Abstract
In the study described here, we have taken steps to characterize the YjeE protein, an Escherichia coli protein of unknown function that is essential for bacterial viability. YjeE represents a protein family whose members are broadly conserved in bacteria, absent from eukaryotes and contain both Walker A and B motifs, characteristic of P-loop ATPases. We have revisited the dispensability of the yjeE gene in E. coli and describe efforts to probe the function of the YjeE protein with in vitro biochemistry. We have looked critically for ATPase activity in the recombinant E. coli protein and have made vigilant use of site-directed variants in the Walker A [K41A (Lys41→Ala) and T42A] and putative Walker B (D80Q) motifs. We noted that any hydrolysis of ATP by the wild-type E. coli protein might be attributed to background ATPase, since it was not appreciably different from that of the variants. To overcome potential contaminants, we turned to crystalline pure YjeE protein from Haemophilus influenzae that was found to hydrolyse ATP at a slow rate (kcat=1 h−1). We have also shown high-affinity binding to YjeE by ADP using equilibrium dialysis (Kd=32 μM) and by fluorescence resonance energy transfer from a conserved tryptophan in YjeE to a fluorescent derivative of ADP, 2′-/3′-O-(N-methylanthraniloyl)adenosine 5′-O-diphosphate (Kd=8 μM). Walker motif variants were notably impaired for ADP binding and T42A and D80Q mutations in yjeE were incapable of complementing the yjeE deletion strain.
Keywords: ADP binding, ATPase, Escherichia coli, in vivo complementation, YjeE protein
Abbreviations: Amp, ampicillin; AMPPNP, adenosine 5′-(β,γ-imido)triphosphate; DTT, dithiothreitol; FRET, fluorescence resonance energy transfer; IPTG, isopropyl β-D-thiogalactopyranoside; KAN, kanamycin; LB, Luria–Bertani; MANT-ADP, 2′-/3′-O-(N-methylanthraniloyl)adenosine 5′-O-diphosphate
INTRODUCTION
Since the first report of a complete genome sequence for a free-living organism (Haemophilus influenzae) in 1995 [1], researchers have been faced with the daunting realization that approx. one-third of the genes in any microbe encode proteins of unknown function [2]. Similar estimates have been made for the human genome [3,4], and there is now widespread agreement that the next challenge faced by researchers is the assignment of function for uncharted genes. Although we might have expected the uncharacterized fraction of genomes to encode auxiliary and otherwise superfluous functions, it has become clear from recent studies that many proteins of unknown function are highly conserved and perform essential tasks. Mutagenesis experiments in a variety of bacteria have suggested that hundreds of proteins are indispensable for the growth of any given organism [5–11]. In contrast, fewer than 30 proteins have been exploited commercially as targets for antibacterial drugs [12].
Proteins of unknown function have particular novelty and promise as new targets in antibacterial research; however, the idiosyncratic nature of the physiology and biochemistry associated with any given gene product profoundly limits genome-scale approaches to understand its function. As such, a knowledge of the functions of uncharacterized proteins will be most significantly advanced one gene/protein at a time through concerted investigations of phenotype and biochemistry.
In the study described here, we have taken steps to understand the function of YjeE, a protein identified as essential for cell viability in a gene dispensability study of 27 uncharacterized Escherichia coli genes [13]. YjeE represents a protein family whose members are broadly conserved in bacteria and absent from eukaryotes. A detailed examination of the sequence alignment of this family, performed during the construction of the Clusters of Orthologous Groups of Proteins database [2], revealed all the diagnostic motifs of the P-loop ATPases [14]. More recently, the co-structure of the YjeE protein from H. influenzae in complex with ADP was reported along with a prediction based on the genome context that this protein could be an ATPase involved in cell-wall synthesis [15].
In the present study, we have revisited the dispensability of yjeE with the construction of a conditional mutant in E. coli and describe biochemical efforts to probe the function of YjeE. We have looked critically for ATPase activity in the recombinant protein from E. coli and have made use of crystallization methods [15] to purify H. influenzae YjeE to homogeneity to characterize unambiguously a very slow ATPase activity resident in YjeE. We have shown high-affinity binding of ADP by YjeE using equilibrium dialysis and FRET (fluorescence resonance energy transfer) with a fluorescent derivative of ADP, MANT-ADP [2′-/3′-O-(N-methylanthraniloyl)adenosine 5′-O-diphosphate]. An in vivo complementation assay of YjeE function is also presented, where Walker box variants were found to be incapable of supporting cell growth.
EXPERIMENTAL
Materials
E. coli strains were grown in a rich LB (Luria–Bertani) medium. Restriction enzymes and Vent polymerase were purchased from New England Biolabs (Beverly, MA, U.S.A.). DTT (dithiothreitol), Tris, Amp (ampicillin), IPTG (isopropyl β-D-thiogalactopyranoside) and urea were obtained from BioShop Canada Inc. (Burlington, ON, Canada); MgCl2 was obtained from BDH Inc. (Toronto, ON, Canada). MANT-ADP was purchased from Molecular Probes (Eugene, OR, U.S.A.). Tritium-labelled ADP ([2,8-3H]ADP) and the Western chemiluminescence reagent were obtained from PerkinElmer Life Sciences (Boston, MA, U.S.A.). Donkey anti-rabbit antibody conjugated to horseradish peroxidase was purchased from Jackson Immunoresearch Laboratories (West Grove, PA, U.S.A.). Nitrocellulose membranes (0.45 μm) were obtained from Bio-Rad Laboratories (Hercules, CA, U.S.A.). Triethylamine (99%) was purchased from Anachemia Canada Inc. (Montreal, PQ, Canada). All other compounds were from Sigma (Oakville, ON, Canada).
Dispensability of yjeE
To analyse the dispensability of yjeE, a gene knockout procedure similar to that described previously [16,17] was implemented using wild-type E. coli MG1655. A strain positive for chromosomal integration of yjeE and a KAN (kanamycin) resistance cassette at the araBAD locus were selected and named EB437 (MG1655 araBAD::yjeE kan). This strain, diploid for yjeE, allowed for the control of YjeE expression using the araBAD promoter. The complementing copy of yjeE was under the control of a consensus ribosome-binding site [18]. The gene replacement procedure was conducted in strain EB437 using a chloramphenicol resistance cassette (promoter to stop codon). PCR confirmed the deletion of yjeE and the resulting strain was named EB445 (MG1655 araBAD::yjeE kan yjeE::cat). A growth analysis in the presence and absence of arabinose was performed to assess the inducer dependence of the yjeE deletion strain in liquid media. The strains EB437 and EB445 were grown overnight on LB/KAN and LB/KAN/arabinose plates respectively. One colony from each plate was used to inoculate an overnight liquid culture. On the following day, 10 μl of each of the overnight cultures was added to 5 ml of fresh media and grown until the cells reached an absorbance A600 of 0.3–0.5. The samples were diluted to an A600 of 0.1, and 1 ml of the strain EB445 was used to inoculate 100 ml of LB/KAN/arabinose (0.2, 0.02 and 0%). Additionally, 1 ml of EB437 was used to inoculate 100 ml of LB/KAN with and without arabinose. The samples were incubated at 37 °C with shaking at 250 rev./min for 480 min. Every hour, the A600 for a 0.5 ml sample was read.
Cloning and purification of YjeE
Recombinant E. coli and H. influenzae YjeE proteins were purified after expression in E. coli. E. coli yjeE was amplified by PCR from E. coli MG1655 chromosomal DNA using Vent polymerase and the primers P1 (5′-G GGGACAAGTTTCTACAAAAAAGCAGGCTTAATGAATCGAGTAATTCCGCTCCCT-3′) and P2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCATTAACCGGCTAAACGCGCCAGCAACAATT-3′). The PCR-amplified product flanked by ‘attB’ recombination sites (in bold-face) was cloned using the Gateway™ phage lambda site-specific recombination system (Invitrogen Canada, Burlington, ON, Canada). Subsequently, according to the manufacturer's instructions, the yjeE gene was moved into the pDEST17 expression vector designed to generate an N-terminal His-tagged recombinant protein. The resulting YjeE expression plasmid was used to transform competent E. coli BL21 (DE3) cells for expression purposes. Three different variants of YjeE [K41A (Lys41→Ala), T42A and D80Q] were constructed with the Quik Change™ site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) using the pDEST17-YjeE expression vector as template. The sequences of the overexpression constructs for YjeE and the three variants were confirmed by sequencing (MOBIX; McMaster University). In a typical YjeE purification, 4×1 litre of LB broth containing 50 μM Amp were inoculated at a ratio of 1:100 with an overnight culture of E. coli BL21(DE3)/pDEST17-YjeE prepared from a freshly streaked plate. Cultures were incubated at 37 °C with shaking at 250 rev./min until the cell density reached an A600 of approx. 0.5, induced with 1 mM IPTG and incubated with shaking at 250 rev./min for 3 h at 37 °C. Cells were harvested by centrifugation (15 min at 4000 g) and resuspended in 20 mM sodium phosphate (pH 7.5), 500 mM NaCl and 0.5 mM PMSF (buffer A). After resuspension, cells were disrupted by three passages through a French Press at 69000 kPa, and the lysate was clarified by centrifugation (30 min at 20000 g). YjeE was purified by nickel chelate chromatography using a 4 ml HiTrap affinity column (Amersham Biosciences, Baie d'Urfé, PQ, Canada), all at 4 °C. The column was washed with buffer A containing 20 mM imidazole and eluted with a linear imidazole gradient of 20–300 mM. Fractions of the eluate were analysed by SDS/PAGE, and those containing pure His-tagged YjeE were pooled and concentrated using an Amicon 8200 stirred cell concentrator and PM10 membranes (Fisher Scientific, Nepean, ON, Canada) and then buffer-exchanged to 20 mM Tris (pH 7.5), 100 mM NaCl, 0.5 mM EDTA and 1 mM DTT, using a Superdex S200 (Amersham Biosciences) gel-filtration column (1.6 cm×64 cm). The pooled fractions were then chromatographed over a Q-Sepharose Fast Flow (Amersham Biosciences) anion-exchange column (2.6 cm×10 cm) and washed with 200 ml of 20 mM Tris (pH 7.5) and 1 mM DTT. Pure His-tagged YjeE was eluted with a linear gradient of 0–1 M NaCl in 900 ml. Fractions containing pure YjeE were pooled and dialysed against 20 mM Tris (pH 7.5) and 2 mM DTT. Using this method, approx. 80 mg of pure YjeE was obtained.
The expression clone for YjeE from H. influenzae [15] was kindly provided by G. Gilliland (University of Maryland). Overexpression, purification and crystallization procedures were as described previously [15]. We grew crystals of YjeE in hanging drops from 10 mg/ml protein solution in 0.1 M Tris/HCl (pH 7.0), 1.4 M ammonium sulphate and 2% (w/v) poly(ethylene glycol) 400. The crystals were grown for 3 days and the corresponding drops were pooled, washed several times with the crystallization buffer and dissolved in 10 mM Hepes (pH 8.0), 1 mM EDTA, 0.5 mM DTT and 150 mM NaCl. Pure E. coli and H. influenzae YjeE proteins were frozen in small aliquots and stored at –80 °C. Protein concentration was determined by the method of Gill and von Hippel [19].
YjeE enzyme assay
A high-sensitivity HPLC assay was developed to monitor slow ATP hydrolysis by purified YjeE. Production of ADP was monitored by HPLC (Waters 600 pump and controller, Waters 486 tunable absorbance detector, Waters 717 autosampler; Waters, Milford, MA, U.S.A.) using a 4.5 mm×50 mm WP QUAT strong ion-exchange column (J. T. Baker, Phillipsburg, NJ, U.S.A.). ATP and ADP were separated with a 12 ml gradient of triethylamine bicarbonate from 20 to 500 mM, using a flow rate of 2 ml/min. The progress of the reaction was monitored by absorbance at 259 nm and the amount of ADP produced was determined by integration of the resolved peaks using the HPLC software (Millenium; Waters). The reaction was performed at room temperature (23 °C) in a mixture containing an appropriate amount of ATP in 50 mM Hepes (pH 7.5), 2 mM DTT and 10 mM MgCl2, with a final addition of YjeE to start the reaction. After incubating for an appropriate time period, the reactions were stopped by adding 2 vol. of 8 M urea (5.3 M final concentration). After completion of the reaction, the mixture was filtered through a 5 kDa MWCO Amicon Ultrafree-MC filter (Millipore, Bedford, MA, U.S.A.) to remove the protein before loading on to the HPLC column.
Complementation experiments with the yjeE deletion strain
The expression vectors for wild-type yjeE and three site-directed mutants therein (see above) were transformed into the yjeE deletion strain EB445 and evaluated for growth in the presence and absence of arabinose on LB/KAN/Amp. To determine the relative expression levels of these strains in the absence of arabinose, we transformed these plasmids into the diploid yjeE complementation strain EB437 for analysis by Western blotting. (Two of these mutants did not support the growth of EB445 in the absence of arabinose.) E. coli lysates were prepared for this analysis by growing each strain in 200 ml of LB medium at 37 °C with shaking at 250 rev./min to an A600 of approx. 1. Cells were then harvested by centrifugation, resuspended in 4 ml of lysis buffer (50 mM Hepes, pH 7.5, 5 mM EDTA, 0.1 mg/ml DNase, 0.1 mg/ml RNase and 0.5 mM PMSF), disrupted by three passages through a French Press at 69000 kPa, and the lysates were clarified by centrifugation (30 min at 100000 g). The lysates were then dialysed overnight against a 50 mM Hepes (pH 7.5) buffer before analysis. Western-blot analysis was performed with a rabbit polyclonal anti-YjeE antibody raised by Cocalico Biologicals (Reamstown, PA, U.S.A.). Anti-YjeE antibodies were affinity-purified using purified YjeE coupled with a bisacrylamide/azlactone co-polymer (Ultralink Biosupport Medium) as described by the manufacturer (Pierce Chemical, Rockville, IL, U.S.A.).
FRET experiments
The active site of YjeE was probed by FRET from a conserved tryptophan residue to the ADP analogue, MANT-ADP. Fluorescence spectra were measured with a PTI spectrofluorimeter (Photon Technology International, London, ON, Canada). The sample buffer (20 mM Tris, pH 7.5, 10 mM MgCl2 and 2 mM DTT) was thoroughly degassed by bubbling with nitrogen gas for at least 1 h. The excitation wavelength was either 356 nm for direct excitation of the MANT-ADP or 295 nm for FRET experiments. The emission spectra were recorded between 370 and 450 nm. Spectra of buffer alone, buffer and protein and buffer with MANT-ADP were determined to correct for the contribution of Raman scattering and unbound MANT-ADP. Excitation and emission slit widths of 6 nm were used in all experiments.
FRET was used to determine the dissociation constants of MANT-ADP for wild-type YjeE and three variants from the blank-corrected emission intensities at 440 nm (excitation at 295 nm). Increasing amounts of MANT-ADP (2–35 μM) were added to a constant amount of YjeE (2 μM) and the change in fluorescence intensity was monitored (ΔF440). In competition experiments, the loss of FRET from YjeE to MANT-ADP was monitored with the addition of the following nucleotides: AMP, ADP, ATP, AMPPNP [adenosine 5′-(β,γ-imido)triphosphate], CTP, CDP, CMP, GMP, GDP, GTP, UMP, UDP and UTP. Data were fitted to a hyperbolic function to determine the dissociation constants for MANT-ADP (Kd) and the unmodified nucleotides (the apparent dissociation constant Kapp).
Equilibrium dialysis
Equilibrium dialysis was used to establish the stoichiometry and affinity of ADP for wild-type YjeE and the K41A variant. A 20-cell equilibrium dialyser (Spectrum Laboratories, Rancho Dominquez, CA, U.S.A.) was used with Spectra-Pro semi-permeable membranes (Spectrum), dividing each cell into two chambers (molecular mass cut-off, 6–8 kDa). A 200 μl volume of YjeE (33 μM) or the K41A variant (100 μM), dialysed previously in 20 mM Tris (pH 8.0), 2 mM DTT and 10 mM MgCl2, was deposited in one chamber of the divided cell. In the other chamber was an equal volume of ADP solution (20 mM Tris, pH 8.0, 2 mm DTT and 10 mM MgCl2, supplemented with 0.2 μCi of [2,8-3H]ADP) ranging in concentration from 2 to 500 μM ADP and 20 to 2000 μM ADP for wild-type YjeE and the K41A variant respectively. The cells were rotated at 15 rev./min at room temperature for a time period necessary to achieve equilibrium of ADP across the membrane in a control cell (2 h). Subsequently, duplicate 40 μl aliquots were removed from each chamber and applied to filter paper and allowed to dry before scintillation counting. These measurements allowed the calculation of the ADP concentration in the ADP chamber (free ADP) and in the protein chamber (free ADP and protein-bound ADP).
RESULTS
Conditional complementation of E. coli yjeE deletion
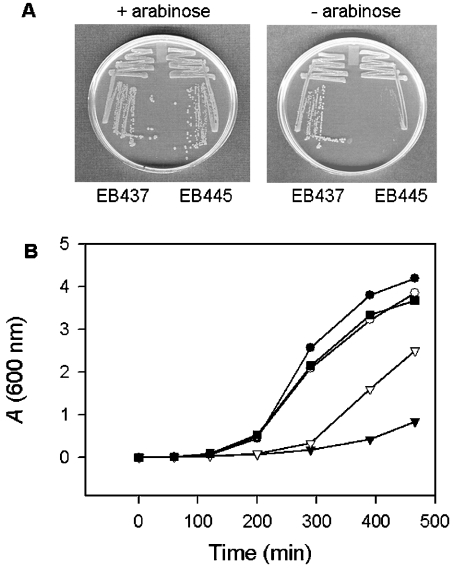
Figure 1 illustrates the growth defect in our conditional yjeE mutant (EB445). In this mutant, yjeE has been deleted from its wild-type location and a rescue copy placed under control of the arabinose promoter on the chromosome of E. coli (MG1655 araBAD::yjeE kan yjeE::cat). Also shown is a control strain, EB437, which is diploid for yjeE (MG1655 araBAD::yjeE kan). As is evident from Figure 1(A), the yjeE deletion strain (EB445) was able to grow well on rich, solid media in the presence of arabinose with wild-type colony morphology; however, on the removal of arabinose, there was a drastic decrease in growth and a loss of the ability to form single colonies. The diploid strain (EB437) was able to grow equally well both in the presence and absence of an inducer.
Figure 1. Conditional complementation of the yjeE deletion strain.
(A) Arabinose dependence of the yjeE deletion strain EB445. The yjeE deletion strain (EB445) and the diploid strain (EB437) were plated in the presence and absence of arabinose and grown overnight at 37 °C. (B) Growth of yjeE-depleted cells in liquid media. Cells (EB445) were grown overnight on LB/KAN/arabinose plates and used to inoculate LB/KAN media with 0.2% (■), 0.02% (▽) and no arabinose (▼). The diploid strain (EB437) was similarly grown in the presence of 0.2% arabinose (●) and no arabinose (○). Growth was followed at 37 °C for 8 h.
Figure 1(B) shows a kinetic growth analysis of the yjeE deletion strain (EB445), where growth in liquid media was monitored at various arabinose concentrations and the diploid strain (EB437) was used as a control. At 0.2% arabinose, the deletion strain exhibited the same lag phase as the control strain and grew at a similar rate during the exponential phase, reaching the same final cell density as the diploid strain. As the level of inducer was decreased to 0.02%, an increase in the lag phase was observed in the deletion strain along with a decrease in the growth rate during the exponential phase. In the absence of an inducer, there was almost no growth during the time course of the experiment. Indeed, that the deletion mutant showed some growth at high inoculum on solid media and over an extended period of time in liquid culture in the absence of an inducer probably arises from the residual gene expression in the absence of an inducer. These results indicate that the ability of E. coli to grow is related to the expression of YjeE in the cell. Taken together, the conditional growth experiments using solid and liquid media provide the first clear evidence that yjeE is essential for the growth of E. coli.
Purification of YjeE
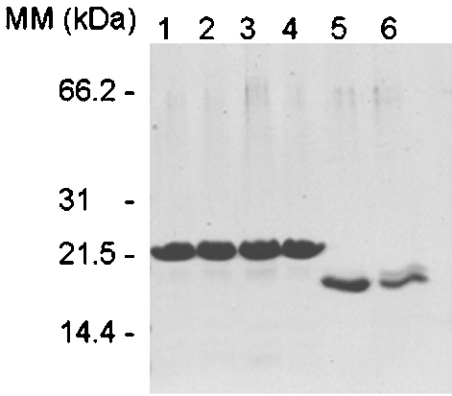
Figure 2 shows an SDS/PAGE analysis of purified recombinant E. coli YjeE, three variants thereof (K41A, T42A and D80Q) and H. influenzae YjeE. The level of expression of all these proteins was high, yielding, for example, approx. 20 mg of pure protein per litre of culture for E. coli YjeE. The E. coli proteins run with apparent molecular masses that are slightly larger than their true molecular masses. Nonetheless, electrospray ionization MS of wild-type E. coli YjeE yielded a molecular mass for the His-tagged protein of 19246 Da (results not shown), in agreement with the mass calculated from the sequence of 19380 or 19249 Da with cleavage of the N-terminal methionine residue. H. influenzae YjeE, which was purified with the benefit of a cleavable His tag [15], migrated consistent with the calculated mass (18107 Da) for the thrombin-processed protein. The purity of all these affinity-purified proteins was very high as evidenced by SDS/PAGE, but particularly noteworthy was our use of protein crystallization to purify the H. influenzae YjeE protein to homogeneity. No detectable difference in purity, however, was observed between the YjeE preparations before and after crystallization.
Figure 2. SDS/PAGE (15% gel) showing purified YjeE and its variants.
Lane 1, wild-type E. coli YjeE; lane 2, E. coli K41A variant; lane 3, E. coli T42A variant; lane 4, E. coli D80Q variant; lanes 5 and 6, purified recombinant H. influenzae YjeE before and after crystallization respectively.
Enzymic characterization of YjeE
We used a high-sensitivity HPLC-based assay to monitor the production of ADP over time to evaluate the slow ATPase activity of YjeE. Repeated analysis with several different preparations of wild-type E. coli YjeE revealed a very low ATPase activity in the range of 10 h−1 (results not shown). We were, nevertheless, concerned that an ATPase contaminant with high activity present in undetectable amounts in the pure preparation could account for such low activities. To test this hypothesis, three YjeE variants were created. Two mutations in the Walker A motif were performed, K41A and T42A, and one mutation in the putative Walker B motif, D80Q. Similar mutations to these have been shown to alter significantly the activity of other ATP-hydrolysing proteins [20–22]. Compared with wild-type YjeE (kcat=12 h−1, kcat/Km=8.5 mM−1·h−1), however, there was little impact due to K41A (kcat=8.3 h−1, kcat/Km=14 mM−1·h−1), T42A (kcat=4.8 h−1, kcat/Km=10 mM−1·h−1) and D80Q (kcat=2.6 h−1, kcat/Km=5 mM−1·h−1) substitutions (Table 1).
Table 1. Characterization of steady-state kinetics of ATP hydrolysis by the YjeE protein.
| Enzyme | Km (mM) | kcat (h−1) | kcat/Km (mM−1·h−1) |
|---|---|---|---|
| Wild-type E. coli YjeE | 1.4 | 12 | 8.5 |
| K41A E. coli YjeE | 0.6 | 8.3 | 14 |
| T42A E. coli YjeE | 0.4 | 4.8 | 10 |
| D80Q E. coli YjeE | 0.5 | 2.6 | 5 |
| Crystalline pure H. influenzae YjeE | 0.8 | 1.2 | 1.5 |
| H. influenzae YjeE before crystallization | 0.7 | 1.1 | 1.6 |
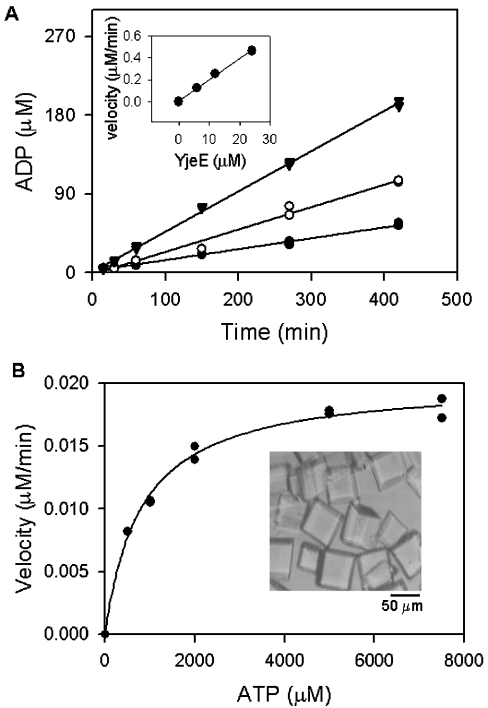
Concerned about the possibility that a contaminating activity might be responsible for the ATPase activity observed for E. coli YjeE and its variants, we turned to H. influenzae YjeE for which crystallization conditions have been determined [15]. Crystals of purified H. influenzae YjeE were grown, washed extensively and redissolved before testing for ATPase activity. Figure 3 shows our kinetic analysis of that preparation using the high-sensitivity HPLC assay. The production of ADP by crystalline pure YjeE correlated with both time and the amount of protein added, yielding a turnover number at saturating ATP of 1.2 h−1 (Figure 3A). Analysis of the dependence of this activity on ATP yielded a Km=0.8 mM for ATP (Figure 3B), allowing the calculation of a catalytic efficiency kcat/Km of 1.6 mM−1·h−1. It is also noteworthy that affinity-purified, but not crystallized, H. influenzae YjeE protein showed kinetic parameters (kcat=1.1 h−1, kcat/Km=1.6 mM−1·h−1) that were in agreement with those of the crystalline pure enzyme. Altogether, our results provide unambiguous evidence for a slow but detectable ATP hydrolysis activity in isolated YjeE protein.
Figure 3. ATPase activity of crystalline pure H. influenzae YjeE.
Crystals of YjeE (B, inset) were prepared and dissolved as described (in the Experimental section) and ATPase activity was determined. (A) YjeE at concentrations of 6 μM (●), 12 μM (○) and 24 μM (▼) was incubated in the presence of 6 mM ATP and 10 mM MgCl2 (reaction volume, 100 μl) and 10 μl fractions were sampled at the times indicated and quenched by the addition of urea to 6 M. The amount of ADP produced was determined by HPLC as described in the Experimental section. The inset shows the initial velocity of the reaction plotted as a function of enzyme concentration; the slope represents the turnover of the enzyme under these conditions (1.2 h−1). (B) Dependence of the reaction velocity on ATP concentration. Reactions (20 μl) containing ATP (0.2–7.5 mM), YjeE (12 μM) and 10 mM MgCl2 were quenched after 7 h and analysed by HPLC to determine the rates. The data were fitted to the Michaelis–Menten equation v=kcat[E][S]/(Km+[S]) using Sigma Plot 2000 (SPSS Science, Chicago, IL, U.S.A.) and Km and kcat values were calculated to be 800 μM and 1.2 h−1 respectively.
It is worth noting that, in the course of their structural determination of H. influenzae YjeE, Teplyakov et al. [15] reported that YjeE-ADP co-crystals were produced in crystallization trials with either ADP or ATP, implying an intrinsic ATPase. Indeed, these authors reported an ATPase activity of 25 h−1 for the purified enzyme, which is 20-fold more than that reported in the present study.
MANT-ADP as a probe of the active site of YjeE
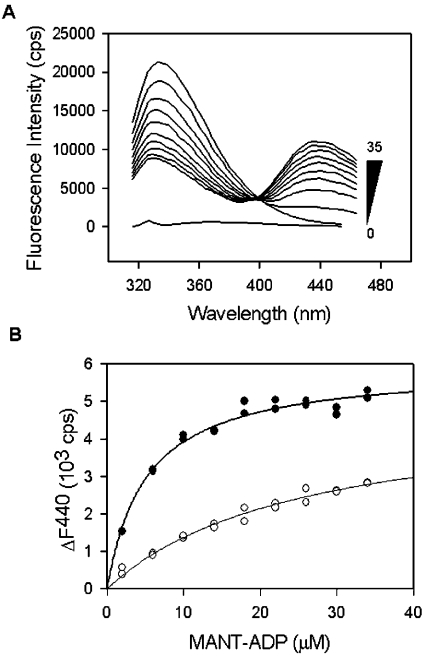
The protein family represented by YjeE has a single conserved tryptophan residue (position 109 in the E. coli sequence) that, as evident from the crystal structure of the H. influenzae enzyme [15], is proximal to the ADP binding site. Indeed, the indole nitrogen of the tryptophan residue is only 7 Å from the β-phosphate and 15 Å from the 2′ oxygen of ADP. Such a distance is appropriate for the observation of FRET [23] from the tryptophan to MANT-ADP on binding of the modified nucleotide to the YjeE protein. Accordingly, excitation of YjeE at 295 nm in the presence of MANT-ADP was found to decrease the intrinsic protein fluorescence emission intensity (peak at 330 nm) and increase the emission intensity associated with MANT-ADP (peak at 440 nm) through FRET. The foster distance R0 was calculated to be 3.1 nm using the method described previously [23]. Figure 4(A) shows the fluorescence emission spectra of the E. coli YjeE protein in the presence of increasing concentrations of MANT-ADP (0–35 μM). We observed no evidence of FRET in the absence of added MgCl2 (results not shown). The FRET signal was completely abolished when a variant (W109A) lacking tryptophan was used (results not shown), indicating that Trp109 is responsible for the totality of the transfer.
Figure 4. Determination of Kd for wild-type YjeE and the K41A variant.
(A) Emission spectra collected on incubation of recombinant E. coli YjeE protein (2 μM) with increasing amounts of MANT-ADP (0–35 μM) as indicated (excitation was at 295 nm). The spectrum shown at the bottom corresponds to that of buffer alone. (B) Change of fluorescence emission intensity at 440 nm for wild-type YjeE (●) and the K41A variant (○) as a function of MANT-ADP concentration. The spectra of the protein alone and MANT-ADP alone at each concentration were subtracted from the spectra of MANT-ADP in the presence of protein, and the change of fluorescence at 440 nm (ΔF440) was plotted as a function of the total MANT-ADP concentration. Fluorescence is expressed in terms of counts/s (cps). The data were fitted to a hyperbolic function to determine Kd values of 8 and 65 μM for wild-type YjeE and the K41A variant respectively.
The Kd for MANT-ADP to wild-type YjeE and the three variants were measured using the FRET signal between Trp109 and MANT-ADP. Figure 4(B) shows the results of this analysis for wild-type E. coli YjeE and the K41A variant, where the measured Kd values were 8 and 65 μM respectively (Table 2). No binding could be detected by FRET for the variants T42A and D80Q at the highest concentration (35 μM) of MANT-ADP used (results not shown). Wild-type H. influenzae YjeE was found to have a Kd=18 μM for MANT-ADP (results not shown). In these experiments, important controls were performed, which are not shown. For example, very little protein fluorescence is detected at 440 nm; however, changes in the fluorescence at 440 nm were corrected for the minor contribution of protein fluorescence. Furthermore, the fluorescence of MANT-ADP in the absence of YjeE was found to increase linearly with the concentration (results not shown), indicating that the curvature in the plots of Figure 4 is not due to saturation of the emission detector.
Table 2. Kd values for nucleotide binding by the YjeE protein.
| Kd (μM) | ||
|---|---|---|
| Enzyme | FRET (MANT-ADP) | Equilibrium dialysis (ADP) |
| E. coli YjeE | 8±0.9 | 32±7.6 |
| K41A E. coli YjeE | 65±3.8 | 211±66 |
| T42A E. coli YjeE | >100 | Not determined |
| D80Q E. coli YjeE | >100 | Not determined |
| H. influenzae YjeE | 18±0.4 | Not determined |
Competition for MANT-ADP binding to YjeE
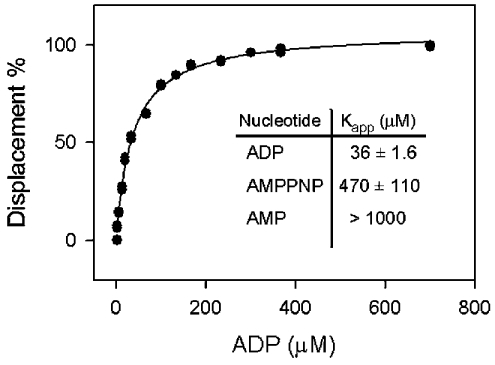
FRET from YjeE to MANT-ADP proved to be a valuable reporter for the binding of unlabelled nucleotides to YjeE. Figure 5 presents the saturation curve for the displacement of MANT-ADP from YjeE with the addition of unlabelled ADP to yield a Kapp value of 36 μM. The non-hydrolysable ATP analogue AMPPNP had a Kapp value of 470 μM, whereas little or no binding was observed for AMP, GDP, GMP, GTP, CMP, CDP, CTP, UMP, UDP or UTP (results not shown).
Figure 5. Displacement of MANT-ADP bound to YjeE by various nucleotides.
The graph depicts the displacement of the MANT-ADP by unmodified ADP as followed by a decrease in the FRET signal associated with the binding of the MANT-ADP (10 μM) to YjeE (6 μM). The data were fitted to a hyperbolic function to determine the Kapp. The inset summarizes the Kapp measured for ADP, AMPPNP and AMP. No displacement was detected when GMP, GDP, GTP, CMP, CDP, CTP, UMP, UDP or UTP was used as a competitor.
Equilibrium dialysis with labelled ADP
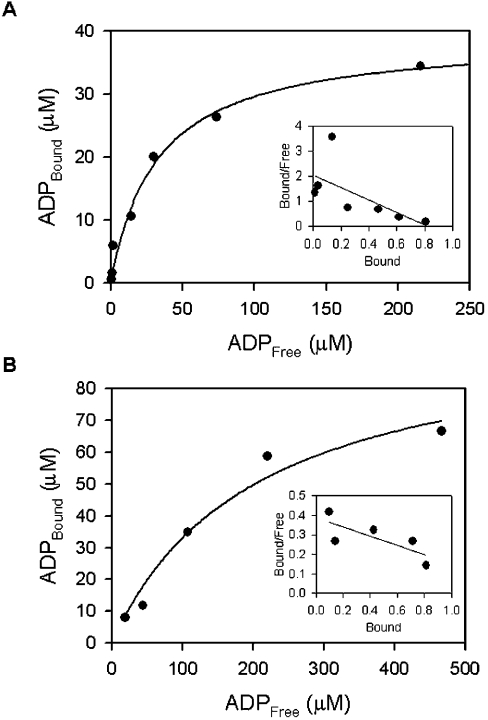
The affinities of ADP for wild-type E. coli YjeE and the K41A variant were determined using equilibrium dialysis (Figure 6). The hyperbolic plot and Scatchard analysis revealed stoichiometric binding of ADP to YjeE and a Kd=32 μM for the interaction. Similarly, the K41A variant was found to exhibit saturable and stoichiometric binding of ADP with Kd=211 μM (Table 2).
Figure 6. Determination of Kd for ADP bound to wild-type YjeE (A) and K41A variant (B) using equilibrium dialysis.
The data are presented in hyperbolic form, where [ADP]Bound is plotted versus [ADP]Free. Equilibration was attained as described in the Experimental section under the following conditions: 20 mM Tris, 2 mM DTT, 10 mM MgCl2 (pH 7.5) and 33 μM wild-type YjeE and 100 μM K41A variant respectively. The insets show Scatchard plots where the [ADP]Bound/[ADPFree] ratio is plotted as a function of [ADP]Bound. The stoichiometry of binding shows that about one molecule of ADP is bound per molecule of protein. The Kd values for ADP bound to wild-type YjeE and K41A variant were calculated to be 32 and 211 μM respectively.
Complementation of the yjeE deletion strain using the three YjeE variants
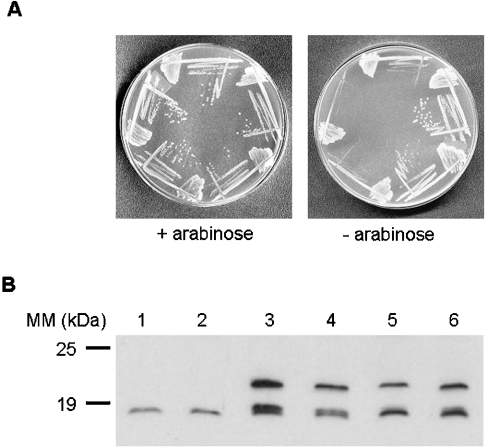
We tested the ability of the Walker box mutants (K41A, T42A and D80Q) in YjeE to complement the yjeE depletion strain. The Gateway expression clones encoding His-tagged versions of wild-type YjeE and three variants of YjeE were transformed into the yjeE deletion strain EB445 (MG1655 araBAD::yjeE kan yjeE::cat). Figure 7 shows the growth of the resulting strains in the presence and absence of arabinose, as well as the results of Western blotting with anti-YjeE polyclonal antibody to establish expression levels of these clones relative to wild-type E. coli. Wild-type yjeE and the K41A mutant were able to complement the yjeE deletion strain, whereas the T42A and D80Q mutants were not able to rescue the defect (Figure 7A). Apart from insights into the relative importance of these residues to YjeE function, there are a couple of interesting conclusions from this experiment. The first is that His-tagged YjeE is able to function effectively in vivo. The second is that His-tagged wild-type YjeE and variants were produced from the pDEST17 in the absence of T7 RNA polymerase (not present in MG1655) and the inducer for this expression system (IPTG). Indeed, the expression levels of His-tagged wild-type YjeE and its variants were similar to one another and to YjeE present in wild-type E. coli (Figure 7B).
Figure 7. In vivo complementation of yjeE deletion strain with wild-type and variants of YjeE.
(A) Strains were grown on LB/CM/arabinose plates (left panel) and LB/CM plates (right panel) at 37 °C overnight. Clockwise from top: strain EB445 (yjeE deletion strain), EB445 with pDEST17-WTYjeE, EB445 with pDEST17-K41AYjeE, EB445 with pDEST17-T42AYjeE and EB445 with pDEST17-D80QYjeE. (B) Western blotting using a YjeE polyclonal antibody to confirm expression of YjeE from the pDEST17 plasmids. Cell lysates (approx. 100 μg) were subjected to SDS/PAGE (15% gel) and electroblotted on to a nitrocellulose membrane. The blot was treated with YjeE polyclonal antibody and donkey anti-rabbit antibody conjugated to horseradish peroxidase. The signal was developed using the Western Exposure Chemiluminescent Detection System and imaged on a chemiluminescence film with a 2 min exposure. Lane 1, wild-type E. coli MG1655; lane 2, EB437 (diploid strain); lane 3, EB437/pDEST17-WTYjeE; lane 4, EB437/pDEST17-K41AYjeE; lane 5, EB437/pDEST17-T42AYjeE; lane 6, EB437/pDEST17-D80QYjeE. Evident from the blot are both His-tagged (expressed from pDEST17) and untagged YjeE migrating with apparent molecular masses of 19.2 and 18.1 kDa respectively.
DISCUSSION
We describe here the indispensability of the yjeE gene in E. coli, through deletion of yjeE and conditional rescue with a complementing copy of the gene that is controlled by the arabinose promoter. The essential nature of yjeE was previously noted in a 27-gene dispensability study by Freiberg et al. [13]. Our results are, nevertheless, the first growth data presented on this mutant. It is also noteworthy that, in [13], conditional expression was from a temperature-sensitive plasmid, where growth was reportedly halted at 43 °C. One ambiguity surrounding the use of temperature-sensitive defects is rooted in the possibility that lethality is due to unusual cell physiology at heat-shock growth temperatures [24]. Our growth data at 37 °C provide unequivocal proof that yjeE is indispensable to the growth of E. coli. Similarly, the conditional mutant generated in this work will be useful for a variety of purposes, including further phenotypic characterization of the cells depleted of YjeE.
We found a low ATPase activity for E. coli YjeE protein (12 h−1) and made use of site-directed mutagenesis to create Walker A and B variants in YjeE. We had hoped that these substitutions would significantly decrease the activity of YjeE so that we could rule out the presence of a contaminating ATPase. The activity of these variants was, however, not convincingly affected (kcat=8.3, 4.8 and 2.6 h−1 for K41A, T42A and D80Q respectively). Since these substitutions were chosen for their impact on chemical steps in the catalysis of other P-loop ATPases [20–22], it remains a possibility that other steps, substrate binding or product release, are overwhelmingly rate-limiting in YjeE. This possibility is reinforced by the finding that our preparation of H. influenzae YjeE showed the same activity before and after further purification to homogeneity by crystallization. Indeed, we crystallized and thoroughly washed H. influenzae YjeE protein crystals in an effort to test for intrinsic ATPase unambiguously. Time, enzyme and substrate dependence of the ATP hydrolysis activity associated with this preparation unequivocally establishes YjeE as an ATPase (kcat=1.2 h−1).
We have shown high-affinity and stoichiometric binding of ADP by YjeE using equilibrium dialysis (Kd=32 μM) and FRET with the fluorescent derivative of ADP, MANT-ADP (Kd=8 μM). The latter technique has provided a convenient surrogate ligand-binding assay to allow us to reveal that YjeE is overwhelmingly selective for ADP compared with other nucleotides. Interestingly, YjeE may have a much lower affinity for the substrate. The FRET assay revealed an affinity of 470 μM for AMPPNP, a non-hydrolysable analogue of ATP, and the substrate dependence of the ATPase activity demonstrated a Km of 800 μM for ATP.
While the site-directed mutagenesis studies were ambiguous with respect to the ATPase activity of E. coli YjeE, ADP binding was significantly impaired by all of the substitutions. K41A YjeE showed a 7–8-fold decrease in affinity for ADP and MANT-ADP, whereas the T42A and D80Q variants had no measurable affinity for the latter dinucleotide. Indeed, the ADP-binding data were in accordance with our complementation study, where only wild-type yjeE and the K41A mutant were able to complement the yjeE deletion strain in vivo. Probably, the residual function of the K41A variant is manifest in its measurable ADP-binding activity. It is also noteworthy that the K41A variant retained the highest level of ATPase activity.
Our mutational results are largely in accordance with the structural data reported recently [15]. In the nucleotide-binding site, the P-loop Thr42, through its hydroxy group, interacts directly with a magnesium molecule, which is presumed to stabilize the negative charges on the phosphate. The conserved aspartate (Asp80) also interacts with this cation through a water molecule. This water molecule may well be a nucleophile in position to attack the γ-phosphate of ATP. Lys41 is in hydrogen-bonding proximity to the β-phosphate ADP and may also interact with the γ-phosphate during catalysis as has been proposed for other ATPases [21,25]. It is therefore somewhat surprising that the loss of this interaction results in only a minor but measurable loss of function. Apart from the Walker A and B motifs, the recently solved crystal structure of YjeE from H. influenzae shows the highest similarity to a subset of P-loop ATPases including F1-ATPases, helicases, DNA-binding proteins and AAA proteins, but also has a unique topology. The bound ADP, for example, was found in a syn conformation, which is unusual in known ATPases with the exception of AAA proteins [15]. Taken together, this information suggests that this protein probably represents a separate family of P-loop proteins with distinct features.
The low ATPase activity of YjeE in isolation and its apparently high affinity for product suggests a role for this protein in energy or signal transduction. ATPases of this variety, such as DNA-binding proteins [26,27] and molecular chaperones [26,27], typically have a very low ATPase activity that is difficult to measure in isolation but is potentiated by interactions with partner macromolecules. Our findings to date for YjeE are consistent with such a paradigm and have positioned us for further investigations to reveal additional molecular interactions of YjeE.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (grant number MOP-64292) and by a Canada Research Chair in Microbial Biochemistry (to E.D.B.). We thank E. Koonin of the National Center for Biotechnology Information in Maryland for engaging discussions regarding the sequence and probable functions of YjeE.
References
- 1.Fleischmann R. D., Adams M. D., White O., Clayton R. A., Kirkness E. F., Kerlavage A. R., Bult C. J., Tomb J. F., Dougherty B. A., Merrick J. M. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Tatusov R. L., Galperin M. Y., Natale D. A., Koonin E. V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., Smith H. O., Yandell M., Evans C. A., Holt R. A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Badarinarayana V., Estep P. W., 3rd, Shendure J., Edwards J., Tavazoie S., Lam F., Church G. M. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 2001;19:1060–1065. doi: 10.1038/nbt1101-1060. [DOI] [PubMed] [Google Scholar]
- 6.Akerley B. J., Rubin E. J., Novick V. L., Amaya K., Judson N., Mekalanos J. J. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U.S.A. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutchison C. A. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 8.Sassetti C. M., Boyd D. H., Rubin E. J. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsyth R. A., Haselbeck R. J., Ohlsen K. L., Yamamoto R. T., Xu H., Trawick J. D., Wall D., Wang L., Brown-Driver V., Froelich J. M., et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes S. Y., Scholle M. D., D'Souza M., Bernal A., Baev M. V., Farrell M., Kurnasov O. V., Daugherty M. D., Mseeh F., Polanuyer B. M., et al. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 2002;184:4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi K., Ehrlich S. D., Albertini A., Amati G., Andersen K. K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P., et al. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haselbeck R., Wall D., Jiang B., Ketela T., Zyskind J., Bussey H., Foulkes J. G., Roemer T. Comprehensive essential gene identification as a platform for novel anti-infective drug discovery. Curr. Pharm. Des. 2002;8:1155–1172. doi: 10.2174/1381612023394818. [DOI] [PubMed] [Google Scholar]
- 13.Freiberg C., Wieland B., Spaltmann F., Ehlert K., Brotz H., Labischinski H. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J. Mol. Microbiol. Biotechnol. 2001;3:483–489. [PubMed] [Google Scholar]
- 14.Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teplyakov A., Obmolova G., Tordova M., Thanki N., Bonander N., Eisenstein E., Howard A. J., Gilliland G. L. Crystal structure of the YjeE protein from Haemophilus influenzae: a putative ATPase involved in cell wall synthesis. Proteins. 2002;48:220–206. doi: 10.1002/prot.10114. [DOI] [PubMed] [Google Scholar]
- 16.Campbell T. L., Brown E. D. Characterization of the depletion of 2-C-methyl-D-erythritol-2,4-cyclodiphosphate synthase in Escherichia coli and Bacillus subtilis. J. Bacteriol. 2002;184:5609–5618. doi: 10.1128/JB.184.20.5609-5618.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko K. A., Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vellanoweth R. L., Rabinowitz J. C. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 19.Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 20.Sato K., Mori H., Yoshida M., Mizushima S. Characterization of a potential catalytic residue, Asp-133, in the high affinity ATP-binding site of Escherichia coli SecA, translocation ATPase. J. Biol. Chem. 1996;271:17439–17444. doi: 10.1074/jbc.271.29.17439. [DOI] [PubMed] [Google Scholar]
- 21.Deyrup A. T., Krishnan S., Cockburn B. N., Schwartz N. B. Deletion and site-directed mutagenesis of the ATP-binding motif (P-loop) in the bifunctional murine ATP-sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. J. Biol. Chem. 1998;273:9450–9456. doi: 10.1074/jbc.273.16.9450. [DOI] [PubMed] [Google Scholar]
- 22.Zhou T., Rosen B. P. Asp45 is a Mg2+ ligand in the ArsA ATPase. J. Biol. Chem. 1999;274:13854–13858. doi: 10.1074/jbc.274.20.13854. [DOI] [PubMed] [Google Scholar]
- 23.Lakowicz J. R. New York: Plenum Press; 1999. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 24.Yura T., Nagai H., Mori H. Regulation of the heat-shock response in bacteria. Annu. Rev. Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 25.Fisher A. J., Smith C. A., Thoden J. B., Smith R., Sutoh K., Holden H. M., Rayment I. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 26.Vernos I., Karsenti E. Motors involved in spindle assembly and chromosome segregation. Curr. Opin. Cell Biol. 1996;8:4–9. doi: 10.1016/s0955-0674(96)80041-x. [DOI] [PubMed] [Google Scholar]
- 27.Yang W. Structure and function of mismatch repair proteins. Mutat. Res. 2000;460:245–256. doi: 10.1016/s0921-8777(00)00030-6. [DOI] [PubMed] [Google Scholar]