Abstract
Our previous studies have demonstrated de novo haem biosynthesis in the malarial parasite (Plasmodium falciparum and P. berghei). It has also been shown that the first enzyme of the pathway is the parasite genome-coded ALA (δ-aminolaevulinate) synthase localized in the parasite mitochondrion, whereas the second enzyme, ALAD (ALA dehydratase), is accounted for by two species: one species imported from the host red blood cell into the parasite cytosol and another parasite genome-coded species in the apicoplast. In the present study, specific antibodies have been raised to PfFC (parasite genome-coded ferrochelatase), the terminal enzyme of the haem-biosynthetic pathway, using recombinant truncated protein. With the use of these antibodies as well as those against the hFC (host red cell ferrochelatase) and other marker proteins, immunofluorescence studies were performed. The results reveal that P. falciparum in culture manifests a broad distribution of hFC and a localized distribution of PfFC in the parasite. However, PfFC is not localized to the parasite mitochondrion. Immunoelectron-microscopy studies reveal that PfFC is indeed localized to the apicoplast, whereas hFC is distributed in the parasite cytoplasm. These results on the localization of PfFC are unexpected and are at variance with theoretical predictions based on leader sequence analysis. Biochemical studies using the parasite cytosolic and organellar fractions reveal that the cytosol containing hFC accounts for 80% of FC enzymic activity, whereas the organellar fraction containing PfFC accounts for the remaining 20%. Interestingly, both the isolated cytosolic and organellar fractions are capable of independent haem synthesis in vitro from [4-14C]ALA, with the cytosol being three times more efficient compared with the organellar fraction. With [2-14C]glycine, most of the haem is synthesized in the organellar fraction. Thus haem is synthesized in two independent compartments: in the cytosol, using the imported host enzymes, and in the organellar fractions, using the parasite genome-coded enzymes.
Keywords: apicoplast, ferrochelatase, haem synthesis, localization, mitochondrion, Plasmodium falciparum
Abbreviations: ALA, δ-aminolaevulinate; ALAD, ALA dehydratase; ALAS, ALA synthase; FC, ferrochelatase; hALAD, host red cell ALAD; hFC, host red cell FC; hsp, heat-shock protein; PfALAD, ALAD from P. falciparum; PfALAS, ALAS from P. falciparum; PfFC, FC from P. falciparum; PfΔFC, truncated recombinant PfFC; PfNT1, nucleotide transporter from P. falciparum; PfPP2C1, protein phosphatase from P. falciparum; TRITC, tetramethylrhodamine β-isothiocyanate
INTRODUCTION
In earlier studies, we have shown that the malarial parasite synthesizes haem de novo, despite acquiring haem from the host red cell haemoglobin in the intra-erythrocytic stage. Inhibition of the de novo haem pathway leads to the death of the parasite and is therefore a drug target [1,2]. Further studies have highlighted the complexities involved in parasite haem biosynthesis. The first enzyme, ALAS [ALA (δ-aminolaevulinate) synthase] encoded by the parasite genome (PfALAS), is localized in the mitochondrion and is functional [3,4]. We have shown that the second enzyme of the pathway, ALAD (ALA dehydratase), is imported by the parasite from the host red cell (hALAD) and is functional [2,5]. However, the Plasmodium falciparum genome sequence reveals that it can code for all the enzymes of the haem-biosynthetic pathway [6], except for uroporphyrinogen-III synthase that is yet to be annotated. Sato and Wilson [7] have shown that the parasite gene-coded ALAD (PfALAD) cDNA can complement a haem B mutant (ALAD−) of Escherichia coli, indicating that the cDNA codes for a functional enzyme. It has been suggested that PfALAD may account for de novo haem synthesis by the parasite rather than the imported enzyme. On the basis of phylogenetic analysis, Sato et al. [8] have predicted that PfALAD protein would be similar to the Mg2+-requiring plant plastid ALAD, rather than the Zn2+-requiring mammalian ALAD. In a recent study [9], we have shown that, while PfALAD is targeted to the apicoplast and the recombinant enzyme has an alkaline optimum pH, like plant ALADs, it is very similar to the enzyme species from Pseudomonas aeruginosa in manifesting metal-independent enzymic activity, although stimulated by Mg2+ to an extent of 20–30%. However, PfALAD accounts only for approx. 10% of the total ALAD enzymic activity of the parasite, the remainder being accounted for by the imported host enzyme. These results have led to the suggestion that PfALAD may only account for haem synthesis in the apicoplast, and an additional machinery involving the PfALAS in the mitochondrion and imported enzymes in the cytosol may be necessary, since the parasite cell has a single apicoplast, unlike plants with numerous chloroplasts per cell.
The ultimate site of haem synthesis in a cell would be decided by the site of localization of FC (ferrochelatase), the terminal enzyme of the pathway. FCs of animals and fungi are localized at the inner membrane of mitochondria [10], whereas in plants it was reported that there are two isoforms, one exclusively targeted to chloroplasts and the other targeted to both mitochondria and chloroplasts [11]. However, a recent study in cucumber has led to the conclusion that both the isoforms are targeted to chloroplasts [12]. Sato and Wilson [13] have cloned the PfFC (FC from P. falciparum) cDNA and shown that it can successfully rescue an FC-null mutant of E. coli, establishing that the cDNA codes for a functional enzyme. Since PfFC seems to lack the bipartite N-terminal sequence, necessary to target parasite proteins to the apicoplast [14], Sato and Wilson [13] have suggested that PfFC is unlikely to be a plastid protein, but may be localized in the mitochondrion.
In this paper, the localization of FC in P. falciparum cultured in human red blood cells has been studied. For this purpose, PfFC-specific antibodies have been raised in mice using PfΔFC (recombinant truncated PfFC). With the use of these as well as antibodies to the hFC (host red cell FC), the pattern of localization of both the enzyme species has been studied using immunofluorescence. The possible co-localization of PfFC with parasite mitochondrial markers such as PfHsp60 (where Hsp stands for heat-shock protein) and MitoTracker dye has been investigated. Further studies of the localization of PfFC and hFC in the parasite were carried out by immunoelectron microscopy. Organellar fractions containing the apicoplast and mitochondria as well as the cytosolic fraction were used for the biochemical analysis of FC localization by Western-blot analysis and enzyme assay. Finally, these isolated fractions have been assessed independently for the potential to synthesize haem from the precursors [2-14C]glycine and [4-14C]ALA. On the basis of these and earlier results, the unique features of haem synthesis in the malarial parasite utilizing the imported host enzymes and the parasite genome-coded enzymes have been discussed.
EXPERIMENTAL
Materials
4-14C-labelled ALA and 59FeCl3 were purchased from NEN Life Science Products. [2-14C]Glycine and [1-14C]glycine were purchased from BRIT (Mumbai, India). Antibodies to hFC, parasite nucleotide transporter (PfNT1) and parasite protein phosphatase (PfPP2C1) were gifts from Dr S. Taketani (Kansai Medical University, Osaka, Japan), Dr N. Rager (Oregon Health Science University, Portland, OR, U.S.A.) and Dr C. Ben Mamoun (University of Connecticut, Farmington, CT, U.S.A.) respectively. PfHsp60 cDNA was a gift from Dr Nirbhay Kumar (Johns Hopkins School of Public Health, Baltimore, MD, U.S.A.). This cDNA was expressed in E. coli using the expression vector pRSETA (Invitrogen). Antibodies were raised to the purified His-tagged protein (PfHsp60) in rabbits.
A truncated PfFC protein (PfΔFC) was obtained from the 3′ portion of PfFC that was amplified with PCR primers to give a 750 bp product. The primer set used was FC 5′-AAGGATCCAATGAGATTTGATATTTTT and FC 3′-AATGAATTCTTACACCCATCCTATTAT. The sequence of the 750 bp product was identical with that reported by Sato and Wilson [13]. This fragment was cloned in pRSETA and the protein was expressed as a histidine-tagged protein by isopropyl β-D-thiogalactoside induction. The protein was purified using an Ni2+-nitrilotriacetate column, and the band on SDS/PAGE (10% gel) was eluted and characterized as PfFC by MALDI (matrix-assisted laser-desorption ionization) analysis and peptide sequencing. This truncated protein was injected into mice with a primary dose of 50 μg and three boosters with 20 μg/mouse at 21-day intervals. The animals were bled 1 week after injecting each booster and IgG was prepared from sera.
Immunofluorescence microscopy
To find the localization of FC in P. falciparum, smears from the culture were fixed in ice-cold acetone/methanol (70:30). After air drying and blocking non-specific sites with 2% (w/v) BSA in PBS, a primary antibody (dilutions: hFC, 1:100; PfΔFC, 1:50; PfHsp60, 1:100) was added to the coverslip and incubated for 3 h. After washing with PBS, the respective secondary goat antiprimary antibodies conjugated to FITC/TRITC (tetramethylrhodamine β-isothiocyanate) were added at 1:200 dilution. After incubation for 45 min, the coverslips were washed with PBS and mounted on a glass slip using Slow Fade from Molecular Probes. Co-localization of PfFC with MitoTracker Red, CM-H2Xros, a mitochondrion-specific dye [15], was also studied. For this purpose, 1.5 ml of parasite culture was incubated with the dye (250 nM) for 30 min. The cells were then washed with complete RPMI 1640 and a smear was prepared on coverslips, fixed and processed for antibody treatment as described above. A multipurpose fluorescence microscope (Axioskop; Carl Zeiss) was used for most of the studies. Images were acquired using 100× oil immersion objective with the help of Axiovision image acquisition software.
Immunoelectron microscopy
The parasite-infected red blood cell pellet was washed three times with PBS and then resuspended in 1% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde in phosphate buffer (pH 7.1) for 3–4 h. Immunostaining was then performed by the method described by PELCO International using the LR white resin. Ultra-thin sections (100 nm) from the embedded block were mounted on nickel grids. All incubations were performed in blocking buffers containing 2% BSA and 1% (v/v) goat serum. Anti-human FC antibodies (rabbit, 1:50) or anti-PfFC antibodies (mouse, 1:20) were used, and incubation was performed for 2–3 h. Goat anti-rabbit IgG or goat anti-mouse IgG coupled with 10 or 20 nm gold particles was used as the secondary antibody at 1:50 dilution and incubated for 1 h. The grids were post-fixed with 1% glutaraldehyde in PBS, washed with distilled water, subjected to silver enhancement and then counter-stained with uranyl acetate in 90% (v/v) methanol. Sections were viewed using a JEOL (X-100) transmission electron microscope.
Parasite organellar and cytosolic preparations
P. falciparum culture was maintained on human O+ red cells by the candle jar method [16]. The cells were pelleted down and the parasites, essentially at the trophozoite stage, were isolated by treatment with 0.15% (w/v) saponin [17] or osmotic lysis [18]. For the latter procedure, packed infected red blood cells were treated with 40 vol. of cold 10 mM potassium phosphate buffer (pH 7.4), mixed thoroughly and kept on ice for 10 min. The parasites released after the lysis of red cells were pelleted down and washed once with the same buffer and three times with PBS. With both procedures, the final washes tested negative for haemoglobin or erythroid ALAD, which were used as red cell cytoplasmic markers.
The parasite pellet was lysed in 10 mM phosphate buffer (pH 7.4), containing 0.1% (w/v) Triton X-100 for 10 min in ice. The suspension was spun down at 800 g for 5 min and then at 12000 g for 30 min. The supernatant after centrifugation at 100000 g was used as the cytosolic fraction and did not contain any detectable red cell haemoglobin. The 12000 g pellet was used as the organellar fraction. The pellet was suspended in 10 mM potassium phosphate buffer (pH 7.4), containing 1% Triton X-100, and incubated for 30 min. It was centrifuged at 12000 g for 30 min, and the supernatant was used as the solubilized organellar fraction.
Western-blot analysis of the organellar and cytosolic fractions
The respective fractions were analysed by SDS/PAGE (10% gel) and the proteins were transferred on to nitrocellulose membranes for immunoblot analysis. Antibodies to PfNT1, PfPP2C, PfHsp60, PfALAD, PfFC and hFC were used at appropriate dilutions as primary antibodies. Secondary goat antibodies conjugated to alkaline phosphatase were used to visualize the specific protein bands.
Synthesis of PfFC by the parasite
P. falciparum culture (1.5 ml, 10% parasitaemia) was incubated with 75 μCi of [35S]methionine for 8 h and the parasite pellet was isolated and fractionated into organellar and cytosolic fractions. The cytosol and solubilized organellar preparations were precleared with normal mouse serum and then incubated with an anti-PfΔFC antibody (mouse, 1:100) for 3 h at 4 °C. This was followed by the addition of Protein A–Sepharose beads. The beads were recovered by centrifugation, washed with RIPA buffer [15 mM NaCl, 50 mM Tris (pH 8.0), 0.1% SDS, 1% Triton X-100 and 0.5% deoxycholate] and subjected to SDS/PAGE (10% gel) and autoradiography. Northern-blot analysis was also performed with parasite RNA using labelled PfΔFC cDNA as the probe.
Haem synthesis by the cytosolic and organellar fractions
The parasite cytosolic and organellar (solubilized) fractions obtained from 6 ml of P. falciparum were incubated with 5 μCi of [2-14C]glycine or [1-14C]glycine or 1 μCi of [4-14C]ALA in a total volume of 100 μl for 8 h at 37 °C. Haem was extracted with an ethyl acetate/acetic acid (4:1) mixture, and the ethyl acetate layer was washed twice with water, 1.5 M HCl and water and used for radioactivity measurement [2].
Partial purification of red blood cell and P. falciparum FC
FC, both human and P. falciparum, was purified by the method described in [19] but with slight modifications. Briefly, human red blood cells and P. falciparum pellet were lysed with ten packed cell volumes of hypo-osmotic lysis buffer and kept at 4 °C for 1 h. The membrane fraction was separated from the cytosolic fraction by using a high-speed spin at 100000 g for 1 h. hFC enzymic activity was detected in both the cytosolic and membrane fractions of red blood cells. However, the membrane being devoid of haemoglobin, the pellet was used as the starting material for the purpose of purification. Membrane pellet was solubilized in ten pellet volumes of solubilization buffer [20 mM Tris/HCl (pH 8.1), 1% (w/v) sodium cholate, 1.0 M KCl, 1 mM sodium dithionite and 0.1% (w/v) protease inhibitor cocktail] by incubating at 4 °C for 1 h, followed by sonication (five pulses in Vibra cell sonicator, 3 s on and 2 s off). The membrane fraction was centrifuged at 100000 g for 1 h at 4 °C. The supernatant was the solubilized membrane fraction. This fraction was subjected to a 0–50% ammonium sulphate cut and centrifuged at 10000 g for 15 min. The resulting pellet was resuspended in equilibration buffer [20 mM Tris/HCl (pH 8.1), 1% Triton X-100, 0.5 M KCl, 1 mM sodium dithionite and protease inhibitor cocktail]. This fraction was loaded on to a 1.0 ml Pharmacia Hi Trap Blue affinity column, previously equilibrated with 10 ml of the equilibration buffer. The column was washed with 3–5 column volumes of the equilibration buffer, followed by 3–5 ml of wash I [20 mM Tris/HCl (pH 8.1), 1% Triton X-100, 1.0 M KCl, 1.0 mM sodium dithionite and 0.1% (v/v) protease inhibitor cocktail] and, finally, with 3–5 ml of wash II [20 mM Tris/HCl (pH 8.1), 0.5% sodium cholate, 1.0 M KCl, 1.0 mM sodium dithionite and protease inhibitor cocktail]. FC was eluted from the column with 5 ml of elution buffer [20 mM Tris/HCl (pH 8.1), 1% sodium cholate, 1.5 M KCl, 1.0 mM sodium dithionite and 0.1% (v/v) protease inhibitor cocktail].
Assay of FC enzyme
This was carried out [2] in a total volume of 100 μl, containing 20 mM Tris (pH 7.5), 100 μM protoporphyrin IX, 1 mM 2-mercaptoethanol, 2.5 μCi of 59FeCl3 and the parasite cytosolic or solubilized organellar fraction. The mixture was incubated at 37 °C for 1 h in a nitrogen atmosphere. Haem was extracted and radioactivity was measured as described above.
RESULTS
Cloning and expression of PFΔFC and its antibody specificity
Full-length PfFC cDNA was constructed using primers on the basis of the open reading frame finder analysis of the gene sequence in Plasmo DB database and the cDNA sequence published by Sato and Wilson [13]. However, in the 1050 bp cDNA component of the sequence published by Sato and Wilson [13], two insertions, 63 and 41 bp, were found in the present study. Sato and Wilson [13] have also found two cDNAs, with and without the 63 bp insertion. The cDNA with the 63 bp insertion was considered to be an alternatively spliced product causing a frameshift with unknown function. The cDNA without the 63 bp insertion was found to be functional in complementation assays with an FC-null mutant of E. coli. Sato and Wilson [13] have not reported the 41 bp insertion in the cDNA that is shown as the terminal part of intron 4 as per PlasmoDB annotation. The reason for the differences is not clear. RNA from P. falciparum in culture was used for reverse transcriptase–PCR amplification in the present study, whereas Sato and Wilson [13] have used cDNA libraries for the purpose.
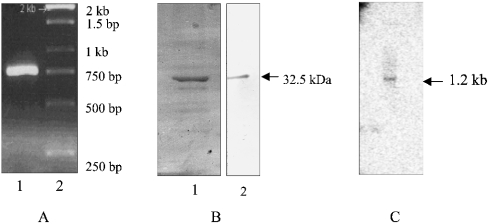
However, the 750 bp 3′-fragment obtained by PCR amplification (Figure 1A) was identical in sequence with that reported by Sato and Wilson [13]. This fragment was used to express the truncated protein (32.5 kDa) that reacted with the His-tagged antibody (Figure 1B). The protein band eluted from the gel was subjected to tryptic digestion and analysed using Ettan MALDI–ToF (time-of-flight) Pro (Amersham Biosciences) and Proteometrics LLC software; the sequence of one of the peptides (VIGGDLYPFFCIES) that led to the identification of the protein as truncated PfFC (PfΔFC) was also analysed. Antibodies to PfΔFC were raised in mice. Northern-blot analysis of RNA probed with PfΔFC-cDNA revealed a band at 1.2 kb, indicating that the gene is transcribed in P. falciparum (Figure 1C).
Figure 1. Preparation of PfΔFC.
RNA from the parasite was subjected to reverse transcriptase–PCR amplification with FC-specific primers, and the full-length cDNA obtained was sequenced. (A) Lanes: 1, PCR-amplified 750 bp fragment; 2, standards (1 kb ladder). (B) SDS/PAGE analysis of the C-terminal 32.5 kDa protein fragment expressed in E. coli and purified on an Ni2+-nitrilotriacetate column: Lanes: 1, Coomassie stain; 2, Western-blot analysis with anti-histidine tag antibodies. (C) Northern-blot analysis of parasite RNA with labelled PfΔFC cDNA showing an RNA band at 1.2 kb.
Origin and localization of FC in the parasite
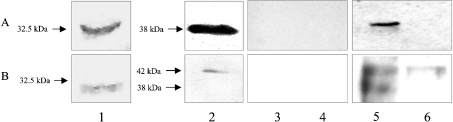
To examine the presence of PfFC and hFC in the infected red cell, the specificities of the respective antibodies were first examined. The results presented in Figure 2(A) reveal that the PfΔFC antibody shows up the PfΔFC (32.5 kDa) used to raise the antibody in Western-blot analysis. It also interacts with the partially purified PfFC from the parasite (38 kDa) and an identical band shows up with the parasite lysate. However, the red cell lysate does not show any cross-reacting protein. Figure 2(B) reveals that the hFC antibody (obtained as a gift) cross-reacts with PfΔFC and also shows up the partially purified hFC protein (42 kDa) from the red cells. It shows up an identical band with red blood cell lysate. However, the parasite lysate gives two bands of 42 and 38 kDa, the former corresponding to hFC and the latter corresponding to PfFC. Thus it appears that, whereas PfΔFC antibodies are specific to PfFC, hFC antibodies react with both hFC and PfFC. Preimmune sera from mice and rabbit, the animals in which the antibodies were raised, do not show any cross-reacting band in Western-blot analysis. These results suggest that the malarial parasite contains FC species of host origin as well as the parasite-genome-coded, endogenously synthesized enzyme.
Figure 2. Specificity of anti-FC antibodies.
Western-blot analysis was performed with antibodies against PfΔFC and hFC using purified FC preparations as well as parasite and red cell lysates. (A) Lanes: 1, 2, 5 and 6, Western-blot analysis with PfΔFC antibodies. (B) Lanes: 1, 2, 5 and 6, Western-blot analysis with hFC antibodies; 1, PfΔFC; 2, partially purified PfFC (top) and hFC (bottom); 3 and 5, parasite lysate; 4 and 6, red blood cell lysate; 3 and 4, preimmune sera from mice (top) and rabbit (bottom) controls.
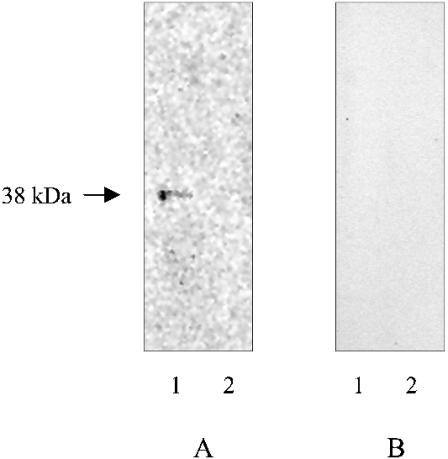
Studies on FC localization were performed using labelled protein synthesis and immunoprecipitation with PfFC-specific antibodies, and by immunofluorescence and immunoelectron microscopy of P. falciparum in culture. When the parasite proteins are labelled with [35S]methionine in a culture of P. falciparum, a band of 38 kDa is immunoprecipitated from the organellar fraction, but not the cytosolic fractions, with PfΔFC antibodies (Figure 3).
Figure 3. Immunoprecipitation and SDS/PAGE analysis of [35S]methionine-labelled P. falciparum in culture.
(A) PfΔFC antibodies. (B) Preimmune sera; 1, organellar fraction; 2, cytosolic fraction.
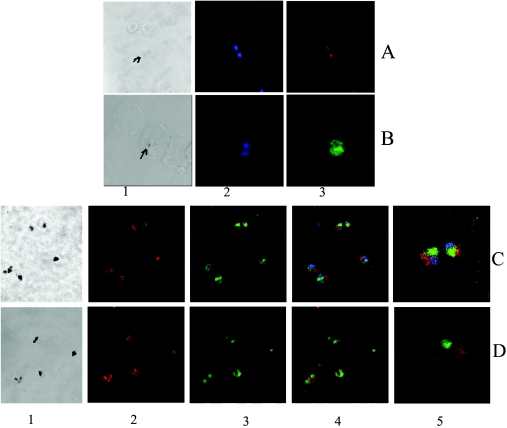
Immunofluorescence studies reveal a compact localization of PfFC in the parasite with PfΔFC antibodies and TRITC-labelled secondary antibodies in the infected red cells (Figure 4A). With hFC antibodies, the green fluorescence obtained with FITC-labelled secondary antibodies in the infected red cell covers almost the entire parasite (Figure 4B). This signal is higher in the parasite than the weaker diffused fluorescence seen in the surrounding red cells, where haemoglobin may have a quenching effect. Figures 4(C) and 4(D) give the co-localization patterns of PfFC with PfHsp60 and MitoTracker respectively. It is clearly seen that the green fluorescence due to PfFC (FITC-conjugated secondary antibody) does not co-localize with the red fluorescence due to PfHsp60 (TRITC-conjugated secondary antibody). The latter is an established Plasmodium mitochondrial marker [20]. Again, PfFC fluorescence does not co-localize with that of the MitoTracker, a mitochondrion-specific dye [15]. These results suggest the possibility that at least the bulk of the PfFC is not present in the parasite mitochondrion.
Figure 4. Immunofluorescence analysis of FC localization in P. falciparum-infected red cells.
(A) Smear analysis with PfΔFC antibodies followed by TRITC-conjugated (red) secondary antibodies; 1, bright field (the arrow indicates hemozoin pigment); 2, Hoescht stain indicating two parasites; 3, localized red fluorescence due to PfFC in the parasite. (B) Smear analysis with hFC antibodies followed by FITC-conjugated (green) secondary antibodies; 1, bright field (the arrow indicates hemozoin pigment); 2, Hoescht stain indicating two parasites; 3, broad green fluorescence covering the entire parasite cytoplasm due to hFC. (C) Analysis of the co-localization of PfFC with PfHsp60 using FITC-conjugated (green) secondary antibodies for the former and TRITC-conjugated (red) secondary antibodies for the latter; 1, bright field (dark spots represent the hemozoin pigment); 2, red fluorescence in the parasite due to PfHsp60; 3, green fluorescence in the parasite due to PfFC; 4, merge of 2 and 3 along with Hoescht stain (blue) showing lack of co-localization; 5, projection of two parasites showing lack of co-localization. (D) Analysis of co-localization of PfFC with MitoTracker; 1, bright field (dark spots represent the hemozoin pigment); 2, red fluorescence in the parasite due to MitoTracker; 3, green fluorescence in the parasite due to PfFC; 4, merge of 2 and 3 showing lack of co-localization; 5, projection of a single parasite showing lack of co-localization.
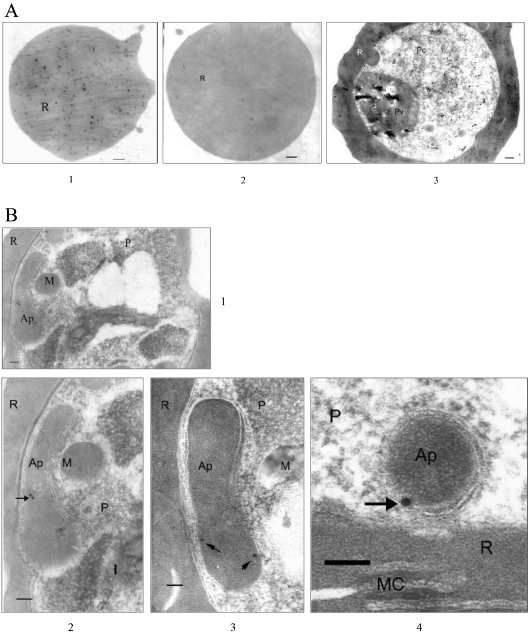
Immunoelectron-microscopy studies were performed with hFC and PfFC antibodies using gold-conjugated secondary antibodies. First of all, hFC antibodies give extensive signals with the uninfected red cells, whereas the PfΔFC antibodies do not show any signal (Figure 5A). Preimmune sera do not show any signal (results not shown). These results confirm the presence of significant amounts of FC in the red blood cell, in addition to confirming the specificity of PfΔFC antibodies. The results presented in Figure 5(A) also reveal that, with the infected red cell using hFC antibodies, FC signals are seen all over the red cell cytoplasm and parasite. However, PfΔFC antibodies give sparse and discrete localization of gold particles in typical multimembrane apicoplast structures, but not elsewhere in the infected cell (Figures 5B1–5B4). Interestingly, signals are seen close to the membranes, suggesting that PfFC may be localized to a subcompartment in the apicoplast. Even though hFC antibodies recognize PfFC, the extensive localization in the parasite cytoplasm seen in immunofluorescence and immunoelectron-microscopy studies using these antibodies is essentially due to imported hFC, since the PfFC signal specifically detected by PfΔFC antibodies is sparse and confined to the apicoplast.
Figure 5. Immunoelectron-microscopic analysis of FC localization in parasite-infected red cells.
The secondary antibodies were conjugated to 10 or 20 nm gold particles. (A) 1 and 3, uninfected and infected red cells probed with hFC antibodies respectively; 2, uninfected red cell probed with PfΔFC antibodies. (B) Three different fields showing discrete gold particles in typical apicoplast structures with multiple membranes. 1, the whole parasite; 2 (enlarged part from 1) and 3, elongating apicoplasts in a later-stage trophozoite; 4, a circular apicoplast in an early-stage tropozoite. The signals are localized close to the membrane. Scale bar, 100 μm. AP, apicoplast; M, mitochondrion; R, red cell; MC, Maurer's cleft; P, parasite; Pv, parasite food vacuole; Pc, parasite cytosol.
Haem synthesis by the organellar fraction
As already mentioned, studies in this laboratory have shown that the malaria parasite imports hALAD (host red cell ALAD) and perhaps the subsequent enzymes of the pathway [2,5,9]. Studies have also shown that PfALAS and PfALAD are present in the parasite mitochondrion [4] and apicoplast [9] respectively. The present study reveals that, whereas hFC is present significantly in the parasite cytoplasm, PfFC is present in the apicoplast. It is thus of interest to examine whether the organellar fraction containing the apicoplast and mitochondrion is capable of independent haem synthesis with enzymes encoded by the parasite genome. It has been reported that the apicoplast and mitochondrion are in close proximity to each other in the malarial parasite ([21] and Figure 5). Similarly, the parasite cytosol containing the imported host enzymes may also be capable of independent haem synthesis.
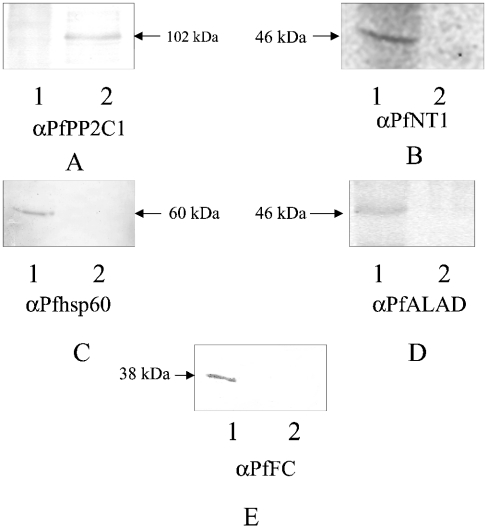
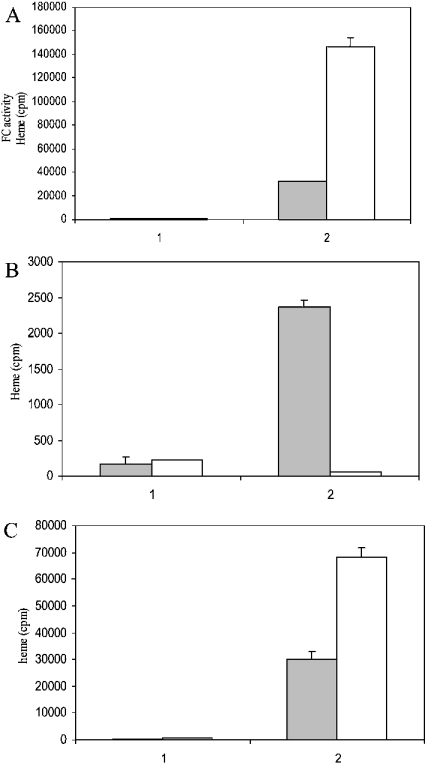
Western-blot analysis of the parasite organellar and cytosolic fractions reveals that the former is positive for PfNT1 (membrane marker) [22], PfHsp60 (mitochondrion marker) [20], PfALAD [9] and PfFC (apicoplast markers). The parasite cytosol is negative for all these markers and red cell haemoglobin (as assessed by Western-blot analysis), but is positive for PfPP2C1 (cytoplasmic marker) [23] (Figures 6A–6E). The cytosol is also positive for hFC (Figure 2). Thus the parasite cytosol is devoid of detectable parasite membrane proteins or red cell components as contaminants, but contains imported host enzymes of the haem-biosynthetic pathway [2,5,9]. Interestingly, FC enzyme assay reveals that the parasite cytosol manifests five times more activity when compared with the solubilized organellar fractions (Figure 7A). Total haem synthesis was measured in these fractions independently with two precursors, and interesting differences were seen. With [2-14C]glycine, the substrate for the first enzyme ALAS, the entire haem synthesis is almost fully accounted for by the solubilized organellar fraction (Figure 7B). With [4-14C]ALA, the substrate for ALAD, the cytosolic fraction accounts for nearly three times more haem synthesis when compared with the solubilized organellar fraction (Figure 7C). No incorporation into haem is seen with [1-14C]glycine, validating our measurement of the radioactivity incorporated into haem using the specific precursors.
Figure 6. Western-blot analysis of the parasite organellar and cytosolic fractions.
Antibodies to (A) PfPP2C1, (B) PfNT1, (C) PfHsp60, (D) PfALAD and (E) PfFC were used. 1, Organellar fraction; 2, cytosolic fraction. The total parasite pellet isolated from 2 ml of culture (10% parasitaemia) was fractionated into organellar and cytosolic fractions and the entire fractions were analysed by SDS/PAGE for Western-blot analysis.
Figure 7. Incorporation of [2-14C]glycine and [4-14C]ALA into haem and the assay of FC enzymic activity in the organellar and cytosolic fractions.
The cytosolic (shaded bars) or solubilized organellar (empty bars) fractions were incubated with [2-14C]glycine or [4-14C]ALA to measure incorporation into haem. FC enzymic activity was assayed in these fractions using 59FeCl3 and protoporphyrin as substrates. Results are expressed in terms of radioactivity in haem for the total cytosolic or solubilized organellar fraction obtained from 6 ml of parasite culture. (A) FC enzymic activity; 1, heat-denatured parasite fractions (control); 2, total enzymic activity. (B) [2-14C]Glycine incorporation into haem; 1, [1-14C]glycine incorporation (control); 2, [2-14C]glycine incorporation. (C) [4-14C]ALA incorporation into haem; 1, heat-denatured parasite fractions (control); 2, native parasite fractions; white bar, cytosol; shaded bar, solubilized organellar fraction.
DISCUSSION
In the present study, both hFC and PfFC have been detected in P. falciparum cultured in red blood cells. It is interesting to note that hFC, which is essentially a mitochondrial enzyme [10], is present in the red blood cell that is devoid of mitochondria. Although the nucleus is extruded during red cell maturation, mitochondria are disintegrated in the reticulocytes, involving the processes of autophagy and cytoplasmic degradation [24–26]. It appears that a fraction of the FC released in the process is still present in the red blood cell and acts as a source for import into the intra-erythrocytic parasite. However, the activity of ALAS, which is also a mitochondrial enzyme, is very low in the red cell and the host enzyme could not be detected in the parasite [2]. It has also been demonstrated that the red cells used to culture P. falciparum (98–99% red blood cells) are capable of haem synthesis using [4-14C]ALA as the substrate, indicating the presence of all subsequent enzymes of the pathway, including FC [2]. In the present study, hFC has been purified from human red blood cells, and its presence in the red blood cell and parasite has been demonstrated by immunofluorescence, immunoelectron microscopy and Western-blot analysis using specific antibodies.
It was predicted previously [6,12] that PfFC would be targeted to the mitochondrion rather than the apicoplast, since the protein sequence does not possess a discernible bipartite signal peptide and transit peptide sequence, considered essential for apicoplast targeting [13,27]. Surprisingly, the present study reveals that PfFC is not detectable in the parasite mitochondrion, but is present in the apicoplast. There are other examples such as triosephosphate isomerase and glycerol phosphate dehydrogenase that are consi- dered as apicoplast proteins, although not recognized as such by automated tools [28]. Application of PlasmoAP [29], a rule-based predictor for the apicoplast targeting of peptides within P. falciparum, suggests that only two enzymes of the haem-biosynthetic pathway, PfALAD and porphobilinogen deaminase, satisfy all the five sequence-based criteria for apicoplast localiz ation. Interestingly, PfALAS also scores fully in this prediction for apicoplast targeting, but with the verdict that it does not have a good signal sequence and seems to have an apicoplast-targeting sequence. However, it has already been demonstrated that PfALAS is in the parasite mitochondrion [4]. It is also possible that the targeting signal is buried elsewhere in some of the apicoplast-targeted proteins, or an apicoplast carrier protein may function to ferry the proteins that do not possess a bipartite signal. Since the isolated parasite organellar fraction is capable of independent haem synthesis without the involvement of cytosol, it is a distinct possibility that the apicoplast has the entire machinery for haem synthesis from ALA and the mitochondrion may only have the PfALAS. This would obviate shuttling of pathway intermediates back and forth between the mitochondrion and the apicoplast, with four membranes to traverse. The source of ALA for apicoplast haem synthesis is an open question. It has been suggested that ALA from mitochondria can get into the apicoplast [8], since the two organelles are seen in close proximity to each other [21]. However, plastids do have an independent machinery to make ALA from glutamic acid [30]. This pathway in the apicoplast has not been seriously investigated, since [2-14C]glycine (shemin pathway) is utilized for haem synthesis in the parasite and PfALAS is localized in the mitochondrion [1,2,4].
The present study indicates that the parasite makes haem in the cytosol utilizing imported host enzymes. Our previous studies have shown that hALAD is imported into the parasite cytoplasm and is functional in haem synthesis [2,5,9]. The present study also reveals that hFC is present significantly in the parasite cytoplasm and is active. hFC enzymic activity in the parasite cytosol is almost five times greater than the PfFC activity in the organellar fraction. This is based on the assumption that the assay conditions are similar for both hFC and PfFC. The fact that the isolated parasite cytosol is capable of making haem from ALA establishes that it has all the enzymes of the pathway. With [4-14C]ALA, the cytosol makes almost three times more haem when compared with the organellar fraction. However, with [2-14C]glycine, the isolated cytosol is unable to make haem since PfALAS is the only source for ALA and it is localized in the mitochondrion. In the normal pathway of haem biosynthesis in liver in vivo, the mitochondrion provides the ALA that is subsequently converted into porphyrins in the cytosol. However, the last two or three enzymes of the pathway in plants or liver are again localized in the mitochondrion, and the substrates have to shuttle back into the organelle to make haem [10,30,31]. For the parasite, the mitochondrion may serve as a source of ALA for cytoplasmic haem synthesis in vivo; however, the parasite is capable of completing haem synthesis in the cytosol using imported mature ALAD and FC and perhaps other intermediate host enzymes of the pathway. The ability of the cytosolic and organellar fractions to perform independent haem synthesis with their own complement of enzymes of the pathway and without any cross-over contribution from one fraction to another suggests that there could be two independent pools of haem and therefore of haemoproteins. Considering that the apicoplast PfALAD and PfFC account for only 10–20% of the total activities of these enzymes in the parasite ([9] and the present study), it appears that the apicoplast may only account for a minor proportion of haem synthesis by the parasite. As already stated, an additional site of haem synthesis is perhaps needed, since unlike plants that have numerous plastids, the parasite cell has only a single apicoplast. Although the apicoplast makes only 10–20% of the total haem synthesized in the parasite, it could still be functionally important for parasite survival.
Acknowledgments
This work was supported by a grant from the Department of Science and Technology and Department of Biotechnology, New Delhi.
References
- 1.Surolia N., Padmanaban G. De novo biosynthesis of haem offers a new chemotherapeutic target in the human malarial parasite. Biochem. Biophys. Res. Commun. 1992;187:744–750. doi: 10.1016/0006-291x(92)91258-r. [DOI] [PubMed] [Google Scholar]
- 2.Bonday Z. Q., Taketani S., Gupta P. D., Padmanaban G. Haem biosynthesis by the malarial parasite – import of δ-aminolaevulinate dehydratase from the host red cell. J. Biol. Chem. 1997;272:21839–21846. doi: 10.1074/jbc.272.35.21839. [DOI] [PubMed] [Google Scholar]
- 3.Wilson C. M., Smith A. B., Baylon R. V. Characterization of the δ-aminolaevulinate synthase gene homologue from P. falciparum. Mol. Biochem. Parasitol. 1996;75:271–276. doi: 10.1016/0166-6851(95)02531-6. [DOI] [PubMed] [Google Scholar]
- 4.Varadharajan S., Dhanasekaran S., Rangarajan P. N., Padmanaban G. Involvement of δ-aminolaevulinate synthase encoded by the parasite gene in de novo haem synthesis by Plasmodium falciparum. Biochem. J. 2002;367:321–327. doi: 10.1042/BJ20020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonday Z. Q., Dhanasekaran S., Rangarajan P. N., Padmanaban G. Import of host delta-aminolaevulinate dehydratase into the malarial parasite. Identification of a new drug target. Nat. Med. 2000;6:898–903. doi: 10.1038/78659. [DOI] [PubMed] [Google Scholar]
- 6.Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson K. E., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature (London) 2002;419:391–398. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato S., Wilson R. J. M. The genome of Plasmodium falciparum encodes an active δ-aminolaevulinic acid dehydratase. Curr. Genet. 2002;40:391–398. doi: 10.1007/s00294-002-0273-3. [DOI] [PubMed] [Google Scholar]
- 8.Sato S., Tews I., Wilson R. J. M. Impact of a plastid bearing endosymbiont on apicomplexan genomes. Int. J. Parasitol. 2000;30:427–439. doi: 10.1016/s0020-7519(99)00185-x. [DOI] [PubMed] [Google Scholar]
- 9.Dhanasekaran S., Chandra N. R., Sagar B. K. C., Rangarajan P. N., Padmanaban G. δ-Aminolaevulinic acid dehydratase from Plasmodium falciparum – indigenous vs imported. J. Biol. Chem. 2004;279:6934–6942. doi: 10.1074/jbc.M311409200. [DOI] [PubMed] [Google Scholar]
- 10.Harbin B. M., Dailey H. A. Orientation of ferrochelatase in bovine liver mitochondria. Biochemistry. 1985;24:366–370. doi: 10.1021/bi00323a019. [DOI] [PubMed] [Google Scholar]
- 11.Chow J. S., Singh D. P., Walker A. R., Smith A. G. Two different genes encode ferrochelatase in Arabidopsis: mapping, expression and cellular targeting of the precursor proteins. Plant J. 1998;15:531–541. doi: 10.1046/j.1365-313x.1998.00235.x. [DOI] [PubMed] [Google Scholar]
- 12.Masuda T., Suzuki T., Shimada H., Ohta H., Takaniya K. Subcellular localization of two types of ferrochelatase in cucumber. Planta. 2003;217:602–609. doi: 10.1007/s00425-003-1019-2. [DOI] [PubMed] [Google Scholar]
- 13.Sato S., Wilson R. J. M. Proteobacteria-like ferrochelatase in the malaria parasite. Curr. Genet. 2003;42:292–300. doi: 10.1007/s00294-002-0360-5. [DOI] [PubMed] [Google Scholar]
- 14.Waller R. F., Keeling P. J., Donald R. G., Striepen B., Handman E., Lang-Unnasch N., Cowman A. F., Besra G. S., Roos D. S., McFadden G. I. Nuclear-encoded proteins target to the plastid of Toxoplasma gondii and Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L. B. Fluorescent labeling of mitochondria. Methods Cell Biol. 1989;30:103–123. doi: 10.1016/s0091-679x(08)60190-9. [DOI] [PubMed] [Google Scholar]
- 16.Trager W., Jensen J. B. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 17.Fitch C. D., Chevli R., Banyal H., Phillips G., Pfaller M. A., Krogstad D. J. C. Lysis of Plasmodium falciparum by ferriprotoporphyrin IX and a chloroquine-ferriprotoporphyrin IX complex. Antimicrob. Agents Chemother. 1982;21:819–822. doi: 10.1128/aac.21.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sirawaraporn W. Preparation of crude fractions from host-parasite complex. In: Panyim S., Wilairat P., Yuthawong Y., editors. Genetic Engineering Techniques in Tropical Diseases Research. Bangkok: Mahidol University; 1985. pp. 407–412. [Google Scholar]
- 19.Dailey H. A., Fleming J. E., Harbin B. E. Purification and characterization of mammalian and chicken ferrochelatase. Methods Enzymol. 1986;123:401–408. doi: 10.1016/s0076-6879(86)23049-9. [DOI] [PubMed] [Google Scholar]
- 20.Das A., Syin C., Fujioka H., Zheng H., Goldman N., Aikawa M., Kumar N. Molecular characterization and ultrastructural localization of Plasmodium falciparum Hsp60. Mol. Biochem. Parasitol. 1997;88:95–104. doi: 10.1016/s0166-6851(97)00081-9. [DOI] [PubMed] [Google Scholar]
- 21.Hopkins J., Fowler R., Krishna S., Wilson I., Mitchell G., Bannister L. The plastid in Plasmodium falciparum asexual blood stages: a three dimensional ultrastructural analysis. Protist. 1999;150:283–295. doi: 10.1016/S1434-4610(99)70030-1. [DOI] [PubMed] [Google Scholar]
- 22.Rager N., Ben Mamoun C., Carter N. S., Goldberg D. E., Ullman B. Localization of the Plasmodium falciparum PfNT1 nucleoside transporter to the parasite plasma membrane. J. Biol. Chem. 2001;276:41095–41099. doi: 10.1074/jbc.M107037200. [DOI] [PubMed] [Google Scholar]
- 23.Ben Mamoun C., Sullivan D. J., Banerjee R., Goldberg D. E. Identification and characterization of an unusual double serine/threonine protein phosphatase 2C in the malarial parasite Plasmodium falciparum. J. Biol. Chem. 1998;273:11241–11247. doi: 10.1074/jbc.273.18.11241. [DOI] [PubMed] [Google Scholar]
- 24.Grullich C., Duvoisin R. M., Wiedmann M., van Leyden K. Inhibition of 15-lipoxygenase leads to delayed organelle degradation in the reticulocyte. FEBS Lett. 2001;489:51–54. doi: 10.1016/s0014-5793(01)02080-4. [DOI] [PubMed] [Google Scholar]
- 25.Heynen M. J., Tricot G., Verwilghen R. L. Autophagy of mitochondria in rat bone marrow erythroid cells. Relation to nuclear extrusion. Cell Tissue Res. 1985;239:235–239. doi: 10.1007/BF00214924. [DOI] [PubMed] [Google Scholar]
- 26.Schewe T., Halangk W., Hiebsch C., Papoport S. M. A lipoxygenase in rabbit reticulocytes which attacks phospholipids and intact mitochondria. FEBS Lett. 1975;60:149–152. doi: 10.1016/0014-5793(75)80439-x. [DOI] [PubMed] [Google Scholar]
- 27.Waller R. F., Reed M. B., Cowman A. F., McFadden G. I. Protein targeting to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ralph S. A., Van Dooren G. G., Waller R. F., Crawford M. J., Fraunholz M. J., Foth B. J., Tonkin C. J., Roos D. S., McFadden G. I. Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Microbiol. Rev. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 29.Foth B. J., Ralph S. A., Tonkin C. J., Struck N. S., Fraunholz M., Roos D. S., Cowman A. F., McFadden G. I. Dissecting apicoplast targeting in the malarial parasite Plasmodium falciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- 30.Jordan P. N. Biosynthesis of 5-aminolevulinic acid and its transformation into coporphyrinogen in animals and bacteria. In: Dailey H. A., editor. Biosynthesis of Haem and Chlorophyll. New York: McGraw-Hill; 1990. pp. 55–121. [Google Scholar]
- 31.Papenbrock J., Grimm B. Regulatory network of tetrapyrrole biosynthesis – studies of intracellular signalling involved in metabolic and developmental control of plastids. Planta. 2001;213:667–681. doi: 10.1007/s004250100593. [DOI] [PubMed] [Google Scholar]