Abstract
UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase is a bifunctional enzyme, which initiates and regulates sialic acid biosynthesis. Sialic acids are important compounds of mammalian glycoconjugates, mediating several biological processes, such as cell–cell or cell–matrix interactions. In order to characterize the function of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase, a number of deletion mutants were generated, lacking either parts of the N-terminal epimerase or the C-terminal kinase domain. N-terminal deletion of only 39 amino acids results in a complete loss of epimerase activity. Deletions in the C-terminal part result in a reduction or complete loss of kinase activity, depending on the size of the deletion. Deletions at either the N- or the C-terminus also result in a reduction of the other enzyme activity. These results indicate that a separate expression of both domains is possible, but that a strong intramolecular dependency of the two domains has arisen during evolution of the enzyme. N-terminal, as well as C-terminal, mutants tend to form trimers, in addition to the hexameric structure of the native enzyme. These results and yeast two-hybrid experiments show that structures required for dimerization are localized within the kinase domain, and a potential trimerization site is possibly located in a region between the two domains. In conclusion, our results reveal that the activities, as well as the oligomeric structure, of this bifunctional enzyme seem to be organized and regulated in a complex manner.
Keywords: deletion mutant, domain, sialic acid, UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase, yeast two-hybrid assay
Abbreviations: GlcNAc, N-acetylglucosamine; ManNAc, N-acetylmannosamine; Neu5Ac, N-acetylneuraminic acid
INTRODUCTION
Sialic acids are essential components of glycoconjugates on deuterostomia cells [1]. They have been shown to be involved in many biological processes, such as the formation or masking of recognition determinants [2] and stabilization of glycoprotein structures [3]. Sialic acids also serve as ligands for bacterial adhesins, mediating, for example, the binding of Escherichia coli to its host cells [4]. Viral receptors, such as the haemagglutinin of the influenza virus, play important roles during infection processes [5]. Two major groups of proteins, selectins and siglecs (sialic-acid-binding, immunoglobulin-like lectins), act as sialic-acid-binding lectins in mammalian cell–cell interactions during physiological and pathological processes [6–8].
Neu5Ac (N-acetylneuraminic acid) is the biosynthetic precursor of nearly all of the 50 naturally occurring sialic acids [9]. In mammals, the biosynthesis is initiated by UDP-GlcNAc 2-epimerase (UDP-N-acetylglucosamine 2-epimerase), which synthesizes ManNAc (N-acetylmannosamine) from UDP-GlcNAc [10]. ManNAc is then phosphorylated at C-6 by a specific ManNAc kinase [11,12]. It has been shown that, in mammals, these two enzymes are combined in a single bifunctional enzyme, UDP-GlcNAc 2-epimerase/ManNAc kinase [13,14]. ManNAc 6-phosphate is metabolized further by Neu5Ac-9-phosphate synthase to Neu5Ac 9-phosphate [12,15]. After release of the phosphate, Neu5Ac is activated by CMP-Neu5Ac synthase [16]. The end-product of the biosynthesis, CMP-Neu5Ac, serves as substrate for the sialyltransferases in glycoconjugate biosynthesis [17].
It has been shown that UDP-GlcNAc 2-epimerase is the key enzyme of Neu5Ac biosynthesis, since its activity is feedback-inhibited by CMP-Neu5Ac [18]. In sialuria, a sialic acid storage disorder, free sialic acid accumulates due to a defect in the regulation of UDP-GlcNAc 2-epimerase by CMP-Neu5Ac [19]. More recently, a second disorder, hereditary inclusion body myopathy, has been shown to be associated with point mutations in the UDP-GlcNAc 2-epimerase/ManNAc kinase gene [20]. Furthermore, UDP-GlcNAc 2-epimerase/ManNAc kinase is a major determinant of cell-surface sialylation [21] and a regulator of the function of specific cell-surface-adhesion molecules [22]. A specific knockout of the gene in mice is lethal to the embryo at day 8.5 [23].
UDP-GlcNAc 2-epimerase/ManNAc kinase has been cloned and characterized from rats, mice and humans [14,24,25]. The bifunctional enzyme consists of 722 amino acids and has a molecular mass of 79 kDa. Natively, it assembles as a hexamer, possessing both enzyme activities, but under certain conditions dimeric or trimeric forms have also been observed [13,26]. UDP-GlcNAc 2-epimerase and ManNAc kinase activities have been mapped to different regions of the polypeptide, suggesting two functional domains. The epimerase domain is located at the N-terminus, whereas the kinase domain is found at the C-terminus [26]. In the present study, we analysed the structure and function of the bifunctional enzyme in more detail by the generation of several N- and C-terminal deletion mutants. When one domain was mutated, the other could still be expressed, but not without a drastic decrease in both enzyme activities, indicating a strong mutual adaptation of the two domains during evolution. Furthermore, structures responsible for subunit oligomerization were identified.
MATERIALS AND METHODS
Generation of deletion mutants of UDP-GlcNAc 2-epimerase/ManNAc kinase
C-terminal deletions of UDP-GlcNAc 2-epimerase/ManNAc kinase were made by introducing additional stop codons, using the QuikChange™ site-directed mutagenesis kit (Stratagene). Rat cDNA of UDP-GlcNAc 2-epimerase/ManNAc kinase, cloned into the pFastBacHTA vector [27], was used as template DNA. The primers used are listed in Table 1. Temperature cycling amplification was performed with PfuTurbo® DNA polymerase (Stratagene). Parental template DNA was then digested with DpnI. The synthesized plasmids containing the desired mutation were transformed into supercompetent InvαF' E. coli cells (Invitrogen). All constructs were verified via sequencing of the whole UDP-GlcNAc 2-epimerase/ManNAc kinase gene.
Table 1. Oligonucleotide primers used for construction of deletion mutants of UDP-GlcNAc 2-epimerase/ManNAc kinase.
Primers were used for site-directed mutagenesis, mismatches with the template DNA are underlined.
| Mutant | Primer sequence |
|---|---|
| Δ383–722 | 5′-TTCAAGAGCCATGACAGAAGAAATTCTGC-3′ |
| Δ490–722 | 5′-GCTGCACTCGACCTAGTAGATACAGGAGTGG-3 |
| Δ597–722 | 5′-GGAATGGCCTTGTAGAGGGAAGCAAAGAAGC-3′ |
| Δ697–722 | 5′-GGATGTGGATGTATAGGTTTCAGACTTGG-3′ |
| Δ717–722 | 5′-GGTTCTGGACTACTAGACCCGCAGGATCC-3′ |
| Δ1–359 | 5′-GGTAAACAGTACCCTTGCTCGAGGATATATGGGG-3′ |
For the generation of N-terminal-deletion mutants, Δ1–39 and Δ1–234 fragments of the UDP-GlcNAc 2-epimerase/ManNAc kinase cDNA were excised with the restriction enzymes SstI and NspV respectively. For generation of the cDNA for Δ1–359, an additional XhoI cleavage site was introduced by site-directed mutagenesis before digestion. cDNAs were cloned into the correct pFastBacHT vectors for generation of fusion proteins with N-terminal His6 tags.
Expression of proteins in Sf9 insect cells
Both the wild-type and the deletion mutants of UDP-GlcNAc 2-epimerase/ManNAc kinase were expressed in insect cells using the Bac-to-Bac® System (GibcoBRL) as described previously [27]. In brief, constructs of the deletion mutants were transformed into DH10Bac E. coli cells for transposition into bacmid DNA. Recombinant bacmid DNA was isolated and used for transfection of Sf9 insect cells, which release recombinant baculoviruses containing the modified UDP-GlcNAc 2-epimerase/ManNAc kinase genes. The presence of the respective genes in the baculo-viruses was monitored by PCR. For this purpose, 25 μl of virus solution was mixed with 5 μl of 10% (w/v) SDS and 5 μl of proteinase K (20 mg/ml) and water to give a final volume of 110 μl, before incubation for 30 min at 37 °C. DNA was isolated by phenol/chloroform extraction. After ethanol precipitation, the DNA was resuspended in 5 μl of water and used directly for PCR with the primers 5′-ACCTATAAATATTCCGGATT-3′ and 5′-AAAGCAAGTAAAACCTCTAC-3′. After 35 cycles, the amplified products were analysed by agarose gel electrophoresis.
For overexpression, Sf-900 insect cells with a density of 2×106 cells/ml were infected with recombinant UDP-GlcNAc 2-epimerase/ManNAc kinase baculoviruses at MOI (multiplicity of infection) of 3 and shaken at 27 °C for 48 h. Cells were harvested by centrifugation at 700 g for 5 min at room temperature (20 °C), then resuspended in lysis buffer (10 mM sodium phosphate, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol and 1 mM PMSF; 1 ml per 10 ml of cell culture) and lysed by sonication. After centrifugation at 20000 g for 30 min at 4 °C, the supernatant was used for detection of UDP-GlcNAc 2-epimerase/ManNAc kinase.
Determination of enzymic activities and protein concentration
Before enzyme activity assays, in order to remove protein aggregates >600 kDa, samples of the different mutants were subjected to gel-filtration chromatography (see below). UDP-GlcNAc 2-epimerase activity was then determined with a radiometric [13] or a colorimetric assay [28]. ManNAc kinase activity was assayed radiometrically as described previously [13]. One unit of enzyme activity was defined as the formation of 1 μmol of product per min at 37 °C. Specific activities were expressed as m-units/mg of protein.
To calculate the amount of UDP-GlcNAc 2-epimerase/Man-NAc kinase protein, aliquots were subjected to a SDS/7.5% PAGE using a Mini Protean II system (Bio-Rad). If necessary, proteins were concentrated by acetone precipitation before analysis. For this purpose, 4 vol. of acetone were mixed with the sample and incubated for at least 1 h at −20 °C. Precipitated proteins were centrifuged at 2000 g for 10 min at 4 °C. The pellets were dried and resuspended in sample buffer. Separated proteins were transferred on to a nitrocellulose membrane, and the UDP-GlcNAc 2-epimerase/ManNAc kinase was detected with the monoclonal antibodies H-15 (Santa Cruz) or Penta-His (Qiagen) specific to the His6-tag epitope. Bands were visualized by the ECL® (enhanced chemiluminescence) kit (Amersham Biosciences) and quantified on a LAS-1000 Fuji imager RayTest with the Image Gauge V3.4 program.
Determination of oligomeric structures
The oligomeric state of wild-type and mutated UDP-GlcNAc 2-epimerase/ManNAc kinase was determined with freshly prepared insect cell cytosol by gel filtration on a Superdex® 200 HR 10/30 column (Amersham Biosciences). A buffer containing 10 mM sodium phosphate, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol and 100 mM NaCl was used as eluent. At a flow rate of 0.2 ml/min, fractions of 0.5 ml were collected and assayed for enzyme activities. The column was calibrated with a protein mixture of thyroglobulin (670 kDa), γ-globulin (158 kDa) and ovalbumin (44 kDa).
Generation of yeast two-hybrid constructs and two-hybrid experiments
Rat cDNA of the UDP-GlcNAc 2-epimerase/ManNAc kinase cloned into pFastBacHTA vector served as template for the amplification of cDNAs for expression of the constructs depicted in Figure 6. For amplification of the cDNA of the constructs, primers (Table 2) with the following combinations were used: construct 1, primers 1 and 2; construct 2, primers 1 and 3; construct 3, primers 4 and 2; construct 4, primers 5 and 6; construct 5, primers 1 and 7; construct 6, primers 8 and 9; construct 7, primers 10 and 3; construct 8, primers 4 and 11; construct 9, primers 12 and 13; construct 10, primers 14 and 12; construct 11, primers 1 and 9; construct 12, primers 8 and 3; construct 13, primers 4 and 13; construct 14, primers 12 and 3. The primers introduced additional SalI and NotI restriction sites for cloning the cDNAs as N-terminal fusions with the LexA DNA-binding and the Gal4-activation domain of the yeast two-hybrid vectors pBTM117c and pGAD426 respectively [29]. All constructs were checked by DNA sequencing. LexA DNA-binding domain fusions (‘baits’) were expressed in L40ccua [MATa] and the activation domain fusion proteins (‘prey’) were expressed in yeast strain L40ccα [MATα]. L40ccα clones were mixed with L40ccua clones for interaction mating and grown on YPD [1% (w/v) yeast extract, 2% (w/v) peptone and 2% (w/v) glucose] agar plates for 24 h at 30 °C. Cells were transferred on to SDII agar plates (minimal medium lacking tryptophan and leucine), and diploid cells were grown for 72 h at 30 °C. For the selection of interactions, diploid cells were transferred onto SDIV agar plates (minimal medium lacking tryptophan, leucine, histidine and uracil) with and without nylon membranes, and incubated for 5 days at 30 °C. For the identification of protein–protein interactions, the activity of the (lexAop)4-HIS3 and the (lexAop)8-URA3 reporter genes was assessed via growth on SDVI. The nylon membranes were subjected to a β-galactosidase assay, which enabled the simultaneous examination of the (lexAop)4-HIS3, (lexAop)8-URA3 and the (lexAop)8-lacZ reporters.
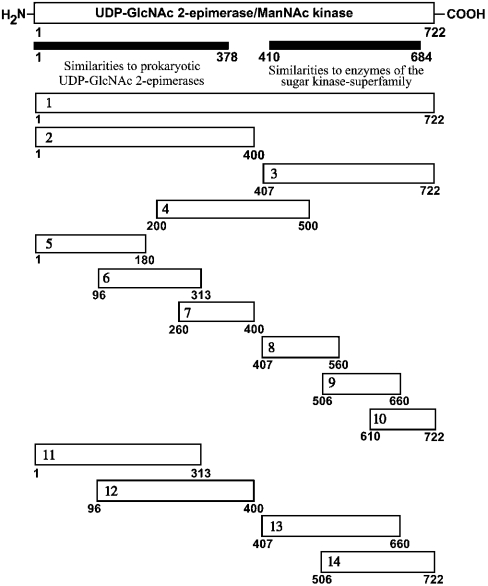
Figure 6. Schematic overview of the constructs of UDP-GlcNAc 2-epimerase/ManNAc kinase generated for the yeast two-hybrid experiments.
cDNAs were generated by PCR and cloned into the bait vector pBTM117c and the prey vector pGAD426 as described in the Materials and methods section.
Table 2. Oligonucleotide primers used for construction of deletion mutants of UDP-GlcNAc 2-epimerase/ManNAc kinase for the yeast two-hybrid experiments.
Primers 1–14 were used for amplification of cDNAs for generation of yeast two-hybrid constructs (homologies with UDP-GlcNAc 2-epimerase/ManNAc kinase sequence are underlined).
| Name | Sequence |
|---|---|
| Primer 1 | 5′-GTCGACGTCGACAATGGAGAAGAACGGGAATAACCGG-3′ |
| Primer 2 | 5′-GCGGCCGCCTAGTGGATTGCGGGCG-3′ |
| Primer 3 | 5′-GCGGCGCGCGGCCGCAATATCCTGAGAGATGTTCT-3′ |
| Primer 4 | 5′-GTCGACGTCGACGAGTGCCTTGGCTGTTGATC-3′ |
| Primer 5 | 5′-GTCGACGTCGACTATTCGGATGTGGCTAGGTTGA-3′ |
| Primer 6 | 5′-GCGGCCGCGAGGTCCACAGAGTTCCACT-3′ |
| Primer 7 | 5′-GCGGCCGCCAAAAGGATGCGGTCGTGGT-3′ |
| Primer 8 | 5′-GTCGACGTCGACGGATGTCCTCAACCGCCTGAAGC-3 |
| Primer 9 | 5′-GCGGCCGCGGGTTCCAAAGGCGCCAACC-3′ |
| Primer 10 | 5′-GTCGACGTCGACGATGGTTCGAGTGATGCGGAAG-3′ |
| Primer 11 | 5′-GCGGCCGCGCTGCCGTGGATCAGCTCGT-3′ |
| Primer 12 | 5′-GTCGACGTCGACACCCGTGTGGGTGGACAACGA-3′ |
| Primer 13 | 5′-GCGGCCGCGCGGCCGCCATAGTGTGGAGGATATTCA-3′ |
| Primer 14 | 5′-GTCGACGTCGACAGTGGAAGGGATGTCAGTGCC-3′ |
RESULTS
Generation of deletion mutants
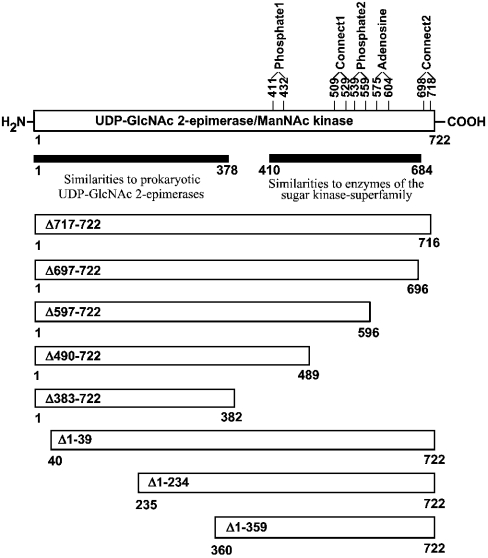
To examine whether the two catalytic functions of the bifunctional enzyme UDP-GlcNAc 2-epimerase/ManNAc kinase can be expressed separately, mutants with different sizes of deletions in the UDP-GlcNAc 2-epimerase and the ManNAc kinase domain were generated (Figure 1). Deletion mutants in the C-terminal ManNAc kinase domain were generated by insertion of earlier stop codons into the UDP-GlcNAc 2-epimerase/ManNAc kinase cDNA. It is well known that sugar kinases contain five subdomains (phosphate1, phosphate2, adenosine, connect1 and connect2) that are responsible for binding distinct parts of the substrate ATP [30]. All subdomains are present in the ManNAc kinase domain (Figure 1) and served as the structural basis for generation of the C-terminal mutants. Mutant Δ717–722 contains all the subdomains, whereas Δ697–722 lacks the connect2 motif. Mutant Δ597–722 additionally lacks the region between connect2 and adenosine, while Δ490–722 lacks four of the five subdomains. Finally, Δ383–722 only consists of the putative UDP-GlcNAc 2-epimerase domain.
Figure 1. Overview of resulting proteins of mutated UDP-GlcNAc 2-epimerase/ManNAc kinase cDNA.
cDNAs for expression were generated by restriction cleavage of different fragments of the coding UDP-GlcNAc 2-epimerase/ManNAc kinase sequence. C-terminal-deletion mutations were generated in the UDP-GlcNAc 2-epimerase/ManNAc kinase cDNA with the QuikChange™ site-directed mutagenesis kit by insertion of additional stop codons.
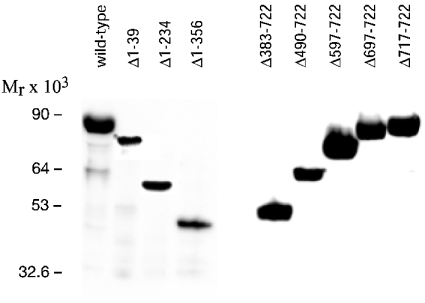
Deletions in the N-terminal UDP-GlcNAc 2-epimerase domain were generated by excising different fragments of the 5′-coding region of the UDP-GlcNAc 2-epimerase/ManNAc kinase cDNA. Since information on potential subdomains of this part of the protein is lacking, naturally occurring restriction sites or restriction sites introduced by site-directed mutagenesis were used. Constructs containing the deleted UDP-GlcNAc 2-epimerase/ManNAc kinase gene were transfected into Sf9 insect cells, which then produced recombinant baculoviruses. The presence of the correct cDNA in the baculoviruses was shown by PCR analysis, followed by sequencing of the PCR product. The recombinant baculoviruses were used for infection of insect cells. The harvested insect cells were lysed, and aliquots were analysed by Western blotting. All deletion mutants of the UDP-GlcNAc 2-epimerase/ManNAc kinase produced overexpressed proteins of the expected size (Figure 2). The wild-type enzyme showed a molecular mass of 85 kDa, which is in agreement with the predicted molecular mass of UDP-GlcNAc 2-epimerase/ManNAc kinase of 79 kDa and the size of the His6 tag of 6 kDa. This result also shows that UDP-GlcNAc 2-epimerase/ManNAc kinase is expressed without glycosylation, in agreement with the cytosolic localization of the enzyme. The sizes of the N-terminal mutant proteins were 79 kDa, 58 kDa and 45 kDa, whereas the sizes of the C-terminal mutant proteins were 85 kDa, 83 kDa, 72 kDa, 61 kDa and 49 kDa. In insect cells, soluble, wild-type UDP-GlcNAc 2-epimerase/ManNAc kinase typically represented 30–50% of the total protein expression [27]. Similar expression levels were observed for the Δ717–722 mutant. But in all other cases between 5% and 30%, and in a few cases <5%, of total protein was soluble (results not shown).
Figure 2. Western blot analysis of the UDP-GlcNAc 2-epimerase/ManNAc kinase deletion mutants expressed in insect cells.
Sf-900 insect cells were infected with recombinant baculoviruses containing the mutated UDP-GlcNAc 2-epimerase/ManNAc kinase gene. After cell lysis, 20 μg of protein were applied to a SDS/7.5% PAGE and analysed by Western blotting using the H-15 antibody.
Enzymic activities of the deletion mutants
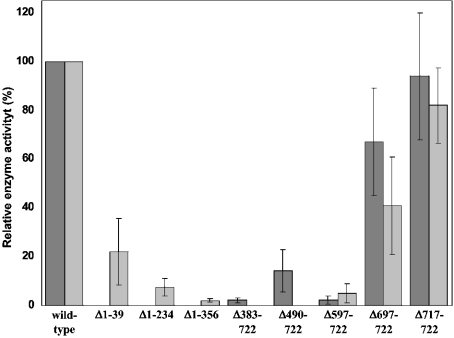
Before the determination of enzymic activities, samples of the different mutants were subjected to gel-filtration chromatography. This method separated aggregates of UDP-GlcNAc 2-epimerase/ManNAc kinase with a size >2000 kDa, which were obtained as by-products of protein expression in insect cells, from fractions with proteins <600 kDa, representing native UDP-GlcNAc 2-epimerase/ManNAc kinase. All fractions were assayed for UDP-GlcNAc 2-epimerase and ManNAc kinase activity. In parallel, the amount of active UDP-GlcNAc 2-epimerase/ManNAc kinase in each fraction was determined by Western blotting. The specific activities of the aggregates were much lower than the activities of the respective native enzyme, as already shown earlier for the wild-type enzyme [27]. Therefore relative specific enzyme activities of the mutants with respect to the wild-type enzyme were determined only for fractions with proteins <600 kDa (Figure 3). All of the N-terminal-deletion mutants display a complete loss of UDP-GlcNAc 2-epimerase activity. The ManNAc kinase activity was also drastically reduced. The Δ1–39 mutant has only 22% ManNAc kinase activity, compared with the wild-type enzyme. The activities of the other mutants are below 10%. These results show that the first 39 amino acids are essential for UDP-GlcNAc 2-epimerase activity.
Figure 3. Relative specific enzyme activities of the UDP-GlcNAc 2-epimerase/ManNAc kinase deletion mutants.
Proteins were expressed in Sf-900 insect cells. After cell lysis and consecutive centrifugation, the supernatant was applied to the Superdex® 200 column in order to remove protein aggregates. Fractions containing the native enzymes with a size <600 kDa were analysed for UDP-GlcNAc 2-epimerase and ManNAc kinase activity using radiometric assays. Relative specific enzyme activities±S.D. (n=4) were calculated from the ratio of measured enzyme activity to the amount of UDP-GlcNAc 2-epimerase/ManNAc kinase detected by Western blotting. The calculated activities represent the average of all fractions from each mutant. Dark grey bars, UDP-GlcNAc 2-epimerase activity; light grey bars, ManNAc kinase activity.
The results for the C-terminal-deletion mutants were more diverse than for the N-terminal-deletion mutants. Loss of the last six amino acids does not markedly influence the ManNAc kinase activity; however, a loss of the next 20 amino acids reduces the kinase activity by 60%. This indicates a significant, but not essential, role for the connect2 subdomain in the ManNAc kinase activity. Deletions of larger parts of the protein then result in further reduction of the kinase activity. The Δ597–722 mutant, containing four of the five ATP-binding subdomains, retains about 5% kinase activity, whereas further deletions result in a total loss of kinase activity. On the other hand, all C-terminal mutants still display UDP-GlcNAc 2-epimerase activity. Loss of the last six and 26 amino acids respectively results in a moderate reduction of epimerase activity. Larger deletions then result in a further significant reduction of enzyme activities, but these mutants still retain epimerase activity, indicating that they still have an enzymically active epimerase domain. Interestingly, the Δ490–722 mutant showed an approx. 4-fold higher UDP-GlcNAc 2-epimerase activity than that of Δ597–722. Probably, the loss of amino acids 490–597 has a positive effect on the stability of some epimerase domain structures essential for the activity.
Finally, all the deletion mutants that still retained some epimerase activity were investigated for inhibition by CMP-Neu5Ac, the feedback inhibitor of the UDP-GlcNAc 2-epimerase. The epimerase activity of all C-terminal mutants was inhibited by 100 μM CMP-Neu5Ac (results not shown). In addition, the fractions after gel-filtration chromatography which contain trimeric protein of the mutants Δ490–722 and Δ383–722 (see below) were analysed. For this oligomeric form, inhibition of epimerase activity by 100 μM CMP-Neu5Ac was also found. These results indicate that all essential structures for inhibition must be located within the epimerase domain and that they are still functional when only trimeric protein is formed. This is in agreement with the postulated binding site for CMP-sialic acid between amino acids 249 and 275 [31].
Oligomeric structure of the deletion mutants
UDP-GlcNAc 2-epimerase/ManNAc kinase assembles natively as a homohexamer of 75 kDa subunits [13], but, under certain conditions, dimeric and trimeric forms were also observed [13,26,28]. Therefore the oligomeric state of the deletion mutants was determined by gel-filtration chromatography. The presence of the respective enzymes was established by detection of ManNAc kinase activity in N-terminal mutants and UDP-GlcNAc 2-epimerase activity in C-terminal mutants (Figures 4 and 5). The presence of UDP-GlcNAc 2-epimerase/ManNAc kinase protein was confirmed by Western blotting using the H-15 antibody (results not shown). Native molecular masses of the deletion mutants were determined by comparison with a molecular-mass standard mixture.
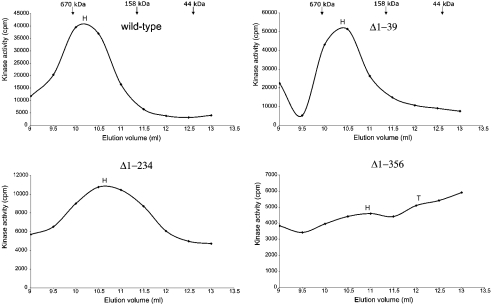
Figure 4. Determination of the oligomeric state of N-terminal-deletion mutants.
Sf-900 insect cells were infected with recombinant baculoviruses, which contain the mutated UDP-GlcNAc 2-epimerase/ManNAc kinase gene. The cytosolic fraction was applied to a gel-filtration column. Enzymic activity in the fractions was detected by the radiometric ManNAc kinase assay and plotted against the elution volume. H and T indicate the elution volumes corresponding to the respective hexamer or trimer of the investigated protein. Since the mutants possessed different activities, different amounts of protein were applied to the column.
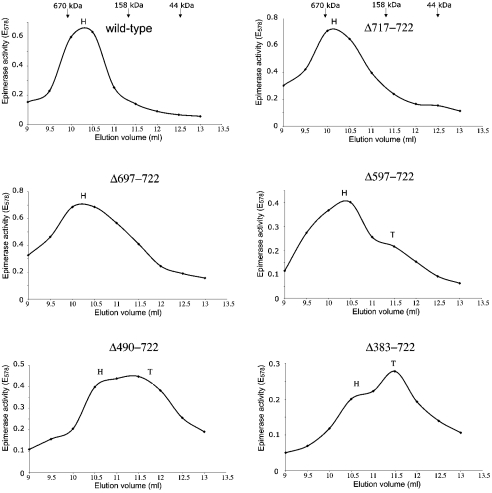
Figure 5. Determination of the oligomeric state of C-terminal-deletion mutants.
Sf-900 insect cells were infected with recombinant baculoviruses, which contain the mutated UDP-GlcNAc 2-epimerase/ManNAc kinase gene. The cytosolic fraction was applied to a gel-filtration column. Enzymic activity in the fractions was detected with the colorimetric UDP-GlcNAc 2-epimerase assay and plotted against the elution volume. H and T indicate the elution volumes corresponding to the respective hexamer or trimer of the investigated protein. Since the mutants possessed different activities, different amounts of protein were applied to the column.
As estimated for a hexamer, the UDP-GlcNAc 2-epimerase/ManNAc kinase wild-type enzyme had a molecular mass of 500 kDa (Figure 4). The N-terminal-deletion mutants Δ1–39, Δ1–234 and Δ1–356 were eluted in fractions corresponding to native molecular masses of 420 kDa, 330 kDa and 290 kDa respectively. This indicates that the deletion mutants also form hexamers. The inserted deletions have no significant influence on the oligomeric structure of the mutated proteins, suggesting that amino acids 1–356 are not involved in the oligomerization process of UDP-GlcNAc 2-epimerase/ManNAc kinase. However, the main elution peaks of the deleted UDP-GlcNAc 2-epimerase/ManNAc kinase proteins were accompanied by a second peak. For the Δ1–356 mutant, the size of this peak was sufficient to calculate its molecular mass as 150 kDa, representing a trimer of the protein.
The C-terminal-deletion mutants Δ717–722, Δ697–722 and Δ597–722 were eluted at volumes corresponding to native molecular masses of 500 kDa, 480 kDa and 410 kDa respectively (Figure 5). Therefore these C-terminal deletion mutants form hexamers comparable with the wild-type enzyme. However, for the Δ597–722 mutant, a small peak corresponding to a trimeric protein is visible. For Δ490–722 and Δ383–722, two peaks are visible, one corresponding to a hexameric protein and another to a trimeric protein (Figure 5). In Δ490–722, the ratio of hexamer to trimer is nearly 1:1, whereas trimers dominate in Δ383–722.
Yeast two-hybrid experiments
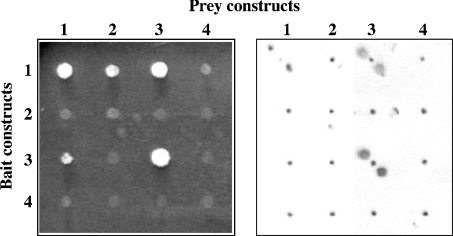
In order to get a more detailed insight into the interactions of epimerase and kinase domains during oligomer formation, we used the yeast two-hybrid system with the complete protein or several deleted variants. A total of 14 different bait and prey constructs were generated (Figure 6) for the pairwise interaction mating in a two-hybrid system. All 154 combinations were assayed for the activation of the HIS3 and URA3 reporter genes via growth on minimal medium (Figure 7). In addition, simultaneous activation of the HIS3, the URA3 and the lacZ reporter genes was examined in a β-galactosidase assay (Figure 7). In both assays, interactions were only found between the constructs 1, 2 and 3, representing the whole enzyme or the complete epimerase and kinase domains respectively (Table 3). Strong interactions, indicated by a strong signal in the β-galactosidase assay, were found for all constructs containing the whole kinase domain in both fusion proteins (i.e. 1 compared with 1, 1 compared with 3, 3 compared with 1, and 3 compared with 3). However, growth on selective medium could also be detected when only the epimerase domain was present in one or both fusion proteins (i.e. 1 compared with 2). Smaller parts of UDP-GlcNAc 2-epimerase/ManNAc kinase did not show any interaction in the yeast two-hybrid assay, indicating that the whole domains are needed for the interactions.
Figure 7. Results of yeast two-hybrid experiment.
Left-hand panel, colony growth on minimal medium. Right-hand panel, β-galactosidase assay. Assays were performed as described in the Materials and methods section. For the β-galactosidase assay, two spots were applied for each sample.
Table 3. Results of the yeast two-hybrid experiments.
Constructs 1–14 (Figure 6) were cloned into the bait vector pBTM117c and into the prey vector pGAD426 and analysed for protein–protein interaction in yeast two-hybrid experiments as described in the Materials and methods section. Interactions were analysed by growth on agar plates and by a β-galacosidase assay. The relative strength of interaction is given as no interaction (−), very weak interaction (−/+), weak interaction (+), strong interaction (++).
| Prey constructs | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bait constructs | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
| 1 | ++ | −/+ | ++ | − | − | − | − | − | − | − | − | − | − | − |
| 2 | − | −/+ | − | − | − | − | − | − | − | − | − | − | − | − |
| 3 | + | − | ++ | − | − | − | − | − | − | − | − | − | − | − |
| 4 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 5 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 6 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 9 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 10 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 11 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 12 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 13 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 14 | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
DISCUSSION
UDP-GlcNAc 2-epimerase/ManNAc kinase is the bifunctional enzyme that catalyses the first two steps of the biosynthesis of sialic acids, and it is also important for pathway regulation. In this work, we investigated the functions of the two different domains of UDP-GlcNAc 2-epimerase/ManNAc kinase by generating several N- and C-terminal-deletion mutants. In a previous study, we inserted different point mutations in the bifunctional enzyme, and were able to show that the epimerase and the kinase of the bifunctional enzyme function independently of each other [26]. We therefore examined whether these two functions can also be separated physically. Our results show that the UDP-GlcNAc 2-epimerase, as well as the ManNAc kinase, can be functionally expressed as separated domains. This indicates that neither domain is essential for the enzyme activity of the other. But the loss of even part of the other domain causes a more or less drastic reduction of the enzyme activity. Structures in each domain are therefore required for optimal enzyme activity of the other domain. Prokaryotic organisms possess monofunctional homologues of the two domains of the mammalian UDP-GlcNAc 2-epimerase/ManNAc kinase [26]. Therefore it is likely that the bifunctional enzyme resulted from the fusion of two formerly separated genes. This gene fusion must have happened early in evolution, since a dissociation of both functional domains is still possible today, although it results in a drastic reduction of the enzyme activities. After gene fusion, mutations in both domains of the bifunctional enzyme resulted in a strong mutual adaptation. Bifunctional enzymes are quite rare in mammalian metabolism, and, to date, only a few cases are known where one or both domains have been separated physically. The functions of the domains of 3′-phosphoadenosine 5′-phosphosulphate synthase can be expressed separately [32], as well as the sulphotransferase domain of heparan sulphate/heparin N-deacetylase/N-sulphotransferase [33].
We have shown that amino acids 1–39 play an important role in UDP-GlcNAc 2-epimerase/ManNAc kinase. Deletion of these amino acids causes a complete loss of the UDP-GlcNAc 2-epimerase activity, suggesting that these amino acids are essential for the epimerase activity. Therefore it can be concluded that some of these amino acids may be part of the active site of the UDP-GlcNAc 2-epimerase or are important for its formation. At the same time, loss of these first 39 amino acids causes a drastic loss of ManNAc kinase activity. As the Δ1–39 mutant still shows kinase activity, these amino acids are not essential for ManNAc kinase activity, although they influence it strongly. Without knowledge of the three-dimensional structure, the possibility that the first 39 amino acids play a direct role in ManNAc kinase activity cannot be excluded, but it is more probable that the epimerase domain stabilizes important structural elements of the kinase domain.
Sugar kinases have five conserved subdomains for binding of ATP [30], which are also found in the ManNAc kinase domain (Figure 1). Loss of the connect2 motive subdomain results in a reduction of ManNAc kinase activity of 60%, indicating that this motif is important, but not essential, for ATP binding. Loss of the amino acids between the connect2 and the adenosine motifs reduced the kinase activity nearly to its detection limit, indicating that these amino acids contribute significantly to the ManNAc kinase activity. These amino acids between the adenosine and the connect2 motifs may be associated with stabilization of catalytically important structures in the ManNAc kinase domain, rather than interacting directly in the catalytic process. Deletion of further motifs results in a complete loss of ManNAc kinase activity. These deletions in the C-terminal part of the enzyme also have an influence on the epimerase domain of the bifunctional enzyme. Indirect allosteric effects on the structure or stability of the protein are thought to be responsible for these effects.
The inserted deletions in the bifunctional UDP-GlcNAc 2-epimerase/ManNAc kinase influence not only the enzyme activities, but also the oligomeric states of the mutated enzymes. The wild-type and all deletion mutants of the UDP-GlcNAc 2-epimerase/ManNAc kinase assemble as hexamers or trimers. Besides detection of trimers by gel filtration in the present and a previous study [26], they were also found by an alternative method. Cross-linking of the enzyme with periodate-oxidized UDP-GlcNAc in a time-dependent manner, followed by SDS/PAGE, revealed formation of cross-linked trimers and hexamers [28], thus complementing the gel-filtration experiments. Strong negative cooperativity was shown earlier for the substrate UDP-GlcNAc, which suggests the existence of more than one active site for UDP-GlcNAc 2-epimerase activity [13]. In the present study, trimers with UDP-GlcNAc 2-epimerase activity were detected, and provide the first structural evidence for more than one active UDP-GlcNAc 2-epimerase site in the hexameric protein.
In its native state, the UDP-GlcNAc 2-epimerase/ManNAc kinase exists as a hexamer. This structure is thought to consist either of a trimer of dimers [13] or a dimer of trimers [28]. N-terminal deletion mutants still assemble as hexamers identical with the wild-type enzyme. This indicates that amino acids of the UDP-GlcNAc 2-epimerase domain have little or no involvement in the oligomerization process. These results are supported by the yeast two-hybrid experiments, showing strong interaction between constructs containing the complete kinase domain, but no or only very weak interactions for epimerase domain constructs. C-terminal deletions of approx. 100 amino acids did not significantly influence the oligomeric structure of the enzyme, as these deletions do not cause a loss of the hexameric structure. Larger deletions obviously resulted in a reduced stability of the hexameric enzyme, as indicated by the occurrence of trimers. Therefore it can be concluded that important structures responsible for the formation of hexamers are located in the amino acid sequence 235–596. In particular, deletion of amino acids 383–596 causes a drastic increase of trimeric structures, suggesting that this region contains important structures for dimerization. A potential subdomain for trimerization is possibly located around amino acids 360–382, a region which cannot be unequivocally assigned to one of the two domains, as all mutated proteins containing this part were found as hexamers or trimers, but not as dimers or monomers.
In the yeast two-hybrid experiments, the complete kinase domain showed strong interactions with itself and the entire bifunctional enzyme. For all other constructs, no interactions were detected, suggesting that essential structures for oligomerization were missing. We note that the constructs expressing amino acid sequences 200–500 and 407–660 did not show interactions, although the gel-filtration experiments suggested that they include structures promoting trimerization and dimerization. Failure to detect these interactions may be attributable to the use of different constructs in the yeast two-hybrid experiments. The respective deletion mutants, which still assemble as hexamers and trimers in gel-filtration experiments, contain the complete epimerase domain, whereas the two-hybrid fragments used are lacking larger parts of it. In order to avoid false-positive signals, the yeast two-hybrid system used here is particularly designed for high stringency, which is achieved by the simultaneous use of three reporter genes and very low protein expression levels. This could also account for the fact that we did not observe interactions with the smaller fragments.
Our results show that the hexameric structure of UDP-GlcNAc 2-epimerase/ManNAc kinase is regulated in a more complex manner by secondary, and possibly also tertiary, structures and cannot be reduced to one simple dimerization or trimerization site. It is also possible that the trimeric structures are artifacts, resulting from the inability of the deletion mutants to form hexamers. In this case, the trimeric structures are not native states of the hexamers. This would also be consistent with the detection of at least small amounts of hexamers in all the deletion mutants and the fact that the quantity of trimers increases with the size of the deletion. On the other hand, trimers were also found after cross-linking the hexameric structure [28], indicating that trimeric structures in general are part of the hexameric protein.
In conclusion, we were able to express the UDP-GlcNAc 2-epimerase and the ManNAc kinase of the bifunctional enzyme separately from each other. Deletions of even small parts of the enzyme result in a significant reduction of the enzyme activities of both domains, showing that they strongly influence each other. Larger N- and C-terminal deletions result in a reduced stability of the hexameric enzyme and the occurrence of trimers of the protein. The oligomerization process leading to hexamer formation seems to involve complex secondary, and possibly also tertiary, structures of the protein.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Bonn, Germany (Grants Lu 799/1-1 and Ho 1959/5-1), the Fonds der Chemischen Industrie, Bonn, Germany and the Sonnenfeld-Stiftung, Berlin, Germany. Work in the AG Neuroproteomics (MDC Berlin) was supported by the NGFN (KB-P4T03). We thank Dr T. A. Scott (Leeds, U.K.) for improving the English style of the manuscript.
References
- 1.Corfield A. P., Schauer R. Occurrence of sialic acids. In: Schauer R., editor. Sialic Acids. Wien: Springer; 1982. pp. 5–50. [Google Scholar]
- 2.Schauer R., Kelm S., Reuter G., Roggentin P., Shaw L. Biochemistry and role of sialic acids. In: Rosenberg A., editor. Biology of the Sialic Acids. New York: Plenum Press; 1995. pp. 7–67. [Google Scholar]
- 3.Rens-Domiano S., Reisine T. Structural analysis and functional role of the carbohydrate component of somatostatin receptors. J. Biol. Chem. 1991;266:20094–20100. [PubMed] [Google Scholar]
- 4.Sakarya S., Öncü S. Bacterial adhesins and the role of sialic acid in bacterial adhesion. Med. Sci. Monit. 2003;9:RA76–RA82. [PubMed] [Google Scholar]
- 5.Suzuki Y., Ito T., Suzuki T., Holland R. E., Jr, Chambers T. M., Kiso M., Ishida H., Kawaoka Y. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 2000;74:11825–11831. doi: 10.1128/jvi.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowe J. B. Glycosyltransferases and glycan structures contributing to the adhesive activities of L-, E- and P-selectin counter-receptors. Biochem. Soc. Symp. 2002;69:33–45. doi: 10.1042/bss0690033. [DOI] [PubMed] [Google Scholar]
- 7.Sawada R., Tsuboi S., Fukuda M. Differential E-selectin-dependent adhesion efficiency in sublines of a human colon cancer exhibiting distinct metastatic potentials. J. Biol. Chem. 1994;269:11425–11431. [PubMed] [Google Scholar]
- 8.Crocker P. R. Siglecs: sialic-acid-binding immunoglobulin-like lectins in cell–cell interactions and signalling. Curr. Opin. Struct. Biol. 2002;12:609–615. doi: 10.1016/s0959-440x(02)00375-5. [DOI] [PubMed] [Google Scholar]
- 9.Angata T., Varki A. Chemical diversity in the sialic acids and related α-keto acids: an evolutionary perspective. Chem. Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 10.Comb D. G., Roseman S. Enzymatic synthesis of N-acetyl-D-mannosamine. Proc. Natl. Acad. Sci. U.S.A. 1958;29:653–654. doi: 10.1016/0006-3002(58)90031-3. [DOI] [PubMed] [Google Scholar]
- 11.Gosh S., Roseman S. Enzymatic phosphorylation of N-acetylmannosamine. Proc. Natl. Acad. Sci. U.S.A. 1961;47:955–958. doi: 10.1073/pnas.47.7.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren L., Felsenfeld H. N-acetylmannosamine-6-phosphate and N-acetylneuraminic acid-9-phosphate as intermediates in sialic acid biosynthesis. Biochem. Biophys. Res. Commun. 1961;5:185–190. doi: 10.1016/0006-291x(61)90107-3. [DOI] [PubMed] [Google Scholar]
- 13.Hinderlich S., Stäsche R., Zeitler R., Reutter W. A bifunctional enzyme catalyzes the two steps in N-acetylneuraminic acid biosynthesis of rat liver: purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J. Biol. Chem. 1997;272:24313–24318. doi: 10.1074/jbc.272.39.24313. [DOI] [PubMed] [Google Scholar]
- 14.Stäsche R., Hinderlich S., Weise C., Effertz K., Lucka L., Moormann P., Reutter W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver: molecular cloning and functional expression of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J. Biol. Chem. 1997;272:24319–24324. doi: 10.1074/jbc.272.39.24319. [DOI] [PubMed] [Google Scholar]
- 15.Chen H., Blume A., Zimmermann-Kordmann M., Reutter W., Hinderlich S. Purification and characterization of N-acetylneuraminic acid-9-phosphate synthase from rat liver. Glycobiology. 2002;12:65–71. doi: 10.1093/glycob/12.2.65. [DOI] [PubMed] [Google Scholar]
- 16.Münster A. K., Eckhardt M., Potvin B., Mühlenhoff M., Stanley P., Gerardy-Schahn R. Mammalian cytidine 5′-monophosphate N-acetylneuraminic acid synthetase: a nuclear protein with evolutionarily conserved structural motifs. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9140–9145. doi: 10.1073/pnas.95.16.9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harduin-Lepers A., Recchi M. A., Delannoy P. 1994, the year of sialyltransferases. Glycobiology. 1995;5:741–758. doi: 10.1093/glycob/5.8.741. [DOI] [PubMed] [Google Scholar]
- 18.Kornfeld S., Kornfeld R., Neufeld E., O'Brien P. J. The feedback control of sugar nucleotide biosynthesis in liver. Proc. Natl. Acad. Sci. U.S.A. 1964;52:371–379. doi: 10.1073/pnas.52.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seppala R., Lehto V. P., Gahl W. A. Mutations in the human UDP-N-acetylglucosamine 2-epimerase gene define the disease sialuria and the allosteric site of the enzyme. Am. J. Hum. Genet. 1999;64:1563–1569. doi: 10.1086/302411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenberg I., Avidan N., Potikha T., Hochner H., Chen M., Olender T., Barash M., Shemesh M., Sadeh M., Grabov-Nardini G., et al. The UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nat. Genet. 2001;29:83–87. doi: 10.1038/ng718. [DOI] [PubMed] [Google Scholar]
- 21.Keppler O. T., Hinderlich S., Langner J., Schwartz-Albiez R., Reutter W., Pawlita M. UDP-GlcNAc 2-epimerase: a regulator of cell surface sialylation. Science. 1999;284:1372–1376. doi: 10.1126/science.284.5418.1372. [DOI] [PubMed] [Google Scholar]
- 22.Horstkorte R., Rau K., Reutter W., Nöhring S., Lucka L. Increased expression of the selectin ligand sialyl-Lewisx by biochemical engineering of sialic acids. Exp. Cell Res. 2004;295:549–554. doi: 10.1016/j.yexcr.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Schwarzkopf M., Knobeloch K. P., Rhode E., Hinderlich S., Wiechens N., Lucka L., Horak I., Reutter W., Horstkorte R. Sialylation is essential for early development in mice. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5267–5270. doi: 10.1073/pnas.072066199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horstkorte R., Nöhring S., Wiechens N., Schwarzkopf M., Danker K., Reutter W., Lucka L. Tissue expression and amino acid sequence of murine UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase. Eur. J. Biochem. 1999;260:923–927. doi: 10.1046/j.1432-1327.1999.00253.x. [DOI] [PubMed] [Google Scholar]
- 25.Lucka L., Krause M., Danker K., Reutter W., Horstkorte R. Primary structure and expression analysis of human UDP-N-acetyl-glucosamine-2-epimerase/N-acetylmannosamine kinase, the bifunctional enzyme in neuraminic acid biosynthesis. FEBS Lett. 1999;454:341–344. doi: 10.1016/s0014-5793(99)00837-6. [DOI] [PubMed] [Google Scholar]
- 26.Effertz K., Hinderlich S., Reutter W. Selective loss of either the epimerase or kinase activity of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase due to site-directed mutagenesis based on sequence alignments. J. Biol. Chem. 1999;274:28771–28778. doi: 10.1074/jbc.274.40.28771. [DOI] [PubMed] [Google Scholar]
- 27.Blume A., Ghaderi D., Liebig V., Hinderlich S., Donner P., Reutter W., Lucka L. UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase, functionally expressed in and purified from Escherichia coli, yeast, and insect cells. Protein Expression Purif. 2004;35:387–396. doi: 10.1016/j.pep.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Blume A., Chen H., Reutter W., Schmidt R. R., Hinderlich S. 2′,3′-Dialdehydo-UDP-N-acetylglucosamine inhibits UDP-N-acetylglucosamine 2-epimerase, the key enzyme of sialic acid biosynthesis. FEBS Lett. 2002;521:127–132. doi: 10.1016/s0014-5793(02)02856-9. [DOI] [PubMed] [Google Scholar]
- 29.Sittler A., Walter S., Wedemeyer N., Hasenbank R., Scherzinger E., Eickhoff H., Bates G. P., Lehrach H., Wanker E. E. SH3GL3 associates with the Huntingtin exon 1 protein and promotes the formation of polygln-containing protein aggregates. Mol. Cell. 1998;2:427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- 30.Bork P., Sander C., Valencia A. An ATPase domain common to procaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U.S.A. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarema K. J., Goon S., Bertozzi C. R. Metabolic selection of glycosylation defects in human cells. Nat. Biotechnol. 2001;19:553–558. doi: 10.1038/89305. [DOI] [PubMed] [Google Scholar]
- 32.Venkatachalam K. V., Akita H., Strott C. A. Molecular cloning, expression, and characterization of human bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase and its functional domains. J. Biol. Chem. 1998;273:19311–19320. doi: 10.1074/jbc.273.30.19311. [DOI] [PubMed] [Google Scholar]
- 33.Berninsone P., Hirschberg C. B. Heparan sulfate/heparin N-deacetylase/N-sulfotransferase: the N-sulfotransferase activity domain is at the carboxyl half of the holoenzyme. J. Biol. Chem. 1998;273:25556–25559. doi: 10.1074/jbc.273.40.25556. [DOI] [PubMed] [Google Scholar]