Abstract
Chicken avidin is a highly popular tool with countless applications in the life sciences. In the present study, an efficient method for producing avidin protein in the periplasmic space of Escherichia coli in the active form is described. Avidin was produced by replacing the native signal sequence of the protein with a bacterial OmpA secretion signal. The yield after a single 2-iminobiotin–agarose affinity purification step was approx. 10 mg/l of virtually pure avidin. Purified avidin had 3.7 free biotin-binding sites per tetramer and showed the same biotin-binding affinity and thermal stability as egg-white avidin. Avidin crystallized under various conditions, which will enable X-ray crystallographic studies. Avidin produced in E. coli lacks the carbohydrate chains of chicken avidin and the absence of glycosylation should decrease the non-specific binding that avidin exhibits towards many materials [Rosebrough and Hartley (1996) J. Nucl. Med. 37, 1380–1384]. The present method provides a feasible and inexpensive alternative for the production of recombinant avidin, avidin mutants and avidin fusion proteins for novel avidin–biotin technology applications.
Keywords: avidin–biotin technology, bacterial, biotin, chicken avidin, expression system, signal peptide
Abbreviations: AVR, avidin-related protein; NS, native avidin signal peptide; OS, signal peptide of OmpA protein; PEG, poly(ethylene glycol)
INTRODUCTION
Chicken avidin and bacterial streptavidin are among the most widely exploited tools in modern biotechnological and biomedical applications [1]. In addition to their high-affinity ligand binding, avidin and streptavidin [referred to as (strept)avidin] owe their popularity to the easy attachment of biotin or its derivatives to almost any biologically useful molecule without compromising the activity of the target. Additionally, the tetrameric nature of (strept)avidin allows for efficient signal amplification. Collectively, the techniques that constitute this methodology are known as (strept)avidin–biotin technology [2].
Both avidin and streptavidin are proteins secreted in their native hosts. Chicken avidin is a minor constituent of egg white (approx. 0.05% of the total protein) [3] and therefore its large-scale purification requires a large number of eggs. Furthermore, wild-type avidin is glycosylated. The production of streptavidin in its native host, Streptomyces avidinii, suffers from a relatively long cultivation time (up to 10 days). In addition, the end-product is usually a mixture of heterogeneous molecules that need further processing to purify biotechnologically compatible core streptavidin [4]. Similarly, avidin suffers from batch-to-batch variations [5]. Therefore it is understandable that much effort has been invested in producing avidin and streptavidin as well as their mutants and fusions in heterologous expression systems.
Production of recombinant streptavidin has been most often reported in bacteria, especially in Escherichia coli [6,7]. The use of cytoplasmic E. coli overexpression systems leads almost always to the accumulation of streptavidin as inclusion bodies, thereby requiring renaturation and further downstream steps to obtain the active protein. In some studies, streptavidin was produced in a soluble form within the periplasmic space of E. coli [8] or within the cytoplasm of baculovirus-infected insect cells [9,10]. For the production of soluble streptavidin, an efficient expression system in Bacillus subtilis has been developed [11,12].
Chicken avidin, being a eukaryotic protein, has proven even more difficult to produce in bacteria when compared with streptavidin. Trials involving cytoplasmic expression in E. coli have produced low yields of the soluble protein [13] or avidin aggregates as inclusion bodies [14]. In contrast, improved yields have been achieved using eukaryotic production systems such as baculovirus-infected insect cells [15], Pichia pastoris [16] and recombinant maize [17]. Insect cell culturing, however, is relatively expensive and requires the use of a special biotin-free medium. On the other hand, the construction of recombinant corn is time-consuming and laborious, rendering it unsuitable for routine use, especially if the aim is to construct and test novel mutated forms of the target protein. Hence, a cheap and easily modifiable expression system for the production of avidin, such as E. coli, is highly desirable.
In the present study, we demonstrate the successful production of soluble active avidin in the periplasmic space of E. coli. The protein produced by this method contained 3.7 free biotin-binding sites per tetramer. To obtain efficient avidin secretion, the bacterial Bordetella avium OmpA secretion signal [18] was cloned in front of the avidin sequence (OS-avidin, where OS represents the signal peptide of OmpA protein), whereas utilization of the natural secretion signal of chicken avidin (NS-avidin, where NS represents native avidin signal peptide) [19] led to poor yields. This novel system provides a fast, easy and cheap alternative for the production of recombinant avidins. The advantage of the system is the fact that the avidin produced lacks the carbohydrate chains that create problems in certain applications of avidin–biotin technology. In addition, the produced avidin crystallized under various conditions, thereby laying an improved foundation for structural studies of avidin in the future.
EXPERIMENTAL
Construction of the expression vector
Avidin cDNA [19] was extended by stepwise elongation PCR [20] to include the OmpA signal peptide-encoding region and attL-recombination sites (Gateway; Invitrogen) in the constructs. The oligonucleotide sequences are shown in Table 1.
Table 1. Oligonucleotides used in the present study.
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| 5′-AVD | CGCTCTGGCGCTTGCCTTCGCCGCCGTTACGGCCTCTGGTGTTGCCTCGGCTCAGACCGTGGCCAGAAAGTGCTCGCTGAC |
| 3′-AVD | TGCTTTCTTATAATGCCAACTTTGTACAAGAAAGCTGGGTATTACTCCTTCTGTGTGCGCAGG |
| 5′-AVD2 | GCTTTTTTATAATGCCAACTTTGTACAAAAAAGCAGGCTATGAACAAACCCTCCAAATTCGCTCTGGCGCTTGCCTTCG |
| 3′-UNIV | CAAATAATGATTTTATTTTGACTGATAGTGACCTGTTCGTTGCAACAAATTGATAAGCAATGCTTTCTTATAATGCCAAC |
| 5′-UNIV | CAAATAATGATTTTATTTTGACTGATAGTGACCTGTTCGTTGCAACAAATTGATAAGCAATGCTTTTTTATAATGCCAACTTTGT |
| 5′-AVD-attl-sig | TTATAATGCCAACTTTGTACAAAAAAGCAGGCTATGGTGCACGCAACCTC |
The expression construct pBVboostFG+OS-AVD: primers 5′-AVD and 3′-AVD were used in the first PCR. The resulting PCR product (488 bp) was extracted from 1% agarose gel and subjected to a second PCR with primers 5′-AVD2 and 3′-UNIV. The PCR product (607 bp) was again isolated from an agarose gel and subjected to a final PCR with primers 5′-UNIV and 3′-UNIV.
The expression construct pBVboostFG+NS-AVD: primers 5′-AVD-attl-sig and 3′-AVD were used in the first PCR. The resulting PCR product (532 bp) was extracted from the agarose gel and subjected to a second PCR with the primers 5′-UNIV and 3′-UNIV.
The resulting PCR products were cloned into pBVboostFG (O. H. Laitinen, K. J. Airenne, V. P. Hytönen, E. Peltomaa, A. J. Mähönen, T. Wirth, M. M. Lind, K. A. Mäkelä, P. I. Toivanen, D. Schenkwein, T. Heikura, H. R. Nordlund, M. S. Kulomaa and S. Ylä-Herttuala, unpublished work) using the Gateway LR-cloning reaction (Invitrogen) and subsequently confirmed by sequencing. In the resulting vectors, the constructs were cloned under the control of the strong T7 promoter [21] that can be induced in T7 system-compatible bacterial strains.
Production
Fresh transformants of E. coli BL21-AI (Invitrogen) harbouring pBVboostFG+OS-AVD or pBVboostFG+NS-AVD were cultured in Luria–Bertani medium supplemented with 7 μg/ml gentamicin (Sigma) at 30 °C (250 rev./min). When the culture density reached an absorbance A595 0.4–0.6, 0.2% L-arabinose was added to induce the avidin expression and the cultivation was continued at 30 °C for an additional 16 h. The cells were collected by centrifugation [1500 g, room temperature (23±1 °C), 10 min] and frozen until analysed.
Protein purification
Bacterial cells (from 100 ml culture) were suspended in 4 ml of GET (0.5 M glucose, 1 mM EDTA and 200 mM Tris, pH 7.4) and 0.15 mg/ml lysozyme was added. The suspension was incubated on ice for 20 min, after which 20 ml of 2 mM EDTA, 150 mM NaCl, 1% (v/v) Triton X-100 and 50 mM Tris, pH 8.0 (HilloI) was added and the suspension was sonicated for 3 min (1 s on/3 s off, 30% amplitude) with a Branson digital sonifier® 450. The resulting crude extract was clarified by centrifugation (15000 g, 4 °C, 15 min). An equal volume of 50 mM sodium carbonate (pH 11) containing 1 M NaCl was added to the supernatant and the pH was adjusted to 10.5 using 1 M NaOH. Affinity chromatography on a 2-iminobiotin column (Affiland, Liège, Belgium) was used to isolate the active avidin, as described previously [15]. The avidin concentration was measured using a molar absorption coefficient of 24280 M−1·cm−1 at 280 nm [22].
Cell fractionation
Bacterial cells suspended in lysozyme-containing GET buffer as described above were centrifuged (15000 g and 5 min) and the periplasmic fraction was obtained from the supernatant [23]. The resulting pellet was suspended in HilloI buffer, sonicated as above and centrifuged (15000 g and 15 min). The cytoplasmic fraction was obtained from the supernatant and the insoluble fraction from the pellet.
Gel-filtration chromatography
The quality of the purified proteins as well as the molecular mass of their tetrameric forms were assayed by FPLC gel filtration using Shimadzu HPLC liquid chromatography equipped with a Superdex 200 HR 10/30 column (Amersham Biosciences, Uppsala, Sweden). The column was calibrated by using the gel-filtration mixture (thyroglobulin, IgG, ovalbumin, myoglobin and vitamin B12; Bio-Rad Laboratories, Hercules, CA, U.S.A.) and BSA (Roche Diagnostics, Mannheim, Germany) as the molecular-mass standards. Sodium carbonate buffer (50 mM, pH 11) with 150 mM NaCl was used as the liquid phase. Protein samples of 5 μg in a volume of 10 μl were used in the analysis.
N-terminal sequencing
Automated N-terminal sequencing of OS-avidin was performed on an Applied Biosystems Procise 494 protein sequencer using Edman degradation chemistry.
MS
Micromass LCT electrospray time-of-flight MS was used for the measurements. A sample of OS-avidin (1 ml, 0.2 mg/ml) was dialysed against water and diluted 1:1 with acetonitrile. The instrument was operated with a source temperature of 100 °C, desolvation temperature of 180 °C, RF lens voltage of 937 V, extraction cone voltage of 6 V, sample cone voltage of 78 V and capillary voltage of 3677 V. The sample was injected at the rate of 25 μl/min.
Biotin-binding analyses
The number of free binding sites in the purified OS-avidin sample was measured as described by Green [24]. For the analysis, 0.25 mM HABA [2-(4′-hydroxyazobenzene) benzoic acid] was added to the protein solution (10 μM) in 50 mM sodium acetate (pH 4.0), and A500 was measured at 23 °C using a Beckman DU640 spectrophotometer. Excess (0.2 mM) biotin was then added to the solution and A500 was again measured. The concentration of biotin-binding sites in the sample was calculated according to the following equation: c (binding sites)=ΔA500/34 mM [24]. The affinity of OS-avidin for 2-iminobiotin was determined with an IAsys optical biosensor as reported previously [25].
The binding of labelled biotin to OS-avidin and control avidin was analysed by a method based on the quenching, owing to the binding to avidin, of the biotin-coupled fluorescent probe ArcDia™ BF560 (ArcDia, Turku, Finland). Measurements were performed using a PerkinElmer LS55 luminometer. Briefly, 50 nM biotin-BF560 was measured in 50 mM sodium phosphate buffer (pH 7) containing 650 mM NaCl using excitation at 560 nm (2.5 nm slit) and the emission was collected at 578 nm (5 nm slit). Continuous stirring was performed throughout the analysis. After recording the emission for 100 s, OS-avidin (or control avidin) was added to a final biotin-binding subunit concentration of 50 nM and the measurements were continued for 500 s. After that, free biotin was added to a final concentration of 5 μM and the signal was measured for 3600 s at room temperature. A one-phase dissociation model was used to analyse the data. The dissociation rate constant kdiss was determined by fitting the following equation to the data: kdisst=ln(B/Bo)+constant. Bo is the maximum binding measured and B is the value determined as a function of time. The first 500 s were omitted from the analysis to prevent the effect of the fast initial phase [22]. Fluorescent biotin release after measurement for 1 h was determined from the actual data.
Crystallization
The initial crystallization conditions for avidin were based either on conditions described earlier for avidins [26,27] or on a sparse matrix protein crystallization screen (Minimal Screen 12 [28,29]). The vapour diffusion method was used employing hanging drops of 2 μl with equal volumes of protein and well solution. For crystallization, purified avidin was concentrated to approx. 0.5 mg/ml with a Centricon Plus-20 concentrator (Millipore) using a buffer containing 50 mM sodium acetate (pH 4) and 20 mM sodium chloride. The protein concentration of the samples was determined by the Bradford assay (Bio-Rad Laboratories) [30]. Within 2 weeks, bar-like crystals with a typical size of 0.4×0.05× 0.05 mm appeared. Crystals were obtained under several conditions using the following reservoir solutions: (i) 0.1 M sodium citrate (pH 4.6) and 15–20% (w/v) PEG 2000 [poly(ethylene glycol)]; (ii) 0.1 M sodium acetate (pH 4.6), 25% PEG 8000 and 0.2 M ammonium acetate; (iii) 0.1 M sodium acetate (pH 5.6), 20% PEG 8000 and 0.2 M ammonium acetate; (iv) 0.1 M Mes (pH 6.6), 20–26% PEG 8000 and 0.2 M magnesium acetate; (v) 0.1 M Hepes (pH 7.5), 10% (v/v) 2-propanol and 18–26% PEG 4000; and (vi) 0.1 M Tris (pH 8.0–8.6) and 20–29% PEG 1000. Diffraction data up to 1.5 Å resolution (1 Å=0.1 nm) were collected at the beam line X13, EMBL/DESY (Deutsches Elektronen Synchrotron), Hamburg, Germany (T. T. Airenne, V. P. Hytönen, T. A. Salminen, H. Kidron, M. S. Johnson and M. S. Kulomaa, unpublished work). Determination and analysis of the crystal structure is currently in progress.
RESULTS AND DISCUSSION
There are two main approaches to the successful production of heterologous proteins in E. coli, each leading to a different outcome: in one case, the expressed proteins are in soluble form and, in the other, in the insoluble form of the so-called inclusion bodies [31,32]. Approaches utilizing inclusion bodies have certain advantages, e.g. protection of the produced proteins from proteolysis and partial purification by centrifugation. However, usually the preferred method is to begin working directly with material that is soluble and active since this is especially beneficial with regard to different high-throughput strategies. Therefore our aim was to adapt the flexible and easily scaleable E. coli expression system to produce a high yield of soluble active chicken avidin. Since previous attempts to produce avidin in the cytoplasm of E. coli have led to the formation of inclusion bodies [14] or low yields of the soluble protein [13], we decided to express avidin within the periplasmic space of E. coli.
Two alternative constructs were created. In the first, the native secretion signal of avidin was retained, whereas in the other, a bacterial signal sequence from the OmpA protein from B. avium [18] was cloned in front of the mature avidin peptide (Figure 1). The latter was performed to study whether it was possible to increase the production and secretion efficiency of avidin by utilizing a genuine bacterial secretion signal. To ensure the correct cleavage of the product, the construct included the first three amino acids of the native OmpA protein. The end-products encoded by these two constructs were named NS-avidin (native signal avidin) and OS-avidin (OmpA signal avidin). Both constructs were cloned under the strong T7 promoter in a novel pBVboostFG vector that allows easy recombinational cloning [40] and simultaneous production of recombinant proteins in bacterial, insect and vertebrate cells (O. H. Laitinen et al., unpublished work).
Figure 1. Schematic representation of the expression cassettes used in the present study.
The upper Figure represents the natural avidin signal construct (NS-avidin) and the lower Figure shows the OmpA signal construct (OS-avidin). The most distal termini correspond to attL nucleic acid sequences that allow recombinational cloning of these constructs to compatible expression vectors [40]. The N-terminal amino acid residues, recognized as signal peptides in their natural context, are underlined and they are followed by the mature avidin sequence (truncated in the Figure). For the OmpA signal, the first three amino acid residues of the mature OmpA protein (QTV) were also included to confirm correct cleavage of the signal.
Utilization of the native secretion signal of avidin produced only modest quantities of NS-avidin within the periplasm and the yield after 2-iminobiotin–agarose affinity purification was less than 1 mg/l. In addition, the E. coli signal peptidase showed poor recognition of the signal peptide of NS-avidin, since the purified preparation contained both the processed and non-processed forms of the protein (Figure 2B). It is a known fact that codon usage, especially at the 5′-end of the transcript, can have a major effect on heterologous protein production and secretion efficiency in E. coli [33]. We found previously that changing the codon usage in the N-terminal part of avidin according to the codon preferences of E. coli had a positive effect on the efficiency of avidin production [13]. Since the NS-avidin secretion signal and the whole avidin coding region of the NS-avidin construct were cloned from native avidin cDNA [19] and therefore do not necessarily follow an optimal E. coli codon usage, this may partially explain the poor production efficacy of NS-avidin. However, codon analysis of the avidin signal sequence did not reveal major bias against the E. coli codon preference (E. coli preference taken from www.kazusa.or.jp [33]). Two leucine-encoding CTC triplets were found in the signal sequence of NS-avidin. The CTC triplets occur rarely in E. coli mRNAs (1.01% of codons). Furthermore, a rarely occurring proline-encoding CCC codon was also found once in the avidin signal sequence (0.55% of codons).
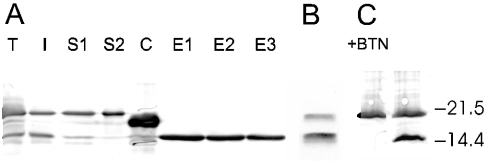
Figure 2. Expression and purification of bacterial avidin.
(A) Immunoblot showing the expression of recombinant OS-avidin within the periplasmic space of E. coli and the purification of the produced protein. Lane T shows a sample of the total E. coli lysate and lane S1 shows the soluble fraction after cell lysis. The upper band represents a non-processed OS-avidin form that still contains a signal peptide, whereas the lower band corresponds to a mature peptide without the signal. Lane I corresponds to the insoluble fraction from cell lysis and lane S2 the flow-through of the soluble fraction after 2-iminobiotin–agarose purification. In lane C, a commercial avidin standard (Belovo) is shown and, in lanes E1–E3, the OS-avidin elution fractions released at pH 4 are shown. As the Figure shows, the processed OS-avidin represents the active form since it is the only form that is not present in the flow-through fraction, and becomes eluted. (B) Eluted fraction from the purification of NS-avidin. The unprocessed and processed forms of avidin are seen. (C) Effect of biotin treatment on the total cell sample heated at 80 °C. Biotin-treated (+BTN) and -untreated samples are shown. Monomeric forms of the processed avidin are not present on the blot in the presence of biotin, whereas the non-processed form stays monomeric with and without biotin. The biotin-stabilized avidin tetramers of the processed form are not visible. Locations of molecular-mass standard bands at 21.5 and 14.4 kDa is shown (Bio-Rad Laboratories).
Another reason for the low expression and lack of secretion efficacy of NS-avidin probably derives from the nature of the avidin signal peptide itself. It is an archetypal eukaryotic signal sequence that does not contain the positively charged residues along its outermost N-terminal region characteristic of prokaryotic signal sequences [34]. The central hydrophobic region of the signal is also occupied almost exclusively by leucine residues (Figure 1) instead of showing an approximately even distribution of alanine and leucine residues, as is typically found in prokaryotic signals [34]. Furthermore, although glycine and serine residues, in positions −3 and −1 upstream of the signal peptidase cleaving site, are small and neutral residues and therefore formally obey the (−3, −1) rule for signal sequences, these positions are almost exclusively occupied by alanine residues in prokaryotic signals [34]. Although it is known that many eukaryotic secretion signals are recognized in prokaryotic organisms, they often lead to poor secretion yield and cleavage accuracy (see [35] and references therein). Considering all these results together, it was hardly surprising that the natural signal sequence of avidin was not efficiently recognized and cleaved in E. coli.
According to our hypothesis, the use of the OmpA signal sequence construct improved the secretion efficiency of OS-avidin by more than one order of magnitude when compared with that of the NS-avidin construct. The typical yield of purified OS-avidin after purification by 2-iminobiotin–agarose affinity chromatography was approx. 10 mg/l (Figures 2A and 3). However, a large fraction of the produced OS-avidin remained unsecreted. It is possible, indeed, that a part of this fraction aggregated into inclusion bodies since the insoluble sample mainly contained the non-processed form (Figure 2A). The OmpA signal peptide was efficiently cleaved from the secreted fraction of OS-avidin, since the purified sample contained only the processed form of the protein. This made the end-product highly homogeneous (Figure 2A). The homogeneity was further confirmed by N-terminal sequencing and MS, which showed that the N-terminal sequence of purified OS-avidin was as expected. The first three residues of the mature OmpA protein were found at the N-terminus and they were followed by the N-terminal sequence of avidin (Table 2). The activity of the processed form was also checked by treating the sample containing whole cells with biotin. Only the processed form withstood the heat treatment as a tetramer after SDS/PAGE analysis similar to egg-white avidin [5], whereas the non-processed form failed to form tetramers (Figure 2C).
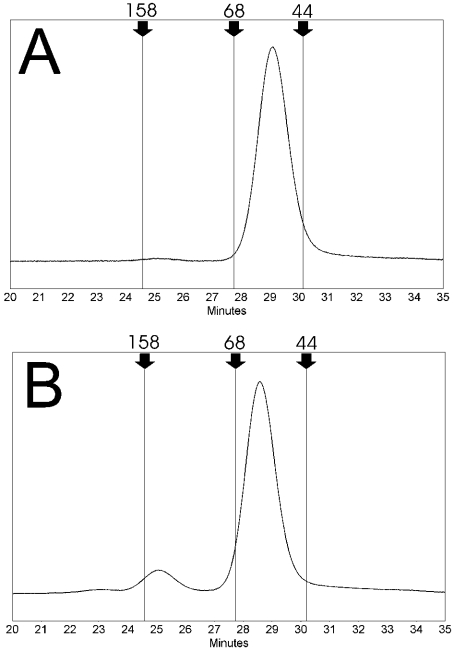
Figure 3. FPLC gel-filtration analysis.
FPLC gel-filtration chromatography profiles of purified recombinant avidin (A) and commercial avidin control (Belovo) (B). The recombinant avidin is seen almost completely as a single sharp peak corresponding to the tetrameric quaternary structure of avidin, whereas a clear higher molecular-mass peak (approx. 25 min) is seen in the chromatogram of control avidin. Elution times of gel-filtration standards are indicated in the chromatograms by arrows: IgG (158 kDa), 24.6 min; BSA (68 kDa), 27.8 min; ovalbumin (44 kDa), 30.1 min.
Table 2. Biochemical properties of bacterial avidin compared with those of avidin isolated from chicken egg white (Belovo).
| OS-avidin | Chicken avidin | Method | |
|---|---|---|---|
| Molecular mass (Da) | 14671.2±0.4 | ∼16500* | ESI–MS |
| Mass of tetramer (kDa) | 52.6 | 59.8 | FPLC gel filtration† |
| Affinity for 2-iminobiotin surface (10−8 M) | 4.3±2.1 | 2.1±0.6 | IAsys optical biosensor‡ |
| No. of biotin-binding sites per tetramer | 3.72±0.05 | 3.44±0.04 | UV/Vis spectroscopy with HABA§ |
| Dissociation rate constant for fluorescent biotin (10−5 s−1) | 1.62±0.23 | 2.26±0.08 | Fluorescence spectroscopy with biotinylated probe |
| Release of biotinylated fluorescent probe in 1 h (%) | 12.4±5.0 | 14.1±1.5 | Fluorescence spectroscopy with biotinylated probe |
| Thermal stability Tr/Tr with biotin ( °C) | 60/90 | 60/90 | SDS/PAGE-based assay∥ |
| N-terminal sequence | QTVARKCSLTGKW | ARKCSLTGKW¶ | N-Terminal sequencing |
The periplasmic production approach offers some clear advantages for proteins such as avidin. It allows the formation of a disulphide bridge and virtually lacks bound biotin, properties that are essential for the full activity of the produced avidin. Consistent with this assumption, OS-avidin purified from the periplasm of E. coli had high specific activity, showing 3.7 free biotin-binding sites per tetramer (Table 2), and this activity was comparable with that of commercial avidin, which in fact has a somewhat lower activity (3.4 free binding sites/tetramer). This result is in good agreement with the report by Shultz et al. [23], who produced streptavidin fused to single-chain antibody fragments in the periplasm of E. coli. Their streptavidin chimaeras had 3.6 free biotin-binding sites per tetramer.
OS-avidin was selected for further analysis, since it was more homogeneous and gave markedly better yields than NS-avidin. The quaternary structure of OS-avidin was studied by FPLC gelfiltration assay. The results showed that OS-avidin was completely tetrameric, just as native avidin (Figure 3). Quenching of the biotinylated fluorescent probe showed that biotin binding to OS-avidin was as tight as for wild-type egg-white avidin (Table 2). Furthermore, the binding affinity of OS-avidin for 2-iminobiotin was similar to the affinity shown by the native protein when studied using an IAsys optical biosensor (Table 2). The purified OS-avidin also tightly bound fluorescent biotin. Both the measured dissociation rate constant (1.62×10−5 s−1) and the total quantity of bound ligand released after measurement for 1 h (12.4%) were close to those measured for control avidin (2.26×10−5 s−1, 14.1%). The heat stability of the purified OS-avidin was studied by an SDS/PAGE-based method [5]. Once again, the ability of OS-avidin to withstand heat, in this analysis, was found to be comparable with that of native avidin (Table 2). On the basis of these biochemical analyses, we can argue that the purified OS-avidin is equal in quality to natural avidin.
Natural chicken avidin has only one post-translational modification, asparagine-linked glycosylation in position 17. The carbohydrate chain is, however, partially responsible for the non-specific binding that hampers the use of avidin in certain applications [36,37,41]. The carbohydrate chain is also processed differently in natural avidins, which furthermore makes native avidin preparations more heterogeneous. Therefore the absence of glycosylation from E. coli is an advantage, especially since it is a known fact that the carbohydrate chain does not affect the biotin-binding activity of avidin [38]. In addition, the use of X-ray crystallography in structure–function studies of proteins often benefits from the absence of carbohydrate units, since they are often more mobile. Determination of the X-ray structure of glycosylated avidin, for example, has been found to be extremely difficult, and well-diffracting avidin crystals have usually been achieved only from deglycosylated avidin preparations [26]. Our periplasmic bacterial expression system provides, therefore, a convenient method by which non-glycosylated avidin mutants of interest can be prepared without the need for any further mutation of the glycosylation site(s) or deglycosylation. The suitability of our novel avidin expression system for crystallization was proven by the crystallization trials. We were able to crystallize OS-avidin under many different conditions over the pH range of 4.6–8.6 (Figure 4). The system also enables the production of non-glycosylated AVRs (avidin-related proteins), an interesting family of highly glycosylated relatives of avidin from chicken [25]. In fact, we have already successfully produced some novel avidin mutants and AVR proteins with this system, obtaining yields similar to those described for OS-avidin (Table 3).
Figure 4. Typical avidin crystals.
Crystals from four different conditions are shown. The following well solutions were used: (A) 0.1 M sodium citrate (pH 4.6) and 15% PEG 2000, (B) 0.1 M Mes (pH 6.6), 26% PEG 8000 and 0.2 M magnesium acetate, (C) 0.1 M Hepes (pH 7.5), 10% 2-propanol and 24% PEG 4000 and (D) 0.1 M Tris (pH 8.6) and 22% PEG 1000. Scale bar, 0.2 mm.
Table 3. Typical yields of avidin proteins produced after affinity purification.
| Produced protein | Ligand in affinity chromatography | Pure protein (mg/l) |
|---|---|---|
| AVD(V37T) | 2-Iminobiotin | 2 |
| AVD(N118L) | Biotin | 10 |
| AVR2 | Biotin | 5 |
| AVR4/5(C122S) | 2-Iminobiotin | 20 |
It is possible to achieve even better expression and secretion yields by optimizing different parameters in the system. Other bacterial signal sequences could be tried or beneficial mutations of the signal could be screened for stronger secretion. It is also possible to modify the codon usage of the whole avidin sequence according to the codon preferences of E. coli. One obvious way to increase protein yields would be to optimize the bioreactor cultivation procedure for synthesizing OS-avidin, instead of using the Erlenmeyer bottle cultivation procedure used here. However, these results prove that it is, in principle, possible to produce chicken avidin within the periplasm of E. coli in an active and soluble form. The produced and purified OS-avidin was highly active, i.e. most of its biotin-binding sites were free, and it showed quality similar to or even better than that of commercial avidin in the SDS/PAGE (only monomers) and FPLC gel-filtration (only tetramers) analyses. By using this novel production system, it should therefore be possible to overcome batch-to-batch variations in avidin preparation that, in some cases, hinder the interpretation of experimental results. Furthermore, no carbohydrate chain is attached to bacterial avidin. It is possible that, in the future, native avidin preparations will be replaced to a large extent by recombinant avidin produced in E. coli.
Acknowledgments
We thank Ms I. Helkala and Ms E. Korhonen for the excellent technical assistance. We thank Professor K. Rissanen and Ms M. Lahtiperä (Department of Chemistry, NanoScience Center, University of Jyväskylä) for MS analysis. This work was supported by the ISB (The National Graduate School in Informational and Structural Biology), the Academy of Finland, the Sigrid Jusélius Foundation and Ark Therapeutics, Ltd.
References
- 1.Wilchek M., Bayer E. Introduction to avidin-biotin technology. Methods Enzymol. 1990;184:5–13. doi: 10.1016/0076-6879(90)84256-g. [DOI] [PubMed] [Google Scholar]
- 2.Wilchek M., Bayer E. A. Foreword and introduction to the book (strept)avidin-biotin system. Biomol. Eng. 1999;16:1–4. doi: 10.1016/s1050-3862(99)00032-7. [DOI] [PubMed] [Google Scholar]
- 3.Green N. M. Avidin and streptavidin. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 4.Bayer E. A., Ben-Hur H., Hiller Y., Wilchek M. Postsecretory modifications of streptavidin. Biochem. J. 1989;259:369–376. doi: 10.1042/bj2590369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer E. A., Ehrlich-Rogozinski S., Wilchek M. Sodium dodecyl sulfate-polyacrylamide gel electrophoretic method for assessing the quaternary state and comparative thermostability of avidin and streptavidin. Electrophoresis. 1996;17:1319–1324. doi: 10.1002/elps.1150170808. [DOI] [PubMed] [Google Scholar]
- 6.Sano T., Cantor C. R. Expression of a cloned streptavidin gene in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1990;87:142–146. doi: 10.1073/pnas.87.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson L. D., Weber P. C. Construction and expression of a synthetic streptavidin-encoding gene in Escherichia coli. Gene. 1993;136:243–246. doi: 10.1016/0378-1119(93)90472-f. [DOI] [PubMed] [Google Scholar]
- 8.Voss S., Skerra A. Mutagenesis of a flexible loop in streptavidin leads to higher affinity for the Strep-tag II peptide and improved performance in recombinant protein purification. Protein Eng. 1997;10:975–982. doi: 10.1093/protein/10.8.975. [DOI] [PubMed] [Google Scholar]
- 9.Karp M., Lindqvist C., Nissinen R., Wahlbeck S., Akerman K., Oker-Blom C. Identification of biotinylated molecules using a baculovirus-expressed luciferase-streptavidin fusion protein. Biotechniques. 1996;20:452–456. 458–459. doi: 10.2144/19962003452. [DOI] [PubMed] [Google Scholar]
- 10.Laitinen O. H., Airenne K. J., Marttila A. T., Kulik T., Porkka E., Bayer E. A., Wilchek M., Kulomaa M. S. Mutation of a critical tryptophan to lysine in avidin or streptavidin may explain why sea urchin fibropellin adopts an avidin-like domain. FEBS Lett. 1999;461:52–58. doi: 10.1016/s0014-5793(99)01423-4. [DOI] [PubMed] [Google Scholar]
- 11.Nagarajan V., Ramaley R., Albertson H., Chen M. Secretion of streptavidin from Bacillus subtilis. Appl. Environ. Microb. 1993;59:3894–3898. doi: 10.1128/aem.59.11.3894-3898.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qureshi M. H., Yeung J. C., Wu S. C., Wong S. L. Development and characterization of a series of soluble tetrameric and monomeric streptavidin muteins with differential biotin binding affinities. J. Biol. Chem. 2001;276:46422–46428. doi: 10.1074/jbc.M107398200. [DOI] [PubMed] [Google Scholar]
- 13.Airenne K. J., Sarkkinen P., Punnonen E.-L., Kulomaa M. S. Production of recombinant avidin in Escherichia coli. Gene. 1994;144:75–80. doi: 10.1016/0378-1119(94)90206-2. [DOI] [PubMed] [Google Scholar]
- 14.Nardone E., Rosano C., Santambrogio P., Curnis F., Corti A., Magni F., Siccardi A. G., Paganelli G., Losso R., Apreda B., et al. Biochemical characterization and crystal structure of a recombinant hen avidin and its acidic mutant expressed in Escherichia coli. Eur. J. Biochem. 1998;256:453–460. doi: 10.1046/j.1432-1327.1998.2560453.x. [DOI] [PubMed] [Google Scholar]
- 15.Airenne K. J., Oker-Blom C., Marjomäki V. S., Bayer E. A., Wilchek M., Kulomaa M. S. Production of biologically active recombinant avidin in baculovirus-infected insect cells. Protein Expr. Purif. 1997;9:100–108. doi: 10.1006/prep.1996.0660. [DOI] [PubMed] [Google Scholar]
- 16.Zocchi A., Jobe A., Neuhaus J.-M., Ward T. Expression and purification of a recombinant avidin with a lowered isoelectric point in Pichia pastoris. Protein Expr. Purif. 2003;32:167–174. doi: 10.1016/j.pep.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Kusnadi A. R., Hood E. E., Witcher D. R., Howard J. A., Nikolov Z. L. Production and purification of two recombinant proteins from transgenic corn. Biotechnol. Prog. 1998;14:149–155. doi: 10.1021/bp970138u. [DOI] [PubMed] [Google Scholar]
- 18.Gentry-Weeks C. R., Hultsch A. L., Kelly S. M., Keith J. M., Curtiss R., III Cloning and sequencing of a gene encoding a 21-kilodalton outer membrane protein from Bordetella avium and expression of the gene in Salmonella typhimurium. J. Bacteriol. 1992;174:7729–7742. doi: 10.1128/jb.174.23.7729-7742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gope M. L., Keinanen R. A., Kristo P. A., Conneely O. M., Beattie W. G., Zarucki-Schulz T., O'Malley B. W., Kulomaa M. S. Molecular cloning of the chicken avidin cDNA. Nucleic Acids Res. 1987;15:3595–3606. doi: 10.1093/nar/15.8.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumder K. Ligation-free gene synthesis by PCR: synthesis and mutagenesis at multiple loci of a chimeric gene encoding OmpA signal peptide and hirudin. Gene. 1992;110:89–94. doi: 10.1016/0378-1119(92)90448-x. [DOI] [PubMed] [Google Scholar]
- 21.Cheetham G. M., Steitz T. A. Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2000;10:117–123. doi: 10.1016/s0959-440x(99)00058-5. [DOI] [PubMed] [Google Scholar]
- 22.Green N. M. Avidin. Adv. Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- 23.Schultz J., Lin Y., Sanderson J., Zuo Y., Stone D., Mallett R., Wilbert S., Axworthy D. A tetravalent single-chain antibody-streptavidin fusion protein for pretargeted lymphoma therapy. Cancer Res. 2000;60:6663–6669. [PubMed] [Google Scholar]
- 24.Green N. M. Spectrophotometric determination of avidin and streptavidin. Methods Enzymol. 1970;18:418–424. [Google Scholar]
- 25.Laitinen O. H., Hytönen V. P., Ahlroth M. K., Pentikäinen O. T., Gallagher C., Nordlund H. R., Ovod V., Marttila A. T., Porkka E., Heino S., et al. Chicken avidin-related proteins show altered biotin-binding and physico-chemical properties as compared with avidin. Biochem. J. 2002;363:609–617. doi: 10.1042/0264-6021:3630609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livnah O., Bayer E. A., Wilchek M., Sussman J. L. Three-dimensional structures of avidin and the avidin-biotin complex. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5076–5080. doi: 10.1073/pnas.90.11.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pazy Y., Eisenberg-Domovich Y., Laitinen O. H., Kulomaa M. S., Bayer E. A., Wilchek M., Livnah O. Dimer-tetramer transition between solution and crystalline states of streptavidin and avidin mutants. J. Bacteriol. 2003;185:4050–4056. doi: 10.1128/JB.185.14.4050-4056.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jancarik J., Scott W. G., Milligan D. L., Koshland D. E., Jr, Kim S. H. Crystallization and preliminary X-ray diffraction study of the ligand-binding domain of the bacterial chemotaxis-mediating aspartate receptor of Salmonella typhimurium. J. Mol. Biol. 1991;221:31–34. doi: 10.1016/0022-2836(91)80198-4. [DOI] [PubMed] [Google Scholar]
- 29.Kimber M. S., Vallee F., Houston S., Necakov A., Skarina T., Evdokimova E., Beasley S., Christendat D., Savchenko A., Arrowsmith C. H., et al. Data mining crystallization databases: knowledge-based approaches to optimize protein crystal screens. Proteins. 2003;51:562–568. doi: 10.1002/prot.10340. [DOI] [PubMed] [Google Scholar]
- 30.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 31.Balbas P. Understanding the art of producing protein and nonprotein molecules in Escherichia coli. Mol. Biotechnol. 2001;19:251–267. doi: 10.1385/MB:19:3:251. [DOI] [PubMed] [Google Scholar]
- 32.Panda A. K. Bioprocessing of therapeutic proteins from the inclusion bodies of Escherichia coli. Adv. Biochem. Eng. Biotechnol. 2003;85:43–93. doi: 10.1007/3-540-36466-8_3. [DOI] [PubMed] [Google Scholar]
- 33.Humphreys D. P., Carrington B., Bowering L. C., Ganesh R., Sehdev M., Smith B. J., King L. M., Reeks D. G., Lawson A., Popplewell A. G. A plasmid system for optimization of Fab' production in Escherichia coli: importance of balance of heavy chain and light chain synthesis. Protein Expr. Purif. 2002;26:309–320. doi: 10.1016/s1046-5928(02)00543-0. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Humphreys D. P., Sehdev M., Chapman A. P., Ganesh R., Smith B. J., King L. M., Glover D. J., Reeks D. G., Stephens P. E. High-level periplasmic expression in Escherichia coli using a eukaryotic signal peptide: importance of codon usage at the 5′ end of the coding sequence. Protein Expr. Purif. 2000;20:252–264. doi: 10.1006/prep.2000.1286. [DOI] [PubMed] [Google Scholar]
- 36.Schechter B., Silberman R., Arnon R., Wilchek M. Tissue distribution of avidin and streptavidin injected to mice. Effect of avidin carbohydrate, streptavidin truncation and exogenous biotin. Eur. J. Biochem. 1990;189:327–331. doi: 10.1111/j.1432-1033.1990.tb15493.x. [DOI] [PubMed] [Google Scholar]
- 37.Yao Z., Zhang M., Sakahara H., Nakamoto Y., Higashi T., Zhao S., Sato N., Arano Y., Konishi J. The relationship of glycosylation and isoelectric point with tumor accumulation of avidin. J. Nucl. Med. 1999;40:479–483. [PubMed] [Google Scholar]
- 38.Hiller Y., Gershoni J. M., Bayer E. A., Wilchek M. Biotin binding to avidin. Oligosaccharide side chain not required for ligand association. Biochem. J. 1987;248:167–171. doi: 10.1042/bj2480167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeLange R. J. Egg white avidin. I. Amino acid composition; sequence of the amino- and carboxyl-terminal cyanogen bromide peptides. J. Biol. Chem. 1970;245:907–916. [PubMed] [Google Scholar]
- 40.Hartley J. L., Temple G. F., Brasch M. A. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosebrough S. F., Hartley D. F. Biochemical modification of streptavidin and avidin: in vitro and in vivo analysis. J. Nucl. Med. 1996;37:1380–1384. [PubMed] [Google Scholar]