Abstract
The pathogenic fungus Cryptococcus neoformans produces an extracellular PLB1 (phospholipase B1), shown previously to be a virulence factor. A novel phospholipase (LPL1) with only LPL (lysophospholipase) and LPTA (transacylase) activities has now been characterized in C. gattii, and found to be a 66-kDa glycoprotein (by SDS/PAGE), with a native molecular mass of 670 kDa. The pI was 6.3, and it was active at high temperatures (to 70 °C), as well as at both acidic and neutral pH values. It was stimulated by calcium and palmitoyl carnitine at pH 7.0, but not at pH 5.0, and palmitoyl lysophosphatidylcholine was the preferred substrate. Sequencing indicated that LPL1 is a novel cryptococcal lysophospholipase, and not the gene product of CnLYSO1 or PLB1. A protein with only LPL and LPTA activities was subsequently isolated from two strains of C. neoformans var. grubii. A PLB1 enzyme was isolated from both C. gattii and a highly virulent strain of C. neoformans var. grubii (H99). In both cases, all three enzyme activities (PLB, LPL and LPTA) were present in one 95–120 kDa glycoprotein (by SDS/PAGE) with pI 3.9–4.3. Characterization of PLB1 from C. gattii showed that it differed from that of C. neoformans in its larger native mass (275 kDa), high PLB activity relative to LPL and LPTA, and preference for saturated lipid substrates. Differences in the properties between the secreted phospholipases of the two cryptococcal species could contribute to phenotypic differences that determine their respective environmental niches and different clinical manifestations.
Keywords: Cryptococcus, phospholipase B1 (PLB1), lysophospholipase 1 (LPL1), secreted glycoprotein, virulence
Abbreviations: DOPC, dioleoyl phosphatidylcholine; DOPE, dioleoyl phosphatidylethanolamine; DOPS, dioleoyl phosphatidylserine; DPPC, dipalmitoyl phosphatidylcholine; IEF, isoelectric focusing; LPL, lysophospholipase; LPTA, LPL transacylase; lyso-PC, lysophosphatidylcholine; lyso-PE, lyso-ethanolamine; lyso-PI, lysophosphatidylinositol; lyso-PS, lysophosphatidylserine; PA, phosphatidic acid; PI, phosphatidylinositol; PLB, phospholipase B; SDA, Sabouraud's dextrose agar
INTRODUCTION
Fungi, such as the basidiomycetous yeast Cryptococcus neoformans, typically cause systemic and potentially fatal disease, especially in immunocompromised hosts. Two species have now been recognised: C. neoformans, which is divided into two varieties, var. neoformans (serotype D) and var. grubii (serotype A), and C. gattii (serotypes B and C). Human disease due to C. gattii is restricted to certain geographic areas, unlike that due to C. neoformans, which has a world-wide distribution. C. gattii is a primary pathogen, affecting immunocompetent hosts almost exclusively, and is virulent in mouse and rat models of cryptococcosis [1,2]. Ecological, genetic and biochemical differences between C. gattii and C. neoformans have also been described [1]. The biological bases of these differences have not been determined.
Enzymes with PLB (phospholipase B, EC 3.1.1.5) activities, but different properties, have been characterized in pathogenic and non-pathogenic fungi [3]. These activities include PLB, which removes both acyl chains from phospholipids, LPL (lysophospholipase), which removes the single acyl chain from lysophospholipids, and LPTA (LPL transacylase), which re-acylates lysophospholipids to form phospholipids. Three proteins with PLB, LPL and LPTA or PLB and LPL activities have been isolated from the non-pathogenic fungus Saccharomyces cerevisiae [4–6]. The PLB1 gene of C. neoformans was proven to be a virulence determinant in a mouse and rabbit model, following gene deletion and reconstitution experiments [7]. The secreted gene product of PLB1 was initially purified from strain BL-1 of C. neoformans var. grubii [8]. This PLB1 protein expressed PLB, LPL and LPTA activities, and was purified as an apparently single acidic glycoprotein with a pH optimum of 4–5 and a molecular mass of 70–90 kDa on SDS/PAGE [8]. We have also cloned a novel LPL gene (CnLYSO1) from C. neoformans var. grubii, which was predicted to code for an extracellular LPL with 21% identity to a human LPL [9].
These findings prompted us to look for phospholipase proteins in C. gattii, where these might differ from those of C. neoformans. We now report the presence of a secreted novel LPL with activity at pH 7.0 in C. gattii, which was also present in two strains of C. neoformans, as well as a secreted PLB1 enzyme with all three activities.
EXPERIMENTAL
Reagents and materials
1-[1-14C]Palmitoyl 2-lyso-PC (1-[1-14C]palmitoyl 2-lysophosphatidylcholine; 56.7 mCi/mmol) and 1,2-di[1-14C]palmitoyl PC (1,2-di[1-14C]palmitoyl phosphatidylcholine; 112 mCi/mmol) were purchased from Amersham Life Science (Little Chalfont, Bucks., U.K.) Carrier lipids, 1-palmitoyl lyso-PC (1-palmitoyl sn-glycero-3-phosphocholine), DPPC (dipalmitoyl phosphatidylcholine) and substances used as substrates were from Sigma (St. Louis, MO, U.S.A.). These included 1-oleoyl lyso-PC (1-oleoyl sn-glycero-3-phosphocholine), 1-palmitoyl lyso-PE (1-palmitoyl sn-glycero-3-phosphoethanolamine), 1-palmitoyl lyso-PI (1-palmitoyl sn-glycero-3-phosphoinositol), DOPC (dioleoyl phosphatidylcholine), DOPS (dioleoyl phosphatidylserine), PI (phosphatidylinositol, from bovine liver), PA (phosphatidic acid), sphingomyelin (from egg yolk), DOPE (dioleoyl phosphatidylethanolamine), triolein and palmitoyl carnitine chloride. Corresponding radiolabelled phospholipids were purchased from Amersham Life Science, with the exception of [palmitoyl-1-14C]-palmitoyl carnitine chloride (40–55 mCi/mmol) which was from Dupont NEN (Boston, MA, U.S.A.). SDA (Sabouraud's dextrose agar) was obtained from Difco Laboratories (Detroit, MI, U.S.A.). Unless specified otherwise, additional reagents were purchased from Sigma and were of analytical grade or the highest available purity.
Cryptococcal strains and culture
Strain TCS-SC1 (serotype B, C. gattii) is an environmental isolate (cultured from Eucalytus teretecornis) from the Westmead Hospital Cryptococcal Collection (T. C. Sorrell and S. C. A. Chen), and strain BL-1 (serotype A, C. neoformans var. grubii) is a clinical isolate from the same collection. The high virulence, high phospholipase-producing strain H99 (serotype A, C. neoformans var. grubii) was a gift from Dr Gary Cox (Department of Medicine, Infectious Disease Division, Duke University Medical Centre, Durham, NC, U.S.A.). All strains were maintained and subcultured on SDA and stored at 4 °C.
Preparation of supernatants
Strains were grown to confluence on 16-cm SDA plates for 72 h at 30 °C in air. Cells were scraped from 80–100 dishes and washed sequentially in saline and harvesting buffer (10 mM imidazole/2 mM CaCl2/2 mM MgCl2/56 mM D-glucose, made up in isotonic saline, pH 5.5) as described previously [8]. For C. gattii, wash steps required centrifugation at 20000 g for 1 h to pellet the cells. Washed cells were resuspended in 50–60 ml of harvesting buffer, and incubated at 37 °C for 24 h. Supernatants were obtained by centrifugation and stored at −70 °C.
Purification of PLB1 protein
C. gattii
The protocol used was based on that reported previously for strain BL-1 [8]. Briefly, the steps involved ammonium sulphate precipitation, hydrophobic chromatography (phenyl-Sepharose CL-4B), anion exchange chromatography (UNO™Q-1), followed by two size-exclusion chromatography steps (Superose 12 HR then Superdex 200 PC). One modification was the substitution of Mes buffer (50 mM Mes/2 mM EDTA, pH 6.3) for 50 mM sodium citrate/2 mM EDTA, pH 6.3, at the first dialysis step, as citrate was found to inhibit C. gattii enzyme activity. Another modification was an increase in the concentration of NaCl from 0.15 M to 0.6 M at the step involving Superose 12 chromatography.
C. neoformans strain H99
This was also carried out as described previously [8], with one modification, which entailed chromatography on a 1 ml HiTrap® Blue Sepharose column (Amersham Biosciences, Sydney, Australia) after passage through the phenyl-Sepharose column. The dialysed phenyl-Sepharose eluate containing activity was concentrated using a 10-kDa cut-off Centriplus™ centrifugal concentrator (Amicon, Millipore Australia Pty. Ltd., North Ryde, NSW, Australia) and loaded on to the Blue Sepharose column equilibrated with 5 vol. of 50 mM Mes/2 mM EDTA buffer, pH 6.3. The PLB1 protein was eluted by a further wash with 5 vol. of the same buffer. Contaminating proteins were retained on the column. The active fractions were then subjected to chromatography on a UNO™Q-1 column, as previously [8], and the PLB1 enzyme eluted between 0.15–0.26 M NaCl. Size-exclusion chromatography was carried out as previously described [8].
Assays for PLB, LPL and LPTA activity
These were carried out by our previously published radiometric assay [8]. Total protein concentration was determined using a Coomassie-Blue-binding assay (Pierce Chemical Co., Rockford, IL, U.S.A.) method using BSA as a standard.
Characterization of enzyme activities
All experiments were performed in duplicate using 0.1–0.2 μg of pure enzyme. DPPC or 1-palmitoyl lyso-PC were used as substrates, unless otherwise specified. Heat-stability was determined by measurement of residual activity at 37 °C after 10 min pre-incubation at various temperatures, unless stated otherwise. The effect of pH was determined over the range pH 3.5–8.5 using an imidazole acetate buffer system (50 mM final concentration).
Modifying agents and cations were added to reaction mixtures at the desired concentration following addition of the substrate. Cation solutions were prepared in deionized water. Other agents were prepared as stock solutions in 50 mM imidazole/acetate buffer, at pH 4–5 or pH 7.0, with the exception of palmitoyl carnitine, which was prepared in methanol. The final concentration of methanol added was <0.1% (v/v) and was also present in the controls.
Determination of released non-esterified fatty acid by colorimetric assay
Where suitable radiolabelled lysophospholipid substrates were not available, reaction products of LPL and LPTA activity were extracted by the method of Bligh and Dyer [10] and evaporated under nitrogen. The relative levels of non-esterified fatty acid in each sample were determined using the acyl-CoA-oxidase system assay kit (Boehringer Mannheim, Mannheim, Germany).
SDS/PAGE and IEF (isoelectric focusing)
Analytical SDS/PAGE was performed by the method of Laemmli [11] under reducing conditions in 4–20% linear gradient polyacrylamide gels (Novex, San Diego, CA, U.S.A.). Gels were stained with Coomassie Brilliant Blue R-250 and silver (Novex Silver Express; Novex) for proteins, and with the glycoprotein staining kit (Pierce). The apparent molecular mass was estimated by SDS/PAGE and gel-filtration chromatography using the Superdex 200 PC 3.2/30 column. In the latter case the column was calibrated with molecular-mass-marker protein standards from Bio-Rad Laboratories (Regent's Park, NSW, Australia) [thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa) and vitamin B-12 (1.35 kDa)].
Two-dimensional chromatography (IEF/PAGE) was carried out using IEF on a non-linear 7-cm Immobiline DryStrip, pH 3.0–10.0, (Amersham Biosciences) using carrier ampholytes of 0.5%, pH 3.0–10.0 (Biolytes; Bio-Rad), for a total of 12 kV/h. Proteins were then run on a 10–20% gradient mini-gel (Bio-Rad) and visualized with Sypro-Ruby or Coomassie Blue R-250.
Amino acid sequencing
The N-terminal amino acid sequence of LPL1 was determined by the method of Matsudaira [12]. Briefly, 15–20 μg of pure protein was separated by SDS/PAGE or IEF/PAGE. The bands/spots of interest were extracted from the gel by mixing overnight in 100 mM sodium acetate/0.1% (w/v) SDS/10 mM dithiothreitol, and passive adsorption on to a PVDF membrane. Sequencing was performed with an ABI model 494 gas-phase sequencer (Applied Biosystems, Foster City, CA, U.S.A.).
Preparation of pivaloyl column
Trimethylacetic acid (pivalic acid; Aldrich, Castle Hill, NSW, Australia) was coupled to EAH (epoxy-aminohexyl)–Sepharose 4B (Amersham Biosciences) using 98% 1-[3-dimethylamino)-propyl]-3-ethylcarbodiimide hydrochloride (Aldrich), using the protocol recommended by Amersham. The washed gel was stored in 20% ethanol at 4 °C.
Chitin deacetylase activity
This was measured by two different colorimetric methods. Firstly, by determining acetic acid production from the substrate hexa-N-acetyl-chitohexaose (Sigma) [13,14], and secondly by measuring the concentration of glucosamine residues produced by the deacetylation reaction [15,16].
RESULTS
Purification of a novel lysophospholipase protein (LPL1) from C. gattii
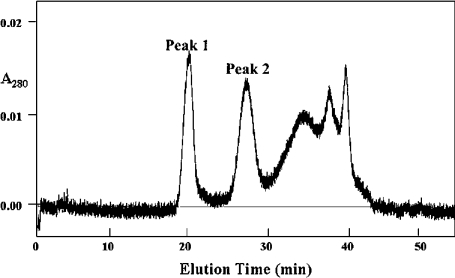
During the Superose chromatography step of the PLB1 purification (see the Experimental section) a second protein peak was detected (Figure 1, peak 1), which eluted before the PLB1 peak (Figure 1, peak 2). Assays of peak 1 revealed no PLB was present, but it contained LPL and LPTA activity (LPTA/LPL ratio, 0.5–0.7).
Figure 1. Separation of LPL1 (peak 1) and PLB1 (peak 2) from C. gattii by Superose 12 HR 10/30 column chromatography.
Freeze-dried active fractions post-UNO™Q-1 chromatography [8] were reconstituted in 50 mM Mes/2 mM EDTA/0.6 M NaCl, pH 6.3, and applied to the Superose column equilibrated in the same buffer. Proteins were eluted at a flow rate of 0.5 ml/min, and fractions were collected and assayed for LPL, LPTA and PLB activities.
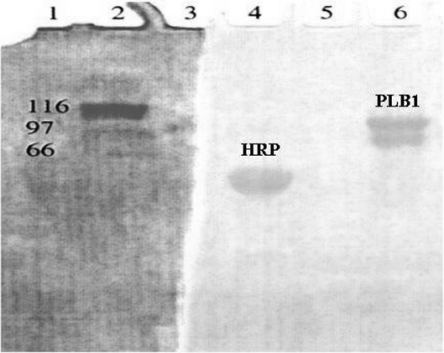
The LPL1 protein occurred on SDS/PAGE as a broad band at approx. 66 kDa which stained for glycoprotein (Figure 2). The molecular mass determined by size-exclusion chromatography was very large (670 kDa), and the isoelectric point on IEF/PAGE was around 6.2. Only one major spot was obtained, suggesting the enzyme was relatively pure (results not shown), and the increase in specific activity above the LPL and LPTA in the crude supernatant was 44-fold. The N-terminal sequence (LSINTPASIV) of the secreted protein was compared with the C. neoformans serotype D database (http://www.TIGR.org), and two good matches were obtained.
Figure 2. SDS/PAGE of purified LPL1 from C. gattii.
The peak 1 protein obtained from the Superose 12 column in Figure 1, containing LPL and LPTA activities, was subjected to SDS/PAGE and stained for carbohydrate, as described in the Experimental section. Horseradish peroxidase (HRP) was used as a positive control for glycoprotein. Molecular-mass markers were stained with Coomassie Blue.
The first matching protein, which contains eight of the ten LSINTPASIV residues (TIGR database search results: 186.m0373 expressed protein, chromosome 2; CNB02410), has a predicted N-terminal leader peptide sequence, indicative of a secretory protein, and contains a lipase-like motif (GXSXG) (these sequences are indicated in bold): MFTKAAIVVALAGTVNAALSINTPASLIECQPAALSWSGGSSTPYYLAVLPGGQVSATALENIDTVDTESYTWTVNLASGTNITIRVTDGSGNIAYSSPVVIQEGSSSSCLTSSSSSSATAAAGSSSGDSSASTTASGSSASSGSSGSSASTTASGSSASASSSSSSSDSGALLTKGNAGVAASLVGIVAAAFAAVA.
The second protein contains nine of the ten LSINTPASIV residues (TIGR database search results: 184.m04411 hypothetical protein, chromosome 4; CND00830) and also has a predicted N-terminal leader sequence (shown in bold type), as follows: MHVSNLVALAALAGSAAALTINTPASIVECQPASITFSDGTSPYILAAIPGGQVSAAAIETINDSLTTSPYTWTVNLAAGTNITLKITDATGTIAYSSPIVVQAGSSQSCINATASTSGLSTAAVTTTASDTSAAGGGAATTSASESSSSSSAAQSSSEASSAAASSSSSASSAASTTAASSTGAASSATSAAATSASGTSGAFANAVVGVPALVAGLFAGVAALL.
Neither protein had significant similarity to the amino acid sequence predicted by the CnLYSO1 gene sequence from serotype D [9], thus we conclude that this gene does not encode LPL1.
Characterization of LPL1
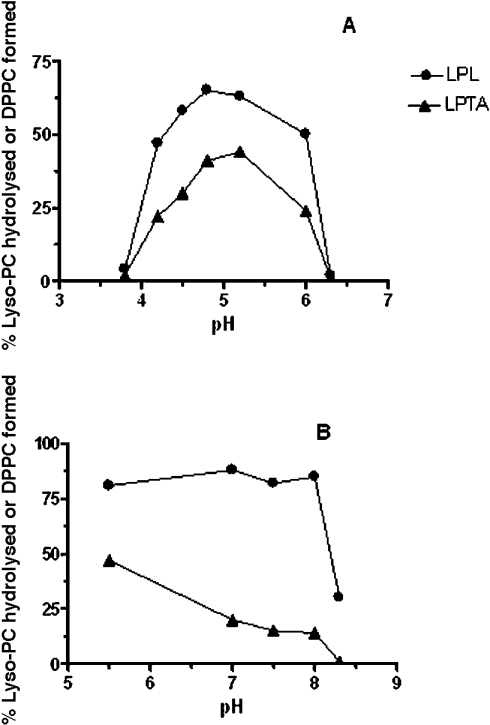
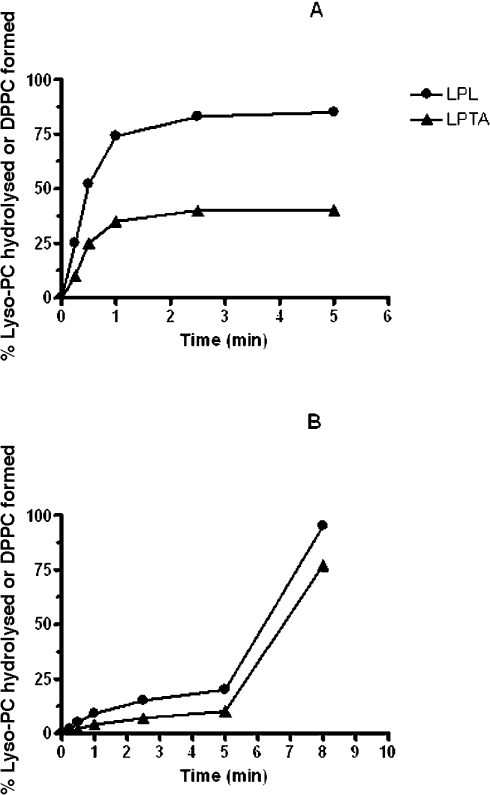
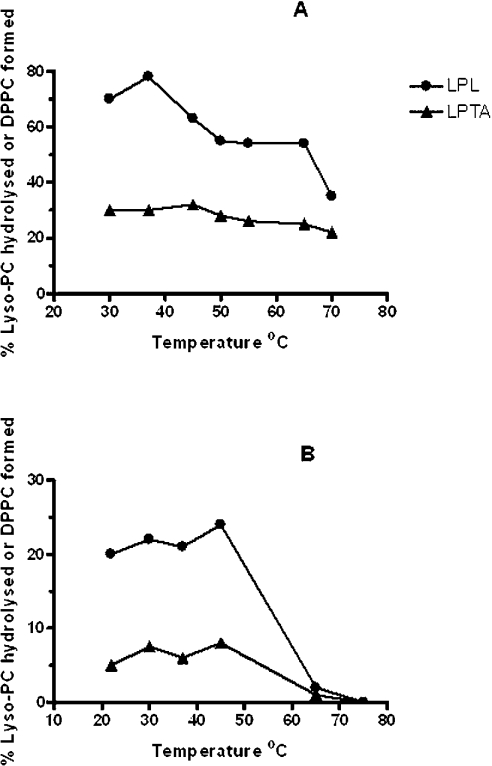
Linearity of activity with enzyme concentration and assay time was observed to 1 μg of protein and 30 s respectively. Using lyso-PC as substrate, 20 μM was the optimal concentration and the pH curves showed clear optima at around pH 5.0 for both LPL and LPTA (Figure 3A). However, activity was also detected at pH 7.0 when the incubation time was extended to 5–7 min. Further experiments showed that this longer incubation time resulted in a dramatic change in response to pH, with a broad optimum from pH 5.5 to 8.0 for LPL (Figure 3B). This effect was due to a lag in the time course of activity at high pH relative to pH 5.0 (Figures 4A and 4B). Another novel feature of this enzyme was its response to assay temperature, which although optimal at 37 °C, displayed considerable activity at 70 °C (Figure 5A). Stability to pre-incubation at high temperatures was also marked, with little change in activity after pre-incubation for 10 min between 37–50 °C, although no activity remained after pre-incubation at 65 °C (Figure 5B).
Figure 3. Effects of pH on LPL1 activities from C. gattii measured using 30 s (A) or 5 min (B) incubation time.
Enzyme activities were measured as described in the Experimental section by radiometric analysis using 1-palmitoyl lyso-PC as substrate. Results are typical of two or more experiments and are expressed as the percentage of lyso-PC hydrolysed (LPL) or percentage conversion to DPPC (LPTA).
Figure 4. Effects of incubation time on LPL1 activities from C. gattii at pH 5.0 (A) and pH 7.0 (B).
Enzyme activities were measured as described in the Experimental section by radiometric analysis using 1-palmitoyl lyso-PC as substrate. Results are typical of two or more experiments and are expressed as the percentage of lyso-PC hydrolysed (LPL) or percentage conversion to DPPC (LPTA).
Figure 5. Effects of temperature on LPL1 activities from C. gattii.
The effects of increasing assay temperatures (incubation time 30 s) are shown (A), and the effects of pre-incubation for 10 min at increasing temperatures before assay at 37 °C for 30 s are shown (B). Enzyme activities were measured as described in the Experimental section by radiometric analysis using 1-palmitoyl lyso-PC as substrate. Results are typical of at least two experiments and are expressed as the percentage of lyso-PC hydrolysed (LPL) or percentage conversion to DPPC (LPTA).
Relative substrate specificities measured at pH 5.0 were: 1-palmitoyl lyso-PC, 100%; 1-oleoyl lyso-PC, 30%; 1-palmitoyl lyso-PE, 20%; lyso-PI, 10%. No activity was detected with lyso-PS, and substrates were not tested at higher pH values. The effects of modifying agents using 1-palmitoyl lyso-PC as substrate are shown in Table 1. In some instances, differences were observed depending on whether the assay was conducted at pH 5.0 for 30 s or pH 7.0 for 7 min. Both calcium and palmitoyl carnitine stimulated activity at pH 7.0, compared with no effect, or strong inhibition, at pH 5.0. Triton X-100 and PA were more inhibitory at pH 7.0, whereas DPPC was less so.
Table 1. Effect of modifying agents on the LPL1 enzyme from C. gattii.
Assays for enzyme activities were performed at pH 5.0 or pH 7.0 using 1-palmitoyl lyso-PC as substrate. The final concentrations of modifiers in the assays are shown in the Table. The results are expressed as the percentage of the control values (100%), which were 37.5±1.9 (pH 5.0) and 2.9±0.5 (pH 7.0) μmol/min per mg of protein. Experiments were performed at least twice and produced similar results.
| Relative LPL activity (%) | ||||
|---|---|---|---|---|
| Metal ion/reagent | Concentration (mM) | pH… | 5.0 | 7.0 |
| Control | 100 | 100 | ||
| CaCl2 | 10 | 96 | 260 | |
| FeCl3 | 10 | 61 | 62 | |
| EGTA | 10 | 98 | 85 | |
| Dithiothreitol | 10 | 122 | 91 | |
| Triton X-100 | 0.1* | 52 | BLD | |
| Carnitine | 10 | 121 | 91 | |
| Palmitoyl carnitine | 0.5 | 10 | 157 | |
| DPPC | 0.5 | 16 | 50 | |
| Sphingomyelin | 0.5 | 79 | 71 | |
| PA | 0.5 | 76 | 34 | |
* Concentration is a percentage (w/v).
Detection of LPL1 in C. neoformans var. grubii
A protein with only LPL and LPTA activities was subsequently isolated from two strains of C. neoformans var. grubii. During purification of PLB1 from strain H99 (see the Experimental section and below), a separate protein was found during ion exchange (UNO™Q-1) chromatography to elute at a higher concentration of NaCl (0.23–0.45 M) than that required for PLB1 (0.15–0.26 M). This protein had only LPL and LPTA activity at pH 4–5 (LPTA/LPL ratio of 0.6) and a trace amount of PLB. The native molecular mass was determined to be 220 kDa by size-exclusion chromatography (Sephadex 200 column), as described in the Experimental section.
A column made of a fatty acid analogue (pivalic acid) immobilized on Sepharose was observed to separate the LPL and LPTA activities from PLB1 produced by C. neoformans strain BL-1. The post-phenyl-Sepharose fraction (see the Experimental section) was dialysed against 50 mM Mes/2 mM EDTA buffer, pH 4.0, and applied to the pivaloyl column equilibrated in the same buffer. LPL and LPTA activities only (LPTA/LPL ratio of 0.5) were detected in the flow-through and wash from the column; the PLB1 protein was eluted in buffer containing 15% (w/v) ammonium sulphate.
Purification of PLB1 from C. gattii
Purification of PLB1 protein from the TCS-SC1 strain of C. gattii produced a double band (90–105 kDa) on SDS/PAGE which stained positive for glycoprotein (Figure 6). The lower of the two bands was identified by amino acid sequencing as chitin deacetylase precursor, a contaminant also sometimes present in serotype A preparations [8]. Because chitin deacetylase has the capacity to remove acetyl groups it was deemed possible that it contributed part of the PLB1 enzyme activity. However, the serotype A plb1 deletion mutant, HCM5, compared with the wild-type H99, had negligible PLB1 enzyme activity [7], but the measured activity of chitin deacetylase was not diminished in HCM5, making its contribution to PLB1 activity unlikely (F. Widmer, unpublished work).
Figure 6. SDS/PAGE of purified PLB1 from C. gattii.
The pure proteins (lane 6) were separated by SDS/PAGE and stained for glycoprotein as described in the Experimental section. Horseradish peroxidase (HRP) was included as a positive control for glycoprotein. The molecular-mass standards were stained by Coomassie Blue.
The C. gattii PLB1 protein had a molecular mass of 275 kDa by size-exclusion chromatography and an acidic isoelectric point (3.9), determined by IEF/PAGE (results not shown). These findings are in agreement with the putative protein sequence derived from the C. gattii PLB1 cDNA [17], which predicted an isoelectric point of 4.12 and a molecular mass of 68.3 kDa. However, the presence of 17 possible N-glycosylation sites and 4 possible O-glycosylation sites suggests that the actual mass of the protein could be as high as that found for the purified protein, and the native molecular mass suggests it consists of two or three subunits. All three enzyme activities (PLB, LPL and LPTA) were present, and their specific activities were increased approx. 100-fold by the purification process.
Characterization of C. gattii PLB1
Linearity of activity with enzyme concentration was maintained to at least 0.2 and 0.1 μg of purified protein for PLB and LPL/LPTA activities, and reaction times were linear to 10 min and 30 s respectively. Optimum substrate concentrations were 500 μM and 40 μM for PLB and LPL/LPTA assays respectively, with excess substrate being strongly inhibitory. All three activities were found to be optimal at 37 °C and pH 4.5–5.0, with no activity detectable at high temperatures or pH 7.0. However, when the incubation time for the LPL/LPTA assays was extended from 30 s to 5–10 min, activity was detected at pH 7.0 (about 5% of that at pH 5.0; Table 2). No PLB activity was detected at pH 7.0.
Table 2. Substrate specificities for the PLB1 enzyme from C. gattii.
PLB and LPL activities were measured by the radiometric and colorimetric assays respectively as described in the Experimental section. The reactions were performed using 500 μM of the various phospholipids at pH 5.0, and 40 μM of the lysophospholipids at pH 5.0 (30 s incubation) and pH 7.0 (5 min incubation). Results are expressed as the percentage of the control value (100%), where this represents a specific activity of 12.5±1.5 μmol/min per mg of protein for PLB (DPPC as substrate) and 82.8±9.4 μmol/min and 3.5±0.3 μmol/min per mg of protein for LPL at pH 5.0 or pH 7.0. BLD, below the limits of detection.
| Relative activity (%) | ||||
|---|---|---|---|---|
| LPL | ||||
| Substrate | PLB | pH… | 5.0 | 7.0 |
| DPPC | 100 | - | - | |
| DOPC | 2 | - | - | |
| DOPE | 11 | - | - | |
| DOPS | BLD | - | - | |
| PA | PA | - | - | |
| PI | BLD | - | - | |
| Sphingomyelin | 3 | - | - | |
| Triolein | BLD | - | - | |
| Palmitoyl carnitine | BLD | - | - | |
| 1-Palmitoyl lyso-PC | - | 100 | 100 | |
| 1-Oleoyl lyso-PC | - | 30 | 667 | |
| Lyso-PI | - | BLD | BLD | |
| 1-Palmitoyl lyso-PE | - | BLD | 100 | |
| 1-Palmitoyl lyso-PS | - | 70 | 100 | |
The substrate specificities for PLB and LPL activities are presented in Table 2 (substrate specificities for LPTA were similar to those of LPL and are not shown). PLB activity was maximal against the fully saturated DPPC, and not significant against DOPS, PA, PI, triolein or sphingomyelin. Interestingly, the substrate preference for LPL activity was pH-dependent, changing from 1-palmitoyl lyso-PC at pH 5.0 (assay time, 2 min) to 1-oleoyl lyso-PC and 1-palmitoyl lyso-PE at pH 7.0 (assay time, 5 min; Table 2).
The effects of modifying agents are shown in Table 3. LPTA activity followed a similar trend to LPL activity (results not shown). None of the activities was stimulated by calcium; all were inhibited by Triton X-100, PA and sphingomyelin. Ferric chloride and carnitine inhibited PLB activity, and the PLB substrate DPPC inhibited LPL activity, whereas the LPL substrate, lyso-PC, slightly stimulated PLB activity. The effects of modifying agents on LPL were the same at pH 5.0 or 7.0, except for palmitoyl carnitine which inhibited at pH 5.0, but strongly stimulated LPL activity at pH 7.0 (Table 3).
Table 3. Effect of modifying agents on the PLB1 enzyme from C. gattii.
Assays for enzyme activities were performed using DPPC at pH 5.0 and 1-palmitoyl lyso-PC at pH 5.0 and pH 7.0 as substrates. The final concentrations of modifiers in the assays are shown in the Table. The results are expressed as the percentage of the control value (100%), where this value was the same as described in Table 2. Experiments were performed at least twice with similar results.
| Relative activity (%) | |||||
|---|---|---|---|---|---|
| LPL | |||||
| Metal ion/reagent | Concentration (mM) | PLB | pH… | 5.0 | 7.0 |
| Control | 100 | 100 | 100 | ||
| CaCl2 | 10 | 78 | 108 | 104 | |
| FeCl3 | 10 | 20 | 83 | 71 | |
| EGTA | 10 | 111 | 99 | 102 | |
| Dithiothreitol | 10 | 89 | 100 | 111 | |
| Triton X-100 | 0.1* | 15 | 9 | 5 | |
| Carnitine | 10 | 62 | 101 | 108 | |
| Palmitoyl carnitine | 1 | 36 | 35 | 239 | |
| DPPC | 0.5 | - | 11 | 18 | |
| Lyso-PC | 0.5 | 132 | - | - | |
| Sphingomyelin | 0.5 | 65 | 55 | 72 | |
| PA | 0.5 | 42 | 8 | 13 | |
* Concentration is a percentage (w/v).
Purification of PLB1 from C. neoformans var. grubii strain H99
H99 is a highly virulent strain of C. neoformans which secretes large amounts of PLB1 activity [18]. Purification produced a glycoprotein with an acidic isoelectric point (approx. 4.3) and molecular mass of approx. 110–120 kDa on IEF/PAGE (results not shown). The contaminant chitin deacetylase was not detected after IEF/PAGE, and was probably removed during purification at the additional Blue Sepharose step. Size-exclusion chromatography (Sephadex 200) revealed a native molecular mass for PLB1 of 110–120 kDa. The H99 enzyme demonstrated all three of the PLB, LPL and LPTA activities (LPL/PLB ratio, 22.5; LPTA/LPL ratio, 0.4). It was not further characterized.
DISCUSSION
In the present study, we have demonstrated the presence of two secreted phospholipase proteins, PLB1 and a novel LPL1, in Cryptococcus gattii, and have confirmed that a protein with the LPL1 enzyme activity is also produced by C. neoformans var. grubii.
We reported previously that the PLB1 genes for all four cryptococcal serotypes have considerable similarity to each other (at least 85%, [17]); however, the PLB1 protein isolated from C. gattii displays some important differences from that of C. neoformans var. grubii strain BL-1 [8]. C. gattii PLB1 is larger (native molecular mass of 275 kDa compared with 160–180 kDa for C. neoformans PLB1). C. neoformans PLB1 had an LPL/PLB ratio of 53, and LPTA/LPL ratio of 0.4–0.5 [8]. For PLB1 from the type strain of C. neoformans var. grubii, H99 (see above), these ratios were 22.5 and 0.3–0.4. In contrast the C. gattii PLB1 enzyme had an LPL/PLB ratio of 6.6 and an LPTA/LPL ratio of 0.3–0.4, which may indicate a predilection for the C. gattii enzyme to degrade phospholipids, rather than lysophospholipids. Although it is possible that more LPL and LPTA were lost during purification of the C. gattii enzyme, this is unlikely, because PLB activity is more labile during purification (L. C. Wright, unpublished work). It has been shown that PLB activity can be lost from the PLB enzyme of Penicillium notatum by proteolysis [19]. Although its mass on SDS/PAGE is smaller than PLB1, LPL1 is unlikely to be a proteolytic product of PLB1, since its native molecular mass (670 kDa) is much larger, and it has different properties (such as pI closer to neutral, stimulation by calcium at pH 7.0).
The PLB1 phospholipid substrate specificities were also very different, with C. gattii PLB1 being specific for DPPC, as opposed to the broad spectrum of phospholipids (including DOPC) attacked by the C. neoformans PLB1 enzyme. Unlike 1-oleoyl lyso-PC for the LPL of C. neoformans PLB1 [8], the LPL substrate preferred by C. gattii PLB1 was the saturated 1-palmitoyl lyso-PC. These observations are of interest, since human infection with C. gattii typically presents with large lung lesions (‘cryptococcomas’) and DPPC is abundant in the surfactant lining the small air spaces of the lung. Furthermore, host PLA (phospholipase A) activity in the lung may generate the potentially toxic pore-forming agent palmitoyl lyso-PC [20], which could be degraded rapidly by cryptococcal PLB. All three of the C. gattii PLB1 activities were less sensitive to inhibition by FeCl3, and were strongly inhibited by Triton X-100, whereas the PLB activity of C. neoformans PLB1 was mildly stimulated by this detergent.
Previously we detected some LPL and LPTA activity (but traces only of PLB) at pH 7.0 in crude cell-free supernatants from C. neoformans serotype D [9] and C. neoformans var. grubii (serotype A, strains H99 and BL1) [18]. We have now shown by purification of PLB1 from C. gattii that part of the activity detected in these crude preparations at a pH close to physiological levels is due to PLB1. The substrate specificity for the LPL and LPTA activities of PLB1 at pH 7.0, in contrast to that at acidic pH, includes unsaturated lysophospholipids and amino-lysophospholipids. This suggests that apart from using host lipids for its own growth, the fungus could protect itself within host tissues and blood, by re-acylating potentially toxic lysophospholipids and fatty acids formed by host PLA activity, and repairing its own damaged membranes. Phagocytes, for example, may attack the fungal membrane outer leaflet (rich in choline-containing lipids), and on activation also produce arachidonic acid [21], which initiates a cascade of fungicidal signals. Removal of this arachidonic acid by fungal re-acylation would be an important immunological silencing mechanism. For cryptococci to escape from within host cells, the inner leaflet (rich in aminophospholipids) would need to be degraded and the breakdown products re-acylated to avoid damage to the fungus.
Most of the LPL and LPTA activity at pH 7 and higher, however, comes from the novel enzyme LPL1. For PLB1, the ratio of LPL activity at pH 5.0 compared with that at pH 7.0 is about 24:1, but with LPL1 this can be as high as 1:1 (see Figure 3). The LPL1 peptide sequence has no similarity to the predicted PLB1 gene product. Furthermore, the two proteins predicted from the cryptococcal database which contained the LPL1 peptide sequence are very small (molecular masses of 24–28 kDa), suggesting that neither is likely to be the LPL1 protein, although one is possibly a lipase. Alternative explanations are possible for this size discrepancy. The annotated cryptococcal genome database we interrogated was from C. neoformans serotype D, not C. gattii. It is possible that a longer sequence will be found when the C. gattii database is completed and the genome subsequently annotated. In addition, LPL1 could be highly glycosylated, such as found for the PLB1 from Saccharomyces cerevisiae [22] or Kluyveromyces lactis [23]. Alternatively, the protein could be made up of a number of tandem repeats of similar domains, such as has been described for rat intestinal PLB/lipase [24], and for which the sequence of only one repeat may be given in the serotype D cryptococcal database. As with PLB1, it is probable that LPL1 is a multimeric protein, with up to ten subunits aggregating to give the high native molecular mass. Slow aggregation at physiological pH would account for the longer incubation time required to reach maximum activity.
Proteins with only LPL and LPTA activity have been purified from other fungi, for example Candida has three secreted calcium-independent LPL activities [25,26] with pH optima around 5.5–6.0, which are inhibited by Triton X-100. Three phospholipases have been characterized in Saccharomyces cerevisiae [5,6], but only the PLB1 and PLB2 gene products have substantial LPL activity. The PLB activity of the product of PLB3 was stimulated by calcium. Interestingly, the yeasts Kluyveromyces lactis [23] and Torulaspora delbrueckii [27] secrete a single PLB enzyme containing PLB, LPL and LPTA activities with two pH optima (2.0–2.5 and 7.5). At alkaline pH the activities all required calcium, and the substrate specificity was different. In contrast to the cryptococcal and other fungal enzymes, Fe3+ stimulated activity at alkaline pH in K. lactis.
Although we have identified a secreted protein with only LPL and LPTA activities in two strains of C. neoformans var. grubii, we cannot be certain that these proteins are identical with LPL1, since they were not sufficiently characterized or sequenced. Similarly, the product of the CnLYSO1 gene described in C. neoformans serotype D has not been characterized. LPL1 may be the source of the residual LPL activity after knock-out of the PLB1 gene ([7], and R. T. Santangelo, unpublished work). It has been proposed that PLB is involved in turnover and remodelling of membrane phospholipids [5], and it has a role in Schizosaccharomyces pombe in signal transduction in response to stress [28]. It seems logical, therefore, that several different phospholipases with different substrate specificities and other characteristics are necessary to survive a wide range of environmental stresses. The fact that LPL1 is active even at very high temperatures could be of advantage to a cryptococcal species such as C. gattii, which is mostly found in tropical and subtropical regions of the world.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia (grants #211040 and #990738). We thank the Australian Proteome Analysis Facility, North Ryde, Australia, for the protein sequencing and two-dimensional gel electrophoresis, and Dr Julie Djordjevic and Dr John Coe for their helpful advice.
References
- 1.Sorrell T. C. Cryptococcus neoformans variety gattii. Med. Mycol. 2001;39:155–168. [PubMed] [Google Scholar]
- 2.Chen S. C. A., Muller M., Zhou J. Z., Wright L. C., Sorrell T. C. Phospholipase activity in Cryptococcus neoformans: a new virulence factor? J. Infect. Dis. 1997;175:414–420. doi: 10.1093/infdis/175.2.414. [DOI] [PubMed] [Google Scholar]
- 3.Ghannoum M. Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 2000;13:122–143. doi: 10.1128/cmr.13.1.122-143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee K., Patton J., Fido M., Hines L., Kohlwein S., Paltauf F., Henry S., Levin D. The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J. Biol. Chem. 1994;269:19725–19730. [PubMed] [Google Scholar]
- 5.Merkel O., Fido M., Mayr J. A., Pruger H., Raab F., Zandonella G., Kohlwein S. D., Paltauf F. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:28121–28127. doi: 10.1074/jbc.274.40.28121. [DOI] [PubMed] [Google Scholar]
- 6.Fyrst H., Oskouian B., Kuypers F. A., Saba J. D. The PLB2 gene of Saccharomyces cerevisiae confers resistance to lysophosphatidylcholine and encodes a phospholipase B/lysophospholipase. Biochemistry. 1999;38:5864–5871. doi: 10.1021/bi9824590. [DOI] [PubMed] [Google Scholar]
- 7.Cox G. M., McDade H. C., Chen S. C. A., Tucker S. C., Gottfredsson M., Wright L. C., Sorrell T. C., Leidich S. D., Casadevall A., Ghannoum M. A., Perfect J. R. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen S. C. A., Wright L. C., Golding J. C., Sorrell T. C. Purification and characterisation of secretory phospholipase B, lysophospholipase-transacylase from a virulent strain of the pathogenic fungus Cryptococcus neoformans. Biochem. J. 2000;347:431–439. doi: 10.1042/0264-6021:3470431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coe J., Wilson C., Sorrell T. C., Latouche N., Wright L. C. Cloning of CnLYSO1, a novel extracellular lysophospholipase of the pathogenic fungus Cryptococcus neoformans. Gene. 2003;316:67–78. doi: 10.1016/s0378-1119(03)00740-6. [DOI] [PubMed] [Google Scholar]
- 10.Bligh E. C., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 13.Kafetzopoulos D., Martinou A., Bouriotis V. Bioconversion of chitin to chitosan: purification and characterization of chitin deacetylase from Mucor rouxii. Proc. Natl. Acad. Sci. U.S.A. 1993;90:2564–2568. doi: 10.1073/pnas.90.7.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao X., Katsumoto T., Onodera K. Purification and characterization of chitin deacetylase from Absidia coerulea. J. Biochem. (Tokyo) 1995;117:257–263. doi: 10.1093/jb/117.2.257. [DOI] [PubMed] [Google Scholar]
- 15.Tokuyasu K., Ohnishini-Kameyama M., Hayashi K. Purification and characterization of extracellular chitin deacetylase from Colletotrichum lindemuthianum. Biosci. Biotechnol. Biochem. 1996;60:1598–1603. doi: 10.1271/bbb.60.1598. [DOI] [PubMed] [Google Scholar]
- 16.Dische Z., Borenfreund E. A spectrophotometric method for the microdetermination of hexosamines. J. Biol. Chem. 1950;184:517–522. [PubMed] [Google Scholar]
- 17.Latouche G. N., Sorrell T. C., Meyer W. Isolation and characterisation of the phospholipase B gene of Cryptococcus neoformans var gattii. FEMS Yeast Res. 2002;2:551–561. doi: 10.1111/j.1567-1364.2002.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 18.Wright L. C., Chen S. C. A., Wilson C. F., Simpanya M. F., Blackstock R., Cox G., Murphy J. W., Sorrell T. C. Strain-dependent effects of environmental signals on the production of extracellular phospholipase by Cryptococcus neoformans. FEMS Microbiol. Lett. 2002;209:175–181. doi: 10.1111/j.1574-6968.2002.tb11128.x. [DOI] [PubMed] [Google Scholar]
- 19.Masuda S., Kitamura N., Saito K. Primary structure of protein moiety of Penicillium notatum phospholipase B deduced from the cDNA. Eur. J. Biochem. 1991;202:783–787. doi: 10.1111/j.1432-1033.1991.tb16433.x. [DOI] [PubMed] [Google Scholar]
- 20.Flieger A., Rydzewski K., Banerji S., Broich M., Heuner K. Cloning and characterization of the gene encoding the major cell-associated phospholipase A of Legionella pneumophila, plaB, exhibiting hemolytic activity. Infect. Immun. 2004;72:2648–2658. doi: 10.1128/IAI.72.5.2648-2658.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castro M., Ralston N. V. C., Morgenthaler T. I., Rohrbach M. S., Limper A. H. Candida albicans stimulates arachidonic acid liberation from alveolar macrophages through α-mannan and β-glucan cell wall components. Infect. Immun. 1994;62:3138–3145. doi: 10.1128/iai.62.8.3138-3145.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witt W., Schweingruber M. E., Mertschung A. Phospholipase B from the plasma membrane of Saccharomyces cerevisiae; separation of two forms with different carbohydrate content. Biochim. Biophys. Acta. 1984;795:108–116. doi: 10.1016/0005-2760(84)90110-3. [DOI] [PubMed] [Google Scholar]
- 23.Oishi H., Morimoto T., Watanabe Y., Tamai Y. Purification and characterization of phospholipase B from Kluyveromyces lactis, and cloning of phospholipase B gene. Biosci. Biotechnol. Biochem. 1999;63:83–90. doi: 10.1271/bbb.63.83. [DOI] [PubMed] [Google Scholar]
- 24.Takemori H., Zolotaryov F. N., Ting L., Urbain T., Komatsubara T., Hatano O., Okamoto M., Tojo H. Identification of functional domains of rat intestinal phospholipase B/lipase. J. Biol. Chem. 1998;273:2222–2231. doi: 10.1074/jbc.273.4.2222. [DOI] [PubMed] [Google Scholar]
- 25.Mirbod F., Banno Y., Ghannoum M. A., Ibraham A. S., Nakashima S., Kitajima Y., Cole G. T., Nozawa Y. Purification and characterization of lysophospholipase-transacylase (h-LPTA) from a highly virulent strain of Candida albicans. Biochim. Biophys. Acta. 1995;1257:181–188. doi: 10.1016/0005-2760(95)00072-k. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi M., Banno Y., Nozawa Y. Secreted Candida albicans phsopholipases: purification and characterization of two forms of lysophospholipase-transacylase. J. Med. Vet. Mycol. 1991;29:193–204. [PubMed] [Google Scholar]
- 27.Kurawabara Y., Maruyama M., Watanabe Y., Tanaka S., Tamai Y. Purification and some properties of membrane-bound phospholipase B from Torulaspora delbrueckii. J. Biochem. 1988;104:236–241. doi: 10.1093/oxfordjournals.jbchem.a122449. [DOI] [PubMed] [Google Scholar]
- 28.Yang P., Du H., Hoffman C. S., Marcus S. The phospholipase B homolog Plb1 is a mediator of osmotic stress response and of nutrient-dependent repression of sexual differentiation in the fission yeast. Mol. Gen. Genomics. 2003;269:116–125. doi: 10.1007/s00438-003-0820-8. [DOI] [PMC free article] [PubMed] [Google Scholar]