Abstract
C4ST-1 (chondroitin 4-sulphotransferase-1) and C6ST-1 (chondroitin 6-sulphotransferase-1) transfer sulphate from PAPS (adenosine 3′-phosphate 5′-phosphosulphate) to positions 4 and 6 respectively of the GalNAc residues of chondroitin. We showed previously that C4ST-1 purified from rat chondrosarcoma and recombinant C4ST-1 both transfer sulphate efficiently to position 4 of the GalNAc residues of DSDS (desulphated dermatan sulphate). We report here the specificity of C4ST-1 and C6ST-1 in terms of uronic acid residue recognition around the GalNAc residue to which sulphate is transferred. When [35S]glycosaminoglycans formed from DSDS after incubation with [35S]PAPS and C4ST-1 were digested with chondroitinase ACII, a major part of the radioactivity was recovered in disaccharide fractions and the remainder distributed to tetrasaccharides and larger fractions, indicating that C4ST-1 mainly transferred sulphate to position 4 of the GalNAc residue located at the GlcA-GalNAc-GlcA sequence. Structural analysis of tetrasaccharide and larger oligosaccharide fractions indicated that C4ST-1 mainly transferred sulphate to the GalNAc residue adjacent to the reducing side of the GlcA residue. On the other hand, when [35S]glycosaminoglycans formed from DSDS after incubation with [35S]PAPS and C6ST-1 were digested with chondroitinase ACII, a major part of the radioactivity was recovered in fractions larger than hexasaccharides, indicating that C6ST-1 transferred sulphate to the GalNAc residues located in the L-iduronic acid-rich region. Structural analysis of the tetrasaccharide and larger oligosaccharide fractions indicated that C6ST-1 showed very little preference for the GalNAc residue neighbouring the GlcA residue. These results indicate that C4ST-1 and C6ST-1 differ from each other in the recognition of uronic acid residues adjacent to the targeted GalNAc residue.
Keywords: chondroitin sulphate–dermatan sulphate hybrid, chondroitin 4-sulphotransferase-1 (C4ST-1), C6ST-1, desulphated dermatan sulphate, GalNAc-4-O-sulphation, GalNAc-6-O-sulphation
Abbreviations: C4ST-1, chondroitin 4-sulphotransferase-1; C6ST-1, chondroitin 6-sulphotransferase-1; CS, chondroitin sulphate; HexA, hexuronic acid; ΔHexA, 4,5-unsaturated HexA; DS, dermatan sulphate; DSDS, desulphated DS; D4ST, dermatan 4-sulphotransferase; GalNAc, N-acetylgalactosamine; GalNAc(4,6-SO4), 4,6-bis-O-sulpho-GalNAc; GalNAc(4SO4), 4-O-sulpho-GalNAc; GalNAc(6SO4), 6-O-sulpho-GalNAc; GlcA, D-glucuronic acid; Di-6S, GlcAβ1-3GalNAc(6SO4); IdoA, L-iduronic acid; PAPS, adenosine 3′-phosphate 5′-phosphosulphate; Tet-40, GlcA-GalNAc(4SO4)-GlcA-GalNAc; Tet-44, GlcA-GalNAc(4SO4)-GlcA-GalNAc(4SO4); Tet-46, GlcA-GalNAc(4SO4)-GlcA-GalNAc(6SO4); Tet-60, GlcA-GalNAc(6SO4)-GlcA-GalNAc; Tet-64, GlcA-GlNAc(6SO4)-GlcA-GalNAc(4SO4); Tet-66, GlcA-GalNAc(6SO4)-GlcA-GalNAc(6SO4); Tri-40, GalNAc(4SO4)-GlcA-GalNAc; Tri-44, GalNAc(4SO4)-GlcA-GalNAc(4SO4); Tri-46, GalNAc(4SO4)-GlcA-GalNAc(6SO4); Tri-60, GalNAc(6SO4)-GlcA-GalNAc; Tri-64, GalNAc(6SO4)-GlcA-GalNAc-(4SO4); Tri-66, GalNAc(6SO4)-GlcA-GalNAc(6SO4)
INTRODUCTION
C4ST-1 (chondroitin 4-sulphotransferase-1) and C6ST-1 (chondroitin 6-sulphotransferase-1) transfer sulphate from PAPS (adenosine 3′-phosphate 5′-phosphosulphate) to positions 4 and 6 respectively of the GalNAc residues of chondroitin. We previously purified these enzymes to homogeneity and cloned the cDNAs on the basis of the amino acid sequences of the purified proteins [1–4]. Purified C4ST-1 [1] as well as the recombinant C4ST-1 [2] were found to sulphate DSDS [desulphated DS (dermatan sulphate)]; however, sulphate was transferred to a glucuronic acid-rich region of DSDS because a major part of the 35S-radioactivity was recovered in ΔDi-4S [2-acetamide-2-deoxy-3-O-(β-D-gluco-4-enepyranosyluronic acid)-4-O-sulpho-D-galactose] when the 35S-labelled glycosaminoglycans were formed from DSDS after the reaction with C4ST-1, and [35S]PAPS were digested with chondroitinase ACII [1,2]. Such a value of specificity for C4ST-1 has also been reported by other groups [5–7]; however, how C4ST-1 can differentiate GlcA-containing repeating units from IdoA (L-iduronic acid)-containing repeating units is still not clear.
CS (chondroitin sulphate)–DS hybrid molecules have been reported to be present in various tissues [8–16]. In these hybrid molecules, GalNAc residues located in GlcA-containing moieties were sulphated at position 4 or 6; however, it is not known how the sulphation pattern of these hybrid molecules is determined. C5 epimerization of chondroitin by a particular fraction from human fibroblasts was stimulated by the presence of a sulphate donor, PAPS [17]. Biosynthesis of DS by cultured fibroblasts was decreased when the concentration of inorganic sulphate in the medium was decreased [18,19]. These observations suggest that C5 epimerization is closely linked to 4-O-sulphation of GalNAc residue. On the other hand, recent studies suggest that, during the biosynthesis of DS, C5 epimerization may occur before the 4-O-sulphation of GalNAc residues, since a microsomal fraction obtained from human fibroblasts was reported to sulphate DSDS more efficiently than chondroitin [20], and unlike C4ST-1, the recombinant D4ST (dermatan 4-sulphotransferase) has been reported to prefer DSDS to chondroitin as the acceptor [6,7]. However, the mechanisms by which C5 epimerization of GlcA residues and 4-O-sulphation of GalNAc residues are regulated are at present largely unknown. To clarify whether C4ST-1 and C6ST-1 may be involved in the biosynthesis of a CS–DS hybrid molecule and whether or not C4ST-1 could participate in the biosynthesis of DS, it is necessary to elucidate how C4ST-1 and C6ST-1 recognize the sequence containing GlcA and IdoA around the GalNAc residue to which sulphate is transferred.
In the present study, we investigated the specificity of C4ST-1 and C6ST-1 in terms of uronic acid residue recognition and found that C4ST-1 is quite different from C6ST-1 in its requirement for the GlcA residue.
EXPERIMENTAL
Materials
The following commercial materials were used. H235SO4 was obtained from PerkinElmer; chondroitinase ACII, chondroitinase ABC, DS from pig skin, ΔDi-0S [2-acetamide-2-deoxy-3-O-(β-D-gluco-4-enepyranosyluronic acid)-D-galactose], ΔDi-6S [2-acetamide-2-deoxy-3-O-(β-D-gluco-4-enepyranosyluronic acid)-6-O-sulpho-D-galactose], ΔDi-4S, ΔDi-2S [2-acetamide-2-deoxy-3-O-(2-O-sulpho-β-D-gluco-4-enepyranosyluronic acid)-D-galactose], ΔDi-diSD [2-acetamide-2-deoxy-3-O-(2-O-sulpho-β-D-gluco-4-enepyranosyluronic acid)-6-O-sulpho-D-galactose], ΔDi-diSB [2-acetamide-2-deoxy-3-O-(2-O-sulpho-β-D-gluco-4-enepyranosyluronic acid)-4-O-sulpho-D-galactose] and ΔDi-diSE [2-acetamide-2-deoxy-3-O-(β-D-gluco-4-enepyranosyluronic acid)-4,6-bis-O-sulpho-D-galactose] were obtained from Seikagaku Corporation (Tokyo, Japan); a Partisil-10 SAX (strong anion exchange) column (4.6 mm×25 cm) was from Whatman (Clifton, NJ, U.S.A.); unlabelled PAPS, GalNAc-4-sulphate, -6-sulphate and -4,6-bissulphate, Di-6S [GlcAβ1-3GalNAc(6SO4)] and β-glucuronidase (bovine liver, type B-3) were from Sigma. Hiload Superdex 30 HR 16/60 and Fast Desalting Column HR 10/10 were from Amersham Biosciences; β-N-acetylhexosaminidase was from New England Biolabs.
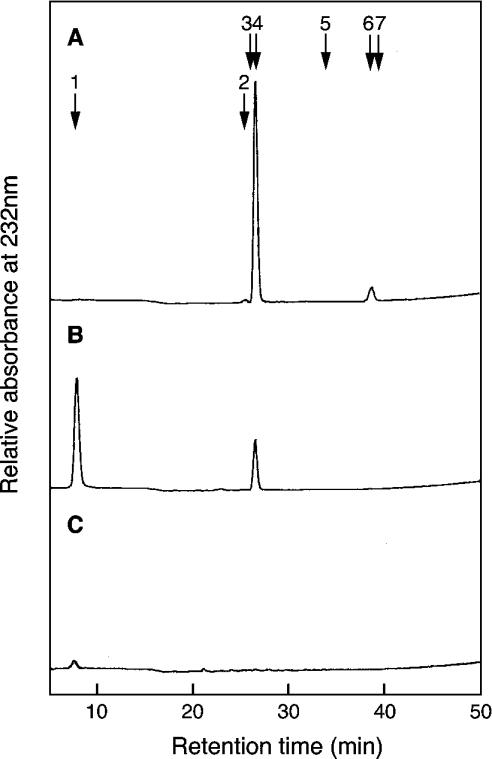
[35S]PAPS was prepared as described in [21]. The disulphated tetrasaccharides GlcA-GalNAc(4SO4)-GlcA-GalNAc(4SO4) (Tet-44), GlcA-GalNAc(6SO4)-GlcA-GalNAc(4SO4) (Tet-64), GlcA-GalNAc(4SO4)-GlcA-GalNAc(6SO4) (Tet-46) and GlcA-GalNAc(6SO4)-GlcA-GalNAc(6SO4) (Tet-66) and the disulphated trisaccharides GalNAc(4SO4)-GlcA-GalNAc(4SO4) (Tri-44) and GalNAc(6SO4)-GlcA-GalNAc(4SO4) (Tri-64), GalNAc-(4SO4)-GlcA-GalNAc(6SO4) (Tri-46) and GalNAc-(6SO4)-GlcA-GalNAc-(6SO4) (Tri-66) were prepared as described in [22]. Two monosulphated tetrasaccharides, GlcA-GalNAc-(4SO4)-GlcA-GalNAc (Tet-40) and GlcA-GalNAc(6SO4)-GlcA-GalNAc (Tet-60), were prepared from Tet-46 and Tet-66 respectively by chondro-6-sulphatase digestion [22]. Two monosulphated trisaccharides, GalNAc(4SO4)-GlcA-GalNAc (Tri-40) and GalNAc(6SO4)-GlcA-GalNAc (Tri-60), were prepared from Tri-46 and Tri-66 respectively by chondro-6-sulphatase digestion [22]. Partially desulphated DS (DSDS) was prepared from pig skin DS by the method of Nagasawa et al. [23]. Solvolysis with 90% (v/v) DMSO was performed at 100 °C for 60 min. When the DSDS was digested with chondroitinase ABC, ΔDi-0S and ΔDi-4S were formed but no ΔDi-diSB was detected (Figure 1B). The degree of desulphation was calculated as 73% from the proportion of ΔDi-0S in the total unsaturated disaccharides formed after chondroitinase ABC digestion (Table 1). The proportion of the GlcA-GalNAc-GlcA sequence in the DSDS was approx. 7% of the total unsulphated repeating disaccharide units as judged from the recovery of ΔDi-0S after digestion with chondroitinase ACII (Figure 1C).
Figure 1. Digestion of DS and DSDS with chondroitinase ABC or ACII.
DS (A) and DSDS (B, C) were digested with chondroitinase ABC (A, B) or ACII (C). The digested materials were separated with a SAX-HPLC. The column was monitored at 232 nm. The arrows indicate the elution position of ΔDi-0S (1), ΔDi-6S (2), ΔDi-2S (3), ΔDi-4S (4), ΔDi-diSD (5), ΔDi-diSB (6) and ΔDi-diSE (7).
Table 1. Disaccharide compositions of DS and DSDS.
Each glycosaminoglycan (15 nmol as galactosamine) was digested with chondroitinase ABC or ACII and subjected to SAX-HPLC. Disaccharide compositions were determined by measuring the absorbance at 232 nm of unsaturated disaccharides as shown in Figure 1. ND, not detected under the conditions used here.
| Glycosaminoglycans | ΔDi-0S | ΔDi-4S | ΔDi-6S | ΔDi-diSB | ΔDi-diSE |
|---|---|---|---|---|---|
| DS | ND | 0.92 | 0.01 | 0.08 | ND |
| DSDS | 0.73 | 0.27 | ND | ND | ND |
Preparation of recombinant human C4ST-1 and C6ST-1
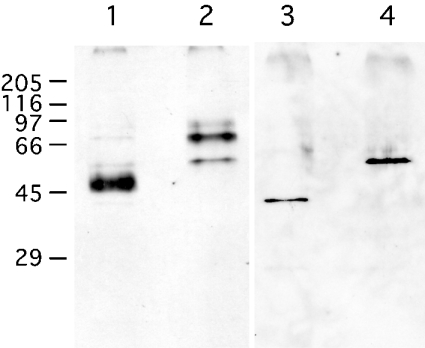
Recombinant human C4ST-1 and C6ST-1 were expressed as fusion proteins with a FLAG peptide and were affinity-purified. A DNA fragment that codes for the full open reading frame of C4ST-1 was amplified by PCR using a plasmid containing the human C4ST-1, pcDNAC4ST-1, as a template [24]. The 5′- and 3′-primers were CGCAAGCTTATGAAGCCAGCGCTGCTGGAA and ACCGTCTAGATTATTCCAATTTCAGGTAGCT respectively. At the 5′-end of the oligonucleotide primers, restriction enzyme recognition sites were introduced: the HindIII site for the sense primer and the XbaI site for the antisense primer. The PCR product was digested with XbaI and HindIII and subcloned into these sites of a pFLAG-CMV-2 plasmid (Sigma). A DNA fragment that codes for the full open reading frame of C6ST-1 was amplified by PCR using a plasmid containing the human C6ST-1, pCNXhC6ST-1, as a template [25]. The 5′- and 3′-primers were CGCAAGCTTATGGAGAAAGGACTCACTTTGC and CAGGAATTCCTACGTGACCCAGAAGGTGCC respectively. At the 5′-end of the oligonucleotide primers, restriction enzyme recognition sites were introduced: the HindIII site for the sense primer and the EcoRI site for the antisense primer. The PCR product was digested with EcoRI and HindIII and subcloned into these sites of a pFLAG-CMV-2 plasmid. The plasmids thus prepared were transfected into COS-7 cells and the fusion proteins produced were extracted as described previously [2]. Cellular extracts from ten 10 cm dishes were applied to an anti-FLAG monoclonal antibody-conjugated agarose column (0.5 ml; Sigma), which was equilibrated either with buffer A [10 mM Tris/HCl, pH 7.2, 20 mM MgCl2, 2 mM CaCl2, 10 mM 2-mercaptoethanol, 0.1% Triton X-100 and 20% (v/v) glycerol] containing 50 mM NaCl for the purification of C4ST-1 or with buffer A containing 150 mM NaCl for the purification of C6ST-1. The absorbed materials were eluted with 1.6 ml of each buffer, containing the FLAG peptide, under the conditions recommended by the manufacturer. The purified proteins were visualized with Western blots, as described below, before or after N-glycosidase F digestion (Figure 2). After N-glycosidase F digestion, a single protein band was detected at the migration position of 43 kDa for C4ST-1 (Figure 2, lane 3) and 56 kDa for C6ST-1 (Figure 2, lane 4). The molecular masses of these proteins agreed well with the molecular masses of 42791 Da for FLAG–C4ST-1 and 55908 Da for FLAG–C6ST-1, as calculated from the cDNAs.
Figure 2. Western-blot analysis of the affinity-purified C4ST-1 and C6ST-1.
The FLAG–C4ST-1 and FLAG–C6ST-1 fusion proteins were extracted from COS-7 cells that were transfected with the respective cDNA and purified with an anti-FLAG monoclonal antibody-conjugated column as described under the Experimental section. The affinity-purified C4ST-1 (lanes 1 and 3) and C6ST-1 (lanes 2 and 4) were detected with anti-FLAG antibody before (lanes 1 and 2) or after (lanes 3 and 4) the N-glycosidase F digestion. Molecular-mass standards were as follows: myosin, 205 kDa; β-galactosidase, 116 kDa; phosphorylase b, 97 kDa; BSA, 66 kDa; egg albumin, 45 kDa; carbonic anhydrase, 29 kDa.
Western-blot analysis
The affinity-purified C4ST-1 and C6ST-1 were precipitated with 10% (w/v) trichloroacetic acid. The precipitates were washed with acetone and digested with recombinant N-glycosidase F (Roche Applied Science, Indianapolis, IN, U.S.A.) using the methods recommended by the manufacturer. After digestion, the samples were separated by SDS/PAGE as described by Laemmli [26]. The separated proteins were electrophoretically transferred on to Hybond ECL® membranes (Amersham Biosciences) and stained with an anti-FLAG M2 monoclonal antibody (Sigma). The blot was developed with a polyclonal anti-mouse IgG antibody coupled with horseradish peroxidase using an ECL® detection kit and a Hyperfilm ECL® (Amersham Biosciences).
Sulphotransferase reaction
The sulphotransferase reaction with C4ST-1 or C6ST-1 was carried out by the method described previously [1,3]. The reaction mixture contained 2.5 μmol of imidazole/HCl (pH 6.8), 10 μg of protamine chloride, 0.1 μmol of dithiothreitol, 25 nmol (as galactosamine) of DSDS, 50 pmol of [35S]PAPS (approx. 5.0×105 c.p.m.) and 2 μl of the recombinant C4ST-1 or C6ST-1 in a final volume of 50 μl. The reaction mixtures were incubated at 37 °C for 20 min and the reaction was stopped by immersing the reaction tubes in a boiling-water bath for 1 min. To prepare the 35S-labelled glycosaminoglycans used for structural analysis, the reaction conditions were modified; 400 pmol of [35S]-PAPS (approx. 8.0×106 c.p.m.) and 10 μl of the recombinant C4ST-1 or C6ST-1 were added to the reaction mixtures and they were incubated for 7.5 h. After the reaction was stopped, the 35S-labelled glycosaminoglycans were precipitated with ethanol, as described previously [3]. The precipitates were dissolved in 50 mM Tris/HCl (pH 8.0) containing 0.5 mg of Actinase (Kaken Seiyaku, Tokyo, Japan) and incubated at 37 °C for 20 h. After digestion with Actinase, the 35S-labelled glycosaminoglycans were isolated by precipitation with ethanol followed by gel chromatography with a Fast Desalting Column, as described previously [3].
Digestion with chondroitinase ACII, chondroitinase ABC, β-N-acetylhexosaminidase and β-glucuronidase
Digestion with chondroitinase ACII was performed for 12 h at 37 °C in a reaction mixture containing, in a final volume of 200 μl, 35S-labelled glycosaminoglycans, 10 μmol of Tris/acetate buffer (pH 7.5), 20 μg of BSA and 100 m-units of chondroitinase ACII. The digestion was continued for a further 12 h with 100 m-units of fresh chondroitinase ACII. Digestion with chondroitinase ABC was performed for 4 h at 37 °C in a reaction mixture containing, in a final volume of 25 μl, 35S-labelled glycosaminoglycans or 35S-labelled oligosaccharides, 1.25 μmol of Tris/acetate buffer (pH 7.5), 2.5 μg of BSA and 50 m-units of chondroitinase ABC. Digestion with β-N-acetylhexosaminidase was performed for 24 h at 37 °C in a reaction mixture containing, in a final volume of 50 μl, 35S-labelled oligosaccharides, 2.5 μmol of sodium acetate buffer (pH 4.5) and 50 units of β-N-acetylhexosaminidase. Digestion with β-glucuronidase was performed for 24 h at 37 °C in a reaction mixture containing, in a final volume of 50 μl, Di-6S (100 nmol as galactosamine), 35S-labelled disaccharides, 2.5 μmol of sodium acetate buffer (pH 4.5), 1 μmol of sodium fluoride and 4000 units of β-glucuronidase.
Removal of unsaturated uronic acid by mercuric acetate
Removal of unsaturated uronic acid was performed as described in [27]. Oligosaccharides containing unsaturated uronic acid were dried and dissolved in 0.5 ml of 35 mM mercuric acetate in 25 mM Tris/25 mM sodium acetate (pH 5.0). The reaction was carried out for 1 h at room temperature (25 °C). After the reaction was completed, the samples were applied to a Dowex 50 (H+) column (bed volume, 1 ml). The column was washed with 3 ml of water. The flow-through fractions and washings were combined and freeze-dried.
Superdex 30 chromatography and HPLC
A Superdex 30 16/60 column was equilibrated with 0.2 M NH4HCO3 and run at a flow rate of 1 ml/min. Fractions (1 ml) were then collected. Separation of the degradation products formed from 35S-labelled glycosaminoglycans and 35S-labelled oligosaccharides was performed by HPLC using a Whatman Partisil-10 SAX column (4.6 mm×25 cm). Unless otherwise stated, the column was developed with 5 mM KH2PO4 for 10 min followed by a linear gradient of 5–500 mM KH2PO4.
RESULTS
Sulphation of DSDS with the recombinant C4ST-1 and C6ST-1
We found previously that the rate of sulphation of DSDS by purified C4ST-1 increased with increase in concentration of protamine up to 0.2 mg/ml [1]; therefore, the rates of sulphation of various glycosaminoglycans by the recombinant C4ST-1 or C6ST-1 were compared at 0.2 mg/ml protamine (Table 2). Both the recombinant C4ST-1 and C6ST-1 transferred sulphate to DSDS at nearly the same rate as the rate of sulphation of chondroitin under the assay conditions. The presence of IdoA residues thus appears to have little effect on the rate of sulphation. CS-A and CS-C were poor acceptors for both the recombinant enzymes. DS was totally inactive as the acceptor for both C4ST-1 and C6ST-1, suggesting that DS contained no unsulphated disaccharide units available to both C4ST-1 and C6ST-1. The disaccharide analysis of DS shown in Figure 1 supports this idea.
Table 2. Sulphation of various glycosaminoglycans with affinity-purified human C4ST-1 and C6ST-1.
Sulphotransferase activities (pmol·min−1·ml−1×10−2) were assayed using various glycosaminoglycans as described in the Experimental section.
| Acceptors | C4ST-1 activity | C6ST-1 activity |
|---|---|---|
| Chondroitin | 2.50 | 6.28 |
| CS-A | 0.04 | 0.39 |
| CS-C | 0.02 | 0.17 |
| DS | 0.00 | 0.00 |
| DSDS | 2.83 | 5.97 |
Chondroitinase ACII digestion of the 35S-labelled glycosaminoglycans formed from DSDS by reaction with the recombinant C4ST-1 or C6ST-1
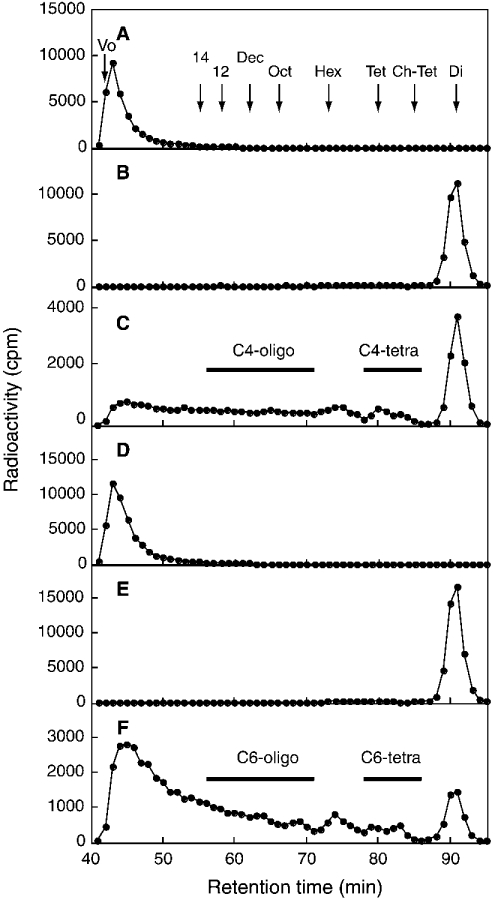
To examine the uroninc acid recognition of C4ST-1 and C6ST-1, we utilized the difference in the specificity between chondroitinase ACII and chondroitinase ABC. GalNAc–GlcA bonds are susceptible to both chondroitinase ACII and chondroitinase ABC, whereas GalNAc–IdoA bonds can be cleaved only by chondroitinase ABC. When [35S]glycosaminoglycans obtained from DSDS after incubation with [35S]PAPS and C4ST-1 were digested with chondroitinase ACII and subjected to Superdex 30 chromatography, 32% of the total radioactivity was recovered in the disaccharide fraction (Figure 3C), whereas the [35S]glycosaminoglycans were quantitatively converted into disaccharide after chondroitinase ABC digestion (Figure 3B). The disaccharide fraction in Figure 3(B) was identified as ΔDi-4S by SAX-HPLC (results not shown). ΔDi-4S should be released from the GlcA-GalNAc(4SO4)-GlcA sequence in the [35S]glycosaminoglycans. Since the proportion of the acceptor sequence GlcA-GalNAc-GlcA in DSDS was only 7% of the total unsulphated repeating disaccharide units (Figure 1), these results indicate that C4ST-1 preferentially transferred sulphate to GalNAc residues located at the GlcA-GalNAc-GlcA sequence in DSDS. Such a specificity of C4ST-1 was observed previously in the C4ST-1 purified from rat chondrosarcoma [1]. On the other hand, when the [35S]glycosaminoglycans obtained from DSDS by incubation with [35S]-PAPS and C6ST-1 were digested with chondroitinase ACII, a major part of the radioactivity was recovered in the fractions larger than hexasaccharides (Figure 3F), whereas the [35S]glycosaminoglycans were also quantitatively converted into disaccharide by chondroitinase ABC digestion (Figure 3E). The disaccharide fraction in Figure 3(E) was identified as ΔDi-6S by SAX-HPLC (results not shown). These results indicate that, unlike C4ST-1, C6ST-1 could transfer sulphate efficiently to the GalNAc residues embedded in the IdoA-rich region.
Figure 3. Superdex 30 chromatography of chondroitinase ACII or ABC digests of 35S-labelled glycosaminoglycans derived from DSDS by incubation with [35S]PAPS and the affinity-purified C4ST-1 or C6ST-1.
The sulphotransferase reaction was carried out as described under the Experimental section. The 35S-labelled glycosaminoglycans formed from DSDS after the reaction with C4ST-1 (A–C) or C6ST-1 (D–F) were applied to the Superdex 30 column before (A, D) or after digestion with chondroitinase ABC (B, E) or ACII (C, F). The arrows indicate the elution position of Blue Dextran (Vo), ΔDi-4S (Di), CS tetrasaccharide (Tet), chondroitin tetrasaccharide (Ch-Tet), CS hexasaccharide (Hex), CS octasaccharide (Oct), CS decasaccharide (Dec), CS dodecasaccharide (12) and CS tetradecasaccharide (14).
Structural analysis of the tetrasaccharide fractions released after chondroitinase ACII digestion
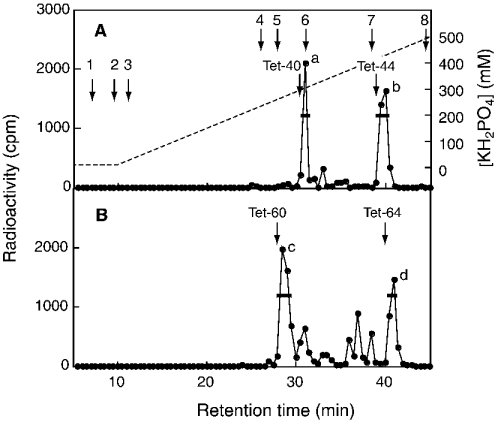
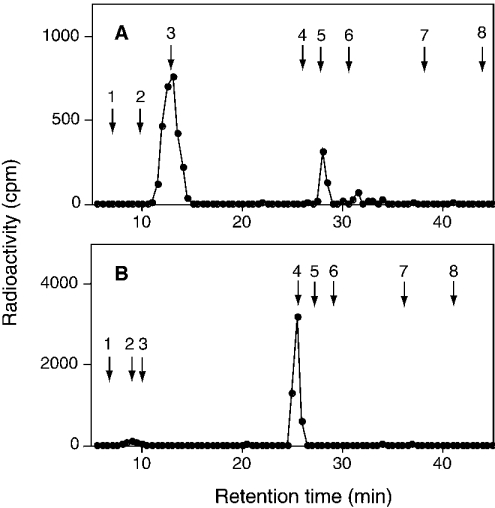
To obtain information about the preference for HexA (hexuronic acid) residues adjacent to the targeted GalNAc residue, we analysed the tetrasaccharide fractions that were supposed to be generated by chondroitinase ACII digestion from a sequence, GlcA1-GalNAc(±SO4)2-IdoA3-GalNAc(±SO4)4-GlcA5-. The tetrasaccharide fraction (C4-tetra) released from the [35S]glycosaminoglycans obtained by incubating with C4ST-1, which is indicated by a horizontal bar in Figure 3(C), was further separated with SAX-HPLC (Figure 4A). Two peaks were obtained from C4-tetra; as judged from the elution positions of Tet-40 and Tet-44, peaks a and b could be assigned to a monosulphated tetrasaccharide and a disulphated tetrasaccharide respectively. To determine the position to which the sulphate was transferred, these tetrasaccharides were subjected to treatment with mercuric acetate, and the resulting trisaccharides were separated with SAX-HPLC; peak e was obtained from peak a and peak f was obtained from peak b (Figure 5). Peak e was eluted near Tri-40 and peak f was eluted near Tri-44 (Figures 5A and 5C), confirming that peaks e and f were a monosulphated trisaccharide and a disulphated trisaccharide respectively. If the radioactive sulphate was transferred to the GalNAc residue located at the non-reducing side of the trisaccharides, the radioactivity should have been released as GalNAc(4SO4) after chondroitinase ABC digestion (see Scheme 1A). Alternatively, if the radioactive sulphate was transferred to the GalNAc residue located at the reducing side of the trisaccharides, the radioactivity should be released as ΔDi-4S. Peaks e and f were digested with chondroitinase ABC and the digestion products were separated with SAX-HPLC. Only GalNAc(4SO4) was released from peak e after chondroitinase ABC digestion (Figure 5B), indicating that sulphate was transferred exclusively to the No. 2 GalNAc residue in the sequence, -GlcA1-GalNAc2-IdoA3-GalNAc4-GlcA5-. On the other hand, GalNAc(4SO4) and a small amount of ΔDi-4S were released from peak f (Figure 5D), indicating that the No. 4 GalNAc residue in the sequence, -GlcA1-GalNAc(4SO4)2-IdoA3-GalNAc4-GlcA5-, could also be sulphated by C4ST-1 at a much slower rate than the No. 2 GalNAc residue in the sequence,-GlcA1-GalNAc2-IdoA3-GalNAc(4SO4)4-GlcA5-. Taken together, C4ST-1 appears to transfer sulphate mainly to the GalNAc residues located at the reducing side of GlcA, and such specificity could be slightly affected by the presence of a sulphate group on the GalNAc residue.
Figure 4. Separation of tetrasaccharide fractions by SAX-HPLC.
The C4-tetra (A) and C6-tetra (B) fractions shown in Figures 3(C) and 3(F) respectively were pooled separately, freeze-dried and applied to SAX-HPLC as described under the Experimental section. The broken line depicts the concentration of KH2PO4. The arrows indicate the elution position of ΔDi-0S (1), GalNAc(6SO4) (2), GalNAc(4SO4) (3), ΔDi-6S (4), ΔDi-4S (5), GalNAc(4,6-SO4) (6), ΔDi-diSD (7) and ΔDi-diSE (8). The elution positions of Tet-40, Tet-44, Tet-60 and Tet-64 are indicated.
Figure 5. Chondroitinase ABC digestion of the trisaccharides derived from C4-tetra after mercuric acetate treatment.
The trisaccharides derived from peak a (A, B) or peak b (C, D) shown in Figure 4(A) were separated with SAX-HPLC before (A, C) or after (B, D) digestion with chondroitinase ABC. The standards were the same as those described in the legend to Figure 4. The elution positions of Tri-40 and Tri-44 are indicated.
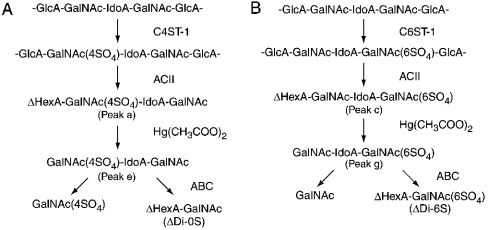
Scheme 1. Structural analysis of monosulphated tetrasaccharides formed after chondroitinase ACII digestion.
35S-labelled glycosaminoglycans formed from DSDS after the reaction with C4ST-1 (A) or C6ST-1 (B) were digested with chondroitinase ACII. The monosulphated tetrasaccharide fractions (peak a in Figure 4A and peak c in Figure 4B) were purified and treated with mercuric acetate. The resulting trisaccharides (peak e in Figure 5A and peak g in Figure 6A) were digested with chondroitinase ABC. GalNAc(4SO4) was obtained from the non-reducing terminal of peak e, and ΔDi-6S was obtained from the reducing terminal of peak g.
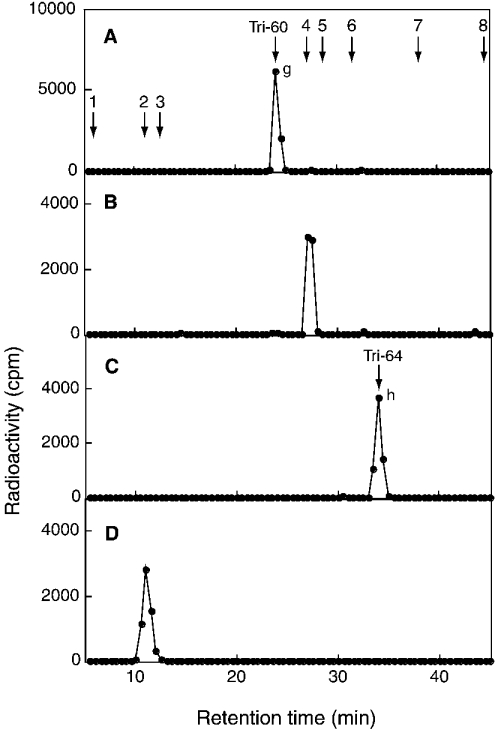
To compare the specificity of C4ST-1 with that of C6ST-1, we analysed the tetrasaccharide fraction (C6-tetra) released from the [35S]glycosaminoglycans obtained by incubation with C6ST-1, which is indicated by a horizontal bar in Figure 3(F). C6-tetra was also proposed to be derived from the sequence -GlcA1-GalNAc-(±SO4)2-IdoA3-GalNAc(±SO4)4-GlcA5-. Two major peaks (peaks c and d) and several minor peaks were obtained from C6-tetra after SAX-HPLC (Figure 4B). As judged from the elution position of Tet-60 and Tet-64, peaks c and d could be assigned to a monosulphated tetrasaccharide and a disulphated tetrasaccharide respectively. Other small peaks seem to represent tetrasaccharides with different sulphation patterns; however, these peaks were not examined further because of their limited amounts. Materials contained in peaks c and d were recovered separately and subjected to treatment with mercuric acetate; peak g was obtained from peak c and peak h was obtained from peak d (Figure 6). Peak g was eluted near Tri-60 and peak h was eluted near Tri-64 (Figures 6A and 6C), confirming that peaks g and h were a monosulphated trisaccharide and a disulphated trisaccharide respectively. When peak g derived from peak c was digested with chondroitinase ABC, ΔDi-6S was exclusively detected (Figure 6B), indicating that sulphate was transferred to the No. 4 GalNAc residue in the sequence, -GlcA1-GalNAc2-IdoA3-GalNAc4-GlcA5- (see Scheme 1B). In contrast, when peak h derived from peak d was digested with chondroitinase ABC, only GalNAc-(6SO4) was obtained (Figure 6D), indicating that C6ST-1 could transfer sulphate to the No. 2 GalNAc residue in the sequence, -GlcA1-GalNAc2-IdoA3-GalNAc(4SO4)4-GlcA5-. These results suggest that the specificity of C6ST-1 appears to depend markedly on the presence or absence of a sulphate group on the GalNAc residue, but not on the HexA type; C6ST-1 could transfer sulphate almost at the same rate to both the GalNAc residue located at the reducing side of IdoA and the GalNAc residue located at the reducing side of GlcA. These results were obtained from an analysis of the two major peaks in Figure 4(B). Structural analysis of the other minor peaks may depict a more complicated picture regarding the specificity of C6ST-1.
Figure 6. Chondroitinase ABC digestion of the trisaccharides derived from C6-tetra after mercuric acetate treatment.
The trisaccharides derived from peak c (A, B) or peak d (C, D) shown in Figure 4(B) were separated with SAX-HPLC before (A, C) or after (B, D) digestion with chondroitinase ABC. The standards were the same as those described in the legend to Figure 4. The elution positions of Tri-60 and Tri-64 are indicated.
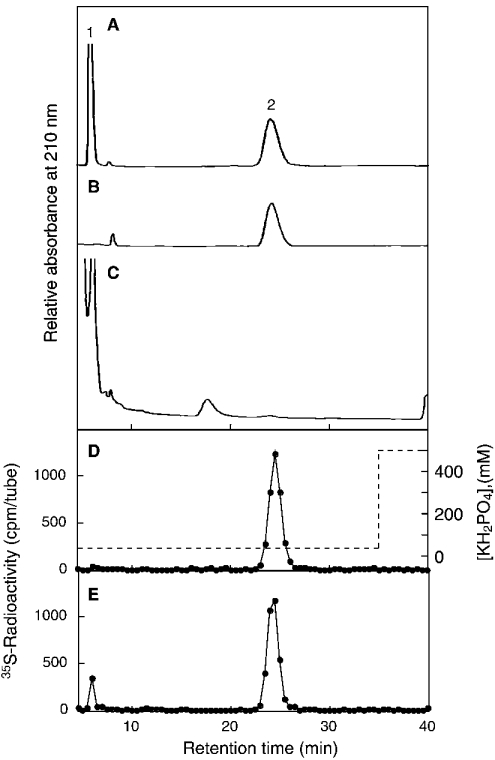
The presence of an IdoA residue in the chondroitinase ACII-resistant oligosaccharides was assumed on the basis of the reported specificity of chondroitinase ACII [28]. We tried to confirm the type of uronic acid residue contained in peak g. When peak g was digested with β-N-acetylhexosaminidase, an 35S-labelled disaccharide (Di-X) was obtained. Di-X was eluted from the Superdex 30 at the position ΔDi-6S. If Di-X contained GlcA, GalNAc(6SO4) would be released after β-glucuronidase digestion, but if Di-X contained IdoA, GalNAc(6SO4) would not be released. 35S-labelled Di-X was mixed with standard Di-6S and subjected to β-glucuronidase digestion. The reaction products were separated by SAX-HPLC (Figure 7). After digestion with β-glucuronidase, Di-6S was completely degraded to yield GalNAc-(6SO4) (Figure 7C). On the other hand, 35S-labelled Di-X remained almost intact (Figure 7E). A small amount of radioactivity was detected at the position of GalNAc(6SO4) after digestion with β-glucuronidase (Figure 7E). It was reported previously that a trace amount of α-L-iduronidase activity was detected in commercially available β-glucuronidase from bovine livers [29]. The low radioactivity of GalNAc(6SO4) may be released by the contaminating α-L-iduronidase. As described above, peak g was degraded by chondroitinase ABC to ΔDi-6S. Taken together, it is concluded that peak g contained IdoA residue. 35S-labelled Di-X was eluted approx. 0.5 min later than Di-6S (Figures 7B and 7D), supporting the idea that Di-X contained an IdoA residue because IdoA-anhydromannitol(6SO4) was eluted later than GlcA-anhydromannitol(6SO4) under the same elution conditions [30].
Figure 7. β-Glucuronidase digestion of a disaccharide (Di-X) derived from peak g after β-N-acetylhexosaminidase digestion.
The monosulphated trisaccharide (peak g in Figure 6A) was digested with β-N-acetylhexosaminidase and separated by Superdex 30 chromatography. The resulting disaccharide (Di-X) was mixed with Di-6S and separated with SAX-HPLC before (B, D) or after (C, E) digestion with β-glucuronidase. The column was eluted with 40 mM KH2PO4 isocratically and monitored by the absorption at 210 nm (A–C) and the 35S-radioactivity (D, E). The broken line depicts the concentration of KH2PO4. The elution profile of standard GalNAc(6SO4) (1) and Di-6S (2) is indicated in (A). A large background before 5 min and a small peak at approx. 17 min in (C) are derived from the β-glucuronidase preparation.
Structural analysis of the higher oligosaccharide fractions
When the 35S-labelled glycosaminoglycans, derived from DSDS after incubation with C4ST-1, were digested with chondroitinase ACII, 68% of the total radioactivity was recovered in the fractions larger than tetrasaccharide (Figure 3C). Since the larger fractions were not obtained after chondroitinase ABC digestion (Figure 3B), these fractions should have been composed of oligosaccharides containing different numbers of IdoA-GalNAc repeating units. The observation that a radioactive sulphate was found in the larger fractions might lead to the tentative proposal that C4ST-1 could also transfer sulphate efficiently to the GalNAc residues existing between IdoA residues; however, this was not the case, as described below. The larger oligosaccharide fractions are supposed to be formed by the eliminative cleavage of the GalNAc–GlcA bond from IdoA-rich regions and, hence, should contain a ΔHexA residue at their non-reducing ends. If C4ST-1 prefers the GalNAc residue located at the reducing side of GlcA as shown in C4-tetra, the radioactive sulphate groups should reside mainly on the penultimate GalNAc residue adjacent to ΔHexA. To address these issues, we investigated the distribution of radioactive sulphate groups incorporated into the higher molecular fractions (C4-oligo) indicated by a horizontal bar in Figure 3(C). As judged from the elution positions of an even number of CS oligosaccharides, C4-oligo seems to contain an octasaccharide to tetradecasaccharide. C4-oligo was subjected to treatment with mercuric acetate and then digested with chondroitinase ABC. The chondroitinase ABC digestion products were separated with SAX-HPLC (Figure 8A). A major part of the radioactivity was recovered in GalNAc(4SO4), and a small amount of radioactivity was detected at ΔDi-4S. These results also indicate that C4ST-1 transferred sulphate mainly to GalNAc residues adjacent to the reducing side of GlcA residues.
Figure 8. Chondroitinase ABC digestion of the oligosaccharides derived from C4- or C6-oligo after mercuric acetate treatment.
Oligosaccharides derived from C4-oligo (A) and C6-oligo (B) after mercuric acetate treatment were separated with SAX-HPLC after digestion with chondroitinase ABC. The standards were the same as those described in the legend to Figure 4.
When the 35S-labelled glycosaminoglycans derived from DSDS after incubation with C6ST-1 were digested with chondroitinase ACII, a major part of the radioactivity was recovered in the fractions larger than tetrasaccharides (Figure 3F). As shown in the analysis of C6-tetra, the C6ST-1 activity seems to be independent of the presence or absence of the GlcA residue; therefore, most of the radioactivity in the larger oligosaccharide fractions is expected to reside in the internal regions. To determine the location of the radioactive sulphate in the oligosaccharide, we analysed the oligosaccharide fractions (C6-oligo) indicated by a horizontal bar in Figure 3(F) by the methods described above. C6-oligo was subjected to treatment with mercuric acetate and then digested with chondroitinase ABC. The chondroitinase ABC digestion products were separated with SAX-HPLC (Figure 8B). The highest radioactivity was recovered in ΔDi-6S and a trace amount was detected at GalNAc(6SO4). These results indicate that C6ST-1 transferred sulphate mainly to the internal GalNAc residue of these oligosaccharides and that C6ST-1 does not require the presence of a GlcA residue adjacent to the non-reducing side of the targeting GalNAc residue.
DISCUSSION
In the present study, we showed that C4ST-1 and C6ST-1 were different from each other not only in the position to which sulphate was transferred, but also in the recognition of the uronic acid residue adjacent to the targeted GalNAc residue. C4ST-1 was found to prefer GalNAc residues located between GlcA residues, and was demonstrated to transfer sulphate mainly to GalNAc residues adjacent to the reducing side of the GlcA residue. In contrast, C6ST-1 did not show any requirement for the presence of GlcA residues.
NMR analysis of the 35S-labelled tetrasaccharides obtained from the 35S-DSDS after chondroitinase ACI showed that D4ST transfers sulphate to the No. 4 GalNAc residue in the sequence of GlcA1-GalNAc2-IdoA3-GalNAc4-GlcA [7]. Such a specificity is quite different from the specificity of C4ST-1 described in the present study and appears to coincide with the observation that a microsomal fraction obtained from fibroblasts was reported to sulphate DSDS more efficiently than chondroitin [20]; nevertheless, the strict specificity of D4ST should be confirmed by determining the position of radioactive sulphate transferred to the acceptor DSDS, since the amount of radioactive sulphate transferred to DSDS through the reaction catalysed by D4ST might be much smaller than the amount of sulphate groups present in the acceptor molecules. C5 epimerization of GlcA residue appears to be closely linked to C4 sulphation of GalNAc residue in the biosynthesis of DS [17–19]. If C4 sulphation of the GalNAc residue occurs after the C5 epimerization of the GlcA residue, C4ST-1 could be hardly involved in the biosynthesis of DS, since C4ST-1 showed very weak activity towards the No. 4 GalNAc residue in the sequence GlcA1-GalNAc2-IdoA3-GalNAc4-GlcA. Instead, C4ST-1 may participate in the biosynthesis of a hybrid sequence, GlcA-GalNAc(4SO4)-IdoA-GalNAc(4SO4) [15]. Genetic approaches will be necessary to clarify the sulphotransferases that are involved in the biosynthesis of DS or CS–DS hybrid structures.
C6ST-1 was found to sulphate the GalNAc residues present in the regions containing contiguous IdoA-GalNAc units; however, glycosaminoglycans having IdoA-GalNAc(6SO4) units are very rare in vertebrate tissues. On the other hand, an ascidian species has been reported to have a DS containing IdoA-GalNAc(6SO4) units [31]. It remains to be determined why the DS containing IdoA-GalNAc(6SO4) units is not synthesized in the vertebrate tissues. CS–DS hybrid molecules from the human meniscus were reported to have GlcA-GalNAc(6SO4) units on the non-reducing side of IdoA-GalNAc(4SO4) units [10]. C6ST-1 may be involved in the synthesis of these hybrid structures since it was demonstrated to transfer sulphate to the No. 2 GalNAc residue in the sequence GlcA1-GalNAc2-IdoA3-GalNAc(4SO4)4-GlcA as shown in Figure 6(D).
We previously found that the ability of chondroitinase ACII to cleave the GalNAc–GlcA bond was not affected by the sulphation pattern on GalNAc residues since GlcA-containing trisaccahrides having sulphate groups on position 4 or 6 of GalNAc residues were all easily degraded by chondroitinase ACII [22]. We also found that a trisaccharide having a sulphate group on position 2 of the GlcA residue was refractory to chondroitinase ACII digestion [22]; therefore, the tetrasaccharides obtained from the 35S-labelled DSDS might have contained GlcA(2SO4) instead of IdoA. However, this was not the case, because only [35S]ΔDi-4S was detected after chondroitinase ABC digestion of the 35S-labelled glycosaminoglycan formed from DSDS (Figure 3B), and the acceptor DSDS did not yield any detectable ΔDi-diSB or ΔDi-2S after chondroitinase ABC digestion (Figure 1B).
Glycosaminoglycans containing IdoA-GalNAc(4,6-SO4) units have been reported in the hag fish notochord [32,33], glomeruli [34], rat mesangial cells [35] and an ascidian species [36]. These glycosaminoglycans may be synthesized by sulphotransferases different from C6ST-1, since C6ST-1 failed to transfer sulphate to position 6 of the GalNAc(4SO4) residues. Instead, GalNAc(4SO4) 6-O-sulphotransferase may be involved in the synthesis of these glycosaminoglycans, since both the squid and human GalNAc-(4SO4) 6-O-sulphotransferases were found to transfer sulphate to DS efficiently [37,38].
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on No. 5801 and on Priority Areas No. 10178102 from the Ministry of Education, Science, Sports and Culture of Japan and by a special research grant from Seikagaku Corporation.
References
- 1.Yamauchi S., Hirahara Y., Usui H., Takeda Y., Hoshino M., Fukuta M., Kimura J. H., Habuchi O. Purification and characterization of chondroitin 4-sulfotransferase from the culture medium of a rat chondrosarcoma cell line. J. Biol. Chem. 1999;274:2456–2463. doi: 10.1074/jbc.274.4.2456. [DOI] [PubMed] [Google Scholar]
- 2.Yamauchi S., Mita S., Matsubara T., Fukuta M., Habuchi H., Kimata K., Habuchi O. Molecular cloning and expression of chondroitin 4-sulfotransferase. J. Biol. Chem. 2000;275:8975–8981. doi: 10.1074/jbc.275.12.8975. [DOI] [PubMed] [Google Scholar]
- 3.Habuchi O., Matsui Y., Kotoya Y., Aoyama Y., Yasuda Y., Noda M. Purification of chondroitin 6-sulfotransferase secreted from cultured chick embryo chondrocytes. J. Biol. Chem. 1993;268:21968–21974. [PubMed] [Google Scholar]
- 4.Fukuta M., Uchimura K., Nakashima K., Kato M., Kimata K., Shinomura T., Habuchi O. Molecular cloning and expression of chick chondrocyte chondroitin 6-sulfotransferase. J. Biol. Chem. 1995;270:18575–18580. doi: 10.1074/jbc.270.31.18575. [DOI] [PubMed] [Google Scholar]
- 5.Hiraoka N., Nakagawa H., Ong E., Akama T. O., Fukuda M. N., Fukuda M. Molecular cloning and expression of two distinct human chondroitin 4-O-sulfotransferases that belong to the HNK-1 sulfotransferase gene family. J. Biol. Chem. 2000;275:20188–20196. doi: 10.1074/jbc.M002443200. [DOI] [PubMed] [Google Scholar]
- 6.Evers M. R., Xia G., Kang H. G., Schachner M., Baenziger J. U. Molecular cloning and characterization of a dermatan-specific N-acetylgalactosamine 4-O-sulfotransferase. J. Biol. Chem. 2001;276:36344–36353. doi: 10.1074/jbc.M105848200. [DOI] [PubMed] [Google Scholar]
- 7.Mikami T., Mizumoto S., Kago N., Kitagawa H., Sugahara K. Specificities of three distinct human chondroitin/dermatan N-acetylgalactosamine 4-O-sulfotransferases demonstrated using partially desulfated dermatan sulfate as an acceptor: implication of differential roles in dermatan sulphate biosynthesis. J. Biol. Chem. 2003;278:36115–36127. doi: 10.1074/jbc.M306044200. [DOI] [PubMed] [Google Scholar]
- 8.Fransson L.-A., Roden L. Structure of dermatan sulfate. II. Characterization of products obtained by hyaluronidase digestion of dermatan sulfate. J. Biol. Chem. 1967;242:4170–4175. [PubMed] [Google Scholar]
- 9.Fransson L.-A. Structure of dermatan sulfate. III. The hybrid structure of dermatan sulfate from umbilical cord. J. Biol. Chem. 1968;243:1504–1510. [PubMed] [Google Scholar]
- 10.Habuchi H., Yamagata T., Iwata H., Suzuki S. The occurrence of a wide variety of dermatan sulfate-chondroitin sulfate copolymers in fibrous cartilage. J. Biol. Chem. 1973;248:6019–6028. [PubMed] [Google Scholar]
- 11.Figura K., Kiowski W., Buddecke E. Studies on the chemistry of arterial wall, XVII. Metabolic characteristics of different types of chondroitin sulfate-dermatan sulfate hybrids in arterial tissue. Hoppe Seylers Z. Physiol. Chem. 1975;356:1517–1525. doi: 10.1515/bchm2.1975.356.2.1517. [DOI] [PubMed] [Google Scholar]
- 12.Blake D. A., Conrad H. E. Hybrid glycosaminoglycans synthesized by monolayers of chick embryo arterial fibroblasts. Biochemistry. 1979;18:5475–5482. doi: 10.1021/bi00591a033. [DOI] [PubMed] [Google Scholar]
- 13.Cheng F., Heinegard D., Malmstrom A., Schmidtchen A., Yoshida K., Fransson L. A. Patterns of uronosyl epimerization and 4-/6-O-sulphation in chondroitin/dermatan sulphate from decorin and biglycan of various bovine tissues. Glycobiology. 1994;4:685–696. doi: 10.1093/glycob/4.5.685. [DOI] [PubMed] [Google Scholar]
- 14.Karamanos N. K., Vanky P., Syrokou A., Hjerpe A. Identity of dermatan and chondroitin sequences in dermatan sulfate chains determined by using fragmentation with chondroitinases and ion-pair high-performance liquid chromatography. Anal. Biochem. 1995;225:220–230. doi: 10.1006/abio.1995.1147. [DOI] [PubMed] [Google Scholar]
- 15.Huckerby T. N., Lauder R. M., Brown G. M., Nieduszynski I. A., Anderson K., Boocock J., Sandall P. L., Weeks S. D. Characterization of oligosaccharides from the chondroitin sulfates. (1)H-NMR and (13)C-NMR studies of reduced disaccharides and tetrasaccharides. Eur. J. Biochem. 2001;268:1181–1189. doi: 10.1046/j.1432-1327.2001.01948.x. [DOI] [PubMed] [Google Scholar]
- 16.Zamfir A., Seidler D. G., Kresse H., Peter-Katalinic J. Structural investigation of chondroitin/dermatan sulfate oligosaccharides from human skin fibroblast decorin. Glycobiology. 2003;13:733–742. doi: 10.1093/glycob/cwg086. [DOI] [PubMed] [Google Scholar]
- 17.Malmstrom A., Fransson L.-A. Biosynthesis of dermatan sulfate. I. Formation of L-iduronic acid residues. J. Biol. Chem. 1975;250:3419–3425. [PubMed] [Google Scholar]
- 18.Silbert J. E., Palmer M. E., Humphries D. E., Silbert C. K. Formation of dermatan sulfate by cultured human skin fibroblasts. Effects of sulfate concentration on proportions of dermatan/chondroitin. J. Biol. Chem. 1986;261:13397–13400. [PubMed] [Google Scholar]
- 19.Silbert C. K., Humphries D. E., Palmer M. E., Silbert J. E. Effects of sulfate deprivation on the production of chondroitin/dermatan sulfate by cultures of skin fibroblasts from normal and diabetic individuals. Arch. Biochem. Biophys. 1991;285:137–141. doi: 10.1016/0003-9861(91)90340-o. [DOI] [PubMed] [Google Scholar]
- 20.Eklund E., Roden L., Malmstrom M., Malmstrom A. Dermatan is a better substrate for 4-O-sulfation than chondroitin: implications in the generation of 4-O-sulfated, L-iduronate-rich galactosaminoglycans. Arch. Biochem. Biophys. 2000;383:171–177. doi: 10.1006/abbi.2000.2043. [DOI] [PubMed] [Google Scholar]
- 21.Delfert D. M., Conrad H. E. Preparation and high-performance liquid chromatography of 3′-phosphoadenosine-5′-phospho[35S]sulfate with a predetermined specific activity. Anal. Biochem. 1985;148:303–310. doi: 10.1016/0003-2697(85)90233-7. [DOI] [PubMed] [Google Scholar]
- 22.Ohtake S., Kimata K., Habuchi O. A unique nonreducing terminal modification of chondroitin sulfate by N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase. J. Biol. Chem. 2003;278:38443–38452. doi: 10.1074/jbc.M306132200. [DOI] [PubMed] [Google Scholar]
- 23.Nagasawa K., Inoue Y., Tokuyasu T. An improved method for the preparation of chondroitin by solvolytic desulfation of chondroitin sulfates. J. Biochem. (Tokyo) 1979;86:1323–1329. doi: 10.1093/oxfordjournals.jbchem.a132648. [DOI] [PubMed] [Google Scholar]
- 24.Okuda T., Mita S., Yamauchi S., Matsubara T., Yagi F., Yamamori D., Fukuta M., Kuroiwa A., Matsuda Y., Habuchi O. Molecular cloning, expression and chromosomal mapping of human chondroitin 4-sulfotransferase, whose expression pattern is different from that of chondroitin 6-sulfotransferase. J. Biochem. (Tokyo) 2000;128:763–770. doi: 10.1093/oxfordjournals.jbchem.a022813. [DOI] [PubMed] [Google Scholar]
- 25.Fukuta M., Kobayashi Y., Uchimura K., Kimata K., Habuchi O. Molecular cloning and expression of human chondroitin 6-sulfotransferase. Biochim. Biophys. Acta. 1998;1399:57–61. doi: 10.1016/s0167-4781(98)00089-x. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Ludwigs U., Elgavish A., Esko J. D., Meezan E., Roden L. Reaction of unsaturated uronic acid residues with mercuric salts. Cleavage of the hyaluronic acid disaccharide 2-acetamido-2-deoxy-3-O-(beta-D-gluco-4-enepyranosyluronic acid)-D-glucose. Biochem. J. 1987;245:795–804. doi: 10.1042/bj2450795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bienkowski M. J., Conrad H. E. Structural characterization of the oligosaccharides formed by depolymerization of heparin with nitrous acid. J. Biol. Chem. 1985;260:356–365. [PubMed] [Google Scholar]
- 29.Kosaka H., Isemura M., Ono T., Nishimura Y., Kato K. Studies on the alpha-L-iduronidase activity of beta-glucuronidase preparations from bovine liver, rat liver, and rat preputial gland. J. Biochem. (Tokyo) 1980;88:69–75. [PubMed] [Google Scholar]
- 30.Hiyama K., Okada S. Crystallization and some properties of chondroitinase from Arthrobacter aurescens. J. Biol. Chem. 1975;250:1824–1828. [PubMed] [Google Scholar]
- 31.Mourao P. A., Pavao M. S. G., Mulloy B., Wait R. Chondroitin ABC lyase digestion of an ascidian dermatan sulfate. Occurrence of unusual 6-O-sulfo-2-acetamido-2-deoxy-3-O-(2-O-sulfo-alpha-L-idopyranosyluronic acid)-beta-D-galactose units. Carbohydr. Res. 1997;300:315–321. doi: 10.1016/s0008-6215(97)00061-x. [DOI] [PubMed] [Google Scholar]
- 32.Anno K., Seno N., Mathews M. B., Yamagata T., Suzuki S. A new dermatan polysulfate, chondroitin sulfate H, from hagfish notochord. Biochim. Biophys. Acta. 1971;237:173–177. doi: 10.1016/0304-4165(71)90046-8. [DOI] [PubMed] [Google Scholar]
- 33.Ueoka C., Nadanaka S., Seno N., Khoo K.-H., Sugahara K. Structural determination of novel tetra- and hexasaccharide sequences isolated from chondroitin sulfate H (oversulfated dermatan sulfate) of hagfish notochord. Glycoconj. J. 1999;16:291–305. doi: 10.1023/a:1007022229813. [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi S., Oguri K., Yaoita E., Kobayashi K., Okayama M. Highly sulfated proteochondroitin sulfates synthesized in vitro by rat glomeruli. Biochim. Biophys. Acta. 1985;841:71–80. doi: 10.1016/0304-4165(85)90275-2. [DOI] [PubMed] [Google Scholar]
- 35.Yaoita E., Oguri K., Okayama E., Kawasaki K., Kobayashi S., Kihara I., Okayama M. Isolation and characterization of proteoglycans synthesized by cultured mesangial cells. J. Biol. Chem. 1990;265:522–531. [PubMed] [Google Scholar]
- 36.Vilela-Silva A. C., Werneck C. C., Valente A. P., Vacquier V. D., Mourao P. A. Embryos of the sea urchin Strongylocentrotus purpuratus synthesize a dermatan sulfate enriched in 4-O- and 6-O-disulfated galactosamine units. Glycobiology. 2001;11:433–440. doi: 10.1093/glycob/11.6.433. [DOI] [PubMed] [Google Scholar]
- 37.Ito Y., Habuchi O. Purification and characterization of N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase from the squid cartilage. J. Biol. Chem. 2000;275:34728–34736. doi: 10.1074/jbc.M909633199. [DOI] [PubMed] [Google Scholar]
- 38.Ohtake S., Ito Y., Fukuta M., Habuchi O. Human N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase cDNA is related to human B cell recombination activating gene-associated gene. J. Biol. Chem. 2001;276:43894–43900. doi: 10.1074/jbc.M104922200. [DOI] [PubMed] [Google Scholar]