Abstract
We detected a protein in rabbit skeletal muscle extracts that was phosphorylated rapidly by SGK1 (serum- and glucocorticoid-induced kinase 1), but not by protein kinase Bα, and identified it as NDRG2 (N-myc downstream-regulated gene 2). SGK1 phosphorylated NDRG2 at Thr330, Ser332 and Thr348 in vitro. All three residues were phosphorylated in skeletal muscle from wild-type mice, but not from mice that do not express SGK1. SGK1 also phosphorylated the related NDRG1 isoform at Thr328, Ser330 and Thr346 (equivalent to Thr330, Ser332 and Thr348 of NDRG2), as well as Thr356 and Thr366. Residues Thr346, Thr356 and Thr366 are located within identical decapeptide sequences GTRSRSHTSE, repeated three times in NDRG1. These threonines were phosphorylated in NDRG1 in the liver, lung, spleen and skeletal muscle of wild-type mice, but not in SGK1−/− mice. Knock-down of SGK1 in HeLa cells using small interfering RNA also suppressed phosphorylation of the threonine residues in the repeat region of NDRG1. The phosphorylation of NDRG1 by SGK1 transformed it into an excellent substrate for GSK3 (glycogen synthase kinase 3), which could then phosphorylate Ser342, Ser352 and Ser362 in the repeat region. Incubation of HeLa cells with the specific GSK3 inhibitor CT 99021 increased the electrophoretic mobility of NDRG1 in HeLa cells, demonstrating that this protein is phosphorylated by GSK3 in cells. Our results identify NDRG1 and NDRG2 as physiological substrates for SGK1, and demonstrate that phosphorylation of NDRG1 by SGK1 primes it for phosphorylation by GSK3.
Keywords: glycogen synthase kinase 3 (GSK3), n-myc downstream-regulated gene (NDRG), p53, phosphorylation, serum- and glucocorticoid-induced kinase 1 (SGK1)
Abbreviations: CDK, cyclin-dependent kinase; DYRK1A, dual-specificity tyrosine phosphorylated and regulated kinase 1A; ENaC, epithelial sodium channel; ERK, extracellular-signal-regulated kinase; FOXO3a, forkhead box O3a; GSK3, glycogen synthase kinase 3; GST, glutathione S-transferase; IGF-1, insulin-like growth factor-1; KESTREL, kinase substrate tracking and elucidation; LDS, lithium dodecyl sulphate; MALDI-TOF, matrix-assisted laser-desorption ionization–time-of-flight; MAPK, mitogen-activated protein kinase; NDRG, n-myc downstream-regulated gene; PDK1, 3-phosphoinositide-dependent kinase 1; PKB, protein kinase B; PKC, protein kinase C; RSK, p90 ribosomal S6 kinase; SAPK, stress-activated protein kinase; S6K, p70 ribosomal S6 kinase; SGK1, serum- and glucocorticoid-induced kinase 1; siRNA, small interfering RNA
INTRODUCTION
SGK1 (serum- and glucocorticoid-induced kinase 1) is an immediate early gene whose level of expression is greatly enhanced within 1 h of exposing most cells to serum, glucocorticoids or other agonists (reviewed in [1,2]). In addition, the activity of SGK1 increases in response to signals that activate phosphoinositide 3-kinase and elevate the intracellular level of PtdIns(3,4,5)P3 (reviewed in [1,2]). This ‘second messenger’ induces the activation of an as yet unidentified protein kinase(s), which phosphorylate(s) the C-terminal hydrophobic motif of SGK1. This creates a docking site for PDK1 (3-phosphoinositide-dependent kinase 1), allowing it to phosphorylate a threonine residue located in the activation loop, which activates SGK1 [3].
SGK1 is a member of the AGC subfamily of protein kinases and most closely resembles PKB (protein kinase B; also called Akt), with 54% identity in the catalytic domain. Based on studies with synthetic peptide substrates, SGK1 was found to have similar specificity requirements to PKB, phosphorylating serine and threonine residues that lie in Arg-Xaa-Arg-Xaa-Xaa-Ser/Thr- motifs [4,5]. SGK1 and PKB have also been shown to phosphorylate the same proteins both in vitro and when overexpressed in cells, such as GSK3 (glycogen synthase kinase 3) [5,6] and the transcription factor FOXO3a (forkhead box O3a; formerly called FKHRL1) [7], suggesting that they might have some physiological substrates in common. However, in embryonic stem cells that express a PDK1 mutant which activates PKB normally but is unable to activate SGK1, IGF-1 (insulin-like growth factor-1)-induced phosphorylation of GSK3 and FOXO3a is not impaired, indicating that SGK1 is not rate limiting for the phosphorylation of these proteins under the conditions tested [3]. Moreover, PKB and SGK must phosphorylate at least some distinct substrates in cells, because the phenotypes of mice that do not express these protein kinases are quite different. For example, mice that do not express PKBβ have impaired insulin-stimulated glucose uptake into muscle and become diabetic as they age [8]. In contrast, mice that do not express SGK1 have an impaired ability to adequately decrease Na+ excretion when dietary NaCl is restricted [9].
SGK1 has been implicated in the activation of a number of ion channels (reviewed in [10]). This is thought to be mediated by the SGK1-catalysed phosphorylation of the protein ubiquitin ligase NEDD4-2, because phosphorylation of NEDD4-2 in vitro and in overexpression studies impairs its ability to ubiquitinate the ENaC (epithelial sodium channel) and target it for degradation, thereby increasing expression of the ENaC at the cell membrane [11,12]. However, definitive evidence that SGK1 is required for the site-specific phosphorylation of endogenous NEDD4-2 in vivo is still lacking. Moreover, the level of ENaC in the apical membrane and collecting ducts of the kidney is only decreased moderately in SGK1−/− mice [9], and there is no impairment of renal water and electrolyte secretion at standard NaCl intake. This suggests that regulation of the channel may be more complex and/or that another SGK isoform [13] or a related protein kinase, such as PKB, may be able to substitute for SGK1, at least partially, if it is not expressed.
The identification of physiological substrates for SGK1 has proved difficult for several reasons; first because potent and selective inhibitors of this enzyme are not yet available, and secondly because mice that do not express SGK1 have only recently been generated [9]. Moreover, searching databases for proteins with Arg-Xaa-Arg-Xaa-Xa-Ser/Thr motifs is of little help because, even if these sites are accessible for phosphorylation in the native proteins, they may be phosphorylated by PKB or other protein kinases with similar specificity determinants, such as isoforms of RSK (p90 ribosomal S6 kinase) and S6K (p70 S6 kinase) [14]. To try to identify novel substrates for SGK1, we therefore decided to adopt the KESTREL (kinase substrate tracking and elucidation) approach [15]. In this method, cell extracts are subjected to ion exchange chromatography, and aliquots of the fractions collected are incubated with Mg[γ-32P]ATP in the absence or presence of two or more closely related protein kinases that have similar substrate specificity requirements in vitro. The aim is to detect proteins that are phosphorylated selectively by just one of these protein kinases and then investigate whether they are bona fide physiological substrates in appropriate follow-up studies. Using this approach, we were able to identify elongation factor 2-kinase as a protein that is inactivated by phosphorylation at Ser359 catalysed by SAPK4 (stress-activated protein kinase 4; also called p38δ), but not by the closely related isoforms SAPK2a/p38α or SAPK3/p38γ [15].
In the present paper, we have identified NDRG2 (n-myc downstream-regulated gene 2) as a protein in muscle extracts that is phosphorylated efficiently by SGK1, but not by PKB, and we go on to show that this protein and the related NDRG1 isoform are indeed physiological substrates for SGK1. In the accompanying paper [16], we use the same approach to identify a new physiological substrate for PKB that is not phosphorylated by SGK1.
MATERIALS AND METHODS
Materials
[γ-32P]ATP, ECL® reagent and materials for protein purification were obtained from Amersham Biosciences (Chalfont St Giles, Bucks., U.K.). Unlabelled ATP and ‘complete EDTA-free protease inhibitor cocktail’ were from Roche Molecular Biochemicals (Lewes, E. Sussex, U.K.), Precision prestained protein molecular mass markers from Bio-Rad (Hemel Hempstead, Herts., U.K.) and cell culture media, precast Bis-Tris SDS/10% polyacrylamide gels, running buffer and transfer buffer were from Invitrogen (Paisley, Scotland, U.K.). Foetal bovine serum was purchased from Cambrex (Wokingham, Surrey, U.K.), ImmobilonP membranes from Millipore (Watford, Herts., U.K.) and LY 294002 from Merck Biosciences (Nottingham, U.K.). Microcystin-LR was obtained from Dr Linda Lawton (Robert Gordon University, Aberdeen, Scotland, U.K.). All peptides were synthesized at the Molecular Recognition Centre, University of Bristol, U.K. All other chemicals were of the highest purity and purchased from Merck (Poole, Dorset, U.K.) or Sigma-Aldrich (Poole, Dorset, U.K.).
Cloning of NDRG1 and NDRG2
NDRG2 (AAL08624) was amplified from IMAGE EST 4215141 with the 5′NDRG2 and 3′NDRG2 oligonucleotides shown below using EXPAND HIFI DNA Polymerase (Roche). The PCR product was cloned into pCR2.1 (Invitrogen) and sequenced by The DNA Sequencing Service (School of Life Sciences, University of Dundee, Dundee, U.K.). The insert was found to contain a deletion, which was corrected by PCR using the oligonucleotides MP290 and MP291 (see below) with the 5′NDRG2 and 3′NDRG2 probes. The new fragment was cloned into pCR2.1 and sequenced. pCR2.1 NDRG2 was digested with BamHI and subcloned into the same site in pGEX6P-1 to produce pGEX6P-1 NDRG2.
NDRG1 (XP_005243) was amplified by reverse transcription–PCR using an Access RT-PCR System (Promega, Southampton, U.K.) from total RNA extracted from HeLa cells with the oligonucleotides 5′NDRG1 and 3′NDRG1. The product was cloned into pCR2.1, sequenced, and the resulting plasmid pCR2.1 NDRG1 was again digested with BamHI and subcloned into the same site in pGEX6P-1 to produce pGEX6P-1 NDRG1.
Oligonucleotides
Oligonucleotide sequences were as follows: 5′NDRG1, GGATCCGCCACCATGGACTACAAGGACGACGATGACAAGTCTCGGGAGATGCAGGATGTAGACC; 3′NDRG1, GGATCCCTAGCAGGAGACCTCCATGGACTTG; 5′NDRG2, GAATTCGCCACCATGGACTACAAGGACGACGATGACAAGGCGGAGCTGCAGGAGGTGCA; 3′NDRG2, GAATTCTCAACAGGAGACCTCCATGGTGTGC; MP290, AGGAGACCAAGCACCTCATGAAGATGCAGTGGTGGAATGTAACTCAAAACTGGATCCCACC; MP291, ACCACTGCATCTTCATGAGGTGCTTGGTCTCCTACCACCAGCATCACAGGGCACC.
Protein expression and purification
His-tagged proteins were expressed in insect Sf21 cells, and GST (glutathione S-transferase) fusion proteins in Escherichia coli. His-tagged PKBα-(118–480) in which Ser473 was mutated to Asp, SGK1-(60–431) in which Ser422 was mutated to Asp and S6K1-(1–421) in which Thr412 was mutated to Glu were purified by chromatography on nickel-nitrilotriacetate–agarose and then maximally activated by phosphorylation with His-tagged PDK1-(52–556). N-terminally His-tagged full-length RSK1-(1–735) was purified and then maximally activated with PDK1 and full-length GST–ERK2 (extracellular-signal-regulated kinase 2). PDK1 binds very tightly to heparin–Sepharose, and was removed from all activated protein kinases by passage through this column. GST–ERK2 was removed by passage through glutathione–agarose. The activated protein kinases were concentrated by repurification on nickel-nitrilotriacetate–agarose, dialysed against 50 mM Tris/HCl, pH 7.5, 270 mM sucrose, 150 mM NaCl, 0.1 mM EGTA, 0.1% (v/v) 2-mercaptoethanol, 0.2 mM PMSF and 1 mM benzamidine, and stored at −80 °C.
GST–NDRG1 and GST–NDRG2 fusion proteins were expressed in E. coli BL21 CodonPlus-RIL (Merck Biosciences) and purified on glutathione–Sepharose. Purified proteins were dialysed against 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.1 mM EGTA, 0.1% (v/v) 2-mercaptoethanol, 0.2 mM PMSF and 1 mM benzamidine, snap frozen in liquid nitrogen and stored at −80 °C.
Antibodies
Antibodies that recognize PKBα and SGK1 were raised against these proteins in sheep. The production of antibodies that recognize SGK1 phosphorylated at Ser422 has been described previously [5]. Antibodies recognizing PKBα phosphorylated at Ser473 were obtained from Cell Signalling Technologies (Hitchin, Herts., U.K.). Antibodies against NDRG1 and NDRG2 were made in sheep using recombinant full-length NDRG1 or a GST-fusion of full-length NDRG2. The antisera were then purified by affinity chromatography on CH-Sepharose to which the proteins had been coupled covalently and, in the case of the NDRG2, the antibodies were passed through GST–Sepharose to remove anti-GST antibodies. Phospho-specific antibodies that recognize NDRG2 phosphorylated at Thr330, Ser332, Thr330 plus Ser332, Thr348, Ser350 and Thr348 plus Ser350 were made against the peptides LSRSRpTASLTSAA, LSRSRTApSLTSAA, LSRSRpTApSLTSAA, GNRSRSRpTLSQSS, GNRSRSRTLpSQSS and GNRSRSRpTLpSQSS respectively (where pT is phosphothreonine and pS is phosphoserine). An antibody that recognizes NDRG1 phosphorylated at Thr346, Thr356 and Thr366 (termed anti-p3xThr) was raised against the nonapeptide RSRSHpTSEG, whose sequence is common to all three sites. The phospho-specific antibodies were purified by affinity chromatography on CH-Sepharose to which the phosphopeptide immunogen had been coupled covalently, and used for immunoblotting in the presence of the unphosphorylated peptide antigens (10 μg/ml) to neutralize any antibodies that recognized the unphosphorylated NDRG1 or NDRG2.
Purification of a specific substrate for SGK1
Rabbit skeletal muscle extracts were prepared as described [17] and passed through a Sephadex G-25 column equilibrated with 30 mM Mops, pH 7.0, 5% (v/v) glycerol, 0.1% (v/v) 2-mercaptoethanol and 0.03% (w/v) Brij 35 (buffer A). This material, containing 2 g of protein, was chromatographed on a 25 ml column of heparin–HP-Sepharose. The protein that did not bind to the column was diluted with 5 vol. of 30 mM Tris/HCl, pH 7.5, 5% (v/v) glycerol, 0.1% (v/v) 2-mercaptoethanol and 0.03% (w/v) Brij 35 (buffer B), passed through a 0.22 μm filter and chromatographed on an 8 ml Source15 Q-Sepharose (HR10/10) equilibrated in buffer B. The column was washed with 40 ml of buffer B, and developed with a 160 ml non-linear gradient from 0 to 1 M NaCl in buffer B. Fractions of 4 ml were collected at a flow rate of 2 ml/min. Aliquots of each fraction were diluted 5-fold into 30 mM Tris/HCl, pH 7.5, 2 mM MgCl2, 10 mM 2-mercaptoethanol, 0.1 mM EGTA and 1.0 μg/ml each of aprotinin and leupeptin. The fractions were then incubated for 4 min at 30 °C at a 6-fold final dilution with 20 nM [γ-32P]ATP (2.5×106 c.p.m.) with or without SGK1 or PKBα (each at 0.3 unit/ml). The reactions were stopped by the addition of 10 μl of 320 mM Tris/HCl, pH 6.8, 8% (w/v) SDS, 20 mM EDTA, 32% (v/v) glycerol, 1.14 M 2-mercaptoethanol and 0.02% (w/v) Bromophenol Blue (SDS sample buffer) heated for 3 min at 100 °C, subjected to SDS/PAGE, electroblotted on to ImmobilonP membranes and autoradiographed to reveal phosphorylated proteins.
Assay of protein kinases
These were assayed at 30 °C as described previously [18,19]. One unit of PKBα, SGK1, RSK1 or S6K1 activity was that amount which catalysed the phosphorylation of 1 nmol of the standard substrate peptide CROSStide (GRPRTSSFAEG) in 1 min [20].
Preparation of cell and tissue extracts
HeLa cells were cultured on 10-cm-diam. dishes in Dulbecco's modified Eagle's medium supplemented with 2 mM glutamine, 10% (w/v) foetal bovine serum, penicillin and streptomycin. HeLa cells were deprived of serum for 4 h. Cells were then treated with or without inhibitors for 1 h, before stimulation for 3 h with 10% (w/v) foetal bovine serum. Cells were lysed in 0.25 ml of 50 mM Tris/HCl (pH 7.5), 1 mM EGTA, 1 mM EDTA, 1% (w/v) Triton X-100, 1 mM sodium orthovanadate, 50 mM NaF, 5 mM sodium pyrophosphate, 0.27 M sucrose, 1 μM microcystin-LR, 0.1% (v/v) 2-mercaptoethanol and complete proteinase inhibitor cocktail. Protein concentrations of lysates were determined using the Bradford method with BSA as standard. Lysates were resuspended in LDS (lithium dodecyl sulphate) sample buffer (Invitrogen) and heated for 10 min at 70 °C prior to SDS/PAGE.
Mice were killed and a variety of tissues were removed, snap frozen in liquid nitrogen, powdered and extracted with the cell lysis buffer described above, then centrifuged at 16000 g for 15 min at 4 °C to pellet insoluble material. The supernatant was decanted, denatured in LDS and used for immunoblotting.
Production and transfection of siRNA (small interfering RNA)
Synthetic sense and antisense oligonucleotides were synthesized by the Oligonucleotide Synthesis Service (School of Life Sciences, University of Dundee, Dundee, U.K.) and amplified with a Silencer siRNA construction kit (Ambion, Huntington, U.K.). The NDRG1 sense oligonuceotide was 5′-AAGTTACTCTGCATTTCTTCCCCTGTCTC-3′, while the antisense oligonucleotide was 5′-AAGGAAGAAATGCAGAGTAACCCTGTCTC-3′. The SGK1 sense oligonuceotide was 5′-AATATTTGTAGCAGCAATGCTCCTGTCTC-3′ and the antisense oligonucleotide was 5′-AAAGCATTGCTGCTACAAATACCTGTCTC-3′. HeLa cells were transfected with 100 nM siRNA duplexes using lipofectAMINE 2000™ (Invitrogen) and cultured for 48 h before stimulation.
GSK3 inhibitors
The Chiron inhibitor CT 99021 (6-{2-[4-(2,4-dichlorophenyl)-5-(4-methyl-1H-imidazol-2-yl)-pyrimidin-2-ylamino]ethylamino}nicotinonitrile) was efficiently synthesized in three steps in 7% overall yield using a convergent approach from 2,4-dichlorobenzoyl chloride and 6-chloronicotinonitrile respectively following standard literature procedures ([21] and references cited therein). The AstraZeneca inhibitor AR-A0144-18 [1-(4-methoxybenzyl)-3-(5-nitrothiazol-2-yl)urea] was prepared in a single step from 5-nitrothiazol-2-ylamine and 1-isocyanoto-4-methoxybenzene in 73% yield following the method of Bhat and coworkers [22]. The GlaxoSmithKline inhibitors SB 415286 and SB 216763 were obtained from Sigma.
RESULTS
SGK1 phosphorylates proteins of 45 kDa and 35 kDa in skeletal muscle extracts
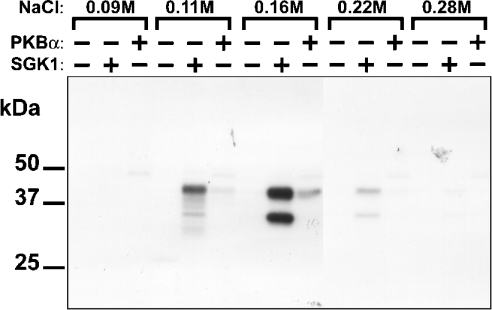
Desalted rabbit skeletal muscle extracts were fractionated on heparin–Sepharose, and the proteins not retained by this column were chromatographed on Source-Q. To identify putative substrates for SGK1 or PKBα, the eluted fractions were incubated with Mg[γ-32P]ATP in the absence or presence of these protein kinases. The reactions were subjected to SDS/PAGE and the phosphorylated proteins visualized by autoradiography. Two proteins of apparent molecular masses 45 kDa and 35 kDa, eluting at 0.16 M NaCl, were phosphorylated strongly by SGK1, but only weakly by PKBα (Figure 1) and therefore merited further investigation.
Figure 1. Identification of 45 kDa and 35 kDa proteins that are phosphor- ylated by SGK1, but only weakly by PKBα.
The proteins not retained on heparin–Sepharose were chromatographed on Source 15 Q-Sepharose, as described in the Materials and methods section. Each fraction was incubated for 4 min at 30 °C with 2 mM MgCl2/20 nM [γ-32P]ATP in the absence (−) or presence (+) of 0.3 unit/ml SGK1 or PKBα, denatured in SDS, subjected to SDS/PAGE, transferred to ImmobilonP membranes and autoradiographed. Two substrates for SGK1 with apparent molecular masses of 45 kDa and 35 kDa, eluting between 0.11 M and 0.22 M NaCl, were detected by autoradiography.
The peak fraction from Source Q contained many proteins, two of which co-migrated with the 32P-labelled bands (Figure 2A). These bands were excised from the gel and tryptic mass fingerprinting revealed that both were the product of NDRG2 (Table 1), suggesting that this protein might be the SGK1 substrate.
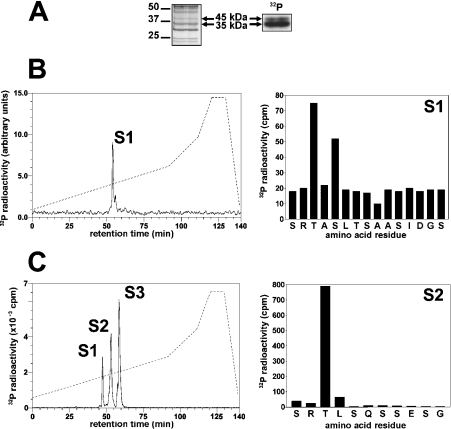
Figure 2. Identification of the residues in NDRG2 that are phosphorylated by SGK1 in vitro.
Partially purified rabbit skeletal muscle NDRG2 was phosphorylated by incubation for 30 min with 10 mM MgCl2/0.1 mM [γ-32P]ATP and 1.0 unit/ml SGK1, denatured in LDS and subjected to SDS/PAGE. (A) The gel was stained with colloidal Coomassie Blue (left panel) or autoradiographed (right panel). (B) The 45 kDa 32P-labelled band from (A) was excised, digested with trypsin and the digest chromatographed on a Vydac C18 column (Separations Group) equilibrated in 0.1% (v/v) trifluoroacetic acid. The column was developed with an acetonitrile gradient (left panel, broken line) at a flow rate of 0.8 ml/min, and fractions of 0.4 ml were collected. The major 32P-labelled peptide S1 (left panel, solid line) was subjected to MS and the sites of phosphorylation identified by solid-phase sequencing (right panel) after coupling the peptide to a Sequalon-AA membrane [23,52]. (C) Bacterially expressed human NDRG2 (1.6 μM) was phosphorylated with SGK1 as in (A) and analysed as in (B).
Table 1. Identification of the 45 and 35 kDa substrates for SGK1 as NDRG2.
The 32P-labelled 35 and 45 kDa substrates (Figure 2A) were excised from the gel, digested with trypsin and analysed on a Perseptive Biosystems Elite STR MALDI-TOF mass spectrometer with saturated α-cyanocinnamic acid as the matrix, as described in [23]. The mass spectrum was acquired in the reflector mode and was internally mass calibrated. The tryptic peptide ions obtained were scanned against the Swiss-Prot and Genpep databases using the MS-FIT program of Protein Prospector. Met-ox, methionine oxidation.
| Mass | Residue no. | ||||
|---|---|---|---|---|---|
| Peptide | Submitted | Matched | Start | End | Peptide |
| 45 kDa | 793.4 | 793.4 | 242 | 247 | DLNFER |
| 1101.5 | 1101.5 | 177 | 185 | GWMDWAAHK | |
| 1117.5 | 1117.5 | 177 | 185 | GWMDWAAHK+1 Met-ox | |
| 1760.0 | 1759.9 | 62 | 76 | RPAILTYHDVGLNYK | |
| 2406.2 | 2406.3 | 155 | 176 | YALNHPDTVEGLVLINIDPNAK | |
| 2582.2 | 2582.3 | 255 | 276 | CPVMLVVECNSKLDPTQTSFLK | |
| 2592.3 | 2592.2 | 292 | 313 | LTEAFKYFLQGMGYMASSCMTR | |
| 35 kDa | 759.4 | 759.4 | 26 | 32 | EAELAAR |
| 793.4 | 793.4 | 242 | 247 | DLNFER | |
| 1101.5 | 1101.5 | 177 | 185 | GWMDWAAHK | |
| 1117.5 | 1117.5 | 177 | 185 | GWMDWAAHK+1 Met-ox | |
| 1149.7 | 1149.6 | 267 | 276 | LDPTQTSFLK | |
| 1760.0 | 1759.9 | 62 | 76 | RPAILTYHDVGLNYK | |
| 2406.3 | 2406.3 | 155 | 176 | YALNHPDTVEGLVLINIDPNAK | |
Identification of the residues in NDRG2 phosphoryated by SGK1
To investigate whether the SGK1 substrate and NDRG2 were the same protein, the partially purified material was phosphorylated as in Figure 2(A), and the 32P-labelled bands were digested with trypsin and chromatographed on a C18 column (see [23] for a detailed description of the methodology used). The 45 kDa (Figure 2B, left panel) and 35 kDa (results not shown) bands both gave rise to one major phosphopeptide S1, whose mass was identical to that of residues 328–343 of murine NDRG2 (SRTASLTSAASIDGSR) plus two phosphate groups. Thus the SGK1 substrate was indeed NDRG2. Solid-phase sequencing identified the sites of phosphorylation as Thr330 and Ser332 (Figure 2B, right panel).
Human GST–NDRG2 could be maximally phosphorylated to 1.5 mol/mol of protein, indicating that at least two sites had been phosphorylated. Chromatography on the C18 column showed three tryptic phosphopeptides, S1, S2 and S3 (Figure 2C, left panel). Peptide S1 comprised residues 328–343 phosphorylated at both Thr330 and Ser332 (results not shown). The elution of this peptide at a slightly lower acetonitrile concentration than the equivalent peptide from rabbit NDRG2 (see above) is explained by the substitution of Ile339 by Val and Ser342 by Asn in the human protein. Peptides S2 and S3 both corresponded to the peptide starting at Ser346 and terminating at the C-terminus of NDRG2. Solid-phase sequencing showed that the site of phosphorylation was Thr348 (Figure 2C, right panel). Partial oxidation of the methionine residue in this peptide may account for its elution at two positions on the C18 column.
Identification of the residues in NDRG2 phosphorylated by protein kinases with similar substrate specificities to SGK1
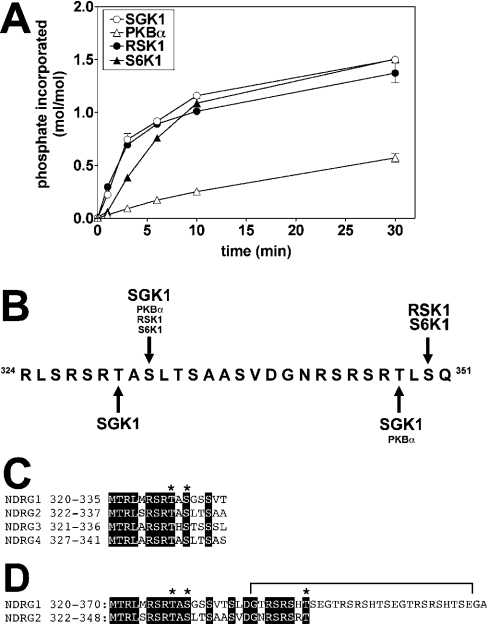
Studies with synthetic peptide substrates have shown that RSK isoforms and S6K1 have similar specificity requirements to PKBα and SGK isoforms [14]; RSK isoforms preferentially phosphorylate Arg/Lys-Xaa-Arg-Xaa-Xaa-Ser- motifs and S6K1 phosphorylates Arg/Lys-Xaa-Arg-Xaa-Xaa-Ser/Thr- motifs [14]. PKBα phosphorylated human GST–NDRG2 at a much lower initial rate than SGK1. However, RSK1 and S6K1 phosphorylated NDRG2 at similar rates to SGK1 when all four kinases were matched for activity towards CROSStide (Figure 3A).
Figure 3. Phosphorylation of NDRG2 by SGK1, PKBα and RSK1.
(A) GST–NDRG2 (0.5 μM) was phosphorylated at 30 °C with the indicated protein kinases (each at 1.0 unit/ml) as in Figure 2(A). Reactions were stopped by the addition of LDS and subjected to SDS/PAGE. The GST–NDRG2 bands were excised from the gel and analysed by Čerenkov counting. (B) Location of the residues on human NDRG2 phosphorylated by SGK1, PKBα, RSK1 and S6K1. (C) Two of the sites in NDRG2 phosphorylated by SGK1 (Thr330 and Ser332) are conserved in the other three NDRG isoforms. Conserved residues are highlighted by a black background, and the sites phosphorylated by SGK1 are marked by asterisks, (D) Residues 320–370 of NDRG1 aligned with residues 322–355 of NDRG2. Conserved residues are highlighted by a black background, and the sites in NDRG2 phosphorylated by SGK1 that are conserved in NDRG1 are marked by asterisks. Residues 339–368 of NDRG1 (bracketed) comprise three decapeptide repeats, whose function is described elswhere in the text.
The sites on human GST–NDRG2 phosphorylated by PKBα, RSK1 and S6K1 were identified as described above for SGK1, and the results are summarized in Figure 3(B). All four kinases were found to phosphorylate NDRG2 at Ser332 in vitro, while PKBα and SGK1 phosphorylated Thr348 and RSK1 and S6K1 phosphorylated Ser350. Thr330, which lies in an ‘atypical’ sequence, was uniquely phosphorylated by SGK1. These sites all lie close to the C-terminus of NDRG2, which terminates at residue 371.
NDRG1 is also phosphorylated by SGK1
Human NDRG2 is one of four members of the NDRG family, the others being NDRG1, NDRG3 and NDRG4. Thr330, Ser332 and the sequences surrounding them are conserved in all four family members (Figure 3C), while Thr348 is conserved in NDRG1 and NDRG2 (Figure 3D). The RSK1 and S6K1 phosphorylation site in NDRG2 is not conserved in the other three isoforms. However, interestingly, NDRG1 contains three copies of the 10-amino-acid repeat sequence GTRSRSHTSE between residues 339 and 369, each of which contains an Arg-Xaa-Arg-Xaa-Xaa-Thr- motif (Figure 3D). The first of these contains Thr346, the residue equivalent to Thr348 of NDRG2. We therefore cloned human NDRG1 as a GST-fusion protein and studied its phosphorylation in vitro.
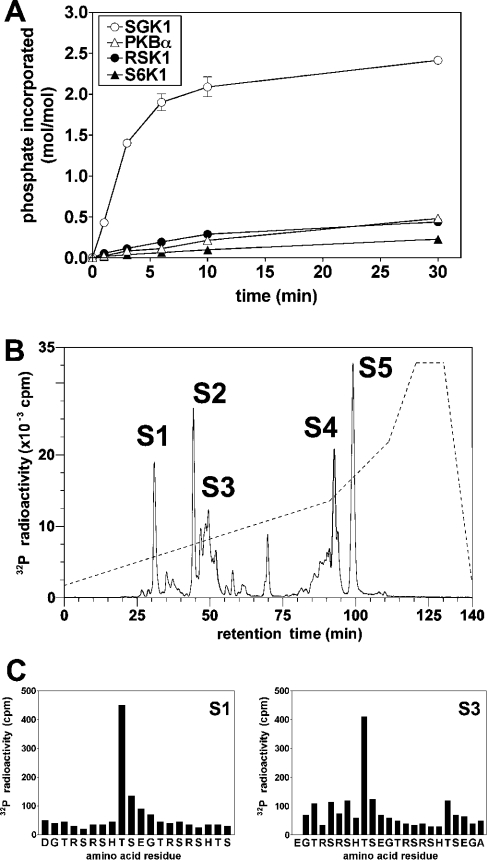
SGK1 phosphorylated NDRG1 far more rapidly and to a much greater extent than did PKBα, RSK1 or S6K1 (Figure 4A). The phosphorylation of NDRG1 by SGK1 approached a plateau at approx. 2.5 mol of phosphate per mol of protein. The maximally phosphorylated 32P-labelled protein was cleaved with N-Asp proteinase, which gave rise to multiple phosphopeptides after chromatography on the C18 column (Figure 4B). Phosphopeptide S1 was identified by MALDI-TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS/MS as the monophosphorylated derivative of the peptide comprising residues 338–357, and was shown by solid-phase sequencing to be phosphorylated at Thr346 (Figure 4C, left panel). Peptide S2 comprised residues 358–372, phosphorylated at Thr366 (results not shown), while the peptides in the S3 region were identified as mono- and di-phosphorylated derivatives of the peptide comprising residues 348–372. The diphosphorylated peptide was phosphorylated at Thr356 and Thr366 (Figure 4C, right panel). The heterogeneous nature of peptide S3 may result from the presence of two different monophosphorylated derivatives and a diphosphorylated derivative and/or variable binding of cations to the three histidine residues in this peptide. The tight binding of this region of NDRG1 to Ni2+ and Cu2+ has been noted previously [24]. Peptides S4 and S5 both comprised the peptide corresponding to residues 303–337 containing one phosphate in which the four methionine residues had become partially oxidized. These peptides were digested with CNBr and subjected to solid-phase sequencing. Some [32P]phosphate was released after the fourth cycle and even more after the sixth cycle, which should correspond to Thr328 and Ser330 respectively if CNBr cleaved after Met324 as expected. Taken together, the results indicate that NDRG1 is phosphorylated by SGK1 at Thr346, Thr356 and Thr366 in the 10-residue repeats, as well as at Thr328 and Ser330, the two residues conserved in all NDRG family members.
Figure 4. Identification of the residues in NDRG1 phosphorylated by SGK1.
(A) GST–NDRG1 (0.5 μM) was phosphorylated at 30 °C with the indicated protein kinases (each at 1.0 unit/ml) as in Figure 2(A). Reactions were stopped by the addition of LDS and subjected to SDS/PAGE. The GST–NDRG1 bands were excised from the gel and analysed by Čerenkov counting. (B) 32P-labelled NDRG1 from the 30 min time point in (A) was digested with N-Asp proteinase and the digest chromatographed on a Vydac C18 column as in Figure 2(B). The five major peptides S1–S5 were then analysed as described in the Results section. (C) Peptides S1 and S3 were subjected to solid-phase sequencing as in Figure 2(C) to identify the sites of phosphorylation.
Generation of antibodies that recognize NDRG1 and NDRG2
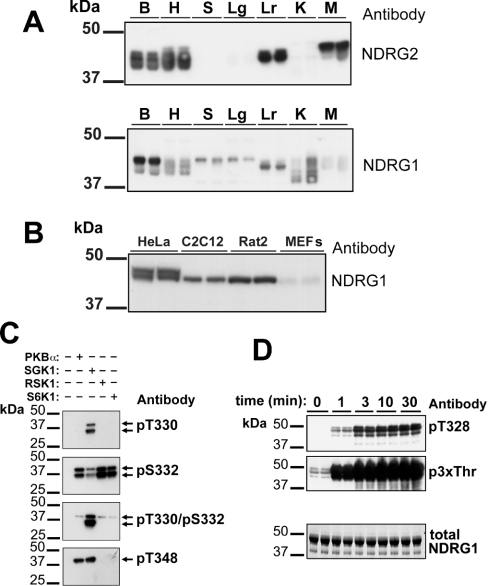
In order to study the phosphorylation of NDRG1 and NDRG2 in vivo, we generated polyclonal antibodies against the bacterially expressed human proteins. These antibodies were used to show that the murine NDRG2 protein is expressed in brain, heart, liver and striated muscles, but not in spleen, lung or kidney. In contrast, the NDRG1 protein is expressed at various levels in all tissues examined (Figure 5A). These results are consistent with the previously reported levels of the mRNAs in these tissues [25]. We also observed that NDRG1 and NDRG2 migrated as multiple bands after SDS/PAGE, which may arise from alternative splicing of the gene in the case of NDRG2 [26] and/or by differential modification of both proteins by phosphorylation and other post-translational mechanisms that may vary from tissue to tissue. NDRG1 migrated as a doublet in HeLa cells, and as single bands in C2C12 myoblasts and Rat2 fibroblasts (but not mouse embryonic fibroblasts) that co-migrated with the lower band of the doublet seen in HeLa cells (Figure 5B). The NDRG2 protein was not detected in any cell line examined (results not shown). The upper band of the doublet in HeLa cells appears to result from phosphorylation alone, because the omission of inhibitors of serine/threonine-specific protein phosphatases from the lysis buffer (NaF, sodium pyrophosphate, microcystin LR) resulted in its disappearance and conversion into the faster migrating band of the doublet (results not shown).
Figure 5. Characterization of antibodies that recognize NDRG1 and NDRG2.
All antibodies were used at a concentration of 1 μg/ml. (A) Extracts from brain (B), heart (H), spleen (S), lung (Lg), liver (Lr) and skeletal muscle (M) (15 μg of protein) were subjected to SDS/PAGE, transferred to ImmobilonP membranes and immunoblotted with either anti-NDRG2 (upper panel) or anti-NDRG1 (lower panel) antibodies using the ECL® detection method. The data show the results with extracts from two individual mice. (B) Lysates (20 μg of protein) from HeLa cells, Rat2 fibroblasts, C2C12 myoblasts and mouse embryonic fibroblasts (MEFs) were immunoblotted with anti-NDRG1 antibody as in (A). (C) Partially purified NDRG2 from rabbit skeletal muscle was maximally phosphorylated with SGK1, PKBα, RSK1 or S6K1 as in Figure 3(A), except that unlabelled ATP was used. Each sample was subjected to SDS/PAGE, transferred to ImmobilonP membranes and immunoblotted as in (A) using the phospho-specific antibodies raised against the sites on NDRG2 phosphorylated by SGK1. (D) Human NDRG1 was phosphorylated with SGK1 for the times indicated as in Figure 3(A), except that unlabelled ATP was used. The samples were electrophoresed and immunoblotted with antibodies that recognize NDRG1 phosphorylated at Thr328 (i.e. the antibody raised against the equivalent site in NDRG2, Thr330; top panel) or at Thr346, Thr356 and Thr366 (i.e. using the anti-p3xThr antibody; middle panel). Total NDRG1 was detected by staining with Coomassie Blue (bottom panel).
To investigate the phosphorylation of NDRG2 in vivo, we raised phosphorylation site-specific antibodies that recognized this protein only when it was phosphorylated at Thr330, Ser332, Thr330 plus Ser332, or Thr348. To examine the specificities of these antibodies, partially purified rabbit skeletal muscle NDRG2 was phosphorylated in vitro with SGK1, PKBα, RSK1 or S6K1 (Figure 5C). Consistent with the sequencing data presented above and summarized in Figure 3(B), phosphorylation of NDRG2 at Thr330 or Thr330 plus Ser332 was detected with the anti-pThr330 antibody only after phosphorylation by SGK1, while phosphorylation at Ser332 was detected with the anti-pSer332 antibody after phosphorylation by any of the four protein kinases tested. The weaker signal obtained with the anti-pSer332 antibody after phosphorylation with SGK1 may be explained by the failure of this antibody to recognize NDRG2 phosphorylated at both Thr330 and Ser332. Similarly, phosphorylation at Thr348 was detected after phosphorylation by SGK1 and PKBα, but not after phosphorylation by RSK1 and S6K1, as expected. Only the fulllength 45 kDa form of NDRG2 could be phosphorylated at Thr348, suggesting that the 35 kDa species detected in the initial KESTREL screen (Figure 1) is C-terminally truncated, since it lacks the epitope recognized by this antibody.
The amino acid sequence surrounding Thr328 and Ser330 of NDRG1 is very similar to that surrounding Thr330 and Ser332 of NDRG2 and, for this reason, NDRG1 phosphorylated at Thr328 was also recognized by the phospho-specific antibody that recognizes pThr330 of NDRG2 (Figure 5D, upper panel). We also raised an antibody (termed anti-p3xThr) that recognizes all three phosphothreonines (Thr346, Thr356 and Thr366) present in the repeated motif of NDRG1 (Figure 5D, middle panel).
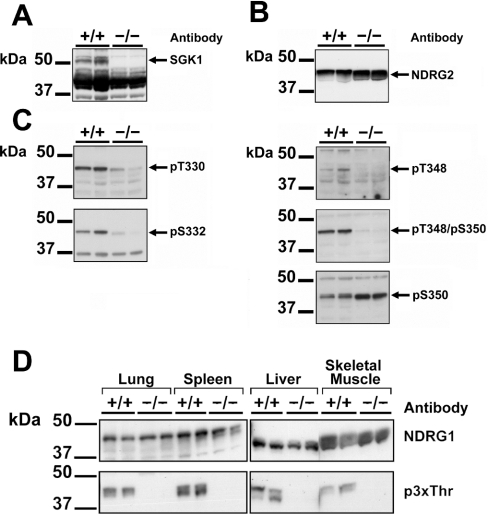
Phosphorylation of NDRG2 is severely impaired in the skeletal muscle of SGK1−/− mice
Since NDRG2 is highly expressed in striated muscles, we examined the phosphorylation of this protein in skeletal muscle of wild-type mice (SGK1+/+) and homozygous SGK1 knockout mice (SGK1−/−) that do not express this protein kinase (Figure 6A). The expression of NDRG2 protein was similar in skeletal muscle from wild-type and knockout mice (Figure 6B), but the phosphorylation at Thr330 and Ser332 was greatly reduced in the latter, and almost no phosphorylation at Thr348 could be detected with either the anti-pThr348 or a further antibody that recognizes NDRG2 phosphorylated at both Thr348 and Ser350 (Figure 6C). In contrast, there was no decrease in the phosphorylation of Ser350 (Figure 6C), which is not phosphorylated by SGK1 in vitro (Figure 3B). The apparent increase in phosphorylation of Ser350 in the SGK1−/− muscle may be explained by decreased phosphorylation of Thr348 in these mice, allowing improved recognition by the anti-pSer350 antibody.
Figure 6. Phosphorylation of NDRG2 and NDRG1 is impaired in the tissues of SGK1−/− mice.
(A) Skeletal muscle extracts (30 μg of protein) prepared from two individual SGK1−/− mice and two individual wild-type SGK1+/+ mice were subjected to SDS/PAGE, transferred to ImmobilonP membranes and immunoblotted with an antibody raised against the SGK1 protein. (B) As in (A), except that the membranes were immunoblotted with anti-NDRG2 antibody. (C) As in (A), except that the membranes were immunoblotted with the five phospho-specific antibodies indicated. (D) Tissue extracts (30 μg of protein) prepared from two individual SGK1−/− mice and two individual wild-type SGK1+/+ mice were immunoblotted as in (A) using antibodies raised against the NDRG1 protein (upper panel) or the anti-p3xThr antibody raised against the phosphorylated decapeptide repeat in NDRG1 (lower panel).
Taken together, these data establish that SGK1 expression and activity is required for the phosphorylation of NDRG2 at Thr330, Ser332 and Thr348 in skeletal muscle.
Phosphorylation of NDRG1 is severely impaired in SGK1−/− mice
We next examined the phosphorylation of NDRG1 in extracts prepared from several tissues of SGK+/+ and SGK−/− mice. The anti-p3xThr antibody detected phosphorylation of the C-terminal region of NDRG1 in skeletal muscle, spleen, liver and lung of SGK+/+ mice, which was totally absent in the same tissue extracts prepared from SGK1−/− mice. In contrast, the total level of NDRG1 protein was similar in the tissues from wild-type and knockout animals. These results establish that NDRG1 is also a physiological substrate for SGK1 (Figure 6D).
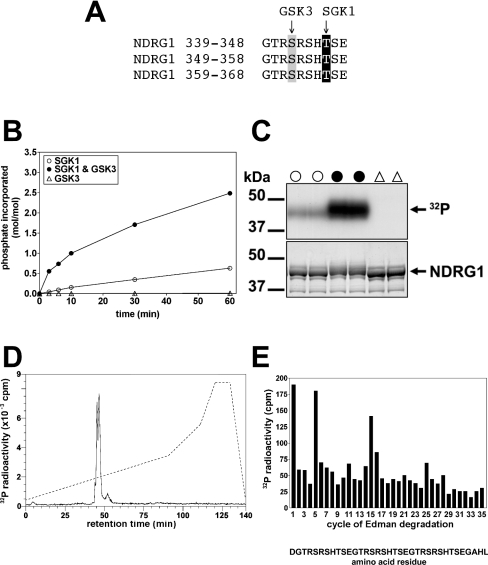
NDRG1 is a substrate for GSK3
The three C-terminal repeats of NDRG1, GTRSRSHTSE, possess a serine residue four amino acids N-terminal to the threonine residues phosphorylated by SGK1 (Figure 7A). Since the consensus sequence for phosphorylation by GSK3 is Ser/Thr-Xaa-Xaa-Xaa-pSer/pThr [27], this suggested that GSK3 might be capable of phosphorylating each serine residue provided that the threonine residues had already been phosphorylated (Figure 7A). We tested this possibility in vitro, and showed that bacterially expressed NDRG1 indeed became an excellent substrate for GSK3, with phosphorylation by this protein kinase approaching 3 mol per mol, after prior phosphorylation of the three threonines by SGK1 (Figure 7B). Phosphorylation by SGK1 in vitro caused a small decrease in the electrophoretic mobility of NDRG1, while the combined phosphorylation with SGK1 plus GSK3 induced a much larger decrease in mobility. Similar observations were made after phosphorylation of NDRG2 in vitro by SGK1 and GSK3 (results not shown).
Figure 7. Phosphorylation of NDRG1 by SGK1 primes it for phosphorylation by GSK3.
(A) Amino acid sequence of the decapeptide repeats in NDRG1 showing the sites phosphorylated by SGK1 (highlighted in black) and GSK3 (highlighted in grey). (B) Recombinant human NDRG1 was phosphorylated for 60 min in the absence (△) or presence (○, ●) of 1.0 unit/ml SGK1 as in Figure 3(A), except that unlabelled ATP was used. The reactions were then incubated for the times indicated in the presence of 1.0 unit/ml GSK3 (●, △) or the absence of this protein kinase (○) and Mg[γ-32P]ATP. The samples were then analysed as in Figure 3(A). (C) Two aliquots of each of the 60 min time point samples from (B) were subjected to SDS/PAGE, then autoradiographed (upper panel) or stained with Coomassie Blue (lower panel). The symbols above the lanes correspond to those in (B). (D) NDRG1 that had been maximally phosphorylated by GSK3 as in (B) was digested with N-Asp proteinase and chromatographed on a Vydac C18 column as described in the legend to Figure 2(B). The solid line shows the 32P-radioactivity and the broken line the acetonitrile gradient. (E) The 32P-labelled peak from (D) was subjected to solid-phase sequencing as in Figure 2(B) to identify the sites phosphorylated by GSK3. The interpretation of the results is described in the text.
NDRG1 was maximally phosphorylated by GSK3 (Figure 7C), digested with N-Asp proteinase and chromatographed on a Vydac C18 column. All of the 32P-radioactivity eluted as a closely spaced doublet (Figure 7D). MALDI-TOF MS analysis of each fraction within the radioactive peaks revealed that they all comprised residues 338–372 in different states of phosphorylation. The earliest eluting fractions contained the hexaphosphorylated peptide (molecular mass 4287.4 Da) and later eluting fractions contained pentaphosphorylated (4207.4 Da) and tetraphosphorylated (4127.4 Da) peptides. Solid-phase sequencing confirmed that GSK3 had phosphorylated all three serines in the repeat motif, i.e. Ser342, Ser352 and Ser362 (Figure 7E). The release of 32P-radioactivity at the first cycle of Edman degradation resulted from coupling of the peptide through the side-chain carboxylate of N-terminal Asp without any other anchoring [23].
Comparison of the potency and specificity of several small-molecule inhibitors of GSK3
Several compounds have been developed recently that are reported to be potent and relatively selective inhibitors of GSK3. In order to decide which of these compounds would be most suitable to examine whether NDRG1 was phosphorylated by GSK3 in cells, we made a side-by-side comparison of the effects of these compounds on a panel of 30 protein kinases (Table 2) [18,19]. We also determined IC50 values with the protein kinases that were inhibited most potently (Table 3). CT 99021 [28] was the most potent and selective of these compounds. It inhibited GSK3 350-fold more potently than CDK2 (cyclin-dependent kinase 2) and did not significantly affect any other protein kinase in the panel. AR-A0144-18 [21] was slightly less potent, but equally selective. SB 216763 and SB 415286 [29] inhibited GSK3 with similar potency to AR-A0144-18, but also inhibited CDK2 and DYRK1A (dual-specificity tyrosine phosphorylated and regulated kinase 1A) at slightly higher concentrations. RSK1 and PKCα (protein kinase Cα) were inhibited significantly. We therefore selected CT 99021 and AR-A0144-18 for the studies described below.
Table 2. Comparison of the specificities of four GSK3 inhibitors.
Assays for each protein kinase were carried out at the ATP concentration specified, which approximates the Km value for each kinase. Each protein kinase used was expressed, purified and assayed as described previously [18,19]. The results shown are percentage activity compared with DMSO-only controls for each enzyme tested, and are means of duplicate determinations. Similar results were obtained in another independent experiment. Abbreviations: MKK, MAPK kinase; JNK, c-Jun N-terminal kinase; MAPKAP-K; MAPK-activated protein kinase; MSK, mitogen- and stress-activated kinase; PRAK, p38-regulated/activated kinase; PKA, cAMP-dependent protein kinase; ROCK, Rho-dependent kinase; AMPK, AMP-activated protein kinase; CHK, checkpoint kinase; PHK, phosphorylase kinase; LCK, lymphocyte kinase; CSK; C-terminal Src kinase; NEK, NIMA-related protein kinase.
| Activity (%) | |||||||
|---|---|---|---|---|---|---|---|
| Protein kinase | ATP (μM) | CT99021 (1 μM) | CT99021 (10 μM) | AR-AO144-18 (1 μM) | AR-AO144-18 (10 μM) | SB 216763 (10 μM) | SB 415286 (10 μM) |
| MKK1 | 5 | 102±3 | 67±4 | 84±8 | 84±8 | 66±4 | 28±8 |
| ERK2 | 50 | 89±3 | 90±4 | 100±7 | 103±5 | 82±5 | 75±9 |
| JNK | 20 | 78±5 | 73±1 | 96±7 | 80±4 | 95±9 | 96±8 |
| SAPK2a/p38α | 50 | 100±0 | 93±1 | 100±8 | 111±9 | 87±2 | 86±5 |
| SAPK2b/p38β2 | 20 | 80±5 | 91±5 | 95±3 | 99±2 | 79±2 | 89±3 |
| SAPK3/p38γ | 5 | 92±2 | 97±0 | 99±3 | 89±8 | 65±2 | 81±1 |
| SAPK4/p38δ | 5 | 86±5 | 84±3 | 91±5 | 86±4 | 81±5 | 92±2 |
| RSK1 | 50 | 91±0 | 80±5 | 109±2 | 108±5 | 31±3 | 14±4 |
| MAPKAP-K2 | 20 | 90±1 | 85±5 | 87±5 | 83±1 | 89±5 | 83±4 |
| MSK1 | 20 | 99±0 | 89±3 | 104±3 | 98±4 | 58±9 | 89±4 |
| PRAK | 20 | 82±1 | 72±7 | 92±6 | 100±3 | 79±1 | 87±7 |
| PKA | 20 | 87±5 | 98±3 | 89±7 | 101±7 | 91±0 | 94±2 |
| PKC | 20 | 90±4 | 85±7 | 93±2 | 90±5 | 30±7 | 30±0 |
| PDK1 | 20 | 91±4 | 88±8 | 105±0 | 105±8 | 85±3 | 74±4 |
| PKB | 5 | 94±5 | 67±0 | 95±6 | 81±4 | 53±1 | 79±2 |
| SGK | 20 | 101±8 | 59±7 | 94±8 | 79±0 | 82±4 | 45±4 |
| S6K1 | 20 | 123±3 | 111±4 | 97±5 | 119±5 | 109±5 | 119±1 |
| GSK3b | 5 | 3±0 | 1±1 | 6±1 | 1±1 | 1±0 | 1±0 |
| ROCK-II | 20 | 96±1 | 82±5 | 102±1 | 84±6 | 76±5 | 78±5 |
| AMPK | 50 | 90±8 | 71±2 | 96±4 | 89±1 | 64±3 | 21±0 |
| CHK1 | 20 | 90±3 | 82±2 | 110±8 | 107±4 | 49±5 | 53±7 |
| CK2 | 5 | 96±6 | 74±4 | 86±5 | 87±3 | 90±0 | 85±7 |
| PHK | 50 | 96±3 | 86±1 | 73±3 | 75±8 | 77±77 | 84±0 |
| LCK | 50 | 88±2 | 83±4 | 94±2 | 95±6 | 64±3 | 65±5 |
| CSK | 20 | 88±5 | 96±4 | 107±5 | 101±1 | 96±4 | 88±4 |
| CDK2–cyclin A | 20 | 78±9 | 13±8 | 86±3 | 45±1 | 14±2 | 6±2 |
| CK1 | 20 | 90±7 | 89±8 | 95±7 | 91±9 | 84±4 | 78±6 |
| DYRK1a | 50 | 87±3 | 81±1 | 91±8 | 78±3 | 6±0 | 8±2 |
| NEK6 | 50 | 89±2 | 98±6 | 103±1 | 91±3 | 91±3 | 93±4 |
| NEK2a | 50 | 109±3 | 84±6 | 90±1 | 93±6 | 91±1 | 111±6 |
Table 3. Concentrations of CT 99021, AR-AO144-18, SB 216763 and SB 415286 required for 50% inhibition of GSK3, CDK2 and DYRK1A at the ATP concentrations specified.
| Inhibitor | ATP (μM) | Kinase | IC50 (μM) |
|---|---|---|---|
| CT 99021 | 5 | GSK3β | 0.04 |
| AR-A0144-18 | 5 | GSK3β | 0.14 |
| SB 216763 | 5 | GSK3β | 0.10 |
| SB 415286 | 5 | GSK3β | 0.20 |
| CT 99021 | 20 | CDK2 | 1.40 |
| AR-A0144-18 | 20 | CDK2 | 6.90 |
| SB 216763 | 20 | CDK2 | 0.95 |
| SB 415286 | 20 | CDK2 | 0.80 |
| SB 216763 | 50 | DYRK1A | 0.80 |
| SB 415286 | 50 | DYRK1A | 0.90 |
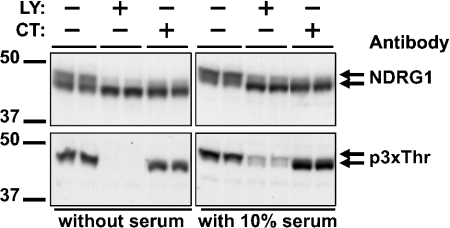
Inhibitors of GSK3 and phosphoinositide 3-kinase suppress the phosphorylation of NDRG1 in HeLa cells
In HeLa cells deprived of serum for 4 h, NDRG1 migrated as a doublet (Figure 8, upper panels). Stimulation with serum increased the proportion of NDRG1 in the upper band of the doublet, and only the upper band was phosphorylated on the threonine residues in the C-terminal repeats (Figure 8, lower panel). Phosphorylation of the C-terminal repeats was prevented by incubation of the cells with the phosphoinositide 3-kinase inhibitor LY 294002, which suppresses the phosphorylation and hence the activity of SGK1 (see Introduction), but not by incubation of the cells with PD 184352, which suppresses activation of the classical MAPK (mitogen-activated protein kinase) cascade [18,30] and hence the activation of RSK isoforms, or with rapamycin, which inhibits the protein kinase mTOR (mammalian target of rapamycin) and hence the activation of S6K1 (results not shown). After incubation of the cells with 2 μM CT 99021 (Figure 8, upper panel) or AR-A0144-18 (results not shown), NDRG1 no longer migrated as a doublet, with nearly all the material now migrating in the position of the lower band, although phosphorylation of the threonine residues in the C-terminal repeats was unaffected. The results indicate that phosphorylation by both SGK and GSK3 is required for a large decrease in the electrophoretic mobility of NDRG1. These results demonstrate that NDRG1 is phosphorylated by GSK3 in HeLa cells, presumably at the three serine residues in the C-terminal repeats. Thus the C-terminal region of NDRG1 is likely to be phosphorylated at up to eight sites, five catalysed by SGK1 (Thr328, Ser330, Thr346, Thr356 and Thr366) and three by GSK3 (Ser342, Ser352 and Ser362).
Figure 8. Effects of GSK3 and phosphoinositide 3-kinase inhibitors on the phosphorylation of NDRG1 in HeLa cells.
HeLa cells were deprived of serum for 4 h, incubated for 1 h with (+) or without (−) 2 μM CT 99021 (CT) or 50 μM LY 294002 (LY) and then for 3 h with or without 10% (v/v) serum in the continued absence or presence of the inhibitor. The phosphorylation state of NDRG1 was analysed after subjecting 20 μg of cell lysate protein to SDS/PAGE followed by transfer to ImmobilonP membranes and immunoblotting with the anti-NDRG1 or anti-p3xThr antibodies.
Knock-down of SGK1 protein expression levels by siRNA ablates phosphorylation of NDRG1 in HeLa cells
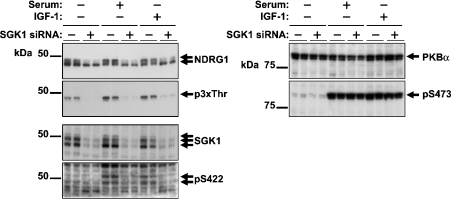
In order to provide further evidence that the threonines in the C-terminal repeats were phosphorylated by SGK1 in HeLa cells, we used siRNA directed towards SGK1 to reduce the expression of this protein kinase. Cells transfected with siRNA against SGK1 showed greatly reduced expression of this protein, but no reduction in NDRG1, whereas cells transfected with siRNA against NDRG1 showed reduced expression of NDRG1 protein, but no reduction in SGK1 (results not shown). Treatment of HeLa cells with the SGK1 siRNA did not reduce the total amount of PKB or its ability to become phosphorylated at Ser473 in response to serum or IGF-1 (Figure 9). Moreover, as a result of the SGK1 knock-down, the phosphorylation of NDRG1 detected by the anti-p3xThr antibody was greatly reduced and, as a consequence, the upper band of the doublet detected with the anti-NDRG1 antibody disappeared (Figure 9). We conclude from the siRNA data that SGK1, and not PKBα, is largely responsible for phosphorylation of the C-terminal threonine residues in NDRG1, under the conditions studied.
Figure 9. Knock-down of SGK1 expression by siRNA in HeLa cells inhibits phosphorylation of the decapeptide repeat region of NDRG1.
HeLa cells were transiently transfected with synthetic siRNA duplexes targeted towards SGK1 or NDRG1 (see the Materials and methods section) and incubated for 48 h. They were then deprived of serum for 4 h and stimulated for 3 h with or without 10% (v/v) serum or 25 ng/ml IGF-1. Aliquots of 20 μg of cell lysate protein were subjected to SDS/PAGE and, after transfer to ImmobilonP membranes, immunoblotted with anti-NDRG1, anti-p3xThr, anti-SGK1, anti-pSer422, anti-PKB and anti-pSer473 antibodies.
DISCUSSION
The work described in this paper has confirmed the power of the KESTREL technique [15] for identifying physiological substrates of protein kinases. Using this method we detected NDRG2 as a protein that is phosphorylated much more rapidly by SGK1 than by PKBα (Figures 1 and 3), two protein kinases with similar substrate specificity requirements, and this led us to discover that this is also true for the NDRG1 isoform (Figure 4). The residues on NDRG1 and NDRG2 that are phosphorylated by SGK1 in vitro were also phosphorylated on the endogenous proteins present in tissue extracts of wild-type mice, but not in extracts from mice that do not express SGK1 (Figure 6). Moreover, knock-down of SGK1 in HeLa cells, which did not affect the level of PKB or its ability to be phosphorylated in response to serum or IGF-1, greatly reduced the phosphorylation of NDRG1 at the sites that are targeted by SGK1 (Figure 9). These results provide strong evidence that both NDRG isoforms are indeed physiological substrates for SGK1.
While the present paper was in preparation, another laboratory reported that NDRG2 was phosphorylated by PKBα in vitro and when overexpressed in cells, and that the major site of phosphorylation was Thr348 [31]. The phosphorylation of NDRG2 that had been overexpressed in C2C12 cells was stimulated by insulin and suppressed by pharmacological inhibition of phosphoinositide 3-kinase, which would be equally consistent with phosphorylation by PKBα or SGK1. Although we cannot exclude the possibility that PKBα may be able to phosphorylate endogenous NDRG2 in cells under conditions that we have not studied, our results point to SGK1 being the relevant protein kinase, and suggest that the overexpression of PKBα may be mimicking effects that are carried out by SGK1 under physiological conditions.
Burchfield et al. [31] reported that PKCθ phosphorylated NDRG2 predominantly at Ser332 and that the overexpression of PKCθ suppressed the phosphorylation of overexpressed NDRG2 at Thr348, as judged by immunoblotting with a ‘pan-PKB substrate’ antibody. Similar results were obtained when cells overexpressing NDRG2 were exposed to PMA. These authors suggested that the phosphorylation of Ser332 may induce a conformational change that prevents PKBα from phosphorylating Thr348. However, it is also possible that the phosphorylation of Ser350 occurs under these conditions and prevents the recognition of Thr348 by the phospho-specific antibody used. PMA is known to activate the classical MAPK cascade and hence induces the activation of RSK isoforms, which we have shown to phosphorylate NDRG2 at Ser350 in vitro (Figure 3B). Suppression of recognition by a phosphospecific antibody as a result of phosphorylation of a nearby site is a problem that is encountered frequently (e.g. [32,33]). The sequence surrounding Thr330 of SGK1 does not conform to the normal consensus sequence for this protein kinase and would therefore not have been detected by Burchfield et al. [31].
We have also shown that the phosphorylation of NDRG1 by SGK1 at three threonines in vitro transforms it into an excellent substrate for GSK3, allowing the latter to phosphorylate Ser342, Ser352 and Ser362. Moreover, phosphorylation of NDRG1 by GSK3 is physiologically relevant (Figure 8), since incubation of HeLa cells with a highly specific inhibitor of GSK3 caused a marked increase in the electrophoretic mobility of NDRG1. Nearly all of the physiological substrates for GSK3 reported to date are primed by the phosphorylation of a serine residue, rather than a threonine residue [34]. The present results therefore demonstrate that the specific docking site on GSK3 for the priming phosphate [35,36] can accommodate phosphothreonine as well as phosphoserine. Interestingly, we have also identified NDRG2 as a GSK3 substrate in a completely independent KESTREL screen (G. Auld, C. Morris and P. Cohen, unpublished work). Although more work is needed to establish whether NDRG2 is also a physiological substrate for GSK3, this seems likely in view of the number of potential sites on this protein for phosphorylation by this protein kinase. For example, the phosphorylation of Thr330 or Ser332 by SGK1 may prime NDRG2 for phosphorylation by GSK3 at Ser326 and Ser328 respectively, the phosphorylation of Thr348 by SGK1 may prime for phosphorylation at Ser344, while the phosphorylation of Thr350 by RSK or another protein kinase(s) may prime for GSK3-mediated phosphorylation at Ser346 (see Figure 3B).
NDRG1 was initially identified as a protein whose expression is up-regulated by a variety of stress signals [37–39], p53 expression and DNA damage [40], and its expression is inhibited under conditions of cell growth [41]. It was also identified as a protein whose expression is up-regulated in mouse embryos deficient in N-myc [42] and down-regulated in tumours [43]. Its expression is induced by stimuli that promote differentiation in cancer cells [41,44], and it has been reported to be a metastasis suppressor gene [44,45]. NDRG1 has also been reported to be regulated downstream of p53 function during the mitotic spindle checkpoint [46]. It has been reported that NDRG2 is induced by mineralocorticoid hormones, such as aldosterone [47], which also induce the expression of SGK1 [1,2]. However, the precise physiological functions(s) of NDRG1 and NDRG2, and hence the role of phosphorylation, is unknown. Others have reported that NDRG1 shows a cytoplasmic and nuclear localization [48], and more recently that NDRG1 is associated with the mitotic spindle apparatus [46], and we have confirmed both results (not shown). However, we have so far been unable to detect any change in the subcellular distribution of NDRG1 in serum-stimulated HeLa cells pretreated with either LY 294002 or CT 99021.
A mutation in the gene encoding NDRG1 appears to be the cause of Hereditary Motor and Sensory Neuropathy-Lom, also called Charcot–Marie–Tooth disease type 4D [49,50], a hereditary disease that is characterized by muscle weakness, sensory loss and neural deafness, symptoms caused by demyelination of peripheral nerves. Thus NDRG1 may be necessary for axonal survival. Very recently, NDRG1-deficient mice have been generated [51]. The sciatic nerve degenerates in these mice, with demyelination at 5 weeks of age, and the animals show muscle weakness. However, myelination of the Schwann cells in the sciatic nerve is normal after 2 weeks. A more detailed analysis suggested that NDRG1 deficiency leads to Schwann cell dysfunction, suggesting that NDRG1 is essential for maintenance of the myelin sheaths in the peripheral nerves. It will clearly be of interest to examine whether some of the phenotypic effects observed in NDRG1−/− mice are also present in SGK1−/− mice.
Acknowledgments
This study was supported by the U.K. Medical Research Council, The Royal Society, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co., Merck KGaA and Pfizer. We thank the protein production and antibody purification teams in the Division of Signal Transduction Therapy (DSTT) (School of Life Sciences, University of Dundee, U.K.), co-ordinated by Hilary McLauchlan and James Hastie, for expression and purification of enzymes and affinity purification of antibodies. We also thank The DNA Sequencing Service (School of Life Sciences, University of Dundee; www.dnaseq.co.uk) for DNA sequencing.
References
- 1.Lang F., Cohen P. Regulation and physiological roles of serum- and glucocorticoid-induced protein kinase isoforms. Science STKE 2001. 2001;108:RE17. doi: 10.1126/stke.2001.108.re17. [DOI] [PubMed] [Google Scholar]
- 2.Firestone G. L., Giampaolo J. R., O'Keeffe B. A. Stimulus-dependent regulation of serum and glucocorticoid-induced protein kinase (SGK) transcription, sub-cellular location and enzymatic activity. Cell. Physiol. Biochem. 2003;13:1–12. doi: 10.1159/000070244. [DOI] [PubMed] [Google Scholar]
- 3.Collins B. J., Deak M., Arthur J. S. C., Armit L. J., Alessi D. R. In vivo role of the PIF-binding docking site of PDK1 defined by knock-in mutation. EMBO J. 2003;22:4202–4211. doi: 10.1093/emboj/cdg407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alessi D. R., Caudwell F. B., Andjelkovic M., Hemmings B. A., Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP-K1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi T., Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- 6.Park J., Leong M. L., Buse P., Maiyar A. C., Firestone G. L., Hemmings B. A. Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunet A., Park J., Tran H., Hu L. S., Hemmings B. A., Greenberg M. E. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol. Cell. Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae S. S., Cho H., Mu J., Birnbaum M. J. Isoform-specific regulation of insulin-dependent glucose uptake by Akt/protein kinase B. J. Biol. Chem. 2003;278:49530–49536. doi: 10.1074/jbc.M306782200. [DOI] [PubMed] [Google Scholar]
- 9.Wulff P., Vallon V., Huang D. Y., Volkl H., Yu F., Richter K., Jansen M., Schlunz M., Klingel K., Loffing J., et al. Impaired renal Na+ retention in the sgk1-knockout mouse. J. Clin. Invest. 2002;110:1263–1268. doi: 10.1172/JCI15696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang F., Henke G., Embark H. M., Waldegger S., Palmada M., Bohmer C., Vallon V. Regulation of ion channels by the serum and glucocorticoid-inducible kinase; implications for transport, excitability and cell proliferation. Cell. Physiol. Biochem. 2003;13:41–50. doi: 10.1159/000070248. [DOI] [PubMed] [Google Scholar]
- 11.Debonneville C., Flores S. Y., Kamynina E., Plant P. J., Tauxe C., Thomas M. A., Munster C., Chraibi A., Pratt J. H., Horisberger J. D., et al. Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na+ channel cell surface expression. EMBO J. 2001;20:7052–7059. doi: 10.1093/emboj/20.24.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder P. M., Olson D. R., Thomas B. C. Serum and glucocorticoid-regulated kinase modulates Nedd4-2-mediated inhibition of the epithelial Na+ channel. J. Biol. Chem. 2002;277:5–8. doi: 10.1074/jbc.C100623200. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T., Deak M., Morrice N., Cohen P. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced kinase. Biochem. J. 1999;344:189–197. [PMC free article] [PubMed] [Google Scholar]
- 14.Leighton I. A., Dalby K. N., Caudwell F. B., Cohen P. T. W., Cohen P. Comparison of the specifcities of p70 S6 kinase and MAPKAP kinase-1 identifies a relatively specific substrate for p70 S6 kinase: the N-terminal kinase domain of MAPKAP-K1 is essential for peptide phosphorylation. FEBS Lett. 1995;375:289–293. doi: 10.1016/0014-5793(95)01170-j. [DOI] [PubMed] [Google Scholar]
- 15.Knebel A., Morrice N., Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38δ. EMBO J. 2001;20:4360–4369. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray J. T., Campbell D. G., Peggie M., Alfonso M., Cohen P. Identification of filamin C as a new physiological substrate of PKBα using KESTREL. Biochem. J. 2004;384:489–494. doi: 10.1042/BJ20041058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuenda A., Alonso G., Morrice N., Jones M., Meier R., Cohen P., Nebreda A. R. Purification and cloning of SAPKK3, the major activator of RK/p38 in stress and cytokine-stimulated monocytes and epithelial cells. EMBO J. 1996;15:4156–4164. [PMC free article] [PubMed] [Google Scholar]
- 18.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bain J., McLauchlan H., Elliott M., Cohen P. The specificities of protein kinase inhibitors: an update. Biochem. J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature (London) 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 21.Goff D. A., Harrison D. S., Nuss J. M., Ring D. B., Zhou X. A. Inhibitors of glycogen synthase kinase 3. 6,417,185. U.S. Pat. 2002
- 22.Bhat R., Xue Y., Berg S., Hellberg S., Ormo M., Nilsson Y., Radesater A. C., Jerning E., Markgren P. O., Borgegard T., et al. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418. J. Biol. Chem. 2003;278:45937–45945. doi: 10.1074/jbc.M306268200. [DOI] [PubMed] [Google Scholar]
- 23.Campbell D. G., Morrice N. Identification of protein phosphorylation sites by a combination of mass spectrometry and solid phase Edman sequencing. J. Biomol. Techn. 2002;13:119–130. [PMC free article] [PubMed] [Google Scholar]
- 24.Zoroddu M. A., Kowalik-Jankowska T., Kozlowski H., Salnikow K., Costa M. Ni(II) and Cu(II) binding with a 14 amino acid sequence of Cap43 protein, TRSRSHTSEGTRSR. J. Inorg. Biochem. 2001;85:47–54. doi: 10.1016/s0162-0134(00)00204-x. [DOI] [PubMed] [Google Scholar]
- 25.Zhou R. H., Kokame K., Tsukamoto Y., Yutani C., Kato H., Miyata T. Characterization of the human NDRG gene family: a newly identified member, NDRG4, is specifically expressed in brain and heart. Genomics. 2001;73:86–97. doi: 10.1006/geno.2000.6496. [DOI] [PubMed] [Google Scholar]
- 26.Qu X., Zhai Y., Wei H., Zhang C., Xing G., Yu Y., He F. Characterization and expression of three novel differentiation-related genes belonging to the human NDRG gene family. Mol. Cell. Biochem. 2002;229:35–44. doi: 10.1023/a:1017934810825. [DOI] [PubMed] [Google Scholar]
- 27.Fiol C. J., Mahrenholz A. M., Wang Y., Roeske R. W., Roach P. J. Formation of protein kinase recognition sites by covalent modification of the substrate; molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase-3. J. Biol. Chem. 1987;262:14042–14048. [PubMed] [Google Scholar]
- 28.Ring D. B., Johnson K. W., Henricksen E. J., Nuss J. M., Goff D., Kinnick T. R., Ma S. T., Reeder J. W., Samuels I., Slabiak T., et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52:588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- 29.Coghlan M. P., Culbert A. A., Cross D. A., Corcoran S. L., Yates J. W., Pearce N. J., Rausch O. L., Murphy G. J., Carter P. S., Roxbee Cox L., Mills D., et al. Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem. Biol. 2000;7:793–803. doi: 10.1016/s1074-5521(00)00025-9. [DOI] [PubMed] [Google Scholar]
- 30.Seebolt-Leopold J. S., Dudley D. T., Herrera R., Van Becelaere K., Wiland A., Gowan R. C., Tecle H., Barrett S. D., Bridges A., Przybranowski S., et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 31.Burchfield J. G., Lennard A. J., Narasimhan S. N., Hughes W. E., Wasinger V. C., Corthals G. L., Okuda T., Kondoh H., Biden T. J., Schmitz-Peiffer C. Akt mediates insulin-stimulated phosphorylation of Ndrg2 – evidence for crosstalk with protein kinase C theta. J. Biol. Chem. 2004;279:18623–18632. doi: 10.1074/jbc.M401504200. [DOI] [PubMed] [Google Scholar]
- 32.Woods Y. L., Cohen P., Becker W., Jakes R., Goedert M., Wang X., Proud C. G. The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bε at Ser539 and the microtubule-associated protein tau at Thr212: potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem. J. 2001;355:609–615. doi: 10.1042/bj3550609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morton S., Davis R. J., McLaren A., Cohen P. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 2003;22:3876–3886. doi: 10.1093/emboj/cdg388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen P., Goedert M. GSK3 inhibitors: development and potential for the treatment of disease. Nat. Rev. Drug Discov. 2004;3:479–487. doi: 10.1038/nrd1415. [DOI] [PubMed] [Google Scholar]
- 35.Frame S., Cohen P., Biondi R. M. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell. 2001;7:1321–1327. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 36.Dajani R., Fraser E., Roe S. M., Young N., Good V., Dale T. C., Pearl L. H. Crystal structure of glycogen synthase kinase-3β: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 37.Kokame K., Kato H., Miyata T. Homocysteine-respondent genes in vascular endothelial cells identified by differential display analysis. J. Biol. Chem. 1996;271:29659–29665. doi: 10.1074/jbc.271.47.29659. [DOI] [PubMed] [Google Scholar]
- 38.Salnikow K., An W. G., Melillo G., Blagosklonny M. V., Costa M. Nickel-induced transformation shifts the balance between HIF-1 and p53 transcription factors. Carcinogenesis. 1999;20:1819–1823. doi: 10.1093/carcin/20.9.1819. [DOI] [PubMed] [Google Scholar]
- 39.Zhou D., Salnikow K., Costa M. Cap43, a novel gene specifically induced by Ni2+ compounds. Cancer Res. 1998;58:2182–2189. [PubMed] [Google Scholar]
- 40.Kurdistani S. K., Arizti P., Reimer C. L., Sugrue M. M., Aaronson S. A., Lee S. W. Inhibition of tumour cell growth by RTP/rit42 and its responsiveness to p53 and DNA damage. Cancer Res. 1998;58:4439–4444. [PubMed] [Google Scholar]
- 41.Piquemal D., Joulia D., Balaguer P., Basset A., Marti J., Commes T. Differential expression of the RTP/Drg1/Ndr1 gene product in proliferating and growth-arrested cells. Biochim. Biophys. Acta. 1999;1450:364–373. doi: 10.1016/s0167-4889(99)00056-7. [DOI] [PubMed] [Google Scholar]
- 42.Shimono A., Okuda T., Kondoh H. N-myc-dependent repression of Ndr1, a gene identified by direct subtraction of whole mouse embryo cDNAs between wild type and N-Myc mutant. Mech. Dev. 1999;83:39–52. doi: 10.1016/s0925-4773(99)00025-8. [DOI] [PubMed] [Google Scholar]
- 43.Van Belzen N., Dinjens W. N., Diesveld M. P., Groen N. A., van der Made A. C., Nozawa Y., Vliestra R., Trapman J., Bosman F. T. A novel gene which is up-regulated during colon epithelial cell differentiation and down-regulated in colorectal neoplasms. Lab. Invest. 1997;77:85–92. [PubMed] [Google Scholar]
- 44.Bandyopadhyay S., Pai S. K., Gross S. C., Hirota S., Hosobe S., Niura K., Saito K., Commes T., Hayashi S., Watabe M., Watabe K. The Drg-1 gene suppresses tumour metastasis in prostate cancer. Cancer Res. 2003;63:1731–1736. [PubMed] [Google Scholar]
- 45.Guan R. J., Ford H. L., Fu Y., Li Y., Shaw L. M., Pardee A. B. Drg-1 as a differentiation-related, putative metastatic suppressor gene in human colon cancer. Cancer Res. 2000;60:749–755. [PubMed] [Google Scholar]
- 46.Kim K. T., Ongusaha P. P., Hong Y. K., Kurdistani S. K., Nakamura M., Lu K. P., Lee S. W. Function of Drg1/Rit42 in p53-dependent mitotic spindle checkpoint. J. Biol. Chem. 2004;279:38597–38602. doi: 10.1074/jbc.M400781200. [DOI] [PubMed] [Google Scholar]
- 47.Boulkroun S., Fay M., Zennaro M. C., Escoubet B., Jaisser F., Blot-Chabaud M., Farman N., Courtois-Coutry N. Characterization of rat NDRG2 (N-myc downstream regulated gene-2), a novel early mineralocorticoid-specific induced gene. J. Biol. Chem. 2002;277:1506–1515. doi: 10.1074/jbc.M200272200. [DOI] [PubMed] [Google Scholar]
- 48.Lachat P., Shaw P., Gebhard S., van Belzen N., Chaubert P., Bosman F. T. Expression of NDRG1, a differentiation-related gene, in human tissues. Histochem. Cell Biol. 2002;118:399–408. doi: 10.1007/s00418-002-0460-9. [DOI] [PubMed] [Google Scholar]
- 49.Kalaydjieva L., Gresham D., Gooding R., Heather L., Baas F., de Jonge R., Blechschmidt K., Angelicheva D., Chandler D., Worseley P., et al. N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am. J. Hum. Genet. 2000;67:47–58. doi: 10.1086/302978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berger P., Young P., Suter U. Molecular cell biology of Charcot-Marie-Tooth disease. Neurogenetics. 2002;4:1–15. doi: 10.1007/s10048-002-0130-z. [DOI] [PubMed] [Google Scholar]
- 51.Okuda T., Higashi Y., Kokame K., Tanaka C., Kondoh H., Miyata T. Ndrg1-deficient mice exhibit a progressive demyelinating disorder of the peripheral nerves. Mol. Cell. Biol. 2004;24:3949–3956. doi: 10.1128/MCB.24.9.3949-3956.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stokoe D., Campbell D. G., Nakielny S., Hidaka H., Leevers S. J., Marshall C., Cohen P. MAPKAP-K2; a novel protein kinase activated by mitogen-activated protein kinase. EMBO J. 1992;11:3985–3994. doi: 10.1002/j.1460-2075.1992.tb05492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]