Abstract
Purpose
Artificial Intelligence (AI) has become increasingly integrated clinically within neurosurgical oncology. This report reviews the cutting-edge technologies impacting tumor treatment and outcomes.
Methods
A rigorous literature search was performed with the aid of a research librarian to identify key articles referencing AI and related topics (machine learning (ML), computer vision (CV), augmented reality (AR), virtual reality (VR), etc.) for neurosurgical care of brain or spinal tumors.
Results
Treatment of central nervous system (CNS) tumors is being improved through advances across AI—such as AL, CV, and AR/VR. AI aided diagnostic and prognostication tools can influence pre-operative patient experience, while automated tumor segmentation and total resection predictions aid surgical planning. Novel intra-operative tools can rapidly provide histopathologic tumor classification to streamline treatment strategies. Post-operative video analysis, paired with rich surgical simulations, can enhance training feedback and regimens.
Conclusion
While limited generalizability, bias, and patient data security are current concerns, the advent of federated learning, along with growing data consortiums, provides an avenue for increasingly safe, powerful, and effective AI platforms in the future.
Keywords: Artificial intelligence (AI), Machine learning (ML), Computer vision (CV), Augmented / virtual reality (AR/VR), Neurosurgery, Brain tumor
Introduction
Advanced technologies, generally under the umbrella of artificial intelligence (AI), are having a profound impact on the practice of medicine and particularly with neuro-oncological patient care. A brief definition of the different AI components and principals discussed in this review are described below to outline their interconnected relationship (Table 1; Fig. 1). AI is the use of technology to simulate intelligence and human thought [1]. Machine learning (ML) is a subset of AI that uses algorithms to recognize patterns and learn from data [1]. Neural networks, inspired from the brain’s neural structure, are complex interconnected layers of information [1, 2]. Deep learning is a type of ML algorithm that leverages neural networks to iterate its knowledge base and performance [1, 3]. Similar to deep learning, radiomics is another approach that is often used for radiographic analysis. However, it is trained with a pre-defined feature set that generates model variables, and the model can be based in linear regressions or ML algorithms [3]. Computer vision (CV) leverages different imaging hardware (like cameras and microscopes) along with ML and other algorithms to understand and analyze images [1]. Often, CV is used in radiomic feature preparation. Natural language processing (NLP) is a different type of AI that leverages ML (and deep learning) to understand and create text [4]. Augmented reality (AR) superimposes virtual information into the real world, while virtual reality (VR) creates an entirely new world [5]. Importantly, both AR and VR systems utilize AI, including ML and CV, to process and construct these environments [5]. Collectively, these different tools are dramatically improving medicine, especially within neuro-oncology.
Table 1.
Defining relevant AI concepts
| Term | Definition |
|---|---|
| Artificial Intelligence (AI) | The use of technology to simulate intelligence and human thought [1] |
| Machine Learning (ML) | A subset of AI that uses algorithms to recognize patterns and learn from data [1] |
| Deep Learning | A type of ML algorithm based in neural networks to iterate knowledge base and model performance [1, 3] |
| Neural Networks | Complex, interconnected nodes of information and algorithms inspired by the brain’s neural structure [1, 2] |
| Computer Vision (CV) | Computerized image analysis that leverages AI, including ML, along with other image capturing hardware (cameras, microscopes, etc.) [1] |
| Radiomics | An approach to radiographic analysis that harnesses pre-defined feature sets to generate variables, which can power linear regressions or ML models (including deep learning models) [3] |
| Natural Language Processing (NLP) | A type of AI that utilizing ML, including deep learning, to analyze language and create text [4] |
| Augmented Reality (AR) | A process that utilizes AI, including ML and CV, to superimpose virtual information into the real world [5] |
| Virtual Reality (VR) | Like AR, VR also uses ML and CV to create an entirely new, virtual world [5] |
Fig. 1.
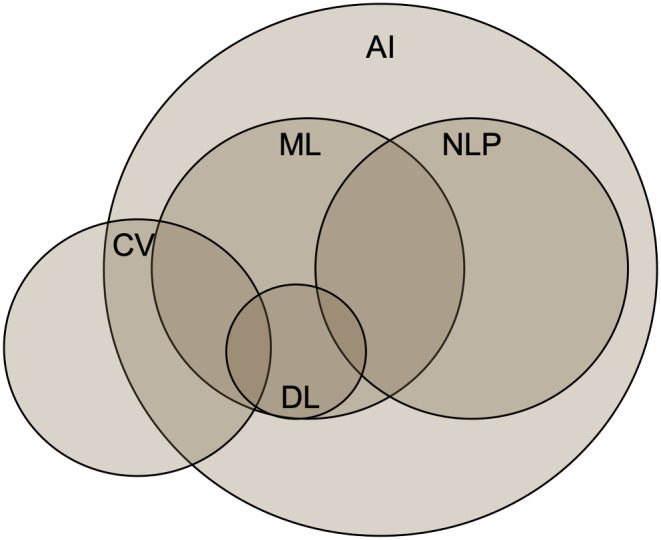
AI is a broad term with many interconnected concepts. While machine learning (ML) and natural language processing (NLP) are separate fields, they both leverage tools from each other, including deep learning (DL). Similarly, computer vision (CV) utilizes ML and DL but also many tools outside of the AI umbrella
The innovative nature of neurosurgery, along with the recent computing advances discussed above, has led to further clinical integration of impactful AI tools. Brain and spinal tumor treatment, in particular, has benefitted from platforms powered by ML, CV, and AR/VR. From pre-operative prognostication and planning to intra-operative pathologic identification and post-operative surgical feedback, AI is enhancing patient care at every step of neuro-oncologic treatment (Fig. 2). Moreover, AR/VR platforms are creating immersive surgical training supplements, with real-time feedback. In this review, we discuss AI tools currently improving the field of surgical neuro-oncology.
Fig. 2.
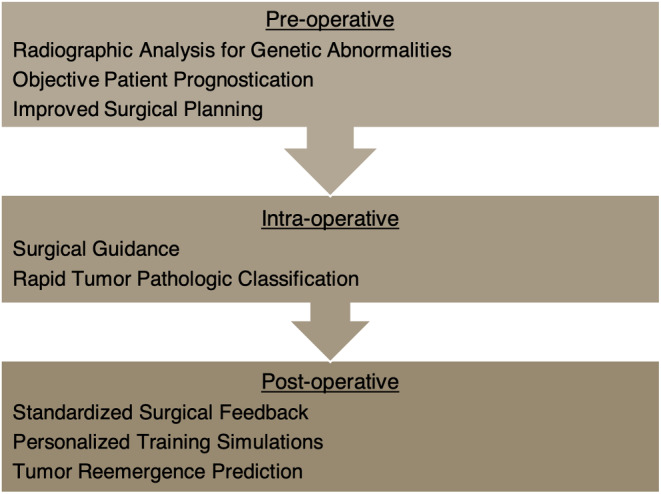
AI technologies influence care in a variety of ways throughout a neuro-oncological patient’s experience, from their pre-operative visits to their surgery and beyond
Preoperative care
A diagnosis of a Central Nervous System (CNS) tumor can be challenging for patients and their families. The initial patient encounter, including obtaining a diagnosis and prognosticating likely outcomes, helps shape the patient’s experience with the healthcare system and can guide their medical decisions. AI is improving this process through harnessing tremendous quantities of data–nearly 1/3 of all data in the world is in the electronic health record (EHR)–and finding associations with pathology and outcomes [6, 7].
Leveraging AI has improved non-invasive, imaging-based diagnosis of CNS tumors, which benefits from rapid, early identification and treatment. Radiomics is a quantitative approach using AI to extract features, or variables, from radiographic images and to use these features to build predictive models [3]. In cranial imaging, multiple studies have demonstrated that radiomic approaches can recognize radiographic abnormalities much more quickly and accurately than radiologists [8–10]. In oncology, these approaches for imaging analysis can identify genetic abnormalities and extract predictive information with a high degree of accuracy [11]. Using this approach, Pease and Gersey et al. employed the Maximum Relevance Minimum Redundancy technique to identify the most relevant glioblastoma features to build a robust predictive pipeline, which estimates MGMT methylation, EGFR amplification, and molecular subgroup [11]. Differentiating tumor pathologies with radiomic models can help predict overall survival and accurately detail tumor genetics [11–13].
While radiomic MRI analysis benefits from expert level feature identification, this process requires a higher time commitment, and deep learning is a more automated approach [11, 14]. Allowing the computer to learn on its own, even identifying features unnoticeable to the human eye, has similarly resulted in powerful pre-operative genetic predictions. Cluceru et al. employed this approach to develop a glioma subtype classifier [15]. This model leveraged traditional T2 images with diffusion weighted images to concurrently evaluate for IDH mutations and 1p19q codeletions [15]. In a similar approach, Shu et al. identified Ki-67 to predict aggressive pituitary adenoma invasiveness [16]. This information can dictate treatment by better outlining available targeted therapies and surgical need. Applying a deep-learning strategy beyond primary tumors, Grossman et al. distinguished between metastasized small cell and non-small cell cancers [17]. Since small cell cancer is usually more aggressive but not typically surgically treated, better differentiating it from non-small cell cancer can vastly alter patient care [17]. Indeed, both radiomics-based and deep learning-based MRI analysis provide a non-invasive, pre-operative characterization of primary and metastasized tumors that leads to better treatment.
Beyond tumor classification, AI tools have led to accurate prognostic models. These models are needed to guide clinical care, appropriately treating those with a chance for favorable outcomes and avoiding unnecessary or painful procedures with little chance for a prolonged quality of life. ML and neural network models–through capturing complex, nonlinear relationships and interaction effects in large datasets–can improve predictions compared to traditional statistical techniques in oncology patients. For example, in brain metastasis patients treated with stereotactic radiosurgery, Oermann et al. demonstrated neural network models with common clinical data such as gender, tumor type, and performance status significantly outperformed regression approaches by nearly ten points in the area under the receiver operating curve [18]. Other models, such as the one developed by Muhlestein et al. to identify high risk patients or those likely to have nonhome discharge, may assist with preoperative counseling and medical optimization [19]. In one of the largest efforts to date, ML models outperformed clinical oncologists predicting 3-month mortality in a multi-institutional cohort of thousands of patients with metastatic cancer [20]. Here, Zachariah et al. used traditional clinical variables easily obtainable in the EHR to build this model with a tree boosting approach [20].
Unfortunately, manual EHR analysis can be tedious. Thus, large language models—using similar techniques to ChatGPT—offer the promise to harness the complex data stored in free text clinical notes. Jiang et al. demonstrated that NLP algorithms could successfully predict a variety of relevant hospital tasks including disposition, comorbidities, and increased length of stay in an “all purpose prediction engine” [21]. Within oncology, Muhlestein et al. and Nunez et al. used this approach to successfully improve nonhome discharge and survival prediction from patient notes [4, 22]. One benefit of these NLP approaches is the relatively simple aspect of data extraction from the EHR [4, 22]. Future approaches will utilize multiple data sources to help guide the patient course from diagnosis to prognosis. For example, Pease and Gersey et al. developed a ‘report card’ that packaged likely diagnoses, genetic alterations, and survival curves in a clinically applicable tool [11]. Applications such as this can provide more information to providers, so that they are best prepared to guide potential conversations with patients and their families, should they seek more information on their clinical course [11]. As data availability increases, these pre-operative tools will lead to objective prognostication and optimal treatment plans that can improve patient care.
Surgical planning
Advances in CV were amongst the first AI methods to be utilized in the clinical space, and these have shown great promise for planning of intervention and following treatment response. This has been especially apparent in stereotactic radiosurgery (SRS) thanks to sophisticated segmentation algorithms such as the U-net [23]. Recently, Lu et al. used computer vision to automatically identify tumor borders in several different pathologies [24]. This improved contouring accuracy—especially for inexperienced clinicians—and improved efficiency by roughly 30%. In a cohort of patients with only brain metastases, Bousabarah et al. had similar segmentation accuracy but model performance was limited somewhat by lesion size [25]. Performing similar to expert clinicians, Hsu et al’s automated ML pipeline was able to track treatment response [26]. Additionally, similar results were obtained from Peng et al. in the pediatric brain tumor population [27]. Using MRIs from pediatric patients with medulloblastoma and high-grade gliomas, the authors were able to achieve excellent segmentation of tumors both pre- and post-operatively. This allowed for less variability in assessing treatment response in this challenging group of patients. For a more thorough discussion of the use of AI in SRS, see the recent review by Lin et al. [28].
The aforementioned progress in tumor segmentation has also greatly aided planning of neurosurgical tumor resections. For example, Musigmann at al manually extracted 107 features from T1 post contrast MRIs and trained a ML model on just these features to predict the likelihood of gross total resection, which can influence the use of surgical adjuncts like 5-aminolevulinic acid or other intra-operative imaging tools [14]. Importantly, the models used had a level of interpretability that allowed the researchers to determine what features were most predictive of gross total resection, which included expected variables such as tumor location and shape [14]. While such studies show the potential for AI to help guide surgical planning, even more exciting work is being done on automating surgery itself. Currently in development by Tucker et al. is one such tool that uses a robotic laser system to resect brain tumor tissue [29]. It makes use of fluorescence to construct an AI model to help guide the laser to areas of tumor tissue while also incorporating computer vision to help the operator optimize areas for resection [29]. Although the development of these systems is still in its infancy, AI methods are set to potentially revolutionize surgical interventions for brain tumors.
Intra-operative pathologic classification
While pre-operative tumor genetic predictions aid surgical planning, these predictions are commonly confirmed intra-operatively. A definitive genetic identification is typically made in collaboration with a neuropathologist, which can be laborious, time-intensive, and subject to variability; however, this information can have a rapid impact on surgical outcomes. For example, optimal tumor resection goals can vary between different spinal cord lesions, specific glioma subtypes, and teratoid rhabdoid tumors, which benefit from definitive tumor identification during surgery [30]. Even since the 1990s, neurosurgical teams have harnessed artificial neural networks to enhance identification speed and objectivity [31]. Two recent applications of neural networks provide substantial improvements to histological and molecular tumor identification.
Leveraging advanced microscopy techniques, Orringer et al. introduced a rapid method for tumor imaging, coined stimulated Raman histology (SRH). This technique, which mimics traditional Hematoxylin and Eosin (H&E) histology without requiring staining, allows for tissue structure visualization at a microscopic level [32]. Hollon et al. furthered this approach to build a novel clinical tool by training a convolutional neural network on SRH tumor data [33]. In less than three minutes, this comprehensive clinical pipeline can provide expert-level tumor analysis of pathologic diagnosis and grade during surgery [32, 33].
Compared to histological analyses, molecular-level tumor classification historically required days to weeks for complete analysis, which can provide critical tumor subtype information, like MGMT methylation or IDH mutations. Nanopore DNA sequencing is a rapid technique that has accelerated DNA methylation profile determination, but unfortunately large nanopore-based datasets are nonexistent [30]. To overcome this limitation, Vermeulen et al. developed stimulated nanopore data to train their neural network classifier, Sturgeon [30]. This approach allows Sturgeon to perform a methylation analysis intra-operatively and classify tumor subtypes in less than 90 min [30].
Analysis pipelines harnessing ML have made histological and molecular-based tumor identification readily available within the typical surgical window. However, neural network-based classification is limited by tissue quality [30, 32, 33]. While these tools are readily compatible in clinical settings, the quantity and purity of tumor in resected tissue samples directly influences performance [30, 32, 33]. With reasonable hardware requirements for clinical implementation, this approach can provide a critical supplement when neuropathologists are not readily available [30, 32, 33]. Ultimately, ML based tumor classification produces an accurate, rapid tumor diagnosis, which can positively direct surgical decision-making and improve patient outcomes.
Surgical skills training and evaluation
Surgical skills are constantly developed and refined through a surgeon’s career. Traditionally, graduated constructive feedback has been the predominant form of learning via a model that is driven by mentored, direct verbal feedback [34–36]. Substantial variability in the effectiveness of this approach is a major limitation to efficient training. The evaluation of surgical skills and improvement of training regimens may be augmented through novel applications of AI, including ML and AR/VR systems. Modern algorithms have emerged as powerful tools that may one day be suited to power personalized training and performance evaluation in surgical practices [37–40]. These technologies can analyze vast datasets of surgical procedures, identify patterns, and optimize training regimens to suit individual learning curves [37–40]. AI-driven simulations provide realistic scenarios, enabling surgeons to hone their skills in a risk-free environment [38]. Additionally, Kiyasseh et al. have used ML algorithms to assess a surgeon’s performance, offering constructive feedback and facilitating continuous improvement [40].
AR/VR systems are only starting to become more available with advanced interfaces that are integrated into surgical fields. By providing a low-risk interface for both the creation and simulation of surgical tasks, VR environments are an increasingly important medium for surgical training. In their systematic review, Chan et al. identified 33 virtual reality systems that simulated multiple surgical procedures while also providing objective feedback, such as force, kinematics, distance to target, and other biologic outcomes (surgical time, blood loss, volume resected) [38]. This automated objective feedback allows for the methodical assessment of skills and evaluation of the skill level between users (i.e. resident versus attending) [38].
Integrating these technologies within surgical training enhances technical skills with the future promising more sophisticated simulations and personalized training modules. The collaboration between medical professionals and technology experts is poised to reshape the paradigm of surgical education, ensuring that practitioners are equipped with the most advanced skills to deliver optimal patient outcomes.
Pitfalls
Despite the promise of AI tools to improve neuro-oncological care and training, they are limited to the effectiveness of their training. A model or platform reflects their underlying data, and unfortunately, many novel AI tools exist with underlying bias [41]. While this bias is commonly related to racial, ethnic, or socioeconomic factors, there is also risk of small or skewed datasets leading to sampling bias [41–43]. Since it is difficult to understand how many machine learning algorithms reach a decision (i.e. they may be considered ‘black boxes’), it can be challenging to ensure algorithms properly reflect the role that social health determinants play in care and outcomes [41, 42]. Though larger, more robust datasets are slowly emerging in neuro-oncology, most models are still reliant on smaller, potentially unbalanced datasets [14–17, 30, 44]. Therefore, creating robust tools can be more challenging, as algorithms can be susceptible to anomalies, which may worsen their generalizability. Data quality, similarly, varies greatly worldwide and is often worse in under-resourced countries, reducing the accuracy of AI tools trained exclusively on higher quality inputs [43]. Continued efforts to debias AI–with bias mitigation preprocessing, with careful external validation, and with larger, more representative datasets–will be crucial for widespread clinical efficacy [41, 42].
Advances in AI are not healthcare exclusive, and innovations have also increased the possibility of exploitation. For example, working backwards from cranial MRIs, Schwarz et al. reconstructed facial images for patient recognition [45, 46]. While ‘de-facing’ methods exist for altering MRIs, they do not completely protect patients and introduce potential artifacts that influence standard MRI analysis [45]. Therefore, it is important for increased discussion on effective patient consent before including their imaging in larger databases. Along with the continued development of more effective de-identification protocols for data, increased data privacy standards are necessary to facilitate patient protection with growing accessibility to patient data.
Future
Data consortiums are a growing avenue for centralized creation and dissemination of large, tumor specific datasets. Multi-institutional consortiums, like ReSPOND (Radiomics Signatures for PrecisiON Diagnostics) for glioblastomas and GLASS (Glioma Longitudinal Analysis) for gliomas, have helped create standardized, comprehensive molecular and imaging datasets [47, 48]. Likewise, the Brain Tumor Image Segmentation (BraTS) Challenge is a global competition that makes large MRI datasets widely available and encourages continuous model improvement and evaluation [44]. However, top performing algorithms tended to be isolated to a particular brain region, tumor size, and/or image quality, indicating there is still substantial room for widely generalizable tumor segmentation models [43, 44]. Indeed, many contextual differences exist across health systems, conditions, and patient populations, which increases the complexity of broadly implemented AI tools. Federated learning–where models are shared and updated at different institutions without centralized sharing of data–has emerged as a paradigm to safely enhance training without increased data risk [49]. This approach helps expose models more broadly to diverse data, and along with expanded centralized datasets, has led to vastly improved generalizability of AI tools [49].
Robust AI systems promise to revolutionize neuro-oncological treatment through accurate patient prognostication, rapid tumor segmentation and histopathologic identification, and real-time objective feedback. With growing datasets and increased collaboration for federated learning, these tools will become more effective and clinically integrated. Even now, re-emergence of glioblastomas following initial treatment is predicted with MRI analysis [50–52]. As these models improve, the capability to predict future tumor locations and emergence will drive improved preemptive care. Ultimately, these toolkits along with other AI based tools will lead to holistic, optimized outcomes and enhance the ability of neurosurgeons to treat patients with these challenging diseases.
Acknowledgements
Thank you to Philip Walker for his guidance during the literature search for this review.
Author contributions
L.B.C. conceptualized the review article idea. C.R.B. performed the literature search. All authors contributed to the structure of the article. C.R.B., M.P., D.P.S., and A.A. initially drafted the article, and C.R.B. prepared the accompanying figures. L.B.C. critically revised the work with C.R.B.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reference list
- 1.Tangsrivimol JA, Schonfeld E, Zhang M, Veeravagu A, Smith TR, Härtl R, Lawton MT, El-Sherbini AH, Prevedello DM, Glicksberg BS, Krittanawong C (2023) Artificial Intelligence in Neurosurgery: a state-of-the-art review from past to Future. Diagnostics (Basel) 13. 10.3390/diagnostics13142429 [DOI] [PMC free article] [PubMed]
- 2.Mofatteh M (2021) Neurosurgery and artificial intelligence. AIMS Neurosci 8:477–495. 10.3934/Neuroscience.2021025 10.3934/Neuroscience.2021025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner MW, Namdar K, Biswas A, Monah S, Khalvati F, Ertl-Wagner BB (2021) Radiomics, machine learning, and artificial intelligence-what the neuroradiologist needs to know. Neuroradiology 63:1957–1967. 10.1007/s00234-021-02813-9 10.1007/s00234-021-02813-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muhlestein WE, Monsour MA, Friedman GN, Zinzuwadia A, Zachariah MA, Coumans JV, Carter BS, Chambless LB (2021) Predicting Discharge Disposition following Meningioma Resection using a multi-institutional Natural Language Processing Model. Neurosurgery 88:838–845. 10.1093/neuros/nyaa585 10.1093/neuros/nyaa585 [DOI] [PubMed] [Google Scholar]
- 5.Kazemzadeh K, Akhlaghdoust M, Zali A (2023) Advances in artificial intelligence, robotics, augmented and virtual reality in neurosurgery. Front Surg 10:1241923. 10.3389/fsurg.2023.1241923 10.3389/fsurg.2023.1241923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faggella D (2018) Where Healthcare’s Big Data Actually Comes From. In: Emerj (ed). Emerj
- 7.Suter-Crazzolara C (2018) Better Patient Outcomes through Mining of Biomedical Big Data. Front ICT 5:30 10.3389/fict.2018.00030 [DOI] [Google Scholar]
- 8.Titano JJ, Badgeley M, Schefflein J, Pain M, Su A, Cai M, Swinburne N, Zech J, Kim J, Bederson J, Mocco J, Drayer B, Lehar J, Cho S, Costa A, Oermann EK (2018) Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat Med 24:1337–1341. 10.1038/s41591-018-0147-y 10.1038/s41591-018-0147-y [DOI] [PubMed] [Google Scholar]
- 9.Chilamkurthy S, Ghosh R, Tanamala S, Biviji M, Campeau NG, Venugopal VK, Mahajan V, Rao P, Warier P (2018) Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet 392:2388–2396. 10.1016/s0140-6736(18)31645-3 10.1016/s0140-6736(18)31645-3 [DOI] [PubMed] [Google Scholar]
- 10.Pease M, Arefan D, Barber J, Yuh E, Puccio A, Hochberger K, Nwachuku E, Roy S, Casillo S, Temkin N, Okonkwo DO, Wu S (2022) Outcome prediction in patients with severe traumatic brain Injury using Deep Learning from Head CT scans. Radiology 304:385–394. 10.1148/radiol.212181 10.1148/radiol.212181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pease M, Gersey ZC, Ak M, Elakkad A, Kotrotsou A, Zenkin S, Elshafeey N, Mamindla P, Kumar VA, Kumar AJ, Colen RR, Zinn PO (2022) Pre-operative MRI radiomics model non-invasively predicts key genomic markers and survival in glioblastoma patients. J Neurooncol 160:253–263. 10.1007/s11060-022-04150-0 10.1007/s11060-022-04150-0 [DOI] [PubMed] [Google Scholar]
- 12.Kumar S, Choudhary S, Jain A, Singh K, Ahmadian A, Bajuri MY (2023) Brain tumor classification using deep neural network and transfer learning. Brain Topogr 36:305–318. 10.1007/s10548-023-00953-0 10.1007/s10548-023-00953-0 [DOI] [PubMed] [Google Scholar]
- 13.Pálsson S, Cerri S, Poulsen HS, Urup T, Law I, Van Leemput K (2022) Predicting survival of glioblastoma from automatic whole-brain and tumor segmentation of MR images. Sci Rep 12:19744. 10.1038/s41598-022-19223-3 10.1038/s41598-022-19223-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musigmann M, Akkurt BH, Krähling H, Brokinkel B, Henssen D, Sartoretti T, Nacul NG, Stummer W, Heindel W, Mannil M (2022) Assessing preoperative risk of STR in skull meningiomas using MR radiomics and machine learning. Sci Rep 12:14043. 10.1038/s41598-022-18458-4 10.1038/s41598-022-18458-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cluceru J, Interian Y, Phillips JJ, Molinaro AM, Luks TL, Alcaide-Leon P, Olson MP, Nair D, LaFontaine M, Shai A, Chunduru P, Pedoia V, Villanueva-Meyer JE, Chang SM, Lupo JM (2022) Improving the noninvasive classification of glioma genetic subtype with deep learning and diffusion-weighted imaging. Neuro Oncol 24:639–652. 10.1093/neuonc/noab238 10.1093/neuonc/noab238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu XJ, Chang H, Wang Q, Chen WG, Zhao K, Li BY, Sun GC, Chen SB, Xu BN (2022) Deep learning model-based approach for preoperative prediction of Ki67 labeling index status in a noninvasive way using magnetic resonance images: a single-center study. Clin Neurol Neurosurg 219:107301. 10.1016/j.clineuro.2022.107301 10.1016/j.clineuro.2022.107301 [DOI] [PubMed] [Google Scholar]
- 17.Grossman R, Haim O, Abramov S, Shofty B, Artzi M (2021) Differentiating small-cell Lung Cancer from Non-small-cell Lung Cancer Brain metastases based on MRI using efficientnet and transfer Learning Approach. Technol Cancer Res Treat 20:15330338211004919. 10.1177/15330338211004919 10.1177/15330338211004919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oermann EK, Kress MA, Collins BT, Collins SP, Morris D, Ahalt SC, Ewend MG (2013) Predicting survival in patients with brain metastases treated with radiosurgery using artificial neural networks. Neurosurgery 72:944–951 discussion 952. 10.1227/NEU.0b013e31828ea04b 10.1227/NEU.0b013e31828ea04b [DOI] [PubMed] [Google Scholar]
- 19.Muhlestein WE, Akagi DS, Kallos JA, Morone PJ, Weaver KD, Thompson RC, Chambless LB (2018) Using a guided machine learning ensemble model to Predict Discharge Disposition following Meningioma Resection. J Neurol Surg B Skull Base 79:123–130. 10.1055/s-0037-1604393 10.1055/s-0037-1604393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zachariah FJ, Rossi LA, Roberts LM, Bosserman LD (2022) Prospective Comparison of Medical oncologists and a machine learning model to Predict 3-Month Mortality in patients with metastatic solid tumors. JAMA Netw Open 5:e2214514. 10.1001/jamanetworkopen.2022.14514 10.1001/jamanetworkopen.2022.14514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang LY, Liu XC, Nejatian NP, Nasir-Moin M, Wang D, Abidin A, Eaton K, Riina HA, Laufer I, Punjabi P, Miceli M, Kim NC, Orillac C, Schnurman Z, Livia C, Weiss H, Kurland D, Neifert S, Dastagirzada Y, Kondziolka D, Cheung ATM, Yang G, Cao M, Flores M, Costa AB, Aphinyanaphongs Y, Cho K, Oermann EK (2023) Health system-scale language models are all-purpose prediction engines. Nature 619:357–362. 10.1038/s41586-023-06160-y 10.1038/s41586-023-06160-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nunez JJ, Leung B, Ho C, Bates AT, Ng RT (2023) Predicting the survival of patients with Cancer from their initial oncology Consultation Document using Natural Language Processing. JAMA Netw Open 6:e230813. 10.1001/jamanetworkopen.2023.0813 10.1001/jamanetworkopen.2023.0813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference. Springer International Publishing, Munich, Germany
- 24.Lu SL, Xiao FR, Cheng JC, Yang WC, Cheng YH, Chang YC, Lin JY, Liang CH, Lu JT, Chen YF, Hsu FM (2021) Randomized multi-reader evaluation of automated detection and segmentation of brain tumors in stereotactic radiosurgery with deep neural networks. Neuro Oncol 23:1560–1568. 10.1093/neuonc/noab071 10.1093/neuonc/noab071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bousabarah K, Ruge M, Brand JS, Hoevels M, Rueß D, Borggrefe J, Große Hokamp N, Visser-Vandewalle V, Maintz D, Treuer H, Kocher M (2020) Deep convolutional neural networks for automated segmentation of brain metastases trained on clinical data. Radiat Oncol 15:87. 10.1186/s13014-020-01514-6 10.1186/s13014-020-01514-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu DG, Ballangrud Å, Prezelski K, Swinburne NC, Young R, Beal K, Deasy JO, Cerviño L, Aristophanous M (2023) Automatically tracking brain metastases after stereotactic radiosurgery. Phys Imaging Radiat Oncol 27:100452. 10.1016/j.phro.2023.100452 10.1016/j.phro.2023.100452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng J, Kim DD, Patel JB, Zeng X, Huang J, Chang K, Xun X, Zhang C, Sollee J, Wu J, Dalal DJ, Feng X, Zhou H, Zhu C, Zou B, Jin K, Wen PY, Boxerman JL, Warren KE, Poussaint TY, States LJ, Kalpathy-Cramer J, Yang L, Huang RY, Bai HX (2022) Deep learning-based automatic tumor burden assessment of pediatric high-grade gliomas, medulloblastomas, and other leptomeningeal seeding tumors. Neuro Oncol 24:289–299. 10.1093/neuonc/noab151 10.1093/neuonc/noab151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin YY, Guo WY, Lu CF, Peng SJ, Wu YT, Lee CC (2023) Application of artificial intelligence to stereotactic radiosurgery for intracranial lesions: detection, segmentation, and outcome prediction. J Neurooncol 161:441–450. 10.1007/s11060-022-04234-x 10.1007/s11060-022-04234-x [DOI] [PubMed] [Google Scholar]
- 29.Tucker M, Ma G, Ross W, Buckland DM, Codd PJ (2021) Creation of an automated fluorescence guided tumor ablation system. IEEE J Transl Eng Health Med 9:4300109. 10.1109/jtehm.2021.3097210 10.1109/jtehm.2021.3097210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vermeulen C, Pagès-Gallego M, Kester L, Kranendonk MEG, Wesseling P, Verburg N, de Witt Hamer P, Kooi EJ, Dankmeijer L, van der Lugt J, van Baarsen K, Hoving EW, Tops BBJ, de Ridder J (2023) Ultra-fast deep-learned CNS tumour classification during surgery. Nature 622:842–849. 10.1038/s41586-023-06615-2 10.1038/s41586-023-06615-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sieben G, Praet M, Roels H, Otte G, Boullart L, Calliauw L (1994) The development of a decision support system for the pathological diagnosis of human cerebral tumours based on a neural network classifier. Acta Neurochir (Wien) 129:193–197. 10.1007/bf01406504 10.1007/bf01406504 [DOI] [PubMed] [Google Scholar]
- 32.Orringer DA, Pandian B, Niknafs YS, Hollon TC, Boyle J, Lewis S, Garrard M, Hervey-Jumper SL, Garton HJL, Maher CO, Heth JA, Sagher O, Wilkinson DA, Snuderl M, Venneti S, Ramkissoon SH, McFadden KA, Fisher-Hubbard A, Lieberman AP, Johnson TD, Xie XS, Trautman JK, Freudiger CW, Camelo-Piragua S (2017) Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng 1. 10.1038/s41551-016-0027 [DOI] [PMC free article] [PubMed]
- 33.Hollon TC, Pandian B, Adapa AR, Urias E, Save AV, Khalsa SSS, Eichberg DG, D’Amico RS, Farooq ZU, Lewis S, Petridis PD, Marie T, Shah AH, Garton HJL, Maher CO, Heth JA, McKean EL, Sullivan SE, Hervey-Jumper SL, Patil PG, Thompson BG, Sagher O, McKhann GM 2nd, Komotar RJ, Ivan ME, Snuderl M, Otten ML, Johnson TD, Sisti MB, Bruce JN, Muraszko KM, Trautman J, Freudiger CW, Canoll P, Lee H, Camelo-Piragua S, Orringer DA (2020) Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med 26:52–58. 10.1038/s41591-019-0715-9 [DOI] [PMC free article] [PubMed]
- 34.Porte MC, Xeroulis G, Reznick RK, Dubrowski A (2007) Verbal feedback from an expert is more effective than self-accessed feedback about motion efficiency in learning new surgical skills. Am J Surg 193:105–110. 10.1016/j.amjsurg.2006.03.016 10.1016/j.amjsurg.2006.03.016 [DOI] [PubMed] [Google Scholar]
- 35.Haglund MM, Cutler AB, Suarez A, Dharmapurikar R, Lad SP, McDaniel KE (2021) The Surgical Autonomy Program: a pilot study of Social Learning Theory Applied to Competency-based Neurosurgical Education. Neurosurgery 88:E345–e350. 10.1093/neuros/nyaa556 10.1093/neuros/nyaa556 [DOI] [PubMed] [Google Scholar]
- 36.Al Fayyadh MJ, Hassan RA, Tran ZK, Kempenich JW, Bunegin L, Dent DL, Willis RE (2017) Immediate Auditory Feedback is Superior to other types of feedback for Basic Surgical skills Acquisition. J Surg Educ 74:e55–e61. 10.1016/j.jsurg.2017.08.005 10.1016/j.jsurg.2017.08.005 [DOI] [PubMed] [Google Scholar]
- 37.Farquharson AL, Cresswell AC, Beard JD, Chan P (2013) Randomized trial of the effect of video feedback on the acquisition of surgical skills. Br J Surg 100:1448–1453. 10.1002/bjs.9237 10.1002/bjs.9237 [DOI] [PubMed] [Google Scholar]
- 38.Chan J, Pangal DJ, Cardinal T, Kugener G, Zhu Y, Roshannai A, Markarian N, Sinha A, Anandkumar A, Hung A, Zada G, Donoho DA (2021) A systematic review of virtual reality for the assessment of technical skills in neurosurgery. Neurosurg Focus 51:E15. 10.3171/2021.5.focus21210 10.3171/2021.5.focus21210 [DOI] [PubMed] [Google Scholar]
- 39.Pangal DJ, Kugener G, Shahrestani S, Attenello F, Zada G, Donoho DA (2021) A guide to annotation of Neurosurgical Intraoperative Video for Machine Learning Analysis and Computer Vision. World Neurosurg 150:26–30. 10.1016/j.wneu.2021.03.022 10.1016/j.wneu.2021.03.022 [DOI] [PubMed] [Google Scholar]
- 40.Kiyasseh D, Ma R, Haque TF, Miles BJ, Wagner C, Donoho DA, Anandkumar A, Hung AJ (2023) A vision transformer for decoding surgeon activity from surgical videos. Nat Biomed Eng 7:780–796. 10.1038/s41551-023-01010-8 10.1038/s41551-023-01010-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J, Galal G, Etemadi M, Vaidyanathan M (2022) Evaluation and mitigation of racial Bias in Clinical Machine Learning models: scoping review. JMIR Med Inf 10:e36388. 10.2196/36388 10.2196/36388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panch T, Mattie H, Atun R (2019) Artificial intelligence and algorithmic bias: implications for health systems. J Glob Health 9:010318. 10.7189/jogh.09.020318 10.7189/jogh.09.020318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adewole M, Rudie JD, Gbdamosi A, Toyobo O, Raymond C, Zhang D, Omidiji O, Akinola R, Suwaid MA, Emegoakor A, Ojo N, Aguh K, Kalaiwo C, Babatunde G, Ogunleye A, Gbadamosi Y, Iorpagher K, Calabrese E, Aboian M, Linguraru M, Albrecht J, Wiestler B, Kofler F, Janas A, LaBella D, Kzerooni AF, Li HB, Iglesias JE, Farahani K, Eddy J, Bergquist T, Chung V, Shinohara RT, Wiggins W, Reitman Z, Wang C, Liu X, Jiang Z, Familiar A, Van Leemput K, Bukas C, Piraud M, Conte GM, Johansson E, Meier Z, Menze BH, Baid U, Bakas S, Dako F, Fatade A, Anazodo UC (2023) The brain tumor segmentation (BraTS) challenge 2023: Glioma Segmentation in Sub-Saharan Africa Patient Population (BraTS-Africa). ArXiv. United States
- 44.Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Ç, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SM, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K (2015) The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 34:1993–2024. 10.1109/tmi.2014.2377694 10.1109/tmi.2014.2377694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarz CG, Kremers WK, Wiste HJ, Gunter JL, Vemuri P, Spychalla AJ, Kantarci K, Schultz AP, Sperling RA, Knopman DS, Petersen RC, Jack CR Jr (2021) Changing the face of neuroimaging research: comparing a new MRI de-facing technique with popular alternatives. NeuroImage 231:117845. 10.1016/j.neuroimage.2021.117845 10.1016/j.neuroimage.2021.117845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwarz CG, Kremers WK, Therneau TM, Sharp RR, Gunter JL, Vemuri P, Arani A, Spychalla AJ, Kantarci K, Knopman DS, Petersen RC, Jack CR Jr (2019) Identification of Anonymous MRI Research participants with Face-Recognition Software. N Engl J Med 381:1684–1686. 10.1056/NEJMc1908881 10.1056/NEJMc1908881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bakas S, Ormond DR, Alfaro-Munoz KD, Smits M, Cooper LAD, Verhaak R, Poisson LM (2020) iGLASS: imaging integration into the Glioma Longitudinal Analysis Consortium. Neuro Oncol 22:1545–1546. 10.1093/neuonc/noaa160 10.1093/neuonc/noaa160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davatzikos C, Barnholtz-Sloan JS, Bakas S, Colen R, Mahajan A, Quintero CB, Capellades Font J, Puig J, Jain R, Sloan AE, Badve C, Marcus DS, Seong Choi Y, Lee SK, Chang JH, Poisson LM, Griffith B, Dicker AP, Flanders AE, Booth TC, Rathore S, Akbari H, Sako C, Bilello M, Shukla G, Fathi Kazerooni A, Brem S, Lustig R, Mohan S, Bagley S, Nasrallah M, O’Rourke DM (2020) AI-based prognostic imaging biomarkers for precision neuro-oncology: the ReSPOND consortium. Neuro Oncol, England, pp 886–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pati S, Baid U, Edwards B, Sheller M, Wang SH, Reina GA, Foley P, Gruzdev A, Karkada D, Davatzikos C, Sako C, Ghodasara S, Bilello M, Mohan S, Vollmuth P, Brugnara G, Preetha CJ, Sahm F, Maier-Hein K, Zenk M, Bendszus M, Wick W, Calabrese E, Rudie J, Villanueva-Meyer J, Cha S, Ingalhalikar M, Jadhav M, Pandey U, Saini J, Garrett J, Larson M, Jeraj R, Currie S, Frood R, Fatania K, Huang RY, Chang K, Bala?a C, Capellades J, Puig J, Trenkler J, Pichler J, Necker G, Haunschmidt A, Meckel S, Shukla G, Liem S, Alexander GS, Lombardo J, Palmer JD, Flanders AE, Dicker AP, Sair HI, Jones CK, Venkataraman A, Jiang M, So TY, Chen C, Heng PA, Dou Q, Kozubek M, Lux F, Mich?lek J, Matula P, Ke?kovsk? M, Kop?ivov? T, Dost?l M, Vyb?hal V, Vogelbaum MA, Mitchell JR, Farinhas J, Maldjian JA, Yogananda CGB, Pinho MC, Reddy D, Holcomb J, Wagner BC, Ellingson BM, Cloughesy TF, Raymond C, Oughourlian T, Hagiwara A, Wang C, To MS, Bhardwaj S, Chong C, Agzarian M, Falc?o AX, Martins SB, Teixeira BCA, Sprenger F, Menotti D, Lucio DR, LaMontagne P, Marcus D, Wiestler B, Kofler F, Ezhov I, Metz M, Jain R, Lee M, Lui YW, McKinley R, Slotboom J, Radojewski P, Meier R, Wiest R, Murcia D, Fu E, Haas R, Thompson J, Ormond DR, Badve C, Sloan AE, Vadmal V, Waite K, Colen RR, Pei L, Ak M, Srinivasan A, Bapuraj JR, Rao A, Wang N, Yoshiaki O, Moritani T, Turk S, Lee J, Prabhudesai S, Mor?n F, Mandel J, Kamnitsas K, Glocker B, Dixon LVM, Williams M, Zampakis P, Panagiotopoulos V, Tsiganos P, Alexiou S, Haliassos I, Zacharaki EI, Moustakas K, Kalogeropoulou C, Kardamakis DM, Choi YS, Lee SK, Chang JH, Ahn SS, Luo B, Poisson L, Wen N, Tiwari P, Verma R, Bareja R, Yadav I, Chen J, Kumar N, Smits M, van der Voort SR, Alafandi A, Incekara F, Wijnenga MMJ, Kapsas G, Gahrmann R, Schouten JW, Dubbink HJ, Vincent A, van den Bent MJ, French PJ, Klein S, Yuan Y, Sharma S, Tseng TC, Adabi S, Niclou SP, Keunen O, Hau AC, Valli?res M, Fortin D, Lepage M, Landman B, Ramadass K, Xu K, Chotai S, Chambless LB, Mistry A, Thompson RC, Gusev Y, Bhuvaneshwar K, Sayah A, Bencheqroun C, Belouali A, Madhavan S, Booth TC, Chelliah A, Modat M, Shuaib H, Dragos C, Abayazeed A, Kolodziej K, Hill M, Abbassy A, Gamal S, Mekhaimar M, Qayati M, Reyes M, Park JE, Yun J, Kim HS, Mahajan A, Muzi M, Benson S, Beets-Tan RGH, Teuwen J, Herrera-Trujillo A, Trujillo M, Escobar W, Abello A, Bernal J, G?mez J, Choi J, Baek S, Kim Y, Ismael H, Allen B, Buatti JM, Kotrotsou A, Li H, Weiss T, Weller M, Bink A, Pouymayou B, Shaykh HF, Saltz J, Prasanna P, Shrestha S, Mani KM, Payne D, Kurc T, Pelaez E, Franco-Maldonado H, Loayza F, Quevedo S, Guevara P, Torche E, Mendoza C, Vera F, R?os E, L?pez E, Velastin SA, Ogbole G, Soneye M, Oyekunle D, Odafe-Oyibotha O, Osobu B, Shu'aibu M, Dorcas A, Dako F, Simpson AL, Hamghalam M, Peoples JJ, Hu R, Tran A, Cutler D, Moraes FY, Boss MA, Gimpel J, Veettil DK, Schmidt K, Bialecki B, Marella S, Price C, Cimino L, Apgar C, Shah P, Menze B, Barnholtz-Sloan JS, Martin J, Bakas S (2022) Federated learning enables big data for rare cancer boundary detection. Nat Commun 13: 7346 10.1038/s41467-022-33407-5
- 50.Metz MC, Molina-Romero M, Lipkova J, Gempt J, Liesche-Starnecker F, Eichinger P, Grundl L, Menze B, Combs SE, Zimmer C, Wiestler B (2020) Predicting Glioblastoma recurrence from preoperative MR Scans using fractional-anisotropy maps with free-water suppression. Cancers (Basel) 12. 10.3390/cancers12030728 [DOI] [PMC free article] [PubMed]
- 51.Cepeda S, Luppino LT, Pérez-Núñez A, Solheim O, García-García S, Velasco-Casares M, Karlberg A, Eikenes L, Sarabia R, Arrese I, Zamora T, Gonzalez P, Jiménez-Roldán L, Kuttner S (2023) Predicting regions of local recurrence in Glioblastomas using Voxel-based Radiomic features of Multiparametric Postoperative MRI. Cancers (Basel) 15. 10.3390/cancers15061894 [DOI] [PMC free article] [PubMed]
- 52.Rathore S, Akbari H, Doshi J, Shukla G, Rozycki M, Bilello M, Lustig R, Davatzikos C (2018) Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: implications for personalized radiotherapy planning. J Med Imaging (Bellingham) 5:021219. 10.1117/1.jmi.5.2.021219 10.1117/1.jmi.5.2.021219 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.


