SUMMARY
The intestinal environment facilitates HIV-1 infection via mechanisms involving the gut-homing vitamin A-derived retinoic acid (RA), which transcriptionally reprograms CD4+ T cells for increased HIV-1 replication/outgrowth. Consistently, colon-infiltrating CD4+ T cells carry replication-competent viral reservoirs in people with HIV-1 (PWH) receiving antiretroviral therapy (ART). Intriguingly, integrative infection in colon macrophages, a pool replenished by monocytes, represents a rare event in ART-treated PWH, thus questioning the effect of RA on macrophages. Here, we demonstrate that RA enhances R5 but not X4 HIV-1 replication in monocyte-derived macrophages (MDMs). RNA sequencing, gene set variation analysis, and HIV interactor NCBI database interrogation reveal RA-mediated transcriptional reprogramming associated with metabolic/inflammatory processes and HIV-1 resistance/dependency factors. Functional validations uncover post-entry mechanisms of RA action including SAMHD1-modulated reverse transcription and CDK9/RNA polymerase II (RNAPII)-dependent transcription under the control of mammalian target of rapamycin (mTOR). These results support a model in which macrophages residing in the intestine of ART-untreated PWH contribute to viral replication/dissemination in an mTOR-sensitive manner.
Graphical abstract
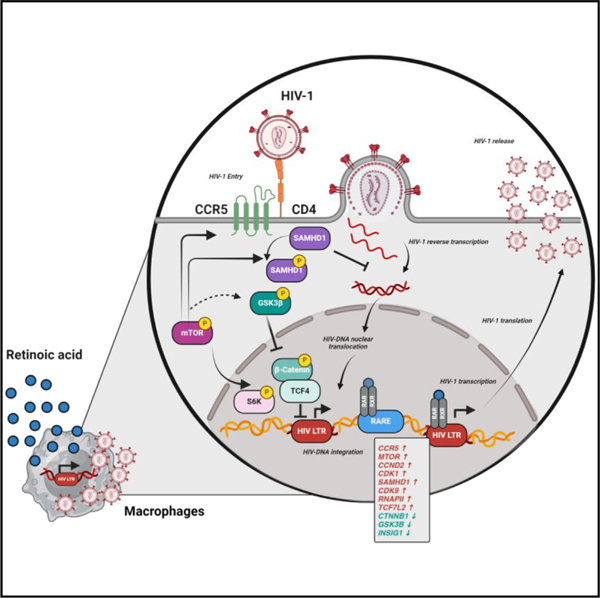
In brief
Dias et al. investigated the effects of retinoic acid (RA), a gut-homing elixir, on macrophages. RA boosts permissiveness to HIV-1 replication by facilitating multiple steps of the viral replication cycle. By analyzing the effect of RA on gene expression, the authors identify new mechanisms of RA action, namely SAMHD1/CDK9/RNAPII-dependent and mTOR-modulated mechanisms.
INTRODUCTION
Antiretroviral therapy (ART) controls HIV-1 replication to undetectable plasma levels but fails to eradicate HIV-1.1,2 The barrier to cure is the persistence of viral reservoirs (VRs) carrying integrated HIV-DNA during ART, thus leading to rapid viral rebound upon treatment interruption in people with HIV (PWH).1,2 The persistence of VRs is well-documented in long-lived memory CD4+ T cells.1,2 Importantly, other immune cells may represent sanctuaries for VRs during ART, such as macrophages residing in deep tissues, which remain poorly investigated due to restricted accessibility for biological samples.3–10
Pioneering studies in the pre-ART era documented HIV-1 infection in tissue macrophages of PWH.11,12 In line with this observation, macrophages express the HIV-1 receptor CD4 and co-receptors CCR5/CXCR4 and support productive HIV-1 infection in vitro.3,4 Other mechanisms by which macrophages become infected with HIV-1 include cell-to-cell transmission or phagocytosis of infected CD4+ T cells.3,8,13 Studies in ART-treated PWH provided evidence that macrophages isolated from liver, brain,8 bronchoalveolar lavage,14–16 duodenum,17 testis,18 and most recently urethra19 carry HIV-1.3 Similarly, studies performed in humanized mice models, especially in myeloid-only models,20 as well as non-human primate simian immunodeficiency virus (SIV)-infection models,21–23 pointed to macrophages as important sites of viral infection/persistence. Finally, by their intrinsic resistance to apoptosis and killing by CD8+ T cells and natural killer (NK) cell killing, HIV-infected macrophages escape from immunological pressure.24,25
The persistence of VRs in macrophages depends on their long-term survival potential and self-renewal capacity in ART-treated PWH, features that vary among macrophage subsets relative to their ontogeny. Tissue-resident macrophages (TRMs) from the brain, liver, lungs, and epidermis derive from embryonic/fetal precursors during intrauterine development and represent long-lived self-renewing TRM.26,27 In contrast, fractions of macrophages from the heart, pancreas, intestine, and dermis derive from bone marrow-derived monocytes and represent short-lived macrophages.26,27 Advances in the past decade demonstrated that long-lived TRM exist in all tissues and their role is mainly in tissue remodeling/homeostasis.26,27 This pool of self-renewing TRM progenitors suffer attrition with aging, as documented in the heart TRMs in mouse28 and human studies,29 thus allowing a niche for pro-inflammatory short-lived monocyte-derived macrophages (MDMs) to infiltrate.30 This evidence points to MDMs as key players in HIV-1 pathogenesis, especially in the context of an aging population of ART-treated PWH.
The intestine is an anatomic site containing the vast majority of immune cells preferentially targeted by HIV-1 for infection and VR persistence during ART.31–34 The intestinal environment is rich in the gut-homing ‘‘elixir’’ retinoic acid (RA), a metabolic derivative of vitamin A, produced by mucosal dendritic cells (DCs) expressing retinaldehyde dehydrogenase (RALDH) activity.35–38 RA binds to the RA receptor alpha (RARα), which forms a heterodimer with retinoid X receptor (RXR).36,38 The RARα/RXR heterodimer undergoes nuclear translocation and binds on RA-responsive elements (RAREs) on the promoter of specific genes,36,38 including the gut-homing integrin beta 7 (ITGB7).35 Importantly, the HIV long terminal repeat (LTR) also contains RAREs,39 suggesting a potential role of RA in HIV-1 transcription. We have previously demonstrated that RA transcriptionally reprograms CD4+ T cells for increased HIV-1 replication/outgrowth.32,40–42 Consistently, our group and others have demonstrated that colon-infiltrating CD4+ T cells carry VRs in PWH receiving ART.32–34,43 Intriguingly, our studies have revealed that integrative infection of colon macrophages, a pool constantly replenished by circulating monocytes,26,27 represents a rare event in ART-treated PWH.43 These findings raised questions on the effect of RA on HIV-1 permissiveness in macrophages from this anatomic site.
In the present study, we explored the effects of all-trans RA (ATRA) on HIV-1 replication in MDMs. Our results support a model in which macrophages in an environment rich in RA, such as the intestine, contribute to primary R5 HIV-1 infection before ART initiation, as well as viral rebound upon ART interruption. Our results also point to mammalian target of rapamycin (mTOR) as a therapeutic target to counteract the effects of ATRA on HIV-1 permissiveness in MDMs. Finally, our results suggest that the rarity of VR detection in colon-infiltrating macrophages of ART-treated PWH43 is likely not due to their resistance to HIV-1 infection but rather explained by the rapid turnover of the intestinal macrophages.26,27
RESULTS
ATRA increases CCR5-tropic HIV-1 replication in macrophages
To investigate the effects of RA on HIV-1 replication in macrophages, MDMs were obtained by culturing highly pure monocytes, isolated by negative selection from peripheral blood mononuclear cells (PBMCs) of HIV-uninfected participants, in the presence of macrophage colony stimulating factor (M-CSF) (Figures 1A and S1A–S1C). MDMs were generated in the presence (ATRA-MDMs) or absence (DMSO-MDMs) of ATRA. One documented action of ATRA is the upregulation of the gut-homing integrin β7 (ITGB7), as we have previously reported.41 In preliminary experiments, ATRA (10 nM) induced a statistically significant increase in ITGB7 expression in MDMs (Figures S2A–S2C). Also, ATRA decreased the expression of its receptor, indicative of a negative feedback, as demonstrated by RT-PCR quantification of RARα and RXR mRNA (Figures S2D and S2E). Similar effects were observed for MDMs generated in the presence or absence of M-CSF, indicative of M-CSF-independent effects of ATRA (Figures S2B–S2E). The optimal concentration of ATRA was identified in dose-response experiments (10, 100, and 1,000 nM), where HIVNL4.3BaL replication was measured by ELISA at days 3–15 post infection and cell viability was observed by flow cytometry (Figures S3A–S3C). Similar dose-response experiments were performed, where cell viability was measured using the lactate dehydrogenase (LDH) assay (Figure S4A) and HIVTHRO DNA integration by PCR (Figure S4B). The ATRA at 10 nM proved to be optimal in terms of boosting HIV-1 replication with no deleterious effects on cell viability (Figures 3 and 4). This concentration is within the range of physiological plasma RA levels (4–14 nM)44,45 and was previously demonstrated by our group to boost HIV-1 replication in CD4+ T cells.32,40–42
Figure 1. ATRA increases CCR5 expression and R5 HIV-1 replication in macrophages.
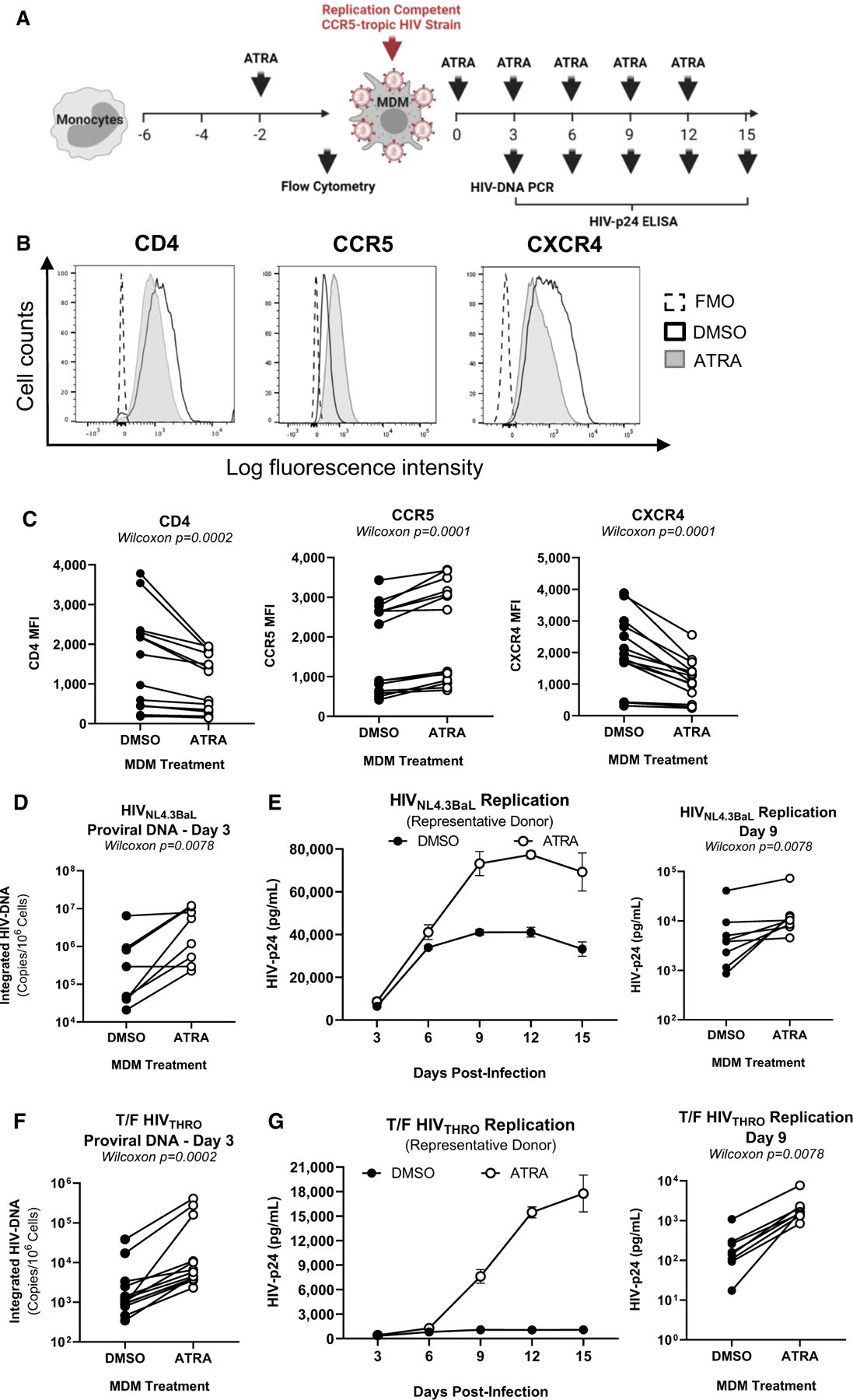
(A) The experimental flowchart. Briefly, monocyte-derived macrophages (MDMs) were obtained by culturing monocytes in medium containing M-CSF (20 ng/mL) for 6 days. MDMs were exposed (ATRA-MDMs) or not (DMSO-MDMs) to ATRA (10 nM) before and after HIV-1 exposure.
(B and C) Prior to HIV-1 exposure, MDMs were analyzed by flow cytometry upon staining with CD4, CCR5, and CXCR4 antibodies. Shown are histograms for CD4, CCR5, and CXCR4 expression on MDMs from one representative donor (B), and statistical analysis of CD4, CCR5, and CXCR4 MFI expression on MDMs from n = 14 participants. In parallel, MDMs were exposed to replication-competent CCR5-tropic HIV-1 strains (HIVNL4.3BaL; T/F HIVTHRO) and cultured in medium containing M-CSF in the presence/absence of ATRA for 15 additional days. Cell-culture supernatants were collected, and fresh medium containing M-CSF and/or ATRA was added every 3 days.
(D–G) MDMs exposed to HIVNL4.3BaL (D and E) and T/F HIVTHRO (F and G) were analyzed for HIV-DNA integration by PCR at day 3 post infection (D and F) and viral replication by HIV-p24 ELISA every 3 days up to 15 days post infection (E and G). Shown are the kinetics of HIV-1 replication in one representative donor (E and G, left panels) and statistical analysis performed at day 9 post infection for n = 8 participants (E and G, middle and right panels). Wilcoxon p values are on the graphs.
Figure 3. ATRA increases HIV-1 replication at post-entry levels before and after integration.
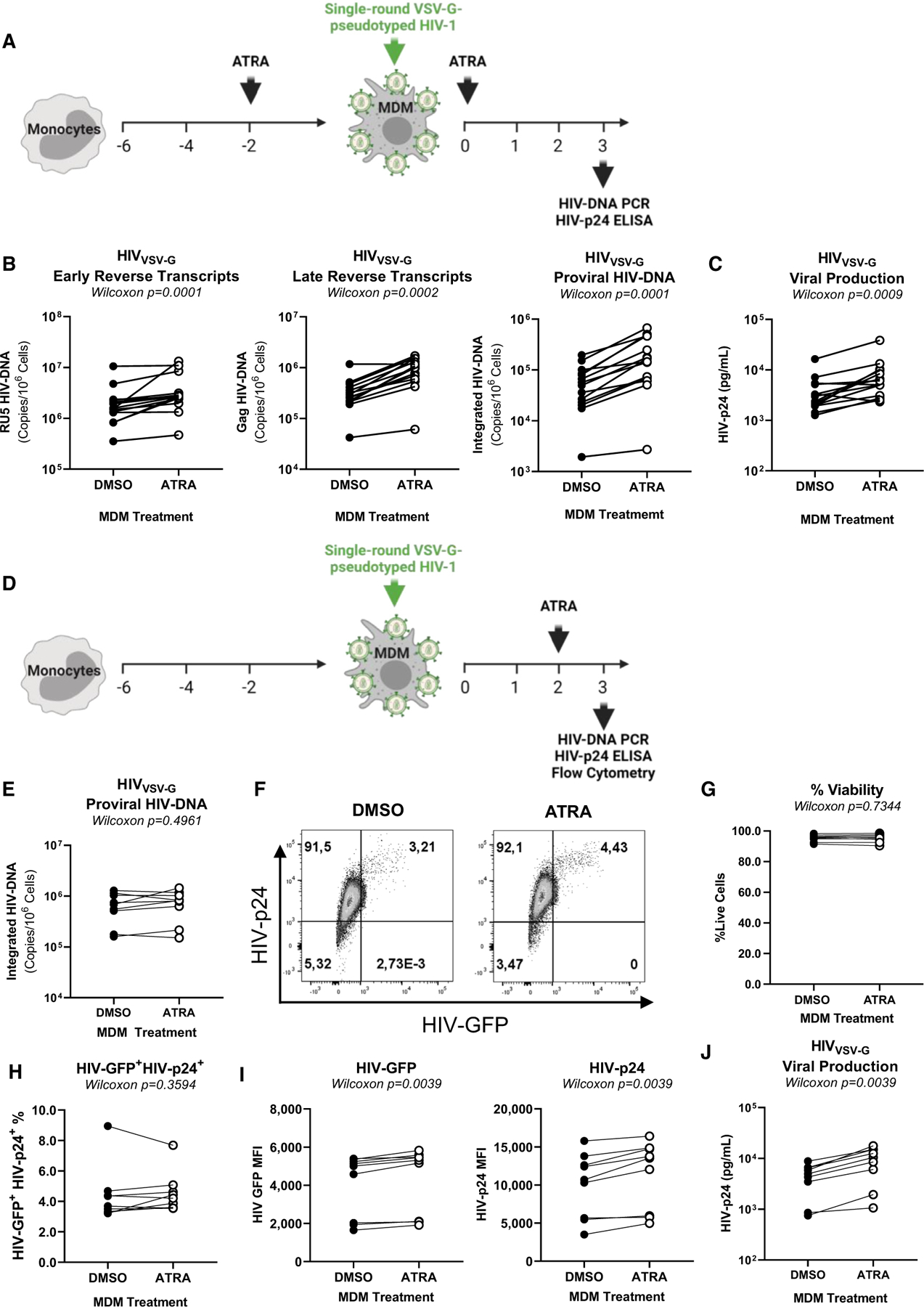
(A) Shown is the experimental flowchart, when MDMs were exposed to single-round VSV-G pseudotyped HIV-1 (HIVVSV-G) and treated with ATRA (10 nM) before and after infection (A–C). HIV-DNA integration and HIV-p24 levels were quantified at day 3 post infection. Shown are the levels of early reverse transcripts (RU5) (B, left panel), late reverse transcripts (Gag) (B, middle panel), and integrated HIV-DNA (B, right panel), as well as (C) HIV-p24 levels in cell-culture supernatants. (D) Shown is the experimental flowchart when MDMs were exposed to ATRA (10 nM) at day 2 post infection. At day 3 post -infection, cells were collected and analyzed for HIV-DNA integration by real-time nested PCR and for GFP and intracellular HIV-p24 expression by flow cytometry. Shown are statistical analysis of HIV-DNA integration (E), GFP, and HIV-p24 co-expression in MDMs from one representative donor (F), as well as statistical analysis of the frequency of viable (G) infected (GFP+HIV-p24+) MDMs (H), the GFP (I, left panel), and HIV-p24 (I, right panel) MFI expression from n = 9 participants. Finally, shown are statistical analysis of HIV-p24 levels in cell-culture supernatants measured by ELISA (J). Experiments were performed on MDMs from n = 14 (A–C) or n = 9 (E–J) HIV-uninfected participants. Wilcoxon p values are indicated on the graphs.
Figure 4. ATRA transcriptionally reprograms MDMs for increased expression of HIV permissiveness transcripts and pathways.
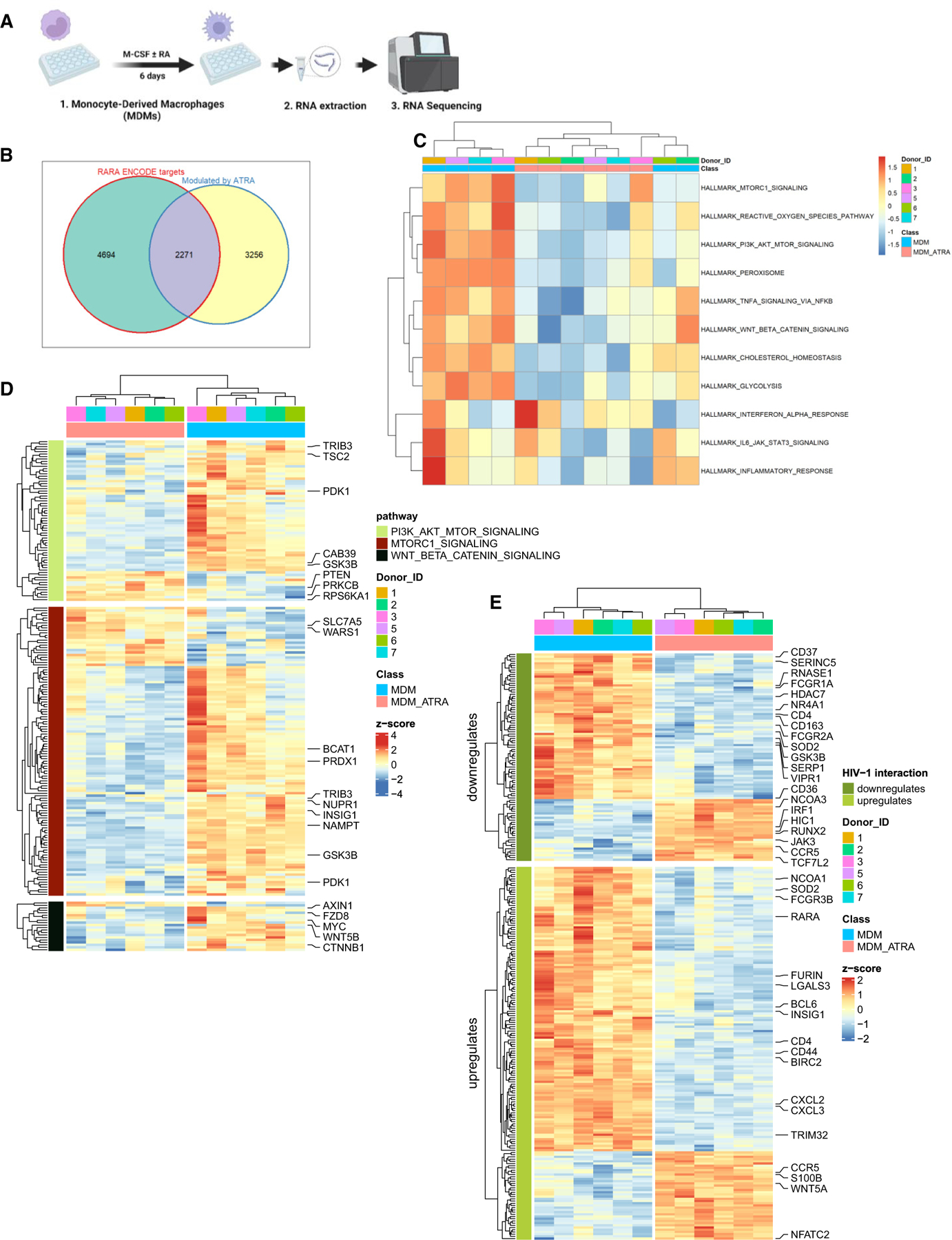
(A) Shown is the experimental flowchart. Briefly, RNA sequencing was performed on total RNA extracted from MDMs of n = 6 HIV-uninfected participants generated in the presence/absence of ATRA (10 nM) before infection.
(B) Differentially expressed genes (DEGs) were analyzed for the presence of RA-responsive elements (RAREs) in their promoters, using the ENCODE bioinformatic tool (https://www.encodeproject.org). This allowed the identification of n = 2,271 DEGs that may represent putative direct RA transcriptional targets in ATRA-treated MDMs.
(C and D) Further, gene set variation analysis (GSVA) was performed to identify signaling pathways modulated by ATRA in MDMs. Heatmaps depict top modulated signaling pathways in ATRA-treated MDMs (C), as well as gene sets (shown are selected transcripts) associated with three top modulated pathways: mTORC1, PI3K AKT mTOR, and Wnt/β-Catenin (D), as well as gene sets and selected transcripts modulated by ATRA in MDMs (p < 0.05; FC cutoff 1.3) matching the lists of genes included on the NCBI HIV interaction database (E). Heatmap cells are scaled by the expression level Z scores for each probe individually. Results from each donor are indicated with a different color code.
Prior to HIV-1 exposure, MDMs were analyzed by flow cytometry for the expression of the HIV-1 receptor CD4 and co-receptors CCR5/CXCR446 (Figure 1B). A significant increase in CCR5 and a decrease in CD4 and CXCR4 mean fluorescence intensity (MFI) expression were observed in ATRA-MDMs versus DMSO-MDMs, raising the possibility that ATRA facilitates CCR5-mediated HIV entry (Figure 1C). To test this possibility, MDMs were exposed to two replication-competent CCR5-tropic HIV-1 strains, HIVNL4.3BaL or transmitted founder (T/F) HIVTHRO. In the case of HIVNL4.3BaL, ATRA significantly increased HIV-DNA integration at day 3 post infection (p = 0.0078) (Figure 1D). Consistently, soluble HIV-p24 levels, indicative of replicative infection, were increased by ATRA, with statistically significant differences observed at day 9 post infection (p = 0.0078) (Figures 1E and S5). Similar results were observed with HIVTHRO, where integrated HIV-DNA levels at day 3 post infection (p = 0.0002) (Figure 1F), as well as soluble HIV-p24 levels at day 9 post infection (p = 0.0078), were significantly increased by ATRA (Figures 1G and S6).
Considering the ATRA-mediated CXCR4 decreases (Figure 1B), MDMs were also exposed to the replication-competent CXCR4-tropic HIVNDK. In contrast to R5 HIV-1, ATRA fails to affect HIVNDK integration/replication (Figures S7A and S7B). Together, these results demonstrate that ATRA renders MDMs highly permissive to productive CCR5 but not CXCR4-tropic HIV-1 infection, possibly as a consequence of a more efficient CCR5-mediated viral entry.
ATRA does not increase the efficiency of CD4/CCR5-mediated HIV-1 entry
To determine the effects of ATRA on CCR5-mediated viral entry, the β-lactamase (Blam) Vpr (Blam-Vpr) assay was performed using the ADA-Env-pseudotyped NL4.3Env−Vpr−/ADA-Env/Blam-Vpr HIV-1 (HIVBlam-ADA) and vesicular stomatitis virus-G (VSV-G)-pseudotyped NL4.3Env−Vpr−/VSV-G/Blam-Vpr HIV-1 (HIVBlam-VSV-G). While HIVBlam-ADA uses CCR5 for entry, the HIVBlam-VSV-G enters cells via the low-density lipoprotein receptor (LDLR).41 HIV-1 entry, reflected by CCF2 cleavage, was measured by flow cytometry (Figure 2A). Maraviroc (MVC) was used to block CCR5-mediated HIV-1 entry. ATRA did not affect the frequency of cleaved CCF2 upon infection with HIVBlam-ADA (Figure 2B), but a statistically significant decrease was observed for MDMs infected with HIVBlam-VSV-G (p = 0.0139) (Figure 2C), raising questions on the effect of ATRA on LDLR expression. As expected, MVC strongly inhibited HIVBlam-ADA entry but had no effect on HIVBlam-VSV-G (Figures 2B and 2C). These results demonstrate that ATRA has no impact on the efficacy of CCR5-mediated HIV-1 entry, indicative that ATRA increases HIV-1 permissiveness in MDMs via post-entry levels.
Figure 2. HIV-1 entry assay in ATRA-treated macrophages.
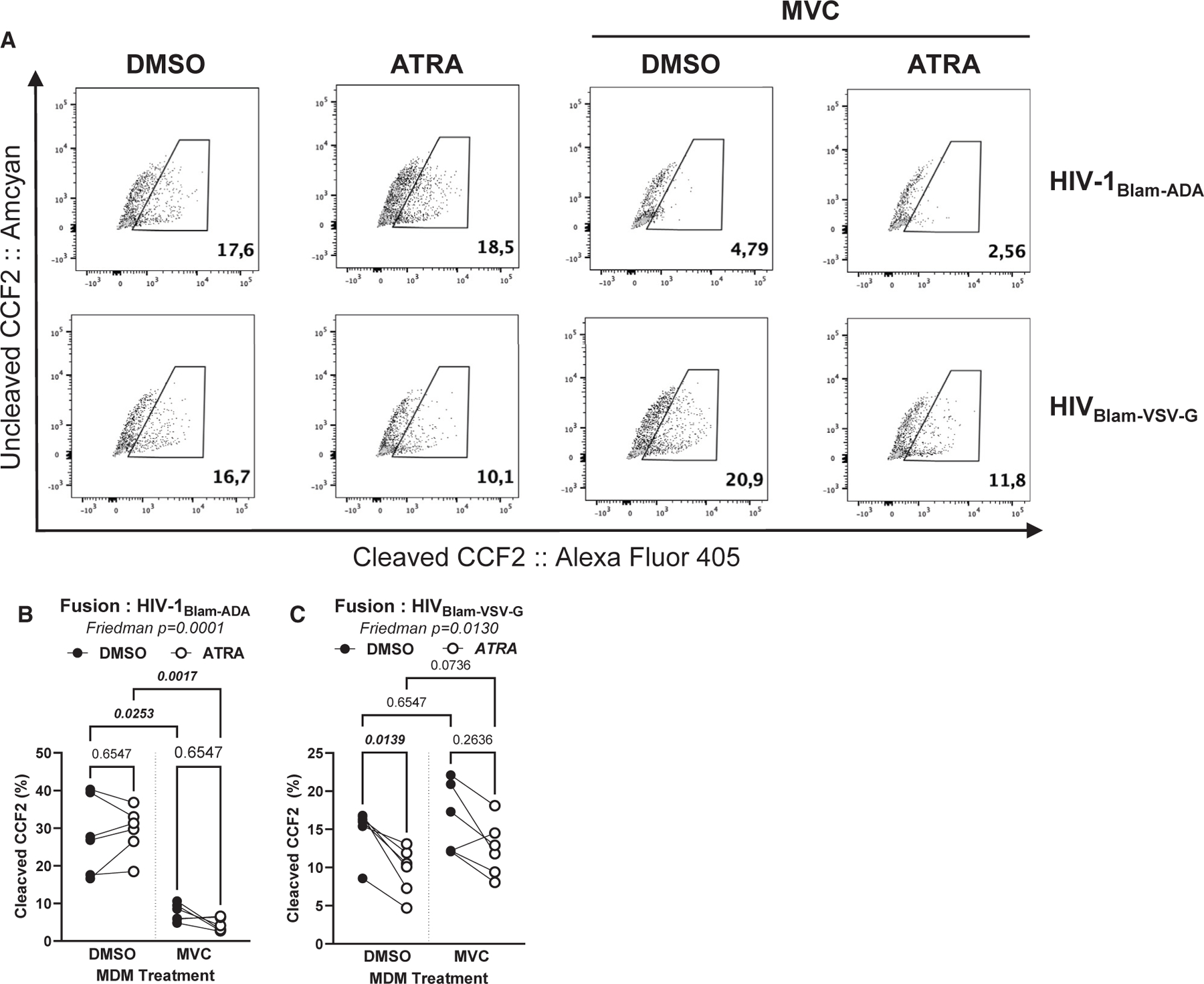
The Blam-Vpr HIV-1 entry assay was performed in DMSO-MDMs and ATRA-MDMs, using the single-round CCR5-tropic ADA-Env HIV-1 (NL4.3Env−Vpr−/ADA-Env/Blam-Vpr; HIVBlam-ADA) or VSV-G-pseudotyped HIV-1 (NL4.3Env−Vpr−/VSV-G-Env/Blam-Vpr; HIVBlam-VSV-G) viruses containing the β-lactamase (Blam)-Vpr protein chimera. MDMs were loaded with CCF2-AM and analyzed by flow cytometry for the change in CCF2 fluorescence from green (520 nm; AmCyan channel; uncleaved) to blue (447 nm; Alexa Fluor 405 channel; cleaved) upon Blam-mediated cleavage. Shown are representative dot plots for cleaved and uncleaved CCF2-AM in DMSO-MDMs versus ATRA-MDMs pretreated or not with MVC (A). Statistical analyses of HIV entry in MDMs exposed to HIVBlam-ADA (B) or HIVBlam-VSV-G (C). Friedman and uncorrected Dunn’s p values are on the graphs.
ATRA increases HIV-1 permissiveness in MDMs at post-entry and post-integration levels
To identify the post-entry steps of the viral replication cycle modulated by ATRA, MDMs were exposed to single-round VSV-G-pseudotyped HIV-1 (HIVVSV-G), a viral construct capable of entering cells via the LDLR.41 In the first set of experiments, MDMs were treated with ATRA before HIVVSV-G exposure (Figure 3A). At day 3 post infection, levels of early (RU5) and late (Gag) reverse transcripts, and integrated HIV-DNA levels (proviral HIV-DNA), were significantly increased by ATRA (Figure 3B), indicative of a higher efficacy in viral reverse transcription and/or integration. Consistently, ATRA significantly increased soluble HIV-p24 levels, indicative of efficient viral release (n = 14; p = 0.0009) (Figure 3C). These results indicate that ATRA acts on the early post-entry steps of the HIV replication cycle, between reverse transcription and integration, thereby leading to efficient subsequent viral production.
Considering the presence of RARE in the HIV-LTR,39 we aimed to study the effects of ATRA in MDMs at post-integration level. In a second set of experiments, MDMs were treated with ATRA at day 2 post infection (Figure 3D), a time when HIV integration is maximal.47 As expected under these conditions, there were no differences in HIV-DNA integration (Figure 3E) or in the viability and the frequency of productively infected GFP+HIV-p24+ cells (Figures 3F–3H) in ATRA-MDMs versus DMSO-MDMs. However, exposure to ATRA post integration significantly increased the MFI for GFP and HIV-p24 expression (p = 0.0039) (Figure 3I), as well as viral release (p = 0.0039) (Figure 3J), indicative of an efficient translation in the presence of ATRA.
ATRA transcriptionally reprograms MDMs for increased HIV-1 permissiveness
To gain insights into molecular mechanisms underlying the effects of ATRA on HIV-1 replication in MDMs, genome-wide RNA sequencing was performed prior to HIV-1 exposure (Figure 4A). Differentially expressed genes (DEGs) were identified based on p values (<0.05), adjusted p values (<0.05), and fold-change (FC, cutoff of 1.3) (Figure S8A), with 1,772 and 2,047 transcripts identified as being upregulated and downregulated, respectively, in ATRA-MDMs versus DMSO-MDMs (Data S1 and S2).
Among the top 50 modulated genes, HIVEP2 (an enhancer of HIV transcription48), IRF1 (a facilitator of HIV replication49), TTC7A (a regulator of HIV transcription50), PHOSPHO1 (a regulator of energy metabolism51), and RUNX2 (a modulator of macrophage differentiation52 and a negative regulator CXCR4 expression53) were upregulated, while HIVEP1 (a negative regulator of nuclear factor κB [NF-κB] activation54) and SLC11A1 (encoding for natural resistance-associated macrophage protein 1, with polymorphism associated with HIV mortality55) were downregulated by ATRA in MDMs (Figure S4B). These transcriptional changes may explain the increased permissiveness to productive R5 HIV-1 replication in ATRA-MDMs with increased viral transcription.
Identification of direct RA target genes
Among DEGs modulated by ATRA in MDMs, 2,271 transcripts expressed RARE in their promoters, as identified using the in silico bioinformatics tool ENCODE search (https://www.encodeproject.org) (Figure 4B; Data S3), indicative that ATRA directly regulates the expression of HIV permissiveness factors in MDMs.
Gene set variation analysis
To extract further meaning from these transcriptional changes, gene set variation analysis (GSVA) (C2, C3, C5, C7, C8, and Hallmark databases; https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) was performed. Top modulated Hallmark pathways are depicted in Figure 4C. Components and regulators of the mTORC1 signaling, PI3K/Akt/mTOR signaling, and Wnt/β-catenin signaling are illustrated in Figure 4D and detailed here below.
mTORC1 signaling
Among transcripts linked to the mTOR pathway, the expression of amino acid transporters such as SLC7A5 and tryptophanyl-tRNA synthetases 1 such as WARS1 are upregulated, suggesting an increase in protein synthesis and metabolic activity (Figure 4D). Moreover, the expression of certain negative regulators of mTOR (e.g., BCAT1, TRIB3, NUPR1, NAMPT, and PRDX1) was downregulated by ATRA in MDMs (Figure 4D). Furthermore, the expression of the insulin-induced gene 1 (INSIG1), a regulator of lipid metabolism and an HIV-1 restriction factor involved in Gag degradation,56 was downregulated by ATRA in MDMs (Figure 4D). Overall, these results are indicative of an mTOR-dependent ATRA-mediated reprogramming of MDMs’ metabolic activity for increased HIV-1 permissiveness.
PIK3-AKT-mTOR signaling
The engagement of the mTOR pathway requires the activation of several kinases such as phosphoinositide 3 kinase (PIK3) and protein kinase B (AKT).57 The class IA PI3K consists of heterodimers of the p110 catalytic (PIK3CA, PI3KCB, or PI3KCD) subunit and p85 regulatory subunit (PIK3R1, PIK3R2, and PIK3R3). Results in Figure 4D depict that the expression of the p85 regulatory sub-units (PI3KR3) was upregulated, while the expression of AKT, encoding for a kinase upstream of mTOR, was downregulated by ATRA in MDMs. The latter coincided with the increased expression of phosphatase and tensin homolog (PTEN) (Figure 4D), a negative regulator of PI3K and AKT.57 The expression of tuberous sclerosis 2 (TSC2), a negative regulator of mTOR,57 was downregulated, while the expression of kinases downstream of mTORC1, such as the ribosomal protein S6 kinase 1 (RPS6KA1),57 was upregulated (Figure 4D), supporting the activation of mTORC1 pathway by ATRA in MDMs. Importantly, S6K has been reported to inhibit the PI3K-AKT-mTOR pathway through a negative feedback loop.57 The latter is consistent with the higher expression of PTEN and lower expression of AKT genes we observed in ATRA-versus DMSO-treated MDMs (Figure 4D). Moreover, the expression of the protein kinase C (PRKCB), another kinase downstream mTORC2,57 was upregulated in ATRA-MDMs versus DMSO-MDMs (Figure 4D), suggesting that the activation of mTORC2 occurs subsequent to AKT activation. Furthermore, the expression of CAB39, responsible for activating AMPK, a kinase that inhibits mTOR activation through sensing of cellular energy (AMP:ATP ratio),57 was also downregulated in ATRA-MDMs compared to DMSO-MDMs (Figure 4D). Overall, these results are indicative of ATRA activating the PI3K-AKT-mTOR pathway in MDMs, thus facilitating HIV-1 replication.
Wnt/β-catenin
The Wnt/β-catenin pathway controls key cellular functions and negatively regulates HIV-1 transcription.58 Results in Figure 4D depict the downregulation of FZD8 (Wnt receptor), AXIN1 (a component of the β-catenin/APC/GSK3β complex), CTNNB1 (β-catenin), and WNT.5B transcripts. Other components/regulators of the Wnt/β-catenin pathway are listed among DEGs (Data S1 and S2). Components of the canonical Wnt/β-catenin pathway were downregulated (i.e., MYC, PDK1, MMP) (Data S2), suggesting the inactivation of this pathway.59 Furthermore, transcripts linked to the non-canonical Wnt/β-catenin pathway were upregulated (i.e., PLC, PRKCB, NFATC2) in ATRA-MDMs versus DMSO-MDMs (Figure 4D; Data S1), pointing to the activation of the non-canonical Wnt/β-catenin pathway. The expression of TCF4/TCF7L2, a gene encoding for the transcription factor TCF4 that inhibits HIV transcription through its binding on the 5′ HIV-LTR promoter upon association with β-catenin,60 was upregulated (Data S1) and transcripts for GSK3B, a serine-threonine kinase that mediates the proteolytic degradation of β-catenin,59 were downregulated by ATRA in MDMs (Figure 4D). While the ATRA-mediated upregulation of TCF4/TCF7L2 and the downregulation of GSK3B transcripts remain intriguing, the downregulation of β-catenin expression is consistent with high levels of HIV-1 replication in ATRA-MDMs.
These results point to profound transcriptional reprogramming mediated by ATRA in MDMs and reveal two druggable pathways, the mTOR and Wnt/beta-catenin pathways, that may play a key role in this process.
ATRA modulates the expression of HIV interactors in MDMs
The NCBI HIV interactor database was interrogated for known HIV permissiveness/restriction factors modulated by ATRA in MDMs (Figure 4E). First, ATRA increased CCR5 and decreased CD4 mRNA expression in MDMs (Figure 4E), consistent with the flow cytometry results in Figures 1B and 1C. Among transcripts known to increase HIV replication, ATRA increased the expression of the transcription factor NFATC2 and coactivator NCOA3 (Figure 4E). In contrast, ATRA decreased expression of BIRC2 (suppressor of HIV transcription),61 SERINC5 (restriction factor for HIV-1 release),62 RNASE1 (inhibitor of HIV-1 production), HDAC7 (suppressor of HIV transcription), and GSK3B (an inducer of zinc-finger antiviral protein [ZAP] functions)63 (Figure 4E). Unexpectedly, genes encoding positive regulators of HIV transcription (TRIM32 and NCOA164) and virion production/release (LGALS3 and FURIN65) were upregulated by ATRA. Furthermore, HIC1 and TCF4/TCF7L2 (Figure 4E), two negative regulators of HIV transcription,66,67 were upregulated by ATRA in MDMs. RT-PCR validations confirmed the increased expression of HIC1 mRNA in ATRA-exposed MDMs (Figure S9A). In contrast, the expression of PPARG, an HIV-1 transcriptional repressor.68 was downregulated in MDMs upon exposure to ATRA (Data S2), as validated by RT-PCR quantification (Figure S9B).
Together, these results reveal a panel of HIV-1 restriction and dependency factors, with a fraction of them carrying putative RARE in their promoters (e.g., SAMHD1, CTNB1, GSK3B, SLC7A5, WARS1, TRIB3, NUPR1, NAMPT, PRDX1, AKT1, PTEN, HIVEP2, HIVEP1, and SERINC5) (Data S3). Some of these transcripts are linked to the mTOR and Wnt/β-catenin pathways. Genes upregulated by ATRA (Figure S1) expressing RARE in their promoters and reported to contribute to HIV-1 replication were identified using another in silico search method on www.genecards.org and included CCR5, MTOR, RPS6KB1, TCF7L2, and INSIG1.
ATRA increases HIV-1 permissiveness in MDMs via mTOR-dependent mechanisms
To explore the role of the mTOR pathway in modulating HIV-1 permissiveness in ATRA-treated MDMs (Figure 4D), the expression of total and phosphorylated mTOR was visualized by western blotting. Results in Figures S10A–S10C demonstrated a similar expression of total and phosphorylated mTOR in ATRA-MDMs and DMSO-MDMs. Exposure to INK128, a documented mTOR inhibitor,41 significantly decreased phosphorylated mTOR expression in DMSO-MDMs but not ATRA-MDMs (Figures S10A–S10C). Nevertheless, the expression of total and phosphorylated S6K, a kinase downstream mTOR indicative of mTOR activity,57 was significantly increased in ATRA-MDMs versus DMSO-MDMs and INK128 strongly reduced phosphorylated S6K expression in ATRA-MDMs (Figures S11A–S11C). This prompted us to explore the contribution of mTOR activation to HIV-1 permissiveness in ATRA-MDMs. The expression of CD4 and CCR5 was significantly reduced on both DMSO-MDMs and ATRA-MDMs, while the CXCR4 expression was increased in DMSO-MDMs upon exposure to INK128 (Figures 5A and 5B). Similarly, exposure to INK128 significantly decreased HIVTHRO integration and replication, mainly in ATRA-MDMs (Figures 5C and 5D). These results demonstrate the capacity of INK128 to act on the mTOR-S6K pathway in ATRA-MDMs and limit CCR5-tropic HIV-1 replication, in part, by decreasing CD4 and CCR5 expression.
Figure 5. mTOR inhibition counteracts the effect of ATRA on CCR5 expression and HIV replication and integration.
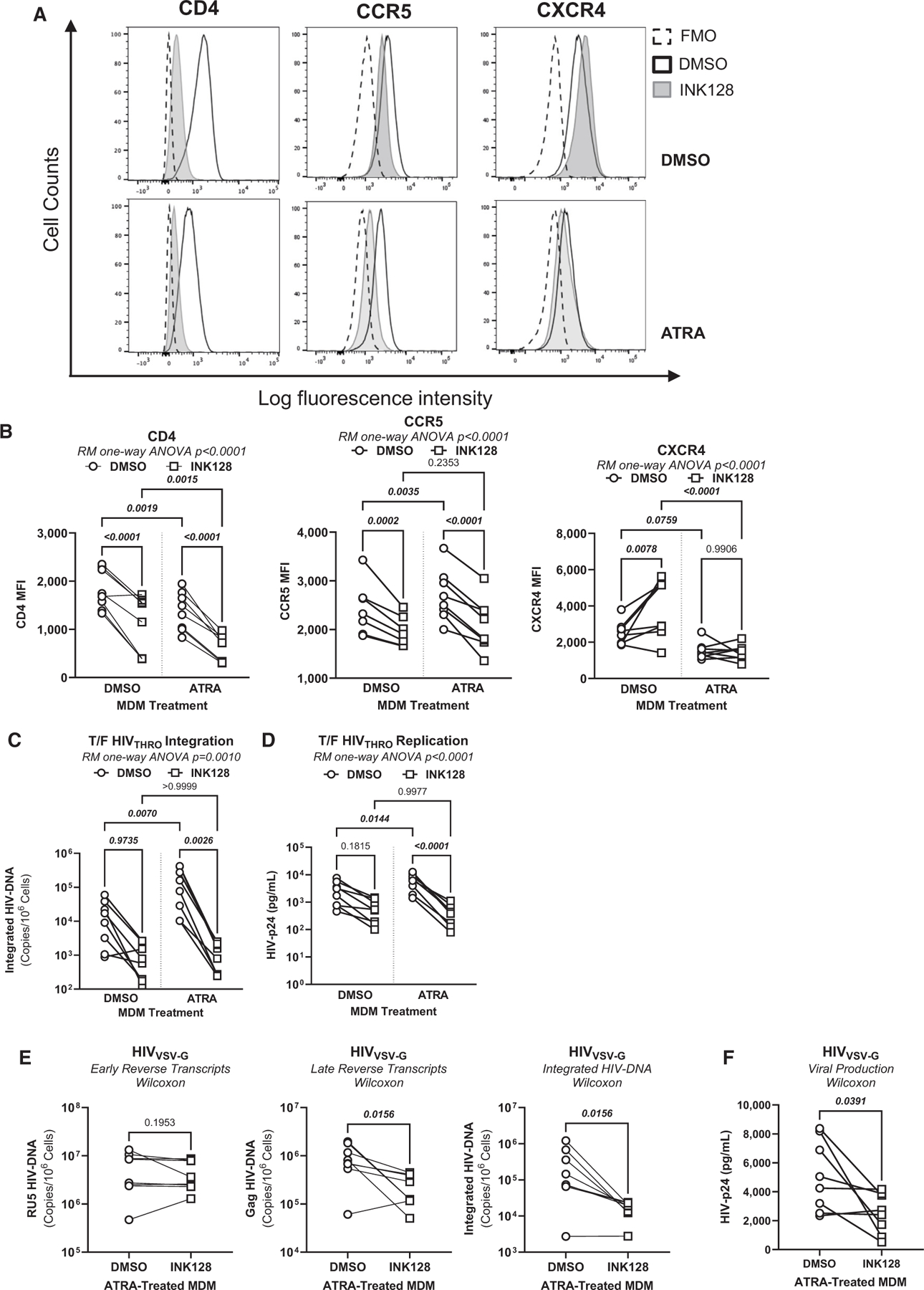
MDMs generated as in Figure 1A were treated with the mTOR inhibitor INK128 (50 nM) 2 days before infection and the day of infection.
(A) The representative flow cytometry histograms of extracellular CD4, CCR5, and CXCR4 expressions.
(B) Statistical analyses of the relative MFI of CD4, CCR5, and CXCR4 expressions.
(C and D) A fraction of MDMs, pretreated or not with INK128, were exposed to T/F HIVTHRO. Cells and cell-culture supernatants were harvested on day 3 post infection for the quantification of integrated HIV-DNA by nested real-time PCR (C) and soluble HIV-p24 by ELISA (D).
(E and F) In parallel, another fraction of ATRA-MDMs and DMSO-MDMs pretreated with INK128 were exposed to HIVVSV-G for 3 days. Shown are the levels of early reverse transcripts (RU5; E, left panel), late reverse transcripts (Gag; E, center panel), integrated HIV-DNA (Alu; E, right panel), as well as the levels of HIV-p24 (F). Experiments were performed on MDMs from n = 8 HIV-uninfected individuals. Repeated measures (RM) one-way ANOVA, Tukey’s test, and Wilcoxon p values are indicated on the graphs.
To identify the viral replication steps targeted by mTOR in ATRA-MDMs, a single-round infection was performed with HIVVSV-G. It is noteworthy that INK128 significantly decreased late reverse transcripts (Gag) and proviral HIV-DNA, but not early reverse transcripts (RU5), indicative of mTOR-dependent mechanisms affecting the completion of reverse transcription and subsequently integration but not the initiation of reverse transcription (Figure 5E). Consistently, results in Figure 5F demonstrated that INK128 also reduced virion release in ATRA-treated MDMs in this single-round model of infection. Together, these results demonstrate that INK128 can counteract the effects of ATRA on CCR5 expression, HIV reverse transcription and integration, and virion release, indicative that ATRA promotes HIV-1 permissiveness in MDMs via mTOR-dependent mechanisms.
ATRA modulates SAMHD1 phosphorylation via mTOR-dependent mechanisms
The mTOR pathway interferes with HIV-1 replication at multiple steps of the viral replication cycle, including reverse transcription.69 One restriction factor originally identified for its capacity to restrict HIV-1 replication in myeloid cells by interfering with the completion of reverse transcription is SAMHD1.70 Interestingly, SAMHD1 mRNA was upregulated by ATRA, in line with the presence of RARE in its promoter, as identified by in silico search using ENCODE (Data S1 and S3). The potential link between mTOR and SAMHD1 activity is also supported by the upregulated expression of CCND2 mRNA (Data S1; Figure 6A) encoding for cyclin D2, a modulator of cell-cycle and SAMHD1 activity.71,72 Indeed, phosphorylation via cyclin-dependent kinases (CDK)1/2 was reported to reduce SAMHD1 capacity to restrict HIV in macrophages.72 Indeed, ATRA significantly increased the expression of CDK1 mRNA (Wilcoxon p = 0.0078) in MDMs (Figure 6B). When ATRA-MDMs were pretreated with INK128, the expression of CCND2 and CDK1 significantly decreased (Figures 6A and 6B). Furthermore, the expression of SAMHD1 mRNA significantly increased (p = 0.0010) upon exposure to ATRA (Figure 6C). In line with our prediction, results in Figures 6D–6F demonstrate increased phosphorylated and total SAMHD1 protein expression in MDMs upon exposure to ATRA. In contrast, exposure to INK128 led to a significant decrease in SAMHD1 phosphorylation, mainly in ATRA-MDMs (Figures 6D–6F), in line with the antiviral activity of INK128 (Figures 5C–5F). These results provide evidence that SAMHD1 mediates HIV restriction in MDMs via mTOR-dependent mechanisms.
Figure 6. ATRA modulates SAMHD1 phosphorylation in a mTOR-dependent manner.
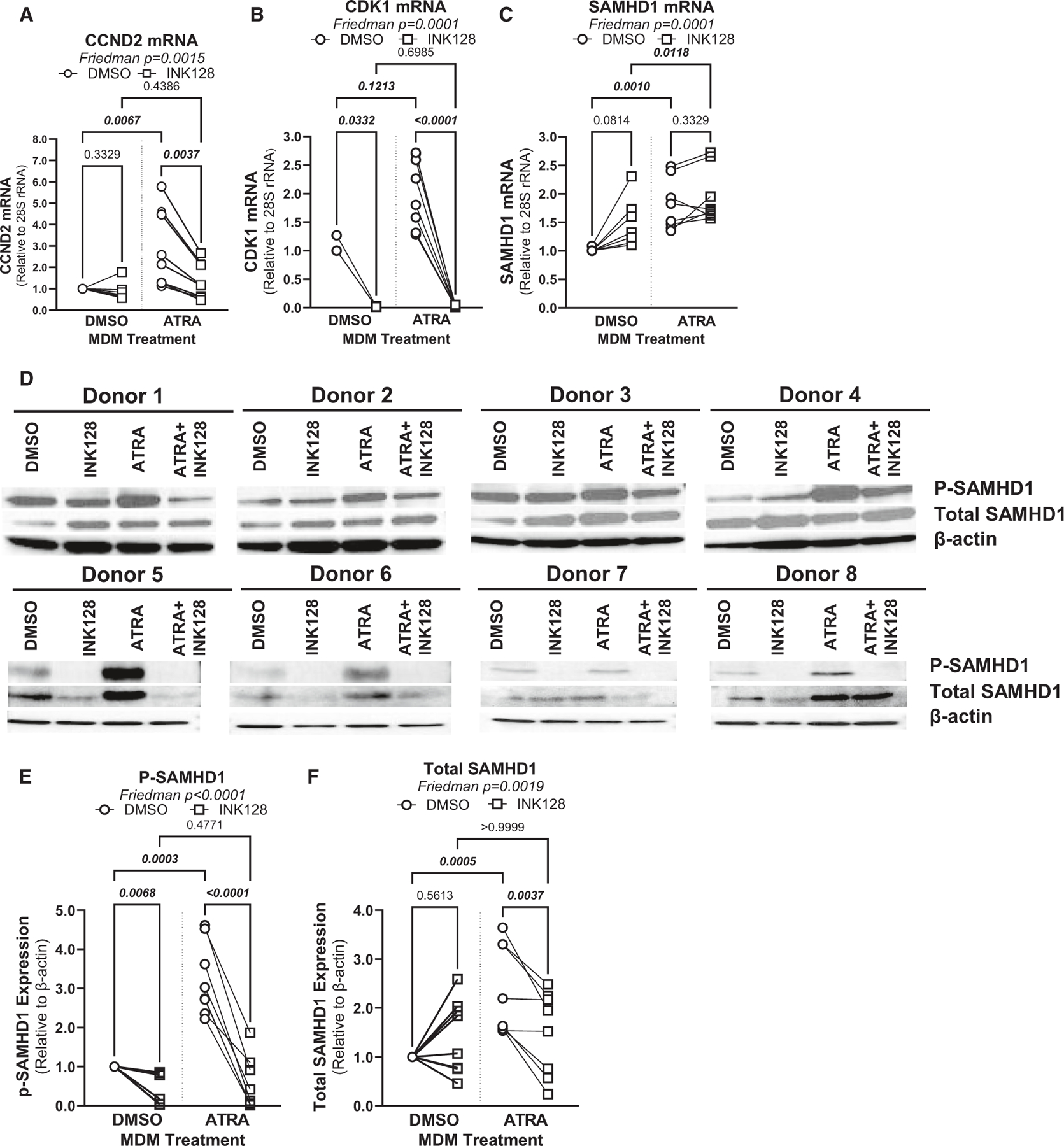
(A–D) MDMs were generated in the presence/absence of ATRA (10 nM) and pretreated or not with INK128 (50 nM), as in Figure 5. Prior to HIV-1 infection, cells were harvested for RT-PCR (A–C) and western blotting investigations (D–F). Shown are mRNA levels of Cyclin D2 (CCND2) (A), CDK1 (B), and SAMHD1 expressions (C), as well as levels of total (molecular weight [MW], 72 kDa) and phosphorylated (MW, 72 kDa) SAMHD1 protein, relative to β-actin (MW, 42 kDa) expression (D) in MDMs from eight different HIV-uninfected donors.
(E and F) Graphs depict total (E) and phosphorylated SAMHD1 protein levels (F) normalized to β-actin expression. Experiments were performed on MDMs from n = 8 HIV-uninfected individuals. Friedman and uncorrected Dunn’s test p values are indicated on the graphs.
The β-catenin/TCF4 pathway limits excessive HIV-1 replication in ATRA-MDMs
RNA sequencing revealed a decreased expression of CTNNB1 (encoding β-catenin) and GSK3B, and an increased expression of TCF7L2 (encoding TCF7L2/TCF4) in ATRA-MDMs versus DMSO-MDMs (Data S1 and S2). Here, RT-PCR and western blotting were used to validate upregulated TCF4 expression (Figures S12A–S12C) as well as the downregulation of CTNNB1/β-catenin (Figures S13A–S13C) and GSK3β (Figures S13D–S13F) at the mRNA and protein level in ATRA-MDMs versus DMSO-MDMs. Then, to explore the role of β-catenin/TCF4 in regulating HIVTHRO replication in ATRA-treated MDMs, ATRA-MDMs were exposed to PRI-724, a potent inhibitor that disrupts the interaction between β-catenin and CBP,73 and PNU-74654, a potent inhibitor that disrupts the interaction between β-catenin and TCF4.74 In preliminary experiments, the optimal concentrations of PNU-724 (0.1 μM) or PNU-74654 (5 μM) were selected based on preliminary dose-response experiments measuring an effect on HIV-1 replication without changes in cell viability. Levels of HIV-DNA integration were further significantly increased in ATRA-MDMs when HIVTHRO infection was performed in the presence of PRI-724 (Figure S13G) or PNU-74654 (Figure S13H). Together, results point to β-catenin/TCF4 as important negative regulators of HIV-1 replication in ATRA-MDMs.
ATRA upregulates HIV-1 transcription via CDK9-dependent and mTOR-modulated mechanisms
Multiple transcripts modulated by ATRA in MDMs point to a direct effect on HIV-1 transcription (Figure 4). To explore this possibility, chromatin immunoprecipitation followed by quantitative PCR assays (ChIP-qPCR) were performed using the monocytic cell line THP1 productively infected with a full-length HIV-1 NL4.3 strain. Importantly, our results showed the recruitment of RARα to the HIV-1 promoter following stimulation with ATRA, together with an increased recruitment of CDK9, as well as of the RNA polymerase II (RNAPII) phosphorylated on serine 2 and 5, thereby demonstrating a direct positive effect of ATRA on HIV-1 transcription (Figure 7A). Consistent with a previous report,75 the recruitment of CDK9 was dependent on mTOR, as demonstrated in ATRA-treated THP-1 cells exposed to the mTOR inhibitor INK128 (Figure 7A). These observations were in line with a robust increase in HIV-1 transcription promoted by ATRA and further inhibited by INK128 (Figure 7B). Together, our results demonstrate the regulation by ATRA of HIV-1 transcription via the direct binding of the receptor RARα to the HIV 5′LTR promoter and the subsequent recruitment of CDK9 and RNAPII.
Figure 7. ATRA promotes HIV-1 transcription via RARα/CDK9/RNAPII-dependent and/or mTOR-dependent mechanisms.
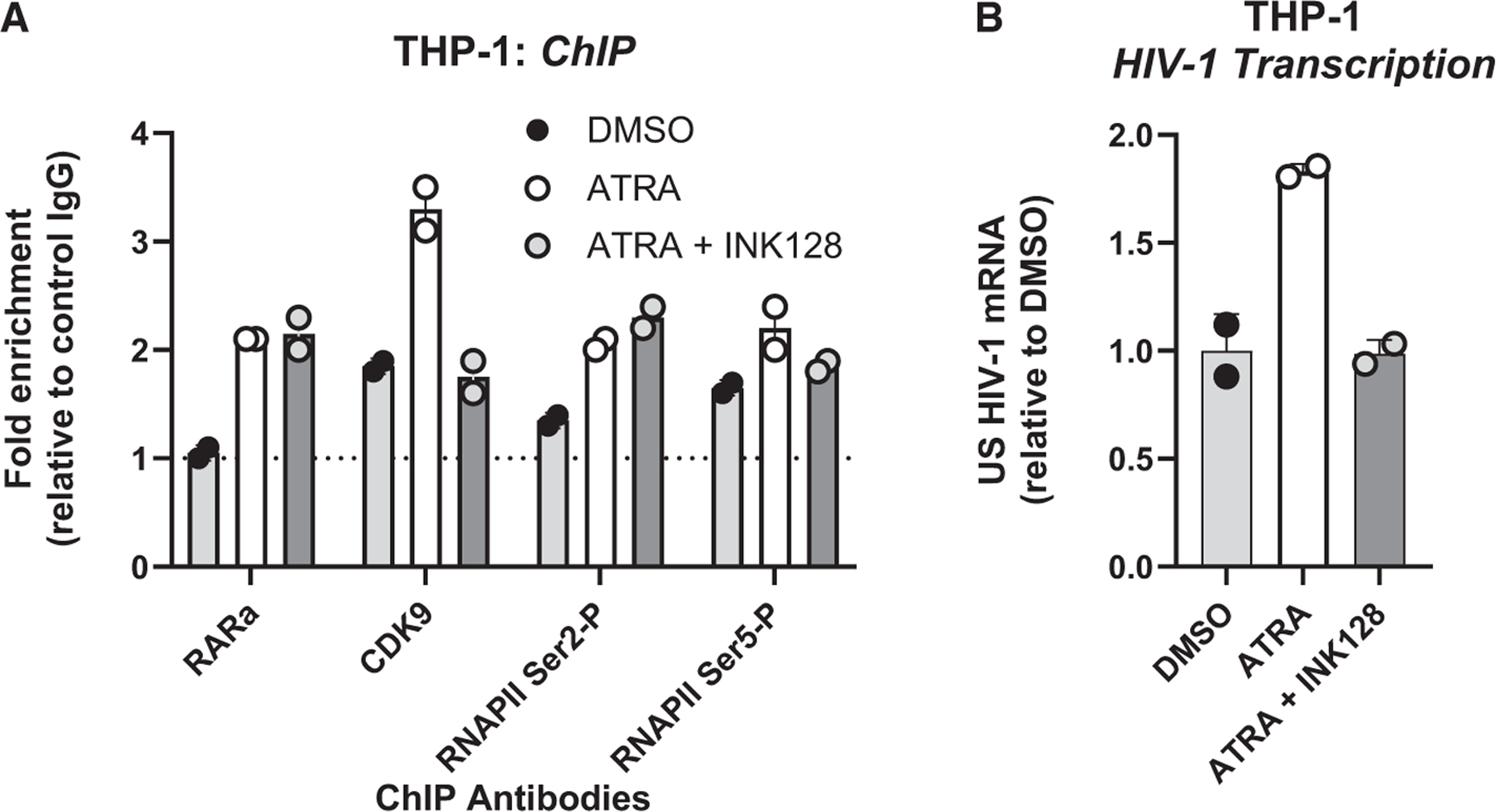
(A) The THP1 monocytic cell line productively infected with a full-length NL4.3 HIV-1 was cultured in the presence/absence of INK128 (50 nM) 1 day before treatment with ATRA (10 nM) for 24 h. Chromatin samples were immunoprecipitated with RARα, CDK9, the phosphorylated RNA polymerase II (RNAPII) on its serine 2 or 5 antibodies, or with rabbit immunoglobulin (Ig) G as negative control. qPCRs were performed with primers amplifying specifically the 5′ LTR HIV-1, in the Nuc-1 region. Fold increases relative to IgG are presented, where fold enrichments for each immunoprecipitated DNA were calculated by the relative standard curve on input DNA. Values represent means of duplicate samples ± SD. One representative experiment out of three is presented.
(B) In parallel, total RNA preparations from THP-1 cells were reverse transcribed. Unspliced HIV-1 RNAs (unspliced transcripts) were quantified by RT-qPCR using GAPDH and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ) as first normalizer and the DMSO-treated condition as second normalizer. Shown are results representative of two different experiments on THP-1 cells. Means from duplicate ± SD are indicated.
DISCUSSION
In this study, we report that ATRA transcriptionally reprograms MDMs for increased permissiveness to HIV-1 replication. Our multiomic approaches elucidated that ATRA acts at post-entry levels, via mechanisms dependent on mTOR, a pathway that regulates the efficacy of reverse transcription via the phosphorylation of SAMHD1, as well as HIV-1 transcription via the recruitment of CDK9 to the HIV-LTR in MDMs. These results support a model in which HIV-1 replicates efficiently in macrophages residing in anatomic sites rich in RA and point to mTOR inhibitors as important regulators of HIV-1 permissiveness in macrophages. These findings are particularly important since our group and others previously reported that mTOR controls HIV replication in CD4+ T cells,76,77 and drugs that are active on both macrophages and CD4+ T cells should receive priority for testing in HIV-1 remission/cure strategies.
Macrophages are major players in host-pathogen interactions. Studies on factors regulating HIV-1 replication in MDMs become highly relevant in the context of premature aging reported in ART-treated PWH,78,79 a process associated with the replacement of long-lived TRMs by short-lived MDMs. Pioneering studies by Poli et al. reported on the proviral effects of RA in macrophages.80 One major finding of our study is that the proviral effects of ATRA in MDMs were mediated by mTOR, a serine/threonine protein kinase that adapts cell metabolism in response to changes in the environment, such as nutrients and growth hormones.57 The current findings are in line with our previous reports that ATRA acts on Th17-polarized CCR6+CD4+ T cells to increase their permissiveness to HIV-1 infection in vitro and enhances viral outgrowth from cells of ART-treated PWH via mTOR-dependent mechanisms.32,40–42 Similarly, other groups have demonstrated the key role played by mTOR in the positive regulation of HIV-1 replication in CD4+ T cells via mechanisms acting on CCR5 expression,81 the deoxynucleoside triphosphate (dNTP) pools required for HIV-1 reverse transcription and acetylcoenzyme A (CoA) required for nuclear transportation of viral products,69 and HIV-1 transcription via the modulation of the CDK9/P-TEFb complex, a component of the viral transcriptional machinery.75 Our findings further support the link between viral replication and host-cell metabolism and point to the use of mTOR inhibitors for HIV-1 treatment. Consistently, our group and others have provided evidence pointing to metformin, an indirect mTOR inhibitor, as an efficient therapeutic strategy for the control of residual HIV-1 transcription and premature aging in ART-treated PWH.82,83
We showed here that ATRA upregulated CCR5, while downregulating CD4 and CXCR4, and preferentially promoted CCR5-tropic HIV-1 replication in MDMs. These results raise new questions on the role of ATRA in the selection of R5 HIV-1 strains during primary infection in MDMs. The fact that ATRA decreased CD4 expression in MDMs without impeding the efficacy of productive HIV-1 infection is consistent with the reported capacity of HIV-1 to enter macrophages with low CD4 requirement.3,4 The upregulation of CCR5 in ATRA-treated MDMs is in line with our in silico findings (ENCODE database interrogation) that RAREs are present in the CCR5 promoter, suggesting that CCR5 is a direct transcriptional target for ATRA. Molecular mechanisms by which the mTOR inhibitor INK128 counteracted the effect of ATRA on CCR5 expression remain to be elucidated. Nevertheless, ATRA did not increase CCR5-mediated HIV-1 entry in MDMs, pointing to the existence of post-entry mechanisms of action. Consistently, single-round infection with VSV-G-pseudotyped HIV-1 demonstrated that ATRA acted at post-entry levels, facilitating reverse transcription and leading to an efficient HIV-DNA integration. Genome-wide RNA sequencing, GSVA, NCBI HIV interactor database search, and the in silico ENCODE search for genes with RARE in their promoters, together with functional validations and pharmacological targeting, allowed us to identify ATRA as an inducer of SAMHD1 phosphorylation, a post-translational modification associated with the loss of its capacity to restrict HIV-1 reverse transcription.84 SAMHD1 was originally identified as a key HIV-1 restriction factor in macrophages via the control of the pool of dNTP essential for reverse transcription.85 In the present study, ATRA-induced SAMHD1 phosphorylation coincided with increased expression of the cyclin CCND2. Other studies have previously reported the role of CDK1/2 in the phosphorylation of SAMHD1.72 We here demonstrate that the mTOR inhibitor INK128 abrogated the ATRA-mediated SAMHD1 phosphorylation. This knowledge adds to the beneficial effects of mTOR inhibitors on controlling HIV-1 infection, mainly in the context where the antiviral features of SAMHD1 are linked to disease progression in PWH.86,87
In the context of single-round infection with VSV-G-pseudotyped HIV-1, MDM exposure to ATRA upon optimal HIV-DNA integration demonstrated an increased intracellular expression of HIV-p24 and an increased virion release in cell-culture supernatants. The latter points to an effect of ATRA on HIV-1 transcription and/or translation. The effect of ATRA on HIV-1 transcription can be explained by the presence of RAREs in the HIV-LTR.39 Indeed, our ChIP-qPCR experiments using the THP-1 myeloid cell line demonstrated the direct binding of RARα onto the HIV-LTR, together with an increased recruitment of CDK9 and RNAPII upon exposure to ATRA. Moreover, ATRA has been reported to activate mTOR/S6K via phosphorylation, a pathway documented to promote HIV-1 transcription.75 Consistently, we demonstrated that the recruitment of CDK9, but not RARα and RNAPII, to the HIV-LTR, as well as HIV-1 transcription, were reduced by the mTOR inhibitor INK128.
In addition to HIV-1 positive regulators modulated by ATRA, our RNA sequencing results originally revealed that ATRA decreased the expression of β-catenin and GSK3β, while increasing the expression of TCF7L2/TCF4 in MDMs. The Wnt/β-catenin pathway represses HIV-1 transcription via mechanisms involving the β-catenin interaction with TCF4 and subsequent binding on HIV-LTR58,60 or the suppression of transcription factors that increase HIV transcription, namely C/EBP and NF-κβ. In the absence of Wnt signaling, GSK3β promotes the proteasomal degradation of β-catenin.59 Therefore, by decreasing the expression of β-catenin and GSK3β, ATRA may facilitate HIV-1 transcription. Nevertheless, we showed that the Wnt/β-catenin/TCF4 pathway remained functional in ATRA-treated MDMs, likely as a mechanism to limit exacerbated HIV-1 replication, as demonstrated by the increased expression of TCF7L2/TCF4 mRNA/protein and the use of the β-catenin inhibitors PRI-742 and PNU-74654. In line with this idea, it has been reported that CD4+ T cells from elite controllers exhibit higher expression/activation of the Wnt/β-catenin pathway compared to ART-treated patients,88 explaining better viral control. Finally, Barbian et al. have demonstrated that Wnt/β-catenin pathway blockade reactivates HIV-1 in primary CD4+ T cells from ART-treated PWH and increases the activity of latency-reversing agents.58
RA is produced from vitamin A, mainly in the intestine, and subsequently stored in different tissues, including the liver, lungs, and bone marrow.38,89,90 In the intestine, RA is produced by dendritic cells91,92 and is reported to represent a gut-homing elixir.35 In line with this evidence, our previous studies revealed an RA signature (CCR5+ITGB7+mTOR+) in CCR6+CD4+ T cells infiltrating the colon but not those circulating in the peripheral blood,41,82 indicative that RA is active in the colon. The role of RA in HIV pathogenesis was recently reviewed,38 and protease inhibitors have been reported to affect the activity of vitamin A metabolizing enzymes, subsequently modulating RA production in PWH.93 It remains unknown whether ART interferes with RA production/storage in tissues and subsequently with tissue-resident macrophage functions in PWH.
In conclusion, our results demonstrate that ATRA transcriptionally reprograms macrophages for increased permissiveness to HIV-1 replication via post-entry and mTOR-dependent mechanisms that involve (1) a reduced SAMHD1-mediated restriction, thus facilitating reverse transcription; (2) a sustained transcription coinciding with the recruitment of RARα, CDK9, and RNAPII to the HIV-LTR and a decrease in the expression/activation of the Wnt/β-catenin pathway; and (3) a more efficient HIV-1 production/release, potentially via a reduced INSIG1-mediated Gag degradation (graphical abstract). These findings point to ATRA-mediated and mTOR-modulated mechanisms of action that may be targeted to interfere with HIV-1 replication in macrophages. Although short-lived MDMs are limited in their capacity to contribute to VR persistence during ART, in the intestinal environment rich in RA, these MDMs likely contribute to the establishment of primary HIV-1 infection and viral rebound upon ART interruption in PWH. Therefore, HIV-1-eradication strategies should consider MDMs as therapeutic targets.
Study limitations
This study has several limitations. The effect of ATRA was studied on bulk MDMs from HIV-uninfected study participants. While the functional characterization of CD4+ T cells carrying VR generated valuable insights into the host-cell determinants associated with integrative HIV-1 infection, such studies are needed for macrophages as well, especially in the context of a tremendous macrophage heterogeneity at single-cell level.94 Trypsin was used to detach macrophages. Although potential deleterious effects of trypsin on surface protein expression cannot be excluded, our flow cytometry results correlated with findings at transcriptional level, indicative that our experimental design does not affect readouts, especially in the presence of proper controls. Although this manuscript provides original evidence that ATRA promotes CCND2 and CDK1 mRNA expression and SAMHD1 phosphorylation, the molecular mechanisms by which SAMHD1-mediated antiviral responses are blunted by ATRA remain to be elucidated. Our transcriptional profiling revealed multiple HIV restriction/dependency factors modulated by ATRA that were not functionally validated herein. For example, the down-modulation of BIRC2, a suppressor of HIV transcription,61 INSIG1, involved in Gag degradation,56 and SERINC5, an HIV release inhibitor counteracted by Nef,62 point to an effect of ATRA on HIV transcription, translation, and viral release, respectively. These mechanisms require further investigation. Studies on the existence of an RA signature in MDMs or TRMs from ART-treated PWH are needed to provide a proof for the clinical relevance of our ex vivo findings. Finally, although our RNA sequencing results revealed downregulation of estrogen receptor 1 (ESR1) by ATRA in MDMs (Data S2), in this study we did not address sex-related differences in the capacity of RA to modulate HIV-1 replication in macrophages. Given the reported interplay between RA and estrogen pathways, such studies are highly relevant to pursue.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Petronela Ancuta (petronela.ancuta@umontreal.ca).
Materials availability
This study did not generate unique reagents.
Data and code availability
RNA-seq data have been deposited at Gene Expression Omnibus (GEO) database under accession GSE226653 and are publicly available as of the date of publication, as listed in the key resources table. Original western blot images have been deposited at Mendeley database (https://data.mendeley.com/datasets/5b38g36z9v/1).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
This paper does not report original code.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal anti-SAMHD1 mouse antibody clone OTI3F5 (30 μL) | Cedarlane | Cat#TA502024S |
| Polyclonal Anti-SAMHD1 (PHOSPHO THR592) (0.02 mg) antibody | Cedarlane | Cat#8005-0.02MG |
| Goat Anti-Mouse IgG (H + L) Secondary Antibody, HRP (1/2000) | Thermo Fisher | Cat#32340; RRID: AB_1185566 |
| Phospho-mTOR (Ser2448) Antibody (1/1000) | Cell Signaling | Cat#2971S; RRID: AB_330970 |
| mTOR (7C10) Rabbit mAb (1/1000) | Cell Signaling | Cat#2983; RRID: AB_2105622 |
| Anti-phospho-ribosomal protein S6 Ser240/Ser244 (1/500) | EMD Millipore | Cat#07-2113 |
| S6 Ribosomal Protein (5G10) Rabbit mAb (1/1000) | Cell Signaling | Cat#2217 |
| TCF4/TCF7L2 (C48H11) Rabbit (1/1000) | Cell Signaling | Cat#2569; RRID: AB_2199816 |
| Monoclonal Anti-β-actin Antibody Produce in Mouse (1/5000) | Sigma | Cat#A5441; RRID: AB_476744 |
| Anti-rabbit IgG HRP-linked Antibody (1/5000) | Cell Signaling | Cat#7074; RRID: AB_2099233 |
| ITGB7 BUV737 (Clone FIB504) | BD | Cat#749664 |
| GSK3B pS9 Antibody, anti-human/mouse/rat REAfinity APC | Miltenyi | Cat#130-106-901; RRID: AB_2651905 |
| Beta Catenin Monoclonal Antibody (15B8), Alexa Fluor 488 | Invitrogen | Cat#53-2567-41; RRID: AB_10807094 |
| Mouse anti-human CD3 Pacific Blue (Clone UCHT1) | BD | Cat#558117; RRID: AB_397038 |
| Mouse anti-human CD4 Alexa Fluor 700 (Clone RPA-T4) | BD | Cat#557922; RRID: AB_396943 |
| Mouse anti-human CD16 Phycoerythrin-Cyanine 7 (Clone 3G8) | BD | Cat#560918; RRID: AB_10563252 |
| Mouse anti-human CD14 APC (Clone M5E2) | BD | Cat#555399; RRID: AB_398596 |
| Anti-human CD1c (BDCA-1) Phycoerythrin (Clone REA694) | Miltenyi | Cat#130-110-536; RRID: AB_2656038 |
| Anti-human HLA-DR Brilliant Violet 785 (Clone L243) | Biolegend | Cat#307642; RRID: AB_2563461 |
| Mouse Anti-human CD195 Fluorescein Isothiocyanate (Clone 2D7/CCR5) | BD | Cat#555992; RRID: AB_396278 |
| CD184 CXCR4 Allophycocyanine (Clone 12G5) | Biolegend | Cat#306510; RRID: AB_314616 |
| HIV-1 core (p24) antigen-RD1 (Clone KC57) | Beckman Coulter | Cat#6604667; RRID: AB_1575989 |
| RARα | Cell signaling | Cat#62294; RRID: AB_2799625 |
| Phospho-Rpb1 CTD (Ser2) | Cell signaling | Cat#13499; RRID: AB_2798238 |
| Phospho-Rpb1 CTD (Ser5) | Cell signaling | Cat#13523; RRID: AB_2798246 |
| RNA polymerase II | Cell signaling | Cat#2629S |
| Rabbit IgG | Cell signaling | Cat#2729; RRID: AB_1031062 |
| CDK9 | Abcam | Cat#ab239364; RRID: AB_3096172 |
| Bacterial and virus strains | ||
| One Shot Stbl3 Chemically Competent E. coli | Life Technologies | Cat# C737303 |
| HIV-1 THRO plasmid (pTHRO.c/2626), subtype B | NIH AIDS Reagent Program (Contribution of Dr. John Kappes and Dr. Christina Ochsenbauer) | Cat#11745 |
| NL4.3BaL HIV plasmid | From Dr. Dana Gabuzda (Dana-Farber Cancer Institute, Boston, MA, USA) | N/A |
| VSV-G Plasmid | National Institution of Health (NIH) | Cat#ARP-4693 |
| NL4.3BaLΔenv GFP | National Institution of Health (NIH) | Cat#ARP-12637 |
| NL4.3Env-Vpr- | From Dr. Eric Cohen | N/A |
| ADA-Env | From Dr. Eric Cohen | N/A |
| Blam-Vpr | From Dr. Eric Cohen | N/A |
| VSV-G-Env | From Dr. Eric Cohen | N/A |
| pNL4.3-IRES-GFP | NIH HIV Reagent Program | Cat#ARP-11349 |
| Biological samples | ||
| Leukapheresis collected from HIV-participants | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium Bicarbonate (NaHCO3) | Sigma | Cat#S5761 |
| Sodium Carbonate (Na2CO3) | Sigma | Cat#223530-500G |
| Thimerosal | Sigma | Cat#T5125-10G |
| Phosphate Buffered Saline (PBS) | Thermo Fisher | Cat#10010023 |
| Tween 20 | Fisher Scientific | Cat#BP337-500 |
| Triton X-100 | Sigma | Cat#X100-500mL |
| Trypan Blue | Thermo Fisher | Cat#15250061 |
| Bovine Serum Albumin (BSA) | BioShop | Cat#ALB001.500 |
| Streptavidin Horseradish Peroxidase (Strep-HRP) | Fisher Scientific | Cat#65R-S104PHRP |
| 3,3′,5,5′-Tetramethylbenzidine (TMB) | Quimigen | Cat#42R-TB10265R-S104PHRP |
| Phosphoric Acid (H3PO4) | Sigma | Cat#PX0996 |
| Nonidet P-40 | Bioshop | Cat#NON505 |
| Tris HCl | BioShop | Cat#TRS002.500 |
| Proteinase K | Fisher Scientific | Cat#25530-015 |
| Molecular Grade Water (H2O) | Wisent | Cat#809-115-CL |
| Guanidine-HC; | Promega | Cat#H5381 |
| Trypsin EDTA with phenol red | Wisent | Cat#075-350 |
| X-tremeGENE HP DNA Transfection Reagent | Roche | Cat#6366244001 |
| Probenecid | Sigma-Aldrich | Cat#P8761 |
| Methanol 99,98% | Fisher Scientific | Cat#BPA4084 |
| ReBlot Plus Strong Antibody Stripping Solution | Sigma | Cat#2504 |
| 4X Laemmli Sample Buffer | Bio-Rad | Cat#1610747 |
| 2-Mercaptoethanol | Sigma | Cat#M6250 |
| Precision Plus Protein Dual Color Standards | Bio-Rad | Cat#1610374 |
| Glycine | BioShop | Cat#GLN001.500 |
| Sodium Chloride | BioShop | Cat#SOD002 |
| 30% Bis-acrylamide Solution | Bioshop | Cat#ACR010.500 |
| UltraPure Tris Buffer | Thermo Fisher | Cat#15504-020 |
| Sodium Dodecyl Sulfate (SDS) | Bio-Rad | Cat#1610302 |
| N,N,N′,N′-Tetramethyl Ethylenediamine (TEMED) | Sigma | Cat#8087420005 |
| Ammonium Persulfate (APS) | Bioshop | Cat#AMP001.100 |
| Radioimmunoprecipitation Assay Buffer (RIPA) 10X | Cell Signaling | Cat#9806S |
| Phosphatase Inhibitor (PhosSTOP) | Roche | Cat#4906845001 |
| Complete, Mini, EDTA-free protease inhibitor | Roche | Cat#11836170001 |
| Formaldehyde solution 37 wt. % in H2O | Sigma | Cat#F1635-500ML |
| Sodium Azide | Bioshop | Cat#SAZ001.250 |
| Fetal Bovine Serum (FBS) | Wisent | Cat#091-150 |
| Dimethyl Sulfoxide (DMSO) | Sigma | Cat#34869-500mL |
| RPMI 1640 Medium (RPMI) | Thermo Fisher | Cat#11875119 |
| All-trans Retinoic Acid (ATRA) | Sigma | Cat#R2625-50MG |
| Sapanisertib (INK128) | Cayman Chemical | Cat#11811-1 |
| PRI-724 (ICG-001) | Selleck | Cat# S8968 |
| PNU-74654 | Selleck | Cat# S8429 |
| Recombinant Human Macrophage Colony Stimulating Factor (M-CSF) | Cedarlane | Cat#216-MC-025 |
| EDTA | Bioshop | Cat#EDT001.1 |
| Phosphate Buffered Saline (PBS) | Thermo Fisher | Cat#10010023 |
| Penicillin/streptomycin (PenStep) | Thermo Fisher | Cat#15140122 |
| Lymphocyte Separation Medium (LSM) | Wisent | Cat#305-010-CL |
| CO2-independent media | Life Technologies | Cat#18045-088 |
| DMEM High glucose, pruvate | Gibco | Cat#11995065 |
| Opti-MEM | Gibco | Cat#31985070 |
| DNAse | Invitrogen | Cat#AM1907 |
| Critical commercial assays | ||
| LDH Assay Kit | Abcam | Cat#ab65393 |
| p24 Enzyme-linked Immunosorbent Assay (ELISA) | Homemade. Hybridome provided by Dr. Michel J. Tremblay95 | N/A |
| QuantiTect SYBR Green RT-PCR Kit | Qiagen | Cat#204243 |
| 10X PCR Buffer/Magnesium Chloride (MgCl2) Buffer/Thermus Aquaticus (TAQ) Polymerase | Thermo Fisher | Cat#18038067 |
| LC480 probe master mix | Roche | Cat#4707494001 |
| QuantiTect SYBR Green RT-PCR Kit | Qiagen | Cat#204243 |
| Deoxynucleoside Triphosphates (dNTP) | Thermo Fisher | Cat#10297018 |
| Promega Wizard Plus DNA Purification System | Fisher Scientific | Cat#PR-A7100 |
| EndoFree Plasmid Maxi Kit | Qiagen | Cat#12362 |
| Memory CD4+ T cell Isolation Kit, human | Miltenyi | Cat#130-091-893 |
| CCF2-AM Substrate and Loading Solution | Life Technologies | Cat#K1032 |
| All Prep DNA/RNA/miRNA Universal Kit | Qiagen | Cat#80224 |
| Detergent Compatible (DC) Protein Assay | Bio-Rad | Cat#5000111 |
| Clarity and Clarity Max ECL Western Blotting Substrates | Bio-Rad | Cat#1705062S |
| Fixation/Permeabilization Kit | BD | Cat#554714 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (405 nm excitation) | Thermo Fisher | Cat#L34957 |
| Pan Monocyte Isolation Kit | Miltenyi | Cat#130-096-537 |
| CHIP assay kit | EMD Millipore | Cat#17-295 |
| Luna® Universal qPCR Master Mix | NEB | Cat# M3003S |
| GeneJET PCR Purification Kit | Thermo Fisher | Cat# K0702 |
| EZ-10 Spin Column Total RNA Miniprep Super Kit | Bio Basic | Cat#BS584 |
| Prime-Script RT reagent kit | TaKaRa | Cat#RR037A |
| Experimental models: Cell lines | ||
| HT-29 | ATCC | Cat#HTB-38 |
| 293T | ATCC | Cat#CRL-3216 |
| ACH-2 | NIH HIV Reagent Program | Cat#ARP-349 |
| THP-1 | NIH HIV Reagent Program | Cat#ARP-9942 |
| Oligonucleotides | ||
| PPARy | Qiagen | GeneGlobal ID: QT00029841 |
| TCF7L2 | Qiagen | GeneGlobal ID: QT00071120 |
| HIC1 | Qiagen | GeneGlobal ID: QT00203175 |
| CTNNB1 | Qiagen | GeneGlobal ID: QT00077882 |
| GSK3β | Qiagen | GeneGlobal ID: QT00057134 |
| CCND2 | Qiagen | GeneGlobal ID: QT00057575 |
| CDK1 | Qiagen | GeneGlobal ID: QT00042672 |
| SAMHD1 | Qiagen | GeneGlobal ID: QT00084322 |
| RARA | Qiagem | GeneGlobal ID: QT00095865 |
| RXRA | Qiagen | GeneGlobal ID:QT00005726 |
| See Table S1: Oligonucleotides | See Table S1: Oligonucleotides | See Table S1: Oligonucleotides |
| Software and algorithms | ||
| FlowJo version 10 | BD | https://www.flowjo.com/ |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/ |
| Image Lab | Bio-Rad | https://www.bio-rad.com/en-ca/product/image-lab-software?ID=KRE6P5E8Z |
| QuantStudio™ Design & Analysis Software | Thermo Fisher | https://www.thermofisher.com/ |
| Other | ||
| MACS LS Columns | Miltenyi | Cat# 130-042-401 |
| Pre-Separation Filters 30 μm | Miltenyi | Cat# 130-041-407 |
| Immobilon-PSQ Polyvinylidene Difluoride (PVDF) | Sigma | Cat#ISEQ00010 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Study participants
Leukapheresis samples were collected from HIV-uninfected individuals [HIV−; n = 22, 19 males and 3 females, with median age of 50 years old (range: 26–71 years old)], as we previously described.41,43,68,96 PBMC were isolated from leukapheresis by gradient centrifugation using the lymphocyte separation medium (Wisent, Saint-Jean-Baptiste/Canada), and preserved frozen in 10% DMSO (SIGMA, St. Louis/United States) in fetal bovine serum (FBS; Wisent, Saint-Jean-Baptiste/Canada) until use.
Ethics statement
A written informed consent following the guidelines of the Declaration of Helsinki and approved by the Institutional Ethics Review Board of the McGill University Health Center (MUHC; Montréal, Québec, Canada) and the Center de Recherche du Center Hospitalier de l’Université de Montréal (CR-CHUM; Montréal, Québec, Canada) was provided, clarified, and signed by all participants in this study.
METHOD DETAILS
Monocyte isolation
PBMCs from HIV− individuals were used to isolate monocytes by negative selection using a pan monocyte magnetic associated cell sorting (MACS) isolation kit (Miltenyi, Bergisch Gladbach/Germany) using a MACS buffer FACS buffer (1X PBS, 10% FBS, 2 mM EDTA). Typically, monocyte purity was >95%, with less than 1% contaminations in CD3+CD4+ T-cells, as determined by flow cytometry upon staining with appropriate Abs (Figure S1), as we previously reported.43
Generation of monocyte-derived macrophages
MDMs were generated by culturing monocytes in 48-well plates (Costar, Arizona/United States) (106 monocytes/well/ml) in the presence of M-CSF for 6 days. The MDMs differentiation media consisted of RPMI-1640 (Thermo Fisher; Waltham/United States), 10% of FBS, 1% penicillin/streptomycin (Thermo Fisher, Waltham/United States), and M-CSF (20 ng/mL; R&D Systems, Minneapolis/United States). Media containing M-CSF was refreshed every 2 days. In parallel, MDMs were generated in the presence of ATRA (Sigma, St. Louis/United States) and exposed or not to the following drugs: INK128 (an MTORC1/2 inhibitor; Cayman Chemical, Ann Arbor/United States) and/or PRI-724 (a Wnt/β-catenin inhibitor; Selleck, Houston/United States) and/or PNU-74654 (a Wnt/β-catenin inhibitor; Selleck, Houston/United States). All drugs were titrated for their effect on MDMs viability and optimal concentrations were used, as indicated in the Figure legends.
Flow cytometry analysis
Surface staining was performed on PBMC and monocytes, as well as on MDMs harvested from 48-well plates using cold 1X PBS. Cells were washed using a FACS buffer (1X PBS, 10% FBS, EDTA 2mM, 0.2% sodium azide) and incubated with the following antibodies against CD3, CD4, CD16, CD14, CD1c, HLA-DR, CD14, CD16, CD195/CCR5, CD184/CXCR4, ITGB7, GSK3β, and β-Catenin (key resources table). Intracellular staining of MDMs infected with HIV-1 in vitro was performed with the HIV-p24 Abs (KC57; Beckman Coulter, Brea/United States) was performed using the BD cytofix/cytoperm fixation/permeabilization solution kit (BD Biosciences, Franklin Lakes/United States) according to the manufacturer’s protocols. To exclude dead cells, the live/dead fixable aqua dead cell stain kit (Thermo Fisher, Waltham/United States) was used. In addition, positive gates were placed according to fluorescence minus one (FMO) strategy, as previously reported.97 Samples were acquired by flow cytometry using the LSRIIA cytometer and the BD FACS Diva software (BD Bioscience, San Jose, CAL; California). Finally, results were analyzed using BD Flowjo (Tree Star, Inc., Ashland, Oregon, USA).
RNA extraction and mRNA expression by real-time RT-PCR
Dual DNA/RNA extraction was performed using the AllPrep DNA/RNA/miRNA universal kit (Qiagen; Hilden/Germany), according to the manufacturer’s protocol. Briefly, RLT plus buffer containing β-mercaptoethanol was added directly onto the MDMs monolayer to lyse the cells. Each lysed sample was transferred onto an AllPrep mini spin column. RNA was eluted at a final volume of 30 μL. The expression of HIC1, TCF4, CCND2, CTNNB1, GSK3β, SAMHD1, CDK1 and PPARγ mRNA was quantified using a one-step SYBR green real-time PCR kit (Qiagen, Hilden/Germany) and the Lightcycler 480 II (Roche, Basel/Switzerland), using QuantiTect primers (Qiagen, Hilden/Germany). The control 28S rRNA primers (were designed as we previously reported41,43,68,96 (key resources table) and purchased from Integrated DNA Technologies (IDT, Newark/United States). Real-time RT-PCR was performed in triplicates on various quantities of total RNA for the quantification of PPARγ (70 ng), HIC1 (50 ng), TCF4 (25 ng), CCND2 (10 ng) CTNNB1 (10 ng), GSK3β (10 ng), SAMHD1 (10 ng), CDK1 (10 ng) mRNA, and 28S rRNA (2 ng). Negative controls (without RNA, without RT enzyme, without mix) were performed for each transcript and the mRNA expression was normalized relative to the 28S rRNA. mRNA expression levels were calculated based on the relative differences (delta CT) between the target gene of interest (PPARγ, HIC1, TCF4, CCND2, CDK1, SAMHD1, CTNNB1, GSK3β) and the control gene (28S rRNA), as we previously reported.41,43,68,96
HIV-1 infection in vitro
The HIV-1 infection in vitro was performed, as we previously reported.41,43,68,96 Briefly, the X-tremeGENE HP DNA transfection reagent (Roche, Basel/Switzerland) was used to generate HIV-1 stocks by transfecting 293T cells with plasmids obtained from the National Institute of Health (NIH) AIDS Research Program. The following molecular clones were used in this study: i) replication-competent R5 NL4.3BaL (HIVNL4.3BaL), ii) T/F THRO (HIVTHRO), iii) replication-competent X4 NDK and iv) replication-defective VSV-G-pseudotyped HIV-1 (HIVVSV-G). The pHEF plasmid Expressing VSV-G (ARP-4693) (contributed by Dr. Lung-Ji Chang) and the HIV plasmid containing the NL4-3 backbone encoding for enhanced green fluorescent protein (EGFP) in place of the Envelope (Env) (NL4.3EGFPΔEnv) (ARP-11100) (contributed by Dr. Haili Zhang, Dr. Yan Zhou and Dr. Robert Siliciano) were obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH. Viral stocks were quantified by HIV-p24 ELISA and titrated on TCR-activated CD4+ T-cells for the identification of optimal infectious concentrations. For infection, MDMs plated in 48-well plates at a density of 106 cells/well were exposed to HIV-1 (30 ng HIV-p24/300 μL/well) and incubated at 37°C for 3 h. Unbound virions were removed by three-times washing with 1 mL media/well. MDMs were further cultured for up to fifteen days in media containing M-CSF (10 ng/mL) in the presence/absence of ATRA and/or other drugs, as indicated in Figure legends. The media containing M-CSF and/or drugs was refreshed every three days post-infection. Cell-culture supernatants were used for HIV-p24 quantification by ELISA. In parallel, MDMs were collected for real-time PCR quantification HIV-DNA levels and flow cytometry analysis of intracellular HIV-p24 expression at day 3 and 15 post-infection, respectively.
HIV-1 entry assay
An HIV-1 entry assay was performed, as described by Cavrois et al.,98 with modifications. Briefly, calcium phosphate was used as a DNA transfection reagent to generate HIV-1 stocks by cotransfecting HEK293T cells with an NL4.3Env−Vpr− proviral construct,99 the Blam-Vpr plasmid98 and either the SVIIIEnvADA99 or SV-CMV-VSV-G99 plasmids in order to generate the R5-tropic ADA-Env NL4.3Env−Vpr−/ADA-Env/Blam-Vpr (HIVADA-Env) or the VSV-G-pseudotyped NL4.3Env−Vpr−/VSV-G-Env/Blam-Vpr (HIVVSV-G) viruses, respectively. Viral stocks were quantified by HIV-p24 ELISA and titrated on TZM-BL cells for the identification of optimal infectious concentrations. MDMs were then loaded with CCF2, a substrate of β-lactamase; thus, CCF2 is cleaved by virion-associated Blam-Vpr transduced in cells susceptible to viral entry. For infection, MDMs plated in 96-well V bottom plates at a density of 105 cells/well were exposed to HIV-1 (50 ng HIV-p24/100 μL/well) and incubated at 37°C for 5 h. Unbound virions were removed by washing with CO2-independent media (Invitrogen) 100 μL media/well. MDMs were resuspended with 100 μL CCF2-AM loading solution/well (Invitrogen, the GeneBlazer kit) for 1 h. Unbound CCF2-AM was removed, and cells incubated for 16 h in the dark, at room temperature. MDMs were later washed and fixed with 2% paraformaldehyde solution, and virus entry measured using a Fortessa (BD Bioscences) flow cytometer. Results were analyzed using the FlowJo software.
Quantification of early HIV-1 reverse transcripts by SYBR green PCR
MDMs were digested directly in 48-well plates in a lysis buffer containing 0.1mM Tris HCl pH 8.0, 0.5% Tween 20 detergent, 10 mg/mL proteinase K (Thermo Fisher, Waltham/United States), and molecular grade water (Wisent, Saint-Jean-Baptiste/Canada) at a concentration of 50,000 cells/15 μL (or 200 μL per 48-well plate). The quantification of different forms of HIV-DNA was performed using nested real-time PCR, relative to CD3 as a housekeeping gene, using primers/SYBR Green as indicated in Table S1, as we previously described.41,43,68,96 Serial dilutions from 3×105 to three ACH-2 T-cells were used as a standard curve for early reverse transcripts quantification. Amplification products from the first PCR reaction were diluted by a factor of ten before adding buffer, primers, and SYBR Green. The limit of detection for this assay is three HIV/CD3 copies per test. All PCR reactions were performed in triplicates.
Quantification of gag and integrated HIV-DNA by nested real-time PCR
MDMs lysates were prepared as described above. The quantification of Gag and integrated HIV-DNA was performed using nested real-time PCR, relative to CD3 as a housekeeping gene, using primers/probes as indicated in Table S1, as we previously described.41,43,68,96 Briefly, late reverse transcripts were quantified using primers directed against Gag HIV-1 regions (45 Amplification cycles), and integrated HIV-DNA levels (45 Amplification cycles) were quantified using primers against the Alu repetitive sequences and HIV-LTR region.41,43,68,96 Serial dilutions from 3×105 to three ACH2 cells were used as a standard curve for late reverse transcripts and HIV-DNA quantification. Amplification products from the first PCR reaction were diluted by a factor of ten before adding buffer, primers, and probes. The limit of detection for this assay is three HIV/CD3 copies per test. All PCR reactions were performed in triplicates.
Quantification of HIV-p24 by ELISA
The HIV-p24 levels were quantified in cell culture supernatants using a homemade ELISA assay, as we previously reported.41,43,68,96
Illumina RNA sequencing and analysis
The RNA sequencing and analysis were performed, as we previously reported.68,96 Briefly, total RNA was extracted from ATRA-MDMs and DMSO-MDMs harvested prior to HIV infection (Figure 1A) using AllPrep DNA/RNA/miRNA universal kit (Qiagen; Hilden/Germany), according to the manufacturer’s protocol. Genome-wide RNA sequencing profiles were generated by Genome Québec (Montreal, Québec, Canada) using the Illumina RNA-Sequencing technology (NovaSeq6000 S4 PE 100bp 25M reads). The paired-end sequencing reads were aligned to coding and non-coding transcripts from Homo Sapiens database GRCh 37 version 75 and quantified with the Kallisto software version 0.44.0. The entire RNA-Sequencing data set and the technical information requested by Minimum Information About a Microarray Experiment (MIAME) are available at the GEO database under accession GSE226653. Statistical analyses were performed using R version 4.21. Differential expression analysis was performed using the limma Bioconductor R package (version 3.52.2) on the log2-counts per million (logCPM) transformed transcript-level and gene-level data. Differentially expressed genes (DEG) were identified based on pp-values (p < 0.05), adjusted pp-values (adj. p < 0.05), and fold-change (FC, cutoff 1.3) (Data S1 and S2). Gene set variation analysis (GSVA; C2, C3, C5, C7, C8, and Hallmark databases; https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) was performed using the GSVA method (package version 1.344.2) on the logCPM data using a Gaussian cumulative distribution function. Finally, genes presenting RARE in their promoters were identified using the ENCODExplorerData (version 0.99.5) and listed in Data S3.
Western blotting
The Western blotting visualisation of proteins was performed, as we previously reported.41,96 Briefly, total lysates of MDMs (3×106 per condition) were generated using the Radio-immunoprecipitation Assay (RIPA) Buffer 1X (Cell Signaling, Danvers/United States) containing phosphatase (PhosSTOP; Roche, Basel/Switzerland) and protease inhibitors (Complete, Mini, EDTA-free protease inhibitor; Roche, Basel/Switzerland). Total protein content from each condition was quantified using DC Protein Assay (Bio-Rad, Hercules/United States) in triplicate samples. SDS-PAGE gel electrophoresis was performed on a gradient polyacrylamide gel for 75 min at 130 V and transferred on immobilon-PSQ polyvinylidene difluoride (PVDF) membranes (Sigma, St. Louis/United States) for 75 min at 100 V. PVDF membranes were blocked with Tris-Buffered Saline (TBS) 0.1% Tween 5% Bovine Serum Albumin (BSA) for 45 min and incubated overnight with primary antibodies against target proteins at 4°C (key resources table). PVDF membranes were washed four times with TBS 0.1% Tween and incubated with HRP-linked secondary Abs (key resources table) for 1 h at room temperature. All Abs were diluted with blocking TBS 0.1% Tween 5% BSA. PVDF membranes were washed four times and proteins were revealed with chemiluminescence western blotting substrates (Bio-Rad, Hercules/United States). PVDF membranes were reused by using a reblot stripping solution (Sigma St. Louis/United States). A Chemidoc imaging system from Bio-Rad was used for chemiluminescence and colorimetric detection to visualize the bands on PVDF membranes and Image lab software (Sigma St. Louis/United States) was used to quantify the band intensity between each condition and each donor.
Lactate dehydrogenase (LDH) cytotoxicity assay
The cytotoxicity of ATRA on MDMs was measured using the LDH cytotoxicity assay (Abcam; Cambridge/United Kingdom), according to the manufacture’s protocol. Briefly, MDMs were placed in a 96-well plate at a concentration of 104 MDMs/well. A portion of MDM were also treated with lysis buffer II/cell lysis solution provided by the kit (positive control). MDM were then incubated at 37°C for 2 days in the presence or the absence of ATRA (10, 100, 1,000 nM). MDM were then centrifuged at 600 g for 10 min and the supernatants were transferred to another 96 well plate and 100 μL of LDH reaction mix was added, mixed, and incubated for 30 min at room temperature. The absorbance of the 96-well plate was read using a plate reader at 490nm. Reference wavelength of 650 nm was used.
Chromatin immunoprecipitation followed by quantitative PCR assay (ChIP-qPCR)
ChIP assays were performed following the ChIP assay kit from EMD Millipore on chromatin preparations of the HIV-1 infected THP-1 monocytic cell line treated or not with ATRA (10 nM) and/or INK128 (50 nM). Briefly, cells were cross-linked for 10 min at room temperature with 1% formaldehyde before lysis followed by chromatin sonication (Bioruptor Plus, Diagenode) to obtain DNA fragments of 200-400bp. Chromatin immunoprecipitations were performed with chromatin from cells and 5 μg of antibodies against either CDK9 (Abcam), RARα, the total RNA polymerase II, the RNA polymerase phosphorylated on serine 2 or serine 5 (Cell Signaling). A Normal Rabbit IgG (Cell Signaling) was used as a negative control. Quantitative real-time PCR reactions were performed using 1/60 of the immunoprecipitated DNA and the Luna Universal qPCRMaster Mix (NEB). The 5′LTR was studied using oligonucleotide primer pairs covering the Nuc-1 region (FW: 5′-TGTGTGCCCGTCTGTTGTGTGA-3′, RV: 5′-TCGGGCGCCACTGCTAGAGA-3′). Relative quantification using the standard curve method was performed and 96-well Optical Reaction plates were read in a QuantStudio3 PCR instrument (Applied Biosystem). Fold enrichments were calculated as fold inductions relative to the values measured with IgG. Primer sequences used for quantification were designed using the software Primer 3.
HIV-1 transcription
Total RNA samples were isolated using the EZ-10 Spin Column Total RNA Miniprep Super Kit (BIO BASIC), according to the manufacturer’s protocol. Following DNAse treatment (Invitrogen), reverse transcription was performed with the Prime-Script RT reagent kit (TaKaRa). Unspliced HIV-1 transcripts were detected using specific primers (FW: 5′- TTCTTCAGAGCAGACCAGAGC-3′, RV: 5′- GCTGCCAAAGAGTGATCTGA-3′). All cDNAs were quantified and normalized to the GAPDH (FW: 5′-GGACCTGACCTGCCGTCTA GAA-3′, RV: 5′- GGTGTCGCTGTTGAAGTCAGAG-3′) and YWHAZ (FW: 5′- ACTTTTGGTACATTGTGGCTTCAA-3′, RV: 5′- CCGCC AGGACAAACCAGTAT-3′) mRNA levels.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Statistical analyses were performed using the GraphPad Prism 9 software (GraphPad Software, Inc.), as we previously reported.41,43,68,96 The Kolmogorov-Smirnov, Shapiro-Wilk, and Anderson-Darling normality tests were applied to orient the use of parametric (normal distribution) versus non-parametric (nonnormal distribution) tests. For comparisons between two matched groups, the non-parametric Wilcoxon test or the parametric t Test were used. For comparisons between more than three groups, the non-parametric the Friedman test with uncorrected Dunn’s post-test or the parametric RM one-way ANOVA with Tukey’s multiple comparison test were used. pP-values <0.05 were considered statistically significant. The pp-values are indicated in all Figures.
Supplementary Material
Highlights.
Retinoic acid (RA) enhances R5 but not X4 HIV-1 replication in macrophages
RA facilitates HIV-1 replication via post-entry mechanisms
RA blunts SAMHD1-mediated HIV-1 restriction via mTOR-modulated mechanisms
RA increases the CDK9/RNAPII-dependent HIV-1 transcription
ACKNOWLEDGMENTS
The authors thank Dr. Dominique Gauchat, Philippe St. Onge, and Dr. Gael Duluth (Flow Cytometry Core Facility, CHUM-Research Center, Montréal, QC, Canada) for expert technical support with polychromatic flow cytometry sorting; Olfa Debbeche and Laurent Knaffo (Biosafety Level 3 Core Facility CHUM-Research Center, Montréal, QC, Canada); Mario Legault (FRQ-S/AIDS and Infectious Diseases Network; Montréal, QC, Canada) for help with ethical approvals and informed consents; and Josée Girouard and Angie Massicotte (McGill University Health Center, Montréal, QC, Canada) for their key contribution to blood collection and clinical information from PWH and uninfected study participants. The authors also thank Dr. Dana Gabuzda (Dana-Farber Cancer Institute, Boston, MA, USA), Dr Roger J. Pomerantz (Thomas Jefferson University, Philadelphia, PA, USA), and Dr. Michel Tremblay (Université Laval, Quebec, QC, Canada) for providing us with VSV-G and HIV plasmids. Finally, the authors acknowledge the key contribution of all study participants for their precious gift of leukapheresis essential for this study.
This work was supported by grants from the Canadian HIV Cure Enterprise Team (CanCURE 1.0) funded by the Canadian Institutes of Health (CIHR) in partnership with CANFAR and IAS (CanCURE 1.0; # HIG-133050 to P.A.) and the Canadian HIV Cure Enterprise Team (CanCURE 2.0) funded by the CIHR (#HB2-164064) to P.A. and E.A.C., as well as by a CIHR project grants to P.A. (PJT #153052; PJT 178127). Core facilities and PWH cohorts were supported by the Fondation du CHUM and the FRQ-S/AIDS and Infectious Diseases Network. C.V.L. acknowledges funding from the Belgian National Fund for Scientific Research (FRS-FNRS, Belgium); ViiV Healthcare; the Fondation Roi Baudouin; and the US National Institutes of Health (NIH) (MDC grant UM1AI164562 co-funded by National Heart, Lung, and Blood Institute, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, National Institute on Drug Abuse, and the National Institute of Allergy and Infectious Diseases). M.B. is funded by the Belgian Fonds pour la formation à la Recherche dans l’Industrie et dans l’Agriculture (FRIA) (FRS-FNRS) Fellowship. A.D. and C.V.L. are Aspirant and Directrice de Recherches of the FRS-FNRS, respectively. The funding institutions played no role in the design, collection, analysis, and interpretation of data. The experimental flow charts in the main figures and the graphical abstract were created with BioRender.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114414.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Siliciano JD, and Siliciano RF (2022). In Vivo Dynamics of the Latent Reservoir for HIV-1: New Insights and Implications for Cure. Annu. Rev. Pathol. 17, 271–294. 10.1146/annurev-pathol-050520-112001. [DOI] [PubMed] [Google Scholar]
- 2.Deeks SG, Archin N, Cannon P, Collins S, Jones RB, de Jong MAWP, Lambotte O, Lamplough R, Ndung’u T, Sugarman J, et al. (2021). Research priorities for an HIV cure: International AIDS Society Global Scientific Strategy 2021. Nat. Med. 27, 2085–2098. 10.1038/s41591-021-01590-5. [DOI] [PubMed] [Google Scholar]
- 3.Sattentau QJ, and Stevenson M (2016). Macrophages and HIV-1: An Unhealthy Constellation. Cell Host Microbe 19, 304–310. 10.1016/j.chom.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton KL, Garcia JV, Clements JE, and Walker BD (2017). HIV Infection of Macrophages: Implications for Pathogenesis and Cure. Pathog. Immun. 2, 179–192. 10.20411/pai.v2i2.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vine EE, Rhodes JW, Warner van Dijk FA, Byrne SN, Bertram KM, Cunningham AL, and Harman AN (2022). HIV transmitting mononuclear phagocytes; integrating the old and new. Mucosal Immunol. 15, 542–550. 10.1038/s41385-022-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendricks CM, Cordeiro T, Gomes AP, and Stevenson M (2021). The Interplay of HIV-1 and Macrophages in Viral Persistence. Front. Microbiol. 12, 646447. 10.3389/fmicb.2021.646447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chitrakar A, Sanz M, Maggirwar SB, and Soriano-Sarabia N (2022). HIV Latency in Myeloid Cells: Challenges for a Cure. Pathogens 11, 611. 10.3390/pathogens11060611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruize Z, and Kootstra NA (2019). The Role of Macrophages in HIV-1 Persistence and Pathogenesis. Front. Microbiol. 10, 2828. 10.3389/fmicb.2019.02828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell BI, Laws EI, and Ndhlovu LC (2019). Impact of Myeloid Reservoirs in HIV Cure Trials. Curr. HIV AIDS Rep. 16, 129–140. 10.1007/s11904-019-00438-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veenhuis RT, Abreu CM, Shirk EN, Gama L, and Clements JE (2021). HIV replication and latency in monocytes and macrophages. Semin. Immunol. 51, 101472. 10.1016/j.smim.2021.101472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Embretson J, Zupancic M, Ribas JL, Burke A, Racz P, Tenner-Racz K, and Haase AT (1993). Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature 362, 359–362. 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 12.Orenstein JM, Fox C, and Wahl SM (1997). Macrophages as a source of HIV during opportunistic infections. Science 276, 1857–1861. 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 13.Dupont M, and Sattentau QJ (2020). Macrophage Cell-Cell Interactions Promoting HIV-1 Infection. Viruses 12, 492. 10.3390/v12050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costiniuk CT, Salahuddin S, Farnos O, Olivenstein R, Pagliuzza A, Orlova M, Schurr E, De Castro C, Bourbeau J, Routy JP, et al. (2018). HIV persistence in mucosal CD4+ T cells within the lungs of adults receiving long-term suppressive antiretroviral therapy. AIDS 32, 2279–2289. 10.1097/QAD.0000000000001962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jambo KC, Banda DH, Kankwatira AM, Sukumar N, Allain TJ, Heyderman RS, Russell DG, and Mwandumba HC (2014). Small alveolar macrophages are infected preferentially by HIV and exhibit impaired phagocytic function. Mucosal Immunol. 7, 1116–1126. 10.1038/mi.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cribbs SK, Lennox J, Caliendo AM, Brown LA, and Guidot DM (2015). Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res. Hum. Retrovir. 31, 64–70. 10.1089/AID.2014.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zalar A, Figueroa MI, Ruibal-Ares B, Baré P, Cahn P, de Bracco M.M.d.E., and Belmonte L. (2010). Macrophage HIV-1 infection in duodenal tissue of patients on long term HAART. Antivir. Res. 87, 269–271. 10.1016/j.antiviral.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Jenabian MA, Costiniuk CT, Mehraj V, Ghazawi FM, Fromentin R, Brousseau J, Brassard P, Bélanger M, Ancuta P, Bendayan R, et al. (2016). Immune tolerance properties of the testicular tissue as a viral sanctuary site in ART-treated HIV-infected adults. AIDS 30, 2777–2786. 10.1097/QAD.0000000000001282. [DOI] [PubMed] [Google Scholar]
- 19.Ganor Y, Real F, Sennepin A, Dutertre CA, Prevedel L, Xu L, Tudor D, Charmeteau B, Couedel-Courteille A, Marion S, et al. (2019). HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat. Microbiol. 4, 633–644. 10.1038/s41564-018-0335-z. [DOI] [PubMed] [Google Scholar]
- 20.Honeycutt JB, Thayer WO, Baker CE, Ribeiro RM, Lada SM, Cao Y, Cleary RA, Hudgens MG, Richman DD, and Garcia JV (2017). HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat. Med. 23, 638–643. 10.1038/nm.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Micci L, Alvarez X, Iriele RI, Ortiz AM, Ryan ES, McGary CS, Deleage C, McAtee BB, He T, Apetrei C, et al. (2014). CD4 depletion in SIV-infected macaques results in macrophage and microglia infection with rapid turnover of infected cells. PLoS Pathog. 10, e1004467. 10.1371/journal.ppat.1004467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernard-Stoecklin S, Gommet C, Corneau AB, Guenounou S, Torres C, Dejucq-Rainsford N, Cosma A, Dereuddre-Bosquet N, and Le Grand R (2013). Semen CD4+ T cells and macrophages are productively infected at all stages of SIV infection in macaques. PLoS Pathog. 9, e1003810. 10.1371/journal.ppat.1003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moeser M, Nielsen JR, and Joseph SB (2020). Macrophage Tropism in Pathogenic HIV-1 and SIV Infections. Viruses 12, 1077. 10.3390/v12101077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clayton KL, Mylvaganam G, Villasmil-Ocando A, Stuart H, Maus MV, Rashidian M, Ploegh HL, and Walker BD (2021). HIV-infected macrophages resist efficient NK cell-mediated killing while preserving inflammatory cytokine responses. Cell Host Microbe 29, 435–447.e9. 10.1016/j.chom.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clayton KL, Collins DR, Lengieza J, Ghebremichael M, Dotiwala F, Lieberman J, and Walker BD (2018). Resistance of HIV-infected macrophages to CD8(+) T lymphocyte-mediated killing drives activation of the immune system. Nat. Immunol. 19, 475–486. 10.1038/s41590-018-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mass E, Nimmerjahn F, Kierdorf K, and Schlitzer A (2023). Tissue-specific macrophages: how they develop and choreograph tissue biology. Nat. Rev. Immunol. 23, 563–579. 10.1038/s41577-023-00848-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ginhoux F, and Guilliams M (2016). Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 44, 439–449. 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Molawi K, Wolf Y, Kandalla PK, Favret J, Hagemeyer N, Frenzel K, Pinto AR, Klapproth K, Henri S, Malissen B, et al. (2014). Progressive replacement of embryo-derived cardiac macrophages with age. J. Exp. Med. 211, 2151–2158. 10.1084/jem.20140639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bajpai G, Schneider C, Wong N, Bredemeyer A, Hulsmans M, Nahrendorf M, Epelman S, Kreisel D, Liu Y, Itoh A, et al. (2018). The human heart contains distinct macrophage subsets with divergent origins and functions. Nat. Med. 24, 1234–1245. 10.1038/s41591-018-0059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Y, Mouton AJ, and Lindsey ML (2018). Cardiac macrophage biology in the steady-state heart, the aging heart, and following myocardial infarction. Transl. Res. 191, 15–28. 10.1016/j.trsl.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busman-Sahay K, Starke CE, Nekorchuk MD, and Estes JD (2021). Eliminating HIV reservoirs for a cure: the issue is in the tissue. Curr. Opin. HIV AIDS 16, 200–208. 10.1097/COH.0000000000000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gosselin A, Wiche Salinas TR, Planas D, Wacleche VS, Zhang Y, Fromentin R, Chomont N, Cohen ÉA, Shacklett B, Mehraj V, et al. (2017). HIV persists in CCR6+CD4+ T cells from colon and blood during antiretroviral therapy. AIDS 31, 35–48. 10.1097/QAD.0000000000001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson JL, Khoury G, Fromentin R, Solomon A, Chomont N, Sinclair E, Milush JM, Hartogensis W, Bacchetti P, Roche M, et al. (2020). Human Immunodeficiency Virus (HIV)-Infected CCR6+ Rectal CD4+ T Cells and HIV Persistence On Antiretroviral Therapy. J. Infect. Dis. 221, 744–755. 10.1093/infdis/jiz509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khoury G, Fromentin R, Solomon A, Hartogensis W, Killian M, Hoh R, Somsouk M, Hunt PW, Girling V, Sinclair E, et al. (2017). Human Immunodeficiency Virus Persistence and T-Cell Activation in Blood, Rectal, and Lymph Node Tissue in Human Immunodeficiency Virus-Infected Individuals Receiving Suppressive Antiretroviral Therapy. J. Infect. Dis. 215, 911–919. 10.1093/infdis/jix039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mora JR, and von Andrian UH (2004). Retinoic acid: an educational “vitamin elixir” for gut-seeking T cells. Immunity 21, 458–460. [DOI] [PubMed] [Google Scholar]
- 36.Hall JA, Grainger JR, Spencer SP, and Belkaid Y (2011). The role of retinoic acid in tolerance and immunity. Immunity 35, 13–22. 10.1016/j.immuni.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manicassamy S, Ravindran R, Deng J, Oluoch H, Denning TL, Kasturi SP, Rosenthal KM, Evavold BD, and Pulendran B (2009). Toll-like receptor 2-dependent induction of vitamin A-metabolizing enzymes in dendritic cells promotes T regulatory responses and inhibits autoimmunity. Nat. Med. 15, 401–409. 10.1038/nm.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sidell N, and Kane MA (2022). Actions of Retinoic Acid in the Pathophysiology of HIV Infection. Nutrients 14, 1611. 10.3390/nu14081611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MO, Hobbs PD, Zhang XK, Dawson MI, and Pfahl M (1994). A synthetic retinoid antagonist inhibits the human immunodeficiency virus type 1 promoter. Proc. Natl. Acad. Sci. USA 91, 5632–5636. 10.1073/pnas.91.12.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monteiro P, Gosselin A, Wacleche VS, El-Far M, Said EA, Kared H, Grandvaux N, Boulassel MR, Routy JP, and Ancuta P (2011). Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin β7. J. Immunol. 186, 4618–4630. 10.4049/jimmunol.1004151. [DOI] [PubMed] [Google Scholar]
- 41.Planas D, Zhang Y, Monteiro P, Goulet JP, Gosselin A, Grandvaux N, Hope TJ, Fassati A, Routy JP, and Ancuta P (2017). HIV-1 selectively targets gut-homing CCR6+CD4+ T cells via mTOR-dependent mechanisms. JCI Insight 2, e93230. 10.1172/jci.insight.93230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Planas D, Raymond Marchand L, Massanella M, Chen H, Wacleche VS, Gosselin A, Goulet JP, Filion M, Routy JP, et al. (2020). Improving HIV Outgrowth by Optimizing Cell-Culture Conditions and Supplementing With all-trans Retinoic Acid. Front. Microbiol. 11, 902. 10.3389/fmicb.2020.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cattin A, Wiche Salinas TR, Gosselin A, Planas D, Shacklett B, Cohen EA, Ghali MP, Routy JP, and Ancuta P (2019). HIV-1 is rarely detected in blood and colon myeloid cells during viral-suppressive antiretroviral therapy. AIDS 33, 1293–1306. 10.1097/QAD.0000000000002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Leenheer AP, Lambert WE, and Claeys I (1982). All-trans-retinoic acid: measurement of reference values in human serum by high performance liquid chromatography. J. Lipid Res. 23, 1362–1367. [PubMed] [Google Scholar]
- 45.Eckhoff C, and Nau H (1990). Identification and quantitation of all-trans- and 13-cis-retinoic acid and 13-cis-4-oxoretinoic acid in human plasma. J. Lipid Res. 31, 1445–1454. [PubMed] [Google Scholar]
- 46.Gorry PR, and Ancuta P (2011). Coreceptors and HIV-1 pathogenesis. Curr. HIV AIDS Rep. 8, 45–53. 10.1007/s11904-010-0069-x. [DOI] [PubMed] [Google Scholar]
- 47.Wiche Salinas TR, Zhang Y, Sarnello D, Zhyvoloup A, Marchand LR, Fert A, Planas D, Lodha M, Chatterjee D, Karwacz K, et al. (2021). Th17 cell master transcription factor RORC2 regulates HIV-1 gene expression and viral outgrowth. Proc. Natl. Acad. Sci. USA 118, e2105927118. 10.1073/pnas.2105927118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomura N, Zhao MJ, Nagase T, Maekawa T, Ishizaki R, Tabata S, and Ishii S (1991). HIV-EP2, a new member of the gene family encoding the human immunodeficiency virus type 1 enhancer-binding protein. Comparison with HIV-EP1/PRDII-BF1/MBP-1. J. Biol. Chem. 266, 8590–8594. [PubMed] [Google Scholar]
- 49.Sivro A, Su RC, Plummer FA, and Ball TB (2013). HIV and interferon regulatory factor 1: a story of manipulation and control. AIDS Res. Hum. Retrovir. 29, 1428–1433. 10.1089/AID.2013.0098. [DOI] [PubMed] [Google Scholar]
- 50.El-Daher MT, Cagnard N, Gil M, Da Cruz MC, Leveau C, Sepulveda F, Zarhrate M, Tores F, Legoix P, Baulande S, et al. (2018). Tetratricopeptide repeat domain 7A is a nuclear factor that modulates transcription and chromatin structure. Cell Discov. 4, 61. 10.1038/s41421-018-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Wu Y, and Jiang M (2022). The emerging roles of PHOSPHO1 and its regulated phospholipid homeostasis in metabolic disorders. Front. Physiol. 13, 935195. 10.3389/fphys.2022.935195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Himes SR, Cronau S, Mulford C, and Hume DA (2005). The Runx1 transcription factor controls CSF-1-dependent and -independent growth and survival of macrophages. Oncogene 24, 5278–5286. 10.1038/sj.onc.1208657. [DOI] [PubMed] [Google Scholar]
- 53.Chopin M, Preston SP, Lun ATL, Tellier J, Smyth GK, Pellegrini M, Belz GT, Corcoran LM, Visvader JE, Wu L, and Nutt SL (2016). RUNX2 Mediates Plasmacytoid Dendritic Cell Egress from the Bone Marrow and Controls Viral Immunity. Cell Rep. 15, 866–878. 10.1016/j.celrep.2016.03.066. [DOI] [PubMed] [Google Scholar]
- 54.Matsumoto H, Scicluna BP, Jim KK, Falahi F, Qin W, Gürkan B, Malmström E, Meijer MT, Butler JM, Khan HN, et al. (2021). HIVEP1 Is a Negative Regulator of NF-κB That Inhibits Systemic Inflammation in Sepsis. Front. Immunol. 12, 744358. 10.3389/fimmu.2021.744358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDermid JM, and Prentice AM (2006). Iron and infection: effects of host iron status and the iron-regulatory genes haptoglobin and NRAMP1 (SLC11A1) on host-pathogen interactions in tuberculosis and HIV. Clin. Sci. 110, 503–524. 10.1042/CS20050273. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Lu J, Ma J, and Liu X (2019). Insulin-induced gene 1 (INSIG1) inhibits HIV-1 production by degrading Gag via activity of the ubiquitin ligase TRC8. J. Biol. Chem. 294, 2046–2059. 10.1074/jbc.RA118.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Battaglioni S, Benjamin D, Wälchli M, Maier T, and Hall MN (2022). mTOR substrate phosphorylation in growth control. Cell 185, 1814–1836. 10.1016/j.cell.2022.04.013. [DOI] [PubMed] [Google Scholar]
- 58.Barbian HJ, Seaton MS, Narasipura SD, Wallace J, Rajan R, Sha BE, and Al-Harthi L (2022). β-catenin regulates HIV latency and modulates HIV reactivation. PLoS Pathog. 18, e1010354. 10.1371/journal.ppat.1010354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lecarpentier Y, Schussler O, Hébert JL, and Vallée A (2019). Multiple Targets of the Canonical WNT/β-Catenin Signaling in Cancers. Front. Oncol. 9, 1248. 10.3389/fonc.2019.01248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aljawai Y, Richards MH, Seaton MS, Narasipura SD, and Al-Harthi L (2014). β-Catenin/TCF-4 signaling regulates susceptibility of macrophages and resistance of monocytes to HIV-1 productive infection. Curr. HIV Res. 12, 164–173. 10.2174/1570162x12666140526122249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pache L, Dutra MS, Spivak AM, Marlett JM, Murry JP, Hwang Y, Maestre AM, Manganaro L, Vamos M, Teriete P, et al. (2015). BIRC2/cIAP1 Is a Negative Regulator of HIV-1 Transcription and Can Be Targeted by Smac Mimetics to Promote Reversal of Viral Latency. Cell Host Microbe 18, 345–353. 10.1016/j.chom.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Firrito C, Bertelli C, Vanzo T, Chande A, and Pizzato M (2018). SERINC5 as a New Restriction Factor for Human Immunodeficiency Virus and Murine Leukemia Virus. Annu. Rev. Virol. 5, 323–340. 10.1146/annurev-virology-092917-043308. [DOI] [PubMed] [Google Scholar]
- 63.Sun L, Lv F, Guo X, and Gao G (2012). Glycogen synthase kinase 3b (GSK3b) modulates antiviral activity of zinc-finger antiviral protein (ZAP). J. Biol. Chem. 287, 22882–22888. 10.1074/jbc.M111.306373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Recio JA, Martínez de la Mata J, Martín-Nieto J, and Aranda A (2000). Retinoic acid stimulates HIV-1 transcription in human neuroblastoma SH-SY5Y cells. FEBS Lett. 469, 118–122. 10.1016/s0014-5793(00)01249-7. [DOI] [PubMed] [Google Scholar]
- 65.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, and Garten W (1992). Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360, 358–361. 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 66.Henderson LJ, Narasipura SD, Adarichev V, Kashanchi F, and Al-Harthi L (2012). Identification of novel T cell factor 4 (TCF-4) binding sites on the HIV long terminal repeat which associate with TCF-4, beta-catenin, and SMAR1 to repress HIV transcription. J. Virol. 86, 9495–9503. 10.1128/JVI.00486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Douce V, Forouzanfar F, Eilebrecht S, Van Driessche B, Ait-Ammar A, Verdikt R, Kurashige Y, Marban C, Gautier V, Candolfi E, et al. (2016). HIC1 controls cellular- and HIV-1-gene transcription via interactions with CTIP2 and HMGA1. Sci. Rep. 6, 34920. 10.1038/srep34920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Planas D, Fert A, Zhang Y, Goulet JP, Richard J, Finzi A, Ruiz MJ, Marchand LR, Chatterjee D, Chen H, et al. (2020). Pharmacological Inhibition of PPARy Boosts HIV Reactivation and Th17 Effector Functions, While Preventing Progeny Virion Release and de novo Infection. Pathog. Immun. 5, 177–239. 10.20411/pai.v5i1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor HE, Calantone N, Lichon D, Hudson H, Clerc I, Campbell EM, and D’Aquila RT (2020). mTOR Overcomes Multiple Metabolic Restrictions to Enable HIV-1 Reverse Transcription and Intracellular Transport. Cell Rep. 31, 107810. 10.1016/j.celrep.2020.107810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S, Schwartz O, and Benkirane M (2011). SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657. 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Badia R, Pujantell M, Riveira-Muñoz E, Puig T, Torres-Torronteras J, Martí R, Clotet B, Ampudia RM, Vives-Pi M, Esté JA, and Ballana E (2016). The G1/S Specific Cyclin D2 Is a Regulator of HIV-1 Restriction in Non-proliferating Cells. PLoS Pathog. 12, e1005829. 10.1371/journal.ppat.1005829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kueck T, Cassella E, Holler J, Kim B, and Bieniasz PD (2018). The aryl hydrocarbon receptor and interferon gamma generate antiviral states via transcriptional repression. Elife 7, e38867. 10.7554/eLife.38867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mavigner M, Zanoni M, Tharp GK, Habib J, Mattingly CR, Lichterfeld M, Nega MT, Vanderford TH, Bosinger SE, and Chahroudi A (2019). Pharmacological Modulation of the Wnt/β-Catenin Pathway Inhibits Proliferation and Promotes Differentiation of Long-Lived Memory CD4+ T Cells in Antiretroviral Therapy-Suppressed Simian Immunodeficiency Virus-Infected Macaques. J. Virol. 94, e01094–19. 10.1128/JVI.01094-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Albalawi YA, Narasipura SD, and Al-Harthi L (2022). Wnt/β-Catenin Protects Lymphocytes from HIV-Mediated Apoptosis via Induction of Bcl-xL. Viruses 14, 1469. 10.3390/v14071469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Besnard E, Hakre S, Kampmann M, Lim HW, Hosmane NN, Martin A, Bassik MC, Verschueren E, Battivelli E, Chan J, et al. (2016). The mTOR Complex Controls HIV Latency. Cell Host Microbe 20, 785–797. 10.1016/j.chom.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fert A, Raymond Marchand L, Wiche Salinas TR, and Ancuta P (2022). Targeting Th17 cells in HIV-1 remission/cure interventions. Trends Immunol. 43, 580–594. 10.1016/j.it.2022.04.013. [DOI] [PubMed] [Google Scholar]
- 77.Planas D, Routy JP, and Ancuta P (2019). New Th17-specific therapeutic strategies for HIV remission. Curr. Opin. HIV AIDS 14, 85–92. 10.1097/COH.0000000000000522. [DOI] [PubMed] [Google Scholar]
- 78.Rodes B, Cadinanos J, Esteban-Cantos A, Rodriguez-Centeno J, and Arribas JR (2022). Ageing with HIV: Challenges and biomarkers. EBioMedicine 77, 103896. 10.1016/j.ebiom.2022.103896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akusjarvi SS, and Neogi U (2023). Biological Aging in People Living with HIV on Successful Antiretroviral Therapy: Do They Age Faster? Curr. HIV AIDS Rep. 20, 42–50. 10.1007/s11904-023-00646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Poli G, Kinter AL, Justement JS, Bressler P, Kehrl JH, and Fauci AS (1992). Retinoic acid mimics transforming growth factor beta in the regulation of human immunodeficiency virus expression in monocytic cells. Proc. Natl. Acad. Sci. USA 89, 2689–2693. 10.1073/pnas.89.7.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heredia A, Le N, Gartenhaus RB, Sausville E, Medina-Moreno S, Zapata JC, Davis C, Gallo RC, and Redfield RR (2015). Targeting of mTOR catalytic site inhibits multiple steps of the HIV-1 lifecycle and suppresses HIV-1 viremia in humanized mice. Proc. Natl. Acad. Sci. USA 112, 9412–9417. 10.1073/pnas.1511144112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Planas D, Pagliuzza A, Ponte R, Fert A, Marchand LR, Massanella M, Gosselin A, Mehraj V, Dupuy FP, Isnard S, et al. (2021). LILAC pilot study: Effects of metformin on mTOR activation and HIV reservoir persistence during antiretroviral therapy. EBioMedicine 65, 103270. 10.1016/j.ebiom.2021.103270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Corley MJ, Pang APS, Shikuma CM, and Ndhlovu LC (2024). Cell-type specific impact of metformin on monocyte epigenetic age reversal in virally suppressed older people living with HIV. Aging Cell 23, e13926. 10.1111/acel.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cribier A, Descours B, Valadão ALC, Laguette N, and Benkirane M (2013). Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3, 1036–1043. 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 85.Simon V, Bloch N, and Landau NR (2015). Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat. Immunol. 16, 546–553. 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin-Gayo E, Buzon MJ, Ouyang Z, Hickman T, Cronin J, Pimenova D, Walker BD, Lichterfeld M, and Yu XG (2015). Potent Cell-Intrinsic Immune Responses in Dendritic Cells Facilitate HIV-1-Specific T Cell Immunity in HIV-1 Elite Controllers. PLoS Pathog. 11, e1004930. 10.1371/journal.ppat.1004930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiselinova M, De Spiegelaere W, Buzon MJ, Malatinkova E, Lichterfeld M, and Vandekerckhove L (2016). Integrated and Total HIV-1 DNA Predict Ex Vivo Viral Outgrowth. PLoS Pathog. 12, e1005472. 10.1371/journal.ppat.1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fonseca SG, Procopio FA, Goulet JP, Yassine-Diab B, Ancuta P, and Sékaly RP (2011). Unique features of memory T cells in HIV elite controllers: a systems biology perspective. Curr. Opin. HIV AIDS 6, 188–196. 10.1097/COH.0b013e32834589a1. [DOI] [PubMed] [Google Scholar]
- 89.Vilhais-Neto GC, and Pourquié O (2008). Retinoic acid. Curr. Biol. 18, R191–R192. 10.1016/j.cub.2007.12.042. [DOI] [PubMed] [Google Scholar]
- 90.Bhat PV (1997). Tissue concentrations of retinol, retinyl esters, and retinoic acid in vitamin A deficient rats administered a single dose of radioactive retinol. Can. J. Physiol. Pharmacol. 75, 74–77. [PubMed] [Google Scholar]
- 91.Stagg AJ (2018). Intestinal Dendritic Cells in Health and Gut Inflammation. Front. Immunol. 9, 2883. 10.3389/fimmu.2018.02883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Agace WW, and Persson EK (2012). How vitamin A metabolizing dendritic cells are generated in the gut mucosa. Trends Immunol. 33, 42–48. 10.1016/j.it.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 93.Toma E, Devost D, Chow Lan N, and Bhat PV (2001). HIV-protease inhibitors alter retinoic acid synthesis. AIDS 15, 1979–1984. 10.1097/00002030-200110190-00010. [DOI] [PubMed] [Google Scholar]
- 94.Bassler K, Schulte-Schrepping J, Warnat-Herresthal S, Aschenbrenner AC, and Schultze JL (2019). The Myeloid Cell Compartment-Cell by Cell. Annu. Rev. Immunol. 37, 269–293. 10.1146/annurev-immunol-042718-041728. [DOI] [PubMed] [Google Scholar]
- 95.Bounou S, Leclerc JE, and Tremblay MJ (2002). Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4(+)-T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J Virol 76, 1004–1014. 10.1128/jvi.76.3.1004-1014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chatterjee D, Zhang Y, Ngassaki-Yoka CD, Dutilleul A, Khalfi S, Hernalsteens O, Wiche Salinas TR, Dias J, Chen H, Smail Y, et al. (2023). Identification of aryl hydrocarbon receptor as a barrier to HIV-1 infection and outgrowth in CD4(+) T cells. Cell Rep. 42, 112634. 10.1016/j.celrep.2023.112634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roederer M (2002). Compensation in flow cytometry. Curr. Protoc. Cytom. Chapter 1, Unit 1.14. 10.1002/0471142956.cy0114s22. [DOI] [PubMed] [Google Scholar]
- 98.Cavrois M, Neidleman J, and Greene WC (2014). HIV-1 Fusion Assay. Bio. Protoc. 4, e1212. 10.21769/bioprotoc.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lodge R, Ferreira Barbosa JA, Lombard-Vadnais F, Gilmore JC, Deshiere A, Gosselin A, Wiche Salinas TR, Bego MG, Power C, Routy JP, et al. (2017). Host MicroRNAs-221 and −222 Inhibit HIV-1 Entry in Macrophages by Targeting the CD4 Viral Receptor. Cell Rep. 21, 141–153. 10.1016/j.celrep.2017.09.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at Gene Expression Omnibus (GEO) database under accession GSE226653 and are publicly available as of the date of publication, as listed in the key resources table. Original western blot images have been deposited at Mendeley database (https://data.mendeley.com/datasets/5b38g36z9v/1).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
This paper does not report original code.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal anti-SAMHD1 mouse antibody clone OTI3F5 (30 μL) | Cedarlane | Cat#TA502024S |
| Polyclonal Anti-SAMHD1 (PHOSPHO THR592) (0.02 mg) antibody | Cedarlane | Cat#8005-0.02MG |
| Goat Anti-Mouse IgG (H + L) Secondary Antibody, HRP (1/2000) | Thermo Fisher | Cat#32340; RRID: AB_1185566 |
| Phospho-mTOR (Ser2448) Antibody (1/1000) | Cell Signaling | Cat#2971S; RRID: AB_330970 |
| mTOR (7C10) Rabbit mAb (1/1000) | Cell Signaling | Cat#2983; RRID: AB_2105622 |
| Anti-phospho-ribosomal protein S6 Ser240/Ser244 (1/500) | EMD Millipore | Cat#07-2113 |
| S6 Ribosomal Protein (5G10) Rabbit mAb (1/1000) | Cell Signaling | Cat#2217 |
| TCF4/TCF7L2 (C48H11) Rabbit (1/1000) | Cell Signaling | Cat#2569; RRID: AB_2199816 |
| Monoclonal Anti-β-actin Antibody Produce in Mouse (1/5000) | Sigma | Cat#A5441; RRID: AB_476744 |
| Anti-rabbit IgG HRP-linked Antibody (1/5000) | Cell Signaling | Cat#7074; RRID: AB_2099233 |
| ITGB7 BUV737 (Clone FIB504) | BD | Cat#749664 |
| GSK3B pS9 Antibody, anti-human/mouse/rat REAfinity APC | Miltenyi | Cat#130-106-901; RRID: AB_2651905 |
| Beta Catenin Monoclonal Antibody (15B8), Alexa Fluor 488 | Invitrogen | Cat#53-2567-41; RRID: AB_10807094 |
| Mouse anti-human CD3 Pacific Blue (Clone UCHT1) | BD | Cat#558117; RRID: AB_397038 |
| Mouse anti-human CD4 Alexa Fluor 700 (Clone RPA-T4) | BD | Cat#557922; RRID: AB_396943 |
| Mouse anti-human CD16 Phycoerythrin-Cyanine 7 (Clone 3G8) | BD | Cat#560918; RRID: AB_10563252 |
| Mouse anti-human CD14 APC (Clone M5E2) | BD | Cat#555399; RRID: AB_398596 |
| Anti-human CD1c (BDCA-1) Phycoerythrin (Clone REA694) | Miltenyi | Cat#130-110-536; RRID: AB_2656038 |
| Anti-human HLA-DR Brilliant Violet 785 (Clone L243) | Biolegend | Cat#307642; RRID: AB_2563461 |
| Mouse Anti-human CD195 Fluorescein Isothiocyanate (Clone 2D7/CCR5) | BD | Cat#555992; RRID: AB_396278 |
| CD184 CXCR4 Allophycocyanine (Clone 12G5) | Biolegend | Cat#306510; RRID: AB_314616 |
| HIV-1 core (p24) antigen-RD1 (Clone KC57) | Beckman Coulter | Cat#6604667; RRID: AB_1575989 |
| RARα | Cell signaling | Cat#62294; RRID: AB_2799625 |
| Phospho-Rpb1 CTD (Ser2) | Cell signaling | Cat#13499; RRID: AB_2798238 |
| Phospho-Rpb1 CTD (Ser5) | Cell signaling | Cat#13523; RRID: AB_2798246 |
| RNA polymerase II | Cell signaling | Cat#2629S |
| Rabbit IgG | Cell signaling | Cat#2729; RRID: AB_1031062 |
| CDK9 | Abcam | Cat#ab239364; RRID: AB_3096172 |
| Bacterial and virus strains | ||
| One Shot Stbl3 Chemically Competent E. coli | Life Technologies | Cat# C737303 |
| HIV-1 THRO plasmid (pTHRO.c/2626), subtype B | NIH AIDS Reagent Program (Contribution of Dr. John Kappes and Dr. Christina Ochsenbauer) | Cat#11745 |
| NL4.3BaL HIV plasmid | From Dr. Dana Gabuzda (Dana-Farber Cancer Institute, Boston, MA, USA) | N/A |
| VSV-G Plasmid | National Institution of Health (NIH) | Cat#ARP-4693 |
| NL4.3BaLΔenv GFP | National Institution of Health (NIH) | Cat#ARP-12637 |
| NL4.3Env-Vpr- | From Dr. Eric Cohen | N/A |
| ADA-Env | From Dr. Eric Cohen | N/A |
| Blam-Vpr | From Dr. Eric Cohen | N/A |
| VSV-G-Env | From Dr. Eric Cohen | N/A |
| pNL4.3-IRES-GFP | NIH HIV Reagent Program | Cat#ARP-11349 |
| Biological samples | ||
| Leukapheresis collected from HIV-participants | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium Bicarbonate (NaHCO3) | Sigma | Cat#S5761 |
| Sodium Carbonate (Na2CO3) | Sigma | Cat#223530-500G |
| Thimerosal | Sigma | Cat#T5125-10G |
| Phosphate Buffered Saline (PBS) | Thermo Fisher | Cat#10010023 |
| Tween 20 | Fisher Scientific | Cat#BP337-500 |
| Triton X-100 | Sigma | Cat#X100-500mL |
| Trypan Blue | Thermo Fisher | Cat#15250061 |
| Bovine Serum Albumin (BSA) | BioShop | Cat#ALB001.500 |
| Streptavidin Horseradish Peroxidase (Strep-HRP) | Fisher Scientific | Cat#65R-S104PHRP |
| 3,3′,5,5′-Tetramethylbenzidine (TMB) | Quimigen | Cat#42R-TB10265R-S104PHRP |
| Phosphoric Acid (H3PO4) | Sigma | Cat#PX0996 |
| Nonidet P-40 | Bioshop | Cat#NON505 |
| Tris HCl | BioShop | Cat#TRS002.500 |
| Proteinase K | Fisher Scientific | Cat#25530-015 |
| Molecular Grade Water (H2O) | Wisent | Cat#809-115-CL |
| Guanidine-HC; | Promega | Cat#H5381 |
| Trypsin EDTA with phenol red | Wisent | Cat#075-350 |
| X-tremeGENE HP DNA Transfection Reagent | Roche | Cat#6366244001 |
| Probenecid | Sigma-Aldrich | Cat#P8761 |
| Methanol 99,98% | Fisher Scientific | Cat#BPA4084 |
| ReBlot Plus Strong Antibody Stripping Solution | Sigma | Cat#2504 |
| 4X Laemmli Sample Buffer | Bio-Rad | Cat#1610747 |
| 2-Mercaptoethanol | Sigma | Cat#M6250 |
| Precision Plus Protein Dual Color Standards | Bio-Rad | Cat#1610374 |
| Glycine | BioShop | Cat#GLN001.500 |
| Sodium Chloride | BioShop | Cat#SOD002 |
| 30% Bis-acrylamide Solution | Bioshop | Cat#ACR010.500 |
| UltraPure Tris Buffer | Thermo Fisher | Cat#15504-020 |
| Sodium Dodecyl Sulfate (SDS) | Bio-Rad | Cat#1610302 |
| N,N,N′,N′-Tetramethyl Ethylenediamine (TEMED) | Sigma | Cat#8087420005 |
| Ammonium Persulfate (APS) | Bioshop | Cat#AMP001.100 |
| Radioimmunoprecipitation Assay Buffer (RIPA) 10X | Cell Signaling | Cat#9806S |
| Phosphatase Inhibitor (PhosSTOP) | Roche | Cat#4906845001 |
| Complete, Mini, EDTA-free protease inhibitor | Roche | Cat#11836170001 |
| Formaldehyde solution 37 wt. % in H2O | Sigma | Cat#F1635-500ML |
| Sodium Azide | Bioshop | Cat#SAZ001.250 |
| Fetal Bovine Serum (FBS) | Wisent | Cat#091-150 |
| Dimethyl Sulfoxide (DMSO) | Sigma | Cat#34869-500mL |
| RPMI 1640 Medium (RPMI) | Thermo Fisher | Cat#11875119 |
| All-trans Retinoic Acid (ATRA) | Sigma | Cat#R2625-50MG |
| Sapanisertib (INK128) | Cayman Chemical | Cat#11811-1 |
| PRI-724 (ICG-001) | Selleck | Cat# S8968 |
| PNU-74654 | Selleck | Cat# S8429 |
| Recombinant Human Macrophage Colony Stimulating Factor (M-CSF) | Cedarlane | Cat#216-MC-025 |
| EDTA | Bioshop | Cat#EDT001.1 |
| Phosphate Buffered Saline (PBS) | Thermo Fisher | Cat#10010023 |
| Penicillin/streptomycin (PenStep) | Thermo Fisher | Cat#15140122 |
| Lymphocyte Separation Medium (LSM) | Wisent | Cat#305-010-CL |
| CO2-independent media | Life Technologies | Cat#18045-088 |
| DMEM High glucose, pruvate | Gibco | Cat#11995065 |
| Opti-MEM | Gibco | Cat#31985070 |
| DNAse | Invitrogen | Cat#AM1907 |
| Critical commercial assays | ||
| LDH Assay Kit | Abcam | Cat#ab65393 |
| p24 Enzyme-linked Immunosorbent Assay (ELISA) | Homemade. Hybridome provided by Dr. Michel J. Tremblay95 | N/A |
| QuantiTect SYBR Green RT-PCR Kit | Qiagen | Cat#204243 |
| 10X PCR Buffer/Magnesium Chloride (MgCl2) Buffer/Thermus Aquaticus (TAQ) Polymerase | Thermo Fisher | Cat#18038067 |
| LC480 probe master mix | Roche | Cat#4707494001 |
| QuantiTect SYBR Green RT-PCR Kit | Qiagen | Cat#204243 |
| Deoxynucleoside Triphosphates (dNTP) | Thermo Fisher | Cat#10297018 |
| Promega Wizard Plus DNA Purification System | Fisher Scientific | Cat#PR-A7100 |
| EndoFree Plasmid Maxi Kit | Qiagen | Cat#12362 |
| Memory CD4+ T cell Isolation Kit, human | Miltenyi | Cat#130-091-893 |
| CCF2-AM Substrate and Loading Solution | Life Technologies | Cat#K1032 |
| All Prep DNA/RNA/miRNA Universal Kit | Qiagen | Cat#80224 |
| Detergent Compatible (DC) Protein Assay | Bio-Rad | Cat#5000111 |
| Clarity and Clarity Max ECL Western Blotting Substrates | Bio-Rad | Cat#1705062S |
| Fixation/Permeabilization Kit | BD | Cat#554714 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (405 nm excitation) | Thermo Fisher | Cat#L34957 |
| Pan Monocyte Isolation Kit | Miltenyi | Cat#130-096-537 |
| CHIP assay kit | EMD Millipore | Cat#17-295 |
| Luna® Universal qPCR Master Mix | NEB | Cat# M3003S |
| GeneJET PCR Purification Kit | Thermo Fisher | Cat# K0702 |
| EZ-10 Spin Column Total RNA Miniprep Super Kit | Bio Basic | Cat#BS584 |
| Prime-Script RT reagent kit | TaKaRa | Cat#RR037A |
| Experimental models: Cell lines | ||
| HT-29 | ATCC | Cat#HTB-38 |
| 293T | ATCC | Cat#CRL-3216 |
| ACH-2 | NIH HIV Reagent Program | Cat#ARP-349 |
| THP-1 | NIH HIV Reagent Program | Cat#ARP-9942 |
| Oligonucleotides | ||
| PPARy | Qiagen | GeneGlobal ID: QT00029841 |
| TCF7L2 | Qiagen | GeneGlobal ID: QT00071120 |
| HIC1 | Qiagen | GeneGlobal ID: QT00203175 |
| CTNNB1 | Qiagen | GeneGlobal ID: QT00077882 |
| GSK3β | Qiagen | GeneGlobal ID: QT00057134 |
| CCND2 | Qiagen | GeneGlobal ID: QT00057575 |
| CDK1 | Qiagen | GeneGlobal ID: QT00042672 |
| SAMHD1 | Qiagen | GeneGlobal ID: QT00084322 |
| RARA | Qiagem | GeneGlobal ID: QT00095865 |
| RXRA | Qiagen | GeneGlobal ID:QT00005726 |
| See Table S1: Oligonucleotides | See Table S1: Oligonucleotides | See Table S1: Oligonucleotides |
| Software and algorithms | ||
| FlowJo version 10 | BD | https://www.flowjo.com/ |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/ |
| Image Lab | Bio-Rad | https://www.bio-rad.com/en-ca/product/image-lab-software?ID=KRE6P5E8Z |
| QuantStudio™ Design & Analysis Software | Thermo Fisher | https://www.thermofisher.com/ |
| Other | ||
| MACS LS Columns | Miltenyi | Cat# 130-042-401 |
| Pre-Separation Filters 30 μm | Miltenyi | Cat# 130-041-407 |
| Immobilon-PSQ Polyvinylidene Difluoride (PVDF) | Sigma | Cat#ISEQ00010 |


