Abstract
We detected a protein in rabbit skeletal muscle extracts that was phosphorylated rapidly by PKBα (protein kinase Bα), but not by SGK1 (serum- and glucocorticoid-induced kinase 1), and identified it as the cytoskeletal protein FLNc (filamin C). PKBα phosphorylated FLNc at Ser2213 in vitro, which lies in an insert not present in the FLNa and FLNb isoforms. Ser2213 became phosphorylated when C2C12 myoblasts were stimulated with insulin or epidermal growth factor, and phosphorylation was prevented by low concentrations of wortmannin, at which it is a relatively specific inhibitor of phosphoinositide 3-kinase. PD 184352 [an inhibitor of the classical MAPK (mitogen-activated protein kinase) cascade] and/or rapamycin [an inhibitor of mTOR (mammalian target of rapamycin)] had no effect. Insulin also induced the phosphorylation of FLNc at Ser2213 in cardiac muscle in vivo, but not in cardiac muscle that does not express PDK1 (3-phosphoinositide-dependent kinase 1), the upstream activator of PKB. These results identify the muscle-specific isoform FLNc as a new physiological substrate for PKB.
Keywords: cytoskeleton, filamin, kinase substrate tracking and elucidation (KESTREL), phosphorylation, protein kinase B (PKB)
Abbreviations: EGF, epidermal growth factor; ERK, extracellular-signal-regulated kinase; FLNa, FLNb and FLNc, filamin isoforms A, B and C respectively; GST, glutathione S-transferase; KESTREL, kinase substrate tracking and elucidation; MAPK, mitogen-activated protein kinase; MKK, MAPK kinase; mTOR, mammalian target of rapamycin; NDRG, n-myc downstream-regulated gene; PI 3-kinase, phosphoinositide 3-kinase; PDK1, 3-phosphoinositide-dependent kinase 1; PH, pleckstrin homology; PKB, protein kinase B; RSK, p90 ribosomal S6 kinase; S6K, p70 ribosomal S6 kinase; SAPK, stress-activated protein kinase; SGK1, serum and glucocorticoid-induced kinase 1; SHIP, Src homology 2-interacting protein
INTRODUCTION
Signals that activate PI 3-kinase (phosphoinositide 3-kinase) and elevate the intracellular concentration of PtdIns(3,4,5)P3 trigger the activation of several protein kinases, including PKB (protein kinase B; also called Akt). PtdIns(3,4,5)P3 binds to the PH (pleckstrin homology) domains of PKB and PDK1 (3-phosphoinositide-dependent protein kinase 1), co-localizing them at the plasma membrane. This allows PDK1 to activate PKB by phosphorylation of Thr308 (reviewed in [1]).
PKB appears to play a central role in mediating many of the actions of signals that elevate PtdIns(3,4,5)P3, including many of the metabolic actions of insulin and the anti-apoptotic effects of survival factors (reviewed in [2]). However, although a number of physiological substrates for PKB have been identified, many more remain to be discovered. For example, mice that do not express the β-isoform of PKB are unable to recruit the glucose transporter GLUT4 to the plasma membrane or to stimulate glucose uptake in response to insulin [3], but the substrate(s) for PKBβ that might mediate this effect are unknown.
PKB isoforms belong to the AGC subfamily of protein kinases, and are most similar to SGK (serum- and glucocorticoid-induced protein kinase) isoforms [4–6], with which they share 55% identity in the catalytic domain. Based on studies with synthetic peptide substrates, PKB was found to phosphorylate serine and threonine residues that lie in Arg-Xaa-Arg-Xaa-Xaa-Ser/Thr- motifs [7]. However, several other protein kinases, including SGK [4,6], RSK (p90 ribosomal S6 kinase) and S6K (p70 S6 ribosomal kinase) isoforms [8], have similar substrate specificities in vitro, making it difficult to predict substrates for PKB simply by searching protein sequence databases with this motif.
In order to try and identify new physiological substrates for PKB, we therefore decided to adopt the KESTREL (kinase substrate tracking and elucidation) approach [9]. In this method, cell extracts are subjected to ion exchange chromatography, and aliquots of the fractions collected are incubated with MgATP in the absence or presence of closely related protein kinases with similar specificity requirements. The aim is to detect proteins that are phosphorylated rather selectively by just one of these kinases and then investigate whether such proteins are bona fide physiological substrates in appropriate follow-up studies. Using this approach, we have identified elongation factor 2-kinase as a protein that is phosphorylated at Ser359 and inactivated by SAPK4 (stress-activated protein kinase 4; also called p38δ), but not by the closely related isoforms SAPK2a/p38α or SAPK3/p38γ [9].
In the preceding paper [10], we used KESTREL to identify NDRG2 (n-myc downstream-regulated gene 2) as a protein that is phosphorylated much more rapidly by SGK1 than by PKBα. Here we identify the muscle-specific filamin isoform FLNc (filamin C) as a protein that is phosphorylated efficiently by PKBα, but not by SGK1. We go on to show that FLNc is indeed a new physiological substrate for PKB.
MATERIALS AND METHODS
Materials
Human insulin was purchased from Roche Molecular Biochemicals (Lewes, E. Sussex, U.K.), EGF (epidermal growth factor) and precast Tris/acetate SDS/3–8%-polyacrylamide gels from Invitrogen (Paisley, Scotland, U.K.), and wortmannin and rapamycin from Merck Biosciences (Nottingham, U.K.). PD 184352 [11] was obtained by chemical synthesis, while peptides were synthesized by Dr Graham Bloomberg (University of Bristol, Bristol, U.K.).
Antibodies
Antibodies that recognize PKB phosphorylated on Ser473 were raised in sheep. Antibodies that recognize PKB phosphorylated on Thr308 and those that recognize ERK1 (extracellular-signal-regulated kinase 1) and ERK2 phosphorylated at their pThr-Glu-pTyr motifs (where pThr and pTyr are phosphothreonine and phosphotyrosine respectively) were purchased from Cell Signalling (Hitchin, Herts., U.K.). Antibodies that recognize ribosomal protein S6 phosphorylated at Ser235 were obtained from Upstate (Milton Keynes, U.K.). Antibodies against human FLNc were raised in a rabbit against the peptide SKTRGGETKREVRVEEST, corresponding to residues 2160–2177. A phospho-specific antibody that recognizes FLNc phosphorylated at Ser2213 was raised in sheep against the phosphopeptide GRERLGpSFGSITR (where pS is phosphoserine), corresponding to residues 2207–2219 of human FLNc. Each peptide was coupled separately to keyhole limpet haemocyanin and BSA, and the conjugates were mixed before injection at Diagnostics Scotland (Edinburgh, U.K.). Both antisera were affinity purified on CH-Sepharose to which the relevant peptide antigen had been coupled covalently. The phosphospecific antibody that recognizes Ser2213 was used for immunoblotting in the presence of the unphosphorylated peptide antigen (10 μg/ml) to neutralize any antibodies that recognized unphosphorylated FLNc.
Cloning and expression of GST (glutathione S-transferase)–FLNc-(1915–2446)
The DNA coding for amino acids 1915–2446 of human FLNc (XP_045856) was amplified with the Expand HiFidelity PCR System (Roche) from IMAGE EST 4121595 using oligonucleotides MP339 (GAATTCAAGCACATCCCGGGGAGCCC) and MP340 (GCGGCCGCTTACGGATGGTGTGCTTGTCACTGTCC). This produced a fragment containing 5′ EcoRI and 3′ NotI restriction enzyme sites. The PCR product was cloned into pCR2.1 (Invitrogen), sequenced and subcloned into pGEX6P-1.
GST–FLNc-(1915–2446) was expressed in Escherichia coli BL21 pLysS (Merck Biosciences) and purified on glutathione–Sepharose. The purified protein was dialysed against 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.1 mM EGTA, 0.1% (v/v) 2-mercaptoethanol, 0.2 mM PMSF and 1 mM benzamidine, snap frozen in liquid nitrogen and stored at −80 °C.
Preparation and chromatography of human skeletal muscle tissue extracts
Human soleus muscle was minced and homogenized with 5 vol. of 4 mM EDTA, 4 mM EGTA, 1 mM benzamidine, 0.2 mM PMSF and 0.1% (v/v) 2-mercaptoethanol. The suspension was centrifuged at 2500 g for 45 min at 4 °C, the floating fat pellet was removed and the supernatant was decanted through glass wool. The extract was centrifuged at 25000 g for 20 min at 4 °C, then the supernatant was passed through Sephadex G-25 equilibrated in 30 mM Mops, pH 7.0, 5% (v/v) glycerol, 0.1% (v/v) 2-mercaptoethanol and 0.03% (w/v) Brij 35 (buffer A), and this material (750 mg of protein) was chromatographed on a 25 ml column of heparin–HP-Sepharose. The column was washed with 125 ml of buffer A and the column developed with a 500 ml non-linear gradient from 0 to 1 M NaCl in buffer A. Fractions of 12.5 ml were collected at a flow rate of 2 ml/min. Aliquots of each fraction were diluted 5-fold into 30 mM Tris/HCl, pH 7.5, 2 mM MgCl2, 10 mM 2-mercaptoethanol, 0.1 mM EGTA, 1.0 μg/ml aprotinin and 1.0 μg/ml leupeptin. A 25 μl aliquot was then incubated for 4 min at 30 °C with 5 μl of 20 nM [γ-32P]ATP (2.5×106 c.p.m.) with or without PKBα or SGK1 (0.3 unit/ml). The reactions were stopped by the addition of 10 μl of 320 mM Tris/HCl, pH 6.8, 8% (w/v) SDS, 20 mM EDTA, 32% (v/v) glycerol, 1.14 M 2-mercaptoethanol and 0.02% (w/v) Bromophenol Blue (SDS sample buffer), heated for 3 min at 100 °C, subjected to SDS/PAGE, electroblotted on to ImmobilonP membranes and autoradiographed to reveal phosphorylated proteins.
Assay of protein kinases
These were assayed at 30 °C as described previously [12,13]. One unit of PKBα or SGK1 activity was that amount which catalysed the phosphorylation of 1 nmol of the standard substrate peptide CROSStide (GRPRTSSFAEG) in 1 min [14].
Cell culture, and preparation of cell and mouse tissue extracts
C2C12 myoblasts were cultured on 15 cm-diameter dishes in Dulbecco's modified Eagle's medium supplemented with 2 mM glutamine, 10% (v/v) foetal bovine serum, penicillin and streptomycin. Cells were lysed in 0.5 ml of lysis buffer, comprising 50 mM Tris/HCl (pH 7.5), 1 mM EGTA, 1 mM EDTA, 1% (w/v) Triton-X 100, 1 mM sodium orthovanadate, 50 mM NaF, 5 mM sodium pyrophosphate, 0.27 M sucrose, 1 μM microcystin-LR, 0.1% (v/v) 2-mercaptoethanol and ‘complete’ proteinase inhibitor cocktail. Mice were killed and cardiac muscle was removed, snap frozen in liquid nitrogen, powdered and extracted with the lysis buffer described above, then centrifuged at 16000 g for 15 min at 4 °C to pellet insoluble material. Protein concentrations were determined by the Bradford method using BSA as standard. Lysates were denatured in lithium dodecyl sulphate and heated for 10 min at 70 °C before being subjected to SDS/PAGE.
RESULTS
Identification of a high-molecular-mass protein phosphorylated by PKBα in skeletal muscle extracts
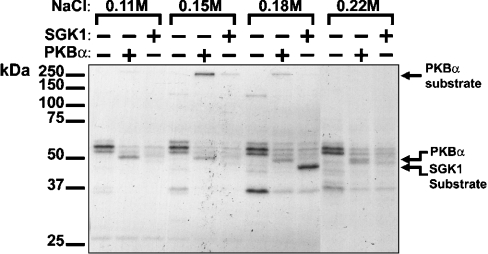
Desalted human skeletal muscle extracts were chromatographed on heparin–Sepharose and the fractions collected were phosphorylated with PKBα or SGK1 as described in the Materials and methods section. A protein was identified of apparent molecular mass >250 kDa, eluting at 0.15 M NaCl, that was phosphorylated by PKBα, but more weakly by SGK1 (Figure 1), and therefore merited further investigation.
Figure 1. Detection of a >250 kDa PKBα substrate in human skeletal muscle.
A skeletal muscle extract (750 mg of protein) was chromatographed on a heparin–Sepharose column, and aliquots of each fraction were incubated with 2 mM MgCl2 and 20 nM [γ-32P]ATP in the absence or presence of 0.3 unit/ml PKBα or SGK1. After SDS/PAGE and autoradiography, a >250 kDa protein, eluting at 0.15 M NaCl, was detected that was phosphorylated by PKBα, but only weakly by SGK1. The position of PKBα, which phosphorylates itself, is indicated. Autophosphorylation of SGK1 is negligible under the conditions used, but the fraction eluting at 0.18 M NaCl contains a protein that is phosphorylated by SGK1, but not by PKB.
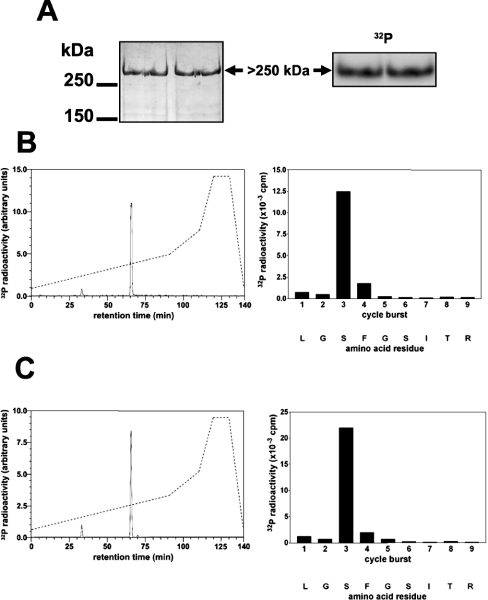
The peak fraction from heparin–Sepharose contained many proteins, but only one co-migrated on a 3–8% gradient gel with the unusually high molecular mass of the 32P-labelled band (Figure 2A). This band was excised from the gel, and tryptic mass fingerprinting revealed that it was FLNc, the muscle-specific isoform of filamin (Table 1), suggesting that this protein might be the PKB substrate.
Figure 2. Identification of the residue in FLNc phosphorylated by SGK1 in vitro.
The partially purified PKBα substrate from Figure 1 was phosphorylated by incubation for 30 min with 10 mM MgCl2/0.1 mM [γ-32P]ATP (106 c.p.m./nmol) and 1.0 unit/ml PKBα, denatured in lithium dodecyl sulphate and subjected to SDS/PAGE. (A) The gel was stained with colloidal Coomassie Blue (left panel) or autoradiographed (right panel). (B) The >250 kDa 32P-labelled band from (A) was excised, digested with trypsin and the digest chromatographed on a Vydac C18 column (Separations Group) equilibrated in 0.1% (v/v) trifluoroacetic acid. The column was developed with an acetonitrile gradient (broken line) at a flow rate of 0.8 ml/min, and fractions of 0.4 ml were collected. The major 32P-labelled peptide P1 (solid line in left panel) was subjected to MS and the sites of phosphorylation were identified by solid-phase sequencing (right panel) after coupling the peptide to a Sequalon-AA membrane [33]. (C) Same as (B), except that GST–FLNc-(1915–2446) was used instead of partially purified human FLNc.
Table 1. Identification of the >250 kDa substrate of PKBα as the muscle-specific isoform of filamin.
The 32P-labelled 250 kDa substrate (see Figure 2A) was excised from the gel, digested with trypsin and analysed on a Perseptive Biosystems Elite STR MALDI-TOF (matrix-assisted laser-desorption–time-of-flight) mass spectrometer with saturated α-cyanocinnamic acid as the matrix, as described in [28]. The mass spectrum was acquired in the reflector mode and was internally mass calibrated. The tryptic peptide ions obtained were scanned against the Swiss-Prot and Genpep databases using the MS-FIT program of Protein Prospector.
| Mass | Residue no. | |||
|---|---|---|---|---|
| Submitted | Matched | Start | End | Peptide sequence |
| 977.5 | 977.5 | 1519 | 1526 | YADQEVPR |
| 1057.6 | 1057.5 | 992 | 1002 | GAGGQGQLDVR |
| 1180.6 | 1180.6 | 756 | 766 | VNVGEGSHPER |
| 1188.6 | 1188.6 | 2609 | 2620 | GPGLSQAFVGQK |
| 1226.8 | 1226.8 | 70 | 80 | LIALLEVLSQK |
| 1230.7 | 1230.7 | 1147 | 1157 | ATIRPVFDPSK |
| 1267.6 | 1267.6 | 1268 | 1278 | EVTTEFTVDAR |
| 1298.7 | 1298.7 | 173 | 183 | VPQLPITNFNR |
| 1303.7 | 1303.7 | 1255 | 1267 | VSGPGVEPHGVLR |
| 1346.8 | 1346.7 | 350 | 361 | VTVLFAGQNIER |
| 1386.7 | 1386.7 | 2544 | 2556 | YGGPQHIVGSPFK |
| 1400.7 | 1400.6 | 1616 | 1627 | YGGDEIPYSPFR |
| 1489.8 | 1489.8 | 2294 | 2307 | GVAGVPAEFSIWTR |
| 1505.8 | 1505.8 | 978 | 991 | VAVGQEQAFSVNTR |
| 1518.8 | 1518.8 | 97 | 109 | LENVSVALEFLER |
| 1533.8 | 1533.8 | 1936 | 1949 | ITESDLSQLTASIR |
| 1565.7 | 1565.7 | 1461 | 1474 | VPQTFTVDCSQAGR |
| 1571.8 | 1571.9 | 114 | 128 | LVSIDSKAIVDGNLK |
| 1571.8 | 1571.8 | 2390 | 2404 | VNQPASFAVQLNGAR |
| 1595.9 | 1595.9 | 545 | 558 | YVVTITWGGYAIPR |
| 1601.8 | 1601.8 | 559 | 573 | SPFEVQVSPEAGVQK |
| 1823.9 | 1823.9 | 2228 | 2244 | VEAAEIVEGEDSAYSVR |
| 1923.9 | 1924.0 | 1639 | 1657 | CLVTVSIGGHGLGACLGPR |
| 1941.8 | 1941.9 | 462 | 478 | SPFPVHVSEACNPNACR |
| 2002.1 | 2002.1 | 1475 | 1494 | APLQVAVLGPTGVAEPVEVR |
| 2006.0 | 2006.1 | 1009 | 1027 | RPIPCKLEPGGGAEAQAVR |
| 2006.0 | 2006.0 | 2017 | 2034 | GLSEGHTFQVAEFIVDTR |
| 2324.1 | 2324.1 | 2174 | 2195 | VEESTQVGGDPFPAVFGDFLGR |
| 2363.1 | 2363.1 | 1807 | 1827 | YDGNHIPGSPLQFYVDAINSR |
| 2527.2 | 2527.2 | 2354 | 2376 | FNDEHIPDSPFVVPVASLSDDAR |
| 2729.2 | 2729.3 | 2411 | 2434 | VHTPSGAVEECYVSELDSDKHTIR |
Identification of the residue in FLNc phosphorylated by PKB
To investigate whether the PKB substrate and FLNc were the same protein, the partially purified material was phosphorylated as in Figure 2(A), and the 32P-labelled band was digested with trypsin and chromatographed on a C18 column. One major peptide was detected (Figure 2B, left panel), whose molecular mass corresponded to that of a peptide comprising residues 2205–2217 of FLNc plus one phosphate group. Solid-phase sequencing (Figure 2B, right panel) identified the site of phosphorylation as Ser2213, establishing that the 32P-labelled protein was indeed FLNc. This residue lies in an optimal consensus sequence for PKB (RERLGSF). We also phosphorylated a fragment of FLNc, expressed as a GST fusion protein, and showed that it was again phosphorylated specifically at Ser2213 (Figure 2C). This fragment was phosphorylated equally well by PKBα and PKBβ when the two protein kinases were matched for activity towards CROSStide (results not shown).
Generation of antibodies that recognize unphosphorylated and phosphorylated FLNc
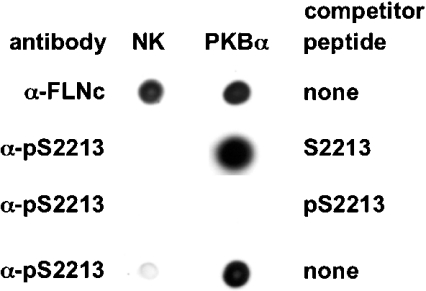
To examine whether Ser2213 of FLNc becomes phosphorylated in cells, we raised antibodies that recognize FLNc only when it is phosphorylated at Ser2213 (anti-pSer2213), as well as antibodies that recognize phosphorylated and unphosphorylated FLNc equally well (anti-FLNc) (Figure 3). The latter antibody was raised against a peptide sequence, conserved in mouse and human FLNc, that is present within the unique insert region of FLNc, but not in the closely related FLNa and FLNb isoforms. The peptide sequence to which the anti-pSer2213 antibody was raised was also conserved in mouse and human FLNc. This antibody recognized GST–FLNc only after phosphorylation by PKBα (Figure 3), and recognition was blocked by incubation with the phosphopeptide immunogen, but not with the unphosphorylated form of this peptide. These antibodies were then used to study the phosphorylation of FLNc by PKBα in cells.
Figure 3. Characterization of antibodies that recognize FLNc.
Bacterially expressed FLNc (residues 1915–2446) was left unphosphorylated (no kinase; NK) or maximally phosphorylated with PKBα. Aliquots (50 ng) were spotted on to a nitrocellulose membrane (Schleicher and Schuell, London, U.K.) and immunoblotted using the anti-pSer2213 antibody (α-pS2213) or an antibody that recognizes phosphorylated and unphosphorylated FLNc equally well (α-FLNc) in the presence or absence (‘none’) of the competitor peptides shown. The sequences of these peptides are given in the Materials and methods section. The prefix p denotes a phosphorylated peptide.
Insulin and EGF stimulate phosphorylation of FLNc on Ser2213 in C2C12 myoblasts
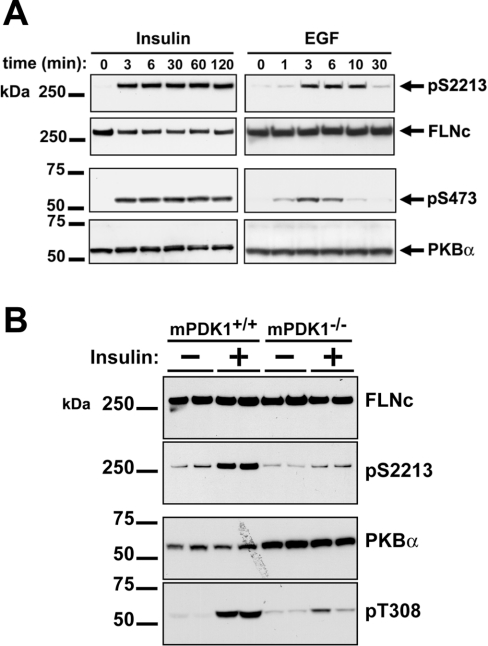
Insulin induced rapid phosphorylation of FLNc at Ser2213 that was maximal after a few minutes and was sustained for at least 2 h. The phosphorylation of FLNc paralleled the activation of PKBα, as assessed by the phosphorylation of Ser473 of PKBα (Figure 4A). The same result was obtained if the extracts were immunoblotted directly (Figure 4A) or if FLNc was immunoprecipitated first (results not shown).
Figure 4. Insulin and EGF induce the phosphorylation of FLNc at Ser2213 in C2C12 cells.
(A) Murine C2C12 myoblasts were deprived of serum for 12 h, then stimulated with 250 ng/ml insulin or 20 ng/ml EGF for the times indicated. The cells were lysed and the lysates subjected to SDS/PAGE followed by transfer to an ImmobilonP membrane. The membranes were then probed with the anti-pSer2213 and anti-FLNc antibodies, and with further antibodies that recognize PKBα phosphorylated at Ser473 (p473) and the phosphorylated and unphosphorylated forms of PKBα equally well (PKBα). (B) Cardiac muscle extracts (10 μg of protein) were prepared from either wild-type or PDK1 conditional knockout duplicate individual mice that had been fasted for 12 h before injection with or without insulin (10 m-units/g) for 20 min. Extracts were subjected to SDS/PAGE, transferred to ImmobilonP membranes and immunoblotted with antibodies as in (A), except that phosphorylation of PKBα was detected with an antibody that recognized PKBα phosphorylated at Thr308 (pT308).
EGF also induced a rapid, but transient, phosphorylation of FLNc at Ser2213, with maximal phosphorylation at 3–10 min. The phosphorylation and subsequent dephosphorylation of FLNc lagged slightly behind the activation and inactivation of PKBα, consistent with FLNc being a substrate of PKB (Figure 4A).
Phosphorylation of FLNc at Ser2213 is severely impaired in cardiac muscle from PDK1−/− mice
Since FLNc is highly expressed in striated muscles, we analysed the phosphorylation of this protein in the cardiac muscle of both wild-type mice (mPDK1+/+) and ‘conditional knockout’ mice (mPDK1−/−) that no longer express PDK1 (3-phosphoinositide-dependent kinase 1), which activates PKB by phosphorylating it on Thr308 [15]. Mice were starved for 12 h before injection with insulin, and cardiac muscle was removed immediately or after 20 min of insulin stimulation. Immunoblot analysis showed that the expression of FLNc was similar in cardiac muscle from wild-type and conditional knockout mice (Figure 4B). In wild-type tissues, FLNc had low basal phosphorylation on Ser2213, which was increased after 20 min of insulin stimulation. In contrast, the basal phosphorylation of Ser2213 in cardiac muscle of the mPDK1−/− mice was not increased by insulin (Figure 4B). The insulin-stimulated phosphorylation of Ser2213 in wild-type cardiac muscle was correlated with activation of PKB, as determined by phosphorylation of Thr308 (Figure 4B).
The phosphorylation of FLNc at Ser2213 is prevented by inhibition of PI 3-kinase
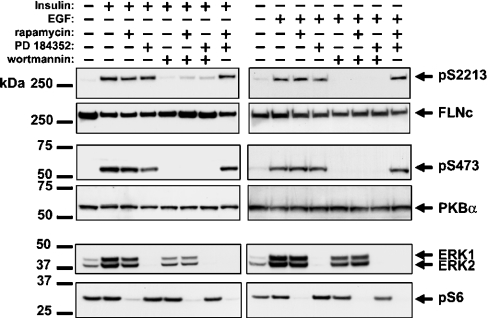
To investigate the signalling pathway involved in the insulin- and EGF-induced phosphorylation of FLNc at Ser2213, we first incubated C2C12 myoblasts in the absence or presence of inhibitors of different signalling pathways, prior to stimulation with these agonists. These experiments showed that the insulin- or EGF-induced phosphorylation of FLNc at Ser2213 and the phosphorylation of PKB at Ser473 were completely prevented by wortmannin, an inhibitor of PI 3-kinase (Figure 5). In contrast, the phosphorylation of FLNc at Ser2213 was unaffected by PD 184352, an inhibitor of the classical MAPK (mitogen-activated protein kinase) cascade and/or by rapamycin, an inhibitor of the protein kinase mTOR (mammalian target of rapamycin). However, as expected, PD 184352 prevented the activation of ERK1 and ERK2, the MAPKs of the classical MAPK cascade, while rapamycin prevented the phosphorylation of ribosomal protein S6, a substrate of S6K which is a downstream component of the mTOR pathway (Figure 5).
Figure 5. Effects of inhibitors of signalling pathways on the insulin- and EGF-stimulated phosphorylation of FLNc on Ser2213 in C2C12 cells.
The experiments were carried out as in Figure 4(A), except that, prior to stimulation with insulin for 3 min or EGF for 6 min, the cells were incubated for 1 h without (−) or with (+) 50 nM rapamycin, 2 μM PD 184352 or 100 nM wortmannin. The membranes were immunoblotted with the antibodies described in the legend to Figure 4, as well as with antibodies that recognize the active, phosphorylated forms of ERK1 and ERK2 and ribososomal protein S6 phosphorylated at Ser235 (pS6).
DISCUSSION
In this paper, we have used the KESTREL method to identify FLNc as a new physiological substrate for PKB that is specific to striated muscle. PKB phosphorylated FLNc at Ser2213 in vitro, and at a much higher rate than did SGK. Moreover, Ser2213 became phosphorylated in C2C12 myoblasts in response to insulin or EGF, with phosphorylation and dephosphorylation correlating with the activation and inactivation respectively of PKB. The phosphorylation of FLNc, like that of PKB, was prevented by the PI 3-kinase inhibitor wortmannin. The phosphorylation of Ser2213 was not suppressed by rapamycin or PD 184352, which prevent activation of S6K1/S6K2 and RSK isoforms respectively, protein kinases that have similar substrate preferences to PKB, and are also activated in response to insulin or EGF. Consistent with this finding, PMA, a potent activator of the classical MAPK cascade and hence the activation of RSK isoforms, did not induce the phosphorylation of FLNc at Ser2213 (results not shown). The present study, in conjunction with the results described in the preceding paper [10], in which NDRG1 and NDRG2 were identified as physiological substrates for SGK, again illustrates the power of the KESTREL method for identifying physiological substrates of closely related protein kinases with similar substrate preferences.
Filamin, which was first identified over 25 years ago [16], is expressed in cells and tissues as three highly related isoforms, termed FLNa, FLNb and FLNc. It is an actin-binding protein that is thought to stabilize three-dimensional networks of actin filaments, linking them to cell membranes. However, it may also act as a scaffolding protein, tethering components of signalling pathways to enhance their activation by particular agonists. For example, FLNa has been reported to bind to MKK4 [MAPK kinase 4, also called SEK1 (SAPK/ERK kinase 1)], one of the activators of JNK (c-Jun N-terminal kinase), and to be required for the tumour necrosis factor α-induced activation of MKK4 in melanoma cells [17]. FLNa has also been reported to bind to SHIP2 (Src homology 2-interacting protein 2) [18], a lipid phosphatase that converts PtdIns(3,4,5)P3 into PtdIns(3,4)P2 and is involved in modulating signalling through PI 3-kinase.
FLNc, the muscle-specific isoform of filamin, contains a 78-amino-acid residue insert that is not present in either FLNa or FLNb, and which is located between the 19th and 20th immunoglobulin-like repeats, of which 24 are present in all three isoforms [19]. Strikingly, it is within this region unique to FLNc that the PKB-phosphorylation site Ser2213 is situated. In contrast, FLNa is not phosphorylated by PKB [20]. In skeletal muscle, FLNc is associated with the Z-disc of the myofibrillar apparatus and binds directly to the Z-disc proteins FATZ [21] and myotilin [22,23]. It also interacts with sarcoglycans γ and δ, which are components of the dystrophin–dystroglycan complex at the sarcolemma [24]. Such a localization at regions where the sarcolemma interacts with myofibrils suggests that FLNc, like other filamin isoforms, may be involved in assembling signalling complexes near the cell membrane. It is therefore of considerable interest that LL5β, a large 160 kDa protein containing a PtdIns(3,4,5)P3-binding PH domain at its C-terminus, has been found to interact with FLNc [25].
Although FLNc is a muscle-specific protein, it is also expressed in some cell lines and, in such cells, agonists that activate PI 3-kinase cause LL5β to translocate to the cell periphery. This is presumably a consequence of the interaction of its PH domain with membrane-associated PtdIns(3,4,5)P3, because translocation did not occur in mutants of LL5β in which the PH domain was deleted. However, incubation of the cells with inhibitors of PI 3-kinase causes LL5β to relocate to an unidentified intracellular vesicular compartment. In COS7 cells co-transfected with LL5β and FLNc, treatment with wortmannin caused FLNc to partially redistribute to this compartment. In cells not treated with PI 3-kinase inhibitors, or in cells transfected with FLNc alone, FLNc adopted a punctate distribution that was insensitive to wortmannin. Interestingly, a similar relocalization to the intracellular vesicular compartment occurred in cells co-transfected with FLNc and a mutant of LL5β that lacks the C-terminal PH domain [25]. This indicates that wortmannin does not induce relocalization by preventing the interaction of PtdIns(3,4,5)P3 with the PH domain of LL5β, but by inhibiting a distinct PtdIns(3,4,5)P3-dependent mechanism. It will clearly be of great interest to investigate whether inhibition of the PKB-catalysed phosphorylation of FLNc at Ser2213 underlies this effect.
We have carried out several experiments to address the function of phosphorylation of FLNc on Ser2213. First, we examined the localization of FLNc in MRC5 fibroblasts, which express this protein, but could not detect any change in FLNc localization or its co-localization with actin stress fibres after stimulation with insulin in the presence or absence of wortmannin (results not shown). Since SHIP2 is reported to associate with FLNa, we also analysed whether it interacted with FLNc, but were unable to detect any SHIP2 activity associated with FLNc immunoprecipitates from control or insulin-stimulated C2C12 cells (results not shown). The muscle-specific proteinase calpain III regulates the ability of FLNc to interact with sarcoglycans γ and δ by cleaving FLNc during calpain-mediated remodelling of cytoskeleton–membrane interactions, including myoblast fusion and muscle repair [26]. FLNa becomes resistant to cleavage by calpain upon phosphorylation by cAMP-dependent protein kinase [27]. We immunopreciptated FLNc from control or insulin-stimulated C2C12 cells, but the rate and extent of proteolytic cleavage that occurred upon incubation with recombinant calpain II in vitro were unaltered (results not shown). In summary, our current data provided no evidence that the phosphorylation of Ser2213 affects the localization of FLNc, its interaction with SHIP2 or its susceptibility to cleavage of calpain.
Finally, it should be mentioned that FLNc is likely to be phosphorylated at sites additional to Ser2213. The C-terminal region in FLNa, which is conserved in FLNc, is reported to be phosphorylated in vitro by several protein kinases, including cAMP-dependent protein kinase [27,28], calcium/calmodulin-dependent protein kinase II [29], protein kinase C [30,31] and PAK1 (p21-activated kinase 1) [32]. Although RSK isoforms do not phosphorylate FLNc at Ser2213, RSK2 has been reported to phosphorylate FLNa at Ser2152, and mutation of this site to alanine blocked the EGF-stimulated phosphorylation of FLNa [20]. The multisite phosphorylation of FLNc and other filamin isoforms may therefore influence their interaction with other proteins and their ability to modulate signal transduction in a complex manner.
Acknowledgments
We thank Mr Amar Jain (Ninewells Hospital, Dundee, U.K.) for providing resected human muscle. We also thank Dr Dario Alessi (MRC Protein Phosphorylation Unit, University of Dundee) for providing the conditional PDK1 knockout mice that do not express PDK1 in striated muscle. This study was supported by the U.K. Medical Research Council, The Royal Society, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co., Merck KGaA and Pfizer. We thank the Protein Production and Antibody Purification Teams (co-ordinated by Dr Hilary McLaughlin and Dr James Hastie) in the Division of Signal Transduction Therapy (School of Life Sciences, University of Dundee) for PKBα, PKBβ, SGK1 and the anti-filamin antibodies. DNA sequencing was performed by The Sequencing Service (School of Life Sciences, University of Dundee; www.dnaseq.co.uk).
References
- 1.Mora A., Komander D., van Aalten D. M. F., Alessi D. R. PDK1, the master regulator of AGC kinase signal transduction. Semin. Dev. Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 2.Brazil D. P., Hemmings B. A. Ten years of protein kinase B: a hard AKT to follow. Trends Biochem. Sci. 2001;26:657–666. doi: 10.1016/s0968-0004(01)01958-2. [DOI] [PubMed] [Google Scholar]
- 3.Bae S. S., Cho H., Mu J., Birnbaum M. J. Isoform-specific regulation of insulin-dependent glucose uptake by Akt/protein kinase B. J. Biol. Chem. 2003;278:49530–49536. doi: 10.1074/jbc.M306782200. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi T., Deak M., Morrice N., Cohen P. Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem. J. 1999;344:189–197. [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi T., Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- 6.Park J., Leong M. L., Buse P., Maiyar A. C., Firestone G. L., Hemmings B. A. Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alessi D. R., Caudwell F. B., Andjelkovic M., Hemmings B. A., Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 8.Leighton I. A., Dalby K. N., Caudwell F. B., Cohen P. T., Cohen P. Comparison of the specificities of p70 S6 kinase and MAPKAP kinase-1 identifies a relatively specific substrate for p70 S6 kinase: the N-terminal kinase domain of MAPKAP kinase-1 is essential for peptide phosphorylation. FEBS Lett. 1995;375:289–293. doi: 10.1016/0014-5793(95)01170-j. [DOI] [PubMed] [Google Scholar]
- 9.Knebel A., Haydon C. E., Morrice N., Cohen P. Stress-induced regulation of eukaryotic elongation factor 2 kinase by SB 203580-sensitive and -insensitive pathways. Biochem. J. 2002;367:525–532. doi: 10.1042/BJ20020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray J. T., Campbell D. G., Morrice N., Auld G. A., Shpiro N., Marquez R., Peggie M., Bain J., Bloomberg G. B., Grahammer F., et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebolt-Leopold J. S., Dudley D. T., Herrera R., Van Becelaere K., Wiland A., Gowan R. C., Tecle H., Barrett S. D., Bridges A., Przybranowski S., et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 12.Bain J., McLauchlan H., Elliott M., Cohen P. The specificities of protein kinase inhibitors: an update. Biochem. J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature (London) 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 15.Mora A., Davies A. M., Bertrand L., Sharif I., Budas G. R., Jovanovic S., Mouton V., Kahn C. R., Lucocq J. M., Gray G. A., et al. Deficiency of PDK1 in cardiac muscle results in heart failure and increased sensitivity to hypoxia. EMBO J. 2003;22:4666–4676. doi: 10.1093/emboj/cdg469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallach D., Davies P. J., Pastan I. Cyclic AMP-dependent phosphorylation of filamin in mammalian smooth muscle. J. Biol. Chem. 1978;253:4739–4745. [PubMed] [Google Scholar]
- 17.Marti A., Luo Z., Cunningham C., Ohta Y., Hartwig J., Stossel T. P., Kyriakis J. M., Avruch J. Actin-binding protein-280 binds the stress-activated protein kinase (SAPK) activator SEK-1 and is required for tumor necrosis factor-alpha activation of SAPK in melanoma cells. J. Biol. Chem. 1997;272:2620–2628. doi: 10.1074/jbc.272.5.2620. [DOI] [PubMed] [Google Scholar]
- 18.Dyson J. M., Munday A. D., Kong A. M., Huysmans R. D., Matzaris M., Layton M. J., Nandurkar H. H., Berndt M. C., Mitchell C. A. SHIP-2 forms a tetrameric complex with filamin, actin, and GPIb-IX-V: localization of SHIP-2 to the activated platelet actin cytoskeleton. Blood. 2003;102:940–948. doi: 10.1182/blood-2002-09-2897. [DOI] [PubMed] [Google Scholar]
- 19.Thompson T. G., Chan Y. M., Hack A. A., Brosius M., Rajala M., Lidov H. G., McNally E. M., Watkins S., Kunkel L. M. Filamin 2 (FLN2): A muscle-specific sarcoglycan interacting protein. J. Cell Biol. 2000;148:115–126. doi: 10.1083/jcb.148.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo M. S., Ohta Y., Rabinovitz I., Stossel T. P., Blenis J. Ribosomal S6 kinase (RSK) regulates phosphorylation of filamin A on an important regulatory site. Mol. Cell. Biol. 2004;24:3025–3035. doi: 10.1128/MCB.24.7.3025-3035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faulkner G., Pallavicini A., Comelli A., Salamon M., Bortoletto G., Ievolella C., Trevisan S., Kojic S., Dalla Vecchia F., et al. FATZ, a filamin-, actinin-, and telethonin-binding protein of the Z-disc of skeletal muscle. J. Biol. Chem. 2000;275:41234–41242. doi: 10.1074/jbc.M007493200. [DOI] [PubMed] [Google Scholar]
- 22.van der Ven P. F., Wiesner S., Salmikangas P., Auerbach D., Himmel M., Kempa S., Hayess K., Pacholsky D., Taivainen A., Schroder R., et al. Indications for a novel muscular dystrophy pathway. Gamma-filamin, the muscle-specific filamin isoform, interacts with myotilin. J. Cell Biol. 2000;151:235–248. doi: 10.1083/jcb.151.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salmikangas P., van der Ven P. F., Lalowski M., Taivainen A., Zhao F., Suila H., Schroder R., Lappalainen P., Furst D. O., Carpen O. Myotilin, the limb-girdle muscular dystrophy 1A (LGMD1A) protein, cross-links actin filaments and controls sarcomere assembly. Hum. Mol. Genet. 2003;12:189–203. doi: 10.1093/hmg/ddg020. [DOI] [PubMed] [Google Scholar]
- 24.Bonnemann C. G., Thompson T. G., van der Ven P. F., Goebel H. H., Warlo I., Vollmers B., Reimann J., Herms J., Gautel M., Takada F., et al. Filamin C accumulation is a strong but nonspecific immunohistochemical marker of core formation in muscle. J. Neurol. Sci. 2003;206:71–78. doi: 10.1016/s0022-510x(02)00341-6. [DOI] [PubMed] [Google Scholar]
- 25.Paranavitane V., Coadwell W. J., Eguinoa A., Hawkins P. T., Stephens L. LL5beta is a phosphatidylinositol (3,4,5)-trisphosphate sensor that can bind the cytoskeletal adaptor, gamma-filamin. J. Biol. Chem. 2003;278:1328–1335. doi: 10.1074/jbc.M208352200. [DOI] [PubMed] [Google Scholar]
- 26.Guyon J. R., Kudryashova E., Potts A., Dalkilic I., Brosius M. A., Thompson T. G., Beckmann J. S., Kunkel L. M., Spencer M. J. Calpain 3 cleaves filamin C and regulates its ability to interact with gamma- and delta-sarcoglycans. Muscle Nerve. 2003;28:472–483. doi: 10.1002/mus.10465. [DOI] [PubMed] [Google Scholar]
- 27.Chen M., Stracher A. In situ phosphorylation of platelet actin-binding protein by cAMP-dependent protein kinase stabilizes it against proteolysis by calpain. J. Biol. Chem. 1989;264:14282–14289. [PubMed] [Google Scholar]
- 28.Jay D., Garcia E. J., Lara J. E., Medina M. A., de la Luz Ibarra M. Determination of a cAMP-dependent protein kinase phosphorylation site in the C-terminal region of human endothelial actin-binding protein. Arch. Biochem. Biophys. 2000;377:80–84. doi: 10.1006/abbi.2000.1762. [DOI] [PubMed] [Google Scholar]
- 29.Wu M. P., Jay D., Stracher A. Existence of multiple phosphorylated forms of human platelet actin binding protein. Cell. Mol. Biol. Res. 1994;40:351–357. [PubMed] [Google Scholar]
- 30.Kawamoto S., Hidaka H. Ca2+-activated, phospholipid-dependent protein kinase catalyzes the phosphorylation of actin-binding proteins. Biochem. Biophys. Res. Commun. 1984;118:736–742. doi: 10.1016/0006-291x(84)91456-6. [DOI] [PubMed] [Google Scholar]
- 31.Tigges U., Koch B., Wissing J., Jockusch B. M., Ziegler W. H. The F-actin cross-linking and focal adhesion protein filamin A is a ligand and in vivo substrate for protein kinase C alpha. J. Biol. Chem. 2003;278:23561–23569. doi: 10.1074/jbc.M302302200. [DOI] [PubMed] [Google Scholar]
- 32.Vadlamudi R. K., Li F., Adam L., Nguyen D., Ohta Y., Stossel T. P., Kumar R. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat. Cell Biol. 2002;4:681–690. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- 33.Campbell D. G., Morrice N. Identification of protein phosphorylation sites by a combination of mass spectrometry and solid phase Edman sequencing. J. Biomol. Tech. 2002;13:119–130. [PMC free article] [PubMed] [Google Scholar]