREB. After 24 h of transfection, the mRNA levels of NALP1 were determined by real-time PCR. The histogram shows the means±S.D. for triplicate analyses.
To determine whether CREB mediated the expression of NALP1, we transfected TF1 cells with A-CREB, a dominant negative inhibitor that functions by preventing DNA binding of CREB in a dimerization domain-dependent fashion [16]. Figure 4(C) shows that transfection with A-CREB, but not with vector alone, led to a significant reduction (more than 3-fold) in the mRNA levels of NALP1 as assessed by real-time PCR.
NALP1 promoter responds to modulators of CREB activity
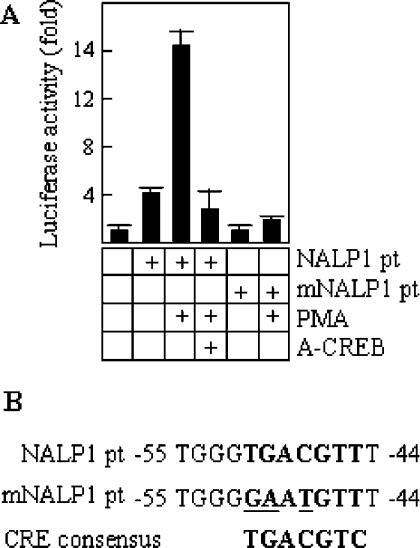
To study the promoter activity of the CRE sequence, a 297 bp fragment from the promoter region of NALP1 (NALP1pt), containing the CRE consensus site, was cloned into a promoterless luciferase vector (NALP1pt-luciferase), and this construct was transiently transfected into TF1 cells. Figure 5(A) shows a representative experiment. The levels of luciferase activity detected in cells stimulated with PMA were significantly higher than those observed in unstimulated cells (approx. 3.5-fold). However, this activation was abolished in the presence of A-CREB, suggesting that it is dependent on endogenous CREB. To further clarify the functional specificity of the CRE site contained in the NALP1pt fragment, a mutagenesis analysis was performed. We mutated three bases within the sequence motif (Figure 5B) and transfected TF1 cells with an NALP1pt mutant luciferase vector. As shown in Figure 5(A), there were no significant differences between the PMA-stimulated and the unstimulated cells, and in both conditions the level of luciferase activity was similar to that observed in non-transfected control cells. Thus these data show the importance of the CRE consensus site in the transcriptional activation of NALP1 gene, and suggest a low, if any, contribution of other putative binding sequences, at least in the cell system used here.
Figure 5. Transcriptional regulation of CRE-containing NALP1 promoter.
(A) Cells were transfected with a wild-type NALP1 promoter fragment (NALP1pt) either alone or with A-CREB, and a mutant NALP1 promoter [mNALP1pt; the mutation is shown in (B)], in the presence of a constitutive β-galactosidase reporter vector (pRSV-β-gal). Cells were induced with 50 ng/ml PMA for 6 h or left untreated. Units of luciferase activity were normalized based on values of pRSV-β-gal activity to control for transfection efficiency. The results are expressed as the mean±S.D. for triplicate cultures.
DISCUSSION
NALP1 is a member of the NALP protein family that shows a domain structure similar to that found in Nod family members [1]. Studies using genetically modified cells have revealed that Nod1 and Nod2 activate NF-κB, a transcription factor that plays a central role in inflammatory responses and innate immunity [8]. In the present study we show that NALP1 is upregulated during the myelomonocytic differentiation of CD34+ progenitor cells. This expression pattern is similar to that described previously for Nod2 [15]. Furthermore, NALP1 has been shown to be expressed in peripheral blood mononuclear leucocytes and polymorphonuclear cells [2,3]. We have extended these results, demonstrating that NALP1 is mainly expressed in T-lymphocytes, granulocytes and monocytes. However, we found an apparent discrepancy in NALP1 expression between granulocytes and monocytes when comparing cells obtained from peripheral blood or in vitro-derived CD34+ progenitors. A likely explanation is that CD34+ cells have been differentiated under the selective pressure of individual factors (i.e., G-CSF or M-CSF), whereas peripheral blood cells have matured under different external stimuli, including interactions with stromal or other haematopoietic cells, and with an array of growth and differentiation factors. This may lead to differences in the expression pattern of genes targeted by the affected signalling pathways. In line with this, differences in the expression of Nod2 between CD15+ and CD14+ subpopulations were much higher than those found between CD34-derived granulocytes and monocytes [15]. Recent studies have shown that NALP1 protein forms a large signal-induced multiprotein complex that comprises caspase-1, caspase-5 and Pycard/Asc, which plays a crucial role in the processing of pro-IL-1β [1,4]. Constitutive expression of IL-1β has been detected in many forms of leukaemia, and, once released, it can operate in an autocrine or paracrine manner to induce the proliferation of leukaemic cells [23]. Interestingly, we found that the levels of NALP1 mRNA varied significantly among patients with acute myeloid or lymphocytic leukaemia, although we could not establish a clear correlation between NALP1 expression and disease outcome, because of the lack of clinical data in most of the samples analysed. Of note, two major splice variants of NALP1 exist, one of which does not contain an LRR. In line with this, it has been described that a Nod2 variant lacking part of the LRRs, shows a defective response to bacterial products, and is associated with susceptibility to a chronic inflammatory bowel disease [10]. We showed that some of the bone marrow samples from leukaemia patients mainly express the LRR-deficient isoform of NALP1. Thus, it is tempting to speculate that this NALP1 isoform is defective in sensing the signals triggered by external stimuli, including pro-inflammatory molecules, and consequently leukaemic cells carrying this variant would be less efficient in the production of IL-1β. Further studies will be needed to determine the contribution of NALP1 isoforms to the clinical course of leukaemia.
Based on the differential expression of NALP1 in different haematopoietic cell systems, we focussed on the transcriptional regulation of this gene, and found that CREB may be the major transcriptional inducer of NALP1 in myeloid leukaemia cells.
Activation of CREB by phosphorylation has been shown to upregulate the expression of the pro-inflammatory cytokine IL-6 [24], and to induce the transcriptional activation of cyclo-oxygenase-2, which plays a critical role in human inflammatory disorders [25]. Thus, as described for NALP1, CREB is also implicated in the regulation of inflammatory responses.
CREB responds to a diverse array of extracellular signals that activate a variety of protein kinases, including PKA (protein kinase A), MAPKs (mitogen-activated protein kinases), CaMKs (Ca2+/calmodulin-dependent protein kinases) and PKC [26,27]. It has been shown that IL-3 and GM-CSF (granulocyte/macrophage/monocyte colony-stimulating factor) induce phosphorylation of CREB in TF1 cells through PKC, indicating that, at least in myeloid cells, PKC plays a major role in CREB activation [21]. Consistently, we have shown that treatment of leukaemic TF1 cells with the PKC activator PMA induces CREB phosphorylation and upregulates NALP1 mRNA, and that blockade of PKC or CREB activity reduces the expression of NALP1. In addition, db-cAMP also induces NALP1 and phospho-CREB in TF1. This cAMP analogue activates PKA, a key effector of cAMP-mediated signal transduction [28]. Thus it is likely that PKA also contributes to NALP1 expression. Nevertheless, it remains to be elucidated whether the PKC–CREB pathway regulates the expression of NALP1 in more physiologically relevant models, including primary leukaemia cells.
In conclusion, we describe in the present study that CREB activates transcription of NALP1 through signalling pathways dependent, at least in part, on PKC in a myeloid leukaemia cell line. This transcriptional mechanism may be involved in the regulation of myeloid cell responses to pro-inflammatory stimuli.
Acknowledgments
This work was supported by a grant from ‘Fundacion Marcelino Botin’ (proyecto Terapia Genica) to J.L.F.-L., and by grant C03/10 from ‘Fondo de Investigacion Sanitaria’ (programa RTICCC) to J.L.F.-L. and F.P. We are grateful to David Ginty for providing the A-CREB plasmid.
References
- 1.Tschopp J., Martinon F., Burns K. NALPs: a novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 2003;4:95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- 2.Hlaing T., Guo R. F., Dilley K. A., Loussia J. M., Morrish T. A., Shi M. M., Vincenz C., Ward P. A. Molecular cloning and characterization of DEFCAP-L and -S, two isoforms of a novel member of the mammalian Ced-4 family of apoptosis proteins. J. Biol. Chem. 2001;276:9230–9238. doi: 10.1074/jbc.M009853200. [DOI] [PubMed] [Google Scholar]
- 3.Chu Z. L., Pio F., Xie Z., Welsh K., Krajewska M., Krajewski S., Godzik A., Reed J. C. A novel enhancer of the Apaf1 apoptosome involved in cytochrome c-dependent caspase activation and apoptosis. J. Biol. Chem. 2001;276:9239–9245. doi: 10.1074/jbc.M006309200. [DOI] [PubMed] [Google Scholar]
- 4.Martinon F., Burns K., Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 5.Harton J. A., Linhoff M. W., Zhang J., Ting J. P. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J. Immunol. 2002;169:4088–4093. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- 6.Inohara N., Ogura Y., Chen F. F., Muto A., Nunez G. Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J. Biol. Chem. 2001;276:2551–2554. doi: 10.1074/jbc.M009728200. [DOI] [PubMed] [Google Scholar]
- 7.Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 8.Silverman N., Maniatis T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 9.Wang C. Y., Mayo M. W., Korneluk R. G., Goeddel D. V., Baldwin A. S., Jr NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 10.Ogura Y., Bonen D., Inohara N., Nicolae D., Chen F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R., et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 11.Hugot J., Chamaillard M., Zouali H., Lesage S., Cezard J., Belaiche J., Almer S., Tysk C., O'Morain C., Gassull M., et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 12.Miceli-Richard C., Lesage S., Rybojad M., Prieur A., Manouvrier-Hanu S., Hafner R., Chamaillard M., Zouali H., Thomas G., Hugot J. CARD15 mutations in Blau syndrome. Nat. Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 13.Dode C., Le Du N., Cuisset L., Letourneur F., Berthelot J. M., Vaudour G., Meyrier A., Watts R. A., Scott D. G., Nicholls A., et al. New mutations of CIAS1 that are responsible for Muckle–Wells syndrome and familial cold urticaria: a novel mutation underlies both syndromes. Am. J. Hum. Genet. 2002;70:1498–1506. doi: 10.1086/340786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robles A. I., Bemmels N. A., Foraker A. B., Harris C. C. APAF-1 is a transcriptional target of p53 in DNA damage-induced apoptosis. Cancer Res. 2001;61:6660–6664. [PubMed] [Google Scholar]
- 15.Gutierrez O., Pipaon C., Inohara N., Fontalba A., Ogura Y., Prosper F., Nunez G., Fernandez-Luna J. L. Induction of Nod2 in myelomonocytic and intestinal epithelial cells via nuclear factor-κB activation. J. Biol. Chem. 2002;277:41701–41705. doi: 10.1074/jbc.M206473200. [DOI] [PubMed] [Google Scholar]
- 16.Riccio A., Ahn S., Davenport C. M., Blendy J. A., Ginty D. D. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science. 1999;286:2358–2361. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 17.Benito A., Silva M., Grillot D., Nunez G., Fernandez-Luna J. Apoptosis induced by erythroid differentiation of human leukemia cell lines is inhibited by Bcl-XL. Blood. 1996;87:3837–3843. [PubMed] [Google Scholar]
- 18.Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-κB. J. Biol. Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 19.Sanz C., Benito A., Silva M., Albella B., Richard C., Segovia J. C., Insunza A., Bueren J. A., Fernandez-Luna J. L. The expression of Bcl-x is downregulated during differentiation of human hematopoietic progenitor cells along the granulocyte but not the monocyte/macrophage lineage. Blood. 1997;89:3199–3204. [PubMed] [Google Scholar]
- 20.Ji L., Mochon E., Arcinas M., Boxer L. M. CREB proteins function as positive regulators of the translocated bcl-2 allele in t(14;18) lymphomas. J. Biol. Chem. 1996;271:22687–22691. doi: 10.1074/jbc.271.37.22687. [DOI] [PubMed] [Google Scholar]
- 21.Gubina E., Luo X., Kwon E., Sakamoto K., Shi Y. F., Mufson R. A. βc cytokine receptor-induced stimulation of cAMP response element binding protein phosphorylation requires protein kinase C in myeloid cells: a novel cytokine signal transduction cascade. J. Immunol. 2001;167:4303–4310. doi: 10.4049/jimmunol.167.8.4303. [DOI] [PubMed] [Google Scholar]
- 22.Boer A. K., Drayer A. L., Rui H., Vellenga E. Prostaglandin-E2 enhances EPO-mediated STAT5 transcriptional activity by serine phosphorylation of CREB. Blood. 2002;100:467–473. doi: 10.1182/blood.v100.2.467. [DOI] [PubMed] [Google Scholar]
- 23.Beaupre D. M., Talpaz M., Marini F. C., 3rd, Cristiano R. J., Roth J. A., Estrov Z., Albitar M., Freedman M. H., Kurzrock R. Autocrine interleukin-1β production in leukemia: evidence for the involvement of mutated RAS. Cancer Res. 1999;59:2971–2980. [PubMed] [Google Scholar]
- 24.Sitaraman S. V., Merlin D., Wang L., Wong M., Gewirtz A. T., Si-Tahar M., Madara J. L. Neutrophil-epithelial crosstalk at the intestinal lumenal surface mediated by reciprocal secretion of adenosine and IL-6. J. Clin. Invest. 2001;107:861–869. doi: 10.1172/JCI11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schroer K., Zhu Y., Saunders M. A., Deng W. G., Xu X. M., Meyer-Kirchrath J., Wu K. K. Obligatory role of cyclic adenosine monophosphate response element in cyclooxygenase-2 promoter induction and feedback regulation by inflammatory mediators. Circulation. 2002;105:2760–2765. doi: 10.1161/01.cir.0000018127.10968.34. [DOI] [PubMed] [Google Scholar]
- 26.Shaywitz A. J., Greenberg M. E. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 27.Xie H., Rothstein T. L. Protein kinase C mediates activation of nuclear cAMP response element-binding protein (CREB) in B lymphocytes stimulated through surface Ig. J. Immunol. 1995;154:1717–1723. [PubMed] [Google Scholar]
- 28.Meinkoth J. L., Alberts A. S., Went W., Fantozzi D., Taylor S. S., Hagiwara M., Montminy M., Feramisco J. R. Signal transduction through the cAMP-dependent protein kinase. Mol. Cell. Biochem. 1993;127–128:179–186. doi: 10.1007/BF01076769. [DOI] [PubMed] [Google Scholar]