Abstract
AMPK (AMP-activated protein kinase) responds to intracellular ATP depletion, while PPARα (peroxisome proliferator-activated receptor α) induces the expression of genes coding for enzymes and proteins involved in increasing cellular ATP yields. PPARα-mediated transcription is shown here to be co-activated by the α subunit of AMPK, as well as by kinase-deficient (Thr172Ala) and kinase-less (Asp157Ala, Asp139Ala) mutants of AMPKα. The Ser452Ala mutant of mPPARα mutated in its putative consensus AMPKα phosphorylation site is similarly co-activated by AMPKα. AMPKα or its kinase-less mutants bind to PPARα; binding is increased by MgATP, to a lesser extent by MgADP, but not at all by AMP or ZMP [AICAR (5-aminoimidazole-4-carboxamide ribonucleoside) monophosphate]. ATP-activated binding of AMPKα to PPARα is mediated primarily by the C-terminal regulatory domain of AMPKα. PPARα co-activation by AMPKα may, however, require its secondary interaction with the N-terminal catalytic domain of AMPKα, independently of its kinase activity. While AMPK catalytic activity is activated by AICAR, PPARα co-activation and PPARα-controlled transcription are robustly inhibited by AICAR, with concomitant translocation of nuclear AMPKα or its kinase-less mutants to the cytosol. In conclusion, AMPKα, independently of its kinase activity, co-activates PPARα both in primary rat hepatocytes and in PPARα-transfected cells. The kinase and transcriptional co-activation modes of AMPKα are both regulated by the cellular ATP/AMP ratio. Co-activation of PPARα by AMPKα may transcriptionally complement AMPK in maintaining cellular ATP status.
Keywords: 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR), AMP-activated protein kinase (AMPK), ATP/AMP ratio, nuclear translocation, peroxisome proliferator-activated receptor α (PPARα), transcription
Abbreviations: AICAR, 5-aminoimidazole-4-carboxamide ribonucleoside; AMPK, AMP-activated protein kinase; AOX, peroxisomal acyl-CoA oxidase; CAT, chloramphenicol acetyltransferase; DMEM, Dulbecco's modified Eagle's medium; GFP, green fluorescent protein; GST, glutathione S-transferase; HNF, hepatocyte nuclear factor; LBD, ligand-binding domain; PGC-1, PPARγ co-activator-1; PPAR, peroxisome proliferator-activated receptor; PPRE, peroxisome proliferator-responsive element; RT-PCR, reverse transcription–PCR; RXR, retinoid X receptor; ZMP, AICAR monophosphate; the prefixes h, m and r denote human, mouse and rat respectively
INTRODUCTION
Peroxisome proliferators (e.g. hypolipidaemic fibrate drugs, natural and xenobiotic long-chain mono- or di-carboxylic acids) induce in rodents a dramatic increase in the expression of genes coding for proteins and enzymes involved in the peroxisomal, mitochondrial and microsomal α-, β- and ω-oxidation of fatty acids, with a concomitant increase in ATP yield (reviewed in [1]). Activation of gene expression by peroxisome proliferators results from binding of the PPARα (peroxisome proliferator-activated receptor α)/RXRα (retinoid X receptor) heterodimer to cognate DR-1 (direct repeat-1) response elements [peroxisome proliferator-responsive element (PPRE)] in the promoters of relevant genes, leading to activation of transcription. PPARα activation by peroxisome proliferators is modulated further by PPARα phosphorylation by protein kinase A and other putative kinases [2,3].
AMPK (AMP-activated protein kinase; reviewed in [4,5]) serves as an intracellular ‘stress gauge’ which responds to intracellular ATP depletion induced by heat shock, hypoxia, prolonged exercise, inhibitors of the tricarboxylic acid cycle and uncouplers of oxidative phosphorylation [6]. Phosphorylation of target proteins by activated AMPK inhibits ATP-consuming anabolic pathways such as the synthesis of fatty acids and sterols, and activates ATP-generating catabolic pathways such as fatty acid oxidation. Depletion of intracellular ATP is transduced to AMPK by an increase in intracellular AMP as a result of the rapid equilibrium between ATP and AMP catalysed by adenylate kinase. AMPK is activated by direct AMP binding [7] as well as by AMP activation of an upstream AMPK kinase, recently identified as the serine/threonine kinase LKB1 (also known as STK11) [8], which phosphorylates the α subunit of AMPK at Thr172 [9,10]. Activation of AMPKα catalytic activity by AMP is mimicked by the 5-monophosphate metabolite (ZMP) of AICAR (5-aminoimidazole-4-carboxamide ribonucleoside) [11]. Active AMPK is composed of a heterotrimeric complex consisting of a catalytic α1 or α2 subunit together with the regulatory β (β1/β2) and γ (γ1/γ2/γ3) subunits that are required for stabilizing the α subunit and for forming a fully active AMPK complex [12]. Intracellular AMPK consists of a heterogeneous population of complexes composed of different α, β and γ subunit isoforms. The functional roles of each of these complexes are still unknown.
The present study has been initiated in order to search for putative concerted action of AMPK and PPARα in maintaining intracellular ATP status. As PPARα activates the transcription of genes coding for enzymes and proteins involved in increasing cellular ATP yields, modulation of PPARα transcriptional activity by AMPK could link PPARα-responsive genes to ATP depletion sensed by AMPK. Thus PPARα activation by AMPK could transcriptionally complement AMPK effects mediated by direct modulation of the catalytic activity of ATP-generating/consuming enzymes.
EXPERIMENTAL
Plasmid constructs
The pSG5-PPAR expression plasmid for mPPARα (mouse PPARα) [13] was from S. Green (AstaZeneca, Alderley Park, Cheshire, U.K.). The GAL4-mPPARα(LBD) construct (where LBD denotes ligand-binding domain) was prepared by subcloning the ScaI/StuI sequence of pSG5-mPPARα (amino acids 156–468) into pCMV-GAL4. mPPARα(S452A) was prepared by PCR amplification of the SphI/BamHI sequence of mPPARα, using the sense primer 5′-CGGAGCATGCGCAGCTCGTACAGGTCATCAAGAAGACCGAGCGCA-3′ and the antisense primer 5′-CGCGGATCCTCAGTACATGCTTCTGTAGA-3′. This mutated sequence was then ligated to a BamHI/SphI fragment of mPPARα. The product obtained after ligation was cloned into the BamHI site of pSG5.
hAMPKα2 (human hAMPKα2) cDNA cloned into Bluescript (a gift from N. H. Sarkar, Institute for Molecular Medicine and Genetics, Augusta, GA, U.S.A.) was cut by HindIII and XbaI and cloned into pcDNA3. rAMPKα1 (rat AMPKα1) cDNA was prepared from rat liver total RNA by RT-PCR (reverse transcription–PCR), using the sense primer 5′-GAATTCATGGCCGAGAAGCAGAAGCACGAC-3′ and the antisense primer 5′-GCGCTCTAGATTACTGTGCAAGAATTTTAATTAG-3′. The in- sert was cloned into the EcoRI/XbaI site of pcDNA3. hAMPKβ1 cDNA was prepared from HepG2 total RNA by RT-PCR using the sense primer 5′-ATTGGATCCCGCCGTCCGCCTTCCCTGTGT-3′ and the antisense primer 5′-ATCAGATCTTGGCCACCATCGCCCCCAGC-3′. The phosphorylated insert was cloned into the EcoRV site of pcDNA3. rAMPKγ1 cDNA was prepared from rat liver total RNA by RT-PCR using the sense primer 5′-GCTTTCCAAGCTGAGGAACTG-3′ and the antisense primer 5′-TCCGTTCTCTCAGGATTCCAT-3′. The phosphorylated insert was cloned into the EcoRV site of pcDNA3.
The site-specific rAMPKα1(T172A), hAMPKα2(T172A), hAMPKα2(D139A) and hAMPKα2(D157A) mutants were prepared by the Kunkel method of mutagenesis [14], using the synthetic oligonucleotides 5′-ATAATTGGGCGAGCCACAGCTGGCTCTTAAAAATTCACC-3′, 5′-ATAATTTGGAGATCCGCAGCTGGCTCTCAGAAATTCAAC-3′, 5′-ATTCTCTGGTTTCAGAGCTCGATGAACAACCAT-3′ and 5′-ATTAGATAATCCGAAAGCGGCTATCTTGGCGTTCATGTGTGCATCCAA-3′ respectively. The hAMPKα2(L204YAAA) mutant was prepared by the Kunkel method [14], using the primer 5′-AAATGGGAGGGTGCCGCATGCAGCAGCATACAAGATAAC-3′. cDNA encoding hAMPKα2-(1–312) was prepared by PCR using pcDNA3-hAMPKα2 as template, the sense primer for T7, and the antisense primer 5′-CGCGGATCCTTAATATAAACTGTTCATTAC-3′. The PCR product was cloned into the HindIII/BamHI site of pcDNA3. cDNA encoding hAMPKα2-(313–552) was prepared by PCR using pcDNA3-hAMPKα2 as template, the sense primer 5′-CCCAAGCTTACCATGAGTGGTGACCCTCAA-3′ and the antisense primer 5′-CGCGGATCCTCAACGGGCTAAAGTAGT-3′. The PCR fragment was cloned into a HindIII/BamHI site of pcDNA3. cDNA encoding hAMPKα2-(398–552) was prepared by PCR using pcDNA3-hAMPKα2 as template, the sense primer 5′-CCCAAGCTTACCATGAAAAAAGCCAAGTGGCGT-3′ and the antisense primer 5′-CGCGGATCCTCAACGGGCTAAAGTAGT-3′. The PCR fragment was cloned into a HindIII/BamHI site of pcDNA3. cDNA encoding hAMPKα2-(357–552) was prepared by PCR using pcDNA3-hAMPKα2 as template, the sense primer 5′-AAGCTTACCATGCATATTCCCCCAGGC-3′ and the antisense primer 5′-CGCGGATCCTCAACGGGCTAAAGTAGT-3′. The PCR fragment was cloned into a HindIII/BamHI site of pcDNA3. cDNA encoding hAMPKα2-(357–552)(P361A, P365A, P367A) was prepared by PCR using pcDNA3-hAMPKα2 as template, the sense primer 5′-AAGCTTACCATGCATATTCCCGCAGGCCTGAAAGCTCATGCAGAAAGGATGCCA-3′ and the antisense primer 5′-CGCGGATCCTCAACGGGCTAAAGTAGT-3′. The PCR fragment was cloned into a HindIII/BamHI site of pcDNA3.
The FLAG-hAMPKα2 plasmid was prepared by cloning in-frame an AscI/XbaI hAMPKα2 restriction fragment from Bluescript-hAMPKα2 into the pFlag-CMV-2 expression vector (Eastman Kodak Co.). The GFP-rAMPKα1 plasmid (where GFP is green fluorescent protein) was prepared by cloning in-frame a BstXI/XbaI rAMPKα1 fragment from pcDNA3-rAMPKα1 into the SmaI restriction site of the pEGFP-C2 expression vector (Clontech). The GFP-hAMPKα2 plasmid was prepared by cloning in-frame an AscI/XbaI hAMPKα2 restriction fragment into the SmaI restriction site of the pEGFP-C1 expression vector (Clontech). The GFP-hAMPKα2(D157A) plasmid was prepared by cloning in-frame an AscI/XbaI hAMPKα2(D157A) restriction fragment into the SmaI restriction site of the pEGFP-C1 expression vector (Clontech). The GFP-mPPARα plasmid was prepared by cloning in-frame a BamHI restriction fragment from pSG5-mPPARα into the BamHI restriction site of the pEGFP-C1 expression vector (Clontech). GST-mPPARα(LBD) (where GST is glutathione S-transferase) was prepared by cloning the ScaI/StuI fragment from pSG5-mPPARα into the SmaI restriction site of pGEX-1T (Pharmacia). The reporter plasmid containing the 1.3 kb Δ(−472 to −129) AOX (peroxisomal acyl-CoA oxidase) promoter linked to CAT (chloramphenicol acetyltransferase) [15] was from T. Osumi (Graduate School of Science, Himeji Institute of Technology, Kamigori, Hyogo, Japan). The pG5-CAT reporter plasmid consists of five copies of the GAL4 binding site upstream of the adenovirus E1b promoter [16]. The GFP-human p53 plasmid was from Y. Haupt (School of Pharmacy, Hadassah Medical School, The Hebrew University, Jerusalem, Israel).
RNA extraction and Northern blot
Total RNA was prepared using the EZ-RNA extraction kit (Biological Industries, Beit Haemek, Israel). Peroxisomal AOX mRNA was analysed by Northern blot hybridization using a 1756 bp AOX cDNA prepared by RT-PCR from rat total RNA using the sense primer 5′-CTGAACGACCCAGACTTC-3′ and the antisense primer 5′-CCATCATAGCGGCCAAGA-3′. Blots were visualized and quantified by phosphor-imaging.
Cell lines and transfection
COS-7, HeLa and 293 cells were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) fetal calf serum. Cultured cells were transfected by the calcium phosphate precipitation method with the indicated reporter [rAOX(PPRE)-CAT (5.0 μg) or pG5-CAT (3.0 μg)] and the indicated expression plasmids, followed by incubating the cells for 24 h with the indicated peroxisome proliferators added in DMSO solutions. The β-galactosidase expression plasmid pCMV-βgal added to each calcium phosphate precipitate served as an internal control for transfection. Presented transfection experiments are representative of 3–5 similar experiments.
Primary culture of rat hepatocytes
Rat hepatocytes were prepared as described by Berry et al. [17]. Viability was evaluated by exclusion of erythrosin B, and amounted to >90%. Cells were plated on rat tail collagen gel [18], and grown overnight in DMEM containing 10% (v/v) fetal calf serum, 100 μ-units of insulin/ml, 1 μM dexamethasone, 100 μg/ml streptomycin sulphate and 100 μg/ml penicillin-G, followed by incubating the cells with additions as indicated.
Protein–protein interaction in vitro
35S-labelled proteins were prepared using the in vitro TNT/T7-coupled transcription/translation system (Promega). The GST–mPPARα(LBD) protein was produced in Escherichia coli BL21 bacteria after induction with 0.2 mM isopropyl β-D-thiogalactoside for 3 h at 30 °C. Bacterial cells were harvested, resuspended in lysis buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, 1.4 mM KH2PO4, 1 mM PMSF, 1 mM benzamidine, 10 μg/ml leupeptin and 10 μg/ml aprotinin) and mildly sonicated. Triton X-100 was then added to a final concentration of 1% (v/v). After centrifugation to remove cell debris, dithiothreitol was added to a final concentration of 20 mM, followed by glutathione–agarose beads (Sigma) equilibrated with lysis buffer. The GST–mPPARα(LBD) protein was allowed to bind to the beads for 30 min at 4 °C under constant rotation. Tethered proteins were washed three times with lysis buffer and resuspended in the same buffer. For the pull-down assay, 2 μg of GST–mPPARα(LBD) tethered to glutathione beads was resuspended in 200 μl of pull-down buffer (10 mM Hepes/NaOH, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol, 100 mM NaCl, 10% glycerol, 0.1% Nonidet P40) followed by addition of 35S-labelled proteins and other additions as indicated. Expression of AMPKβ and AMPKγ in reticulocytes was determined by synthesizing the respective 35S-labelled proteins. After incubating the reaction mixture for 2 h at 4 °C with constant rotation, the beads were washed three times with pull-down buffer without glycerol. Bound proteins were eluted by boiling the sample in SDS buffer for 3 min and subjected to SDS/PAGE analysis.
Protein–protein interaction in vivo
At 48 h following transfection with expression plasmids for mPPARα and FLAG–AMPKα, cells were lysed in lysis buffer (25 mM Hepes, pH 7.5, 1% Nonidet P40, 70 mM KCl, 20 mM NaF, 1 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 0.2 mM EGTA, 2 mM vanadate) for 30 min on ice. Insoluble material was removed by centrifugation at 14000 g for 15 min at 4 °C. Expression of FLAG–AMPKα was determined by Western blotting of cell lysates using anti-FLAG M2 monoclonal antiserum. PPARα was immunoprecipitated by incubating the lysate for 2 h on ice with anti-PPARα polyclonal antibody raised in rabbits against the 16 C-terminal amino acids of rPPARα [19] (a gift from M. Dauca, Faculty of Sciences, University Henri Poincare-Nancy 1, Vandoeuvre-les-Nancy, France). The lysate was added to 6.5 mg of Sepharose–Protein A (Sigma) and incubated for 90 min at 4 °C under constant rotation. The Sepharose–Protein A beads were rinsed three times with lysis buffer without protease inhibitors and the bound proteins were eluted by boiling in SDS sample buffer for 3 min. Immunoprecipitated proteins were then separated by SDS/PAGE and analysed by Western blotting using anti-FLAG M2 monoclonal antibody (Sigma).
Nuclear/cytosolic localization of AMPKα
INS cells were cultured on glass coverslips at 37 °C and 5% CO2 in RPMI 1640 containing 25 mM glucose, 1 mM sodium pyruvate, 10 mM Hepes, 50 μM 2-mercaptoethanol and 10% (v/v) fetal calf serum with additions as indicated. HeLa cells were cultured on glass coverslips at 37 °C and 5% CO2 in DMEM containing 10% fetal calf serum with additions as indicated. Following treatment, the cells were washed three times in PBS (containing the respective additions) and fixed for 10 min at room temperature with 4% (v/v) paraformaldehyde in PBS. Following fixation, cells were washed three times in PBS and permeabilized by incubating them for 20 min at room temperature in PBS containing 1% Nonidet P40. Cells were then washed again three times in PBS and incubated for 30 min at room temperature with PBS containing 2% (v/v) horse serum to block non-specific binding sites. The coverslips were then incubated with sheep anti-AMPKα1, sheep anti-AMPKα2, rabbit anti-PPARα or mouse anti-p53 for 3 h as indicated. After extensive washing in PBS, cells were incubated for 1 h with the appropriate secondary antibody, washed again in PBS and mounted on slides using H-1000 mounting medium for fluorescence (Vector Laboratories). The slides were then sealed with nail polish and stored at 4 °C until analysed by confocal microscopy.
RESULTS
AMPK activates PPARα-dependent transcription
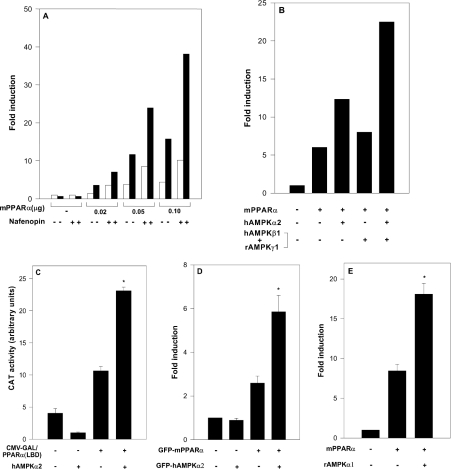
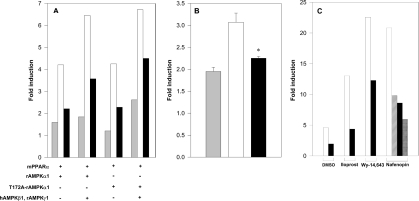
The effects of AMPK on transcriptional activation mediated by PPARα were studied in a variety of cells (293, COS-7 and HeLa) transfected with a reporter plasmid consisting of the AOX gene enhancer (PPRE) linked to the CAT gene [rAOX(PPRE)-CAT] [15], and with expression plasmids for mPPARα and for the α1 or α2 subunits of AMPK (rAMPKα1 or hAMPKα2). Transfected cells were incubated in the absence or presence of added peroxisome proliferators (e.g. nafenopin). As shown in Figure 1(A), PPARα-dependent CAT expression was activated by co-transfected hAMPKα2, within a range of transfected mPPARα levels, in both the presence and the absence of added nafenopin. Activation of PPARα by hAMPKα2 was amplified further by transfected β and γ subunits of AMPK (Figure 1B), in line with their reported effect of stabilizing the α subunit and in forming the fully active AMPK complex [12]. PPARα activation by hAMPKα2 did not require full-length PPARα, but was mediated by the LBD of mPPAR. Thus CAT expression induced by a chimaeric recombinant protein consisting of the DNA-binding domain of GAL4 fused to the LBD of mPPARα was activated by co-transfected hAMPKα2 (Figure 1C). Activation by AMPKα was similarly observed upon using expression plasmids for GFP–mPPARα and GFP–hAMPKα2 (Figure 1D), thus making it possible to study the nuclear localization of the two interacting proteins (see below). PPARα activation by AMPKα was not specific for the α2 isoform, but was also observed with transfected rAMPKα1 (Figure 1E). Hence mPPARα is activated by each of the two α isoforms of AMPK, and activation is further amplified by the β and γ subunits.
Figure 1. Activation of PPARα by AMPK.
Cells were transfected with expression plasmids for mPPARα and AMPK subunits as described in the Experimental section. Fold induction represents CAT activity (means±S.E.M. for triplicate plates, or means of duplicate plates differing by no more than 10%) normalized by CMV-β-galactosidase and further by CAT activity of pSG5-transfected cells. (A) 293 cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and with the expression plasmid for mPPARα (0.02–0.1 μg) as indicated, and co-transfected with the expression plasmid for hAMPKα2 (2.0 μg) (■) or the control plasmid pcDNA3 (□). Following transfection, cells were incubated in the absence or presence of 20 μM nafenopin as indicated. (B) 293 cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and the expression plasmid for mPPARα (0.1 μg), and co-transfected with expression plasmids for hAMPKα2 (2.0 μg), hAMPKβ1 (1.0 μg) and/or rAMPKγ1 (1.0 μg) as indicated. Following transfection, cells were incubated in the presence of 20 μM nafenopin. (C) HeLa cells were transfected with the reporter plasmid pG5-CAT and the expression plasmid for GAL4 (0.4 μg) or GAL4-mPPARα(LBD) (0.4 μg), and co-transfected with the expression plasmid for hAMPKα2 (2.0 μg) as indicated. Following transfection, cells were incubated in the presence of 100 μM nafenopin. (D) HeLa cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and co-transfected with the expression plasmid for GFP–mPPARα (0.02 μg) with or without the expression plasmid for GFP–hAMPKα2 (2.0 μg) as indicated. Following transfection, cells were incubated in the presence of 10 μM nafenopin. (E) 293 cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and co-transfected with the expression plasmids for mPPARα (0.05 μg) and/or rAMPKα1 (2.0 μg) as indicated. Following transfection, cells were incubated in the presence of 10 μM nafenopin. Significance of differences: *P<0.05 compared with no added AMPKα.
PPARα activation by transfected AMPK does not involve its kinase activity
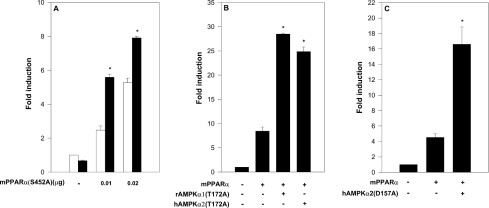
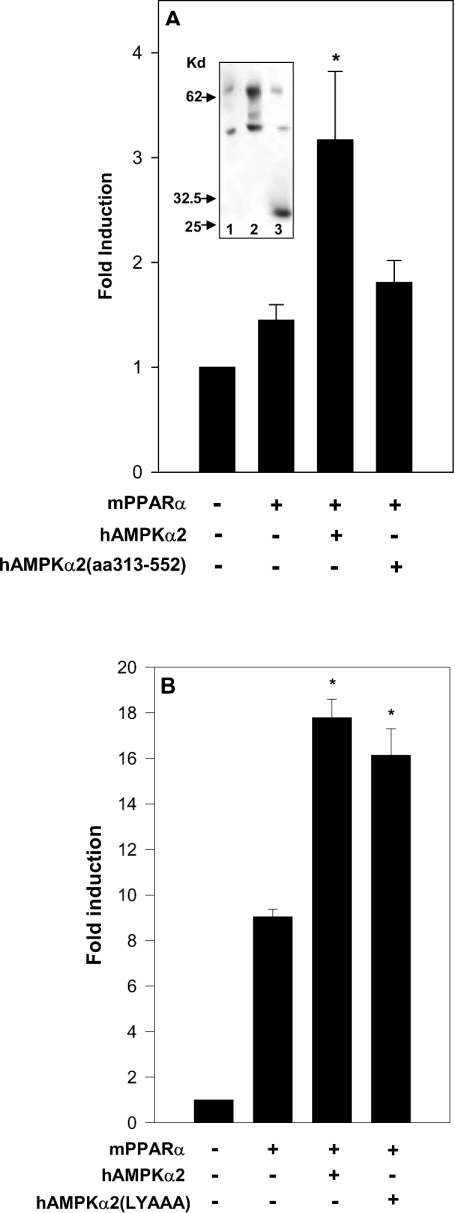
Putative phosphorylation of mPPARα by AMPK was evaluated by co-transfecting cells with hAMPKα2 and with mPPARα mutated in its putative consensus AMPKα phosphorylation site [20]. This mPPARα site consists of the amino acid sequence IKKTES452DAAL, and is the only potential AMPK consensus site of mPPARα. The mPPARα(S452A) mutant was activated by hAMPKα2 (Figure 2A) similarly to wild-type mPPARα (Figure 1A), thus implying that activation was not accounted for by phosphorylation of the AMPK consensus site of PPARα.
Figure 2. Activation of PPARα by AMPKα is independent of its kinase activity.
Cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and co-transfected with the indicated expression plasmids for mPPARα and AMPKα as described in the Experimental section. Fold induction represents CAT activity (means±S.E.M. for triplicate plates, or means of duplicate plates differing by no more than 10%) normalized by CMV-β-galactosidase and normalized further by CAT activity of pSG5-transfected cells. (A) 293 cells were transfected with the expression plasmid for mPPARα(S452A) (0–0.02 μg) as indicated, in the absence (□) or presence (■) of transfected hAMPKα2 (2.0 μg). Following transfection, cells were incubated in presence of 10 μM nafenopin. Significance: *P<0.05 compared with no added AMPKα. (B) 293 cells were transfected with the expression plasmid for mPPARα (0.05 μg), and co-transfected with expression plasmids for rAMPKα1(T172A) (2.0 μg) or hAMPKα2(T172A) (2.0 μg) as indicated. Following transfection, cells were incubated in the presence of 10 μM nafenopin. Significance: *P<0.05 compared with no added rAMPKα(T172A). (C) COS-7 cells were transfected with the expression plasmid for hAMPKα2(D157A) (2.0 μg) as indicated. Following transfection, cells were incubated in the presence of 10 μM nafenopin. Significance: *P<0.05 compared with no added hAMPKα2(D157A).
The role played by the kinase activity of AMPK in PPARα activation was evaluated further by site-directed mutagenesis of Thr172 or Asp157 of AMPKα, which are essential for AMPK catalytic activity [8,10]. As shown in Figures 2(B) and 2(C), the kinase-deficient mutants rAMPKα1(T172A) and hAMPKα2(T172A) and the kinase-less mutant hAMPKα2(D157A) were as efficient as the respective wild-type AMPKα (Figure 1) in activating mPPARα transcriptional activity. The kinase-less hAMPKα2(D139A) mutant was similarly effective (results not shown). Hence PPARα activation by transfected AMPKα is independent of the kinase activity of AMPK.
The α subunit of AMPK binds to PPARα
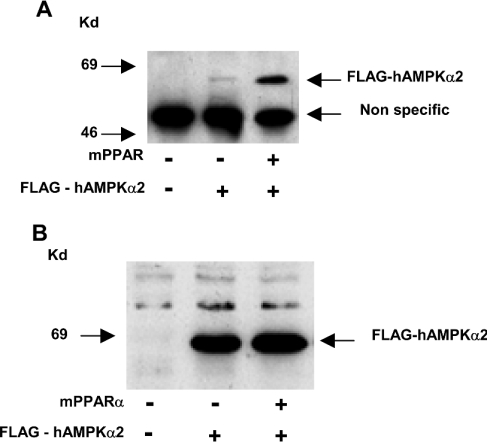
Binding of the α subunit of AMPK to PPARα was verified in 293 cells co-transfected with mPPARα together with FLAG–hAMPKα2. Putative mPPARα–hAMPKα2 complexes immunoprecipitated with anti-mPPARα antisera were characterized by anti-FLAG antisera. As shown in Figure 3, the mPPARα–hAMPKα2 complex was formed in vivo.
Figure 3. PPARα–AMPKα interaction in vivo.
293 cells were transfected with expression plasmids for mPPARα (0.2 μg) and FLAG–hAMPKα2 (1.0 μg) as indicated. (A) mPPARα complexes were immunoprecipitated with anti-mPPARα antiserum, immunoprecipitates were resolved by SDS/PAGE and FLAG–hAMPKα2 complexes were characterized by Western blotting using anti-FLAG M2 monoclonal antiserum as described in the Experimental section. (B) Expression of FLAG–hAMPKα2 was monitored by Western blotting of cell lysates using anti-FLAG antisera. Kd, kDa.
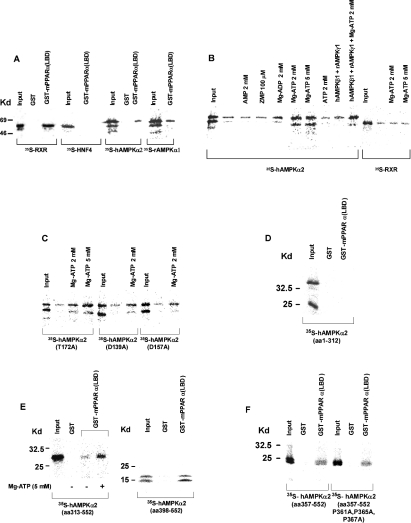
Binding of the α subunit of AMPK to PPARα was verified further by pulling down the AMPKα subunit via its affinity binding to the chimaeric GST–PPARα recombinant protein tethered to glutathione–agarose (Figure 4). As shown in Figure 4(A), [35S]methionine-labelled rAMPKα1 or hAMPKα2 bound to GST–mPPARα(LBD) (but not to the GST recombinant) with a yield similar to RXRα (used as a positive control). Binding was specific for the AMPKα moiety, as [35S]methionine-labelled HNF4α (hepatocyte nuclear factor 4α), used as negative control [21], was not pulled down by the chimaeric GST–mPPARα(LBD) recombinant. Binding affinities for the interaction of PPARα with the α1 and α2 isoforms of AMPK were verified by titrating the respective α-subunits in pull-down assays, and were found to be similar for the two AMPKα isoforms. Binding was unaffected by AMPKβ, AMPKγ (Figure 4B) or peroxisome proliferators (results not shown) added to the pull-down mixture.
Figure 4. PPARα–AMPKα interaction in vitro.
GST or GST–mPPARα(LBD) was tethered to glutathione–agarose beads and incubated in the presence of the indicated 35S-labelled proteins as described in the Experimental section. Input represents 20% of the respective 35S-labelled proteins subjected to pull-down. (A) Interaction of PPARα with hAMPKα2 and rAMPKα1. (B) Activation by MgATP of the PPARα–hAMPKα2 interaction. (C) PPARα interaction with hAMPKα2 mutants. (D) PPARα interaction with AMPKα2-(1–312). (E) PPARα interaction with hAMPKα2-(313–552) and hAMPKα2-(398–552). (F) PPARα interaction with hAMPKα2-(357–552) and hAMPKα2-(357–552)(P361A, P365A, P367A). Kd, kDa.
Binding of the hAMPKα2 subunit to PPARα was activated by MgATP, within the range of its physiological concentrations, less by MgADP, but not by AMP, ZMP or free ATP (Figure 4B). Activation by MgATP was not competed out by increasing concentrations of AMP or ZMP, and binding was inhibited by excess of free Mg (results not shown). Activation by MgATP was unaffected by added AMPKβ or AMPKγ proteins to the pull-down mixture. MgATP activation of AMPKα binding to PPARα was specific, as binding of RXRα remained unaffected by MgATP (Figure 4B). Furthermore, similar to wild-type hAMPKα2, the kinase-less hAMPKα2(D157A) and hAMPKα2(D139A) mutants and the kinase-deficient hAMPKα2(T172A) mutant bound to mPPARα, and binding was activated by MgATP (Figure 4C), thus indicating that the effect of MgATP did not involve the kinase activity of AMPK.
Putative AMPKα2 domains involved in the PPARα–AMPKα interaction consist of the LXXLL motif (aa 204–208) and the proline-rich domain (aa 360–383) of AMPKα. The LXXLL and proline-rich sequences are common recognition motifs involved in the binding of co-activators to their cognate nuclear receptors [22,23]. AMPKα domains involved in PPARα binding were verified by evaluating the interaction of PPARα with either the catalytic (aa 1–312) or the regulatory (aa 313–552) domain of hAMPKα2 using pull-down assays. hAMPKα2-(1–312) failed to bind GST–PPARα(LBD) (Figure 4D). In contrast, hAMPKα2-(313–552) did bind mPPARα(LBD), and binding was activated by MgATP (Figure 4E), thus indicating that PPARα–AMPKα2 interaction and its activation by MgATP are accounted for primarily by the regulatory AMPKα2 domain, independently of the catalytic AMPKα domain and its ATP-binding site GXGXXG. The putative contribution made by the proline-rich domain of hAMPKα2-(360–383) to the PPARα–hAMPKα2 interaction was verified by mutating proline residues 361, 365 and 367 to alanine. Binding of the regulatory hAMPKα2 domain to PPARα was unaffected by mutating the proline-rich subdomain (Figure 4F). Furthermore, hAMPKα2-(398–552), which lacks the proline-rich domain, still bound to mPPARα (Figure 4E), thus indicating that the proline-rich domain is not involved in binding of AMPKα to PPARα. It is worth noting that the shorter form of AMPKα observed in pull-down experiments (Figures 4A–4C) is presumably due to AMPKα being translated at two different start codons, rather than representing a degradation product. Indeed, the production of two AMPKα2 isoforms during in vitro transcription/translation has been reported previously by Sarkar and co-workers [24]. The shorter protein may result from an in-frame ATG codon with a Kozak consensus sequence at nucleotide 339, corresponding to Met93 [24].
The putative contribution made by the L204YALL motif [25] to the PPARα–AMPKα interaction was verified using the L204YAAA mutant of full-length hAMPKα2. The mutant was found to bind to tethered mPPARα(LBD) with 50% yield as compared with wild-type hAMPKα2 (results not shown), thus indicating that the PPARα–AMPKα interaction, mediated primarily by the C-terminal regulatory domain of AMPKα, may be complemented by the LXXLL motif of the catalytic domain of AMPKα.
Binding of mPPARα to the regulatory domain of hAMPKα2 was not sufficient for transcriptional co-activation of mPPARα by hAMPKα2. Thus, in contrast with mPPARα activation by full-length hAMPKα2, the truncated regulatory domain of hAMPKα2 did not activate mPPARα in transfection assays (Figure 5A), in spite of its similar expression level to full-length AMPKα (Figure 5A, inset). Hence mPPARα co-activation initiated by its binding to the regulatory domain of hAMPKα2 may require a secondary interaction with the catalytic domain of hAMPKα2. The putative role of the LYALL motif of the catalytic domain of hAMPKα2 in co-activating mPPARα was verified by evaluating mPPARα activation by the hAMPKα2(L204YAAA) mutant in transfection assays. As shown in Figure 5(B), mPPARα co-activation by the hAMPKα2(L204YAAA) mutant was similar to that by wild-type hAMPKα2, thus implicating other catalytic subdomains of hAMPKα2 as being involved in mPPARα co-activation.
Figure 5. PPARα co-activation by AMPKα is independent of the LXXLL domain of AMPKα.
(A) 293 cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and the expression plamid for RXRα (0.03 μg), and co-transfected with the expression plasmid for mPPARα (0.03 μg), hAMPKα2 (5.0 μg) or hAMPKα2-(313–552) (5.0 μg) as indicated. Following transfection, cells were incubated for 17 h in the presence of 20 μM nafenopin. Fold induction represents CAT activity (means±S.E.M. of duplicate plates differing by no more than 10% from five independent experiments) normalized by CMV-β-galactosidase and further normalized by CAT activity of pSG5-transfected cells. Significance: *P<0.05 compared with no added hAMPKα2. Expression of hAMPKα2 and hAMPKα2-(313–552) was verified by Western blot analysis of the cell lysate using sheep anti-hAMPKα2 antibody (inset): lane 1, pcDNA3 (5.0 μg); lane 2, hAMPKα2 (5.0 μg); lane 3, hAMPKα2-(313–552) (5.0 μg). Kd, kDa. (B) COS-7 cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and the expression plasmid for mPPARα (0.05 μg), and co-transfected with hAMPKα2 (2.0 μg) or hAMPKα2(L204YAAA) (2.0 μg) as indicated. Following transfection, cells were incubated in the presence of 10 μM nafenopin. Fold induction represents CAT activity (means±S.E.M. for triplicate plates or means of duplicate plates differing by no more than 10%) normalized by CMV-β-galactosidase and further normalized by CAT activity of pSG5-transfected cells. Significance: *P<0.05 compared with no added AMPKα.
Inhibition by AICAR of transcriptional co-activation by AMPKα
PPARα co-activation by AMPKα was verified further by evaluating the effects of AICAR in modulating the transcriptional activity of PPARα by transfected and endogenous AMPKα. Since AICAR activates AMPK catalytic activity [11], and as PPARα co-activation by transfected AMPKα was independent of its kinase activity (Figure 2), AICAR was expected to be without effect on PPARα co-activation by transfected or endogenous AMPKα. Surprisingly, however, AICAR acted as a potent inhibitor of PPARα transactivation by transfected AMPKα (Figure 6A). Furthermore, AICAR inhibition of PPARα activation by AMPKα was also observed with mPPARα(S452A), which is mutated in its AMPK consensus site (results not shown), with the kinase-deficient mutants rAMPKα1(T172A) (Figure 6A) and hAMPKα2(T172A) (results not shown), and with the kinase-less mutant hAMPKα2(D157A) (Figure 6B), thus indicating that inhibition by AICAR of PPARα activation was independent of the catalytic activity of transfected AMPKα.
Figure 6. Inhibition of PPARα by AICAR.
Cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and co-transfected with the indicated expression plasmids for mPPARα and AMPKα mutants as described in the Experimental section. Fold induction represents CAT activity (means+S.E.M. for triplicate plates or means of duplicate plates differing by no more than 10%) normalized by CMV-β-galactosidase and further normalized by CAT activity of pSG5-transfected cells. (A) 293 cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and the expression plasmids for mPPARα (0.1 μg) or mPPARα(S452A) (0.1 μg), and co-transfected with expression plasmids for rAMPKα1 (1.0 μg), rAMPKα1(T172A) (1.0 μg) and/or hAMPKβ1 (1.0 μg) plus rAMPKγ1 (1.0 μg) as indicated. CAT activity was determined in cells incubated overnight in the absence of added ligand (grey bars), as well as in cells incubated for an additional 7 h with 30 μM nafenopin in the absence (□) or presence (■) of added 250 μM AICAR. Note the nafenopin-dependent activity during the 7 h incubation period in the absence and presence of AICAR. (B) 293 cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and the expression plasmid for mPPARα (0.02 μg), and co-transfected with expression plasmid for hAMPKα2(D157A) (2.0 μg). CAT activity was determined in cells incubated overnight in the absence of added ligand (grey bar), as well as in cells incubated for an additional 6 h with 10 μM nafenopin in the absence (□) or presence (■) of added 500 μM AICAR. Note the nafenopin-dependent activity during the 6 h incubation period in the absence and presence of AICAR. Significance: *P<0.05 compared with no added AICAR. (C) COS-7 cells were transfected with the reporter plasmid rAOX(PPRE)-CAT and the expression plasmid for mPPARα (0.05 μg). CAT activity was determined in cells incubated overnight in the absence of added ligands and incubated for an additional 7 h in the presence of DMSO vehicle, 50 nM iloprost, 10 μM Wy-14,643 or 10 μM nafenopin as indicated, in the absence (white bars) or presence of 200 (hatched bars), 250 (black bars) or 600 (cross-hatched bars) μM AICAR.
Similar to AICAR inhibition of PPARα co-activation by transfected AMPKα, AICAR inhibited dose-dependently the transcriptional activity of wild-type PPARα (Figure 6C) or PPARα(S452A) (results not shown) in the absence of overexpressed AMPKα and independently of the peroxisome proliferators used for PPARα activation.
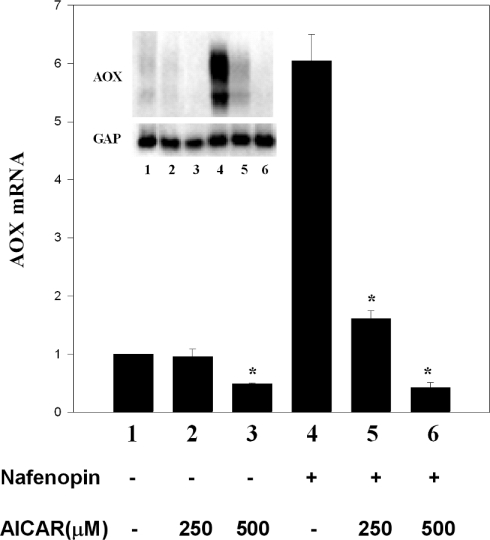
AICAR inhibition of PPARα transcriptional activity was verified further in primary rat hepatocytes. As shown in Figure 7, basal endogenous AOX mRNA, presumably induced by an endogenous PPARα ligand, and its induction by added nafenopin were markedly inhibited by AICAR, in line with AICAR inhibition of PPARα transcriptional activity in transfection assays (Figure 6). Inhibition of PPARα-controlled transcription by AICAR was specific, as expression of glyceraldehyde-3-phosphate dehydrogenase remained unaffected by AICAR under conditions of complete suppression of nafenopin-induced peroxisomal AOX.
Figure 7. Inhibition by AICAR of the expression of PPARα-responsive genes.
Rat primary hepatocytes were prepared and cultured in triplicate on collagen gels as described in the Experimental section. Overnight-cultured cells were treated with 250 μM or 500 μM AICAR for 1 h and then incubated for a further 24 h in the absence or presence of 30 μM nafenopin as indicated. Peroxisomal AOX was probed using a 1756 bp peroxisomal AOX cDNA. Bars represent the AOX transcript normalized by glyceraldehyde-3-phosphate dehydrogenase (GAP) mRNA. Values are means±S.E.M. of triplicate plates; *P<0.05 compared with no added AICAR. The results are representative of three similar experiments.
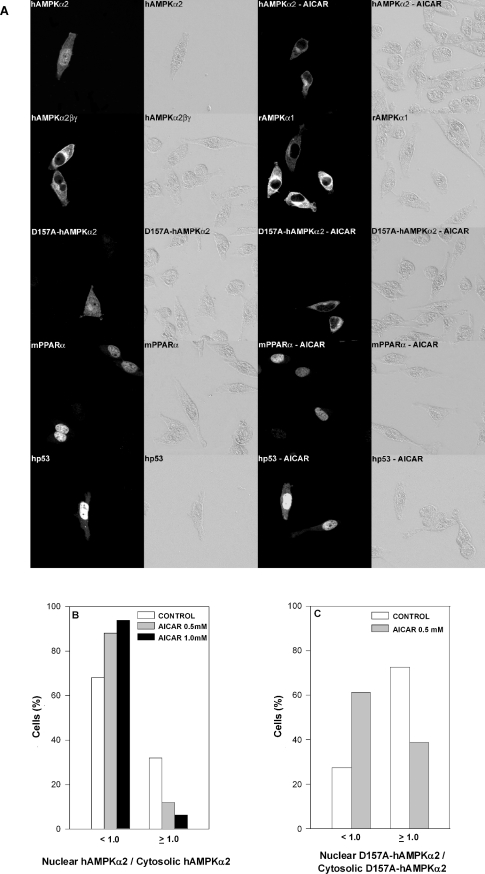
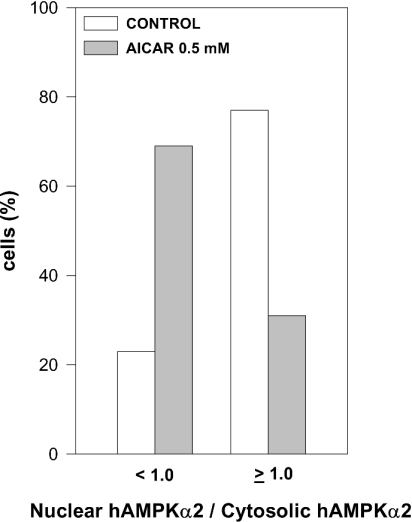
Inhibition by AICAR of PPARα activation by AMPKα was analysed further by evaluating the effects of AICAR on the nuclear localization of AMPKα or PPARα. Non-transfected INS cells or HeLa cells transfected with either rAMPKα1 or hAMPKα2 were incubated in the presence or absence of AICAR and were then reacted with anti-AMPKα1 or anti-AMPKα2 antibodies respectively. The nuclear/cytosolic distributions of transfected (Figure 8) and endogenous (Figure 9) AMPKα as a function of added AICAR were analysed by confocal microscopy. In line with a previous report [26], AMPKα2 was found to be significantly enriched in the nuclei of HeLa cells as compared with AMPKα1 (Figure 8A). Added AICAR resulted in the displacement of nuclear AMPKα2 to the cytosol, and hence a significant loss of nuclear AMPKα2 (Figures 8A and 8B). Nuclear AMPKα2 was similarly displaced by co-transfecting the cells with hAMPKβ1 and rAMPKγ1 (Figure 8A). Displacement of nuclear AMPKα2 by AICAR was independent of the kinase activity of transfected AMPKα2, as the transfected hAMPKα2(D157A) kinase-less mutant was displaced by AICAR similarly to wild-type AMPKα2 (Figures 8A and 8C). Displacement of nuclear AMPKα2 by AICAR was specific, as nuclear mPPARα or human p53 (Figure 8A), as well as the total cellular amount of PPARα (results not shown), remained unaffected by added AICAR. Displacement of nuclear AMPKα2 by AICAR was not limited to transfected AMPKα, but was similarly observed for endogenous nuclear AMPKα (Figure 9). Hence AICAR inhibition of PPARα transcriptional activity and co-activation by AMPKα may be accounted for by the displacement of nuclear AMPKα.
Figure 8. Displacement by AICAR of transfected nuclear AMPKα.
(A) HeLa cells were cultured on glass coverslips and transfected overnight with expression plasmids for GFP–rAMPKα1 (6.0 μg), GFP–hAMPKα2 (6.0 μg), hAMPKβ1 (4.0 μg), rAMPKγ1 (4.0 μg), GFP–hAMPKα2(D157A) (6.0 μg), GFP–mPPARα (4.0 μg) or GFP–human p53 (2.0 μg) as indicated. Following transfection, cells were incubated for 6 h in the absence or presence of 0.5 mM AICAR as indicated, fixed, permeabilized, further incubated with the respective antibody and analysed by bright-field illumination (right panel of each pair) or by confocal microscopy (left panel of each pair) as described in the Experimental section. Representative micrographs are shown. (B, C) HeLa cells were transfected overnight with expression plasmids for GFP–hAMPKα2 (6.0 μg) (B) or GFP–hAMPKα2(D157A) (6.0 μg) (C) Following transfection, cells were incubated for 6 h in the absence or presence of 0.5 or 1.0 mM AICAR as indicated. Following treatment, cells were fixed, permeabilized, further incubated with anti-AMPKα2 antibody and analysed by confocal microscopy as described in the Experimental section. The ratio of nuclear hAMPKα2/cytosolic hAMPKα2 (B) or of nuclear hAMPKα2(D157A)/cytosolic hAMPKα2(D157A) (C) was determined for 100–200 cells.
Figure 9. Displacement by AICAR of endogenous nuclear AMPKα.
INS cells were cultured on glass coverslips and incubated for 4 h in the absence or presence of 0.5 mM AICAR. Following treatment, cells were fixed, permeabilized, further incubated with anti-AMPKα2 antibody and analysed by confocal microscopy as described in the Experimental section. The ratio of nuclear hAMPKα2/cytosolic hAMPKα2 was determined for 100 cells.
DISCUSSION
The present study reports a novel role for the α subunit of AMPK in activating PPARα. In light of the established kinase activity of AMPK, PPARα activation by AMPK was expected essentially to reflect modulation of its trancriptional activity by AMPK-catalysed phosphorylation. Surprisingly, however, activation of PPARα transcriptional activity by AMPK is independent of its kinase activity, but is accounted for by transcriptional co-activation of PPARα mediated by a direct physical PPARα–AMPKα association. Evidence in support of this conclusion is as follows. (a) Kinase-less (Asp157Ala, Asp139Ala) and kinase-deficient (Thr172Ala) AMPKα mutants were as effective co-activators as wild-type AMPKα. (b) Transcriptional co-activation by AMPKα was maintained when the consensus AMPKα phosphorylation site of PPARα was mutated. (c) PPARα transcriptional activity and its co-activation by AMPKα were essentially eliminated by an activator (e.g. AICAR) of the kinase activity of AMPK. AICAR inhibition of co-activation by AMPKα was similarly effective when using the kinase-less AMPKα(D157A) or the kinase-deficient AMPKα(T172A) mutants instead of wild-type AMPKα. (d) Wild-type AMPKα or its kinase-less mutants bound PPARα directly, both in vitro and in vivo. Binding is mediated primarily by the regulatory domain of AMPKα, independently of the catalytic domain. Following PPARα binding to AMPKα, co-activation of PPARα may require a secondary interaction with the LXXLL motif of the catalytic domain (aa 1–312) of AMPKα, independently of its kinase activity. The extent of transcriptional co-activation of mPPARα by AMPKα is similar to that reported previously for CBP [cAMP response element-binding protein (CREB)-binding protein]/p300 or PGC-1 (PPARγ co-activator-1) [27,28]. Interestingly, similarly to PGC-1 [29], AMPKα contains a C-terminal serine/arginine-rich domain (aa 407–552 in AMPKα2) which may interact with the C-terminal domain of RNA polymerase II [30], thus indicating a possible interaction between PPARα and RNA polymerase II mediated by AMPKα.
Similarly to other transcriptional co-activators, co-activation by AMPKα may prove not to be limited to PPARα, but to be shared by other nuclear receptors yet to be identified. It is worth noting, however, that cellular conditions favouring kinase-independent transcriptional co-activation by nuclear AMPKα appear to differ from those favouring its kinase activity. Thus the kinase activity is dependent on the association of AMPKα with its β and γ subunits [12] and is activated by AMP or AICAR/ZMP [11]. In contrast with the kinase activity, nuclear localization of AMPKα is not favoured by its association with the β and γ subunits or by AICAR (Figures 8 and 9). Moreover, the interaction of AMPKα with PPARα is activated by ATP (Figure 3), while its kinase activity is inhibited [7] by ATP. Hence the AMPKα subunit may operate in its kinase and transcriptional co-activation modes as a function of its association with β and γ subunits, modulation of its conformation by the cellular energy charge (i.e. ATP/AMP ratio) or its recruitment by cognate nuclear receptors. The duality of AMPKα as a kinase and as a transcriptional co-activator, taken together with the displacement of nuclear AMPKα by AICAR, may imply that reported effects of AICAR or energy charge on transcription [31–36] may not only implicate the kinase activity of AMPK but may also be transduced by phosphorylation-independent interference with transcriptional co-activation of cognate nuclear receptors by AMPKα.
The mode of action of AICAR in displacing nuclear AMPK remains to be investigated. It is worth noting, however, that displacement of nuclear AMPKα by AICAR was observed with wild-type AMPKα as well as with its kinase-less mutant (Figure 8). Similarly, AICAR inhibited PPARα transcriptional activity with both wild-type PPARα (Figure 6) and mPPARα(S452A), which is mutated in its AMPK consensus site. Likewise, AICAR inhibition of PPARα co-activation by AMPKα was observed with wild-type AMPKα as well as with kinase-deficient or kinase-less mutants of AMPKα (Figure 6). Hence displacement of nuclear AMPKα by AICAR and AICAR inhibition of PPARα co-activation by AMPKα and of its transcriptional activity may be independent of the kinase activity of AMPKα. Kinase-independent effects of AICAR may involve its interaction with nucleoside transporters and/or adenosine receptors [37,38].
Transcriptional co-activation of PPARα by AMPKα may complement AMPK in maintaining cellular ATP status, by linking the transcription of PPARα-responsive genes involved in generating ATP (e.g. genes coding for mitochondrial and peroxisomal β-oxidation of fatty acids) with cellular ATP status sensed by AMPK. Since the kinase and co-activation modes of AMPK are regulated in opposite directions by the ATP/AMP ratio, maintenance of the cellular ATP status by kinase-independent co-activation of PPARα-responsive genes takes turns with the effects of AMPK mediated by the phosphorylation of target enzymes.
Acknowledgments
We thank G. Hardie for critical suggestions, as well as for his gift of anti-AMPKα antibodies. We also thank M. Dauca and Y. Haupt for anti-mPPARα and anti-p53 antibodies respectively, N.H. Sarkar for the gift of hAMPKα2 cDNA, and T. Osumi for the AOX-CAT reporter plasmid.
References
- 1.Desvergne B., Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 2.Lazennec G., Canaple L., Saugy D., Wahli W. Activation of peroxisome proliferator-activated receptors (PPARs) by their ligands and protein kinase A activators. Mol. Endocrinol. 2000;14:1962–1975. doi: 10.1210/mend.14.12.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Latruffe N., Malki M. C., Nicolas-Frances V., Clemencet M. C., Jannin B., Berlot J. P. Regulation of the peroxisomal beta-oxidation-dependent pathway by peroxisome proliferator-activated receptor alpha and kinases. Biochem. Pharmacol. 2000;60:1027–1032. doi: 10.1016/s0006-2952(00)00416-0. [DOI] [PubMed] [Google Scholar]
- 4.Hardie D. G., Carling D., Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 5.Kemp B. E., Mitchelhill K. I., Stapleton D., Michell B. J., Chen Z. P., Witters L. A. Dealing with energy demand: the AMP-activated protein kinase. Trends Biochem. Sci. 1999;24:22–25. doi: 10.1016/s0968-0004(98)01340-1. [DOI] [PubMed] [Google Scholar]
- 6.Hardie D. G., Salt I. P., Hawley S. A., Davies S. P. AMP-activated protein kinase: an ultrasensitive system for monitoring cellular energy charge. Biochem. J. 1999;338:717–722. [PMC free article] [PubMed] [Google Scholar]
- 7.Hawley S. A., Selbert M. A., Goldstein E. G., Edelman A. M., Carling D., Hardie D. G. 5′-AMP activates the AMP-activated protein kinase cascade, and Ca2+/calmodulin activates the calmodulin-dependent protein kinase I cascade, via three independent mechanisms. J. Biol. Chem. 1995;270:27186–27191. doi: 10.1074/jbc.270.45.27186. [DOI] [PubMed] [Google Scholar]
- 8.Woods A., Johnstone S. R., Dickerson K., Leiper F. C., Fryer L. G., Neumann D., Schlattner U., Wallimann T., Carlson M., Carling D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 9.Crute B. E., Seefeld K., Gamble J., Kemp B. E., Witters L. A. Functional domains of the alpha1 catalytic subunit of the AMP-activated protein kinase. J. Biol. Chem. 1998;273:35347–35354. doi: 10.1074/jbc.273.52.35347. [DOI] [PubMed] [Google Scholar]
- 10.Stein S. C., Woods A., Jones N., Davison M. D., Carling D. The regulation of AMP-activated protein kinase by phosphorylation. Biochem. J. 2000;345:437–443. [PMC free article] [PubMed] [Google Scholar]
- 11.Corton J. M., Gillespie J. G., Hawley S. A., Hardie D. G. 5-Aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem. 1995;229:558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- 12.Dyck J. R. B., Gao G., Widmer J., Stapleton D., Fernandez C. S., Kemp B. E., Witters L. A. Regulation of 5′-AMP-activated protein kinase activity by the noncatalytic beta and gamma subunits. J. Biol. Chem. 1996;271:17798–17803. doi: 10.1074/jbc.271.30.17798. [DOI] [PubMed] [Google Scholar]
- 13.Issemann I., Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature (London) 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 14.Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 15.Osumi T., Wen J. K., Hashimoto T. Two cis-acting regulatory sequences in the peroxisome proliferator-responsive enhancer region of rat acyl-CoA oxidase gene. Biochem. Biophys. Res. Commun. 1991;175:866–871. doi: 10.1016/0006-291x(91)91645-s. [DOI] [PubMed] [Google Scholar]
- 16.Lille J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature (London) 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 17.Berry M. N., Edwards A. M., Barrit G. J. Amsterdam: Elsevier Science Publishers; 1991. High-yield preparation of isolated hepatocytes from rat liver. Laboratory Techniques in Biochemistry and Molecular Biology, vol. 21; pp. 15–58. [Google Scholar]
- 18.Michalopoulos G., Pitot H. C. Primary culture of parenchymal liver cells on collagen membranes. Morphological and biochemical observations. Exp. Cell Res. 1975;94:70–78. doi: 10.1016/0014-4827(75)90532-7. [DOI] [PubMed] [Google Scholar]
- 19.Braissant O., Foufelle F., Scotto C., Dauca M., Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology (Baltimore) 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 20.Ching Y. P., Davies S. P., Hardie D. G. Analysis of the specificity of the AMP-activated protein kinase by site-directed mutagenesis of bacterially expressed 3-hydroxy 3-methylglutaryl-CoA reductase, using a single primer variant of the unique-site-elimination method. Eur. J. Biochem. 1996;237:800–808. doi: 10.1111/j.1432-1033.1996.0800p.x. [DOI] [PubMed] [Google Scholar]
- 21.Hertz R., Bishara-Shieban J., Bar-Tana J. Mode of action of peroxisome proliferators as hypolipidemic drugs. Suppression of apolipoprotein C-III. J. Biol. Chem. 1995;270:13470–13475. doi: 10.1074/jbc.270.22.13470. [DOI] [PubMed] [Google Scholar]
- 22.Savkur R. S., Burris T. P. The coactivator LXXLL nuclear receptor recognition motif. J. Pept. Res. 2004;63:207–212. doi: 10.1111/j.1399-3011.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhou D., Quach K. M., Yang C., Lee S. Y., Pohajdak B., Chen S. PNRC: a proline-rich nuclear receptor coregulatory protein that modulates transcriptional activation of multiple nuclear receptors including orphan receptors SF1 (steroidogenic factor 1) and ERRalpha1 (estrogen related receptor alpha-1) Mol. Endocrinol. 2000;14:986–998. doi: 10.1210/mend.14.7.0480. [DOI] [PubMed] [Google Scholar]
- 24.Aquan K., Scott J., See C. G., Sarkar N. H. Characterization and chromosomal localization of the human homologue of a rat AMP-activated protein kinase-encoding gene: a major regulator of lipid metabolism in mammals. Gene. 1994;149:345–350. doi: 10.1016/0378-1119(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 25.Chen J. D. Steroid/nuclear receptor coactivators. Vitam. Horm. 2000;58:391–448. doi: 10.1016/s0083-6729(00)58032-7. [DOI] [PubMed] [Google Scholar]
- 26.Salt I., Celler J. W., Hawley S. A., Prescott A., Woods A., Carling D., Hardie D. G. AMP-activated protein kinase: greater AMP dependence, and preferential nuclear localization, of complexes containing the α2 isoform. Biochem. J. 1998;334:177–187. doi: 10.1042/bj3340177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dowell P., Ishmael J. E., Avram D., Peterson V. J., Nevrivy D. J., Leid M. p300 functions as a coactivator for the peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 1997;272:33435–33443. doi: 10.1074/jbc.272.52.33435. [DOI] [PubMed] [Google Scholar]
- 28.Vega R. B., Huss J. M., Kelly D. P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 30.Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 31.Foretz M., Carling D., Guichard C., Ferré P., Foufelle F. AMP-activated protein kinase inhibits the glucose-activated expression of fatty acid synthase gene in rat hepatocytes. J. Biol. Chem. 1998;273:14767–14771. doi: 10.1074/jbc.273.24.14767. [DOI] [PubMed] [Google Scholar]
- 32.Leclerc I., Kahn A., Doiron B. The 5′-AMP-activated protein kinase inhibits the transcriptional stimulation by glucose in liver cells, acting through the glucose response complex. FEBS Lett. 1998;431:180–184. doi: 10.1016/s0014-5793(98)00745-5. [DOI] [PubMed] [Google Scholar]
- 33.Xavier G., Leclerc I., Salt I. P., Doiron B., Hardie D. G., Kahn A., Rutter G. A. Role of AMP-activated protein kinase in the regulation by glucose of islet beta cell gene expression. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4023–4028. doi: 10.1073/pnas.97.8.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lochhead P. A., Salt I. P., Walker K. S., Hardie D. G., Sutherland C. 5-Aminoimidazole-4-carboxamide riboside mimics the effects of insulin on the expression of the 2 key gluconeogenic genes PEPCK and glucose-6-phosphatase. Diabetes. 2000;49:896–903. doi: 10.2337/diabetes.49.6.896. [DOI] [PubMed] [Google Scholar]
- 35.Leclerc I., Lenzner C., Gourdon L., Vaulont S., Kahn A., Viollet B. Hepatocyte nuclear factor-4alpha involved in type 1 maturity-onset diabetes of the young is a novel target of AMP-activated protein kinase. Diabetes. 2001;50:1515–1521. doi: 10.2337/diabetes.50.7.1515. [DOI] [PubMed] [Google Scholar]
- 36.Habinowski S. A., Witters L. A. The effects of AICAR on adipocyte differentiation of 3T3-L1 cells. Biochem. Biophys. Res. Commun. 2001;286:852–856. doi: 10.1006/bbrc.2001.5484. [DOI] [PubMed] [Google Scholar]
- 37.Gadalla A. E., Pearson T., Currie A. J., Dale N., Hawley S. A., Sheehan M., Hirst W., Michel A. D., Randall A., Hardie D. G., Frenguelli B. G. AICA riboside both activates AMP-activated protein kinase and competes with adenosine for the nucleoside transporter in the CA1 region of the rat hippocampus. J. Neurochem. 2004;88:1272–1282. doi: 10.1046/j.1471-4159.2003.02253.x. [DOI] [PubMed] [Google Scholar]
- 38.Fryer L. G. D., Parbu-Patel A., Carling D. Protein kinase inhibitors block the stimulation of the AMP-activated protein kinase by 5-amino-4-imidazolecarboxamide riboside. FEBS Lett. 2002;531:189–192. doi: 10.1016/s0014-5793(02)03501-9. [DOI] [PubMed] [Google Scholar]