Abstract
We have examined the role of a salt bridge between Lys242 and Glu246 in loop L4 of procaspase 3 and of mature caspase 3, and we show that the interactions are required for stabilizing the active site. Replacing either of the residues with an alanine residue results in a complete loss of procaspase 3 activity. Although both mutants are active in the context of the mature caspase 3, the mutations result in an increase in Km and a decrease in kcat when compared with the wild-type caspase 3. In addition, the mutations result in an increase in the pKa value associated with a change in kcat with pH, but does not affect the transition observed for Km versus pH. The mutations also affect the accessibility of the active-site solvent as measured by tryptophan fluorescence emission in the presence of quenching agents and as a function of pH. We show that, as the pH is lowered, the (pro)caspase dissociates, and the mutations increase the pH-dependent instability of the dimer. Overall, the results suggest that the contacts lost in the procaspase as a result of replacing Lys242 and Glu246 are compensated partially in the mature caspase as a result of new contacts that are known to form on zymogen processing.
Keywords: active site, apoptosis, caspase, dimerization, fluorescence quenching, stability
Abbreviations: DTT, dithiothreitol; procaspase 3 (D3A), procaspase 3 (D9A, D28A, D175A)
INTRODUCTION
A major question in studies of programmed cell death is why the executioner procaspases are inactive in the cell. The recently identified structures of procaspase 7 [1,2], the only procaspase for which structural data are currently available, show that the substrate-binding pocket, called loop L3 (see Figure 1), is unravelled and extends away from the active site. As a result, the procaspase 7 structure is incompatible with substrate binding and catalysis. However, it has been shown by Nicholson and co-workers [3] that an uncleavable variant of procaspase 3 does contain catalytic activity. In this mutant, the three processing sites were removed, but the active-site catalytic residues were intact. In addition, we have shown that the binding of uncleavable procaspase 3 to the substrate is similar to that of the mature caspase 3, but the catalytic efficiency is significantly lower (approx. 200-fold) when compared with the mature caspase [4]. Taken together, these results are not compatible with an unravelled solvent-exposed substrate-binding pocket in procaspase 3.
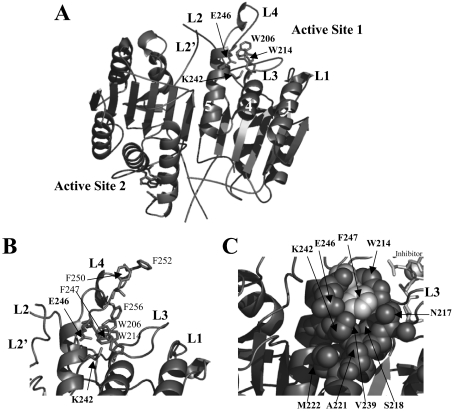
Figure 1. Structure of mature caspase 3.
(A) Mature caspase 3 (PDB entry 1CP3) generated with PyMOL (Delano Scientific, San Carlos, CA, U.S.A.). The positions of Lys242 and Glu246, helices 1, 4 and 5 and the active-site tryptophan residues (Trp206 and Trp214) are indicated. (B) Phenylalanine residues in active-site loop L4 are highlighted. (C) Space-filling model highlighting residues surrounding the Lys242–Glu246 salt bridge. For clarity, the inhibitor was removed from the structures in (A, B).
Nicholson and co-workers [3] showed that a tri-aspartate ‘safety-catch’ (Asp179–Asp181) in the intersubunit linker is important for maintaining the procaspase dormancy [3]. It was further shown that the linker undergoes a pH-dependent conformational change that exposes the Asp175 cleavage site for processing. In addition to the ‘safety-catch’ tri-aspartate described by these authors [3], we showed that the propeptide and the active-site loop L4 also undergo pH-dependent conformational changes [4]. The length of the loop L4 is conserved within, but not between, the three caspase subfamilies [5], with the caspase 3 subfamily containing the longest L4 loops. These amino acids, in part, determine the specificity of the S4 subsite, thus defining the specificity of the caspase.
The C-terminus of helix 5, which is located at the base of loop L4, contains Lys242. The aliphatic side chain is buried by several residues on helix 5 and the adjacent helix 4 (Figure 1). In addition, the charged Nζ atom of Lys242 interacts with the carboxylate of Glu246 (Figure 1). On the basis of the structure of caspase 3 [6,7], the cluster of hydrophobic interactions and charge–charge interactions are predicted to stabilize the conformation of loop L4. However, since Glu246 is not conserved in procaspase 7, it is not known whether these interactions are important for procaspase 3 or whether the same contacts occur in the zymogen. To examine this, we mutated the two residues, Lys242 and Glu246, individually to alanine. We show that the mutations abrogate activity in the procaspase, and whereas the mature caspase mutants are active, their activities are altered compared with the activity of the wild-type caspase 3. Neither mutation affects the dimeric properties of the procaspases at pH 7, but the mutations may affect the formation of the loop bundle between loops L2, L4 and L2′, which stabilizes the active-site structure. Altogether, the results demonstrate that the Lys242–Glu246 interactions are important for maintaining the correct conformation of loop L4 in the procaspase. In the mature caspase, the loss of the interactions is compensated partially, and this is probably due to new interactions that form as a result of processing and subsequent formation of the loop bundle.
EXPERIMENTAL
Materials
Acrylamide, ampicillin, antifoam-C, BSA, CHAPS, citric acid, DEAE-Sepharose, DMSO, DTT (dithiothreitol), EDTA, isopropyl β-D-thiogalactoside, nickel sulphate, Pipes, PMSF, potassium iodide, monobasic and dibasic potassium phosphate, Sephacryl-S100, sodium bicarbonate, sodium citrate (dihydrate), tosyl-lysylchloromethane (‘TLCK’) and tosylphenylalanylchloromethane (‘TPCK’) were obtained from Sigma. Imidazole and urea were from ICN. Sodium chloride, Tris, tryptone and yeast extract were obtained from Fisher (Pittsburgh, PA, U.S.A.), and sucrose was from Mallinckrodt (Phillipsburg, NJ, U.S.A.). Hepes was from Acros (Geel, Belgium), and ultrapure urea was from Nacalai Tesque (Kyoto, Japan). Ac-DEVD-AFC (N-acetyl-Asp-Glu-Val-Asp-(7-amino-4-trifluoromethylcoumarin) and Z-VAD-FMK (benzyloxycarbonyl-valylalanyl-DL-aspartylfluoromethane) were obtained from Calbiochem (San Diego, CA, U.S.A.), His-bind resin was from Novagen, restriction enzymes, Deep Vent polymerase and DpnI were from New England Biolabs, and MgCl2 and dNTPs used for site-directed mutagenesis were from Roche.
Mutagenesis
Mutants were made via site-directed mutagenesis using Deep Vent polymerase and the templates pHC332 [8] and pHC33209 [4], which introduced the mutations in the background of wild-type procaspase 3 or procaspase 3 (D3A)2 [procaspase 3 (D9A, D28A, D175A)2] respectively. Glu246 was mutated to an alanine residue in both templates using primer 1 (GTGGCAACAGCATTTGAGTCCTTTTCC) and primer 2 (GGAAAAGGACTCAAATGCTGTTGCCAC). Lys242 was mutated to an alanine residue in both templates using primer 3 (CCCGGGTTAACCGGGCGGTGGCAACAGAATTT) and primer 4 (AAATTCTGTTGCCACCGCCCGGTTAACCCGGG). There were no new restriction sites introduced with the mutations. The changed bases are shown in boldface. The resulting plasmids were sequenced (both DNA strands) to confirm the mutations.
Protein purification
Escherichia coli BL21(DE3) LysS cells were transformed with plasmid pHC33214 [procaspase 3 (E246A, Glu246→A)], pHC33216 [procaspase 3 (D3A, E246A)], pHC33238 [procaspase 3 (K242A)] or pHC33240 [procaspase 3 (D3A, K242A)]. One should note that the mature caspases are purified after the expression of the active procaspases, which autoprocess in E. coli. Cultures were grown in Fernbach flasks containing 1 litre of Luria–Bertani medium at 37 °C. When the cultures reached an absorbance A600∼1.2, isopropyl β-D-thiogalactoside was added to a final concentration of 0.8 mM to induce protein expression, and the cells were incubated at 25 °C for 4–6 h. The cells were harvested, and the proteins were purified as described in [4].
Size-exclusion chromatography
In separate experiments, proteins were dialysed for >8 h at 25 °C either in 5 mM phosphate buffer at pH 7.0 or 6.0 or in 5 mM citrate buffer at pH 5.0 or 4.0. The protein was then diluted to a concentration of 10 μM, and 5 μl was loaded on to an Agilent G250 Sephacryl HPLC sizing column using an Agilent Bioanalyzer (model 8423) coupled with an Essense HPLC workstation (Lab Alliance, State College, PA, U.S.A.). The column had been pre-equilibrated with the dialysis buffer, and the flow rate was 1 ml/min. A214 was measured. The elution of caspase proteins was compared with that of BSA. The BSA dimer (132 kDa) was eluted at 9.6 ml, whereas the monomer (66 kDa) was eluted at 13.4 ml. The elution volume of BSA did not change over the pH range 4–7.
Enzymic activity assays
Activities of the mutants were determined as described previously [4,9] using the fluorescent substrate Ac-DEVD-AFC. The steady-state parameters kcat and Km were determined as described previously [4] and are presented in Table 1.
Table 1. Catalytic parameters of the caspase 3 loop L4 mutants at pH 7.5.
| pH dependence (pKa) | |||||
|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (M−1·s−1) | kcat | 1/Km | |
| Caspase 3 | 2.2±0.5 | 0.4±0.05 | 1.8×105 | 6.0±0.1 | 6.7±0.1 |
| Caspase 3 (K242A) | 13±2 | 0.04±0.01 | 3.1×103 | 6.3±0.1 | 6.5±0.1 |
| Caspase 3 (E246A) | 23±2 | 0.3±0.02 | 1.3×104 | 6.5±0.1 | 6.5±0.1 |
| Procaspase 3 (D3A) | 3.5±0.8 | 0.003±1.4×10−4 | 8.6×102 | ||
The initial velocity was determined over the pH range 5–10, whereas the catalytic parameters kcat and Km were determined over the pH range 5–9 as described in [4,10]. The following buffers were used: 50 mM citrate (pH 3.0–6.2), 20 mM Pipes (pH 6.1–7.5), 50 mM Tris/HCl (pH 7.2–9.0) and 50 mM sodium bicarbonate (pH 9.2–10.6). All experiments were performed at 25 °C, and all buffers contained NaCl, CHAPS, sucrose and DTT as described in [4]. The data were analysed as described in [4] to determine the pKa values.
Quenching of tryptophan fluorescence emission
Assays were performed in 20 mM citrate (pH 3.0–6.0), 50 mM potassium phosphate (pH 6.0–8.0) or 50 mM Tris/HCl (pH 7.2–9.5). All the buffers contained 1 mM DTT. Protein concentration was 2 μM. Stock solutions of KI (2 M), CsCl (2 M) or acrylamide (5 M) were prepared in the respective buffers and then added to final concentrations of 0–0.3 M (KI or CsCl) or 0–0.8 M (acrylamide). After each addition of the quencher, the samples were incubated at 25 °C. The samples were excited at 295 nm, and fluorescence emission was measured at 345 nm (PTI C61 spectrofluorimeter; Photon Technology International, Lawrenceville, NJ, U.S.A.). All data were corrected for background signal. The data were analysed as described in [4] to determine the Stern–Volmer quenching constant KSV at each pH. The KSV values, determined over the pH range of 3–9, were plotted against pH values as shown in the Figures, and the data were fitted as described by Bose et al. [4] to determine the apparent pKa of each transition.
Fluorescence emission versus pH
Proteins were dialysed against a buffer of 20 mM citrate, pH 6.2 (for experiments between pH 2.5 and 6.2), or a buffer of 20 mM phosphate, pH 9.0 (for experiments between pH 6.0 and 9.0). All buffers contained 1 mM DTT. After dilution into each buffer, the proteins (1 μM) were incubated for at least 1 h at 25 °C. In separate experiments, the equilibration time was determined to be between 15 and 30 min. Samples were excited at 280 nm, and fluorescence emission was monitored from 305 to 400 nm. All data were corrected for background signal. The average emission wavelength (〈λ〉) was calculated as described in [11]. Plots of 〈λ〉 versus pH were then fitted as described previously to determine the apparent pKa of each transition [4]. To examine reversibility, stock proteins were initially dialysed at pH 3 in citrate buffer. The proteins were then diluted into each pH buffer, and the fluorescence emission was monitored as described above. From these experiments, it was found that the results were the same regardless of whether the stock protein was dialysed initially at pH 9 or 3, thus demonstrating reversibility of the transitions shown in Figure 5.
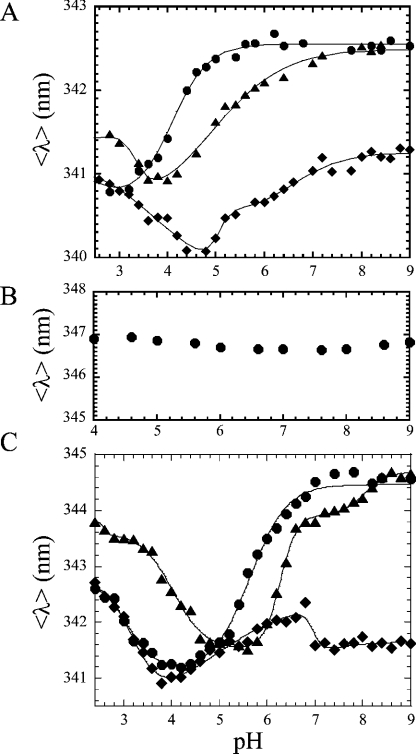
Figure 5. Fluorescence emission versus pH.
(A) The average emission wavelength 〈λ〉 was measured over the pH range 9–2.4 for procaspase 3 (D3A) (●), procaspase 3 (D3A, K242A) (◆) and procaspase 3 (D3A, E246A) (▲). (B) 〈λ〉 versus pH for procaspase 3 (D3A) in 8 M urea-containing buffer. (C) 〈λ〉 versus pH for caspase 3 (●), caspase 3 (K242A) (◆) and caspase 3 (E246A) (▲). The solid lines in (A, C) represent fits of the data as described in the text. The pKa values determined from the fits are given in Table 3.
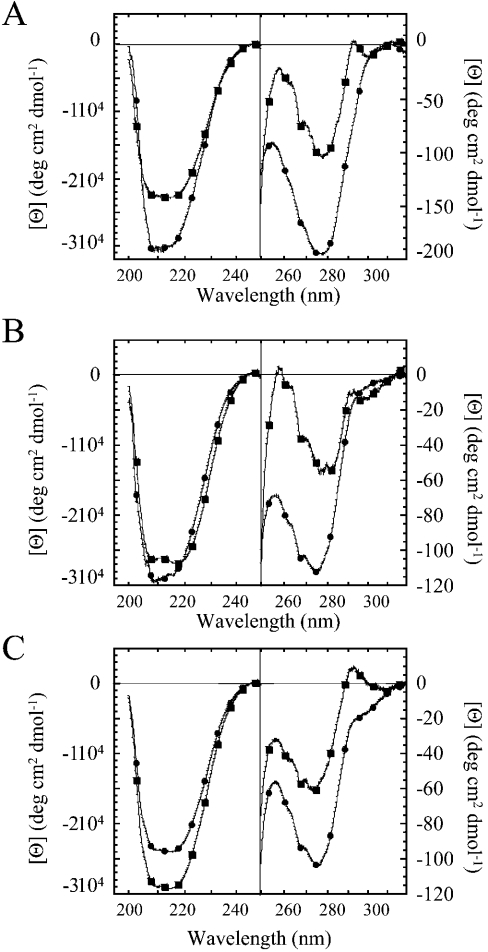
CD spectroscopy
CD spectra were obtained with a Jasco J600A spectropolarimeter (Jasco, Hachioji City, Tokyo, Japan). The protein concentrations for both far- and near-UV CD data were approx. 20 μM, and the samples were in a buffer composed of 50 mM potassium phosphate (pH 7.5) and 1 mM DTT. The path length of the CD cells was either 0.1 cm (far-UV) or 1 cm (near-UV). All experiments were performed at 25 °C, and the spectra were corrected for background signal.
Analytical ultracentrifugation
Sedimentation equilibrium experiments were performed as described in [8,12]. The proteins were in a buffer composed of 20 mM phosphate (pH 7.2) and 1 mM DTT, and experiments were performed at 20 °C. The experimental data were fitted globally using the ORIGIN version [13] of the NONLIN [14] algorithm supplied by Beckman (Fullerton, CA, U.S.A.).
RESULTS
General considerations
Two mutations, K242A and E246A, were obtained depending on the two backgrounds. First, the mutations were introduced into the wild-type procaspase 3. This background provides a means to examine the mutations in the mature caspase 3, because the procaspase autoprocesses when expressed in E. coli. To examine the procaspases, the mutations were introduced into the background of D3A2. As described previously, the three processing sites (Asp9, Asp28 and Asp175) were removed in the D3A mutant, providing a catalytically active yet uncleavable procaspase [4].
We analysed the proteins by size-exclusion chromatography to determine whether the oligomeric properties changed as a result of the mutations. The elution profiles were analysed at several pH values and are described in detail below (see Figure 6); however, at pH 7, wild-type caspase 3, procaspase 3 (D3A), caspase 3 (K242A), procaspase 3 (D3A, K242A) and procaspase 3 (D3A, E246A) were eluted as a single peak at a volume similar to that of the BSA monomer (66 kDa), demonstrating that the proteins are dimers under these conditions (results not shown). The calculated molecular mass of the procaspase 3 monomer was 32.642 kDa. This correlates with our previous results using analytical ultracentrifugation, where we estimated the Kd for dimer dissociation of procaspase 3 (C163S) to be less than 50 nM at pH 7.2 [8]. In contrast, two peaks are observed for caspase 3 (E246A) during elution from the sizing column (see Figure 6C). The first peak is consistent with that of the dimer (59.054 kDa), and the second peak is consistent with that of the monomer (29.527 kDa). Based on the areas under the peaks, we estimate that approx. 10% of caspase 3 (E246A) is monomeric at pH 7. We also analysed the proteins using sedimentation equilibrium studies (results not shown), and the results showed that all of the proteins were dimeric at pH 7, with the exception of caspase 3 (E246A). For that variant, the data were best described by a monomer–dimer equilibrium model with an equilibrium dissociation constant of approx. 6 μM. Overall, the results show that the K242A mutation does not affect the stability of the dimer at pH 7, whereas the E246A mutation decreases dimer stability in the caspase but not in the procaspase.
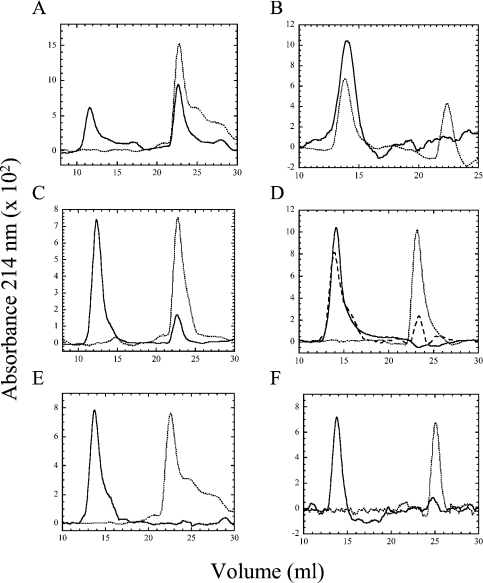
Figure 6. Size-exclusion chromatography versus pH.
(A) Wild-type caspase 3 at pH 5 (——) or pH 4 (- - - -). (B) Procaspase 3 (D3A) at pH 7 (——) or pH 4 (- - - -). (C) Caspase 3 (E246A) at pH 7 (——) or pH 6 (- - - -). (D) Procaspase 3 (D3A, E246A) at pH 6 (——), pH 5 (long broken line) or pH 4 (short broken line). (E) Caspase 3 (K242A) at pH 5 (——) or pH 4 (- - - -). (F) Procaspase 3 (D3A, K242) at pH 5 (——) or pH 4 (- - - -). The monomer of BSA (66 kDa) was eluted at 13.3 ml, and the elution volume did not vary with pH. The dimeric (pro)caspase was eluted at 12–14 ml, whereas the monomeric (pro)caspase was eluted at 23–25 ml.
Enzymic assays
For each mutant, we examined the initial velocity over a range of substrate concentrations, and the results are shown in Figure 2(A), where data for the mature caspase mutants are compared with those for caspase 3 at pH 7.5. Steady-state parameters Km and kcat, determined from the data in Figure 2(A), are presented in Table 1. Results for procaspase 3 (D3A) and mature caspase 3, summarized in Table 1, were described previously [4] and are shown for comparison. Both the procaspase mutants (D3A, K242A and D3A, E246A) were mostly inactive relative to procaspase 3 (D3A) (results not shown), so the steady-state parameters could not be determined for these mutants. Because of the low activity of procaspase 3 (D3A) (Table 1), we assessed the sensitivity of the enzymic assay by examining the initial velocity over a wide range of protein concentrations. The results showed that one can measure approx. 50-fold less enzymic activity than that observed for procaspase 3 (D3A). Thus the mutations in the procaspases result in a decrease in activity of >50-fold compared with that of the procaspase (or 10000-fold compared with that of the mature caspase 3).
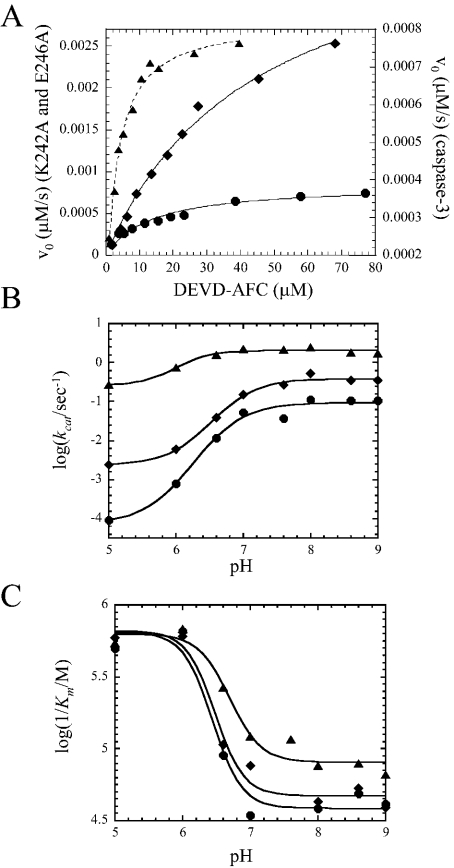
Figure 2. Catalytic parameters of caspase 3 mutants.
(A) Initial velocities of caspase 3 (▲), caspase 3 (K242A) (●) and caspase 3 (E246A) (◆) versus substrate concentration. (B) log (kcat) versus pH for caspase 3 (▲), caspase 3 (K242A) (●) and caspase 3 (E246A) (◆). (C) log (1/Km) versus pH for caspase 3 (▲), caspase 3 (K242A) (●) and caspase 3 (E246A) (◆). The lines represent fits of the data as described in the text. The steady-state parameters Km and kcat (A) and the pKa values of the transitions (B, C) are given in Table 1.
In contrast with the procaspases, both mature caspase 3 mutants were active. As shown in Table 1, the K242A mutation affects both kcat and Km values, where kcat was 10-fold lower and Km was approx. 6-fold higher for the mutant. This results in a specificity constant kcat/Km for caspase 3 (K242A) that is 60-fold lower compared with that for wild-type caspase 3. At pH 7.5, the caspase 3 (E246A) mutant demonstrated a kcat value approximately equal to that of the wild-type caspase 3 (kcat=0.3 s−1), although the Km was significantly higher (23 μM versus 2 μM respectively). This results in a specificity constant that is 14-fold lower than that of wild-type caspase 3. We note that the activity studies were performed with protein concentrations in the low nanomolar range, whereas the oligomeric properties of the proteins were determined in the low micromolar range. It is possible that the low activity of the K242A mutant is due to the formation of the inactive monomers at low protein concentrations. However, one assumes that the presence of the substrate shifts the equilibrium towards the dimeric state and promotes the formation of a competent active state, as was described previously [15,16].
We determined the steady-state parameters over the pH range 5–9, and the results are shown in Figures 2(B) and 2(C). Similar to the mature caspase 3, the catalytic efficiency kcat of caspase 3 (K242A) and that of caspase 3 (E246A) were constant at pH values between 7 and 9 and decreased at lower pH, with an overall larger decrease in kcat value for the mutants. In addition, the pKa values that describe the transitions were higher for the mutants compared with those of wild-type caspase 3 (Table 1). These transitions probably represent conformational changes in the protein that affect the catalytic efficiency rather than protonation of the catalytic cysteine residue at lower pH. As described previously for the unrelated cysteine protease papain [17], it is believed that one form of the enzyme, the thiolate-imidazolium form, is active. Previously, the pKa value for the catalytic cysteine residue of caspase 3 has been suggested to be approx. 6.4 [4,10]. If it is true that the transition represents a protein conformational change, then the results show that the effect is modified by the mutations. The data presented in Figure 2(C) show that Km values, presented as log(1/Km) in the Figure, is constant at pH values higher than 7. This is true also for both the mutants, although the Km values are consistently higher than those of wild-type caspase 3. For all three proteins, the Km values decreased between pH 7 and 6, and the pKa values that describe the transitions (∼6.5) are similar for the three proteins (Table 1). Whereas the pKa values are close to those for the catalytic Cys163 [4,10], it is not clear whether the transition represents the protonation of Cys163 or a conformational change that affects Km. Regardless of these results, the data show that the mutations do not affect the transition. For the mutants, it is worth noting that the same pKa values describe the transitions in kcat (Figure 2B) and in Km (Figure 2C) values. At present, it is not known whether there are two protonation events that affect the two different steps in the enzymic reaction or whether a single protonation event affects both steps. Although the range of pH values is not sufficiently broad to study the ionization of the catalytic histidine, the data shown in Figure 2(C) suggest that the mutations do not affect the ionization of the catalytic cysteine residue. We note that this interpretation should be viewed with some caution, however, since the protonation of Cys163 may be coupled with conformational changes in the protein. As described below, we show that both the mutations alter the protein conformation.
CD spectra
To assess the secondary and tertiary structures of the mutants, we examined the far- and near-UV CD spectra, and the results are compared with those of procaspase 3 (D3A) and of mature caspase 3 (Figure 3). In general, the far-UV CD spectra are similar for the proteins and show that the mutations do not disrupt the overall fold. It is interesting to note that, for procaspase 3 (D3A) (Figure 3A) and procaspase 3 (D3A, K242A) (Figure 3B), the signal increases (i.e. becomes more negative) on maturation. This is not true for the E246A mutant (Figure 3C). At this point, it is not clear whether these results are due to a change in secondary structure after maturation or due to changes in the tertiary structure, which may also affect the signal in this region. Both processes are known to occur after maturation [1,2,4]. We note, however, that the results are not explained by the loss of the propeptide on maturation, since it was observed previously that the signal of procaspase 3 decreases somewhat in the ‘pro-less’ variant of procaspase 3 when compared with the full-length procaspase [8].
Figure 3. CD spectra of caspase mutants.
Far- and near-UV CD spectra of (A) caspase 3 and procaspase 3 (D3A), (B) caspase 3 (K242A) and procaspase 3 (D3A, K242A) and (C) caspase 3 (E246A) and procaspase 3 (D3A, K242A). (A–C) ●, caspase; ■, procaspase.
The near-UV CD spectrum of procaspase 3 (D3A) (Figure 3A) demonstrates maxima at 258 and 292 nm and a minimum at 278 nm. On processing to the mature caspase 3, the near-UV CD signals increase. These changes are consistent with loop movements that occur on maturation (see [18] for a review), which affect the two active-site tryptophan residues (292 nm) and the phenylalanine residues (258 nm). The procaspase 3 monomer contains 15 phenylalanine residues, four of which reside in loop L4 (Figure 1), which is known to undergo a conformational change on maturation [18].
The near-UV CD spectra of the mutant mature caspases (Figures 3B and 3C) are similar to that of the wild-type caspase 3. In all cases, there is an increase in signal on maturation, as observed with the wild-type caspase 3. The mutants demonstrate minima at 275 nm and maxima at 258 nm, although the signals were lower than those described in Figure 3(A). Compared with procaspase 3 (D3A) (Figure 3A), however, the maximum at 292 nm was not present in procaspase 3 (D3A, K242A) (Figure 3B).
Overall, the results show that the proteins are folded with well-packed tertiary structures. The changes in the signal at 292 nm for procaspase 3 (D3A, K242A) suggest an alteration in the environment of Trp206 and/or Trp214 in the active site and are consistent with a change in the enzymic activity described above and the fluorescence emission properties described below.
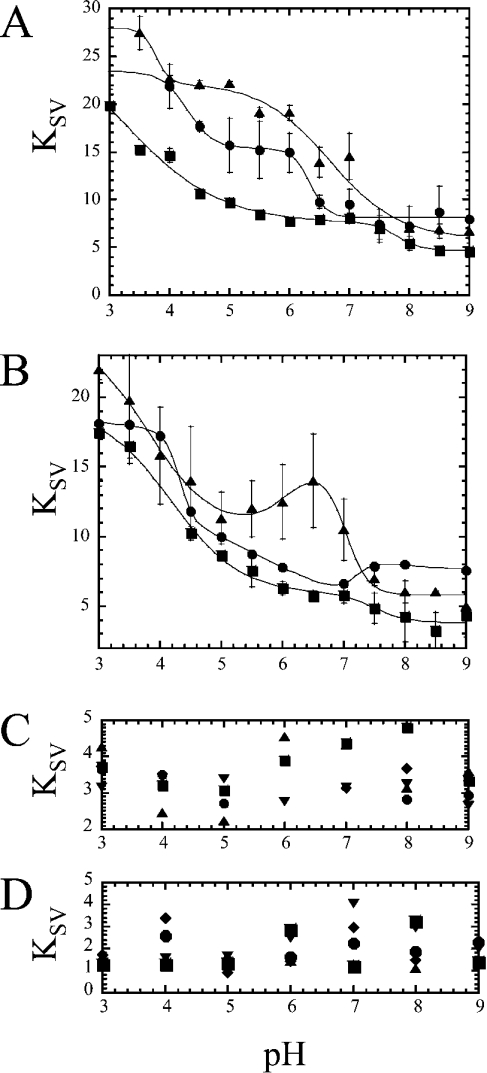
Quenching studies
To examine further the environments of the two active-site tryptophan residues, we examined the accessibility to quenching by iodide. We have shown that, in procaspase 3, the KSV increases as the pH is decreased from pH 9 to 3 [4]. The higher quenching constant indicates a higher accessibility of the KI quencher to the tryptophan residues. Results for procaspase 3 (D3A) (see Figure 4A) are described by two transitions, with pKa values of 6.3 and 4.3 respectively (Table 2). Further studies with acrylamide and CsCl demonstrate a low accessibility to these two quenchers and no pH dependence to quenching (Figures 4C and 4D). Together, the results suggest that the titration of one or more acidic groups and/or histidyl residues affects the electrostatic environment of the tryptophan residues, allowing greater access to the negatively charged iodide at low pH.
Figure 4. Quenching versus pH.
(A) The KSV value was determined for KI quenching over the pH range 9–3 as described in the text for procaspase 3 (D3A) (●), procaspase 3 (D3A, K242A) (■) and procaspase 3 (D3A, E246A) (▲). (B) KSV values versus pH for KI quenching of caspase 3 (●), caspase 3 (K242A) (■) and caspase 3 (E246A) (▲). The solid lines represent fits of the data as described in the text. The pKa values determined from the fits are given in Table 2. (C) KSV from acrylamide; (D) CsCl quenching versus pH of procaspase 3 (D3A) (●), caspase 3 (E246A) (▲), procaspase 3 (D3A, E246A) (■), caspase 3 (K242A) (▼) and procaspase 3 (D3A, K242A) (◆). Error bars show the S.D. determined from the results of four independent experiments.
Table 2. pKa values determined from iodide quenching studies.
| pKa | Caspase 3 | E246A | K242A | D3A | D3A, E246A | D3A, K242A |
|---|---|---|---|---|---|---|
| (1) | 4.3±0.1 | 3.8±0.2 | 4.2±0.2 | 4.3±0.1 | 3.8±0.1 | 3.3±1 |
| (2) | 5.6±0.4 | 6.1±0.6 | ||||
| (3) | 7.2±0.1 | 7.0±0.1 | 7.5±0.3 | 6.3±0.2 | 6.7±0.2 | 7.8±0.2 |
Similar experiments were performed with the K242A and E246A procaspases (Figure 4A) and, as with procaspase 3, the mutants are described by two transitions. At high pH (pH>8), the tryptophan residues appear to be equally accessible to the quencher (KSV∼6), whereas the proteins show differences as the pH is decreased. Procaspase 3 (D3A, E246A) demonstrates a plateau between pH 6 and 4, where KSV is significantly higher than that of procaspase 3 (D3A) or of procaspase 3 (D3A, K242A) (Figure 4A). The pKa of the transition is 6.7 (Table 2). There is a further increase in KSV below pH 4, with a pKa∼3.8. In contrast, the results for procaspase 3 (D3A, K242A) show that the accessibility to the iodide quencher is lower than that for other procaspases. The pKa values that describe the two transitions are approx. 3.3 and 7.8 respectively (Table 2). As with procaspase 3, further studies with acrylamide and CsCl demonstrate a low accessibility to these two quenchers and no pH dependence on quenching (Figures 4C and 4D). Although it is not yet clear what groups are involved in these transitions, the results show that the removal of the two charges affects the pKa values of both transitions as well as the accessibility of the negatively charged iodide. Overall, the results suggest that Glu246 and Lys242 are involved, either directly or indirectly, in maintaining the electrostatic environment of the tryptophan residues in the procaspase active site. Removal of the negative charge allows for increased accessibility to the negatively charged iodide, whereas removal of the positive charge results in a decrease in iodide accessibility.
The iodide quenching of mature caspase 3 against pH values can be described by three transitions (Figure 4B and Table 2). The first transition, between pH 3 and 4.5 (pKa=4.3), results in a decrease in KSV values, as does the second transition, between pH 4.5 and 7 (pKa=5.6). The third transition, between pH 7 and 9 (pKa=7.2), results in a small increase in KSV. Caspase 3 (E246A) is also characterized by three transitions (Figure 4B). In contrast with caspase 3, however, the second transition (pKa=6.1) results in an increase in KSV values, whereas the third transition (pKa=7.0) results in a decrease in KSV values. Although the pKa values are not significantly different from those of caspase 3 (Table 2), the E246A mutant has higher iodide accessibility below pH∼7.5, as was observed with the procaspase form of this mutant (Figure 4A). The increase in iodide accessibility between pH 7 and 6 for this mutant is probably coupled with the dissociation of the heterotetramer, as described above. This issue is discussed in more detail below (Figure 6). For the K242A mutant, the results are adequately described by two transitions, with pKa values of 4.2 and 7.5 (Table 2). Thus the second transition observed with caspase 3 and caspase 3 (E246A) is not observed in this mutant. As with the procaspase form, the K242A mutant has lower accessibility to iodide (below pH∼7.5). Overall, these results again suggest that both the charged groups influence the electrostatic environment of the active site.
We also performed quenching studies with acrylamide and CsCl. As with the other proteins, the mature caspases demonstrate low accessibility to these two quenchers and no pH dependence to quenching (Figures 4C and 4D).
Average emission wavelength versus pH
The average emission wavelength, after excitation at 280 nm, measures the global contribution of all aromatic residues to the fluorescence emission. We have shown that the average emission wavelength 〈λ〉 for caspase 3 reaches a plateau between pH 7 and 9, where 〈λ〉=344.6 nm [4], whereas that of procaspase 3 (D3A) is 342.5 nm (Figure 5A). This shows that, on maturation, a red-shift in fluorescence emission occurs, which is consistent with a more solvent-accessible active site, as described previously [4]. As the pH is decreased, the procaspase undergoes two transitions, with pKa values of 4.2 and <3 respectively (Figure 5A and Table 3).
Table 3. pKa values determined from average emission wavelength studies.
| pKa | Caspase 3 | E246A | K242A | D3A | D3A, E246A | D3A, K242A |
|---|---|---|---|---|---|---|
| (1) | 3.0±0.2 | <3 | 3.2±0.1 | <3 | 3.3±0.2 | 3.8±0.6 |
| (2) | 5.7±0.1 | 4.0±0.1 | 4.5±0.7 | 4.2±0.1 | 5.2±0.4 | 5.1±0.1 |
| (3) | 6.3±0.1 | 6.9±0.1 | 6.5±0.1 | |||
| (4) | 8.0±0.1 |
The fluorescence emission profiles of the mutants are significantly different from that of procaspase 3 (D3A) (Figure 5A). In procaspase 3 (D3A, E246A), the 〈λ〉 reaches a plateau at higher pH similar to that of the control (342.5 nm), and two transitions are observed as the pH is lowered. The first transition, between pH 7 and 4 (pKa=5.2; Table 3), represents a decrease in 〈λ〉 similar to that of the control (341 nm), although the pKa value is significantly higher (5.2 versus 4.2 respectively). A second transition, between pH 4 and 3 (pKa=3.3), results in an increase in 〈λ〉 (341.5 nm). This transition is probably the same, but with a higher pKa value, similar to that observed in procaspase 3 (D3A), where pKa<3.
For the second mutant, procaspase 3 (D3A, K242A) has a significantly blue-shifted fluorescence emission when compared with procaspase 3 (D3A), where 〈λ〉 reaches a plateau at 341.3 nm for pH>8. The results are described by three transitions with pKa=6.5, 5.1 and 3.8 respectively (Table 3). Overall, the results obtained for procaspases show that the mutations affect the pKa of the lowest transition, that a new transition appears between pH 5 and 7 for procaspase 3 (D3A, K242A), and that the fluorescence emission of procaspase 3 (D3A, K242A) is significantly blue-shifted at higher pH relative to that of procaspase 3 (D3A).
For comparison, we collected data for procaspase 3 (D3A) that had been incubated in 8 M-urea-containing buffer (Figure 5B). We have shown that the procaspase is unfolded under these conditions, and the aromatic residues are exposed to the solvent [19]. The results show that, on unfolding, the average emission wavelength increased to 347 nm and that there was no pH dependence to this value. This demonstrates that the blue-shifted transitions described in Figure 5(A) result from a more hydrophobic and probably less solvent-accessible environment of the aromatic residues as the pH is lowered. Although the nature of these transitions remain under investigation, the results show that the active-site tryptophan residues of the native proteins are not completely solvent-exposed, even at low pH.
The mature caspase variants are also significantly different in this assay when compared with the wild-type caspase 3 (Figure 5C). The results show that caspase 3 (E246A) has a 〈λ〉 value similar to that of caspase 3, but only at pH >8. A transition occurs between pH 8.5 and 6.5 (pKa=8.0; Table 3) and results in a plateau of 344 nm. A second transition occurs between pH 6.5 and 5.5 (pKa=6.3), which results in a significant decrease in 〈λ〉 value (to 341.5 nm). Below pH 5, two transitions occur that result in an increase in 〈λ〉 value, with pKa values of 4.0 and <3 respectively. The first transition (pKa=4.0) is probably the same as that observed in caspase 3 (pKa=3.0), but with a higher pKa value. The data show that both forms of this mutant (pro- and mature) attain the same emission maximum as the controls at higher pH values, although the mutation affects the pKa values of the transitions.
For caspase 3 (K242A), the results at low pH are similar to those of wild-type caspase 3, where 〈λ〉 increases from 341 to approx. 343 nm below pH 4 (Figure 5C). However, the mutant varies significantly at higher pH, where it is observed that 〈λ〉 is significantly blue-shifted at pH >7 when compared with either wild-type caspase 3 or caspase 3 (E246A). The results are best described by three transitions with pKa values of 6.9, approx. 4.5 and 3.2 respectively (Table 3). Thus, at pH >7, there is little difference in the fluorescence emission for the two forms of this mutant (cf. Figures 5A and 5B), in contrast with the wild-type caspase 3 and the E246A variant. Below, we will correlate these pH-dependent transitions with the oligomeric properties of the proteins.
Size-exclusion chromatography versus pH
To examine the potential effects of pH on the oligomeric properties of the proteins, we performed size-exclusion chromatography experiments over the pH range of 7–4. In these experiments, approx. 10 μM protein was loaded on to the column. This concentration is consistent with the fluorescence emission and quenching studies described above. As shown previously [8] and described above, procaspase 3 is a dimer at pH 7, and we have estimated the Kd value for dimer dissociation to be below 50 nM. At pH 7 and 6, caspase 3 was eluted as a single peak, consistent with the approx. 60 kDa heterotetramer (results not shown). It was observed that from pH 7 to 5, the heterotetramer was eluted between 11.6 and 12.2 ml. At pH 5, however, two peaks were observed in the elution profile (Figure 6A); the first peak is that of the heterotetramer, whereas the second peak is consistent with the heterodimer (30 kDa). Whereas the baselines are noisy due to the low protein concentration, we estimate that approx. 60% of the protein is monomeric at pH 5. At pH 4, a single peak is observed at approx. 23 ml, demonstrating that the heterodimer has completely dissociated under these conditions. Two smaller peaks are observed at 25 and 28 ml that probably represent the dissociation of the heterodimer into the individual large and small subunits. For procaspase 3 (D3A), a single peak is observed at pH 7 to 5, consistent with the mass of the homodimer (64 kDa). The homodimer of procaspase 3 was eluted between 14.0 and 14.3 ml (Figure 6B), although it is not clear why the procaspase 3 is eluted at a slightly higher elution volume when compared with the mature caspase. At pH 4, two peaks are observed in the elution profile, consistent with that of the dimer (14 ml) and monomer (23 ml). Approx. 40% of the protein is monomeric under these conditions.
For caspase 3 (E246A), two peaks were observed at pH 7, consistent with the presence of a heterotetramer and a heterodimer (Figure 6C), and we estimate that approx. 10% of the protein is heterodimeric under these conditions. As described above, the presence of the heterodimer was confirmed by analytical ultracentrifugation. At pH 6, no heterotetramer is observed, and the protein is eluted as the heterodimer. In contrast, the monomer of the E246A variant of the procaspase was not observed at pH 7 or 6, and approx. 25% of the protein was monomeric at pH 5 (Figure 6D). It was observed that the protein was completely monomeric at pH 4. For caspase 3 (K242A), the protein is a heterotetramer from pH 7 to 5 (Figure 6E). In contrast with the wild-type caspase, no heterodimer is observed at pH 5, whereas the profiles are nearly identical at pH 4. That is, the K242A heterodimer is eluted at 23 ml, and the putative large- and small-subunit minor peaks are observed at 25 and 28 ml respectively. For the procaspase K242A variant (Figure 6F), the protein is a homodimer at pH >5 and a monomer at pH 4. Overall, the data show that the E246A mutation destabilizes the dimer, and the effect is larger in the mature caspase. In contrast, the heterotetramer appears to be more stable in the mature K242A variant, but the homodimer of the procaspase is somewhat less stable when compared with the controls. Unfortunately, it has not been possible to examine the monomer–dimer equilibrium quantitatively at lower pH values using sedimentation equilibrium experiments, as the monomer (or heterodimer) precipitates slowly under the conditions of the experiment (pH 4.0–5.0, >20 μM protein, 20 °C). However, the data shown in Figure 6, as well as our previous data [8], demonstrate that the equilibrium dissociation constant changes from the low nanomolar range at pH 7.2 to the low micromolar range, or higher, at pH 4. For caspase 3 (E246A), dissociation occurs between pH 7 and 6.
DISCUSSION
We have examined the role of a salt bridge between Lys242 and Glu246, located near the base of the active-site loop L4, and we show that the ionic interactions between the two residues are important for maintaining the active-site environment. While removal of either amino acid abrogates activity in the procaspase, the mutations affect the activity of the mature caspase as well. Both the steady-state parameters, kcat and Km, are affected by the mutations. At pH 7.5, where maximum activity is observed for caspase 3, the specificity constants for the mutants, kcat/Km, are between 14- and 60-fold lower than that of caspase 3. Overall, the data show that the values of Km are consistently higher in the mutants, whereas the catalytic efficiencies are consistently lower. The K242A mutation has a stronger effect on the activity, due to a 10-fold decrease in kcat.
Several assays show that Lys242 and Glu246, either directly or indirectly, affect the electrostatic environment of the active site. First, the pKa values associated with changes in kcat versus pH are increased in both mutants. Secondly, a change in accessibility to the negatively charged iodide quencher is observed. Finally, fluorescence emission studies showed that caspase 3 (E246A) undergoes a transition between pH 6 and 8 that results in a red-shift in fluorescence emission.
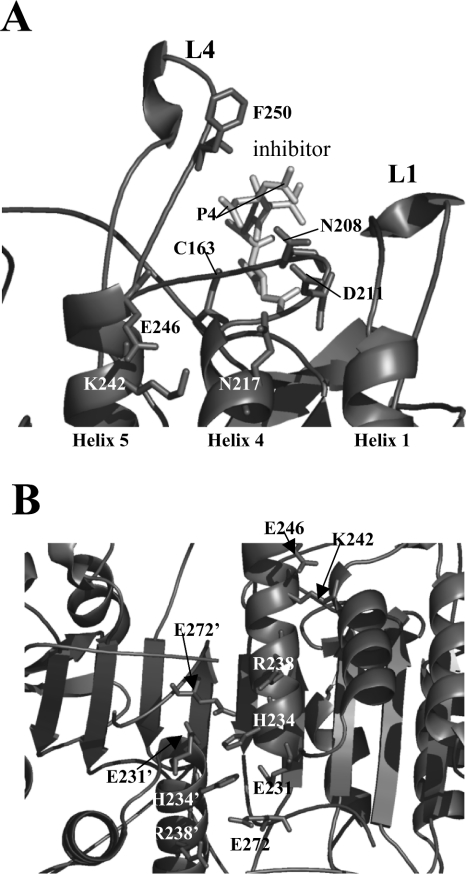
It is well known that pKa values of titratable groups are influenced by the electrostatic field [20,21]. In the active sites of enzymes, the local hydrogen-bonding network and the solvent accessibility will influence the pKa values of the catalytic groups. It is unlikely that Glu246 or Lys242 directly influence the active-site hydrogen-bonding network, since both residues are approx. 19 Å (1 Å=0.1 nm) from the catalytic Cys163. However, indirect effects due to the observed conformational changes can explain the changes in kcat and Km values. For example, if the position of helix 4 is altered due to loss of the Lys242–Glu246 salt bridge, then the position of loop L3, the substrate binding loop, may be affected. Residues Ser209, Lys210 and Asp211, near helix 4, form hydrogen bonds with the side chain Oδ1 atom of Asn208 (loop L3), which in turn hydrogen-bonds with the P4 site of the substrate [7] (Figure 7A). In addition, Ser209 forms additional hydrogen bonds with the substrate. Asp211 is only 3.6 Å from Asn217, located on helix 4 (see Figure 7A), which forms part of the surface surrounding Lys242 (see Figure 1). Thus disruption of the Lys242–Glu246 salt bridge in helix 5 may alter the position of helix 4 and hence affect the hydrogen-bonding network of the S4 subsite. Alternatively, changes in the stability or dynamics of loop L4 may also explain the changes observed in the catalytic parameters. For example, the amide backbone of Phe250 in loop L4 (see Figure 7A) hydrogen-bonds with the P4 carboxylate side chain [7]. Thus changes in the stability of loop L4 may disrupt these interactions. Further studies are required to determine the specific reasons for the changes in the enzymic activity.
Figure 7. Relative position of Lys242–Glu246 salt bridge to the active site and to putative stabilizing ionic interactions.
(A) The relative position of the Lys242–Glu246 salt bridge to the caspase 3 active site. Residues Asn208, Asp211 and Asn217 are described in the text, and the catalytic Cys163 is indicated. (B) Ionic interactions across the dimer interface suggested to stabilize the dimer. A prime (′) indicates a residue from the second heterodimer. The structures were generated using PDB entry 1CP3 and PyMOL (Delano Scientific).
In addition to affecting the enzymic activity, the mutations affected the stability of the dimer. Whereas all proteins were dimeric at pH 7, it was observed that the E246A mutation in the context of the mature caspase destabilized the protein such that the heterodimer was present at pH 7 (∼10%), and the heterotetramer was completely dissociated at pH 6. For caspase 3, heterotetramer dissociation occurred over the pH range of 6–4, whereas the procaspase homodimer dissociated between pH 5 and 4. This was true also for both the K242A variants. In each case, the dissociation of the dimer correlates with a blue-shift in fluorescence emission. While we do not know the groups responsible for the transitions in fluorescence emission, the data suggest that the maturation of the procaspase results in a protein that is more sensitive to pH-dependent dimer dissociation. It is worth noting that while dimer dissociation is accompanied by a blue-shifted fluorescence emission profile, dissociation also leads to an increase in accessibility to the iodide quencher. By utilizing limited V8 proteolysis assays of procaspase 3 (D3A), we showed previously that at least two conformational changes occurred between pH 7 and 4 [4]. At pH 5, the prodomain is more solvent-exposed, and active-site loop L4 undergoes a pH-dependent conformational change that exposes Glu248/Asp253 to rapid cleavage, relative to the accessibility at pH 7. The second conformational change occurs between pH 5 and 4, where it was observed that, at pH 4, the protein was significantly less sensitive to V8 protease than it was at pH 5. Cleavage was observed only in loop L4 at Asp248/Asp253. From the data presented here, we now know that the second transition correlates with dimer dissociation. Thus the picture of the procaspase monomer that emerges from these combined studies is one in which the structure is less accessible to V8 proteolysis, and the tryptophan residues are in a more hydrophobic environment, and exhibit blue-shifted fluorescence emission, yet are more accessible to the iodide quencher. This may be explained by titration of the negative charges at a lower pH close to the tryptophan side chains. We note that, in terms of the fluorescence properties, the procaspase 3 monomer at pH 4 resembles the monomeric folding intermediate that has been described at pH 7.2 [19]. Finally, after dimer dissociation, further transitions are observed at lower pH that result in a red-shift in fluorescence emission. The size-exclusion chromatography data suggest that this may result from dissociation of the subunits in the heterodimer (or a similar unfolding of the procaspase monomer).
In caspase 3, several ion pairs at the N-terminus of helix 5 interact across the dimer interface and are predicted to stabilize the dimer. From one monomer, Glu231, His234, Arg238 and Glu272 form a network of charge–charge interactions with the same residues from the second monomer (Figure 7B). The first three residues reside on the same face of helix 5, whereas Glu272 is contributed by the C-terminus of strand 8. Lys242 resides on the same face of helix 5 as these charged residues, so it is conceivable that the loss of the Lys242–Glu246 salt bridge disrupts, or alters, the interactions of these other ion pairs across the dimer interface. A similar series of polar interactions are observed in procaspase 7 [1,2] as well as other caspases [15,22,23], suggesting that this effect may be observed in other caspases also. Although the residues are not strictly conserved in other caspases, interactions similar to those of Lys242–Glu246 found in caspase 3 may result in similar properties in other caspases. That is, the stability of loop L4 and the C-terminal region of helix 5 may affect the dimeric properties of the caspase by influencing the interactions among the polar groups at the N-terminus of helix 5. For example, we note that Lys242 is conserved in caspase 6 and CED-3 (cell-death determining-3), whereas, in caspase 7, arginine residue is found in the same position [5]. However, Glu246 is not conserved. Thus, in caspase 7, Arg218 (comparable with Lys242 in caspase 3) interacts with Glu201 from helix 4. The Glu201–Arg218 interactions in caspase 7 are predicted to impart similar features to the Lys242–Glu246 interactions in caspase 3.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM065970) and by the North Carolina Agricultural Research Service. We thank Dr M. Goshe for use of the HPLC system and G. Respet for enzyme studies.
References
- 1.Chai J., Wu Q., Shiozaki E., Srinivasula S. M., Alnemri E. S., Shi Y. Crystal structure of a procaspase 7 zymogen: mechanisms of activation and substrate binding. Cell (Cambridge, Mass.) 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 2.Riedl S. J., Fuentes-Prior P., Renatus M., Kairies N., Krapp S., Huber R., Salvesen G. S., Bode W. Structural basis for the activation of human procaspase 7. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14790–14795. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy S., Bayly C. I., Gareau Y., Houtzager V. M., Kargman S., Keen S. L. C., Rowland K., Seiden I. M., Thornberry N. A., Nicholson D. W. Maintenance of caspase 3 proenzyme dormancy by an intrinsic ‘safety catch’ regulatory tripeptide. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6132–6137. doi: 10.1073/pnas.111085198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bose K., Pop C., Feeney B., Clark A. C. An uncleavable procaspase 3 mutant has a lower catalytic efficiency but an active site similar to that of mature caspase 3. Biochemistry. 2003;42:12298–12310. doi: 10.1021/bi034998x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Earnshaw W. C., Martins L. M., Kaufmann S. H. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 6.Mittl P. R. E., DiMarco S., Krebs J. F., Bai X., Karanewsky D. S., Priestle J. P., Tomaselli K. J., Grutter M. G. Structure of recombinant human CPP32 in complex with the tetrapeptide acetyl-Asp-Val-Ala-Asp fluoromethyl ketone. J. Biol. Chem. 1997;272:6539–6547. doi: 10.1074/jbc.272.10.6539. [DOI] [PubMed] [Google Scholar]
- 7.Rotonda J., Nicholson D. W., Fazil K. M., Gallant M., Gareau Y., Labelle M., Peterson E. P., Rasper D. M., Ruel R., Vaillancourt J. P., et al. The three-dimensional structure of apopain/CPP32, a key mediator of apoptosis. Nat. Struct. Biol. 1996;3:619–625. doi: 10.1038/nsb0796-619. [DOI] [PubMed] [Google Scholar]
- 8.Pop C., Chen Y.-R., Smith B., Bose K., Bobay B., Tripathy A., Franzen S., Clark A. C. Removal of the pro-domain does not affect the conformation of the procaspase 3 dimer. Biochemistry. 2001;40:14224–14235. doi: 10.1021/bi011037e. [DOI] [PubMed] [Google Scholar]
- 9.Stennicke H. R., Salvesen G. S. Caspases: preparation and characterization. Methods. 1999;17:313–319. doi: 10.1006/meth.1999.0745. [DOI] [PubMed] [Google Scholar]
- 10.Stennicke H. R., Salvesen G. S. Biochemical characteristics of caspases-3, -6, -7, and -8. J. Biol. Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 11.Royer C. A., Mann C. J., Matthews C. R. Resolution of the fluorescence equilibrium unfolding profile of trp aporepressor using single tryptophan mutants. Protein Sci. 1993;2:1844–1852. doi: 10.1002/pro.5560021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pop C., Feeney B., Tripathy A., Clark A. C. Mutations in the procaspase 3 dimer interface affect the activity of the zymogen. Biochemistry. 2003;42:12311–12320. doi: 10.1021/bi034999p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedegs J., Sarrafzadeh S., Lear J. D., McRorie D. K. Extensions to commercial graphics packages for customization of analysis of analytical ultracentrifuge data. In: Shuster T., Laue T. M., editors. Modern Analytical Ultracentrifugation: Acquisition and Interpretation of Data for Biological and Synthetic Polymer Systems. Boston: Birkhauser; 1994. pp. 227–244. [Google Scholar]
- 14.Johnson M. L., Correira J. J., Yphantis D. A., Halvorson H. R. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., Salvesen G. S. Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talanian R. V., Dang L. C., Ferenz C. R., Hackett M. C., Mankovich J. A., Welch J. P., Wong W. W., Brady K. D. Stability and oligomeric equilibria of refolded interleukin-1β converting enzyme. J. Biol. Chem. 1996;271:21853–21858. doi: 10.1074/jbc.271.36.21853. [DOI] [PubMed] [Google Scholar]
- 17.Lewis S. D., Johnson F. A., Shafer J. A. Effect of cysteine-25 on the ionization of histidine-159 in papain as determined by proton nuclear magnetic resonance spectroscopy. Evidence for a His-159-Cys-25 ion pair and its possible role in catalysis. Biochemistry. 1981;20:48–51. doi: 10.1021/bi00504a009. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 19.Bose K., Clark A. C. Dimeric procaspase 3 unfolds via a four-state equilibrium process. Biochemistry. 2001;40:14236–14242. doi: 10.1021/bi0110387. [DOI] [PubMed] [Google Scholar]
- 20.Pickersgill R. W., Goodenough P. W., Sumner I. G., Collins M. E. The electrostatic fields in the active-site clefts of actinidin and papain. Biochem. J. 1988;254:235–238. doi: 10.1042/bj2540235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perutz M. F. Electrostatic effects in proteins. Science. 1978;201:1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- 22.Wilson K. P., Black J.-A. F., Thomson J. A., Kim E. E., Griffith J. P., Navia M. A., Murcko M. A., Chambers S. P., Aldape R. A., Raybuck S. A., et al. Structure and mechanism of interleukin-1β converting enzyme. Nature (London) 1994;370:270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 23.Watt W., Koeplinger K. A., Mildner A. M., Heinrikson R. L., Tomasselli A. G., Watenpaugh K. D. The atomic-resolution structure of human caspase 8, a key activator of apoptosis. Structure. 1999;7:1135–1143. doi: 10.1016/s0969-2126(99)80180-4. [DOI] [PubMed] [Google Scholar]