Abstract
Marine microalgae such as Pavlova and Isochrysis produce abundant amounts of the ω3-PUFAs (polyunsaturated fatty acids), EPA (eicosapentaenoic acid, 20:5n–3) and DHA (docosahexaenoic acid, 22:6n–3). The pathway leading to the conversion of EPA into DHA in these lower eukaryotes is not well established although it is predicted to involve an elongation step, catalysed by an elongating enzyme complex, leading to the conversion of EPA into ω3-DPA (ω–3-docosapentaenoic acid, 22:5n–3); followed by a desaturation step, catalysed by a Δ4-desaturase, which results in the conversion of DPA into DHA. To date, the enzymes involved in the elongation of EPA have not been identified from any lower eukaryote. In the present study, we describe the identification of microalgal genes involved in the two-step conversion of EPA into DHA. By expressed sequence tag analysis, a gene (pavELO) encoding a novel elongase was identified from Pavlova, which catalysed the conversion of EPA into ω3-DPA in yeast. Unlike any previously identified elongase from higher or lower eukaryotes, this enzyme displayed unique substrate specificity for both n–6 and n–3 C20-PUFA substrates, with no activity towards any C18- or C22-PUFA substrates. In addition, a novel Δ4-desaturase gene (IgD4) was isolated from Isochrysis, which was capable of converting ω3-DPA into DHA, as well as adrenic acid (22:4n–6) into ω6-DPA. Yeast co-expression studies, with pavELO and IgD4, revealed that these genes were capable of functioning together to carry out the two-step conversion of EPA into DHA.
Keywords: docosahexaenoic acid, eicosapentaenoic acid, Isochrysis, microalgae, Pavlova, polyunsaturated fatty acid
Abbreviations: ADA, adrenic acid; ARA, arachidonic acid; DHA, docosahexaenoic acid; DOB, dropout broth; DPA, docosapentaenoic acid; EPA, eicosapentaenoic acid; EST, expressed sequence tag; FAME, fatty acid methyl esters; PKS, polyketide synthase; PUFA, polyunsaturated fatty acid; RACE, rapid amplification of cDNA ends
INTRODUCTION
The long-chain ω3-PUFA (ω3-polyunsaturated fatty acid), DHA (docosahexaenoic acid, 22:6n–3), is an essential component of cell membranes, and is found in high proportions in neuronal membranes and external segments of photoreceptors in the retina [1]. Clinical studies have indicated that DHA is vital for proper visual and neurological development in infants [2,3]. DHA deficiency has been associated with cognitive decline and the onset of Alzheimer's disease in adults [4]. In addition, DHA consumption has been shown to benefit patients with chronic conditions, such as hypertension, coronary heart disease, depression and diabetes [5]. In humans, and particularly in infants, DHA is not produced in adequate amounts to meet metabolic demands of the body and, thus, must be obtained from dietary sources.
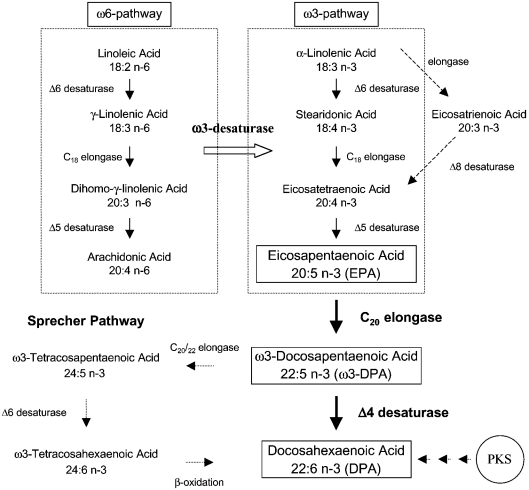
The major dietary sources of DHA are oils from marine fish and microalgae. Fish obtain most of their ω3-PUFAs by consumption of marine microalgae [6], which are considered to be the primary producers of these long-chain ω3-PUFAs. Some of these microalgae are currently exploited for the commercial production of DHA-enriched oils [7]. Thus there is a growing interest in elucidating the DHA biosynthetic pathway in these organisms. In most eukaryotes, the pathway leading to the biosynthesis of DHA begins with the C18-PUFA, α-linolenic acid (18:3n–3) and involves an alternating series of desaturations and elongations (Figure 1). With a few exceptions, both higher and lower eukaryotes share a common pathway leading up to the production of the C20-PUFA, EPA (eicosapentaenoic acid, 20:5n–3) (Figure 1) [8]. However, these organisms are supposed to differ in their mode of converting EPA into DHA (Figure 1). In mammals, the conversion of EPA into DHA is complex, and occurs through the Sprecher pathway, which involves two elongations, a desaturation and a β-oxidation step (Figure 1) [9,10]. On the other hand, in lower eukaryotes, it is predicted that the conversion of EPA into DHA occurs through the elongation of EPA to ω3-DPA (ω3-docosapenatenoic acid, 22:5n–3), which is then acted on by a Δ4-desaturase to form DHA (Figure 1) [11]. Recently, an alternative DHA biosynthesis pathway was identified in a marine protist, Schizochytrium [12], which is similar to the PKS (polyketide synthase) pathway involved in PUFA biosynthesis in prokaryotes [13]. The significance of this PKS pathway in DHA biosynthesis in marine microalgae is unknown.
Figure 1. Alternative DHA biosynthesis routes in higher and lower eukaryotes.
The ω3-pathway leading to EPA production is common to most eukaryotes. A few lower eukaryotes can use an alternative pathway for EPA production (dashed arrows). In addition, in some lower eukaryotes, the ω6-intermediates may be converted into ω3-fatty acids through an ω3-desaturase, leading to EPA production. The conversion of EPA into DHA in lower eukaryotes is a two-step process (dark arrows, boxed text in bold), whereas the conversion of EPA into DHA in higher eukaryotes (mammals) occurs through the Sprecher pathway (broken arrows). An alternative PKS pathway is supposed to function in DHA production in some lower eukaryotes (short broken arrows).
To date, all the enzymes (desaturases and elongases) involved in the production of EPA have been identified from various lower eukaryotic species [8,14]. In addition, Δ4-desaturase genes have been identified from a few organisms [11,15,16]. What remains elusive is the enzyme involved in the elongation of EPA in the pathway leading to DHA production. In general, elongation of fatty acids is catalysed by a multienzyme ‘elongating enzyme complex’. Within this complex, the condensing enzyme (elongase) is the most significant because it catalyses the rate-limiting condensation step in the four-step reaction and determines the substrate specificity of the entire complex [17,18]. A few lower eukaryotic PUFA elongases have been identified, but these are specific for 18-carbon chain length PUFA substrates (C18-PUFAs), with no activity towards C20-PUFAs such as EPA [19–21]. Thus the identification of a C20-PUFA elongase will provide the final link in elucidating the desaturase–elongase pathway involved in DHA biosynthesis in most DHA-producing lower eukaryotes.
In the present study, we set out to identify the genes involved in the final conversion of EPA into DHA in marine microalgae. Two organisms, Pavlova and Isochrysis, were selected for this study because they produce substantial amounts of EPA and DHA [22,23]. EST (expressed sequence tag) analysis of these two organisms resulted in the identification of two novel genes involved in DHA production through the desaturase–elongase pathway. pavELO, which was identified from Pavlova, was found to encode the elusive C20-PUFA elongase that displayed activity towards EPA, but not towards any of the C18- or C22-PUFA substrates tested. This is the first reported elongase with demonstrated specificity for C20-PUFAs. In addition, we identified a novel Δ4-desaturase gene (IgD4) from Isochrysis that catalysed the conversion of ω3-DPA into DHA. These two genes were further evaluated in a heterologous host for their ability to function together in the conversion of EPA into DHA.
EXPERIMENTAL
Strains and growth conditions
Frozen pellets of Pavlova sp. CCMP459 and Isochrysis galbana CCMP1323 were obtained from Provasoli-Guillard National Center for Culture of Marine Phytoplankton (CCMP, West Boothbay Harbor, ME, U.S.A.). For expression studies, the Saccharomyces cerevisiae strains used were SC334 (matα pep4-3 prb1-1122 ura3-52 leu2-3, 112 reg1-501 gal1) [24] or YPH499 (ura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1) (Stratagene, La Jolla, CA, U.S.A.). SC334 was grown either in YPD (yeast extract/peptone/dextrose) media or in selective media DOB (dropout broth) lacking leucine (DOB–Leu) or lacking uracil (DOB–Ura) (Qbiogene, Carlsbad, CA, U.S.A.). YPH499 was cultivated on YPAD (yeast extract/peptone/adenine hemisulphate/dextrose) medium or in the selective DOB media [dextrose-free +2% (w/v) galactose] lacking uracil or DOB media (dextrose-free +2% (w/v) raffinose) lacking leucine and uracil (Qbiogene).
cDNA library construction
To isolate total RNA, frozen cells were crushed in liquid nitrogen, incubated at 55 °C for 3 min in RLT lysis buffer that contains guanidine thioisocyanate, which was provided with the RNeasy Maxi kit (Qiagen, Chatsworth, CA, U.S.A.) and the lysate was homogenized using a Qiashredder column (Qiagen). RNA was then isolated using the RNeasy Maxi kit (Qiagen) according to the manufacturer's instruction. mRNA was isolated from total RNA using an oligo(dT)–cellulose resin, which was then used to synthesize double-stranded cDNA using the SuperScript plasmid system for cDNA synthesis (Invitrogen, Carlsbad, CA, U.S.A.). The Pavlova cDNA was directionally cloned (5′-SalI–3′-NotI) into pSport1 vector to generate a library containing approx. 6.1×105 clones/ml, with an average insert size of approx. 1.2 kb. The I. galbana library that was directionally cloned (5′-NotI–3′-EcoRI) into pBluescript II KS (+) vector (Stratagene) contained approx. 9.4×107 clones/ml, each with an average insert size of approx. 1.3 kb. The libraries were randomly sequenced from the 5′-end using the T7 promoter primer. Sequencing was performed using the ABI BigDye sequencing kit (Applied Biosystems, Foster City, CA, U.S.A.) and the MegaBase Capillary DNA sequencer (Amersham Biosciences, Piscataway, NJ, U.S.A.).
Identification and cloning of a putative elongase from Pavlova sp.
Random sequencing of 2000 clones of the Pavlova cDNA library resulted in the identification of one unique elongase-like fragment, obtained by the alignment of two overlapping clones of 500 bp each. Each clone demonstrated sequence homology to known PUFA elongases, as revealed by BLAST analysis against known sequences in the public domain (GenBank®). Taken together, these two overlapping clones were identified at frequencies of 0.25%. One of these clones contained the putative ‘ATG’ start codon of the gene and was used to design the primer RO1327 (5′-TGCCCATGATGTTGGCCGCAGGCTATCTTCTAGTG-3′). The full-length putative elongase gene was isolated by PCR using RACE (rapid amplification of cDNA ends)-ready cDNA as the template. This cDNA was prepared using the GeneRacer™ kit and Superscript II™ enzyme (Invitrogen), according to the manufacturer's instructions. The primers used in this PCR included 50 pmol of primer RO1327 and 30 pmol of the GeneRacer™ 3′-primer (5′-GCTGTCAACGATACGCTACGTAACG-3′). The PCR amplification was performed using Platinum Taq DNA polymerase (Invitrogen) in 50 μl total volume containing 2 μl of the RACE-ready cDNA, PCR buffer containing 20 mM Tris/HCl, pH 8.4, 50 mM KCl (final concentration), 200 μM each of deoxyribonucleotide triphosphate, 1.5 mM MgSO4 and 0.5 μl of Platinum Taq (HF) DNA polymerase. Amplification was performed as follows: initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 45 s, 55 °C for 30 s, 68 °C for 2 min; the reaction was terminated at 4 °C. The approx. 1.2 kb PCR-amplified fragment thus generated was gel-purified, cloned into the PCR-blunt vector (Invitrogen) and sequenced. Multiple sequence alignment was performed using AlignX, a modified ClustalW algorithm (VectorNTI; InforMax, Bethesda, MD, U.S.A.). This full-length gene was designated PavELO, and subcloned into the EcoRI site of the pYX242 yeast expression vector (Novagen, Madison, WI, U.S.A.) to generate a construct labelled pRPL6-B2.
Identification and cloning of a putative Δ4-desaturase from I. galbana
Clones (2000) of the I. galbana cDNA library were subjected to random sequencing that resulted in the identification of four unique desaturase-like fragments, identified based on the presence of the conserved histidine-box motifs. These desaturase-like fragments were identified at a frequency over the range 0.25–0.5%. One fragment of approx. 647 bp was found to share approx. 30% amino acid sequence identity with known front-end desaturases and this fragment was further pursued. To obtain the 3′-end of this gene, PCR amplification of the cDNA library was performed using the vector primer RO899 (5′-AGCGGATAACAATTTCACACAGGAAACAGC-3′) and the gene-specific primer RO1270 (5′-CACCTGGCTCGAGTCGACGATGATGG-3′). PCR amplification was performed in 50 μl total volume containing 1 μl of the cDNA library template, PCR buffer containing 20 mM Tris/HCl, pH 8.4, 50 mM KCl (final concentration), 200 μM of each deoxyribonucleotide triphosphate, 10 pmol of each primer, 1.5 mM MgSO4 and 0.5 μl of Platinum Taq (HF) DNA polymerase (Invitrogen). Amplification was performed as follows: initial melt at 94 °C for 2 min, followed by 5 cycles of 94 °C for 30 s, 72 °C for 3 min; 10 cycles of 94 °C for 30 s, 70 °C for 30 s, 72 °C for 3 min; 20 cycles of 94 °C for 30 s, 68 °C for 30 s, 72 °C for 3 min; and a final extension at 72 °C for 10 min. This amplification did not result in any PCR bands probably due to the low copy numbers of the transcript in the library. Thus a second round of PCR amplification was performed using 2 μl of the original PCR as template. The set-up of the PCR was identical with that described above. Amplification was performed as follows: initial denaturation at 94 °C for 3 min, followed by 30 cycles of 94 °C for 45 s, 55 °C for 30 s, 68 °C for 2 min; the reaction terminated at 4 °C. A PCR band was thus obtained which was gel-purified, cloned into PCR-Blunt vector (Invitrogen) and sequenced. This fragment was found to contain the putative 3′-end of the gene including the ‘TAA’ stop codon and the poly(A)+ tail. To isolate the 5′-end of this gene, RACE-ready cDNA was prepared and used as a template for the PCRs. PCR was then performed using 30 pmol of GeneRacer™ 5′-primer (5′-CGACTGGAGCACGAGGACACTGA-3′) in combination with 10 pmol of the gene-specific primer RO1286 (5′-CGTCCCGGTGCAATAGAAGGTGAG-3′), using conditions similar to those used to isolate the 3′-end of this gene. Bands thus obtained were gel-purified, cloned and sequenced.
For the full-length gene isolation, both genomic DNA and RACE-derived cDNA were used as templates in PCRs containing the primer RO 1400 (5′-TCAACAGAATTCATGTGCAACGCG-GCGCAGGTCGAGACGCAG-3′), which contained an EcoRI cloning site (underlined) along with the ‘ATG’ start site (bold), and RO 1401 (5′-AAAAGAAAGCTTTTAGTCCGCCTTGACCGTGTCGACCAAAGC-3′), which contained a HindIII cloning site (underlined) along with the ‘TAA’ stop site (bold). PCR amplification was performed using Advantage-GC cDNA polymerase (ClonTech, Palo Alto, CA, U.S.A.) in a 50 μl of total volume containing 1 μl of the RACE-cDNA or 2 μl of genomic DNA, and a PCR buffer [40 mM tricine/KOH, pH 9.2, 15 mM potassium acetate (final concentration), 3.5 mM magnesium acetate, 5% (v/v) DMSO, 3.75 μg/ml BSA, 200 μM each deoxyribonucleotide triphosphate, 1 M GC-melt and 1 μl of Advantage-GC cDNA polymerase]. The amplification conditions included an initial denaturation at 94 °C for 1 min, followed by 30 cycles of 94 °C for 30 s and 68 °C for 3 min, and a final extension at 68 °C for 5 min. A 1.35 kb band thus obtained corresponded to the full-length gene and was designated IgD4. This band was gel-purified, and cloned into the EcoRI–HindIII sites of the pYX242 yeast expression vector (Novagen). This construct was labelled pRIG6 and was transformed into yeast SC334 for expression studies. Multiple sequence alignment of the full-length sequence was performed using AlignX, a modified ClustalW algorithm (VectorNTI; InforMax). A phylogenetic tree was constructed using the neighbour-joining method [25].
Functional expression of pavELO and IgD4 in S. cerevisiae
S. cerevisiae (SC334 or YPH499) was transformed with either pRPL6-B2 containing pavELO cloned into pYX242 or pRIG6 containing IgD4 cloned into pYX242. Transformation was performed using the Alkali-Cation Yeast Transformation kit (Q-biogene, Carlsbad, CA, U.S.A.). Transformants were selected for leucine auxotrophy on media lacking leucine. To characterize enzyme specificity, transformants were grown at 20–24 °C for 44–48 h in DOB–Leu medium containing 25 or 50 μM of various exogenously supplied fatty acid substrates. The host strain transformed with the pYX242 vector alone was used as a negative control in all experiments. At least three independent analyses were performed on these transgenic yeast cultures.
Co-expression of pavELO and IgD4 in S. cerevisiae
To co-express pavELO and IgD4, IgD4 was subcloned into the pESC-Ura vector (Stategene). IgD4 was first released from pRIG6 by digestion with EcoRI and EcoRV, and this insert was subcloned into the EcoRI–SacI restriction site (multiple cloning site 1) of pESC-Ura vector generating construct pRIG9. pRIG9 and pRPL6-B2 were co-transformed into S. cerevisiae strain YPH499 as described previously, and transformants were selected for both uracil and leucine auxotrophy. To characterize enzyme specificity, transformants were grown at 20 °C in the presence of 100 μM exogenously supplied EPA, in the selective DOB media (dextrose-free) containing raffinose. After 24 h of growth, 2% galactose was added to the cultures to induce expression of IgD4, and cultures were grown for an additional 48 h at 20 °C. Cells were then harvested and processed for total lipid analysis. The experimental controls consisted of S. cerevisiae (YPH499) transformed with the two vectors pYX242 and pESC-Ura or S. cerevisiae (YPH499) transformed with the individual plasmids pRIG9 or pRPL6-B2.
Fatty acid analysis
Recombinant yeast cells were washed in deionized water, vortex-mixed in 15 ml of methanol and incubated for 1–2 h at room temperature (24 °C) after the addition of approx. 100 μg of tride-canoin and 29 ml of chloroform. Lipids from the mixture were then separated and extracted in the lower chloroform layer using a separatory funnel. These segregated lipids were filtered through a Whatman filter with 1 g of anhydrous sodium sulphate to remove particulates and residual water. The organic solvents were evaporated to dryness at 40 °C under a stream of nitrogen and the extracted lipids were saponified with 2 ml of 0.5 M KOH in methanol. The saponified samples were heated to 95–100 °C for 30 min and cooled to room temperature. For fatty acid methylation, approx. 2 ml of 14% boron trifluoride in methanol was added, the mixture was heated to 95–100 °C for 30 min, cooled to room temperature, and 2 ml of water and 1 ml of hexane were added to extract the FAME (fatty acid methyl esters). FAME were analysed by GC as described previously [26]. Fatty acids were identified based on the retention time in comparison with authentic standards of long-chain PUFAs, and their identity further verified by GC–MS [26]. The rate of conversion of substrate into product (conversion rate=[product]/[product+substrate]) was calculated to reflect the enzymic activity. This value reflects the overall rate of conversion of total substrate into total product formed, and does not distinguish between activities on esterified versus non-esterified substrates.
RESULTS
Identification of a putative PUFA elongase gene from Pavlova sp.
Marine microalgal species such as Isochrysis and Pavlova represent good candidates for the isolation of genes involved in DHA production because of the high amounts of ω3-PUFAs they produce. To identify these genes, EST libraries from these organisms were constructed, randomly sequenced and analysed for sequence homology to genes present in the public domain (GenBank®). This random sequencing approach resulted in the identification of a cDNA clone with sequence homology to known PUFA elongase genes. The full-length gene, corresponding to this open reading frame, was obtained by RACE-PCR, and this gene was designated pavELO. This gene was 834 bp in length and encoded a protein of 277 amino acids. Comparative sequence analysis revealed that pavELO shared higher overall amino acid sequence identity with mammalian PUFA elongases when compared with C18-PUFA elongases from lower eukaryotes such as Mortierella alpina (GLELO, accession no. AAF70417) (Figure 2). The predicted protein shared greatest sequence homology with mammalian PUFA elongases that recognize C20- and C22-PUFA substrates (C20/C22-PUFA elongases), displaying approx. 35% amino acid sequence identity with the mouse C20/C22-PUFA elongase (mElovl2, accession no. NP_062296) and the human C20/C22-PUFA elongase (hELOVL2, accession no. NP_060240) (Figure 2). Similar to other PUFA elongases, this protein contained several of the conserved motifs characteristic of the PUFA elongase family which included the single conserved histidine box, as well as the 17 invariant amino acid residues that are conserved in the ELO family proteins [14] (Figure 2). Hydropathy analysis revealed that pavELO is highly hydrophobic and is predicted to contain six transmembrane domains, characteristic of other PUFA elongases [19,21,27].
Figure 2. Sequence comparison of PUFA elongases.
The putative amino acid sequences of elongases from Pavlova (pavELO, accession no. AY630573), human (hELOVL2, accession no. NP_060240), mouse (hElovl2, accession no. NP_062296) and M. alpina (GLELO, accession no. AAF70417) were aligned using AlignX (VectorNTI; InforMax). Identical amino acids shared by at least three of the sequences are boxed and shaded. *, the 17 residues that are conserved across the PUFA elongases [14]. The conserved histidine box (HXXHH) is underlined. The three pavELO residues, which caused a decrease in enzymic activity when mutated are indicated by ‘@’ above them.
Characterization of pavELO in S. cerevisiae
To characterize the enzymic activity of pavELO, the plasmid pRPL6-B2 containing pavELO downstream of the constitutive TPI promotor of pYX242, was transformed into S. cerevisiae. Since S. cerevisiae does not produce any endogenous PUFAs, it serves as a suitable expression host for characterizing enzymes involved in the PUFA production. Also, previous studies have revealed that expression of the elongase component (condensing enzyme) alone is sufficient to allow elongation of long-chain PUFAs in yeast [19,21,27], indicating that most PUFA elongases can interact and function with the endogenous reductases and dehydratase of the yeast elongation complex to perform the complete elongation of fatty acids.
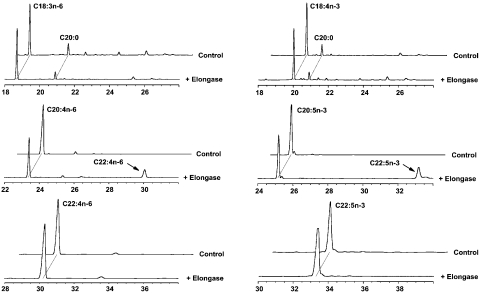
To determine the substrate specificity of pavELO, recombinant yeast expressing pavELO were grown in the presence of various exogenous fatty acid substrates, e.g. C18-PUFAs such as linolenic acid (18:2n–6), γ-linolenic acid (18:3n–6), α-linolenic acid (18:3n–3) and stearidonic acid (18:4n–3); C20-PUFAs such as dihomo-γ-linolenic acid (20:3n–6), ARA (arachidonic acid, 20:4n–6), eicosatetraenoic acid (20:4n–3) and EPA; C22-PUFAs such as ADA (adrenic acid, 22:4n–6) and ω–3-DPA; a saturated fatty acid, arachidic acid (20:0) and a mono-unsaturated fatty acid, eicosenoic acid (20:1). After 48 h of growth in the presence of these substrates, total lipids from these recombinant clones were extracted and analysed by GLC. Results from these studies revealed that pavELO did not recognize the saturated fatty acid or the mono-unsaturated fatty acid substrate that was tested (results not shown). In addition, pavELO did not elongate any of the C18-PUFAs or the C22-PUFA substrates (Figure 3). However, this enzyme did recognize and elongate the C20-PUFA substrate EPA, converting approx. 20% of this into ω3-DPA (Figure 3). In addition, pavELO also elongated the corresponding n–6 C20-PUFA substrate, ARA, converting it into ADA (22:4n–6) (Figure 3). This indicated that pavELO encoded an elongase that was specific for the C20-PUFAs, EPA and ARA, and may function in DHA biosynthesis in Pavlova. Control yeast cultures that were transformed with the vector alone did not demonstrate any elongation activity (Figure 3) confirming that pavELO was responsible for the demonstrated PUFA elongation activity observed in the recombinant yeast.
Figure 3. GLC analysis of FAME from recombinant yeast expressing pavELO.
Recombinant yeast cultures expressing pavELO or the vector (pYX242) alone were grown in the presence of 50 μM of the following substrates added individually to the culture: γ-linolenic acid (18:3n–6), stearidonic acid (18:4n–3), ARA (20:4n–6), EPA (20:5n–3), ADA (22:4n–6) and ω3-DPA (22:5n–3). ω3-DPA or ADA formed by the elongation of EPA or ARA respectively are indicated by arrows. The x-axis represents retention time (min) and y-axis represents degree of response.
During the isolation of the pavELO gene by PCR, we also identified two mutant clones. These mutations, which were probably generated during the PCR process, resulted in a single residue change (G171→A171) in one clone, and a double mutation in the second clone (N57→D57 and R249→C249). These two mutants were tested for elongation activity in yeast to determine the importance of these residues for enzyme functionality (results not shown). The first clone containing the single G171→A171 mutation demonstrated a complete loss of elongation activity indicating that Gly-171 is critical for enzyme functionality. This is not surprising since Gly-171 lies in proximity to the conserved histidine box and the 17 invariant residues that are conserved throughout PUFA elongase family of proteins (Figure 2) [14]. Gly-171 also appears to be conserved in the mammalian C20/22-PUFA elongases (Figure 2). The second clone containing the double mutation (N57→D57 and R249→C249) displayed a 90% loss in elongating activity when compared with the activity of the original pavELO enzyme, indicating the importance of these residues for enzyme functionality. Despite a drastic reduction in activity, this mutant enzyme exhibited the same substrate specificity as the native pavELO did for the C20-PUFAs, EPA and ARA. This suggests that the mutated regions, N57 and R249, by themselves do not determine the substrate specificity of this elongase.
Identification and characterization of the Isochrysis Δ4-desaturase
Random sequencing of the EST library from Isochrysis resulted in the identification of four unique desaturase-like fragments. Full-length sequences of all the four fragments were obtained by RACE-PCR amplification and expressed in yeast, but details of only the one sequence that demonstrated Δ4-desaturase activity has been described in the present study. This gene, designated as IgD4, was 1302 bp long and encoded a protein of 433 amino acids. A homology search revealed that the predicted protein encoded by IgD4 shared <31% sequence identity with known Δ4-, Δ5- and Δ6-desaturases, thus making it difficult to predict the regioselectivity of this desaturase. Similar to all front-end desaturases, this encoded protein contained a cytochrome b5 domain at the N-terminus, and the three conserved histidine-rich motifs that are known to be essential for the enzymic activity of membrane-bound desaturases [29].
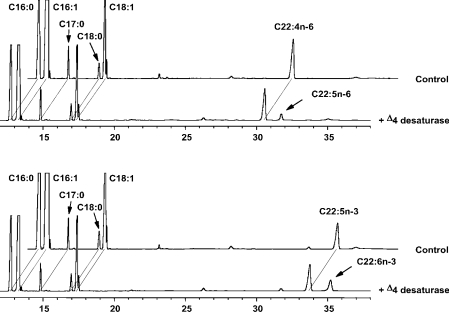
To characterize the enzymic activity of IgD4, the clone pRIG6 that consisted of IgD4 cloned into vector pYX242, was expressed in S. cerevisiae (SC334). Transformants were grown in the presence of various exogenous PUFA substrates, which were taken up by the host, and the corresponding fatty acid products formed were analysed by GLC. Figure 4 demonstrates that IgD4 was capable of recognizing ω3-DPA as a substrate, converting approx. 28% of it into DHA, indicating that the gene encoded an enzyme with Δ4-desaturase activity. In addition, IgD4 also recognized the n–6 substrate, ADA (22:4n–6), converting approx. 13% of it into ω6-DPA (22:5n–6) (Figure 4). This enzyme did not act on any of the C18-PUFAs or the C20-PUFAs tested, indicating that it did not exhibit Δ6-, Δ5- or Δ8-desaturase activity (results not shown). These results confirmed that IgD4 encodes a Δ4-desaturase that probably functions in DHA biosynthesis of I. galbana. No background substrate conversion was detected in recombinant cells containing the vector alone (Figure 4).
Figure 4. GLC analysis of FAME from recombinant yeast expressing IgD4.
Recombinant yeasts expressing IgD4 or vector alone were grown in the presence of 50 μM of the n–6 PUFA, ADA (22:4n–6) or the n–3 PUFA, ω3-DPA (22:5n–3). The corresponding n–6 product, 22:5n–6, and the n–3 product, DHA (22:6n–3), formed by the Δ4-desaturase activity of IgD4, are indicated by arrows. The x-axis represents retention time (min) and y-axis represents degree of response.
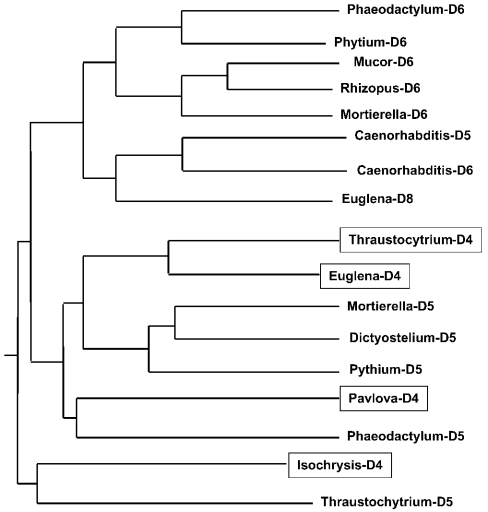
Comparative sequence analysis revealed that IgD4 shared only approx. 30% sequence identity with known Δ4-desaturases from Thraustochytrium [11], Euglena [15] and Pavlova [16]. The histidine box 2 of the deduced polypeptide of IgD4 contains an HXXHH motif (Figure 5) instead of the extended HXXXHH motif that is characteristic of the other Δ4-desaturases. In addition, the extended domain that exists between the second and third histidine boxes of the Δ4-desaturases from Euglena and Thraustochytrium is not present in IgD4 [11,15]. Phylogenetic analysis revealed that the Isochrysis Δ4-desaturase forms a distinct cluster from that of the other known Δ4-desaturases from Euglena (accession no. AY278558), Thraustochytrium (accession no. AF489589) and Pavlova (accession no. AY332747) (Figure 6).
Figure 5. Sequence alignment of Δ4-desaturases.
The putative amino acid sequences of Δ4-desaturases from Isochrysis (IgD4, accession no. AY630574), Pavlova (Pldes1, accession no. AY332747), Euglena (EgD4, accession no. AY278558) and Thraustochytrium (FAD4, accession no. AF489589) were aligned using AlignX (VectorNTI; InforMax). Identical amino acids shared by at least three of the sequences are boxed and shaded. The three conserved histidine box regions are underlined. The second histidine box region of IgD4 does not contain the extended HXXXHH motif shared by the other Δ4-desaturases. The microalgal Δ4-desaturases lack the extended domain that exists between the second and third histidine boxes of the other two Δ4-desaturases.
Figure 6. Phylogenetic analysis of IgD4 and other front-end desaturases.
The phylogenetic tree was created using the neighbour-joining method [25]. Sequences used for the analysis were Δ4-desaturases (boxed): Euglena gracilis (AY278558), I. galbana (AY630574), Pavlova lutheri (AY332747), Thraustochytrium sp. (AF489589); Δ5-desaturases: Caenorhabditis elegans (AAC95143), Dictyostelium discoideum (BAA37090), M. alpina (AAC72755), Phaeodactylum tricornutum (AY082392), Phytium irregulare (AAL13311), Thraustochytrium sp. (AAM09687); Δ8-desaturase: E. gracilis (AAD45877); Δ6-desaturases: C. elegans (AAC15586), M. alpina (AAF08685), Mucor circinelloides (BAB69055), P. tricornutum (AY082393), P. irregulare (AAL13310) and Rhizopus sp. (AAP83964).
Production of DHA in S. cerevisiae
To determine if the Pavlova C20-elongase could function with a Δ4-desaturase in the production of DHA, IgD4 was subcloned into pESC-Ura, a yeast vector that was compatible with the pYX242 vector which contained pavELO. Both these genes were then co-expressed in S. cerevisiae that was grown in the presence of exogenous EPA or ARA, followed by total lipid analysis of the recombinant cells. From Table 1, it is apparent that the recombinant cells expressing pavELO and IgD4 were capable of taking up the exogenous EPA and converting it into DHA. DHA accumulated approx. 3.8% of the total fatty acids confirming that the biosynthetic pathway from EPA to DHA had been successfully reconstructed in yeast. Approx. 24% of EPA that was taken up by the cell was converted into product (ω3-DPA + DHA), and approx. 55% of the ω3-DPA formed by the action of pavELO was converted into DHA by IgD4. Similarly, when the n–6 substrate ARA was added to the culture, ADA (22:4n–6) and ω–6-DPA (22:5n–6) were formed (results not shown), confirming that these proteins were also active on substrates in the n–6 PUFA pathway. Thus this C20-PUFA elongase was capable of functioning in unison with a Δ4-desaturase to catalyse the two-step conversion of EPA into DHA.
Table 1. Conversion of EPA into DHA in yeast.
Yeast strain YPH499 was transformed with two constructs (pYX242+pavELO and pESC-Ura+IgD4) or with the two empty vectors (pYX242+pESC-Ura), and grown in the presence of exogenous EPA. The values shown are means±S.D. of the fatty acid composition for three independent cultures. Fatty acid values correspond to the percentage of total fatty acids.
| Percentage of total fatty acids | ||
|---|---|---|
| Fatty acid | Empty vectors | pavELO+IgD4 |
| 16:0 | 16.01±1.52 | 17.78±0.36 |
| 16:1Δ9 | 30.26±0.94 | 22.70±0.68 |
| 18:0 | 5.33±1.42 | 5.84±0.23 |
| 18:1Δ9 | 22.78±2.9 | 25.56±1.77 |
| 18:2Δ9,12 | − | − |
| 18:3Δ9,12,15 | − | − |
| 18:4Δ6,9,12,15 | − | − |
| 20:4Δ8,11,14,17 | − | − |
| 20:5Δ5,8,11,14,17 | 25.62±5.9 | 21.20±1.06 |
| 22:5Δ7,10,13,16,19 | − | 3.11±0.97 |
| 22:6Δ4,7,10,13,16,19 | − | 3.81±0.35 |
DISCUSSION
Microalgae such as Pavlova and Isochrysis are commonly used as feed in the aquaculture industry because of the considerable amounts of EPA and DHA they produce [30,31]. In these lower eukaryotes, the desaturase–elongase pathway is supposed to be involved in DHA production, and a C20-PUFA elongase in conjunction with a Δ4-desaturase is proposed to catalyse the final conversion of EPA into DHA [11] (Figure 1). Neither one of these enzymes has been identified from Isochrysis to confirm the existence of this pathway in this organism. A Δ4-desaturase gene was recently identified from Pavlova [16], which supports the proposed two-step conversion of EPA into DHA in this organism. However, the missing link in this pathway is the predicted C20-PUFA elongase that elongates EPA. In fact, the gene encoding a C20-PUFA elongase has not been identified from any lower eukaryotes to date, although other PUFA elongases have been identified [19,21,32].
To identify the genes involved in the DHA production in marine microalgae, we performed EST sequence analysis of actively growing DHA-producing cultures of Pavlova and Isochrysis. This search resulted in the identification of the elusive C20-PUFA elongase from Pavlova (pavELO), which demonstrated a specificity for elongating C20-PUFA substrates (i.e. EPA). Although we did not identify this elongase homologue from Isochrysis, we did identify a novel Δ4-desaturase gene from this organism that was capable of catalysing the final conversion of ω3-DPA into DHA in yeast. Recently, an alternative PKS pathway for DHA production was identified in a marine protist, Schizochytrium, and the EST analysis revealed that these PKS gene transcripts were very abundant in this organism [12]. Our sequencing efforts did not reveal the presence of genes with similarity to any of the known PUFA–PKS pathway genes from Schizochytrium [12] or from any prokaryote [13,33,34] in Isochrysis or Pavlova. On the other hand, a number of other genes involved in EPA production were identified from both organisms (S. L. Pereira, unpublished work), including Δ5-desaturase genes, as well as the previously identified C18-PUFA elongase from Isochrysis [32]. This implies that the desaturase–elongase pathway is the major route for the DHA production in both these microalgae. pavELO is the first PUFA elongase to be identified with specificity for C20-PUFA substrates such as EPA. All previously characterized PUFA elongases from lower eukaryotes were found to act on C18-PUFA substrates only [19,21,32]. Although there are known mammalian elongases (e.g. ELOVL2) that recognize C20-PUFA substrates, these tend to display promiscuous substrate specificity, elongating C22-PUFA substrates as well [14,27,28]. The pavELO-encoded enzyme shares structural similarities with other PUFA elongases in that it is highly hydrophobic, contains several transmembrane domains and contains a single conserved histidine box (HXXHH). Although the exact function of the single histidine box in PUFA elongases is unknown, has been proposed to be essential for elongase activity [35].
Expression of pavELO in yeast led to the elongation of exogenously supplied C20-PUFA substrates, which are not normally recognized by the endogenous yeast elongation system (Figure 3). This implies that pavELO encodes a component of the C20-PUFA-‘elongating enzyme complex’ from Pavlova that determines substrate specificity. The ‘elongating enzyme complex’ is a multienzyme complex composed of a condensing enzyme (elongase), a β-keto acyl reductase, a dehydratase and an enoyl reductase [36]. Previous studies in plants and mammals have revealed that the ‘elongase’ component determines the substrate specificity of the complex [17,18]. Although pavELO directs the substrate specificity of the elongation complex, it remains to be determined if this enzyme catalyses the initial condensation reaction in the four-step elongation process. The demonstrated activity of pavELO in yeast also indicates that this enzyme can interact with the other components of the yeast ‘elongating enzyme complex’ to perform the complete elongation reaction. The remaining enzymes of the elongation enzyme complex are supposed to be promiscuous and shared by different elongating systems [17,36].
The precise biochemical characterization of the PUFA elongase family of proteins has been slow. The regions essential for enzymic activity of these enzymes and the determinants of substrate specificity are largely unknown. This is due to the inherent problems associated with purification and handling of these hydrophobic integral membrane proteins. However, some structure–function predictions can be made by comparative sequence analysis of known PUFA elongases. Sequence alignments of all known PUFA elongases have revealed the presence of 17 invariant amino acids that are conserved in all these proteins and, hence, predicted to be critical for enzymic activity [14]. In the present study, by analysing some mutants, we have identified a few additional residues in pavELO (e.g. Gly-171, Asn-57 and Arg-249), which appear to be important for the proper functioning of this protein (Figure 2). Further site-directed mutagenesis studies can be performed to test the structure–function prediction, and thus further the characterization of this family of enzymes. In addition, the identification of pavELO expands the functional repertoire of PUFA elongases, since it is the first elongase with demonstrated substrate specificity for C20-PUFAs. This offers the opportunity to identify regions that determine substrate specificity by comparative sequence analysis with known C18-PUFA elongases and the promiscuous mammalian PUFA elongases. Results from these studies may eventually enable the manipulation of substrate specificity and activity of these enzymes to fit various biotechnology needs.
During the course of EST sequencing, we also identified IgD4, a novel Δ4-desaturase gene from Isochrysis, which is assumed to catalyse the final step in DHA biosynthesis in Isochrysis (Figure 1). Sequence comparison revealed that this new Δ4-desaturase shares low sequence identity with other known Δ4-desaturases (Figure 5). Phylogenetic analysis of various front-end desaturases from lower eukaryotes revealed that in contrast with the Δ6-desaturases, the Δ4- and Δ5-desaturases tend to cluster together, implying a common phylogenetic ancestor (Figure 6). These two classes of desaturases probably arose by a gene duplication event that may have occurred subsequent to the divergence of the Δ6-desaturases. Although IgD4 shares common structural characteristics with other front-end desaturases, such as the three conserved histidine-box motifs and a cytochrome b5 domain at its N-terminus, it does not contain the prolonged amino acid stretch between the second and the third histidine box as seen in other Δ4-desaturases [11,15]. This finding challenges the suggestion that this extended region contributes to the Δ4-regiospecificity of this class of enzymes [15].
With the exception of the C20-PUFA elongase, all the other genes involved in the DHA biosynthesis were previously identified [11,19,26,37]. The identification of pavELO provides the final link in the DHA biosynthesis pathway. Previous studies have demonstrated the possibility of reconstructing the EPA biosynthesis pathway in a heterologous host by co-expressing genes encoding a Δ6-desaturase, a Δ5-desaturase and a C18-PUFA elongase in yeast [20,38,39]. In the present study, we have taken a step further and demonstrated the possibility of converting EPA into DHA in a heterologous host by co-expressing pavELO together with a IgD4 (Table 1). These results support the possibility of reconstituting the entire DHA metabolic pathway in a heterologous host, thus providing a biotechnological tool for synthesizing DHA in alternative sources.
Microalgae that produce abundant amounts of EPA and DHA are widely used as a direct or indirect feed source in aquaculture to feed fish, crustaceans, bivalves etc. [40]. In addition to enhancing the growth of some of these species, these microalgae also serve as an enrichment source of ω3-PUFAs in farmed fish [41]. Thus understanding the PUFA metabolism in these organisms is an area of major focus in the aquaculture arena. The identification of the C20-PUFA elongase gene and the Δ4-desaturase gene from such microalgae provides the opportunity to study DHA biosynthesis at a genetic level, and to uncover the mechanisms involved in the regulation of DHA production at the metabolic level. These studies could eventually enable the manipulation of these organisms so as to enhance the production of DHA.
References
- 1.Neuringer M. Infant vision and retinal function in studies of dietary long-chain polyunsaturated fatty acids: methods, results, and implications. Am. J. Clin. Nutr. 2000;71:256S–267S. doi: 10.1093/ajcn/71.1.256S. [DOI] [PubMed] [Google Scholar]
- 2.Crawford M. A., Costeloe K., Ghebremeskel K., Phylactos A., Skirvin L., Stacey F. Are deficits of arachidonic and docosahexaenoic acids responsible for the neural and vascular complications of preterm babies? Am. J. Clin. Nutr. 1997;66:1032S–1041S. doi: 10.1093/ajcn/66.4.1032S. [DOI] [PubMed] [Google Scholar]
- 3.Nettleton J. A. Are n-3 fatty acids essential nutrients for fetal and infant development? J. Am. Diet. Assoc. 1993;93:58–64. doi: 10.1016/0002-8223(93)92132-h. [DOI] [PubMed] [Google Scholar]
- 4.Tully A. M., Roche H. M., Doyle R., Fallon C., Bruce I., Lawlor B., Coakley D., Gibney M. J. Low serum cholesteryl ester-docosahexaenoic acid levels in Alzheimer's disease: a case-control study. Br. J. Nutr. 2003;89:483–489. doi: 10.1079/BJN2002804. [DOI] [PubMed] [Google Scholar]
- 5.Horrocks L. A., Yeo Y. K. Health benefits of docosahexaenoic acid (DHA) Pharmacol. Res. 1999;40:211–225. doi: 10.1006/phrs.1999.0495. [DOI] [PubMed] [Google Scholar]
- 6.DePauw N., Personne G. Micro-algal biotechnology. In: Borowitzka M. A., Borowitzka J. L., editors. Microalgal Biotechnology. Cambridge: Cambridge University Press; 1988. p. 197. [Google Scholar]
- 7.Kyle D. J., Arterburn L. M. Single cell oil sources of docosahexaenoic acid: clinical studies. World Rev. Nutr. Diet. 1998;83:116–131. doi: 10.1159/000059656. [DOI] [PubMed] [Google Scholar]
- 8.Pereira S. L., Leonard A. E., Mukerji P. Recent advances in the study of fatty acid desaturases from animals and lower eukaryotes. Prostaglandins Leukot. Essent. Fatty Acids. 2003;68:97–106. doi: 10.1016/s0952-3278(02)00259-4. [DOI] [PubMed] [Google Scholar]
- 9.Voss A., Reinhart M., Sankarappa S., Sprecher H. The metabolism of 7,10,13,16,19-docosapentaenoic acid to 4,7,10,13,16,19-docosahexaenoic acid in rat liver is independent of a Δ4-desaturase. J. Biol. Chem. 1991;266:19995–20000. [PubMed] [Google Scholar]
- 10.Sprecher H., Chen Q., Yin F. Q. Regulation of the biosynthesis of 22:5n−6 and 22:6n−3: a complex intracellular process. Lipids. 1999;34:S153–S156. doi: 10.1007/BF02562271. [DOI] [PubMed] [Google Scholar]
- 11.Qiu X., Hong H., MacKenzie S. L. Identification of a Δ4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexaenoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J. Biol. Chem. 2001;276:31561–31566. doi: 10.1074/jbc.M102971200. [DOI] [PubMed] [Google Scholar]
- 12.Metz J. G., Roessler P., Facciotti D., Levering C., Dittrich F., Lassner M., Valentine R., Lardizabal K., Domergue F., Yamada A., et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. 2001;293:290–293. doi: 10.1126/science.1059593. [DOI] [PubMed] [Google Scholar]
- 13.Yazawa K. Production of eicosapentaenoic acid from marine bacteria. Lipids. 1996;31:S297–S300. doi: 10.1007/BF02637095. [DOI] [PubMed] [Google Scholar]
- 14.Leonard A. E., Pereira S. L., Sprecher H., Huang Y. S. Elongation of long-chain fatty acids. Prog. Lipid Res. 2004;43:36–54. doi: 10.1016/s0163-7827(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 15.Meyer A., Cirpus P., Ott C., Schlecker R., Zahringer U., Heinz E. Biosynthesis of docosahexaenoic acid in Euglena gracilis: biochemical and molecular evidence for the involvement of a Δ4-fatty acyl group desaturase. Biochemistry. 2003;42:9779–9788. doi: 10.1021/bi034731y. [DOI] [PubMed] [Google Scholar]
- 16.Tonon T., Harvey D., Larson T. R., Graham I. A. Identification of a very long chain polyunsaturated fatty acid Δ4-desaturase from the microalga Pavlova lutheri. FEBS Lett. 2003;553:440–444. doi: 10.1016/s0014-5793(03)01078-0. [DOI] [PubMed] [Google Scholar]
- 17.Millar A. A., Kunst L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997;12:121–131. doi: 10.1046/j.1365-313x.1997.12010121.x. [DOI] [PubMed] [Google Scholar]
- 18.Moon Y. A., Shah N. A., Mohapatra S., Warrington J. A., Horton J. D. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 2001;276:45358–45366. doi: 10.1074/jbc.M108413200. [DOI] [PubMed] [Google Scholar]
- 19.Parker-Barnes J. M., Das T., Bobik E., Leonard A. E., Thurmond J. M., Chaung L. T., Huang Y. S., Mukerji P. Identification and characterization of an enzyme involved in the elongation of n−6 and n−3 polyunsaturated fatty acids. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8284–8289. doi: 10.1073/pnas.97.15.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaudoin F., Michaelson L. V., Hey S. J., Lewis M. J., Shewry P. R., Sayanova O., Napier J. A. Heterologous reconstitution in yeast of the polyunsaturated fatty acid biosynthetic pathway. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6421–6426. doi: 10.1073/pnas.110140197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zank T. K., Zahringer U., Beckmann C., Pohnert G., Boland W., Holtorf H., Reski R., Lerchl J., Heinz E. Cloning and functional characterization of an enzyme involved in the elongation of Δ6-polyunsaturated fatty acids from the moss Physcomitrella patens. Plant J. 2002;31:255–268. doi: 10.1046/j.1365-313x.2002.01354.x. [DOI] [PubMed] [Google Scholar]
- 22.Meireles L. A., Guedes A. C., Malcata F. X. Lipid class composition of the microalga Pavlova lutheri: eicosapentaenoic and docosahexaenoic acids. J. Agric. Food Chem. 2003;51:2237–2241. doi: 10.1021/jf025952y. [DOI] [PubMed] [Google Scholar]
- 23.Yongmanitchai W., Ward O. P. Omega-3 fatty acids: alternative sources of production. Proc. Biochem. 1989;24:117–125. [Google Scholar]
- 24.Hovland P., Flick J., Johnston M., Sclafani R. A. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene. 1989;83:57–64. doi: 10.1016/0378-1119(89)90403-4. [DOI] [PubMed] [Google Scholar]
- 25.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Knutzon D. S., Thurmond J. M., Huang Y. S., Chaudhary S., Bobik E. G., Chan G. M., Kirchner S. J., Mukerji P. Identification of Δ5-desaturase from Mortierella alpina by heterologous expression in Bakers' yeast and canola. J. Biol. Chem. 1998;273:29360–29366. doi: 10.1074/jbc.273.45.29360. [DOI] [PubMed] [Google Scholar]
- 27.Leonard A. E., Bobik E. G., Dorado J., Kroeger P. E., Chuang L. T., Thurmond J. M., Parker-Barnes J. M., Das T., Huang Y. S., Mukerji P. Cloning of a human cDNA encoding a novel enzyme involved in the elongation of long-chain polyunsaturated fatty acids. Biochem. J. 2000;350:765–770. [PMC free article] [PubMed] [Google Scholar]
- 28.Leonard A. E., Kelder B., Bobik E. G., Chuang L. T., Lewis C. J., Kopchick J. J., Mukerji P., Huang Y. S. Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids. 2002;37:733–740. doi: 10.1007/s11745-002-0955-6. [DOI] [PubMed] [Google Scholar]
- 29.Shanklin J., Whittle E., Fox B. G. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 30.Volkman J. K., Dunstan G. A., Jeffrey S. W., Kearney P. S. Fatty acids from microalgae of the genus Pavlova. Phytochemistry. 1991;30:1855–1859. [Google Scholar]
- 31.Burgess J. G., Iwamoto K., Miura Y., Takano H., Matsunaga T. An optical-fiber photobioreactor for enhanced production of the marine unicellular alga isochrysis aff. galbana t-iso (utex-lb-2307) rich in docosahexaenoic acid. Appl. Microbiol. Biotechnol. 1993;39:456–459. [Google Scholar]
- 32.Qi B., Beaudoin F., Fraser T., Stobart A. K., Napier J. A., Lazarus C. M. Identification of a cDNA encoding a novel C18-Δ9 polyunsaturated fatty acid-specific elongating activity from the docosahexaenoic acid (DHA)-producing microalga, Isochrysis galbana. FEBS Lett. 2002;510:159–165. doi: 10.1016/s0014-5793(01)03247-1. [DOI] [PubMed] [Google Scholar]
- 33.Morita N., Ueno A., Tanaka M., Ohgiya S., Hoshino T., Kawasaki K., Yumoto I., Ishizaki K., Okuyama H. Cloning and sequencing of clustered genes involved in fatty acid biosynthesis from the docosahexaenoic acid producing bacteria, Vibrio marinus strain MP1. Biotechnol. Lett. 1999;21:641–646. [Google Scholar]
- 34.Allen E. E., Bartlett D. H. Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology. 2002;148:1903–1913. doi: 10.1099/00221287-148-6-1903. [DOI] [PubMed] [Google Scholar]
- 35.Qi B., Fraser T. C., Bleakley C. L., Shaw E. M., Stobart A. K., Lazarus C. M. The variant ‘his-box’ of the C18-delta9-PUFA-specific elongase IgASE1 from Isochrysis galbana is essential for optimum enzyme activity. FEBS Lett. 2003;547:137–139. doi: 10.1016/s0014-5793(03)00676-8. [DOI] [PubMed] [Google Scholar]
- 36.Cinti D. L., Cook L., Nagi M. N., Suneja S. K. The fatty acid chain elongation system of mammalian endoplasmic reticulum. Prog. Lipid Res. 1992;31:1–51. doi: 10.1016/0163-7827(92)90014-a. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y. S., Chaudhary S., Thurmond J. M., Bobik E. G., Jr, Yuan L., Chan G. M., Kirchner S. J., Mukerji P., Knutzon D. S. Cloning of δ12- and δ6-desaturases from Mortierella alpina and recombinant production of γ-linolenic acid in Saccharomyces cerevisiae. Lipids. 1999;34:649–659. doi: 10.1007/s11745-999-0410-8. [DOI] [PubMed] [Google Scholar]
- 38.Domergue F., Abbadi A., Ott C., Zank T. K., Zahringer U., Heinz E. Acyl carriers used as substrates by the desaturases and elongases involved in very long-chain polyunsaturated fatty acids biosynthesis reconstituted in yeast. J. Biol. Chem. 2003;278:35115–35126. doi: 10.1074/jbc.M305990200. [DOI] [PubMed] [Google Scholar]
- 39.Qi B., Fraser T., Mugford S., Dobson G., Sayanova O., Butler J., Napier J. A., Stobart A. K., Lazarus C. M. Production of very long chain polyunsaturated omega-3 and omega-6 fatty acids in plants. Nat. Biotechnol. 2004;22:739–745. doi: 10.1038/nbt972. [DOI] [PubMed] [Google Scholar]
- 40.Borowitzka M. A. Microalgae for aquaculture: opportunities and constraints. J. Appl. Phycol. 1997;9:393–401. [Google Scholar]
- 41.Ackmanm R. G. Fatty acid composition in fish oils. In: Barlow S. M., Stanby E., editors. Nutritional Evaluation of Long-chain Fatty Acids in Fish Oil. London: Academic Press; 1982. pp. 25–88. [Google Scholar]