Abstract
UMP kinase catalyses the phosphorylation of UMP by ATP to yield UDP and ADP. In prokaryotes, the reaction is carried out by a hexameric enzyme, activated by GTP and inhibited by UTP. In the present study, Streptococcus pneumoniae UMP kinase was studied as a target for antibacterial research and its interest was confirmed by the demonstration of the essentiality of the gene for cell growth. In the presence of MnCl2 or MgCl2, the saturation kinetics of recombinant purified UMP kinase was hyperbolic for UMP (Km=0.1 mM) and sigmoidal for ATP (the substrate concentration at half-saturation S0.5=9.4±0.7 mM and n=1.9±0.1 in the presence of MgCl2). GTP increased the affinity for ATP and decreased the Hill coefficient (n). UTP decreased the affinity for ATP and only slightly increased the Hill coefficient. The kcat (175±13 s−1 in the presence of MgCl2) was not affected by the addition of GTP or UTP, whose binding site was shown to be different from the active site. The hydrodynamic radius of the protein similarly decreased in the presence of ATP or GTP. There was a shift in the pH dependence of the activity when the ATP concentration was switched from low to high. These results support the hypothesis of an allosteric transition from a conformation with low affinity for ATP to a form with high affinity, which would be induced by the presence of ATP or GTP.
Keywords: cmk, GTP, pyrH, Streptococcus pneumoniae UMP kinase, UTP
Abbreviations: DLS, dynamic light scattering; DSC, differential scanning calorimetry; NMP kinase, nucleoside monophosphate kinase
INTRODUCTION
The de novo biosynthesis of pyrimidine nucleotides are key metabolic pathways, that are conserved in almost all prokaryotic and eukaryotic organisms. The first nucleotide synthesized by the pathway is UMP, which is converted into UDP by a UMP kinase activity. In eukaryotes, the reaction is performed by a non-specific UMP-CMP kinase [1], a member of the NMP (nucleoside monophosphate) kinase family (see [2] for a review), the best example of which is adenylate kinase [3]. In prokaryotes, UMP kinase is encoded by pyrH, and the CMP kinase activity, encoded by cmk, is due to a separate enzyme, a member of the NMP kinase/adenylate kinase family [4]. In Bacillus subtilis and Escherichia coli, CMP kinase possesses a very weak activity when UMP is used as a substrate, with a catalytic efficiency 1000- and 3000-fold lower than that for CMP substrate respectively [5]. In B. subtilis, pyrH is not essential [6], whereas cmk is crucial for cell growth [7] and the UMP-converting activity of CMP kinase was proposed to complement the null-pyrH mutation [5]. In E. coli, overexpression of CMP kinase can suppress some pyrH mutations [8]; nevertheless, pyrH was demonstrated to be essential for growth in both E. coli [9] and Haemophilus influenzae [10]. In addition, the role of UMP kinase is more complex than simply converting UMP into UDP. In E. coli, the enzyme modulates the carAB gene by acting in a complex nucleoprotein structure required for pyrimidine-specific repression of carP1 promoter activity [11] and it also has a putative role in cell division [8], which might explain its localization near the bacterial membranes [12].
The pyrH gene product is highly conserved among bacteria and does not display significant sequence similarities to eukaryotic UMP kinases or other enzymes of the adenylate kinase family [13]. Prokaryotic UMP kinase is instead more related by primary sequence comparison to the aspartokinase family [13]. In addition, molecular modelling studies predict that the E. coli UMP kinase structure shares similarities with carbamate kinases and acetylglutamate kinases [14]. To date, only UMP kinases from E. coli and B. subtilis have been overexpressed, purified and characterized [12,13,15–17]. Both proteins are inhibited by UTP and activated by GTP. They display hyperbolic saturation kinetics with both substrates and, under some conditions, inhibition by excess UMP. They react to UTP or GTP by a change in kcat, with the enzyme from E. coli being particularly sensitive to inhibition by UTP, whereas the enzyme from B. subtilis is more responsive to activation by GTP [13,17].
The essentiality of the pyrH gene in E. coli and H. influenzae, its high conservation among bacteria and its structural divergence from the human counterpart [18] make UMP kinase an attractive enzyme for developing new antibacterials. However, since pyrH of B. subtilis was shown to be non-essential, due to the UMP kinase activity of the cmk gene, the consideration of bacterial UMP kinase as a potential target for antibacterials requires further investigation regarding the essentiality of the pyrH gene in at least one Gram-positive pathogenic species of clinical relevance.
In the present study, we have studied UMP kinase from Streptococcus pneumoniae, a Gram-positive human pathogen that causes life-threatening invasive diseases. First, we have shown its essentiality for cell survival. We have purified the recombinant protein and characterized its kinetics and conformational properties. We have demonstrated that the effectors GTP and UTP did not bind at the active site and the protein underwent significant conformational changes on their binding. Finally, UMP kinase from S. pneumoniae exhibited a unique property, namely co-operativity, for the ATP substrate.
EXPERIMENTAL
Bacterial strains and growth conditions
S. pneumoniae R800 [19] was kindly provided by J.-P. Claverys (UMR 5100, Université Paul Sabatier, Toulouse, France). S. pneumoniae was grown at 37 °C with 5% CO2 in BHI (brain heart infusion; Difco). Epicurian Coli™ XL1-Blue cells (Stratagene) were used as the host for plasmid construction and BL21 (DE3) (Stratagene) was used as the host for recombinant protein expression. Transformation of S. pneumoniae R800 was performed as described by Alloing et al. [20].
DNA manipulation
S. pneumoniae chromosomal DNA was prepared from mid-exponential-phase cultures using the Genomic-tip 500/G extraction kit (Qiagen) according to the manufacturer's instructions. E. coli plasmid DNA was extracted by the alkaline-SDS lysis method with the HiSpeed plasmid midi kit (Qiagen). PCRs were performed using the High-fidelity ampliTac enzyme (Boehringer), and DNA fragments were extracted from agarose gels using the Nucleospin extract kit (BD Biosciences). Restriction enzymes were used as recommended by the manufacturer.
Cloning and expression of the S. pneumoniae UMP kinase
The sequence of the pyrH gene was obtained from the TIGR database, after a BLAST analysis [21,22] with the E. coli pyrH gene. The full-length pyrH coding region was amplified from S. pneumoniae R800 chromosomal DNA with primers pyrHup (5′-GTGCATATGGCGAATCCCAAGTATAAACGT-3′) and pyr-Hdown (5′-TCTGGATCCTTCCTTTTCTTCGATATTATT-3′). The PCR fragment was digested with NdeI and BamHI, whose sites were included in the pyrHup and pyrHdown primers respectively. The purified digested PCR fragment was then ligated with the pET11a expression vector, previously digested with NdeI and BamHI, resulting in the pETpyrH plasmid. The sequence of the cloned gene was 100% identical with the pyrH gene of S. pneumoniae R6 [23]. It harboured one difference as compared with the TIGR strain at 661 bp (T→C), leading to a serine residue in the R800 strain instead of a proline. However, this position is not highly conserved and a serine residue is also found in the sequence of Streptococcus mutans UA159. S. pneumoniae UMP kinase contains 245 amino acids and shares 62.9% identity and 77.1% similarity with the protein of B. subtilis 168 and shares 47.2% identity and 63.3% similarity with E. coli. The pETpyrH plasmid was further introduced into E. coli BL21 (DE3) for the expression of S. pneumoniae UMP kinase.
Deletion experiments of pyrH in S. pneumoniae by insertion of a kanamycin resistance cassette
Two fragments corresponding to the 5′- and 3′-ends of the pyrH gene were PCR-amplified from S. pneumoniae R800 chromosomal DNA. The primers pyrHEup (5′-ATAGAATTCTGGCGAATCCCAAGTATAAAC-3′) and pyrHKdown (5′-CCCGGTACCGTTGCAATGAATCTGCCATCACAA-3′) were used for the 5′-fragment (282 bp), and pyrHBup (5′-TTGGGATCCGAATTGGTTCACCATACTTCT-3′) and pyrHSdown (5′-CTGGCATGCCGATATTTTCACCAAATACGA-3′) were used for the 3′-fragment (302 bp). The 5′- and 3′-PCR-generated fragments were then cloned in pUC19, at the EcoRI–KpnI and BamHI–SphI sites respectively. The resulting pyrH gene was thus devoid of a 118 bp central fragment. A kanamycin cassette (aphA-3) devoid of a promoter-terminator (kindly provided by P. Trieu-Cuot, Institut Pasteur, Paris, France) [24], flanked by KpnI and BamHI, was inserted into the corresponding sites between the two pyrH fragments, resulting in the pPyrGR plasmid. Subsequently, the cmk gene from B. subtilis 168 was amplified using the primers BScmk-S5 (5′-TTCGGATCCTAAATAGATGAGGAGAAAAGAGCAAGGAGA-3′) and BScmk-R (5′-TTGGGATCCTCAGCGGGACTTTTGCTCCACAGC-3′). The cmk gene was then cloned at the BamHI site of pPyrGR, immediately downstream of the aphA-3 cassette. After transformation of E. coli, the clones were checked for correct orientation of the B. subtilis cmk gene. The resulting plasmids, pPKCm and pPyrGR (Figure 1), were then used to transform S. pneumoniae. Transformants were selected by plating on media containing 1 mg/ml kanamycin. Analysis of recombinants for correct insertion was performed by PCR and Southern-blot analysis.
Figure 1. Gene-knockout strategy of the pyrH locus.
(A) pPyrGR and pPKCm are the suicide vectors designed for the insertion at the pyrH site of S. pneumoniae R800, through homologous recombination. The B. subtilis cmk gene (Bscmk) encodes CMP kinase, which also has UMP kinase activity. (B) After the double-crossover events leading to the expected gene replacements, cell viability on kanamycin medium was obtained only when cells were transformed with pPKCm but never with pPyrGR.
Expression and purification of recombinant S. pneumoniae UMP kinase
The expression medium was prepared as described by Riesenberg et al. [25]. Ampicillin was added to a final concentration of 200 mg/l. The cells were cultivated in a 10 litres stirred bioreactor (Type ISF207; INFORS AG, Bottmingen, Switzerland) fully controlled by IRIS software (INFORS Control). The initial culture conditions were as follows: 6 litres working volume, 2 litres/min air flow rate, 200 rev./min stirrer speed, 37 °C, inoculation with 2 ml of a frozen cell suspension at an attenuance (D600) of 2 and dissolved oxygen maintained at 50% by controlling the stirrer speed and air flow. When the culture reached a D600 of 12, the temperature was lowered to 25 °C and protein expression was induced by the addition of 1 mM isopropyl β-D-thiogalactosidase. After 13 h of induction, cells were collected by centrifugation for 30 min at 5000 g, leading to the recovery of 500 g of cell paste. Expression was checked by SDS/PAGE [26] and 90% of the UMP kinase was found in a soluble form.
The cell pellet (84 g) was suspended in 210 ml of 50 mM Tris and 10 mM MgCl2, pH 8.0 (buffer A). Cells were disrupted by sonication using a 19 mm probe and an ultrasonics liquid processor VCX 600 Watt (Sonics & Materials, Newtown, CT, U.S.A.) for 10 min while maintaining the temperature below 15 °C. The lysate was centrifuged at 186000 g for 180 min. The supernatant was applied to a Q Sepharose HP column XK50/10 (Amersham Biosciences) equilibrated with buffer A. After washing, proteins were eluted using a 0–0.5 M NaCl linear gradient for 45 min at a flow rate of 30 ml/min. Fractions containing UMP kinase were pooled and diluted in 4 vol. of 1.5 M (NH4)2SO4 in buffer A. The resulting solution was applied to a phenyl-Sepharose HP column XK50/20 (Amersham Biosciences) equilibrated with 1.2 M (NH4)2SO4 in buffer A. Proteins were eluted using a 1.2–0 M linear gradient of (NH4)2SO4 in buffer A for 45 min at a flow rate of 30 ml/min. The eluted protein was applied to a Superdex 200 column BPG100/700 equilibrated with 50 mM Tris (pH 8.0) and 16% glycerol at a flow rate of 40 ml/min. S. pneumoniae UMP kinase (946 mg) was obtained with a purity higher than 95% as seen by Coomassie Blue staining on SDS/PAGE (Figure 2). Protein concentration was 1.5 mg/ml as measured by the method of Bradford [27] or by UV spectrophotometry using a molar absorption coefficient of 12090 M−1·cm−1 at 280 nm. The N-terminal amino acid sequence was determined by a protein sequencer (Procise 492 cLC; Applied Biosystems). The protein was analysed by HPLC on a Vydac C4 (2.1 mm×150 mm) column using the HP1100 system from Agilent Technologies (Palo Alto, CA, U.S.A.). Elution was performed with a gradient of acetonitrile in 0.1% trifluoroacetic acid. Peaks were detected at 280 nm and analysed on an LCQ mass spectrometer (Thermo Electron S. A., Courtaboeuf, France) fitted with a Finnigan ESI (electrospray ionization) source. All experiments were performed in the positive mode. MS and N-terminal sequencing indicated a molecular mass of 26298 Da and loss of the N-terminal methionine. The enzyme was stored as aliquots and lost less than 10% activity on 12 months storage at −80 °C or during 6 h at 0 °C.
Figure 2. SDS/PAGE of fractions obtained during the purification of UMP kinase from S. pneumoniae.

Lane 1, standard proteins: phosphorylase b (97000 Da), BSA (66000 Da), ovalbumin (45000 Da), carbonic anhydrase (30000 Da), soya-bean trypsin inhibitor (20100 Da) and lysozyme (14400 Da). Lane 2, bacterial extract. Lane 3, Q Sepharose HP chromatography. Lane 4, phenyl-Sepharose HP chromatography. Lane 5, Superdex 200 chromatography. The amount of protein deposited on gel was 5 μg except for the bacterial extract, which was 10 μg.
Determination of the relative molecular mass
Gel filtration was performed on a Superdex 200 3.2/30 column (Amersham Biosciences) in 20 mM PBS (pH 7.4) and 0.15 M NaCl at 40 μl/min, with detection at 280 nm and standard proteins: aldolase (158000 Da), BSA (67000 Da), ovalbumin (43000 Da), chymotrypsinogen A (25000) and RNase A (13700 Da).
UMP kinase assay and kinetic analysis
Nucleotides, phosphoenolpyruvate, NADH, rabbit muscle pyruvate kinase and lactate dehydrogenase were purchased from Sigma. UMP kinase activity was determined at 37 °C by coupling with pyruvate kinase and lactate dehydrogenase and measurement of NADH consumption at 340 nm in a Spectramax 96-well microtitre plate reader (Molecular Devices). The reaction volume was 200 μl. The reaction buffer was 100 mM Bis-Tris and 50 mM KCl and, unless otherwise stated at pH 7.5. For the experiment at pH 8.5, Tris buffer was used. BSA was added to a final concentration of 0.1 mg/ml and DMSO was added to 2.5%. For the coupling, 2.5 mM phosphoenolpyruvate, 0.6 mM NADH, pyruvate kinase corresponding to an activity of 5 μmol−1·min−1·ml−1 and lactate dehydrogenase corresponding to 12.5 μmol−1·min−1·ml−1 were used. Pyruvate kinase activity was always at least 200 times higher than UMP kinase activity. No increase in activity was observed if NDP kinase was added to the assays. ATP, MnCl2 or MgCl2, UMP, GTP and UTP were added at various concentrations for testing UMP kinase activity. MnCl2 solutions were freshly prepared. The rate units are defined as μmol of UMP kinase substrate consumed per min, which corresponded to the consumption of 2 μmol of NADH/min. The reaction was started by the addition of UMP kinase, the final concentration of which varied from 37.5 to 100 ng/ml in different experiments.
Kinetics results were fitted to the following equations, using the non-linear regression module of the Statistica software (Statsoft):
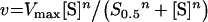 |
(1) |
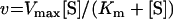 |
(2) |
 |
(3) |
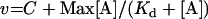 |
(4) |
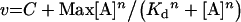 |
(5) |
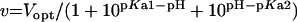 |
(6) |
where v is the steady-state velocity, Vmax and Vopt the maximal rates, Max the plateau rate, C a constant corresponding to the rate in the absence of allosteric effector, [S] the substrate concentration (ATP or UMP), [A] the allosteric effector concentration (GTP or UTP), Km the Michaelis–Menten constant, S0.5 the substrate concentration at half-saturation, Kd the apparent dissociation constant and n the Hill number or a co-operativity index.
DSC (differential scanning calorimetry)
The thermal stability of UMP kinase was determined by DSC, using a WP DSC apparatus (MicroCal, Northampton, MA, U.S.A.), over the temperature range of 15–85 °C at a rate of 60 °C/h. Protein solutions were dialysed against 20 mM Tris and 5 mM MgCl2 (pH 6.5), which was also used in the reference cell. Samples containing 1.8 mg/ml UMP kinase and various concentrations of substrate or effector were carefully degassed and pre-equilibrated for 15 min at 15 °C. Plots of absorbed heat as a function of temperature were analysed with the Origin software provided by MicroCal.
DLS (dynamic light scattering)
Measurements were performed with an Autosizer 4700 (MALVERN) instrument equipped with an argon ion laser operating at 488 nm and 40 mW. The diffused light was collected at a scattering angle of 90°. Measurements were performed with the detector aperture set at 0.2 mm. Five individual runs were taken at 25 °C. The protein was extensively dialysed against 100 mM Tris and 5 mM MgCl2 (pH 6.5). The samples were slowly injected into the scattering cell through a Millex GV4 0.22 μm filter. The time-dependent autocorrelation function of the photon current was analysed by the cumulants method. The hydrodynamic radius was calculated from the measured translational diffusion coefficient using the Stokes–Einstein equation, with the assumption that the species was spherical.
RESULTS
Cloning and sequencing of the S. pneumoniae pyrH gene
The pyrH gene was obtained after PCR amplification from S. pneumoniae R800. The residues identified by Bucurenci et al. [16] through a mutational analysis of E. coli UMP kinase are conserved in S. pneumoniae (Figure 3); these residues were demonstrated to be involved in effector binding (Arg-62 and Asp-77 in E. coli), solubility (Asp-159 and Asp-201 in E. coli), catalysis (Asp-146 in E. coli) and UMP binding (Asp-174 in E. coli). The alanine residue at position 94 of the E. coli enzyme, conserved in most Gram-negative bacteria and involved in the regulation of the carAB promoter [11], is absent from the S. pneumoniae sequence. It is substituted by a serine residue, which in turn is conserved in most Gram-positive organisms.
Figure 3. Alignment of the amino acid sequences of UMP kinase from E. coli (P29464 in SwissProt) and S. pneumoniae.
The secondary-structure elements from the E. coli model of Labesse et al. [14] are shown over the alignment. The residues cited in the text are in boldface. Active-site motifs, as predicted in [14], are underlined in the E. coli sequence and noted below the alignment. Hypothetical GTP-binding site and subunit interface are underlined in the S. pneumoniae sequence and noted below the alignment.
Essentiality of the S. pneumoniae pyrH gene
To demonstrate the essentiality of the pyrH gene in S. pneumoniae, the R800 strain was transformed with the non-replicative pPyrGR and pPKCm vectors (Figure 1). The experiments were repeated three times independently. Transformation with pPKCm allowed the selection of kanamycin-resistant clones, whereas no transformants could be selected with pPyrGR. This strongly suggested that pyrH is essential and cannot be deleted unless complemented with the B. subtilis cmk gene present in the integrated pPKCm plasmid. The complementation experiment with B. subtilis cmk and the use of a terminatorless aphA-3 cassette indicated clearly that the essentiality is not due to a polarity effect on the downstream essential frr gene. Thus S. pneumoniae cells share with E. coli, and not with B. subtilis, the property that pyrH is essential and its deletion cannot be rescued by the endogenous cmk. The differences in pyrH essentiality among bacterial species may come from the in vivo level of activity of the endogenous CMP kinase and also from differences in cell regulation.
The pPKCm S. pneumoniae strain, which is deleted in pyrH and complemented by B. subtilis cmk, constitutes an interesting tool for testing the inhibitors of UMP kinase using a cellular assay. Molecules that exhibited an antibacterial effect on the wild-type strain and not on the pPKCm strain would denote specific targeting of UMP kinase.
Requirement for a bivalent cation
The assay for UMP kinase activity was a coupled system with pyruvate kinase and lactate dehydrogenase. Pyruvate kinase activity is dependent on the presence of a bivalent cation, magnesium or manganese being the preferred co-substrate, with a similar efficiency [28]. In the absence of a bivalent cation, the coupled assay could not be used and it was replaced by an HPLC assay, in which the concentrations of substrates and products were directly measured. In this method, UMP kinase was totally inactive in the absence of MgCl2 or MnCl2. Subsequently, the rate of UMP kinase activity as a function of MgCl2 or MnCl2 concentration up to 20 mM was determined at 1 or 2 mM ATP, in the coupled assay. As long as the cation concentration was lower than the ATP concentration, the rate increased sharply. When the cation concentration was greater than the ATP concentration, the rate increased slowly. Consequently, the ATP–Mg or ATP–Mn complex was considered to be the true substrate for the enzyme. A slow increase in activity for a cation/ATP ratio higher than 1 reflected the incomplete complexation of ATP with the cation. As an example, using dissociation constants of 13.7 and 10 μM for Mg-ATP2− and Mn-ATP2− at pH 8, the concentration of the complexes would be 0.89 and 0.90 mM respectively at 1 mM of both ATP and cation [29]. Moreover, the cations could also be activators, an effect that has been described for other kinases such as rabbit muscle pyruvate kinase [28,30,31] or phosphoenolpyruvate carboxykinase [32]. A cation/ATP molar ratio of 2 was selected for all the subsequent kinetic experiments. This ratio was chosen to enable a close approach to ATP saturation by the cation and to limit the risk of precipitation that could be observed at a high concentration of MnCl2. For example, at 1 mM ATP and 2 mM cation, the concentrations of both Mg-ATP2− and Mn-ATP2− would be 0.99 mM at pH 8. Since GTP or UTP also formed complexes with the cation and since it was assumed that all NTPs had a similar affinity for the cation when several NTPs were present in a reaction, the cation concentration was set to twice the sum of the concentrations of NTPs.
Substrate specificity
Role of ATP, UTP and GTP as phosphate donor was tested at 4 mM in the presence of 2 mM UMP, 8 mM MnCl2 or MgCl2 and a high concentration of UMP kinase (100 ng/ml). ATP was clearly the best phosphate donor and displayed activities of 130 and 66 μmol−1·min−1·mg−1 with MnCl2 and MgCl2 respectively. GTP and UTP were extremely poor substrates with an activity below 1 μmol−1·min−1·mg−1 with either cation.
Substrate saturation curves
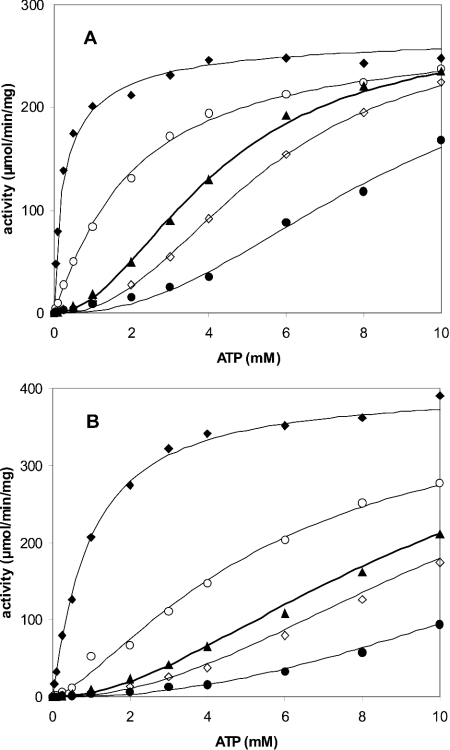
The saturation curves for the substrate ATP were determined at 2 mM UMP in the presence of MnCl2 or MgCl2 (see Figure 4 and Table 1). In the absence of GTP, the curves were sigmoidal. With increase in GTP, the sigmoidicity progressively disappeared. The addition of UTP did not strongly modify the shape of the curve and displaced the position of the inflection point of the sigmoid to higher ATP concentrations. The curves were fitted to eqn (1). For curves with low S0.5, the plateau was clearly defined at the highest concentrations of ATP and the estimation of Vmax was accurate. In contrast, for curves with high S0.5, it was not possible to determine simultaneously and accurately the three parameters Vmax, S0.5 and n. Since we observed that the Vmax parameter was constant for each set of experiments with a cation, a mean value was determined from the curves with a low S0.5. Values of 274±6 and 399±30 μmol−1·min−1·mg−1 (corresponding to kcat=120±3 and 175±13 s−1 per monomer) were obtained for MnCl2 (Figure 4A) and MgCl2 (Figure 4B) respectively. These mean Vmax values were then included in eqn (1) when fitting curves with high S0.5, allowing accurate determination of S0.5 and n. In the absence of any effector, S0.5 was 4.2±0.2 and 9.4±0.7 mM in the presence of MnCl2 and MgCl2 respectively. Whereas the kcat value was 1.5 times higher in the presence of MgCl2, the affinity for ATP was 2.3 times higher in the presence of MnCl2. Finally, n was almost the same for both cations, with values of 2.0±0.1 and 1.9±0.1 for MnCl2 and MgCl2 respectively.
Figure 4. ATP saturation curves of UMP kinase in the presence of MnCl2 (A) or MgCl2 (B).
The concentrations of effectors were 1 mM GTP (◆), 0.1 mM GTP (○), no effector (▲), 0.4 mM UTP (◇) and 2 mM UTP (●). Curves were drawn using kinetic parameters calculated from eqn (1) and displayed in Table 1. UMP concentration was 2 mM and UMP kinase concentration was 37.5 ng/ml. The cation/NTP molar ratio was maintained at 2.
Table 1. S0.5 and n parameters for ATP, calculated from eqn (1).
UMP was 2 mM and UMP kinase was 37.5 ng/ml, with the corresponding Vmax=274±6 and 399±30 μmol·min−1·mg−1 (kcat=120±3 and 175±13 s−1 per monomer of 26298 Da) for MnCl2 and MgCl2 respectively. The cation/NTP molar ratio was maintained at 2. The pH value was 7.5. For the sake of clarity, only five curves are presented in the corresponding Figure 4.
| MnCl2 | MgCl2 | |||
|---|---|---|---|---|
| Effector | S0.5 (mM) | n | S0.5 (mM) | n |
| GTP (mM) | ||||
| 0 | 4.2±0.2 | 2.0±0.1 | 9.4±0.7 | 1.9±0.1 |
| 0.05 | 3.0±0.2 | 1.5±0.1 | 7.7±0.9 | 1.6±0.1 |
| 0.1 | 2.0±0.1 | 1.1±0.1 | 5.7±1.3 | 1.4±0.2 |
| 0.4 | 0.54±0.07 | 0.82±0.06 | 1.8±0.2 | 1.2±0.1 |
| 1 | 0.29±0.05 | 0.75±0.08 | 0.93±0.06 | 1.1±0.1 |
| 2 | 0.19±0.06 | 0.65±0.15 | 0.81±0.14 | 0.92±0.10 |
| UTP (mM) | ||||
| 0 | 4.2±0.2 | 2.0±0.1 | 9.4±0.7 | 1.9±0.1 |
| 0.05 | 4.5±0.2 | 2.0±0.1 | 9.8±1.7 | 2.0±0.2 |
| 0.1 | 4.6±0.2 | 2.2±0.1 | 10±3 | 2.0±0.3 |
| 0.4 | 5.4±0.2 | 2.3±0.1 | 11±2 | 2.1±0.2 |
| 1 | 6.7±0.5 | 2.4±0.2 | 14±5 | 2.2±0.2 |
| 2 | 8.6±2.1 | 2.3±0.4 | 17±14 | 2.2±0.4 |
The addition of GTP abolished the co-operativity. At 2 mM GTP, n was 0.65±0.15 and 0.92±0.10 for MnCl2 and MgCl2 respectively. In parallel, the apparent affinity was increased: at 2 mM GTP, S0.5 was 0.19±0.06 and 0.81±0.14 mM for MnCl2 and MgCl2 respectively. The addition of UTP had an opposite effect on S0.5 and, in parallel, slightly increased the co-operativity. At 2 mM UTP, S0.5 reached 8.6±2.1 and 17±14 mM and n increased to 2.3±0.4 and 2.2±0.4 for MnCl2 and MgCl2 respectively.
The saturation curves with the second substrate, UMP, were not determined at saturating ATP concentrations. High concentrations of ATP were not compatible with solubility. Seven UMP saturation curves were established at different concentrations of ATP in the range 1–5 mM. Both MnCl2 and MgCl2 were tested. All curves were hyperbolic and could be fitted to eqn (2) (results not shown). The Km of UMP was then estimated from a secondary plot of the inverse of the observed Km as a function of the inverse of the ATP concentration. Using a linear fit, Km values for UMP of approx. 0.15 and 0.10 mM were obtained in the presence of MnCl2 and MgCl2 respectively. When GTP or UTP was added at a constant ATP concentration, the curves remained hyperbolic. The apparent Km for UMP slightly decreased and the Vmax increased in the presence of GTP, whereas the reverse phenomena were observed in the presence of UTP.
GTP and UTP saturation curves
The saturation curves of UMP kinase activity with UTP or GTP were obtained at 3 mM ATP, 2 mM UMP and in the presence of MnCl2 or MgCl2. The saturation curves of UTP (Figure 5A) were hyperbolic and the data were fitted to eqn (3). The rates decreased with increase in UTP concentration, and at high concentrations, the activity reached a plateau of low activity, 22.0±1.5 or 8.5±0.7 μmol−1·min−1·ml−1, in the presence of MnCl2 or MgCl2 respectively. This non-null plateau activity could not correspond to a consumption of UTP since UTP was not a good enough substrate (activity less than 1 μmol−1·min−1·ml−1 at 4 mM). Since UMP kinase could not be fully inhibited, it was concluded that UTP did not act by competition at the active site. The fit indicated an apparent Kd of 0.21±0.03 and 0.28±0.10 mM for UTP in the presence of MnCl2 and MgCl2 respectively.
Figure 5. UMP kinase saturation curve by the effector UTP (A) or GTP (B).
The concentrations were 3 mM ATP and 2 mM UMP in the presence of MgCl2 (●) or MnCl2 (□). Curves were drawn using kinetic parameters calculated from eqns (3)–(5); they were hyperbolic except for GTP in the presence of MgCl2, where it was sigmoidal, as shown in the inset. UMP kinase concentration was 75 ng/ml (A) or 37.5 ng/ml (B). The cation/NTP molar ratio was maintained at 2.
The saturation curve of GTP (Figure 5B) was hyperbolic with MnCl2; data were fitted using eqn (4) and the apparent Kd was 0.14±0.02 mM. With MgCl2, a sigmoidal fit was generated using eqn (5), the apparent Kd was 0.21±0.01 mM and the co-operativity index was 2.2±0.2. Additional experiments were performed with different concentrations of ATP (results not shown). When the ATP concentration was decreased, the apparent Kd for GTP and the co-operativity index increased. Despite strong similarities between ATP and GTP in their chemical structure and co-operative kinetics, these two molecules probably do not bind at the same site. Indeed, if GTP activated by mimicking ATP, i.e. through a binding at the ATP site, GTP would be inhibitory at high concentrations. As depicted in Figure 5(B), at 4 mM GTP, which is 40 times the apparent Kd, no significant inhibition was observed. Therefore the strong activation by GTP is probably due to binding at a site distinct from the ATP site.
Effect of pH on the ATP saturation curve
ATP saturation curves were compared at pH 6.5, 7.5 and 8.5 in the presence of MnCl2 (Figure 6A) or MgCl2 (results not shown). Data could be fitted with accuracy to the Hill equation (eqn 1) at pH 6.5 and 7.5. No fit was possible for the data at pH 8.5 with MgCl2 because the measurements only described a reduced part of the curve. The changes in kinetic parameters were qualitatively similar in the experiments with MnCl2 and MgCl2: Vmax increased with pH, whereas the apparent affinity and the co-operativity for ATP decreased with pH. In the presence of MnCl2, for pH values of 6.5, 7.5 and 8.5, Vmax increased from 135±3 to 274±6 and 666±392 μmol−1·min−1·mg−1, S0.5 increased from 1.3±0.1 to 4.2±0.2 and 15±8 mM and n decreased from 2.6±0.3 to 2.0±0.1 and 1.7±0.3 respectively. In the presence of MgCl2, when the pH was shifted from 6.5 to 7.5, Vmax increased from 182±4 to 399±30 μmol−1·min−1·mg−1, S0.5 from 3.2±0.1 to 9.4±0.7 mM and n decreased from 2.8±0.2 to 1.9±0.1. Additional experiments were performed to demonstrate a change in the apparent optimal pH when the ATP concentration was modified. Activity was measured at five different pH values in the range 6.5–8.5 at three ATP concentrations in the presence of MnCl2 (Figure 6B) or MgCl2 (results not shown). Where possible, bell-shaped data were fitted using eqn (6) to determine an optimal pH value and two pKa values. In the presence of MgCl2, when the ATP concentration increased from 2 to 4 and 10 mM, the optimal pH value increased from <6.5 to approx. 6.5 and 7.4, with pKa=6.2±0.1 and 8.6±0.1 at 10 mM ATP. In the presence of MnCl2, for increase in ATP concentration from 2 to 4 and 10 mM, the optimal pH value increased from <6.5 to 7.1 and 7.8, with pKa=6.3±0.2 and 8.0±0.1 at 4 mM ATP and pKa=6.7±0.1 and 8.9±0.2 at 10 mM ATP. Therefore the optimal pH of the reaction increased with increase in ATP concentration and is shifted to slightly higher pH values in the presence of MnCl2 compared with MgCl2.
Figure 6. Influence of pH on (A) ATP saturation curves and (B) the optimal activity at different ATP concentrations, in the presence of MnCl2.
Assays were performed at pH 6.5 (○), pH 7.5 (□) and pH 8.5 (Δ) (A) and 2 mM (●), 4 mM (■) and 10 mM (▲) ATP (B). Curves were drawn using kinetic parameters calculated from eqn (1) (A) and eqn (6) (B). UMP concentration was 2 mM and UMP kinase was 37.5 ng/ml. The MnCl2/ATP molar ratio was maintained at 2.
Protein properties by gel filtration
The enzyme solution at 1.5 mg/ml was submitted to gel filtration at pH 7.4. The protein preparation showed no aggregates. Its major form, representing two-thirds of the protein population, had a molecular mass of 189 kDa. The minor form displayed an apparent mass of 87.5 kDa. Calculations indicated a heptamer for the major form and a trimer for the minor one. Homoheptamers are not frequently observed and the major form may, rather, be a hexamer.
Calculated hydrodynamic radius
The protein preparation was tested by DLS after dialysis in 100 mM Tris and 5 mM MgCl2, pH 6.5 (Table 2). The translational diffusion coefficient was measured in the presence of different ligands and the data were used to calculate the hydrodynamic radius under the assumption of a globular shape. The hydrodynamic radius of UMP kinase was 6.0 nm with a polydispersity of 18%. Addition of 3 mM ATP induced the same effect as 3 mM GTP: the hydrodynamic radius was decreased to 4.8 nm and the polydispersity to 9%, indicating that the protein became more compact in a more homogeneous sample. In contrast, 3 mM UMP or UTP induced a large increase in size and polydispersity. With UMP, the protein solubility was significantly reduced since precipitates were observed in the solution. Because the polydispersity in the preparations containing UTP and UMP was high (44 and 30% respectively), the presence of aggregates was strongly suspected.
Table 2. Hydrodynamic data by DLS.
DT, translational diffusion coefficient; RH, hydrodynamic radius of gyration. UMP kinase content was 2 mg/ml in 5 mM MgCl2 and 100 mM Tris (pH 6.5). Precipitates were observed in the presence of UMP.
| Ligand (3 mM) | DT×108 (cm2/s) | RH (nm) | Polydispersity (%) |
|---|---|---|---|
| None | 41.3±1.5 | 6.0±0.2 | 18±2 |
| ATP | 50.7±0.6 | 4.8±0.1 | 9±2 |
| GTP | 51.1±0.4 | 4.8±0.03 | 9±2 |
| UTP | 22.9±4.0 | 10.9±2.1 | 44±7 |
| UMP | 18.1±0.4 | 13.6±0.2 | 30±2 |
Stability of UMP kinase to thermal denaturation
Changes in Tm (transition midpoint temperature) of the protein were monitored on addition of 3 mM substrates or GTP and UTP effectors (Table 3). Experiments were performed in the presence of 5 mM MgCl2 and at pH 6.5. The DSC profiles showed an endothermic reaction with Tm=50–73 °C. This was followed by an exothermic reaction with an onset temperature in the range 58–75 °C, most probably corresponding to the aggregation of UMP kinase after thermal denaturation. All the ligands induced an increase in Tm. The most significant change was observed with GTP giving a 23 °C shift. ATP induced the lowest Tm shift of 4 °C. A higher ATP concentration of 30 mM was tested to saturate the protein. An increase in Tm of 11 °C was measured. The increases in Tm induced by UTP or UMP binding were similar: 17 or 18 °C respectively. Large increases in Tm obtained with the ligands GTP, UTP and UMP probably indicated extensive conformational changes in the protein. The lowest increase in Tm induced by ATP either corresponds to an absence of conformational change or most probably reflects a lower affinity for ATP in comparison with other ligands.
Table 3. Effect of ligands on the Tm of UMP kinase.
20 mM Tris and 5 mM MgCl2 (pH 6.5) was used except for the 30 mM ATP experiment (100 mM Tris and 30 mM MgCl2, pH 6.5). Precipitates were observed in the presence of UMP.
| Ligand | Tm ( °C) | Increase in Tm ( °C) |
|---|---|---|
| None | 50.0±0.2 | – |
| 3 mM UMP | 67.9±0.8 | 18 |
| 3 mM ATP | 54.3±0.2 | 4 |
| 30 mM ATP | 61.1±0.2 | 11 |
| 3 mM GTP | 73.0±0.4 | 23 |
| 3 mM UTP | 67.1±1.0 | 17 |
DISCUSSION
In the present study, we have demonstrated that the kinetics of S. pneumoniae UMP kinase is Michaelis–Menten type for the substrate UMP and is co-operative for the substrate ATP. We propose a basic model for this enzyme with two forms, R and T, according to allosteric nomenclature [33,34]. R would be the form with a high affinity for ATP and T the form with a low affinity. An allosteric transition would allow the reversible transformation of the T form into the R form and would explain the co-operative effect of ATP. Experiments on the pH dependence of UMP kinase support this model. At low ATP concentrations, the enzyme displayed an optimal pH below 6.5, whereas at high ATP concentrations, the optimal pH was from 7.4 to 7.8. In parallel, apparent pKa values were shifted to higher pH values when the ATP concentration increased. Such a large variation in the optimal pH and pKa values of residues involved in catalysis can be logically related to large conformational changes in the protein. The shift in pH dependence of the activity accompanying an allosteric transition has been described for aspartate transcarbamoylase from E. coli. Its optimal pH is 6.8 at low aspartate concentrations and 8.2 at high aspartate concentrations [35,36].
S. pneumoniae UMP kinase is activated by GTP. Since GTP resembles ATP, it could bind at the ATP site and be the activator at low concentrations by inducing the allosteric transition. Since it is not the inhibitor at high concentrations, we conclude that GTP has a binding site different from the ATP-binding site. GTP strongly decreases the co-operativity for ATP and enhances its affinity. This can indicate that the binding of GTP displaces the equilibrium between the two forms towards the high-affinity R form. UMP kinase is inhibited by UTP. We demonstrated that UTP does not bind at the active site by showing that UTP never completely inhibits the enzyme. UTP decreases the apparent affinity for ATP but only weakly increases the co-operativity. Thus, from the kinetic experiments, we have no clear evidence that UTP might inhibit by inducing the transition to the low-affinity T form. It has been previously shown that some allosteric effectors do not act by shifting the T–R equilibrium, e.g. ATP and aspartate transcarbamoylase from E. coli [37,38].
The enzyme absolutely requires MgCl2 or MnCl2 for activity. The nature of the bivalent cation strongly influences the kinetic parameters and also modifies the apparent optimal pH. With MgCl2, in comparison with MnCl2, the catalytic constant and the affinity for UMP are higher, but the affinities for ATP, GTP and UTP are lower. In addition, some co-operativity for GTP binding is observed solely in the presence of MgCl2. These results suggest that an UMP kinase might contain two metal-binding sites as already known for other kinases [28,30–32]. Nevertheless, further experiments are needed to support this hypothesis.
The oligomerization state of S. pneumoniae UMP kinase was determined by gel filtration. The major form was assigned to a hexamer. A hexamer was also found for E. coli and B. subtilis enzymes, and molecular modelling predicts a three in-plane dimer subunit arrangement [14]. The effect of the binding of the substrates or effectors to the protein was studied by DSC and DLS, at pH 6.5 in the presence of MgCl2. This pH was used to favour the binding of ATP, which displayed a higher affinity for the enzyme at acidic pH. ATP and GTP both strongly decreased the molecular mass of the protein and improved the homogeneity of the sample. Therefore, in the framework of our model, we propose a compact conformation for the high-affinity R form. UTP induced swelling of the protein and, at the same time, increased the polydispersity of the sample. The binding of UTP may induce the protein to adopt a more extended conformation, where exposure of hydrophobic side chains would favour the formation of large polymers or aggregates. Changes in the solubility of the protein have also been described for UMP kinase from E. coli [15,16]. The effect of the substrate UMP on the conformation of S. pneumoniae enzyme is intriguing. The DSC and DLS results are very similar to those obtained with UTP. Binding of UMP at the active site might induce a conformational change similar to that induced by the binding of UTP at a regulatory site. Alternatively, UMP could have a secondary site at the UTP site. UMP would then induce the same conformational change as UTP and it would also probably act as an inhibitor. Serina et al. [15] have described inhibition by excess of UMP with the E. coli UMP kinase, at pH 7.4 and not at pH 6.0. With the S. pneumoniae UMP kinase, no inhibition by excess of UMP was ever obtained either at pH 7.5 or 6.5. Nevertheless, in kinetic experiments, the presence of ATP might alter the binding of UMP at the UTP site by the induction of a structural change. Therefore the binding results by DSC and DLS cannot be directly compared with the kinetic data.
A striking and unique feature of S. pneumoniae UMP kinase kinetics is the co-operativity for ATP, with an extremely low affinity, depicted by S0.5 (the half-saturation constant) of 4.2±0.2 or 9.4±0.7 mM at pH 7.5 in the presence of MnCl2 or MgCl2 respectively. The enzyme is probably poorly active in the absence of GTP under physiological conditions. On the addition of 1 mM GTP, S0.5 is decreased by factors of 14 and 10 and n is decreased by factors of 2.7 and 1.7, in the presence of MnCl2 or MgCl2 respectively. Inhibition by UTP is much less potent compared with the activation by GTP: 1 mM UTP only produced an increase in S0.5 of approx. 1.5 and an increase in n of 1.2 in the presence of either MnCl2 or MgCl2. The B. subtilis and E. coli UMP kinases exhibit very different properties, since Michaelis–Menten kinetics has been described for ATP, with Km=0.9 and 0.12 mM respectively [13,17]. The E. coli enzyme is significantly active in the absence of GTP and is particularly sensitive to inhibition by UTP; the effectors mostly affect the kcat value [13,15]. B. subtilis enzyme is very sensitive to GTP, which induces a large increase in kcat and is minimally affected by UTP [17]. Although UMP kinase from both Gram-positive bacteria have in common a strong sensitivity to activation by GTP, S. pneumoniae enzyme possesses a unique mode of regulation since the allosteric effectors modulate ATP binding and not the kcat.
Labesse et al. [14] have proposed that the overall structure of E. coli UMP kinase is similar to the structure of carbamate kinase from Enterococcus faecalis or Pyrococcus furiosus [39,40]. If their UMP kinase model is valid, then the active site should be similar in E. coli and S. pneumoniae, since the six motifs proposed to participate in the active site are well conserved in S. pneumoniae, with only a few exceptions, for example the absence of a threonine residue in motif 5, numbered 165 in E. coli (Figure 3). Notably, the sequence between residues 100 and 127 in S. pneumoniae, corresponding to the β3 strand and α4 helix in the model, contains a large number of positively charge residues, which might interact with negatively charged groups, possibly phosphates from GTP. Interestingly, this sequence is much better conserved in Gram-positive than in Gram-negative bacteria. Finally, some structural differences between E. coli and S. pneumoniae should be sought at the interface between the subunits of the dimer, since the co-operative signal is probably mediated by the interface. The central region of the interaction between the subunits in E. faecium carbamate kinase is strongly hydrophobic and involves three sulphur-containing residues from the third helix [39]. In E. coli UMP kinase, five methionine residues are present in the corresponding region (Met-71, -79, -81, -86 and -91) and four of them are conserved in S. pneumoniae (Met-68, -78, -83 and -88). In addition, in the periphery of the interaction area in carbamate kinase, α2 helices and β3 strands make contacts through hydrogen bonds. Comparison of the interactions in the α2, α3 and β3 regions in E. coli and S. pneumoniae UMP kinases might be fundamental to understanding the propagation of the allosteric signal.
Acknowledgments
We are particularly grateful to G. Hervé (UMR 7631, Université Pierre et Marie Curie, Paris, France), B. Schoot (Galderma, Sophia Antipolis, France), M. Black, V. Charrier and B. Demers (Aventis Pharma, France) for comments and fruitful discussions. We thank R. Arrebola, B. Genet, H. Rey and L. Chantalat for technical support.
References
- 1.Sugino Y., Teraoka H., Shimono H. Metabolism of deoxyribonucleotides. I. Purification and properties of deoxycytidine monophosphokinase of calf thymus. J. Biol. Chem. 1966;241:961–969. [PubMed] [Google Scholar]
- 2.Yan H., Tsai M.-D. Nucleoside monophosphate kinases: structure, mechanism, and substrate specificity advances. In: Purich D. L., editor. Enzymology and Related Areas of Molecular Biology 73. John Wiley & Sons; 1999. pp. 103–134. [DOI] [PubMed] [Google Scholar]
- 3.Vonrhein C., Schlauderer G. J., Schulz G. E. Movie of the structural changes during a catalytic cycle of nucleoside monophosphate kinases. Structure. 1995;3:483–490. doi: 10.1016/s0969-2126(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 4.Bucurenci N., Sakamoto H., Briozzo P., Palibroda N., Serina L., Sarfati R. S., Labesse G., Briand G., Danchin A., Bârzu O., et al. CMP kinase from Escherichia coli is structurally related to other nucleoside monophosphate kinases. J. Biol. Chem. 1996;271:2856–2862. doi: 10.1074/jbc.271.5.2856. [DOI] [PubMed] [Google Scholar]
- 5.Schultz C. P., Ylisastigui-Pons L., Serina L., Sakamoto H., Mantsch H. H., Neuhard J., Bârzu O., Gilles A.-M. Structural and catalytic properties of CMP kinase from Bacillus subtilis: a comparative analysis with the homologous enzyme from Escherichia coli. Arch. Biochem. Biophys. 1997;340:144–153. doi: 10.1006/abbi.1997.9888. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi K., Ehrlich S. D., Albertini A., Amati G., Andersen K. K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P., et al. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorokin A., Serror P., Pujic P., Azevedo V., Ehrlich S. D. The Bacillus subtilis chromosome region encoding homologues of the Escherichia coli mssA and rpsA gene products. Microbiology. 1995;141:311–319. doi: 10.1099/13500872-141-2-311. [DOI] [PubMed] [Google Scholar]
- 8.Yamanaka K., Ogura T., Koonin E. V., Niki H., Hiraga S. Multicopy suppressors, mssA and mssB, of an smbA mutation of Escherichia coli. Mol. Gen. Genet. 1994;243:9–16. doi: 10.1007/BF00283870. [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka K., Ogura T., Niki H., Hiraga S. Identification and characterization of the smbA gene, a suppressor of the mukB null mutant of Escherichia coli. J. Bacteriol. 1992;174:7517–7526. doi: 10.1128/jb.174.23.7517-7526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akerley B. J., Rubin E. J., Novick V. L., Amaya K., Judson N., Mekalanos J. J. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U.S.A. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kholti A., Charlier D., Gigot D., Huysveld N., Roovers M., Glansdorff N. pyrH-encoded UMP-kinase directly participates in pyrimidine-specific modulation of promoter activity in Escherichia coli. J. Mol. Biol. 1998;280:571–582. doi: 10.1006/jmbi.1998.1910. [DOI] [PubMed] [Google Scholar]
- 12.Landais S., Gounon P., Laurent-Winter C., Mazié J.-C., Danchin A., Bârzu O., Sakamoto H. Immunochemical analysis of UMP kinase from Escherichia coli. J. Bacteriol. 1999;181:833–840. doi: 10.1128/jb.181.3.833-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serina L., Blondin C., Krin E., Sismeiro O., Danchin A., Sakamoto H., Gilles A.-M., Bârzu O. Escherichia coli UMP-kinase, a member of the aspartokinase family, is a hexamer regulated by guanine nucleotides and UTP. Biochemistry. 1995;34:5066–5074. doi: 10.1021/bi00015a018. [DOI] [PubMed] [Google Scholar]
- 14.Labesse G., Bucurenci N., Douguet D., Sakamoto H., Landais S., Gagyi C., Gilles A.-M., Bârzu O. Comparative modelling and immunochemical reactivity of Escherichia coli UMP kinase. Biochem. Biophys. Res. Commun. 2002;294:173–179. doi: 10.1016/S0006-291X(02)00450-3. [DOI] [PubMed] [Google Scholar]
- 15.Serina L., Bucurenci N., Gilles A.-M., Surewicz W. K., Fabian H., Mantsch H. H., Takahashi M., Petrescu I., Batelier G., Bârzu O. Structural properties of UMP-kinase from Escherichia coli: modulation of protein solubility by pH and UTP. Biochemistry. 1996;35:7003–7011. doi: 10.1021/bi960062v. [DOI] [PubMed] [Google Scholar]
- 16.Bucurenci N., Serina L., Zaharia C., Landais S., Danchin A., Bârzu O. Mutational analysis of UMP kinase from Escherichia coli. J. Bacteriol. 1998;180:473–477. doi: 10.1128/jb.180.3.473-477.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagyi C., Bucurenci N., Sirbu O., Labesse G., Ionescu M., Ofiteru A., Assairi L., Landais S., Danchin A., Bârzu O., et al. UMP kinase from the Gram-positive bacterium Bacillus subtilis is strongly dependent on GTP for optimal activity. Eur. J. Biochem. 2003;270:3196–3204. doi: 10.1046/j.1432-1033.2003.03702.x. [DOI] [PubMed] [Google Scholar]
- 18.Van Rompay A. R., Johansson M., Karlsson A. Phosphorylation of deoxycytidine analog monophosphates by UMP-CMP kinase: molecular characterization of the human enzyme. Mol. Pharmacol. 1999;56:562–569. doi: 10.1124/mol.56.3.562. [DOI] [PubMed] [Google Scholar]
- 19.Martin B., Prats H., Claverys J. P. Cloning of the hexA mismatch-repair gene of Streptococcus pneumoniae and identification of the product. Gene. 1985;34:293–303. doi: 10.1016/0378-1119(85)90138-6. [DOI] [PubMed] [Google Scholar]
- 20.Alloing G., de Philip P., Claverys J. P. Three highly homologous membrane-bound lipoproteins participate in oligopeptides transport by the Ami system of the gram-positive Streptococcus pneumoniae. J. Mol. Biol. 1994;241:44–58. doi: 10.1006/jmbi.1994.1472. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoskins J., Alborn W. E., Jr, Arnold J., Blaszczak L. C., Burgett S., DeHoff B. S., Estrem S. T., Fritz L., Fu D. J., Fuller W., et al. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 2001;183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trieu-Cuot P., Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5′-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 25.Riesenberg D., Schulz V., Knorre W. A., Pohl H. D., Korz D., Sanders E. A., Ross A., Deckwer W. D. High cell density cultivation of Escherichia coli at controlled specific growth rate. J. Biotechnol. 1991;20:17–27. doi: 10.1016/0168-1656(91)90032-q. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Baek Y. H., Nowak T. Kinetic evidence for a dual cation role for muscle pyruvate kinase. Arch. Biochem. Biophys. 1982;217:491–497. doi: 10.1016/0003-9861(82)90529-x. [DOI] [PubMed] [Google Scholar]
- 29.O'Sullivan W., Smithers G. W. Stability constants for biologically important metal-ligand complexes. In: Purich D. L., editor. Methods in Enzymology, vol. 63. London: Academic Press; 1979. pp. 294–336. [DOI] [PubMed] [Google Scholar]
- 30.Gupta R. K., Mildvan A. S. Structures of enzyme-bound metal-nucleotide complexes in the phosphoryl transfer reaction of muscle pyruvate kinase. 31P NMR studies with magnesium and kinetic studies with chromium nucleotides. J. Biol. Chem. 1977;252:5967–5976. [PubMed] [Google Scholar]
- 31.Larsen T. M., Benning M. M., Rayment I., Reed G. H. Structure of the bis(Mg2+)-ATP-oxalate complex of the rabbit muscle pyruvate kinase at 2.1 A resolution: ATP binding over a barrel. Biochemistry. 1998;37:6247–6255. doi: 10.1021/bi980243s. [DOI] [PubMed] [Google Scholar]
- 32.Lee M. H., Hebda C. A., Nowak T. The role of cations in avian liver phosphoenolpyruvate carboxykinase catalysis. Activation and regulation. J. Biol. Chem. 1981;256:12793–12801. [PubMed] [Google Scholar]
- 33.Monod J., Changeux J. P., Jacob F. Allosteric proteins and cellular control systems. J. Mol. Biol. 1965;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 34.Monod J., Wyman J., Changeux J. P. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 35.Gerhart J. C., Pardee A. B. Aspartate transcarbamylase, an enzyme designed for feedback inhibition. Fed. Proc., Fed. Am. Soc. Exp. Biol. 1964;23:727–735. [PubMed] [Google Scholar]
- 36.Thiry L., Hervé G. The stimulation of Escherichia coli aspartate transcarbamylase activity by adenosine triphosphate. Relation with the other regulatory conformational changes; a model. J. Mol. Biol. 1978;125:515–534. doi: 10.1016/0022-2836(78)90314-5. [DOI] [PubMed] [Google Scholar]
- 37.Schachman H. K. Can a simple model account for the allosteric transition of aspartate transcarbamoylase? J. Biol. Chem. 1998;263:18583–18586. [PubMed] [Google Scholar]
- 38.Fetler L. P., Tauc P., Hervé G., Moody M. F., Vachette P. X-ray scattering titration of the quaternary structure transition of aspartate transcarbamylase with a bisubstrate analogue: influence of nucleotide effectors. J. Mol. Biol. 1995;251:243–255. doi: 10.1006/jmbi.1995.0432. [DOI] [PubMed] [Google Scholar]
- 39.Marina A., Alzari P. M., Bravo J., Uriarte M., Barcelona B., Fita I., Rubio V. Carbamate kinase: new structural machinery for making carbamoyl phosphate, the common precursor of pyrimidines and arginine. Protein Sci. 1999;8:934–940. doi: 10.1110/ps.8.4.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uriarte M., Marina A., Ramón-Maiques S., Fita I., Rubio V. The carbamoyl-phosphate synthetase of Pyrococcus furiosus is enzymologically and structurally a carbamate kinase. J. Biol. Chem. 1999;274:16295–16303. doi: 10.1074/jbc.274.23.16295. [DOI] [PubMed] [Google Scholar]