Abstract
Existing strategies use bifunctional chimaeras to mediate extracellular protein degradation. However, these strategies rely on specific lysosome-trafficking receptors to facilitate lysosomal delivery, which may raise resistance concerns due to intrinsic cell-to-cell variation in receptor expression and mutations or downregulation of the receptors. Another challenge is establishing a universal platform applicable in multiple scenarios. Here, we develop MONOTAB (MOdified NanOparticle with TArgeting Binders), a plug-and-play monofunctional degradation platform that can drag extracellular targets into lysosomes for degradation. MONOTAB harnesses the inherent lysosome-targeting ability of certain nanoparticles to obviate specific receptor dependency and the hook effect. To achieve high modularity and programmable target specificity, we utilize the streptavidin-biotin interaction to immobilize antibodies or other targeting molecules on nanoparticles, through an antibody mounting approach or by direct binding. Our study reveals that MONOTAB can induce efficient degradation of diverse therapeutic targets, including membrane proteins, secreted proteins, and even extracellular vesicles.
Subject terms: Chemical biology, Biomedical materials, Drug delivery
Existing methods use bifunctional chimaeras for extracellular protein degradation but rely on specific lysosome-targeting receptors, raising resistance issues. Here, the authors develop MONOTAB, a monofunctional platform that degrades extracellular proteins and vesicles without receptor dependency.
Introduction
Targeted protein degradation (TPD) has emerged as a powerful approach for selective elimination of protein targets. The major approaches, such as proteolysis-targeting chimeras (PROTACs)1,2 and molecular glues3, leverage the ubiquitin-proteasome system or the autophagy-lysosome machinery to facilitate protein degradation. Consequently, these platforms primarily target intracellular proteins with cytosolic domains amenable to ligand binding.
Recent advances in lysosome-targeting chimeras (LYTACs)4,5 and related technologies6–12 have provided useful tools for extracellular protein degradation. These approaches involve the use of bifunctional molecules that bridge the extracellular domain of a target protein with lysosome-trafficking receptors (LTRs), leading to endocytosis and subsequent lysosomal degradation of the targeted proteins. Another approach involves bispecific antibody-based platforms13,14 that bring membrane-associated target proteins and transmembrane E3 ubiquitin ligases in close proximity and hence facilitate protein ubiquitination and lysosomal degradation. However, relying on specific surface receptors or E3 ligases for lysosomal delivery can introduce complexities and challenges, especially in therapeutic applications.
First of all, the expression of these surface receptors or E3 ligases can vary significantly among patients and even cells of the same patient. This cell-to-cell variability can confer selection advantages and contribute to drug resistance15,16. Second, many of the receptors or E3 ligases that have been exploited are not essential to cell viability. Cells treated with degraders that utilize non-essential receptors or E3 ligases may select for the loss of the genes encoding those proteins, rendering the degrader ineffective over time17. In addition, bifunctional chimaeras can be confounded by the hook effect due to the differential binding affinities of both sides, which impairs the efficacy of the molecules as a whole to induce an effective ternary complex and can also serve as a potential resistance mechanism18. Apart from the resistance concerns, the complex production process of bifunctional chimaeras, which requires multi-step chemical synthesis or protein recombination, impairs the scalability of these approaches. These limitations underscore the need for a more accessible TPD platform that does not rely on specific surface receptors or E3 ligases for extracellular protein degradation, which, to our knowledge, has not been developed yet.
Many studies have demonstrated that nanomaterials such as polymeric nanoparticles and lipid nanoparticles, without ligand modification, can be easily endocytosed by cells and trafficked to lysosomes19–21. Notably, this lysosome-targeting endocytosis occurs independently of specific surface receptors21. Building upon the inherent propensity of nanomaterials to lysosomal delivery, we develop a plug-and-play TPD platform based on modified nanoparticles with targeting binders (MONOTAB) that can pull extracellular targets into lysosomes for degradation. As a proof of concept, we use Streptavidin-conjugated polystyrene nanoparticles (NPs) as a model chassis. MONOTABs are facilely prepared by sequentially assembling biotinylated anti-IgG (Fc specific) antibodies and targeted antibodies with matching Fc fragments or by directly immobilizing biotinylated targeting binders on the NPs. This modular assembly approach allows for flexibility and versatility in tailoring MONOTAB for different target proteins. We demonstrate the targeting scope of MONOTAB by inducing the degradation of various therapeutically relevant proteins (Fig. 1a), including programmed death-ligand 1 (PD-L1) and matrix metallopeptidase 2 (MMP2). Furthermore, we show that extracellular non-protein targets such as extracellular vesicles (EVs) can also be redirected into lysosomes for degradation using the MONOTAB platform.
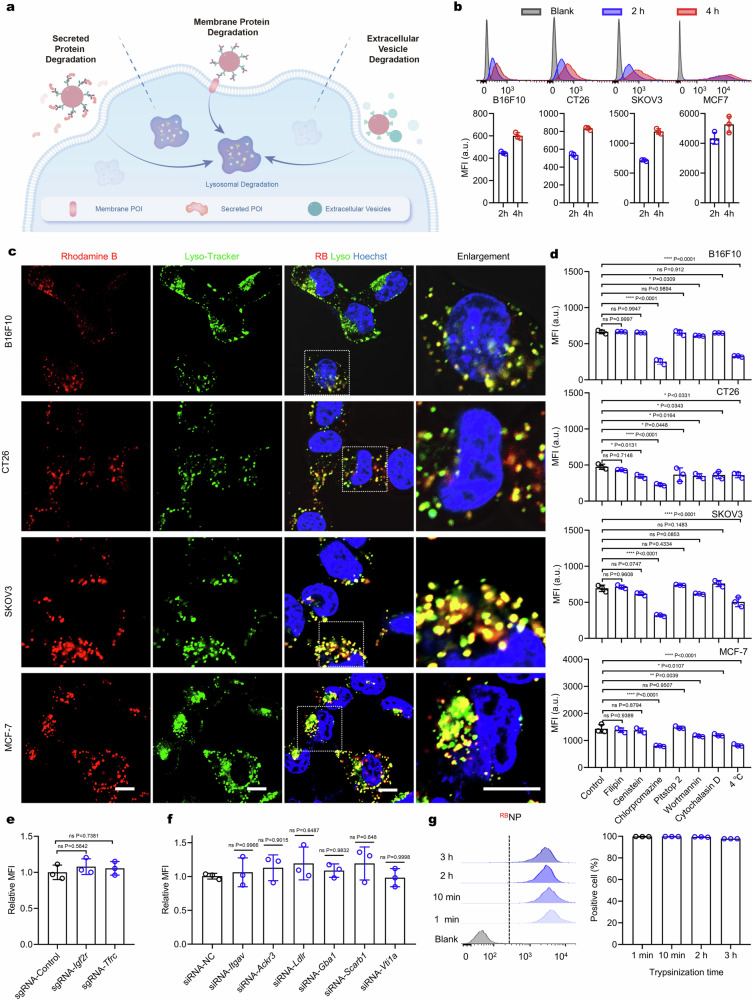
Fig. 1. The concept of MONOTAB based on lysosome-targeting endocytosis of NPs.
a Targeted degradation of extracellular proteins or vesicles mediated by MONOTABs. b Cellular uptake of rhodamine B-labeled NPs (RBNPs) after co-incubation with B16F10 cells for 2 or 4 h (n = 3 biologically independent experiments, each counting 10,000 cells). c Live-cell imaging of different cell lines treated with RBNPs (100 μg mL−1) for 4 h. Scale bar, 10 μm. The images are representative of n = 3 biological replicates. d Effects of endocytic inhibitors on cellular uptake of RBNPs in different cell lines in terms of the RB intensity measured by flow cytometry (n = 3 biologically independent experiments). e and f Cellular uptake of RBNPs (50 μg mL−1) after co-incubation with Igf2r- or Tfrc-KO B16F10 cells, or Itgav-, Ackr3-, Ldlr-, Gba1-, Scarb1- or Vti1a-silencing B16F10 cells for 2 h (n = 3 biologically independent experiments, each counting 10,000 cells). g Cellular uptake of RBNPs (50 μg mL−1) after co-incubation with B16F10 cells for 2 h. Cells were pre-treated with trypsin within 3 h (n = 3 biologically independent experiments, each counting 10,000 cells). Data are presented as mean ± SD where relevant. p values were determined by one-way analysis of variance (ANOVA) with Dunnett’s post hoc test. ns no significance; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Source data are provided as a Source Data file.
Results
Lysosome-targeting endocytosis of NPs
The inherent susceptibility of NPs to lysosome-targeting endocytosis underlies the MONOTAB concept. To validate this rationale, we first explored the lysosomal delivery of NPs. A panel of cells (including B16F10, CT26, SKOV3, and MCF-7) was incubated with rhodamine B-labeled NPs (RBNPs, Supplementary Fig. 1a) for 2 or 4 h, washed and then analyzed by flow cytometry. The results revealed that over 90% of cells in each cell line were fluorescently positive after 4-h incubation (Fig. 1b). Moreover, we confirmed that nearly all the detected NPs were internalized into cells but not absorbed on the membrane via Trypan Blue quenching (Supplementary Fig. 1b and c). The fluorescence intensity measurements of RBNPs in the medium following co-incubation showed that ~22% of the given NPs were internalized within 2 h and ~28% within 4 h (Supplementary Fig. 1d and e). We further examined the subcellular distribution of RBNPs after 4 h incubation using confocal microscopy, which showed high-degree colocalization between the RB signal and lysosomes (Fig. 1c and Supplementary Fig. 1f), with Manders’ colocalization coefficients consistently exceeding 0.8 or even 0.9 (Supplementary Fig. 1g). Notably, despite distinct membrane proteomes among these cell lines22,23, minor variations in NP uptake and lysosomal localization were observed, implying that the lysosome-targeting endocytosis of NPs does not rely on specific surface receptors19.
To identify the critical pathways involved in NP internalization, we next performed a set of endocytosis inhibition experiments. Cells were pre-treated with chemical endocytosis inhibitors or exposed to low temperatures (4 °C), followed by incubation with RBNPs for 1.5 h. Flow cytometry analysis showed that low-temperature treatment significantly inhibited RBNP uptake across all cell lines, pointing to an energy-dependent uptake process. Cholesterol sequestration (filipin)24 or inhibition of clathrin-coated pit (CCP) dynamics (Pitstop 2)25,26 did not influence NP uptake, while the tyrosine kinase inhibitor (genistein)27, actin polymerization inhibitor (cytochalasin D)28,29, and PI3K signaling inhibitor (wortmannin)30,31 showed mild inhibitory effects in a cell line-specific manner (Fig. 1d, Supplementary Fig. 2a and b). Notably, chlorpromazine significantly reduced NP uptake in all cell lines. Since chlorpromazine inhibits clathrin-mediated endocytosis but not specifically21, we used siRNA (small interfering RNA) to knock down clathrin (Supplementary Fig. 2c) to confirm the involvement of clathrin in the internalization process. This knockdown resulted in a significant decrease in NP uptake by ~60% (Supplementary Fig. 2d). These results suggest a major role of clathrin-mediated endocytosis in NP internalization.
To ascertain the independence of NP uptake from specific surface receptors, we screened a panel of typical receptors associated with lysosomal trafficking, including insulin-like growth factor 2 receptor (IGF2R)4, asialoglycoprotein receptor (ASGPR)5,11,12, integrin αvβ3 (ITGAV:ITGB3)7, atypical chemokine receptor 3 (ACKR3/CXCR7)8, scavenger receptor (SCARB1)9, glucagon-like peptide 1 receptor (GLP-1R)32, transferrin receptor (TFRC)33, low-density lipoprotein receptor (LDLR)34, glucosylceramidase beta (GBA)35, and vesicle transport through interaction with t-SNAREs (VTI)36. Since Asgpr and Glp-1r are minimally expressed in B16F10 cells, we focused on the remaining to identify genes for which knockdown may ablate NP uptake. Targeted gene silencing via CRISPR interference (Supplementary Fig. 3a) or RNA interference (Supplementary Fig. 3b) did not impact the internalization of RBNPs (Fig. 1e and f), hinting at the independence of specific receptors. To further confirm this observation, we subjected B16F10 cells to trypsin treatment for 3 h to digest extracellular domains of membrane proteins. Subsequent co-incubation with RBNPs revealed only a mild decrease in the mean fluorescence intensity (MFI) of cells, with no change in the ratio of RB-positive cells (Fig. 1g). In contrast, co-incubation with biotinylated cRGD (biotin-cRGD) and Cy3Avidin (Supplementary Fig. 4), which are internalized via integrin-mediated endocytosis, showed a substantial reduction in the delivery of Cy3Avidin by biotin-cRGD (Supplementary Fig. 4). These results strongly suggest that NP uptake does not rely on specific surface receptors.
Construction of the MONOTAB platform
To ease the preparation of MONOTAB and enable high modularity, we sought to develop an antibody mounting platform for the convenient installation of specific antibodies. Commercially available streptavidin-conjugated anionic NPs were chosen as the chassis, which permits the anchoring of biotinylated anti-IgG (Fc specific) antibodies (αFc) onto the NPs through the streptavidin-biotin interaction. The preparation involves simply mixing the NPs with biotinylated αFc and then centrifugation. As the Fc regions of IgG antibodies from the same host species are identical, the resulting αFc-tethered NPs (αFc-NPs), which serve as the antibody mounting platform, can specifically recognize and immobilize any targeted antibody containing the Fc fragment, allowing for rapid production of diverse MONOTABs towards various targets.
To demonstrate the feasibility of this approach, we incubated NPs with a biotinylated anti-IgG antibody at a 1:1 molar ratio of streptavidin to biotin. Dynamic light scattering (DLS) measurements showed that the average hydrodynamic diameter of the resulting αFc-NPs was ~120 nm, ~20 nm larger than that of the NPs (Supplementary Fig. 5a and b). Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) confirmed the particle size and the uniformly spherical structure of both NPs and αFc-NPs (Supplementary Fig. 5c and d). To verify the presence of αFc on NPs, we ran a reducing SDS-PAGE gel with αFc-NPs, αFc, and NPs (Fig. 2a). After Coomassie blue staining, the αFc-NP lane showed bands at ~17, ~25, ~50, and ~75 kDa (Fig. 2b), which correspond to monomeric streptavidin, the light chain of αFc, the heavy chain of αFc, and monomeric immunoglobulin, respectively, confirming the anchoring of αFc on the NP surface. Furthermore, the αFc-NPs demonstrated excellent stability, as evidenced by the negligible change in particle size over 2-week storage at 4 °C (Supplementary Fig. 5e).
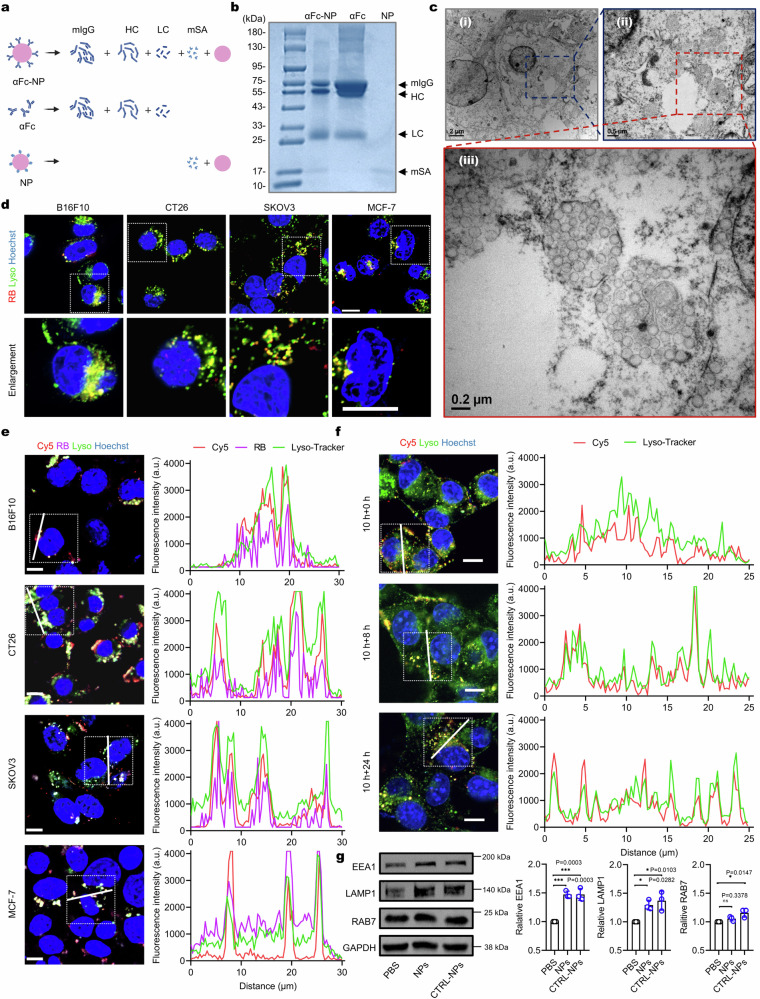
Fig. 2. Antibody anchoring does not alter the lysosome-targeting ability of NPs.
a Putative fragments of αFc-NP, αFc, and NP yielded after protein denaturation with 2-mercaptoethanol. b Reducing SDS-PAGE gel of αFc-NP, αFc, and NP with Coomassie blue staining. The gel images are representative of n = 3 independent replicates. c TEM images of cells incubated with αFc-NP (50 μg mL−1) for 8 h. The TEM images are representative of n = 3 independent replicates. d Live-cell images of B16F10, CT26, SKOV3, or MCF-7 cells treated with αFc-RBNP (100 μg mL−1) for 4 h, respectively. Scale bar, 10 μm. The images are representative of n = 3 biological replicates. e Left panel: live-cell images of different cell lines treated with RB/Cy5CTRL-NPs (50 μg mL−1) for 10 h. Scale bar, 10 μm. Right panel: Fluorescence intensity profiles along the line drawn in the box of the left panel. The images are representative of n = 3 biological replicates. f Left panel: live-cell images of B16F10 cells captured at the indicated time points after incubation with Cy5CTRL-NPs (50 μg mL−1) for 10 h and then cell washing. Scale bar, 10 μm. Right panel: Fluorescence intensity profiles along the line drawn in the box of the left panel. The images are representative of n = 3 biological replicates. g Western blots (left) and quantitative analysis (right) of EEA1, LAMP1, and RAB7 in B16F10 cells after incubation with 50 μg mL−1 CTRL-NPs or NPs for 10 h (n = 3 biologically independent experiments). Data are presented as mean ± SD where relevant. P values were determined by one-way ANOVA with Dunnett’s post hoc test. ns, no significance; *p < 0.05; ***p < 0.001. Source data are provided as a Source Data file.
We next investigated whether the surface anchoring of αFc would alter the biological performance of the NPs, particularly the lysosome-targeting endocytosis. The cell counting kit-8 (CCK8) assay proved the minimal cytotoxicity of both NPs and αFc-NPs to B16F10 cells at serial concentrations ranging from 0.01 ng mL−1 to 100 μg mL−1 (Supplementary Fig. 5f). Flow cytometry analysis showed that the endocytosis in B16F10 cells was concentration-dependent for both NPs and αFc-NPs (Supplementary Fig. 5g), and the uptake efficiency of αFc-RBNP was comparable to that of RBNP after co-incubation for 4 or 8 h, reaching 87% and 96%, respectively (Supplementary Fig. 5h). We examined the cell samples with negative-stain electron microscopy and observed abundant accumulation of spherical nanoparticles within membrane-enclosed compartments (Fig. 2c), which were subsequently identified as lysosomes through live-cell confocal microscopy (Fig. 2d, Supplementary Fig. 5i and j). The endocytosis inhibition experiments indicated that clathrin-mediated endocytosis contributed the most to αFc-NP internalization, which was consistent with the findings of NPs (Supplementary Fig. 5k). These results demonstrate that the inherent lysosome-targeting endocytosis of NPs was not altered by the surface modification with αFc.
We next incubated αFc-NPs with an IgG control antibody produced from the matching host species to generate control MONOTAB (CTRL-NP). At a 1:1 molar ratio of IgG:αFc, the average hydrodynamic diameter of CTRL-NPs was ~30 nm larger than that of αFc-NP, suggesting the successful immobilization of the IgG control onto αFc-NPs (Supplementary Fig. 6a and b). Cryo-TEM confirmed the size, morphology, and well-dispersed state of CTRL-NPs (Supplementary Fig. 6c and d). We then asked if this layer of antibody would influence the uptake efficiency. To this end, we prepared a series of CTRL-NPs with molar ratios of αFc:IgG varying from 1:0.1 to 1:2 (Supplementary Fig. 6e) and then incubated them with different cell lines for 4 h. Flow cytometry analysis showed that the varying αFc:IgG ratios did not impact NP uptake in all cell lines (Supplementary Fig. 6f). Furthermore, we confirmed that the internalization of these CTRL-NPs did not need the interactions between IgG and Fc receptors (FcRs) by using FcR-preblocked B16F10 cells (Supplementary Fig. 6g). To enable more POI binding, we selected the αFc:IgG ratio of 1:2 in the following studies.
Next, we proceeded to investigate whether the CTRL-NPs retained the lysosome-targeting ability. Cy5-labeled IgG (Cy5IgG) was immobilized on the surface of αFc-RBNPs to form RB/Cy5CTRL-NPs. Co-incubation of cells with RB/Cy5CTRL-NPs led to the colocalization of Cy5IgG and RBNPs within the lysosomes (Fig. 2e and Supplementary Fig. 6h). To ensure that the lysosomal delivery of Cy5IgG occurred specifically due to its binding to αFc-NPs, we incubated B16F10 cells with Cy5IgG alone, Cy5IgG plus αFc or NPs, or Cy5IgG plus αFc-NPs (namely Cy5CTRL-NPs). A remarkable 660-fold increase in cellular fluorescence was observed when Cy5IgG was co-incubated with αFc-NPs, whereas no increase was observed when co-incubated with αFc or NPs (Supplementary Fig. 7a, b). Furthermore, even after extended incubation for an additional 8 or 24 h following washing, the Cy5 signal remained localized within the lysosomes (Fig. 2f). These data indicate that NPs could efficiently hijack the tethered IgG into lysosomes and the tethered protein could be stably trapped without lysosomal escape.
Effects of MONOTAB on lysosomal function
After co-incubation with Cy5CTRL-NPs, we observed an unexpected increase in the LysoTracker signal (Supplementary Fig. 7a and c). This observation raises the possibility that MONOTAB might promote lysosomal biogenesis. To test this hypothesis, we examined the expression levels of endo-lysosome markers, including LAMP1 (lysosome), EEA1 (early endosome), and RAB7 (late endosome), and found that all the tested markers were upregulated after the CTRL-NP treatment (Fig. 2g). Immunofluorescence assay further confirmed the increase of LAMP1 (Supplementary Fig. 7d and e). These results are consistent with a previous report indicating that internalization of anionic polystyrene nanoparticles results in activation of the transcription factor EB, a master regulator of lysosome biogenesis, and increased lysosomal degradation capacity37.
As an increase in LysoTracker staining or lysosome markers may also be observed upon lysosomal dysfunction, one may question if the nanoparticles could potentially impair lysosomal health, thereby activating lysosome biogenesis as a compensatory response. To clarify this question, we performed the DQ Green BSA assay to evaluate lysosomal degradation capacity. Untreated B16F10 cells and cells treated with NPs, αFc-NPs, or CTRL-NPs exhibited bright green fluorescence, indicating the effective hydrolysis of the DQ Green BSA into single, dye-labeled peptides by lysosomal proteases. In contrast, no fluorescent signal was observed in cells treated with Bafilomycin A1 (BafA1), an established lysosomal inhibitor (Supplementary Fig. 7f and g). We also examined lysosome membrane stability with acridine orange (AO), a fluorescent dye that emits red fluorescence when protonated in intact lysosomes and green fluorescence when deprotonated in the cytoplasm. Strong green fluorescence was detected in cells treated with chloroquine (CQ, a lysosome-permeability enhancer), while untreated cells and cells treated with NPs, αFc-NPs, or CTRL-NPs exhibited red fluorescence only (Supplementary Fig. 7h and i). These results imply that MONOTAB may promote lysosomal biogenesis without affecting lysosomal health, which promises higher protein degradation potential.
MONOTAB-mediated degradation of membrane-associated protein PD-L1
We next aimed to evaluate the efficacy of the MONOTAB strategy on clinically relevant targets. We first targeted programmed death-ligand 1 (PD-L1), a membrane-associated protein that is often overexpressed on the surface of tumor cells and facilitates their immune evasion38,39. PD-L1-targeted MONOTABs were constructed by incubating αFc-NP with either FITC-labeled or unlabeled anti-PD-L1 antibody (αPD-L1). Co-incubation of cells with FITCαPD-L1-NPs (Fig. 3a) led to lysosomal delivery of FITCαPD-L1 (Fig. 3b and Supplementary Fig. 8a), replicating our findings with CTRL-NPs. We reasoned that αPD-L1-NPs could induce the degradation of PD-L1 by enriching PD-L1 molecules in lysosomes. B16F10 cells were treated with PBS, αPD-L1, αFc-NP, or αPD-L1-NP for 24 h and then assayed for PD-L1 levels. Western blot analysis showed substantial degradation of both total and membrane-associated PD-L1 with 3.3 nM (αPD-L1-equiv. concentration) of αPD-L1-NP (Fig. 3c and d). Immunofluorescence (IF) microscopy revealed nearly complete removal of PD-L1 from cell membranes following the 24-h treatment with αPD-L1-NP, as opposed to the treatments with PBS or αPD-L1 alone (Fig. 3e and Supplementary Fig. 8b). These results highlight the great potential of MONOTAB in inducing robust degradation of membrane proteins.
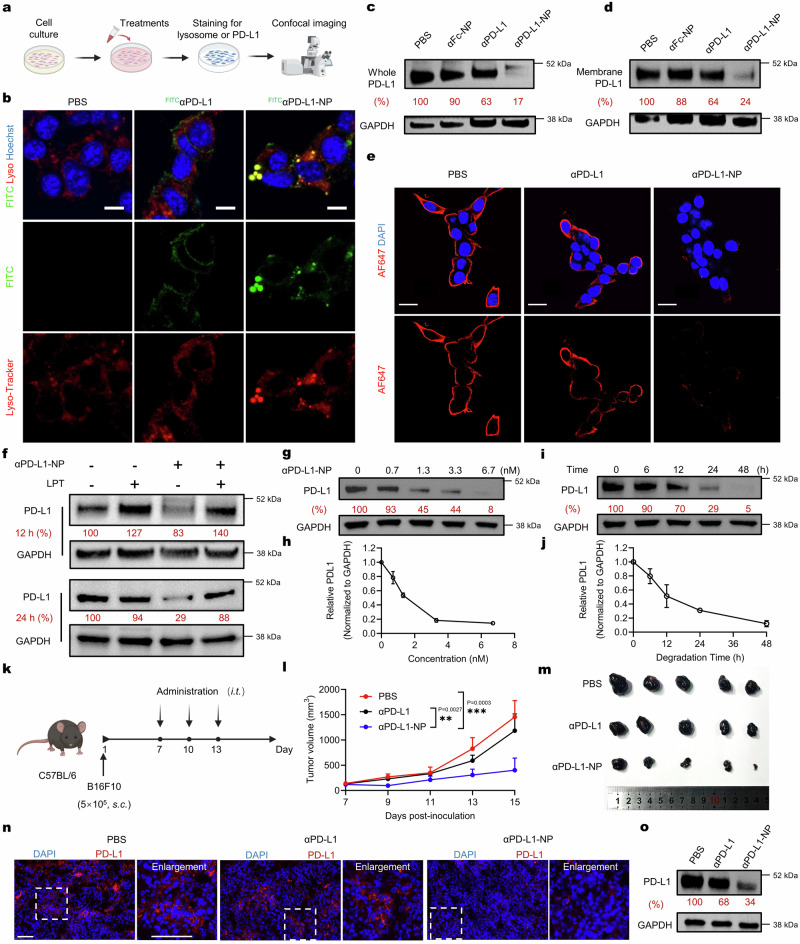
Fig. 3. Degradation of PD-L1 mediated by αPD-L1-NP.
a Schematic illustration of live-cell confocal microscopy assay. b Live-cell images of B16F10 cells treated with FITCαPD-L1 (3.3 nM) or FITCαPD-L1-NP (FITCPD-L1-equiv., 3.3 nM) for 4 h. Scale bar, 10 μm. The images are representative of n = 3 biological replicates. c, d Western blot analysis of total (c) and membrane-associated (d) PD-L1 in B16F10 cells receiving different treatments for 24 h. αPD-L1-equiv. concentration, 3.3 nM; NP-equiv. concentration, 25 μg mL−1. The blots are representative of n = 3 biological replicates. e IF of surface PD-L1 in B16F10 cells treated with αPD-L1 (3.3 nM) or αPD-L1-NP (PD-L1-equiv., 3.3 nM) for 24 h. Scale bar, 20 μm. The IF images are representative of n = 3 biological replicates. f Western blot analysis of B16F10 cells treated with αPD-L1-NP (3.3 nM) for 12 or 24 h in the presence or absence of 0.1 mg mL−1 leupeptin (LPT). The blots are representative of n = 3 biological replicates. g–j Western blot analysis of PD-L1 in B16F10 cells treated with αPD-L1-NP at the indicated concentrations for 24 h (g and h) or at 3.3 nM for the indicated durations (i and j) The blots are representative of n = 3 biological replicates. k–o In vivo antitumor study of αPD-L1-NP in B16F10 tumor-bearing C57BL/6 mice. Mice (n = 5 mice per group) were treated intratumorally (i.t.) with PBS, αPD-L1 (2.0 mg kg−1), or αPD-L1-NP (αPD-L1-equiv. dose, 2.0 mg kg−1) for three times, respectively. k Schematic diagram outlining the experimental design. l Tumor growth curves of mice receiving different treatments. m Image of tumors resected after animal euthanasia. n Immunofluorescence staining of PD-L1 in tumor sections. Scale bar, 100 μm. o Western blot analysis of PD-L1 levels in tumor lysates. The blots are representative of n = 3 biological replicates. All the uncropped blots are included in the Source Data file. Data are presented as mean ± SD. Statistical significance was calculated via one-way ANOVA with Dunnett’s post hoc test. **p < 0.01; ***p < 0.001. Source data are provided as a Source Data file.
To confirm whether the degradation occurs in lysosomes, B16F10 cells were treated with αPD-L1-NPs in the presence or absence of leupeptin (LPT), a commonly used lysosomal protease inhibitor. The LPT treatment significantly diminished the degradation of PD-L1, indicating the involvement of lysosomal proteases in the MONOTAB-mediated degradation (Fig. 3f). Further data of total PD-L1 levels following the 24-h treatment with different concentrations of αPD-L1-NP unveiled a concentration-dependent degradation profile without the hook effect (Fig. 3g and h), echoing the monofunctional modality of MONOTAB. Notably, significant degradation of PD-L1 was already detectable with αPD-L1-NP at a subnanomolar concentration (0.7 nM) and virtually complete elimination was achieved at 6.7 nM. Time-course experiments showed that MONOTAB-mediated PD-L1 degradation occurred in a time-dependent manner, with the levels of PD-L1 persistently decreasing to complete depletion by 48 h (Fig. 3i and j). These results indicate that the MONOTAB approach can efficiently direct surface proteins to lysosomes for degradation.
To validate our approach further, we conducted a comprehensive comparison between MONOTAB and existing approaches using published data on PD-L1 degradation (Supplementary Table 1). Additionally, we benchmarked MONOTAB against two established methods based on bifunctional chimeras, integrin-facilitated lysosomal degradation (IFLD) and GalNAc-LYTAC. PD-L1-targeted BMS-L1-RGD (IFLD type) and αPD-L1-GalNAc (GalNAc-LYTAC type) were synthesized following the described procedures5,7. As opposed to the near-complete degradation of PD-L1 observed with 6.7 nM of αPD-L1-NP, treatment with 50 nM of BMS-L1-RGD for the same duration led to only ~46% of PD-L1 degradation (Supplementary Fig. 8c). On the other hand, to ensure a fair comparison between αPD-L1-NP and αPD-L1-GalNAc, considering GalNAc’s avid binding to ASGPR predominately expressed on hepatocytes, we used Hepa1–6 cells, a murine hepatoma cell line, as the cell model. At a low concentration of 1.3 nM, both αPD-L1-NP and αPD-L1-GalNAc induced similar levels of PD-L1 degradation. However, at higher concentrations, αPD-L1-GalNAc exhibited a typical hook effect due to its bifunctional nature, while αPD-L1-NP caused even more substantial degradation (Supplementary Fig. 8d).
Next, we evaluated the in vivo antitumor effects of αPD-L1-NP. C57BL/6 mice bearing subcutaneous B16F10 tumors were treated with PBS, αPD-L1, or αPD-L1-NP, respectively, and tumor size was measured (Fig. 3k). Compared with the control and αPD-L1 groups, tumor growth was significantly inhibited by the treatment with αPD-L1-NP (Fig. 3l, m and Supplementary Fig. 8e) and no body weight loss was observed during the experiment (Supplementary Fig. 8f). Immunofluorescence analysis of PD-L1 expression in tumor sections revealed a markedly reduced PD-L1 level in the αPD-L1-NP group (Fig. 3n and Supplementary Fig. 8g), which was further corroborated by Western blot analysis (Fig. 3o and Supplementary Fig. 8h). These results underscore the therapeutic potential of MONOTAB in vivo.
MONOTAB-mediated degradation of secreted protein MMP2
Having demonstrated the target scope of MONOTAB towards the membrane-associated protein, we next proceeded to assess its efficacy in degrading secreted proteins. Matrix metalloproteinase 2 (MMP2), which is highly expressed in various tumors and crucial for tumor invasion and metastasis40,41, was chosen as the target protein. The anti-MMP2 MONOTAB (αMMP2-NP) was prepared similarly. B16F10 cells were treated with PBS, αMMP2, or αMMP2-NP for 12 h, and the culture media were then assayed for the MMP2 activity and content (Fig. 4a). Gelatin zymography revealed a substantial decrease in MMP2 activity following the αMMP2-NP treatment. Given that the treatment with αMMP2 also mildly reduced MMP2 activity relative to PBS control, to rule out the possibility that the observed decrease by αMMP2-NPs was exclusively due to the inhibition of MMP2 catalytic activity, we further ran Western blot to quantify the MMP2 content in the media. Similarly, a significant reduction in MMP2 content was observed with αMMP2-NPs. Moreover, αMMP2-NPs substantially elevated the intracellular MMP2 content, especially upon the inhibition of lysosomal proteases with LPT (Fig. 4b), demonstrating the ability of MONOTAB to redirect secreted proteins into lysosomes for degradation. Further analysis of MMP2 activity and content following the treatment with varying concentrations of αMMP2-NP demonstrated a concentration-dependent degradation profile without the hook effect (Fig. 4c). Similar results were observed in other cell lines (Supplementary Fig. 9a and b).
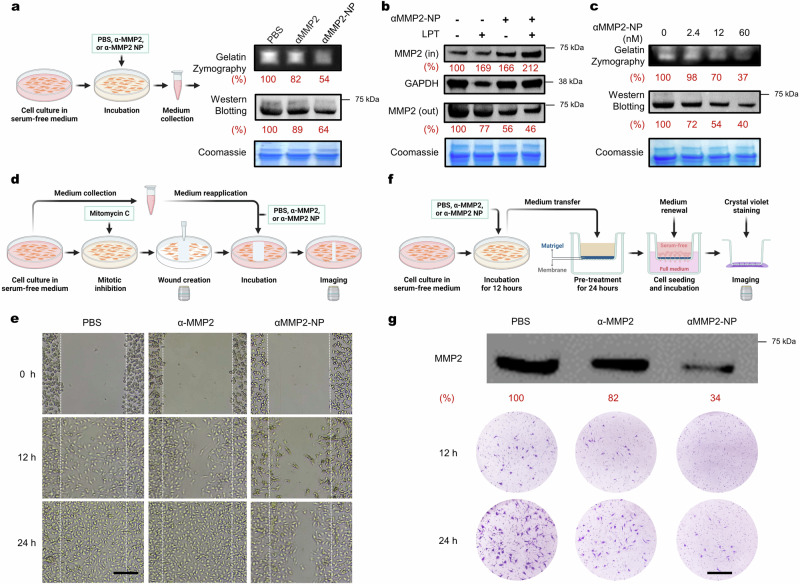
Fig. 4. Degradation of MMP2 mediated by αMMP2-NP.
a Gelatin zymography and Western blot assay of cell culture media for MMP2 activity and content. Media were collected after B16F10 cells were treated with PBS, αMMP2 (12 nM), or αMMP2-NP (α-MMP2-equiv. 12 nM) for 12 h. The gels and blot are representative of n = 3 biological replicates. b Western blot assay of MMP2 inside (IN) or outside (OUT) of B16F10 cells treated with αMMP2-NP (α-MMP2-equiv. 12 nM) for 12 h in the presence or absence of 0.1 mg mL−1 LPT. The gel and blots are representative of n = 3 biological replicates. c MMP2 activity and content in the culture media of B16F10 cells treated with varying concentrations of αMMP2-NP for 12 h. The gels and blot are representative of n = 3 biological replicates. d, e Schematic illustration (d) and results (e) of the wound-healing assay. B16F10 cells were treated with PBS, αMMP2 (12 nM), or αMMP2-NP (α-MMP2-equiv. 12 nM) for 12 h. Scale bar, 200 μm. The images are representative of n = 3 biological replicates. f, g Schematic illustration (f) and results (g) of the transwell cell invasion assay. CT26 cells were treated with PBS, αMMP2 (12 nM), or αMMP2-NP (α-MMP2-equiv. 12 nM). Scale bar, 200 μm. These images are representative of n = 3 biological replicates. All the uncropped gels and blots are included in the Source Data file. Source data are provided as a Source Data file.
Considering that MMP2 degrades a wide range of extracellular matrix components and facilitates cell migration42, we next investigated whether MONOTAB-mediated MMP2 degradation could translate into decreased cell mobility. A wound-healing assay was performed to evaluate the effect of αMMP2-NP on cell migration. B16F10 cells were pre-incubated in serum-free media for 12 h, and the media were collected and re-applied to the cells along with PBS, αMMP2, or αMMP2-NP after wound creation and wash (Fig. 4d). To eliminate the impact of cell proliferation on wound healing, the cells were pre-treated with mitomycin C, a mitotic inhibitor. The αMMP2-NP treatment led to a significantly slower scratch closure rate than observed with αMMP2 or PBS (Fig. 4e), demonstrating the high potency of αMMP2-NP in inhibiting cell mobility.
To further validate our findings, we carried out the cell invasion assay using a transwell apparatus. The serum-free culture media collected after different treatments were incubated in the apical chambers coated with Matrigel for 24 h at 37 °C, followed by cell seeding in the apical chambers with a fresh serum-free medium. A full medium was added to the basolateral chambers (Fig. 4f). Following 12-hour incubation, cells beneath the membrane of the inner chamber were visualized with crystal violet staining. In contrast to PBS or αMMP2, the αMMP2-NP treatment essentially prohibited cell migration towards the basolateral side of the membrane, persisting for at least 24 h (Fig. 4g). This observation was consistent with the results in the wound-healing experiment, further confirming the functional cellular consequences produced by MONOTAB-mediated protein degradation.
MONOTAB-mediated degradation of extracellular vesicles
Inspired by the potent ability of MONOTAB to induce extracellular protein degradation, we next asked whether the target range could be extended to non-protein targets such as extracellular vesicles (EVs). EVs are nanoscale lipid-bound vesicles released by cells and play crucial roles in intercellular signaling and pathological processes. Despite their emerging significance as therapeutic targets, selective degradation of EVs has yet to be achieved. Given the lack of EV-specific proteins for targeting, we explored the possibility of using phosphatidylserine (PS), a molecule commonly exposed on the outer leaflet of EVs but not on viable cells43, as a target for designing the EV-targeted MONOTAB. In light of the strong interaction between PS and Annexin V, we prepared Annexin V-NPs for EV capture by incubating NPs with biotinylated Annexin V. DLS measurement confirmed the binding of EVs to Annexin V-NPs, which resulted in an enlarged particle size (~274 nm) after co-incubation (Supplementary Fig. 10a).
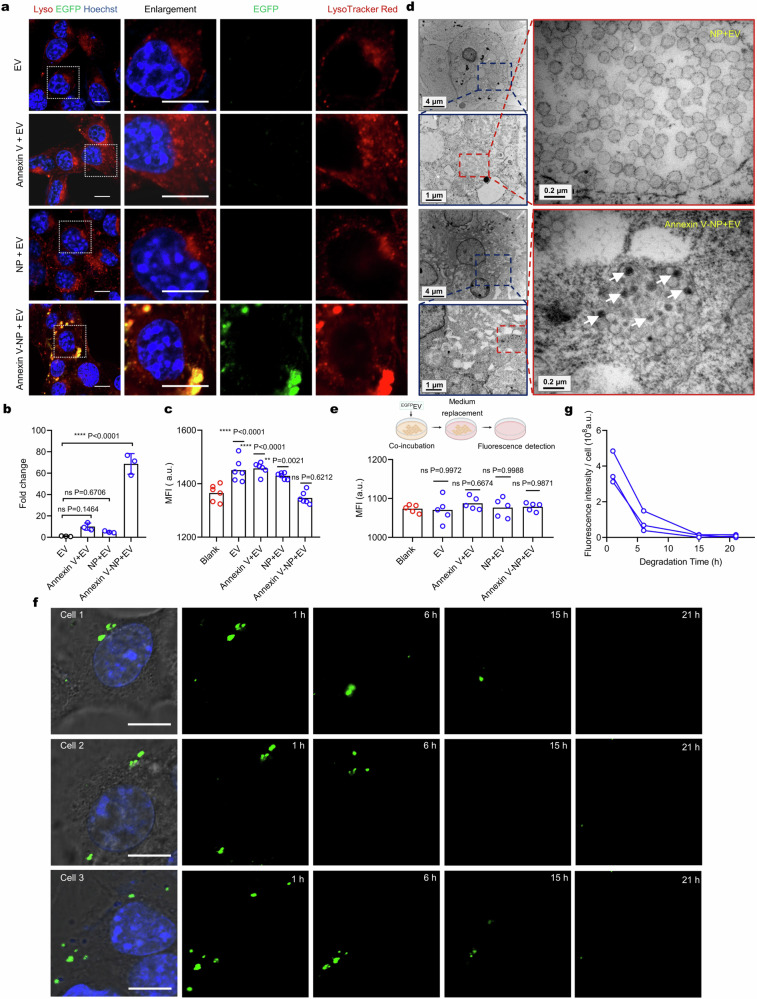
Next, we investigated if Annexin V-NPs could facilitate the uptake of EVs into cells. To ease the detection, we used an ECDHCC1-PalmGRET stable cell line to produce EGFP-labeled EVs (EGFPEVs), which emitted stable fluorescence at pH as low as 5.0 (Supplementary Fig. 10b). Co-incubation of B16F10 cells with Annexin V-NPs and EGFPEVs resulted in a ~70-fold increase in EV uptake compared to EGFPEVs alone, Annexin V plus EGFPEVs, or NPs plus EGFPEVs (Fig. 5a and b). Moreover, co-incubation with Annexin V-NPs significantly reduced the fluorescence intensity of EGFPEVs in the medium (Fig. 5c), approaching the blank level. Colocalization analysis demonstrated that the internalized EVs were transported to lysosomes as well. Similar results were also observed in other cell lines (Supplementary Fig. 10c and d). To ensure that the internalized EVs were indeed trapped in but not fused with lysosomes, we examined the cell samples using negative-stain electron microscopy. As demonstrated in Fig. 5d, treatments with Annexin V-NPs and EGFPEVs showed intact EVs closely associated with NPs within lysosomes. Conversely, when treated with NPs plus EGFPEVs, only NPs were observed within lysosomes. This observation indicates that Annexin V-NPs effectively hijacked EVs into cells and transported them to the lysosomes.
Fig. 5. Degradation of extracellular vesicles mediated by Annexin V-NP.
a Live-cell cell images of B16F10 cells incubated with EGFPEVs, Annexin-V + EGFPEVs, NPs + EGFPEVs, or Annexin-V-NPs + EGFPEVs for 8 h. The concentrations of EV, Annexin-V, and NP were 1.3 × 108 particles mL−1, 0.4, and 50 μg mL−1, respectively. Scale bar, 10 μm. The images are representative of n = 3 biological replicates. b Fold changes in MFI of EGFR signal in a (n = 3 biologically independent experiments). c Fluorescence intensity of EGFPEVs in the medium after co-incubation for 8 h (n = 6 biological replicates). d Representative TEM images of cells incubated with NPs + EVs or Annexin-V-NPs + EVs for 8 h. Scale bars are defined on the panel. White arrow points to the internalized EVs. The images are representative of n = 3 biological replicates. e Fluorescence intensity measurement of exocytosed EGFPEVs in the medium (n = 5 biological replicates). f Real-time tracking of EGFP signal in individual cells after incubation with EVs (1.3 × 108 particles mL−1) and Annexin V-NPs (50 μg mL−1) for 8 h. Scale bar, 10 μm. Cells 1–3 represent randomly tracked n = 3 single cells. g Fluorescence intensity of EGFP signal in individual cells at the indicated time points. Data are presented as mean values ± SD where relevant. Statistical significance was calculated via one-way ANOVA with Dunnett’s post hoc test. ns, no significance; **p < 0.01; ****p < 0.0001. Source data are provided as a Source Data file.
Afterward, we aimed to clarify the fate of the internalized EVs. We first explored the possibility of exocytosis. B16F10 cells treated with EGFPEVs, Annexin-V plus EGFPEVs, NPs plus EGFPEVs, or Annexin-V-NPs plus EGFPEVs were further cultured in fresh serum-free medium. Following incubation for 10 h, the fluorescence intensity of EGFP in the medium was measured, which showed no significant difference across all groups compared to the EGFPEV-free group (Fig. 5e). This suggests that the internalized EVs may not undergo exocytosis. We next asked whether the endocytosed EVs could be degraded. B16F10 cells were incubated with Annexin V-NPs and EGFPEVs for 8 hours, followed by washing for real-time imaging with time-lapse confocal microscopy. We assumed that EGFPEV degradation would be accompanied by the degradation of EGFP, and thus, the diminishment of EGFP signal within the cells could reflect the degree of EV degradation. Live-cell imaging indeed showed a time-dependent decrease in intracellular EGFP signal (Fig. 5f and g), indicating EV degradation. To rule out the contribution of photobleaching, we tracked the EGFP signal in ECDHCC1-PalmGRET cell debris under continuous laser exposure, and only minimal change was detected (Supplementary Fig. 10e and f). These results underscore the potential of using the MONOTAB platform for selective degradation of non-protein targets such as EVs, opening up possibilities for targeting intercellular communication mechanisms mediated by these vesicles.
Discussion
In summary, we have developed MONOTAB as a monofunctional targeted degradation platform that can mediate efficient lysosomal degradation of extracellular targets. Unlike existing strategies that use bifunctional chimaeras to recruit specific LTRs or E3 ligases to trigger degradation, the MONOTAB platform is built on the robust lysosome-targeting ability of nanoparticles. The irrelevance of specific receptors or E3 ligases enables MONOTAB to overcome resistance challenges associated with cellular heterogeneity or genetic mutations. The unique monofunctional modality confers an intrinsic anti-hook effect, which permits more effective degradation and prevents false-negative results that can lead to incorrect dosing decisions in clinical settings. Furthermore, this approach exhibited a considerable ability to promote lysosome biogenesis without impairing lysosomal health. As such, remarkable protein degradation performance was achieved, in which PD-L1 could be virtually eliminated by the targeted MONOTAB at low nanomolar concentrations.
Another key advantage of MONOTAB is the compatibility of commercial molecular binders such as antibodies or biotinylated ligands (via both the antibody mounting platform and the direct streptavidin–biotin interaction), which substantially expands the versatility and modularity. Moreover, compared with other extracellular protein degradation methods that require complex synthesis or protein recombination, the preparation of MONOTAB only needs the simple mixing of solutions followed by centrifugation, which allows rapid generation of diverse MONOTABs towards different targets. We demonstrated the target scope and therapeutic implications of MONOTAB by inducing the degradation of therapeutically relevant targets, including membrane-associated PD-L1, secreted MMP2, and even non-protein targets such as EVs. The ability to degrade EVs is particularly noteworthy, as it opens up possibilities for targeting intercellular communication mechanisms. Beyond this, the platform can also be easily programmed to acquire new capabilities for simultaneously degrading multiple extracellular targets of interest, which is challenging for other approaches.
While the present work utilized polystyrene nanoparticles, the MONOTAB concept can be extended to other lysosome-targeting nanoparticle chassis such as lipid nanoparticles, albumin nanoparticles, and other polymeric nanoparticles. Alternative ligands, such as peptides, nanobodies, aptamers, and small-molecule inhibitors, can also be used instead of antibodies to redirect extracellular proteins to lysosomes, wherein appropriate ligand immobilization strategies (i.e., scFv, SNAP-tag, and DNA self-assembly) would be required to ensure the high modularity. We envision that the MONOTAB platform will provide an effective degradation modality for targeting extracellular proteins and vesicles and hold great promise as a powerful tool for both therapeutic and research applications.
Methods
Reagents
Reagents were purchased from commercial suppliers and used as received. The details of general materials are described in Supplementary Table 2. Antibodies used in this study are listed in Supplementary Table 3.
Characterization of NP, αFc-NP, and CTRL-NP
The hydrodynamic size of NP, αFc-NP, and CTRL-NP (50 μg mL−1) were measured using a Malvern DLS Zetasizer equipped with a 4 mW He-Ne laser at a wavelength of 633 nm at 25 °C. The morphology of NP and αFc-NP was acquired by transmission electron microscopy (TEM, Talos F200X S, Thermo; 50 μg mL−1) and scanning electron microscope (SEM, Scios2 Hivac, Thermo; 500 μg mL−1). The dispersion state of NP, αFc-NP, and CTRL-NP (2.5 mg mL−1) was determined by Cryo-TEM (Talos F200C 200 kV, FEI). The stability of NP and αFc-NP was evaluated by measuring the changes in the particle size after storage at 4 °C for the indicated durations.
SDS–PAGE validation
Streptavidin-modified NPs (0.5 mg), biotinylated IgG (0.02 mg), or αFc-NPs (0.52 mg) were mixed with SDS–PAGE sample loading buffer (5×) and heated at 100 °C for 10 min. After cooling, equal amounts of the sample were loaded on a 12% polyacrylamide gel and separated by SDS–PAGE. The gel was then stained with Coomassie blue (Beyotime) for 2 h at room temperature and destained with H2O. Subsequently, the gel was visualized with a Gel Imaging System (Shenhua Technology, Hangzhou, China).
Cell culture
Cells were cultured at 37 °C and 5% CO2. B16F10, 4T1, HepG2, CT26, SKOV3, and MDAMB231 cells were maintained in RPMI-1640 medium (Thermo Fisher) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Hangzhou Genom). Hela, MCF7, and A549 cells were maintained in DMEM (Thermo Fisher) supplemented with 10% FBS and 1% penicillin/streptomycin. All cell lines were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Science (Shanghai, China).
Cellular uptake analysis of RBNP, αFc-RBNP, or CTRL-RBNP by flow cytometry
Cells were plated in a 12-well plate (100,000 cells per well) one day before the experiment and then incubated with RBNP, αFc-RBNP, or CTRL-RBNP at the indicated concentrations (15, 30, or 75 μg mL−1 for 2 h) or for the indicated durations (100 μg mL−1 for 1, 2, 4, or 8 h). Subsequently, the cells were washed twice with PBS, trypsinized for <1 min, and transferred to Eppendorf tubes. The cells were washed twice with PBS + 0.5% FBS and resuspended in 200 μL of PBS. Flow cytometry was performed on a BD FACSAria III, and FlowJo was used to gate on single cells and live cells for analysis. The experiments were repeated three times independently.
Trypan Blue quenching assay
To distinguish the internalized and surface-bound NPs, a Trypan Blue (TB) quenching assay was performed. Briefly, B16F10 cells were incubated with 50 μg mL−1 of FITCNP at 37 and 4 °C for 1.5 h, respectively, and washed twice with PBS. The cells were trypsinized, collected by centrifugation (300 × g, 3 min), and then incubated with TB (0.01% v/v) for 5 min. After washing with PBS, the cells were analyzed using flow cytometry. The fluorescence of surface-bound FITCNPs would be quenched by TB.
Determination of NP uptake efficiency
B16F10 cells were incubated with RBNP (25 or 50 μg mL−1) for 2 or 4 h. Subsequently, the culture medium was collected, and the fluorescence intensity was measured. The remaining concentration of RBNP was calculated from a standard curve for fluorescence intensity versus RBNP concentration. The NP uptake ratio was calculated accordingly.
Flow cytometry analysis of endocytosis inhibition effects on cellular uptake
Cells were plated in a 12-well plate (100,000 cells per well) one day before the experiment and then treated with filipin (7.5 μM), chlorpromazine (5 μM), Pitstop 2 (20 μM), wortmannin (5 μM), cytochalasin D (5 μM), or genistein (200 μM) for 30 min. Subsequently, the cells were incubated with 50 μg mL−1 of RBNP or αFc-RBNP for 90 min. To analyze the influence of low temperature, the cells were incubated in a 4 °C refrigerator for 30 min and then treated with equal amounts of RBNP or αFc-RBNP for 90 min. After incubation, the cells were washed with PBS three times to remove free RBNP or αFc-RBNP and subjected to flow cytometry analysis.
Generation of Igf2r and Tfrc knockout (KO) cell lines
Igf2r or Tfrc KO B16F10 cells were generated by lentivirus transduction. LentiCRISPR v2 plasmids targeting the coding sequence of Igf2r or Tfrc were constructed based on a lentiviral expression vector Lenti-V2 (Addgene, #52961). Lentiviruses were produced in HEK-293T cells by co-transfecting LentiCRISPR v2 plasmids and their packaging vectors psPAX.2 (Addgene, #12260) and pMD.2G (Addgene, #12259). The virus was collected and filtered through a 0.45 μm syringe filter after 48 h. B16F10 cells were then treated with the filtered viral particles for 48–72 h, and the culture medium was replaced with DMEM supplemented with 10% FBS and 2 μg mL−1 puromycin (Beyotime, China) for Igf2r or Tfrc KO cell screening.
Surface receptor silencing by RNA interference
B16F10 cells were plated in a six-well plate (400,000 cells per well) one day before the experiment and then transfected with 40 nM of each targeted siRNA and RNATransMate (Sangon Biotech, China) according to the manufacturer’s instructions. A non-specific Silencer Select siRNA (NC-siRNA) was used as the negative control.
RT-qPCR
Total RNA was extracted from cell pellets by RNAsimple Total RNA Kit (TianGen, Beijing, China) and reverse-transcribed to cDNA using HyperScript III 1st Strand cDNA Synthesis Kit with gDNA Remover (EnzyArtisan, Shanghai, China) according to the manufacturer's protocols. Quantitative PCR was performed with gene-specific primers on a StepOnePlus Real-time PCR System (Applied Biosystems) using the 2 × S6 Universal SYBR qPCR Mix (EnzyArtisan) to represent the relative expression levels. The following primer pairs were used: human clathrin primer pair (forward: 5’-TGATCGCCATTCTAGCCTTGC-3’; reverse: 5’-CTCCCACCACACGATTTTGCT-3’) and human GAPDH primer pair (forward: 5’-CAGGAGGCATTGCTGATGAT-3’; reverse: 5’-GAAGGCTGGGGCTCATTT-3’). The primers were synthesized by Tsingke (Beijing, China).
Dot blot of CTRL-NPs with varying αFc:IgG ratios
CTRL-NPs were fabricated by using biotinylated goat anti-rabbit IgG-Fc and rabbit IgG. CTRL-NPs with varying αFc:IgG ratios (2 μL, 2.5 mg mL−1) were dropped onto a nitrocellulose membrane and baked in a 37 °C oven for 30 min. The membranes were then blocked with 5% BSA in TBST (137 mM NaCl, 20 mM Tris, 0.1% Tween-20) for 1 h at room temperature and washed three times with TBST. HRP-conjugated goat anti-rabbit IgG(H + L) or donkey anti-goat IgG(H + L) were applied to membranes, respectively, and incubated for 1 hour at room temperature. After washing with TBST, the blots were developed with BeyoECL Star (Beyotime, China) for 2 min and imaged using a Chemiluminescence Imaging System (Shenhua Technology, China). Dot intensities were quantified with ImageJ.
Confocal microscopy for subcellular distribution of RBNP, αFc-RBNP, or Cy5CTRL- RBNP
Cells were seeded into a Nunc glass bottom dish (200,000 cells per dish) one day before the experiment and then incubated with 100 μg mL−1 of RBNP or αFc-RBNP for 4 h, or with 50 μg mL−1 of Cy5CTRL-RBNP for 10 h. Subsequently, the cells were incubated with 2 μM of Hoechst 33342 (Thermo Fisher) for 30 min and 100 nM of LysoTracker Green DND26 (Thermo Fisher) for 15 min. The cells were then washed with PBS three times and imaged using a Nikon AX R confocal microscope. Hoechst 33342 was excited using a 405 nm laser and visualized at 427–475 nm; LysoTracker Green was excited using a 488 nm laser and visualized at 505–544 nm; RB was excited using a 561 nm laser and visualized at 571–625 nm; Cy5 was excited using a 639 nm laser and visualized at 662–737 nm.
Lysosomal health assay
DQ Green BSA assay
B16F10 cells were seeded into a Nunc glass bottom dish (200,000 cells per dish) one day before the experiment and then incubated with 50 μg mL−1 of NP, αFc-NP, or CTRL-NP for 10 h or 100 nM of Bafilomycin A1(Baf A1) for 1 h. The cells were then washed three times with PBS and incubated with 10 μg mL−1 of DQ Green BSA (Share-bio, #D-12050SB) for 10 h, followed by 2 μM of Hoechst 33342 for 30 min. After washing with PBS, the cells were imaged using a Nikon AX R confocal microscope. Hoechst 33342 was excited using a 405 nm laser and visualized at 427–475 nm; FITC was excited using a 488 nm laser and visualized at 505–544 nm.
Acridine orange (AO) test
The lysosomal membrane integrity in living cells was measured using an acridine orange staining kit (Chengong Biotech, #HL10387.1). B16F10 cells were seeded into a Nunc glass bottom dish (200,000 cells per dish) one day before the experiment and then incubated with 50 μg mL−1 of NP, αFc-NP, or CTRL-NP for 10 h or 10 μM of chloroquine (CQ) plus 50 μM of ZnCl2 for 30 min. The cells were then washed with Reagent A from the kit and incubated with 1 mL of Reagent A plus 5 μL of Reagent B for 15 min. Subsequently, the cells were washed twice with Reagent A and imaged using a Nikon AX R confocal microscope. Hoechst 33342 was excited using a 405 nm laser and visualized at 427–475 nm; AO-Green was excited using a 488 nm laser and visualized at 505–544 nm; AO-Orange was excited using a 561 nm laser and visualized at 571–625 nm.
PD-L1 degradation analysis by Western blot
Total protein extraction
Cells were plated in a six-well plate (200,000 cells per well) one day before the experiment and then incubated with αPD-L1-NP or control treatments for the indicated durations. Subsequently, the cells were washed with PBS three times and then lysed with 200 μL of RIPA lysis buffer supplemented with the protease/phosphatase inhibitor cocktail (CST #5872) on ice for 30 min. The lysates were then transferred to Eppendorf tubes and centrifuged at 14,000 × g for 15 min at 4 °C. The supernatant was collected, and the protein concentration was determined using the BCA assay (Beyotime).
Membrane protein extraction
Cells treated as previously described were washed with PBS three times and scraped with a cell scraper. Following centrifugation at 600 × g for 5 min, the supernatant was discarded, and the cell pellet was treated with 1 mL of Reagent A in the Membrane and Cytosol Protein Extraction Kit (Beyotime) supplemented with 1 mM of phenylmethanesulfonyl fluoride (PMSF) on ice for 15 min. Two freeze-thaw cycles were then applied, and the suspension was centrifuged at 700 × g for 10 min at 4 °C. The supernatant was transferred carefully to another Eppendorf tube and then centrifuged at 14,000 × g for 30 min at 4 °C. As much of the supernatant was removed as possible, and the obtained precipitate was treated with 200 μL of Reagent B in the Membrane and Cytosol Protein Extraction Kit. It was then vortexed vigorously and placed on ice for 10 min. Following centrifugation at 14,000 × g for 5 min at 4 °C, the supernatant containing membrane proteins was collected, and the protein concentration was determined using the BCA assay.
Tumor tissue protein extraction
The collected tumor tissues were weighed, cut into pieces, and placed in an EP tube with 4-5 steel balls. Lysis buffer was then added at a ratio of tissue:RIPA:PMSF = 0.1 g:5 mL:50 μL. The EP tube was placed in a ball mill for 30 min (50 Hz) and then kept on ice for 30 min. After centrifugation at 14,000 × g for 10 min at 4 °C, the supernatant was collected, and the protein concentration was determined using the BCA assay.
Western blot
Protein samples were mixed with SDS–PAGE sample loading buffer (5×) and heated at 100 °C for 10 min. After cooling, equal amounts of the sample were resolved on a 12% polyacrylamide gel and then transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore). The membranes were blocked with 5% BSA in TBST for 1 h at room temperature and then incubated with the primary antibody overnight at 4 °C with gentle shaking. After washing with TBST three times, the membrane was incubated with the HRP-conjugated secondary antibody for 1 h at room temperature with gentle shaking and then washed with TBST three times. The blots were developed with BeyoECL Star (Beyotime) for 2 min and imaged using a Chemiluminescence Imaging System (Shenhua Technology, China). Band intensities were quantified with ImageJ.
Animals
Female C57BL/6 mice were purchased from Shanghai Slac Laboratory Animal Co. Ltd. All mice were maintained in a specific pathogen-free (SPF), temperature-controlled (22 ± 1 °C) animal facility on a reverse 12-h light, 12-h dark cycle at Zhejiang University. Food and water were given ad libitum. All animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Zhejiang University (approval number: 20379) in accordance with the institutional guidelines.
In vivo antitumor study
B16F10 cells (5 × 105 cells) in 70 μL of PBS were inoculated subcutaneously into the right flank of the 6–8-week-old female C57BL/6 mice. When the tumor volumes were approximately 100 mm3, mice were randomized into three groups (n = 5 per group) and intratumorally administered with αPD-L1 (2.0 mg kg−1) or αPD-L1-NP (αPD-L1-equiv. concentration, 2.0 mg kg−1; NP concentration, 100 mg kg−1) on a schedule of an injection every 3 days for a total of three injections. Equal volume of PBS was used in the vehicle group. Tumor size and mouse body weight were measured every two days. Tumor volume was calculated based on the formula: tumor volume = length (L) × width (W) × width (W)/2. After animal euthanasia, tumor tissues were resected for further analysis.
MMP2 activity analysis by gelatin zymography
Equal amounts of the sample were mixed with 5× native gel sample loading buffer (Beyotime) and then loaded on a 10% polyacrylamide gel containing 0.1% gelatin. Electrophoresis was carried out at 120 V and 4 °C. After running, the gel was washed with washing buffer (2.5% Triton X-100, 50 mM Tris–HCl, 5 mM CaCl2, 1 μM ZnCl2, pH 7.6) twice for 40 min each at room temperature and then rinse solution (50 mM Tris–HCl, 5 mM CaCl2, 1 μM ZnCl2, pH 7.5) twice for 20 min each. The gel was then incubated in reaction buffer (50 mM Tris–HCl, 150 nM NaCl, 10 mM CaCl2, 1 μM ZnCl2, 0.02% Brij-35, pH 7.5) for 40 h at 37 °C. After incubation, the gel was stained with Coomassie blue for 3 h at room temperature and subsequently destained with destaining solution (5% acetic acid, 10% methanol) until the bands on the gel became clear. The gelatinolytic activity was identified as a clear band on the blue background.
Wound-healing assay
A 0.1% gelatin solution in H2O was added to a 24-well plate (1 mL per well) and incubated at 37 °C for 3 h. The plate was then washed with PBS. Cells were seeded into the plate (100,000 cells per well) and incubated in a full medium. Once the cells reached 90% confluence, the medium was removed, and the cells were incubated in a serum-free medium for 24 h. The medium was collected, and a fresh serum-free medium containing 3 μg mL−1 of mitomycin C was added to each well, followed by a 3-h incubation to halt cell proliferation. The cell layer was scratched with a 20 μL pipette tip and washed with PBS three times. Subsequently, the collected medium was added into the wells along with 12 nM of αMMP2-NP or control treatments. The cells were incubated at 37 °C and imaged at the indicated time points using a SOPTOP XD20 inverted microscope. The wound healing rate was measured using ImageJ. The assay was repeated three times independently.
Cell migration and invasion assay
Cells were plated in a 24-well plate (100,000 cells per well) and incubated in full medium for 24 h. Once the cells reached 90% confluence, the medium was removed, and the cells were cultured in the serum-free medium for an additional 24 h. Then, 12 nM of αMMP2-NP or control treatments were added and incubated for 12 h. The medium was transferred to the gel-coated inserts of Corning Matrigel Invasion Chambers with 8.0 µm PET membrane (#354480) and incubated for 24 h at 37 °C. Subsequently, 5000 cells in serum-free medium were added into the insert chamber. Full medium was added to the lower chamber, and the insert was placed into the wells. After incubation for 12 h, the insert was removed and washed with PBS three times. Then, the cells were fixed with 4% PFA for 15 min, washed with PBS three times, and stained with a 0.1% crystal violet solution for 15 min at room temperature. The staining solution was discarded, and the insert was washed with PBS three times. Cells above the membrane were wiped off with a cotton swab, and the cells beneath the membrane were imaged using a SOPTOP XD20 microscope. The assay was repeated three times independently.
Confocal microscopy of EVs redirected into cells
Cells were seeded into a Nunc glass bottom dish (200,000 cells per dish) one day before the experiment and then incubated with EVs (1.3 × 108 particles mL−1), Annexin-V (0.4 μg mL−1) + EVs, NPs (50 μg mL−1) + EVs, or Annexin-V-NPs (Annexin-V-equiv. 0.4 μg mL−1, NP-equiv. 50 μg mL−1) + EVs for 8 h. Subsequently, the cells were incubated with 2 μM of Hoechst 33342 (Thermo Fisher) for 30 min and 100 nM of LysoTracker Red DND99 (Thermo Fisher) for 15 min. The cells were then washed with PBS three times and imaged with a Nikon AX R confocal microscope. Hoechst 33342 was excited using a 405 nm laser and visualized at 427–475 nm; EGFP was excited using a 488 nm laser and visualized at 505–544 nm; LysoTracker Red was excited using a 561 nm laser and visualized at 571–625 nm.
Confocal microscopy for real-time degradation of intracellular EVs
Cells were seeded into a Nunc glass bottom dish (200,000 cells per dish) one day before the experiment and then incubated with Annexin-V-NPs (50 μg mL−1) + EVs (1.3 × 108 particles mL−1) for 8 h. Subsequently, the cells were incubated with 2 μM of Hoechst 33342 (Thermo Fisher) for 30 min. After washing with PBS three times, the cells in the full medium were imaged using time-lapse confocal microscopy. Hoechst 33342 was excited using a 405 nm laser and visualized at 427–475 nm; EGFP was excited using a 488 nm laser and visualized at 505–544 nm.
Statistical analyses
Experiments were conducted at n = 3. Data are presented as mean ± standard deviation. The significance of the difference between two independent groups was determined using a two-tailed, unpaired Student’s t-test. Multiple groups were compared using one-way ANOVA with Dunnett’s or Tukey’s post hoc accordingly. Manders’ colocalization coefficients and densitometry of blots or gels were calculated by ImageJ. All the statistical analyses were performed using GraphPad Prism 8.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was financially supported by the Fundamental Research Funds for the Central Universities (226-2023-00022) (S.S.), the startup from Zhejiang University (S.S.), the Natural Science Foundation of Zhejiang Province (LR22H160002) (J.L.), the National Natural Science Foundation of China (82172899, 52203193, and 51833008) (J.L., J.X., and Y.S.), the National Key Research and Development Program of China (2021YFA1201200) (Y.S.), and the Open Fund of Zhejiang Provincial Key Laboratory of Pulmonology (J.L.). This work was supported in part by the Dynamic Research Enterprise for Multidisciplinary Engineering Sciences (DREMES) at Zhejiang University and the University of Illinois Urbana Champaign, funded by Zhejiang University (S.S. and J.L.). We thank the Bio-ultrastructure Analysis Lab of Analysis Center of Agrobiology and Environmental Sciences at Zhejiang University for technical support in TEM analyses. We thank Lingyun Wu in the Core Facilities, Zhejiang University School of Medicine, for her technical assistance on Cryo-TEM. Cartoons in Figs. 1a, 2a, 3a, k, 4a, d, f, and 5e were created with BioRender.com, released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Author contributions
S.Y., J.X., J.L., and S.S. conceived the MONOTAB project. J.L., S.S., and Y.S. provided resources. S.Y., Y.W., Q.T., J.F., J.L., and S.S. developed the methodology. S.Y., Y.W., Q.T., Y.Y., Y.G., L.X., S.L., and J.F. performed the experiments. S.Y., Y.W., Q.T., Y.G., S.L., J.X., J.F., J.T., J.L., S.S., and Y.S. analyzed the data. S.Y. and S.S. wrote the manuscript with input from all authors. All authors read and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Data supporting the findings of this study are available in the main manuscript and Supplementary Information. Source Data for the figures in the main text and in the Supplementary Information are provided in the Source Data file. Data are also available from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shasha Yao, Yi Wang, Qian Tang.
Contributor Information
Jiajia Xiang, Email: xiang_jj@zju.edu.cn.
Jian Liu, Email: JianL@intl.zju.edu.cn.
Shiqun Shao, Email: shiqun.shao@zju.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51720-z.
References
- 1.Bekes, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov.21, 181–200 (2022). 10.1038/s41573-021-00371-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang, L. et al. Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy. Nat. Chem. Biol.17, 567–575 (2021). 10.1038/s41589-021-00742-5 [DOI] [PubMed] [Google Scholar]
- 3.Slabicki, M. et al. Small-molecule-induced polymerization triggers degradation of BCL6. Nature588, 164–168 (2020). 10.1038/s41586-020-2925-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banik, S. M. et al. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature584, 291–297 (2020). 10.1038/s41586-020-2545-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn, G. et al. LYTACs that engage the asialoglycoprotein receptor for targeted protein degradation. Nat. Chem. Biol.17, 937–946 (2021). 10.1038/s41589-021-00770-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao, Y. et al. Bispecific aptamer chimeras enable targeted protein degradation on cell membranes. Angew. Chem. Int. Ed. Engl.60, 11267–11271 (2021). 10.1002/anie.202102170 [DOI] [PubMed] [Google Scholar]
- 7.Zheng, J. et al. Bifunctional compounds as molecular degraders for integrin-facilitated targeted protein degradation. J. Am. Chem. Soc.144, 21831–21836 (2022). 10.1021/jacs.2c08367 [DOI] [PubMed] [Google Scholar]
- 8.Pance, K. et al. Modular cytokine receptor-targeting chimeras for targeted degradation of cell surface and extracellular proteins. Nat. Biotechnol.41, 273–281 (2023). 10.1038/s41587-022-01456-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu, C., Wang, W., Wang, Y., Zhang, Y. & Li, J. Dendronized DNA chimeras harness scavenger receptors to degrade cell membrane proteins. Angew. Chem. Int. Ed. Engl.62, e202300694 (2023). 10.1002/anie.202300694 [DOI] [PubMed] [Google Scholar]
- 10.Wu, Y. et al. Aptamer-LYTACs for targeted degradation of extracellular and membrane proteins. Angew. Chem. Int. Ed. Engl.62, e202218106 (2023). 10.1002/anie.202218106 [DOI] [PubMed] [Google Scholar]
- 11.Zhou, Y., Teng, P., Montgomery, N. T., Li, X. & Tang, W. Development of triantennary N-acetylgalactosamine conjugates as degraders for extracellular proteins. ACS Cent. Sci.7, 499–506 (2021). 10.1021/acscentsci.1c00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caianiello, D. F. et al. Bifunctional small molecules that mediate the degradation of extracellular proteins. Nat. Chem. Biol.17, 947–953 (2021). 10.1038/s41589-021-00851-1 [DOI] [PubMed] [Google Scholar]
- 13.Cotton, A. D., Nguyen, D. P., Gramespacher, J. A., Seiple, I. B. & Wells, J. A. Development of antibody-based PROTACs for the degradation of the cell-surface immune checkpoint protein PD-L1. J. Am. Chem. Soc.143, 593–598 (2021). 10.1021/jacs.0c10008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marei, H. et al. Antibody targeting of E3 ubiquitin ligases for receptor degradation. Nature610, 182–189 (2022). 10.1038/s41586-022-05235-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dagogo-Jack, I. & Shaw, A. T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol.15, 81–94 (2018). 10.1038/nrclinonc.2017.166 [DOI] [PubMed] [Google Scholar]
- 16.Jin, H., Wang, L. & Bernards, R. Rational combinations of targeted cancer therapies: background, advances and challenges. Nat. Rev. Drug Discov.22, 213–234 (2023). 10.1038/s41573-022-00615-z [DOI] [PubMed] [Google Scholar]
- 17.Schröder, M. et al. DCAF1-based PROTACs with activity against clinically validated targets overcoming intrinsic- and acquired-degrader resistance. Nat. Commun. 15, 275 (2024) [DOI] [PMC free article] [PubMed]
- 18.Wurz, R. P. et al. Affinity and cooperativity modulate ternary complex formation to drive targeted protein degradation. Nat. Commun.14, 4177 (2023). 10.1038/s41467-023-39904-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donahue, N. D., Acar, H. & Wilhelm, S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv. Drug Deliv. Rev.143, 68–96 (2019). 10.1016/j.addr.2019.04.008 [DOI] [PubMed] [Google Scholar]
- 20.Sousa de Almeida, M. et al. Understanding nanoparticle endocytosis to improve targeting strategies in nanomedicine. Chem. Soc. Rev.50, 5397–5434 (2021). 10.1039/D0CS01127D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rennick, J. J., Johnston, A. P. R. & Parton, R. G. Key principles and methods for studying the endocytosis of biological and nanoparticle therapeutics. Nat. Nanotechnol.16, 266–276 (2021). 10.1038/s41565-021-00858-8 [DOI] [PubMed] [Google Scholar]
- 22.Jang, J. H. & Hanash, S. Profiling of the cell surface proteome. Proteomics3, 1947–1954 (2003). 10.1002/pmic.200300563 [DOI] [PubMed] [Google Scholar]
- 23.Kalxdorf, M. et al. Cell surface thermal proteome profiling tracks perturbations and drug targets on the plasma membrane. Nat. Methods18, 84–91 (2021). 10.1038/s41592-020-01022-1 [DOI] [PubMed] [Google Scholar]
- 24.Chen, Y. et al. Cholesterol sequestration by nystatin enhances the uptake and activity of endostatin in endothelium via regulating distinct endocytic pathways. Blood117, 6392–6403 (2011). 10.1182/blood-2010-12-322867 [DOI] [PubMed] [Google Scholar]
- 25.von Kleist, L. et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell146, 471–484 (2011). 10.1016/j.cell.2011.06.025 [DOI] [PubMed] [Google Scholar]
- 26.Dutta, D., Williamson, C. D., Cole, N. B. & Donaldson, J. G. Pitstop 2 is a potent inhibitor of clathrin-independent endocytosis. PLoS ONE7, e45799 (2012). 10.1371/journal.pone.0045799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandvig, K., Kavaliauskiene, S. & Skotland, T. Clathrin-independent endocytosis: an increasing degree of complexity. Histochem. Cell Biol.150, 107–118 (2018). 10.1007/s00418-018-1678-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casella, J. F., Flanagan, M. D. & Lin, S. Cytochalasin D inhibits actin polymerization and induces depolymerization of actin filaments formed during platelet shape change. Nature293, 302–305 (1981). 10.1038/293302a0 [DOI] [PubMed] [Google Scholar]
- 29.Thiele, L. et al. Evaluation of particle uptake in human blood monocyte-derived cells in vitro. Does phagocytosis activity of dendritic cells measure up with macrophages? J. Control Release76, 59–71 (2001). 10.1016/S0168-3659(01)00412-6 [DOI] [PubMed] [Google Scholar]
- 30.Araki, N., Johnson, M. T. & Swanson, J. A. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J. Cell Biol.135, 1249–1260 (1996). 10.1083/jcb.135.5.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, H.-P. et al. Identification of novel macropinocytosis inhibitors using a rational screen of Food and Drug Administration-approved drugs. Br. J. Pharmacol.175, 3640–3655 (2018). 10.1111/bph.14429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu, L. et al. Conjugation with glucagon like peptide-1 enables targeted protein degradation. Bioorg. Chem.141, 106908 (2023). 10.1016/j.bioorg.2023.106908 [DOI] [PubMed] [Google Scholar]
- 33.Zhang, D. et al. Transferrin receptor targeting chimeras (TransTACs) for membrane protein degradation. Preprint at bioRxiv. 10.1101/2023.1108.1110.552782 (2023).
- 34.Brown, M. S. & Goldstein, J. L. A receptor-mediated pathway for cholesterol homeostasis. Science232, 34–47 (1986). 10.1126/science.3513311 [DOI] [PubMed] [Google Scholar]
- 35.Gehrlein, A. et al. Targeting neuronal lysosomal dysfunction caused by β-glucocerebrosidase deficiency with an enzyme-based brain shuttle construct. Nat. Commun.14, 2057 (2023). 10.1038/s41467-023-37632-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunwar, A. J. et al. Lack of the endosomal SNAREs vti1a and vti1b led to significant impairments in neuronal development. Proc. Natl Acad. Sci. USA108, 2575–2580 (2011). 10.1073/pnas.1013891108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song, W. et al. The autophagic response to polystyrene nanoparticles is mediated by transcription factor EB and depends on surface charge. J. Nanobiotechnol.13, 87 (2015). 10.1186/s12951-015-0149-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mariathasan, S. et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature554, 544–548 (2018). 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong, H. et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med.8, 793–800 (2002). 10.1038/nm730 [DOI] [PubMed] [Google Scholar]
- 40.Sela-Passwell, N. et al. Antibodies targeting the catalytic zinc complex of activated matrix metalloproteinases show therapeutic potential. Nat. Med.18, 143–147 (2011). 10.1038/nm.2582 [DOI] [PubMed] [Google Scholar]
- 41.Orgaz, J. L. et al. Diverse matrix metalloproteinase functions regulate cancer amoeboid migration. Nat. Commun.5, 4255 (2014). 10.1038/ncomms5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarkar, P. et al. Inhibiting matrix metalloproteinase-2 activation by perturbing protein–protein interactions using a cyclic peptide. J. Med. Chem.63, 6979–6990 (2020). 10.1021/acs.jmedchem.0c00180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kira, A. et al. Apoptotic extracellular vesicle formation via local phosphatidylserine exposure drives efficient cell extrusion. Dev. Cell58, 1282–1298.e1287 (2023). 10.1016/j.devcel.2023.05.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available in the main manuscript and Supplementary Information. Source Data for the figures in the main text and in the Supplementary Information are provided in the Source Data file. Data are also available from the corresponding author upon request. Source data are provided with this paper.