Abstract
We analysed key biochemical features that reflect the balance between glycolysis and glucose oxidation in cybrids (cytoplasmic hybrids) harbouring a representative sample of mitochondrial DNA point mutations and deletions. The cybrids analysed had the same 143B cell nuclear background and were isogenic for the mitochondrial background. The 143B cell line and its ρ0 counterpart were used as controls. All cells analysed were in a dynamic state, and cell number, time of plating, culture medium, extracellular volume and time of harvest and assay were strictly controlled. Intra- and extra-cellular lactate and pyruvate levels were measured in homoplasmic wild-type and mutant cells, and correlated with rates of ATP synthesis and O2 consumption. In all mutant cell lines, except those with the T8993C mutation in the ATPase 6 gene, glycolysis was increased even under conditions of low glucose, as demonstrated by increased levels of extracellular lactate and pyruvate. Extracellular lactate levels were strictly and inversely correlated with rates of ATP synthesis and O2 consumption. These results show increased glycolysis and defective oxidative phosphorylation, irrespective of the type or site of the point mutation or deletion in the mitochondrial genome. The different biochemical consequences of the T8993C mutation suggest a uniquely different pathogenic mechanism for this mutation. However, the distinct clinical features associated with some of these mutations still remain to be elucidated.
Keywords: ATP synthesis, cybrid, lactate, mitochondrial DNA mutation, oxygen consumption, pyruvate
Abbreviations: cybrid, cytoplasmic hybrid; DMEM, Dulbecco's modified Eagle's medium; EC, extracellular; IC, intracellular; L/P, lactate-to-pyruvate ratio; M, mutant; MELAS, mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes; MERRF, myoclonus epilepsy with ragged-red fibres; NARP/MILS, neuropathy, ataxia, retinitis pigmentosa/maternal inherited Leigh's syndromes; MIDD, maternally inherited diabetes and deafness; mtDNA, mitochondrial DNA; TCA, trichloracetic acid; WT, wild-type
INTRODUCTION
Mitochondria are the main source of cellular ATP, which is generated by the transfer of electrons along complexes I to IV of the mitochondrial respiratory chain and by the phosphorylation of ADP by complex V (ATP synthase) [1]. In addition to the nuclear genome, cells contain mtDNA (mitochondrial DNA), a double-stranded circular molecule present in multiple copies in each mitochondrion, which encodes for 22 tRNAs, 13 polypeptides and two rRNAs. All 13 polypeptides are components of the respiratory chain. Mutations in the mitochondrial genome have been described in a heterogeneous group of disorders in which the nervous system and skeletal muscle are predominantly, but not exclusively, affected (mitochondrial encephalomyopathies). The main biochemical consequences of pathogenic mtDNA mutations are impaired respiratory capacity and ATP synthesis. Since the oxidative phosphorylation system is controlled by both genomes, genetic mitochondrial diseases can be sporadic, transmitted by Mendelian inheritance (nuclear DNA mutations) or by maternal inheritance (mtDNA mutations). Point mutations in rRNA, tRNA and polypeptide-coding genes are usually maternally inherited, whereas large-scale rearrangements of mtDNA are usually sporadic [2].
The diagnosis of mitochondrial diseases is based on morphological, biochemical and molecular genetic analyses of muscle. Because it is often difficult to obtain enough material to study pathogenic mechanisms, patient-derived cell culture models have been used to establish bioenergetic deficits and to analyse biochemical phenotypes. Even though primary cultures of skin or muscle are appropriate for biochemical and molecular genetic analyses, mtDNA heteroplasmy [i.e. the co-existence of M (mutant) and WT (wild-type) genomes in the same cell] and senescence on prolonged passaging limit their usefulness. Lymphoblastoid cell lines have been useful, but heteroplasmy remains a problem [3]. The cell culture model most extensively used for the study of mitochondrial disorders is the cybrid (cytoplasmic hybrid) system, which was first described in mammalian cells by King and Attardi [4]. Cybrids are generated by fusion of enucleated cytoplasts from patients' cells harbouring mtDNA mutations with cells lacking mtDNA (ρ0), grown under selection and subcloned. This technique permits the analysis of cell lines that are isogenic for mtDNA, have a neutral nuclear background and contain not only homoplasmic WT and homoplasmic M mtDNA, but also cells with different percentages of M molecules from the same patient. Several mtDNA mutations have been studied and characterized using this system [5].
Most point mutations studied by this method are in the tRNA genes. Mutations in the tRNALeu (UUR) gene are found in patients with MELAS (mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes). Cybrids with mutations at nt 3243 and 3271 had decreased complex I activity and defective protein synthesis, resulting in respiratory chain dysfunction [6], although the T3271C mutation was associated with a milder biochemical phenotype [7]. Patients with MERRF (myoclonus epilepsy with ragged-red fibres) have point mutations in the tRNALys gene. Cybrids harbouring the two most frequent MERRF mutations (A8344G and T8356C) show severe reduction in protein synthesis, synthesis of aberrant translation products and defective amino-acylation of the tRNA [8]. Two transitions at the same nucleotide of the ATPase 6 gene (T8993G and T8993C) are associated with the NARP/MILS (neuropathy, ataxia, retinitis pigmentosa/maternal inherited Leigh's syndromes) clinical phenotypes [9,10]. Cybrids homoplasmic for the T8993G mutation showed a residual ATP synthesis capacity of 20% to 35% [11,12]. Cybrids harbouring large-scale mtDNA deletions also had impaired protein synthesis, probably due to the ablation of one or more tRNA genes [13], and resulted in severe respiratory chain deficiency [14,15].
Since mtDNA mutations cause a wide spectrum of clinical and biochemical phenotypes, any understanding of pathogenesis requires that multiple aspects of bioenergetics be analysed in culture models. These include: (i) the end-point of glycolysis at the level of lactate and pyruvate, (ii) the terminal step of oxidative phosphorylation, resulting in ATP synthesis, and (iii) overall respiratory chain function, as reflected by rates of O2 consumption.
Rates of ATP synthesis varied widely in studies of cell lines using different methods [11,16–20], and a few studies of M cybrids reported defective O2 consumption [6,8], but neither the rates of ATP synthesis nor the rates of O2 consumption were correlated with rates of glycolysis or glucose oxidation. Therefore, there is no comprehensive picture of the metabolic profile in cells harbouring mtDNA mutations.
In the present study, we have analysed key biochemical features that reflect the global bioenergetic status of cybrid cell lines harbouring a representative sample of point mutations and rearrangements. We have measured intra- and extra-cellular lactate and pyruvate in homoplasmic WT and homoplasmic M cybrids, and we have correlated these levels with ATP synthesis and the rate of O2 consumption. Both WT and M cybrids had the same 143B cell nuclear background and were isogenic in terms of mitochondrial background (i.e. both WT and M mitochondria came from the same patient). All cells studied were in a dynamic state, where cell number, time of plating, culture components, EC (extracellular) volume, time of harvest and assay were strictly controlled. Our results revealed that all mutations, except for T8993C, increased glycolysis even under conditions of low glucose, as demonstrated by increased EC lactate and pyruvate. We also observed good correlations between EC lactate, mitochondrial ATP synthesis and rates of O2 consumption.
EXPERIMENTAL
Cybrid cell lines and cell culture
The cybrid cell lines used in this study have been described in previous publications (Table 1) [6–11,21–26]. We selected homoplasmic WT and homoplasmic M cell lines for each mutation after RFLP (restriction-fragment-length polymorphism) analysis (point mutations) or Southern blot analysis (deletions). For analysis of ATP synthesis, 1×106 cells were plated in 10 ml of DMEM (Dulbecco's modified Eagle's medium) containing 2 mg/ml glucose, 2.5 mg/ml galactose and 110 μg/ml pyruvate, and were supplemented with 50 μg/ml uridine and 10% fetal bovine serum. Cells growing exponentially in 10 cm2 dishes were harvested at 70–80% confluency and analysed the same day.
Table 1. Homoplasmic cybrid cell lines analysed.
FLP, WS, RN, MM, LB, KB, JCP, DMP, AT and CW refer to patients' cell lines.
| Mutation | Mutation reference | Cybrids used | Cybrids reference | Source of mitochondria |
|---|---|---|---|---|
| MELAS A3243G | [22] | WS 241 (WT) | [6] | Fibroblasts |
| WS 176 (M) | ||||
| RN 236 (WT) | Myoblasts | |||
| RN 164 (M) | ||||
| MELAS T3271C | [23] | MM 71 (WT) | [7] | Fibroblasts |
| MM 37 (M) | ||||
| MM 63 (M) | ||||
| MERRF A8344G | [25] | LB 18 (WT) | [8] | Fibroblasts |
| LB 172 (WT) | ||||
| LB 12 (M) | ||||
| LB 64 (M) | ||||
| MERRF T8356C | [26] | KB 30 (WT) | ||
| KB 42 (M) | [8] | Fibroblasts | ||
| KB 57 (M) | ||||
| KB 106 (M) | ||||
| NARP T8993G | [9] | JCP 213 (WT) | [11] | Platelets |
| JCP 261 (M) | ||||
| JCP 239/Eb13-13 | ||||
| NARP T8993C | [10] | DMP 20/Eb10-21(WT) | Pers.Com. | Platelets |
| DMP 20/Eb10-3 (M) | ||||
| DMP 20/Eb 10-22 (M) | ||||
| AT 153 (WT) | Unpublished work† | Fibroblasts | ||
| AT 101 (M) | ||||
| Deletions | [24] | [21] | ||
| FLP‡ | FLP 6a39.2 (WT) | Fibroblasts | ||
| FLP 6a39.32 (M) | ||||
| CW§ | CW 440-116 (WT) | Fibroblasts | ||
| CW 420-106 (WT) | ||||
| CW 420-115 (M) |
* G. Manfredi, personal communication.
† M. Davidson and M. P. King.
‡ Δ 1.9 kb, nt 7846–9748.
§ Δ 5.8 kb, nt 10155–15945.
For lactate and pyruvate assays, 6×105 cells were plated in 6 cm2 dishes in 5 ml of regular DMEM containing 4.5 mg/ml glucose. After 24 h, the medium was changed to DMEM containing 2 mg/ml glucose, 2.5 mg/ml galactose and 110 μg/ml pyruvate, supplemented with 50 μg/ml uridine and 10% fetal bovine serum. After 48 h in low glucose, the cells were harvested and counted, and lactate and pyruvate levels were measured in the cell pellet (intracellular, IC) and in the medium (EC).
IC and EC lactate and pyruvate measurements
The cell pellet was washed in cold PBS, resuspended in one volume of cold PBS, sonicated with three 15-s pulses, and deproteinized with 3 vol. of 10% TCA (trichloracetic acid). The medium was deproteinized with 4 vol. of 10% TCA. The TCA precipitates from the cell pellet and from the medium were vortex-mixed, left to stand in ice for 5 min and centrifuged. Then the supernatants were used for the assay.
Lactate levels were measured in 0.1 ml of the supernatant spectrophotometrically using the lactate kit UV 826 (Sigma., St. Louis, MO, U.S.A.), together with appropriate blanks containing 0.1 ml of 10% TCA, and standards (metabolite control, Sigma). IC lactate levels were expressed as μmol of lactate per 106 cells. Because cell counts at harvest varied from dish to dish, EC lactate levels in the total volume of the medium (5 ml) were referred to cell counts and expressed as mmol of lactate in medium/106 cells, thus keeping the units of IC and EC lactates uniform.
Pyruvate levels were analysed in 0.25 ml of TCA-treated supernatant from cells (IC) and 0.5 ml of TCA-treated supernatant from medium (EC). The change in absorbance at 340 nm at 37 °C in the presence of NADH and LDH was measured in a Cary UV100 spectrophotometer (Varian Inc., Walnut Creek, CA, U.S.A.) using a pyruvate kit UV-726 (Sigma). IC and EC pyruvate levels were calculated and expressed as above.
ATP synthesis
The rate of ATP synthesis was assayed by incubating cells (5×106/ml) permeabilized by digitonin [27], with 20 mM succinate plus 4 μM rotenone. The reaction, started by addition of 0.5 mM ADP, was stopped after 5 min at 30 °C by addition of about 80% DMSO. The ATP content was measured by the luciferin–luciferase chemiluminescent method [28], and oligomycin sensitivity was determined by pre-incubating the sample with 15 μM oligomycin.
O2 consumption
Homoplasmic WT and M cybrids harbouring the different mtDNA mutations, 143B (ρ+) and 143B206 (ρ0) cells (controls), cultured in 10 cm2 dishes as described above, were treated with trypsin at exponential phase. Aliquots of 5×106 cells were resuspended in 1.5 ml of DMEM without glucose, supplemented with pyruvate, and the rate of O2 consumption of whole cells was recorded for 3 min using a Clark O2 electrode (Hansatech Instruments, King's Lynn, Norfolk, U.K.). All measurements were carried out in triplicate at two different time points.
Citrate synthase
Citrate synthase was assayed in freshly harvested cells by the method of Srère [29]. Cells were resuspended in PBS, lysed by two freeze–thaw cycles, incubated with DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)], acetyl-CoA and oxaloacetate, and the change in absorbance at 412 nm was recorded in a Cary UV100 spectrophotometer. The enzyme activities were normalized to mg of protein in the cell lysate.
RESULTS AND DISCUSSION
The major function of mitochondria is to synthesize ATP from ADP and Pi through the action of the F1Fo-ATP synthase, which transduces the electrochemical gradient energy derived from the oxidation of NADH and FADH2 in the respiratory chain [30]. Therefore, mutations in mtDNA, which encodes the 13 polypeptides of the respiratory chain, are expected to impair energy metabolism. To test this hypothesis, we analysed cybrids with the 143B nuclear background and harbouring the most frequent pathogenic mtDNA point mutations and rearrangements. We compared energy metabolism intermediates and respiratory function in cells repopulated with either homoplasmic WT or M mtDNA molecules from the same patient. The cells analysed were therefore isogenic with respect to both nuclear and mitochondrial genome. We have used the parental 143B cells as WT (ρ+) controls and the ρ0 derivative, 143B206 cells, as negative controls. Cybrid cells are normally grown in medium containing high glucose (4.5 mg/ml). To reveal subtle biochemical defects while maintaining normal cell functions during the period of study, the cells were maintained in medium containing only 2 mg/ml glucose, but supplemented with 2.5 mg/ml galactose as a carbon source for biosynthetic functions. We performed our studies 48 h after fluid change, thus allowing us to examine the products of glycolysis in both the IC and EC compartment at peak growth phase.
The results of the biochemical analyses in cybrids harbouring 0 and 100% M mtDNA, and in ρ0 and ρ+ controls, are presented in Figures 1 and 2, and Tables 2 and 3. The data revealed a wide variation in EC lactate concentration, O2 consumption and ATP synthesis, among the different WT cybrids analysed. This variation, which had been noted by King and Attardi [4] in their original report, is probably due to the different mitochondrial donors of the cybrid lines. King and Attardi [4] showed that the variation in respiratory competence in cybrids obtained from different mitochondrial donors did not correlate with mtDNA copy number. In our present study, we used, as mitochondrial donors, primary cells from patients of very different ages. The aneuploid nature of the parental ρ0 cells may also contribute to these differences. Therefore, we consistently compared data sets from cybrids harbouring WT and M mtDNA from the same patient. Longitudinal comparisons between all WT and all M cybrids would be erroneous and misleading.
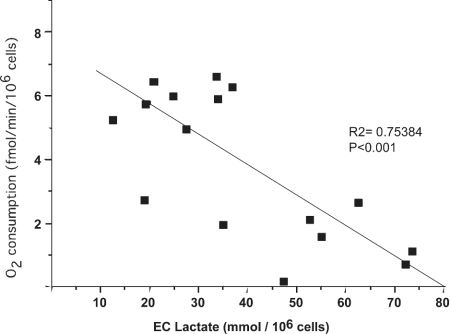
Figure 1. Correlation between EC lactate and O2 consumption in cybrids with homoplasmic WT and homoplasmic mtDNA mutations.
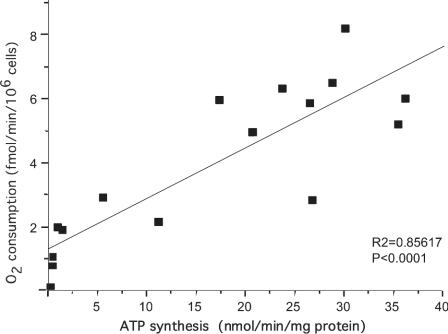
Figure 2. Correlation between ATP synthesis and O2 consumption in cybrids with homoplasmic WT and homoplasmic mtDNA mutations.
Table 2. Lactate and pyruvate levels in cybrids harbouring mtDNA mutations.
The results are expressed as the means±S.D. for each different cell line measured in triplicate. *0.001<P<0.05; **P≤0.001. Units: IC lactate (or pyruvate), μmol of lactate (or pyruvate)/106 cells; EC lactate (or pyruvate), mmol of lactate (or pyruvate) in medium (from 106 cells).
| IC | EC | ||||
|---|---|---|---|---|---|
| Cell line | Lactate | Pyruvate | Lactate | Pyruvate | IC L/P ratio |
| ρ+ (143B) | 15.0±3 | 5.0±1 | 13.52±2.81 | 0.36±0.09 | 3.0 |
| ρ0 (143B206) | 21.0±7* | 16±1** | 47.52±4.4** | 1.44±0.17** | 1.3 |
| MERRF A8344G | |||||
| WT | 11.9±4 | 6.8±5.3 | 33.47±8.73 | 0.49±0.24 | 1.8 |
| M | 31.4±18.0* | 12.8±9.6 | 54.70±11.28** | 0.98±0.33* | 2.6 |
| MERRF T8356C | |||||
| WT | 6.0±4.9 | 9.4±1 | 20.77±2.57 | 0.42±0.09 | 0.6 |
| M | 27.1±16.4 | 18.9±7* | 71.61±24.59* | 1.29±0.49* | 1.4 |
| MELAS A3243G | |||||
| WT | 11.2±3.3 | 11.9±8 | 25.06±9.23 | 0.42±0.22 | 0.9 |
| M | 26.6±13.5* | 16.1±6 | 61.81±21.14* | 1.08±0.35* | 1.7 |
| MELAS T3271C | |||||
| WT | 14.2±4.0 | 9.46±4 | 18.30±5.96 | 0.28±0.033 | 1.5 |
| M | 17.5±4.0 | 12.8±5 | 53.14±12.67* | 0.92±0.42* | 1.4 |
| NARP T8993C | |||||
| WT | 13±9 | 6.3±3 | 19.78±6.01 | 0.31±0.13 | 2.1 |
| M | 12±11 | 8.9±3 | 27.69±6.48* | 0.43±0.25 | 1.3 |
| NARP T8993G | |||||
| WT | 8.8±2.3 | 6.75±1 | 19.94±2.24 | 0.33±0.13 | 1.3 |
| M | 10.2±4.3 | 10.5±4.9 | 33.67±12.02 | 0.62±0.27 | 1.0 |
| Deletions | |||||
| CW (Δ 5.8 kb) | |||||
| WT | 14.0±5.5 | 9.0±4.3 | 44.39±15.67 | 0.71±0.29 | 1.6 |
| M | 23.7±5.4* | 14.1±9.9 | 71.43±22.93 | 1.56±0.61* | 1.7 |
| FLP (Δ 1.9 kb) | |||||
| WT | 11.0±6.6 | 10.4±2.74 | 23.31±5.31 | 0.46±0.14 | 1.1 |
| M | 13.6±3.4 | 24.5±5** | 72.50±12.37** | 1.45±0.46* | 0.6 |
Table 3. Respiratory-chain function in cybrids harbouring mtDNA mutations for each different cell line measured in triplicate.
The results are expressed as the means±S.D. *0.001<P<0.05; **P≤0.001. Units: O2 consumption, fmol of O2 consumed/min per 106 cells; ATP synthesis and citrate synthesis, nmol/min per mg of protein.
| Cell line | O2 consumption | ATP synthesis | Citrate synthase | O2/citrate synthase | ATP/citrate synthase |
|---|---|---|---|---|---|
| ρ+ (143B) | 5.19±0.98 | 35.52±3.00 | 26.0±9.07 | 0.218±0.087 | 1.495±0.63 |
| ρ0 (143B206) | 0.13±0.36** | 0.24±0.07** | 48.4±8.20 | 0.007±0.0001** | 0.005±0.001** |
| MERRF A8344G | |||||
| WT | 6.51±1.93 | 28.86±1.88 | 38.0±10.97 | 0.187±0.022 | 0.918±0.043 |
| M | 1.77±0.97** | 0.897±1.0** | 51.0±20.07 | 0.093±0.033* | 0.015±0.011** |
| MERRF T8356C | |||||
| WT | 6.32±0.71 | 23.815±1.48 | 45.0±5.62 | 0.140±0.019 | 0.562±0.04 |
| M | 0.73±0.34** | 0.173±0.156** | 57±20.23 | 0.016±0.008** | 0.003±0.003** |
| MELAS A3243G | |||||
| WT | 6.01±0.49 | 36.27±3.13 | 28.0±8.76 | 0.236±0.074 | 1.472±0.78 |
| M | 2.76±0.45** | 6.150±4.78* | 55.0±5.57 | 0.052±0.008** | 0.122±0.1 |
| MELAS T3271C | |||||
| WT | 8.20±0.95 | 30.12±2.10 | 24.0±4.42 | 0.342±0.035 | 1.255±0.087 |
| M | 2.18±0.98** | 11.075±0.93** | 43.5±2.12 | 0.050±0.021** | 0.255±0.009** |
| NARP T8993C | |||||
| WT | 5.78±0.86 | 26.59±0.6 | 34.0±15.56 | 0.184±0.050 | 0.967±0.32 |
| M | 4.90±1.29 | 20.67±4.04* | 42.0±16.97 | 0.111±0.051* | 0.540±0.09 |
| NARP T8993G | |||||
| WT | 2.83±1.10 | 26.845±1.10 | 46.0±6.56 | 0.062±0.024 | 0.583±0.023 |
| M | 1.74±0.81* | 1.430±1.12** | 49.0±4.24 | 0.037±0.019* | 0.03±0.02** |
| Deletions | |||||
| CW (Δ 5.8 kb) | |||||
| WT | 7.29±2.31 | 14.625±1.23 | 30.5±0.71 | 0.239±0.080 | 0.48±0.07 |
| M | 1.39±0.16* | 0.82±0.28** | 32.0±1.32 | 0.042±0.004* | 0.03±0.002* |
| FLP (Δ 1.9 kb) | |||||
| WT | 4.65±1.49 | 17.31±1.20 | 32.0±1.76 | 0.145±0.047 | 0.708±0.06 |
| M | 0.67±0.31* | 0.67±1.20** | 37.0±0.98 | 0.018±0.007* | 0.018±0.001** |
Lactate and pyruvate
In normal cells, glycolysis and oxidative metabolism are a continuous process, because pyruvate, the product of one pathway, is the substrate for the other. However, when oxidative metabolism is blocked, pyruvate and lactate are the final products of the glycolytic pathway. Levels of IC lactate were significantly elevated in M cells harbouring the MERRF (A8344G) and the MELAS (A3243G) mutations when compared with WT cells. In the other M cells, including those with NARP, the MELAS T3271C mutation, and the FLP deletion, IC lactate was only slightly elevated or unchanged from baseline. Interestingly, in the 143B206 (ρ0) cells, which are totally glycolytic, the IC lactate was only 40% higher than in the 143B (ρ+) cells. In cybrids harbouring the CW deletion, there was only a slight increase in IC lactate. In cells with the NARP T8993G point mutation, EC lactate was increased 70%, whereas in cells with the NARP T8993C mutation the increase was smaller (40%). The block of glucose oxidation in MERRF (A8344G, T8356C), MELAS (A3243G, T3271C), FLP deletion M clones, and in ρ0 controls is demonstrated by the significantly higher EC lactate and pyruvate levels.
Not surprisingly, the enhanced lactate production seen in cells with mtDNA mutations can be simulated in normal cells by inhibiting mitochondrial respiration with antimycin [31,32]. Accumulation of IC lactate results in abnormally low cytosolic pH, which, in turn, inhibits glycolysis [33]. However, cells in culture seem to maintain IC pH by transporting IC lactate and protons to the EC compartment, a process facilitated by MCTs (monocarboxylate transporters). The rate of lactate transport is determined not only by the kinetics of MCTs, but also by the proton gradient generated by residual mitochondrial respiration and by the volume of the EC space [34]. The large EC space (5 ml) in our tissue culture dishes will both facilitate efflux of lactate from the cells and dilute its EC concentration. Furthermore, under anaerobic conditions in vitro, the lactate formed is utilized continuously as a major respiratory fuel [35–37]. This seems to be different from the in vivo situation in mitochondrial disease patients, in whom lactic acidosis and high CSF (cerebrospinal fluid) lactate are characteristic features. In patients' tissues, cells occupy 85% and EC space only approx. 15% of total volume [38]. This is probably why high intracerebral lactate levels are deleterious to the patients, whereas in cell culture EC lactate diluted in the medium is less toxic.
IC pyruvate levels were significantly elevated in MERRF T8356C M clones and in deleted FLP clones, whereas they were only mildly increased in the M clones with the other mutations. EC pyruvate levels followed the same trend as EC lactate levels, with the exception of the NARP T8993C mutation.
Increase in both lactate and pyruvate in the EC compartment may be deleterious to the cell, as suggested by the observation that levels of ventricular lactate in MELAS patients correlate with clinical severity [39]. Notably, the increase in IC and EC lactate and pyruvate was common to all mutations (with the only exception of NARP T8993C), perhaps because our cellular model is predominantly glycolytic.
In our system, the IC L/P (lactate-to-pyruvate) ratio is not significantly different in WT and M cybrids, which is in contrast with the high L/P ratios observed in primary fibroblast or cardiomyocyte cultures with respiratory chain defects [35,40–42]. This discrepancy may be due to different methodologies for the measurement of lactate and pyruvate or to the different cell types analysed. Our IC lactate and pyruvate values reflect the steady-state levels of a dynamic system, whereas in other systems lactate is measured in cells at a fixed point in time and in the absence of glycolytic substrates.
Citrate synthesis
Since the overall rates of O2 consumption and ATP synthesis depend on the number of mitochondria in the cell, we normalized these values to citrate synthase activity, a nuclear-encoded mitochondrially located enzyme of the tricarboxylic acid cycle. The mean citrate synthase values in M cells (46 nmol/min per mg of protein) are slightly higher than in WT cells (33 nmol/min per mg of protein), perhaps due to compensatory mitochondrial proliferation. When normalized to citrate synthase, the rates of O2 consumption and ATP synthesis are still significantly lower in M cells, except for cybrids harbouring the NARP T8993C mutation (Table 3).
O2 consumption
Rate of O2 consumption by ρ0 cells was only 2.5% of that which occurred in 143B cells. Marked decreases in O2 consumption were observed in the homoplasmic M cell lines compared with the corresponding homoplasmic WT cell lines, with rates of respiration ranging from 11% in MERRF T8356C Ms to 62% in NARP T8993G Ms. As a notable exception, the rate of O2 consumption in cybrids harbouring the NARP T8993C mutation was 85% of the rate in corresponding WT cybrids.
The present study confirmed our previous reports of overall reduction of respiratory capacity in both fibroblasts and cybrids harbouring one or another of the pathogenic mtDNA mutations [6,8,43,44]. In addition, it reveals a close inverse correlation between increased EC lactate levels and rates of O2 consumption (Figure 1). In homoplasmic M A3243G cybrids from patients with MIDD (maternally inherited diabetes and deafness), van den Ouweland et al. [45] found an increased L/P ratio and a residual O2 consumption of 21%, considerably lower than in our cells (46%). Our results are closer to those of King et al. [6], probably because two of our A3243G M lines and one WT cybrid line were also used in that study [6]. On the other hand, our MELAS T3271C M cybrids had a residual respiratory activity of 27%, contrasting with the 62% residual activity in cybrids with the same mutation studied by Koga et al. [7]. This discrepancy is probably due to the low glucose concentration in our culture medium, which enhanced the defect in mitochondrial function.
ATP synthesis
In our assay of ATP synthesis, the contribution of adenylate kinase was reduced to insignificant levels by pre-incubating the sample with a specific inhibitor, diadenosine pentaphosphate. Cells were grown under controlled glycolytic conditions, as described above, and we evaluated the oligomycin-sensitive ATP synthase activity, which ranged between 75% and 90% of the total. The most severe reduction of ATP synthesis was observed in both the A8344G and the T8356C MERRF Ms, followed by the CW and FLP deletion Ms. In these cell lines, the low rates of ATP synthesis showed a good correlation with the reduction of respiratory capacity and the increase of EC lactate and pyruvate (Figures 1 and 2, and Tables 2 and 3). Conversely, in cybrids with MELAS A3243G and the MELAS T3271C mutations, the bioenergetic defects were milder and consistent with previous studies [6,7]. Furthermore, in MELAS cybrids, residual ATP synthase activities (16% for A3243G and 37% for T3271C), were not strictly correlated with rates of O2 consumption, which were lower in T3271C mutants than in A3243G mutants, whereas levels of EC lactate and pyruvate were similar in the two mutations. The reason for this discrepancy is not clear, but perhaps the two mutations affect the subunits of F1Fo-ATPase and the respiratory-chain complexes in different degrees. The severe reduction in ATP synthesis observed in NARP T8993G cybrids (95%) does not correlate with the much milder, though significant, reduction in ADP-dependent respiratory capacity (approx. 40%); the EC accumulation of lactate and pyruvate was also moderate. Therefore, the most important defect observed in NARP T8993G M cybrids was in ATP synthesis [17,19]. The T8993G mutation changes Leu156 to Arg in the F1Fo-ATPase subunit 6, which results in severe reduction of ATP synthesis, but only mild impairment of ADP-stimulated respiration rate, a different enzyme defect from that induced by oligomycin [46]. Interestingly, ATP hydrolysis in the primary fibroblasts from which mitochondria were obtained for the cybrids used in this study was also little affected [17]. In contrast with all other cybrids, cell lines carrying the T8993C mutation (Leu156 to Pro) had only slightly reduced ATP synthesis rate (78% of the control), and no other statistically significant abnormality.
Biochemical analyses not only provide important diagnostic clues for mitochondrial diseases, but can also shed light on pathogenic mechanisms. However, two factors make interpretation of published data in cybrid cells difficult. One is the lack of uniform methods for the study of key points in energy metabolism in cells harbouring pathogenic mtDNA mutations. The other is that most published results were obtained with a single mutation. The availability of cybrids harbouring a representative sample of the main pathogenic mtDNA mutations prompted us to conduct this comparative study. Although we did observe a good correlation among the different biochemical variables, we did not find major differences between cells harbouring different mutations, except for the NARP T8993C. This may be ascribed to the uniform nuclear background (143B) of our cybrids. While the different clinical features that are typically associated with these mtDNA mutations remain largely unexplained, the nuclear background in different tissues may contribute to the tissue-specific phenotypes.
Acknowledgments
This work was supported by grants from the U.S. National Institutes of Health (HD32062 to M. M. D.), and Telethon Fondazione Onlus, Roma (grant GP0280/01 to G.S).
References
- 1.Wallace D. C. Diseases of the mitochondrial DNA. Annu. Rev. Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 2.DiMauro S., Schon E. A. Mitochondrial DNA mutations in human disease. Am. J. Med. Genet. 2001;106:18–26. doi: 10.1002/ajmg.1392. [DOI] [PubMed] [Google Scholar]
- 3.Robinson B. H. Use of fibroblast and lymphoblast cultures for detection of respiratory chain defects. Methods Enzymol. 1996;264:454–464. doi: 10.1016/s0076-6879(96)64041-5. [DOI] [PubMed] [Google Scholar]
- 4.King M. P., Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 5.Schon E. A., DiMauro S. Primary disorders of the mitochondrial DNA and the pathophysiology of mtDNA-related disorders. In: Lemasters J. J., Nieminen A., editors. Mitochondria in Pathogenesis. New York: Kluwer Academic/Plenum Publishers; 2001. pp. 53–80. [Google Scholar]
- 6.King M. P., Koga Y., Davidson M., Schon E. A. Defects in mitochondrial protein synthesis and respiratory chain activity segragate with the tRNALeu(UUR) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. Mol. Cell. Biol. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koga Y., Davidson M., Schon E., King M. Analysis of cybrids harboring MELAS mutations in mitochondrial tRNALeu(UUR) gene. Muscle Nerve (Suppl.) 1995;3:S119–S123. doi: 10.1002/mus.880181424. [DOI] [PubMed] [Google Scholar]
- 8.Masucci J. P., Davidson M., Koga Y., Schon E. A., King M. P. In vitro analysis of mutations causing myoclonus epilepsy with ragged-red fibers in the mitochondrial tRNALys gene: two genotypes produce similar phenotypes. Mol. Cell. Biol. 1995;15:2872–2881. doi: 10.1128/mcb.15.5.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt I. J., Harding A. E., Petty R. K., Morgan Hughes J. A. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- 10.de Vries D., van Engelen B., Gabreels F., Ruitenbeek W., van Oost B. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh's syndrome. Ann. Neurol. 1993;34:410–412. doi: 10.1002/ana.410340319. [DOI] [PubMed] [Google Scholar]
- 11.Manfredi G., Gupta N., Vazquez-Memije M. E., Sadlock J. E., Spinazzola A., De Vivo D. C., Schon E. A. Oligomycin induces a decrease in the cellular content of a pathogenic mutation in the human mitochondrial ATPase 6 gene. J. Biol. Chem. 1999;274:9386–9391. doi: 10.1074/jbc.274.14.9386. [DOI] [PubMed] [Google Scholar]
- 12.Ojaimi J., Pan J., Santra S., Snell W. J., Schon E. A. An algal nucleus-encoded subunit of mitochondrial ATP synthase rescues a defect in the analogous human mitochondrial-encoded subunit. Mol. Biol. Cell. 2002;13:3836–3844. doi: 10.1091/mbc.E02-05-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakase H., Moraes C. T., Rizzuto R., Lombes A., DiMauro S., Schon E. A. Transcription and translation of deleted mitochondrial genomes in Kearns-Sayre syndrome: implications for pathogenesis. Am. J. Hum. Genet. 1990;46:418–427. [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi J. I., Ohta S., Kikuchi A., Takemitsu M., Goto Y. I., Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl. Acad. Sci. U.S.A. 1991;88:1371–1375. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sancho S., Moraes C. T., Tanji K., Miranda A. F. Structural and functional mitochondrial abnormalities associated with high levels of partially deleted mitochondrial DNAs in somatic cell hybrids. Somatic Cell Mol. Genet. 1992;18:431–442. doi: 10.1007/BF01233083. [DOI] [PubMed] [Google Scholar]
- 16.Schon E. A., Santra S., Pallotti F., Girvin M. E. Pathogenesis of primary defects in mitochondrial ATP synthesis. Semin. Cell Dev. Biol. 2001;12:441–448. doi: 10.1006/scdb.2001.0281. [DOI] [PubMed] [Google Scholar]
- 17.Vazquez-Memije M. E., Shanske S., Santorelli F. M., Kranz-Eble P., De Vivo D. C., DiMauro S. Comparative biochemical studies of ATPases in cells from patients with the T8993G or T8993C mitochondrial DNA mutations. J. Inher. Metab. Dis. 1998;21:829–836. doi: 10.1023/a:1005418718299. [DOI] [PubMed] [Google Scholar]
- 18.Garcia J. J., Ogilvie I., Robinson B. H., Capaldi R. A. Structure, functioning, and assembly of the ATP synthase in cells from patients with the T8993G mitochondrial DNA mutation. Comparison with the enzyme in ρ0 cells completely lacking mtDNA. J. Biol. Chem. 2000;275:11075–11081. doi: 10.1074/jbc.275.15.11075. [DOI] [PubMed] [Google Scholar]
- 19.Baracca A., Barogi S., Carelli V., Lenaz G., Solaini G. Catalytic activities of mitochondrial ATP synthase in patients with mitochondrial DNA T8993G mutation in the ATPase 6 gene encoding subunit a. J. Biol. Chem. 2000;275:4177–4182. doi: 10.1074/jbc.275.6.4177. [DOI] [PubMed] [Google Scholar]
- 20.Carelli V., Baracca A., Barogi S., Pallotti F., Valentino M. L., Montagna P., Zeviani M., Pini A., Lenaz G., Baruzzi A., Solaini G. Biochemical–clinical correlation in patients with different loads of the mitochondrial DNA T8993G mutation. Arch. Neurol. 2002;59:264–270. doi: 10.1001/archneur.59.2.264. [DOI] [PubMed] [Google Scholar]
- 21.Davidson M., Zhang L., Schon E., King M. Genetic and functional complementation of deleted mitochondrial DNA. Neurology. 1995;4(Suppl.):A381. [Google Scholar]
- 22.Goto Y., Nonaka I., Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature (London) 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 23.Goto Y., Nonaka I., Horai S. A new mtDNA mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS) Biochim. Biophys. Acta. 1991;1097:238–240. doi: 10.1016/0925-4439(91)90042-8. [DOI] [PubMed] [Google Scholar]
- 24.Moraes C. T., DiMauro S., Zeviani M., Lombes A., Shanske S., Miranda A. F., Nakase H., Bonilla E., Wernec L. C., Servidei S., et al. Mitochondrial DNA deletions in Progressive External Ophthalmoplegia and Kearns–Sayre syndrome. N. Engl. J. Med. 1989;320:1293–1299. doi: 10.1056/NEJM198905183202001. [DOI] [PubMed] [Google Scholar]
- 25.Shoffner J. M., Lott M. T., Lezza A., Seibel P., Ballinger S. W., Wallace D. C. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 26.Silvestri G., Moraes C. T., Shanske S., Oh S. J., DiMauro S. A new mtDNA mutation in the tRNALys gene associated with myoclonic epilepsy and ragged-red fibers (MERRF) Am. J. Hum. Genet. 1992;51:1213–1217. [PMC free article] [PubMed] [Google Scholar]
- 27.Ouhabi R., Boue-Grabot M., Mazat J. P. Mitochondrial ATP synthesis in permeabilized cells: assessment of the ATP/O values in situ. Anal. Biochem. 1998;263:169–175. doi: 10.1006/abio.1998.2776. [DOI] [PubMed] [Google Scholar]
- 28.Stanley P. E., Williams S. G. Use of the liquid scintillation spectrometer for determining adenosine triphosphate by the luciferase enzyme. Anal. Biochem. 1969;29:381–392. doi: 10.1016/0003-2697(69)90323-6. [DOI] [PubMed] [Google Scholar]
- 29.Srère P. Citrate synthase. Methods Enzymol. 1969;264:509–521. [Google Scholar]
- 30.Senior A. E., Nadanaciva S., Weber J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta. 2002;1553:188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 31.D'Aurelio M., Pich M. M., Catani L., Sgarbi G. L., Bovina C., Formiggini G., Castelli G. P., Baum H., Tura S., Lenaz G. Decreased Pasteur effect in platelets of aged individuals. Mech. Ageing Dev. 2001;122:823–833. doi: 10.1016/s0047-6374(01)00239-1. [DOI] [PubMed] [Google Scholar]
- 32.Mancuso M., Filosto M., Bosetti F., Ceravolo R., Rocchi A., Tognoni G., Manca M. L., Solaini G., Siciliano G., Murri L. Decreased platelet cytochrome c oxidase activity is accompanied by increased blood lactate concentration during exercise in patients with Alzheimer's disease. Exp. Neurol. 2003;182:421–426. doi: 10.1016/s0014-4886(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 33.Depre C., Veitch K., Hue L. Role of fructose 2,6-bisphosphate in the control of glycolysis. Stimulation of glycogen synthesis by lactate in the isolated working rat heart. Acta Cardiol. 1993;48:147–164. [PubMed] [Google Scholar]
- 34.Gstraunthaler G., Seppi T., Pfaller W. Impact of culture conditions, culture media volumes, and glucose content on metabolic properties of renal epithelial cell cultures. Are renal cells in tissue culture hypoxic? Cell Physiol. Biochem. 1999;9:150–172. doi: 10.1159/000016312. [DOI] [PubMed] [Google Scholar]
- 35.McKay N. D., Robinson B., Brodie R., Rooke-Allen N. Glucose transport and metabolism in cultured human skin fibroblasts. Biochim. Biophys. Acta. 1983;762:198–204. doi: 10.1016/0167-4889(83)90071-x. [DOI] [PubMed] [Google Scholar]
- 36.Brooks G. A., Dubouchaud H., Brown M., Sicurello J. P., Butz C. E. Role of mitochondrial lactate dehydrogenase and lactate oxidation in the intracellular lactate shuttle. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1129–1134. doi: 10.1073/pnas.96.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks G. A. Lactate shuttles in nature. Biochem. Soc. Trans. 2002;30:258–264. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 38.Dienel G. A., Hertz L. Glucose and lactate metabolism during brain activation. J. Neurosci. Res. 2001;66:824–838. doi: 10.1002/jnr.10079. [DOI] [PubMed] [Google Scholar]
- 39.Kaufmann P., Shungu D., Sano M., Jhung S., Engelstad K., Mitsis E., Mao X., Shanske S., Hirano M., DiMauro S., De Vivo D. Cerebral lactic acidosis correlated with neurological impairment in MELAS. Neurology. 2004;62:1297–302. doi: 10.1212/01.wnl.0000120557.83907.a8. [DOI] [PubMed] [Google Scholar]
- 40.Robinson B. H., McKay N., Goodyer P., Lancaster G. Defective intramitochondrial NADH oxidation in skin fibroblasts from an infant with fatal neonatal lacticacidemia. Am. J. Hum. Genet. 1985;37:938–946. [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson B. H., Glerum D. M., Chow W., Petrova-Benedict R., Lightowlers R., Capaldi R. The use of skin fibroblast cultures in the detection of respiratory chain defects in patients with lacticacidemia. Pediatr. Res. 1990;28:549–555. doi: 10.1203/00006450-199011000-00027. [DOI] [PubMed] [Google Scholar]
- 42.Merante F., Mickle D. A., Weisel R. D., Li R. K., Tumiati L. C., Rao V., Williams W. G., Robinson B. H. Myocardial aerobic metabolism is impaired in a cell culture model of cyanotic heart disease. Am. J. Physiol. 1998;275:H1673–H1681. doi: 10.1152/ajpheart.1998.275.5.H1673. [DOI] [PubMed] [Google Scholar]
- 43.Mariotti C., Tiranti V., Carrara F., Dallapiccola B., DiDonato S., Zeviani M. Defective respiratory capacity and mitochondrial protein synthesis in transformant cybrids harboring the tRNALeu(UUR) mutation associated with maternally inherited myopathy and cardiomyopathy. J. Clin. Invest. 1994;93:1102–1107. doi: 10.1172/JCI117061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.James A. M., Wei Y. H., Pang C. Y., Murphy M. P. Altered mitochondrial function in fibroblasts containing MELAS or MERRF mitochondrial DNA mutations. Biochem. J. 1996;318:401–407. doi: 10.1042/bj3180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Ouweland J. M., Maechler P., Wollheim C. B., Attardi G., Maassen J. A. Functional and morphological abnormalities of mitochondria harbouring the tRNALeu(UUR) mutation in mitochondrial DNA derived from patients with maternally inherited diabetes and deafness (MIDD) and progressive kidney disease. Diabetologia. 1999;42:485–492. doi: 10.1007/s001250051183. [DOI] [PubMed] [Google Scholar]
- 46.Brown G. C., Lakin-Thomas P. L., Brand M. D. Control of respiration and oxidative phosphorylation in isolated rat liver cells. Eur. J. Biochem. 1990;192:355–362. doi: 10.1111/j.1432-1033.1990.tb19234.x. [DOI] [PubMed] [Google Scholar]