Abstract
Ovarian cancer (OC) is a gynecological malignancy that ranks among the most common female cancers worldwide and notably reduces a patient's quality of life. Mitochondrial carrier homology 2 (MTCH2) is a mitochondrial outer membrane protein that serves a regulatory role in mitochondrial metabolism and cell death. The precise contribution and underlying molecular pathways of MTCH2 in the context of OC development is currently unclear. The present study aimed to investigate the roles of MTCH2 in the energy metabolism, cell proliferation and metastatic potential of OC cells and evaluate the regulatory relationship between MTCH2, aminoacyl transfer RNA synthetase-interacting multifunctional protein 2 (AIMP2) and claudin-3. An analysis of 67 patients with high-grade serous OC demonstrated increased expression levels of MTCH2, AIMP2 and claudin-3 in OC tumor tissue samples compared with in corresponding normal tissues adjacent to OC tissue samples. MTCH2 overexpression was significantly associated with the International Federation of Gynecology and Obstetrics stage and tumor differentiation of the OC tumor samples. In vitro experiments using the SK-OV-3 OC cell line demonstrated that MTCH2 exerts a regulatory effect on the cell proliferation, invasion and migratory capabilities of these cells. Knockdown of MTCH2 reduced ATP production, induced mitochondrial dysfunction and promoted cytoskeleton remodeling and apoptosis in SK-OV-3 OC cells. In addition, MTCH2 knockdown downregulated the expression levels of both claudin-3 and AIMP2 proteins. Knockdown of AIMP2 inhibited the regulatory effect of MTCH2. Co-immunoprecipitation experiments demonstrated that MTCH2 interacts with AIMP2 and claudin-3. The present study provides novel insights into the treatment of OC metastasis, as MTCH2 was demonstrated to serve roles in the progression of OC cells through the regulation of claudin-3 via AIMP2, which could provide novel insights into the treatment of ovarian cancer metastasis.
Keywords: mitochondrial carrier homology 2, claudin-3, aminoacyl transfer RNA synthetase-interacting multifunctional protein 2, mitochondrial dysfunction, ovarian cancer
Introduction
According to data from the American Cancer Society, the mortality rate of ovarian cancer (OC) is decreasing, but it remains a common malignant tumor in women (1). OC is typically undetected in patients until the middle or late stages of the disease (2). With the further study of gene targets for cancer therapy, the use of neoadjuvant chemotherapy regimens for patients with OC has the potential to prolong patient survival (3,4). Therefore, the identification of key genes involved in OC progression and understanding of their roles in driving such progression are of importance.
In eukaryotic cells, mitochondria are essential for metabolism and signal transduction (5). Proteins located in the outer mitochondrial membrane (OMM) are essential for the functions of the mitochondria and their connection with the cytoplasm. Therefore, a number of diseases, including certain types of cancer, as well as the aging process, are associated with the pathophysiology of OMM protein malfunction (6). Mitochondrial carrier homology 2 (MTCH2) is a non-classical OMM member within the mitochondrial carrier protein family, and was recently reported to function as an OMM insertion enzyme for tail-anchored proteins (7). Previous studies have demonstrated that MTCH2 serves a role in the regulation of apoptosis and lipid homeostasis (8). Deletion of MTCH2 in mouse embryonic stem cells, fibroblasts and in vivo mouse models has been shown to impede the recruitment of truncated BH3-interacting domain death agonist (tBID) to mitochondria, resulting in reductions in the activation of pro-apoptotic proteins, OMM permeabilization and apoptosis (9). Furthermore, MTCH2 is involved in the maintenance of the normal mitochondrial membrane potential and regulation of mitochondrial dynamics through its enzyme activity; MTCH2 deficiency triggers mitochondrial fragmentation, which contributes to mitochondrial dysfunction (6,10). A previous study on breast cancer suggests that MTCH2 is implicated in the invasion and dissemination of cancer cells, as well as in the metabolic and immune response of these cells (11). MTCH2 knockdown was reported to impair the invasive and migratory capabilities of glioma cells during malignant glioma progression, an effect that was attributed to induced mitochondrial dysfunction characterized by increased mitochondrial oxidative phosphorylation, causing oxidative damage (12). Mitochondrial dysfunction is associated with impaired ATP synthesis and aberrant expression levels of claudins-1, 2 and 7, which compromises the integrity of the intestinal barrier (13).
Claudins are a family of transmembrane tight junction proteins that have a role in size-selective paracellular permeability; claudins-2, 7, 10, 15 and 16 increase cellular cation permeability, whereas claudins 4, 5, 8, 11, 14 and 18 decrease cellular cation permeability (14,15). Claudin-3 is a primary component of cell tight junctions (16,17). Increased expression levels of claudin-3 have been implicated in tumor metastasis and increase the malignant potential of cancer cells (18). Epithelial cell shape changes are promoted through the regulation of claudin-3 expression levels (19). Increased expression levels of claudin-3 have been associated with clinical pathological stage, invasion depth and lymph node metastasis in colorectal cancer (18,20). Increased claudin-3 expression levels were associated with the stage and, lymphatic and distant metastasis of gastric cancer, serving as an independent predictor of poor prognosis of patients with gastric cancer (21). Knockdown of claudin-3 can significantly inhibit cell proliferation and promote cell apoptosis, and therefore has potential as a therapeutic target for OC treatment (22).
Aminoacyl-transfer RNA (tRNA) synthetases (ARSs) are enzymes that ligate their cognate amino acids to tRNAs for protein synthesis (23). The multi-tRNA synthetase complex has also been identified as a macromolecular ARS complex in eukaryotes (24). ARSs usually exist in free form or as multi-tRNA synthetase complexes, which consist of three non-enzymatic ARS interacting multifunctional proteins (AIMP1, AIMP2, and AIMP3) (25). AIMP2 is reported to have anti-proliferative activity and promote cell death as a pro-apoptotic factor (26). Hexokinase 2 (HK2) is a key enzyme in glycolysis that catalyzes glucose phosphorylation and the production of glucose-6-phosphate (27). In previous cancer studies, HK2 increased autophagolysosomal dependent degradation via interactions with AIMP2, which thereby reduced the levels of ionizing radiation-mediated apoptosis and increased the radiation resistance of liver cancer cells (28). Furthermore, AIMP2 has been identified as a potential risk factor in the progression of thyroid papillary carcinoma (29). Using the Gene Expression Profiling Interactive Analysis database, it has been reported that the expression of MTCH2 was associated with AIMP2 (30). Furthermore, claudin-3, as a downstream factor of the Wnt/β-Catenin pathway, was shown to regulate mitochondrial dysfunction (31,32).
Notably, MTCH2, AMIP2 and claudin-3 are associated with the development of various types of tumors. The present study thus aimed to determine the relationship between MTCH2, AMIP2 and claudin-3 expression in OC tissue samples, and investigated the precise role of MTCH2 in the development of OC and potential molecular pathways, thereby providing a novel theoretical basis for the treatment of OC.
Materials and methods
Bioinformatics analyses
The cBioPortal for Cancer Genomics (http://www.cbioportal.org/) was used to analyze the relationship between mutation of the MTCH2 gene in OC and the overall survival time of patients with OC by whole exome sequencing (33). Using the cBioPortal database, OC data from the Pan-Cancer Atlas and the Cancer Genome Atlas were selected for analysis.
Cell culture
Human ovarian carcinoma SK-OV-3 cells were purchased from Shanghai Zhongqiao Xinzhou Biotechnology Co., Ltd. (cat. no. ZQ0074). Cells were cultured in RPMI-1640 medium (cat. no. C11875500BT; Gibco; Thermo Fisher Scientific, Inc.), supplemented with 15% fetal bovine serum (FBS; cat. no. G24-70500; Genial Biologicals, Inc.) and 1% penicillin/streptomycin (cat. no. CA0075, Leagene; Beijing Regen Biotechnology Co., Ltd.) at 37°C with 5% CO2.
Transfection
The pcDNA3.1 cloning vector (cloning sites, NheI and BamHI) was used to generate the MTCH2-overexpression (OE) plasmid, which encodes the MTCH2 transcript (accession no. NM_001317231.2). The plasmid and small interfering (si)RNAs were purchased from Sangon Biotech Co., Ltd. The sequences of MTCH2 and AIMP2 siRNAs used were as follows: MTCH siRNA sense (S), 5′-UCCUCCAACAAUAGGACGAAATT-3′ and anti-sense (AS) 5′-UUUCGUCCUAUUGUUGGAGGATT-3′; AIMP2 siRNA1 S, 5′-GAUGCAGACUUGGAUGUAATT-3′ and AS, 5′-UUACAUCCAAGUCUGCAUCTT-3′; AIMP2 siRNA2 S, 5′-GAUUCACUUUAAUUUGGAATT-3′ and AS, 5′-UUCCAAAUUAAAGUGAAUCTT-3′; AIMP2 siRNA3 S, 5′-AGUGCUUUGGAGAACAGAATT-3′ and AS 5′-UUCUGUUCUCCAAAGCACUTT-3′; and negative control (NC) S, 5′-UUCUCCGAACGUGUCACGUTT-3′ and AS, 5′-ACGUGACACGUUCGGAGAATT-3′. SK-OV-3 cells were seeded into 6-well cell culture plates. The OE transfection plasmid (1.25 µg/ml) and siRNAs (50 pmol/ml) were separately transfected into SK-OV-3 cells using the Lipo8000™ transfection reagent (cat. no. C0533; Beyotime Institute of Biotechnology) at 37°C for 24 h. Co-transfection with AIMP2 siRNA and MTCH2 siRNA or MTCH2-OE was performed to study the downstream mechanism of MTCH2 (34). The control group comprises untransfected cells. The efficiency of the transfection was evaluated through western blotting. After 24 h, cells were collected and used in the follow-up experiments.
Human tissue samples
Tumor tissue samples were collected from high-grade serous ovarian carcinoma (HGSOC). The samples were collected in May 2021. Tumor tissues and normal tissues adjacent to tumor tissues (≥4 cm from the tumor edge) (35) were collected from 67 patients with HGSOC at The Fourth Hospital of Hebei Medical University (Shijiazhuang, China). Patients with a history of chemotherapy or radiotherapy were excluded from the present study. Paired tumor tissues and adjacent normal tissues were collected and stored in liquid nitrogen until further use. The protocol was approved by the Ethics Committee of The Fourth Hospital of Hebei Medical University (approval no. 2020166; March 14 2021; Shijiazhuang, China). All protocols were performed in accordance with the Declaration of Helsinki.
Immunohistochemical staining
OC and adjacent normal tissue samples were fixed using 10% formalin at room temperature for 3 h, embedded in paraffin and sectioned to a 4-µm thickness. After dewaxing with xylene and rehydrated in a decreasing concentration of ethanol, antigen retrieval was performed in a microwave (100°C for 7 min). Endogenous peroxidase activity was quenched with 3% H2O2 for 12 min at 37°C and sections were then blocked with 5% sheep serum (cat. no. ZLI-9056; OriGene Technologies, Inc.) at room temperature for 10 min. After which, the sections were incubated at 4°C overnight with primary antibodies against MTCH2 (1:200; cat. no. 16888-1-AP; Proteintech Group, Inc.) and AIMP2 (1:200; cat no. 10424-1-AP; Proteintech Group, Inc.). The next day, the slides were incubated with secondary antibodies (cat. no. PV-9000; OriGene Technologies, Inc.) at 37°C for 20 min. Staining was performed using 3,3-diaminobenzidine (cat. no. ZLI-9019; OriGene Technologies, Inc.) at room temperature for 5 min and sections were counterstained using hematoxylin for 3 min. A fluorescence microscope used to capture the images. Evaluation of the tissue sample staining intensity was performed as previously described by Wang et al (36). Sections were scored according to the staining intensity as follows: Negative staining, 0; weak positivity, 1; moderate positivity, 2; and strong positivity, 3 points. Further scoring was performed according to the proportion of the section with positive staining as follows: 0–10%, 1; 10–50%, 2; 51–80%, 3; and >80%, 4 points. Subsequently, the two scores were multiplied and the resulting score ranged from 0 to 12. Scores from 0 to 3 were considered negative and scores from 4 to 12 were considered positive. In addition, the mean density was calculated using the following formula: Mean density=IOD/area, where IOD indicates integrated optical density value. The protein expression levels were quantitatively analyzed using Image-Pro Plus software (version 6; Media Cybernetics, Inc.).
Transwell migration assay
Initially, 300 µl Matrigel was used to coat the upper chamber of a Transwell system (24-well plate; pore size, 8.0 µm) at 37°C for 3 h. SK-OV-3 cells were first cultured in serum-free RPMI-1640 medium for 24 h. The next day, SK-OV-3 cells (1×105 cells/ml) were seeded into the upper chambers with serum-free RPMI-1640 medium. RPMI-1640 medium supplemented with 15% FBS and 1% penicillin/streptomycin was added into the bottom of a 24-well plate and placed in an incubator for 48 h (37°C, 5% CO2). Following this, 4% paraformaldehyde (cat. no. DF0135; Leagene; Beijing Regen Biotechnology Co., Ltd.) was used to fix the migrated cells for 10 min and 0.1% crystal violet (cat. no. G1063; Beijing Solarbio Science & Technology Co., Ltd.) was used to stain the cells for 20 min at room temperature. Any residual cells that remained in the upper chambers were gently removed using a cotton swab after staining. A CX31 inverted light microscope (Olympus Corporation) was used to image randomly selected fields of view to evaluate the migrated cells.
Wound healing assay
SK-OV-3 cells (3×105 cells/well) were seeded in 6-well plates and transfected with MTCH2-OE and MTCH2 siRNA for 48 h according to the aforementioned method. When the cell density reached ~90%, 200 µl pipette tips were used to scratch two parallel lines in the central area where the cells grew. PBS was used to wash each well twice, after which, the cells were cultured for 48 h in serum-free RPMI-1640 medium. Images of the scratched area were captured using a CX31 inverted light microscope (Olympus Corporation) at 0 and 24 h. The images were analyzed using the Image-Pro Plus software (version 6; Media Cybernetics, Inc.). The cell migration distance was calculated as follows: (width of the wound at 0 h-width of the wound at 24 h)/width of the wound at 0 h.
Detection of ATP production
ATP content was detected using an ATP fluorescent probe pCMV-AT1.03 (cat. no. D2604, Beyotime Institute of Biotechnology) which allowed ATP levels to be monitored within living single cells in real time. SK-OV-3 cells (2×105 cells/well) were seeded in 12-well plates and cultured at 37°C in 5% CO2 for 24 h. Next, 12 µl pCMV-AT1.03 was added to each well and cultured for 24 h at 37°C. The cells were then imaged at 24-h timepoints using a BX53 fluorescence microscope (Olympus Corporation) and fluorescence intensities were calculated using Image-Pro Plus software (version 6; Media Cybernetics, Inc.).
Mitochondrial activity detection
The mitochondrial network structure was evaluated using Mito-Tracker Red CMXRos (cat. no. C1049B; Beyotime Institute of Biotechnology) according to the manufacturer's instructions. SK-OV-3 cells (1×105 cells/well) were transfected as aforementioned and cultured in 6-well plates for 24 h. The Mito-Tracker Red CMXRos working solution was prepared by adding 1 µl of 200 µM Mito-Tracker Red CMXRos storage solution to 1 ml of cell culture medium (final concentration, 200 nM) mixed well and the cells were then incubated with this working solution for 45 min. The staining solution was then replaced with fresh culture medium prior to imaging. Images were captured of living cells using a fluorescence microscope (BX53; Olympus Corporation) and the fluorescence intensities analyzed using Image-Pro Plus software (version 6, Media Cybernetics).
EdU proliferation assay
The BeyoClick™ EdU-594 Cell Proliferation Assay kit (cat. no. C0078S; Beyotime Institute of Biotechnology) was used to detect the proliferation rate of OC cells, according to the manufacturer's instructions. Briefly, SK-OV-3 cells (1×104 cells) were cultured in a cell incubator at 37°C (5% CO2) for 24 h. Cells were treated with 10 µM of prewarmed EdU working solution at 37°C for 2 h. Then, the cells were fixed with 4% paraformaldehyde for 15 min at room temperature, followed by rinsing three times with PBS solution containing 3% BSA (3 min/wash). The cells were then incubated with PBS containing 0.3%Triton X-100 for 15 min at room temperature. Next, 0.5 ml Click reaction solution was added and incubated in the dark for 30 min at room temperature. The cells were counterstained with DAPI (10 µg/ml) at room temperature for 3 min and imaged using a BX53 fluorescence microscope (Olympus Corporation). The number of positively stained proliferating cells were counted for analysis of proliferation rate.
Caspase-3 activity and annexin V cell apoptosis assay
Apoptosis was detected using Caspase-3 Activity and Apoptosis Detection kit for Live Cells (cat. no. C1077S; Beyotime Institute of Biotechnology) according to the manufacturer's instructions. SK-OV-3 cells were washed with PBS, following which 194 µl annexin V-mCherry Binding Buffer and 5 µl Annexin V-mCherry and 1 µl GreenNuc™ Caspase-3 Substrate (1 mM) were added, gently mixed and incubated at room temperature in the dark for 30 min. Images were captured using a BX53 fluorescence microscope (Olympus Corporation). Annexin V-mCherry labels apoptotic cells with red fluorescence and GreenNuc™ Caspase-3 Substrate labels the nuclei of apoptotic cells with green fluorescence representing caspase-3 activity. The number of apoptotic cells was calculated as follows=(number of apoptotic cells/number of total cells) ×100.
Cytoskeleton remodeling
SK-OV-3 cells (1×105 cells) were spread on slides, fixed with 3.7% formaldehyde in PBS solution for 20 min at room temperature and then washed thrice with PBS containing 0.1% Triton X-100 at room temperature (5 min/wash). Actin-Tracker Green (cat. no. C2201S; Beyotime Institute of Biotechnology) was added dropwise onto the slides and incubated in the dark at room temperature for 60 min. Next, the cells were washed three times with PBS containing 0.1% Triton X-100 at room temperature (5 min/wash), the plates were air-dried, counterstained with DAPI at room temperature for 3 min and images were captured using a BX53 fluorescence microscope (Olympus Corporation). The images were analyzed using the Image-Pro Plus software (version 6; Media Cybernetics, Inc.).
Cellular immunofluorescence
SK-OV-3 cells (2×105 cells) were fixed using 4% paraformaldehyde at room temperature for 10 min, permeabilized with 0.1% Triton-X-100 in PBS and blocked with goat serum (cat. no. ZLI-9056; OriGene Technologies, Inc.). The cells were then incubated with primary antibodies against MTCH2 (1:200; cat no. 16888-1-AP; Proteintech Group, Inc.) and AIMP2 (1:100; cat no. 10424-1-AP; Proteintech Group, Inc.) at 4°C overnight. Next, using dilution buffer (cat. no. ZLI-9030; OriGene Technologies, Inc.) to dilute the antibody, cells were incubated with Alexa Fluor® 488 goat anti-rabbit IgG (1:100; cat. no. ZF-0511; OriGene Technologies, Inc.) secondary antibodies was performed at 37°C for 1 h. PBS was used to wash the section thrice and DAPI was used to stain the nucleus at room temperature for 5 min. Images were captured using a BX53 fluorescence microscope (Olympus Corporation). The fluorescence intensities were analyzed using the Image-Pro Plus software (version 6; Media Cybernetics, Inc.).
Co-immunoprecipitation (co-IP)
The cells were lysed with Lysis Buffer with Protease Inhibitor Cocktail from the IP kit (cat. no. P2179M; Beyotime Institute of Biotechnology). Lysed cells were centrifuged at 13,000 × g for 20 min at 4°C and the supernatant was collected, of which 50 µl was taken for use as the positive control input group. From each sample, 2 µl was used to determine the concentration and the remaining supernatant was stored for subsequent use. The antibodies used for IP were MTCH2 (3 µg; cat no. 16456-1-AP; Proteintech Group, Inc.) and AIMP2 (3 µg; cat. no. 10424-1-AP; Proteintech Group, Inc.) and 3 µg of normal IgG was used as a negative control group. Next, 100 µl Protein A+G magnetic beads (cat. no. P2179M; Beyotime Institute of Biotechnology) were added and incubated with antibodies and IgG on a shaker at 4°C for 2 h. The precipitate was collected by centrifugation at 1,000 × g for 5 min at 4°C. The 500-µl cell lysates were added to protein A+G magnetic beads conjugated with antibody or normal IgG on a shaker at 4°C overnight to bind the proteins to the antibodies. The next day, the samples were centrifuged at 1,000 × g for 5 min at 4°C and the supernatant discarded. The precipitate was washed four times with PBS using 1 ml each time. The precipitate and the input (50 µl of the supernatant after cell lysis) were boiled at 95°C for 5 min with 1× loading buffer (cat. no. ZS306; Beijing Zoman Biotechnology Co., Ltd.). The samples were centrifuged at 1,000 × g for 5 min and the supernatant was used as samples for MTCH2-IP and AIMP2-IP experimental groups for western blotting.
Western blot assay
Total protein was extracted from cells using a Total Protein Extraction kit (cat. no. KGP2100; Nanjing KeyGen Biotech Co., Ltd.). The BCA protein quantification kit (cat. no. PT0001, Leagene; Beijing Regen Biotechnology Co., Ltd.) was used to quantify the concentrations of extracted total proteins. The proteins were then separated by SDS-PAGE on a 9% gel and transferred on to PVDF membranes. The membranes were blocked with 5% milk/TBST (0.05% Tween) for 1 h at room temperature to prevent non-specific binding. The membranes were incubated with MTCH2 (1:1,000; cat. no. 16888-1-AP; Proteintech Group, Inc.), claudin-3 (1:1,000; cat. no. AF0129; Affinity Biosciences), AIMP2 (1:1,000; cat. no. 10424-1-AP; Proteintech Group, Inc.) and β-actin (1:5000, cat. no. AF7018; Affinity Biosciences) overnight at 4°C. Following primary antibody incubation, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (cat. no. ZB-2301; OriGene Technologies, Inc.) at room temperature for 1 h. Protein bands were visualized using electrochemiluminescence solution (cat. no. P1050; Applygen Technologies, Inc.). ImageJ software (version 2.14.0; National Institutes of Health) was used for data analysis.
Statistical analysis
Statistical analysis was conducted using SPSS (version 27; IBM Corp.). Data were presented as mean ± standard deviation. Paired Student's t-test was used to compare two groups. Multiple groups were compared using a one-way ANOVA followed by Bonferroni's post hoc test. Correlation analysis was performed using Pearson correlation test. A survival analysis was performed using Kapan-Meier analysis and log rank test. Contingency tables were assessed using the χ2 test. Data presentation was performed using the GraphPad Prism software (version 8; Dotmatics). P<0.05 was considered to indicate a statistically significant difference.
Results
MTCH2, claudin-3 and AIMP2 were upregulated in OC tumor samples
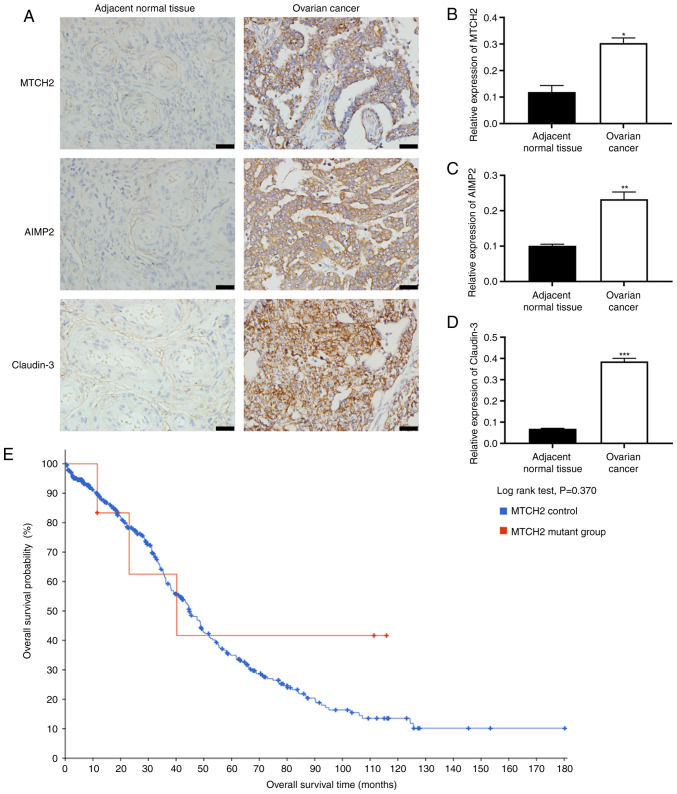
The expression levels of MTCH2, AIMP2 and claudin-3 in OC and adjacent normal tissues were evaluated in tissue samples from 67 patients with OC (median age, 57 years; range, 24-72 years). Immunohistochemical results showed significantly increased expression levels of MTCH2, AIMP2 and claudin-3 in OC tissue samples compared with in adjacent normal tissue samples (Fig. 1A-D), and the tumor samples were scored). According to the immunohistochemical scoring criteria, in OC tissues, 46 cases were positive for MTCH2 and 21 were negative; 42 cases were positive for AIMP2 and 25 were negative; and 43 cases were positive for claudin-3 and 24 were negative (Fig. S1; Table I). Pearson correlation analysis demonstrated a significant positive correlation between MTCH2 expression levels and the expression levels of both AIMP2 and claudin-3 (Table II). Subsequently, the relationship between MTCH2 and the clinicopathological features of patients with OC were assessed, and it was demonstrated that MTCH2 expression levels were significantly associated with International Federation of Gynecology and Obstetrics stage (37) and tumor differentiation (Table III). No significant difference in overall survival time of patients with OC between the MTCH2 mutation group and the MTCH2 control group was detected (Fig. 1E).
Figure 1.
Increased expression levels of MTCH2, AIMP2 and claudin-3 in OC tissue samples. (A) Representative images of immunohistochemical staining of MTCH2, AIMP2 and claudin-3 expression in OC and adjacent normal tissue samples. Scale bar, 40 µm; magnification, 400×. Quantification of the relative expression levels of (B) MTCH2, (C) AIMP2 and (D) claudin-3 in OC and adjacent normal tissue samples. (E) Data from cBioPortal was used to assess the association between MTCH2 control and MTCH2 mutant groups and the overall survival time of patients with OC. *P<0.05, **P<0.01, ***P<0.001. OC, ovarian cancer; MTCH2, mitochondrial carrier homology 2; AIMP2, aminoacyl transfer RNA synthetase-interacting multifunctional protein 2.
Table I.
MTCH2, AIMP2 and claudin-3 expression in adjacent normal tissues and ovarian cancer tissues.
| MTCH2 expression | AIMP2 expression | Claudin-3 expression | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||||
| Tissue | n | - | + | χ2 value | P-value | - | + | χ2 value | P-value | - | + | χ2 value | P-value |
| Normal | 67 | 53 | 14 | 30.905 | <0.001 | 50 | 17 | 18.927 | <0.001 | 47 | 20 | 15.848 | <0.001 |
| Ovarian cancer | 67 | 21 | 46 | 25 | 42 | 24 | 43 | ||||||
MTCH2, mitochondrial carrier homology 2; AIMP2, aminoacyl transfer RNA synthetase-interacting multifunctional protein 2.
Table II.
Correlation of MTCH2 with claudin-3 and AIMP2 expression levels.
| Factors | Pearson correlation | P-value |
|---|---|---|
| MTCH2 and claudin-3 | 0.507 | <0.001 |
| MTCH2 and AIMP2 | 0.287 | 0.019 |
MTCH2, mitochondrial carrier homology 2; AIMP2, aminoacyl transfer RNA synthetase-interacting multifunctional protein 2.
Table III.
Association between MTCH2 expression and clinicopathological features in patients with ovarian cancer.
| Clinical parameters | Total patients, n | MTCH2 positive, n | MTCH2 negative, n | P-value |
|---|---|---|---|---|
| Patients | 67 | 46 | 21 | |
| Age, years | 0.953 | |||
| ≤60 | 45 | 31 | 14 | |
| >60 | 22 | 15 | 7 | |
| Tumor size, cm | 0.831 | |||
| ≤5 | 18 | 12 | 6 | |
| >5 | 49 | 34 | 15 | |
| FIGO stage | 0.014 | |||
| I–II | 16 | 7 | 9 | |
| II–IV | 51 | 39 | 12 | |
| Tumor differentiation | 0.008 | |||
| G1 | 35 | 19 | 16 | |
| G2-G3 | 32 | 27 | 5 | |
| Lymph node metastasis | 0.929 | |||
| Yes | 25 | 17 | 8 | |
| No | 42 | 29 | 13 |
MTCH2, mitochondrial carrier homology 2; FIGO, International Federation of Gynecology and Obstetrics.
MTCH2 overexpression and knockdown regulated OC cell migration and invasion
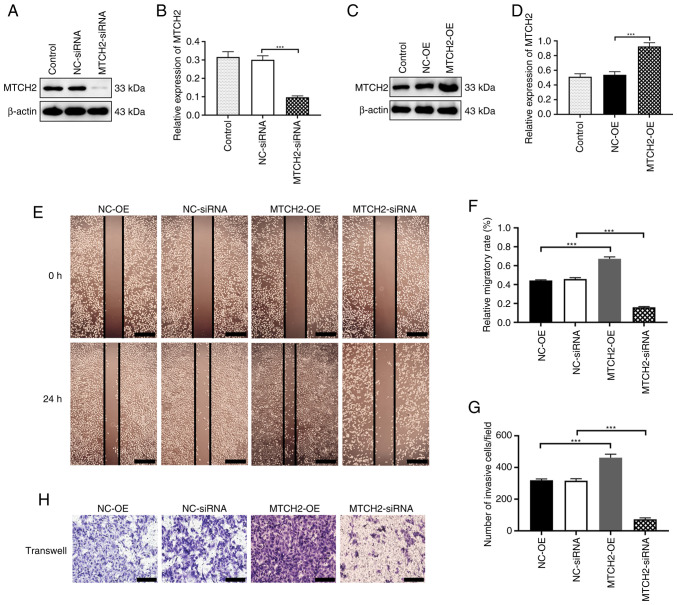
To further explore the role of MTCH2 in OC cells, an MTCH2 overexpression plasmid and MTCH2 siRNA were designed and synthesized. Western blotting was used to confirm the transfection efficiency of the MTCH2-OE plasmid and MTCH2 siRNA in OC cells compared with the negative controls (Fig. 2A-D). The wound healing assay demonstrated that, compared with the NC-OE group, the scratch area of the MTCH2-OE group was significantly reduced at 24 h. By contrast, the scratch area of the MTCH2 siRNA group was significantly larger compared with the NC-siRNA group (Fig. 2E, F). Transwell assays demonstrated that the number of migrating cells in the MTCH2-OE group were significantly increased compared with the NC-OE group (P<0.001), whereas the number of migrating cells were significantly decreased in the MTCH2 siRNA group compared with the NC-siRNA group (Fig. 2G, H).
Figure 2.
MTCH2 effects on cell invasion and migration of SK-OV-3 cells. (A) Representative western blot images and (B) quantification of the validation of MTCH2 siRNA transfection efficiency. (C) Representative western blot images and (D) quantification of the validation of MTCH2-OE transfection efficiency. (E) Representative wound healing assay images of cell migration following knockdown or overexpression of MTCH2 in SK-OV-3 cells and (F) quantification or the relative migratory rate. Scale bar, 400 µm; magnification, 400×. (G) Quantification of the number of invasive cells per field of view and (H) representative Transwell assay images of cell migration following knockdown or overexpression of MTCH2 in SK-OV-3 cells. Scale bar, 100 µm; magnification, 200×. ***P<0.001. MTCH2, mitochondrial carrier homology 2; AIMP2, aminoacyl transfer RNA synthetase-interacting multifunctional protein 2; OE, overexpression; NC, negative control; siRNA, small interfering RNA.
MTCH2 regulated OC cell energy metabolism, cell proliferation and apoptosis
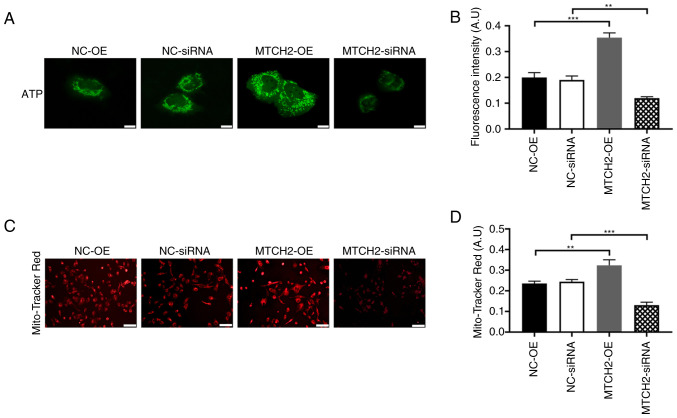
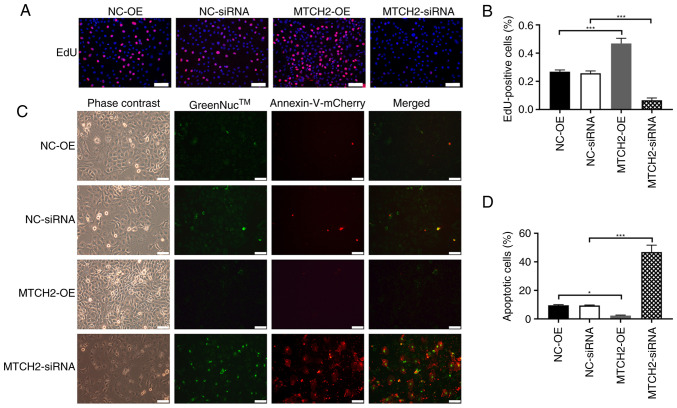
The ATP fluorescence probe assay demonstrated that the fluorescence intensity of the MTCH2-OE group was significantly increased compared with the NC-OE group, which represented increased ATP production in the MTCH2-OE group. By contrast, the fluorescence intensity significantly decreased in the MTCH2 siRNA group compared with the NC-siRNA group (P<0.01; Fig. 3A, B). MitoTracker Red CMXRos staining of mitochondria demonstrated decreased mitochondrial fluorescence in the MTCH2 siRNA group compared with the NC-siRNA group, which indicated mitochondrial dysfunction (Fig. 3C, D). The proliferation of OC cells was assessed through EdU staining. Significantly increased cell proliferation was observed in the MTCH2-OE group compared with the NC-OE group, whereas significantly decreased cell proliferation was demonstrated in the MTCH2 siRNA group compared with the NC-siRNA (Fig. 4A, B). The number of apoptotic cells were significantly decreased in the MTCH2-OE group compared with the NC-OE group, but were significantly increased in the MTCH2 siRNA group compared with the NC-siRNA group (P<0.001; Fig. 4C and D).
Figure 3.
MTCH2 knockdown and overexpression effects mitochondrial function and ATP production in SK-OV-3 cells. (A) Representative images of the ATP fluorescence probe assay in SK-OV-3 cells, indicating ATP production of cells following MTCH2 knockdown or overexpression and (B) quantification of the relative fluorescent intensities. Scale bar, 40 µm; magnification, 400×. (C) Representative images of mitochondrial red fluorescence which indicated mitochondrial function cell following knockdown or overexpression of MTCH2 in SK-OV-3 cells. Scale bar, 100 µm; magnification, 200×. (D) Quantification of the mitochondrial red fluorescence intensities. **P<0.01 and ***P<0.001. MTCH2, mitochondrial carrier homology 2; AIMP2, aminoacyl transfer RNA synthetase-interacting multifunctional protein 2; OE, overexpression; NC, negative control.
Figure 4.
MTCH2 knockdown and overexpression on the proliferation and apoptosis of SK-OV3 cells. (A) Representative images of EdU assay results, which indicated the proliferation of SK-OV-3 cell following knockdown or overexpression of MTCH2. Scale bar, 100 µm; magnification, 200×. (B) Quantification of the EdU-positive cells in these groups. (C) Representative images of GreenNuc™ caspase-3 activity and annexin V cell apoptosis assay in SK-OV-3 cells transfected with MTCH2-OE and MTCH2-siRNA compared with NC-OE and NC-siRNA, respectively and (D) quantification of the apoptotic cells in these groups. Scale bar, 100 µm; magnification, 200×. *P<0.05 and ***P<0.001. EdU, 5-ethynyl-2′-deoxyuridine; MTCH2, mitochondrial carrier homology 2; AIMP2, aminoacyl transfer RNA synthetase-interacting multifunctional protein 2; OE, overexpression; NC, negative control; siRNA, small interfering RNA.
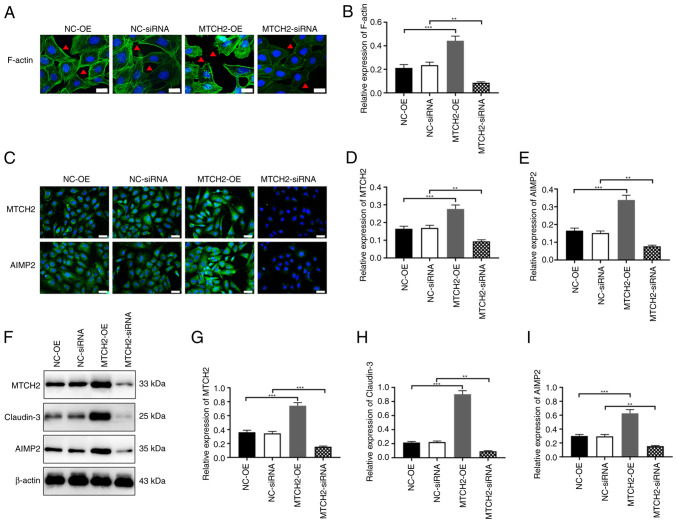
MTCH2 regulated cytoskeletal remodeling and the expression levels of claudin-3 and AIMP2
Transfection with MTCH2-OE in SK-OV-3 cells demonstrated a significant increase in the fluorescence intensity of microfilaments, filamentous feet and pseudopodia compared with the NC-OE group. By contrast, the fluorescence intensity significantly decreased in the MTCH2 siRNA group compared with the NC-siRNA group (Fig. 5A, B). Immunofluorescence and western blot analyses showed that, compared with the NC-OE group, the expression levels of claudin-3 and AIMP2 were significantly increased in the MTCH2-OE group, and were significantly reduced in the MTCH2 siRNA group compared with that in the NC-siRNA group (Fig. 5C-I).
Figure 5.
MTCH2 regulated cytoskeletal remodeling and expression levels of claudin-3 and AIMP2. (A and B) Representative images of the cytoskeleton assay to evaluate the expression of F-actin in SK-OV-3 cells following knockdown or overexpression of MTCH2. The red triangle indicates changes in pseudopodia. Scale bar, 20 µm, magnification, 400×. (C-E) Representative immunofluorescent images to assess the expression levels of MTCH2 and AIMP2 following knockdown or overexpression of MTCH2. Scale bar, 40 µm; magnification, 400×. (F-I) Western blots to assess the expression levels of MTCH2, claudin-3 and AIMP2 in SK-OV-3 cells following knockdown or overexpression of MTCH2. **P<0.01 and ***P<0.001. MTCH2, mitochondrial carrier homology 2; AIMP2, aminoacyl transfer RNA synthetase-interacting multifunctional protein 2; OE, overexpression; NC, negative control.
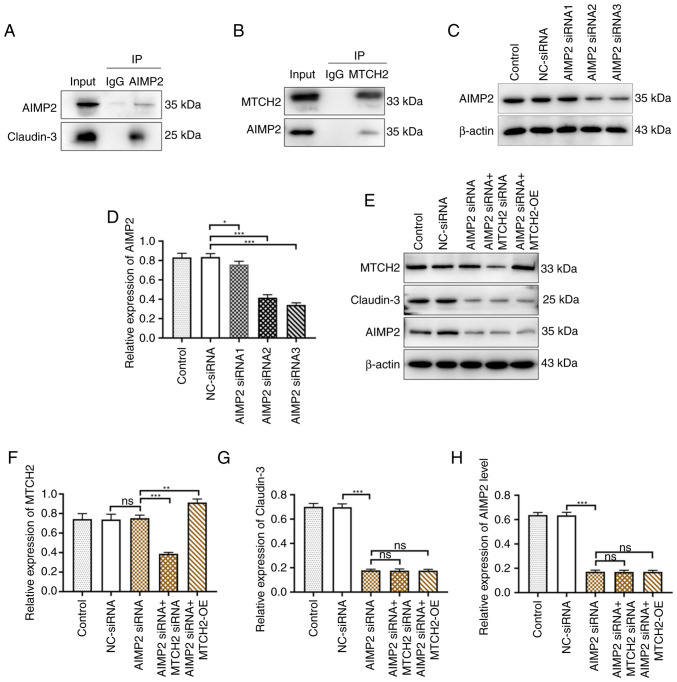
MTCH2 regulated claudin-3 expression levels through AIMP2
To evaluate the pathways involved in MTCH2-mediated OC cell metastasis, a number of AIMP2 siRNA sequences were designed and synthesized. Western blotting was used to assess the transfection efficacy of AIMP2 siRNA. Compared with NC-siRNA, the transfection effects of AIMP2 siRNA2 and AIMP2 siRNA3 were notably better than AIMP2 siRNA1. The results of western blotting revealed that the inhibitory effect of AIMP2 siRNA2 on claudin-3 was markedly stronger than that of AIMP2 siRNA3 therefore, AIMP2 siRNA2 was chosen as the interference sequence for use in subsequent experiments (Figs. 6C and D and S2). The expression levels of claudin-3 were significantly decreased in the AIMP2 siRNA group compared with the NC-siRNA group. However, there was no significant regulatory effect on MTCH2 expression levels upon AIMP2 knockdown by AIMP2 siRNA. Transfection of AIMP2 siRNA with the MTCH2-OE plasmid in OC cells could not restore the expression levels of AIMP2 and claudin-3 to the NC-siRNA group levels, which indicated that knockdown AIMP2 blocked the regulatory effect of MTCH2 on these targets (Figs. 6E-H). Co-immunoprecipitation demonstrated that MTCH2 interacts with both claudin-3 and AIMP2 (Fig. 6A and B).
Figure 6.
MTCH2 regulated claudin-3 expression levels through AIMP2. (A and B) Co-IP assay verified the binding between MTCH2 and AIMP2 in SK-OV-3 cells. (C and D) The efficiency of AIMP2 knockdown was validated using western blotting. (E) The expression levels of (F) MTCH2, (G) claudin-3 and (H) AIMP2 were detected by western blot following the transfection of NC-siRNA, AIMP2 siRNA, AIMP2 siRNA2, AIMP2 siRNA + MTCH2 siRNA and AIMP2 siRNA + MTCH2-OE. *P<0.05, **P<0.01 and ***P<0.001. IP, immunoprecipitation; MTCH2, mitochondrial carrier homology 2; AIMP2, aminoacyl transfer RNA synthetase-interacting multifunctional protein 2; OE, overexpression; NC, negative control; ns, not significant; siRNA, small interfering RNA.
Discussion
Mitochondrial dysfunction can lead to limited ATP production, adversely affecting cellular function; therefore, mitochondrial function has become a focal point of research in cancer therapy (38). Additionally, it has been reported that proteins related to the OMM are upregulated in tumor cells (39). Proteins incorporated into the OMM are essential for mitochondrial function, and a malfunctioning OMM is implicated in the mechanisms underlying cancer metastasis development (40). MTCH2 serves as an insertion enzyme for OMM proteins, which facilitates the entry of various proteins. It is involved in lipid biosynthesis, energy metabolism and mitochondrial division and fusion (6). Claudins are a family of transmembrane tight-linking proteins. Claudin-3 is a highly tissue-specific protein that serves roles in cytoskeletal remodeling and in the early and differential diagnosis in oral squamous cell carcinoma (41). AIMP2 is a scaffold subunit that is part of the polyaminoacyl-tRNA synthase complex and has functions in cellular energy metabolism, protein biosynthesis and apoptosis (28). The present study demonstrated a significant increase in MTCH2, AIMP2 and claudin-3 expression levels in OC tissue samples. Moreover, the expression levels of MTCH2 were significantly positively associated with AIMP2 and claudin-3 expression levels Overexpression of MTCH2 promoted the proliferation, invasion and migration of SK-OV-3 cells. By contrast, knockdown of MTCH2 decreased ATP production in SK-OV-3 cells, induced mitochondrial dysfunction, and promoted cytoskeletal remodeling and apoptosis. The aforementioned results of the present study indicated that MTCH2 knockdown can reduce the protein expression levels of AIMP2 and claudin-3 in OC cells and therefore, could potentially inhibit cancer progression.
It has been reported that MTCH2 is highly expressed in malignant glioma, and its inhibition in glioma cells reduces cell migration and invasion (12). A previous study demonstrated the functional role of MTCH2 in acute myeloid leukemia (AML), reporting that MTCH2 inhibition could reduce tumor growth and induce AML cell differentiation (42). In the present study, increased expression levels of MTCH2 were demonstrated in OC tissues and knockdown of MTCH2 resulted in significantly decreased cell migration and invasion, which suggested that MTCH2 may act as a tumor-promoting factor in the development of OC. It has been reported that MTCH2 is associated with apoptosis involving tBID (43,44). Zhou et al (45) demonstrated that silicon dioxide nanoparticles suppressed the expression of microRNA-450b-3p through the modulation of MTCH2 in spermatocyte cells, which led to mitochondria-activated apoptosis signaling and subsequent apoptosis. In the present study, knockdown of MTCH2 inhibited cell proliferation and promoted apoptosis. Therefore, it could be suggested that MTCH2 represents a key regulatory pathway in OC malignancies.
MTCH2 serves major roles in the biology of neuronal cells as it regulates mitochondrial metabolism, motility, calcium buffering and ATP production (46). The knockdown of MTCH2 in hippocampal neurons results in defects in mitochondrial motility and calcium handling (46). MTCH2 is located in the OMM, where it functions to preserve the structure and function of mitochondria. For instance, the loss of MTCH2 increased the levels of oxidative phosphorylation (47), ATP, NADH and reactive oxygen species (48), markers associated with mitochondrial activity. Additionally, loss of MTCH2 leads to mitochondrial elongation (49), increased mitochondrial size, decreased motility and increased oxidative stress (12). Collectively, this suggests that MTCH2 functions as a central regulator of mitochondrial and overall body energy metabolism. The present study demonstrated that MTCH2 can regulate ATP production and affect mitochondrial function. This further suggests that MTCH2 influences the progression of OC through the modulation of mitochondrial function and energy metabolism. In a previous study of liver cancer, MTCH2 was reported to mediate the signal crosstalk between nucleus and mitochondria, disrupt transport signals and was associated with uncontrolled mitochondrial activity, which led to tumor metabolic reprogramming (50). While the present study substantiates the prevailing understanding of MTCH2 involvement in cancer progression through its modulation of mitochondrial function and energy metabolism, the significance of cytoskeletal rearrangements in the multifaceted landscape of neoplastic transformation was also taken into consideration. The present study enriched the current understanding of the role of MTCH2 in cancer, but also demonstrated the potential of targeting cytoskeleton remodeling as a therapeutic strategy for MTCH2 driven malignant tumors.
Previous research has demonstrated that a cytoskeletal network formed of several unbranched-chain actin filaments surrounds the mitochondria in cells, suggesting a new mechanistic model of mitochondrial activity and cytoskeletal remodeling (51). In the present study, overexpression of MTCH2 promoted the lengthening of pseudopodia, which further demonstrated the role of MTCH2 in promoting the malignant biological behavior of OC cells. It could be suggested that the abnormal expression of MTCH2 may directly or indirectly affect cytoskeletal remodeling and mediate cancer cell metastasis. Transmission electron microscopy could be used to verify this in the future.
Claudin-3 serves a diverse and integral role in maintaining the structural and functional integrity of epithelial cells, governing cell polarity, transcriptional regulation, proliferative capacity, differentiation, metabolic homeostasis and immune-inflammatory responses (52). It is frequently dysregulated in OC, where increased expression levels of claudin-3 have been causally linked to disease progression, the emergence of distant metastases and the acquisition of drug resistance phenotypes (53). Additionally, increased claudin-3 expression levels have been implicated in the malignant transformation of colorectal cancer cells (54). It has been reported that the expression of claudin-3 mRNA and protein is significantly higher in gastric cancer tissues and metastatic tissues than in normal gastric mucosa tissues. (55). Knockdown of claudin-3 inhibited the growth of OC xenografts and promoted benign tumor differentiation (56). Downregulation of Claudin-3 was reported to promote tumor cell apoptosis, inhibit tumor cell proliferation, reduce microvascular density and thus serve an antitumor role (57). Given the selective upregulation of claudin-3 in OC, claudin-3 has emerged as a potential biomarker for early detection (58). The tissue-specificity and intricate molecular signaling networks within cells likely contribute to the biological functions of claudin-3 (59). In the present study, MTCH2 knockdown significantly downregulated claudin-3 expression, which suggested that, in addition to its autonomous oncogenic properties, claudin-3 is under the regulatory control of MTCH2. This finding implicates MTCH2 as a modulator of claudin-3 expression levels and could thereby influence the progression of tumor cells.
AIMP2 has emerged as a potent prognostic marker inversely correlated with patient survival, as dysregulation of AIMP2 expression levels have been reported across a number of types of tumors. Targeted suppression of AIMP2 has been shown to attenuate the migratory capabilities and proliferation of breast cancer cells (60). Moreover, knockdown of AIMP2 expression inhibited EMT, cell invasion, cell migration and tumor growth (55). As a pivotal signaling hub and a robust predictor of adverse outcomes in AML progression (61), AIMP2 has potential as a therapeutic target in cancer treatments. In the present study, MTCH2 knockdown led to downregulation of AIMP2 expression levels. AIMP2 knockout did not affect the expression of MTCH2, but significantly decreased the expression of claudin-3, which blocked the regulation of MTCH2 on mitochondrial function. It could be suggested that AIMP2 may be a downstream factor of MTCH2. Moreover, co-immunoprecipitation data demonstrated a tripartite interaction among these proteins, which suggested that MTCH2 could have a regulatory effect on claudin-3 via AIMP2. It was further demonstrated through knocking down AIMP2 that the interference of AIMP2 siRNA2 on claudin-3 was more obvious than that of AIMP2 siRNA3, which may be due to differences in the interference sequences.
The present study validated the translational relevance of MTCH2 in advanced serous OC tissues and the ovarian endometrioid cancer cell model, represented by the SK-OV-3 cell line. The present study aimed to verify the research value of MTCH2 in HGSOC tissues and the SK-OV-3 cell line. A limitation of the present study was that investigation of MTCH2 functional mechanisms were restricted to the SK-OV-3 model, therefore the results of the present study could not be generalized across different types of OC. Hence, validation using cell lines derived from diverse tissue origins is vital, as it would provide a foundation for future investigation of the role of MTCH2 in other types of OC. As the present experimental results were only confirmed in an in vitro cell model and in vivo animal experiments are currently limited, subcutaneous transplanted tumor models could be used in the future to further investigate the roles of MTCH2 in mechanisms of cancer drug resistance.
The results of the present study indicated that MTCH2 knockdown induced mitochondrial dysfunction, decreased ATP production and promoted apoptosis. MTCH2 regulated claudin-3 through AIMP2 to mediate the progression of OC cells and AIMP2 knockdown inhibited the effect of MTCH2, which indicated that MTCH2 as a novel target for the treatment of OC and provides further insight for the treatment of OC metastasis.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This work was supported by the Project of Hebei Medical Science Research (grant no. 20210129) and the Natural Science Foundation of Hebei Province (grant no. H2020206223).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
GS was responsible for study design. YS and CoL performed the experiments. BS and ChL analyzed the data. JS, PX and ZZ contributed to data interpretation, conceived the study and modified the manuscript. GS and PX confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The protocol was approved by the Ethics Committee of The Fourth Hospital of Hebei Medical University (ethical approval no. 2020166; Shijiazhuang, China). Each patient signed a written informed consent form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Giaquinto AN, Jemal A. Cancer statistics. CA Cancer J Clin. 2024;74:12–49. doi: 10.3322/caac.21820. [DOI] [PubMed] [Google Scholar]
- 2.Xiao Y, Bi M, Guo H, Li M. Multi-omics approaches for biomarker discovery in early ovarian cancer diagnosis. EBioMedicine. 2022;79:104001. doi: 10.1016/j.ebiom.2022.104001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang C, Xia BR, Zhang ZC, Zhang YJ, Lou G, Jin WL. Immunotherapy for ovarian cancer: Adjuvant, combination, and neoadjuvant. Front Immunol. 2020;11:577869. doi: 10.3389/fimmu.2020.577869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsibulak I, Zeimet AG, Marth C. Hopes and failures in front-line ovarian cancer therapy. Crit Rev Oncol Hematol. 2019;143:14–19. doi: 10.1016/j.critrevonc.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Spinelli JB, Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol. 2018;20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guna A, Stevens TA, Inglis AJ, Replogle JM, Esantsi TK, Muthukumar G, Shaffer KCL, Wang ML, Pogson AN, Jones JJ, et al. MTCH2 is a mitochondrial outer membrane protein insertase. Science. 2022;378:317–322. doi: 10.1126/science.add1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman A, Mullokandov M, Zaltsman Y, Regev L, Levin-Zaidman S, Gross A. MTCH2 cooperates with MFN2 and lysophosphatidic acid synthesis to sustain mitochondrial fusion. EMBO Rep. 2023;25:45–67. doi: 10.1038/s44319-023-00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rottiers V, Francisco A, Platov M, Zaltsman Y, Ruggiero A, Lee SS, Gross A, Libert S. MTCH2 is a conserved regulator of lipid homeostasis. Obesity (Silver Spring) 2017;25:616–625. doi: 10.1002/oby.21751. [DOI] [PubMed] [Google Scholar]
- 9.Robinson AJ, Kunji ER, Gross A. Mitochondrial carrier homolog 2 (MTCH2): The recruitment and evolution of a mitochondrial carrier protein to a critical player in apoptosis. Exp Cell Res. 2012;318:1316–1323. doi: 10.1016/j.yexcr.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 10.Manjunath LE, Singh A, Sahoo S, Mishra A, Padmarajan J, Basavaraju CG, Eswarappa SM. Stop codon read-through of mammalian MTCH2 leading to an unstable isoform regulates mitochondrial membrane potential. J Biol Chem. 2020;295:17009–17026. doi: 10.1074/jbc.RA120.014253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kisaki CY, Arcos SSS, Montoni F, Da Silva Santos W, Calacina HM, Lima IF, Cajado-Carvalho D, Ferro ES, Nishiyama-Jr MY, Iwai LK. Bothrops jararaca snake venom modulates key cancer-related proteins in breast tumor cell lines. Toxins (Basel) 2021;13:519. doi: 10.3390/toxins13080519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan Q, Yang W, Zhang S, Li T, Zuo M, Zhou X, Li J, Li M, Xia X, Chen M, Liu Y. Inhibition of mitochondrial carrier homolog 2 (MTCH2) suppresses tumor invasion and enhances sensitivity to temozolomide in malignant glioma. Mol Med. 2021;27:7. doi: 10.1186/s10020-020-00261-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.JanssenDuijghuijsen LM, Grefte S, de Boer VCJ, Zeper L, van Dartel DAM, van Der Stelt I, Bekkenkamp-Grovenstein M, van Norren K, Wichers HJ, Keijer J. Mitochondrial ATP depletion disrupts Caco-2 monolayer integrity and internalizes claudin 7. Front Physiol. 2017;8:794. doi: 10.3389/fphys.2017.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hana C, Dar NN, Venegas MG, Vulfovich M. Claudins in cancer: A current and future therapeutic target. Int J Mol Sci. 2024;25:4634. doi: 10.3390/ijms25094634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zihni C, Mills C, Matter K, Balda MS. Tight junctions: From simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 2016;17:564–580. doi: 10.1038/nrm.2016.80. [DOI] [PubMed] [Google Scholar]
- 16.Lin X, Shang X, Manorek G, Howell SB. Regulation of the epithelial-mesenchymal transition by claudin-3 and claudin-4. PLoS One. 2013;8:e67496. doi: 10.1371/journal.pone.0067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung H, Jun KH, Jung JH, Chin HM, Park WB. The expression of claudin-1, claudin-2, claudin-3, and claudin-4 in gastric cancer tissue. J Surg Res. 2011;167:e185–e191. doi: 10.1016/j.jss.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Li JY, Xie F, Xu XP, Ma JJ, Zhou DC, Liao Y, Tang J, Xie Q, Bai L, Nan QZ. Claudin-3 expression in colorectal carcinoma and its significance. Nan Fang Yi Ke Da Xue Xue Bao. 2017;37:63–67. doi: 10.3969/j.issn.1673-4254.2017.01.11. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senga K, Mostov KE, Mitaka T, Miyajima A, Tanimizu N. Grainyhead-like 2 regulates epithelial morphogenesis by establishing functional tight junctions through the organization of a molecular network among claudin3, claudin4, and Rab25. Mol Biol Cell. 2012;23:2845–2855. doi: 10.1091/mbc.e12-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Zhang W, Li M, Dam J, Huang K, Wang Y, Qiu Z, Sun T, Chen P, Zhang Z, Zhang W. Evaluation of the prognostic relevance of differential claudin gene expression highlights claudin-4 as being suppressed by TGFβ1 inhibitor in colorectal cancer. Front Genet. 2022;13:783016. doi: 10.3389/fgene.2022.783016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Yang X. The expression patterns of tight junction protein claudin-1, −3, and −4 in human gastric neoplasms and adjacent non-neoplastic tissues. Int J Clin Exp Pathol. 2015;8:881–887. [PMC free article] [PubMed] [Google Scholar]
- 22.Oh S, Yang H, Oh HR, Seo MR, Lee CH, Kim YH, Choi JY, Kim NY, Cheon GJ, Kang KW, et al. Visualization of a novel human monoclonal antibody against Claudin-3 for targeting ovarian cancer. Nucl Med Biol. 2022:114–115. 135–142. doi: 10.1016/j.nucmedbio.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Yu YC, Han JM, Kim S. Aminoacyl-tRNA synthetases and amino acid signaling. Biochim Biophys Acta Mol Cell Res. 2021;1868:118889. doi: 10.1016/j.bbamcr.2020.118889. [DOI] [PubMed] [Google Scholar]
- 24.Hyeon DY, Kim JH, Ahn TJ, Cho Y, Hwang D, Kim S. Evolution of the multi-tRNA synthetase complex and its role in cancer. J Biol Chem. 2019;294:5340–5351. doi: 10.1074/jbc.REV118.002958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z, Sun B, Huang S, Yu D, Zhang X. Roles of aminoacyl-tRNA synthetase-interacting multi-functional proteins in physiology and cancer. Cell Death Dis. 2020;11:579. doi: 10.1038/s41419-020-02794-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazaheri M, Yavari M, Marzouni HZ, Stufano A, Lovreglio P, S'amore S, Jahantigh HR. Case report: Mutation in AIMP2/P38, the scaffold for the multi-trna synthetase complex, and association with progressive neurodevelopmental disorders. Front Genet. 2022;13:816987. doi: 10.3389/fgene.2022.816987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu D, Shao F, Bian X, Meng Y, Liang T, Lu Z. The evolving landscape of noncanonical functions of metabolic enzymes in cancer and other pathologies. Cell Metab. 2021;33:33–50. doi: 10.1016/j.cmet.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, Zhan Y, Zhang Y, Zhang Y, Liu Y, Xie Y, Sun Y, Qian J, Ding Y, Ding Y, Fang Y. Hexokinase 2 confers radio-resistance in hepatocellular carcinoma by promoting autophagy-dependent degradation of AIMP2. Cell Death Dis. 2023;14:488. doi: 10.1038/s41419-023-06009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Wang T, Fu G, Xu Y, Zhang N, Han L, Yang M. The allelic regulation of tumor suppressor ADARB2 in papillary thyroid carcinoma. Endocr Relat Cancer. 2023;30:e220189. doi: 10.1530/ERC-22-0189. [DOI] [PubMed] [Google Scholar]
- 30.Yum MK, Kang JS, Lee AE, Jo YW, Seo JY, Kim HA, Kim YY, Seong J, Lee EB, Kim JH, et al. AIMP2 controls intestinal stem cell compartments and tumorigenesis by modulating Wnt/β-catenin signaling. Cancer Res. 2016;76:4559–4568. doi: 10.1158/0008-5472.CAN-15-3357. [DOI] [PubMed] [Google Scholar]
- 31.Miao J, Liu J, Niu J, Zhang Y, Shen W, Luo C, Liu Y, Li C, Li H, Yang P, et al. Wnt/β-catenin/RAS signaling mediates age-related renal fibrosis and is associated with mitochondrial dysfunction. Aging Cell. 2019;18:e13004. doi: 10.1111/acel.13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dias MC, Coisne C, Lazarevic I, Baden P, Hata M, Iwamoto N, Francisco DMF, Vanlandewijck M, He L, Baier FA, et al. Claudin-3-deficient C57BL/6J mice display intact brain barriers. Sci Rep. 2019;9:203. doi: 10.1038/s41598-018-36731-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao L, Liu M, Gao Y, Wei X, Yin Y, Gao L, Zhou R. The long noncoding RNA TARID regulates the CXCL3/ERK/MAPK pathway in trophoblasts and is associated with preeclampsia. Reprod Biol Endocrinol. 2022;20:159. doi: 10.1186/s12958-022-01036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W, Lin J, Huang J, Chen Z, Sheng Q, Yang F, Yang X, Cui X. MicroRNA-409-5p inhibits cell proliferation, and induces G(2)/M phase arrest and apoptosis by targeting DLGAP5 in ovarian cancer cells. Oncol Lett. 2022;24:261. doi: 10.3892/ol.2022.13381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Song X, Cheng G, Zhang J, Dong L, Bai J, Luo D, Xiong Y, Li S, Liu F, et al. The regulatory mechanism and biological significance of mitochondrial calcium uniporter in the migration, invasion, angiogenesis and growth of gastric cancer. Onco Targets Ther. 2020;13:11781–11794. doi: 10.2147/OTT.S262049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berek JS, Renz M, Kehoe S, Kumar L, Friedlander M. Cancer of the ovary, fallopian tube, and peritoneum: 2021 update. Int J Gynaecol Obstet. 2021;155((Suppl 1)):S61–S85. doi: 10.1002/ijgo.13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee C, Park SH, Yoon SK. Genetic mutations affecting mitochondrial function in cancer drug resistance. Genes Genomics. 2023;45:261–270. doi: 10.1007/s13258-022-01359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colpman P, Dasgupta A, Archer SL. The role of mitochondrial dynamics and mitotic fission in regulating the cell cycle in cancer and pulmonary arterial hypertension: Implications for dynamin-related protein 1 and Mitofusin2 in hyperproliferative diseases. Cells. 2023;12:1897. doi: 10.3390/cells12141897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marciniak SJ, Chambers JE, Ron D. Pharmacological targeting of endoplasmic reticulum stress in disease. Nat Rev Drug Discov. 2022;21:115–140. doi: 10.1038/s41573-021-00320-3. [DOI] [PubMed] [Google Scholar]
- 41.Zejc T, Piontek J, Schulzke JD, Fromm M, Ervens J, Rosenthal R. Clinical significance of claudin expression in oral squamous cell carcinoma. Int J Mol Sci. 2022;23:11234. doi: 10.3390/ijms231911234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan DH, Mullokandov M, Wu Y, Voisin V, Gronda M, Hurren R, Wang X, Maclean N, Jeyaraju DV, Jitkova Y, et al. Mitochondrial carrier homolog 2 is necessary for AML survival. Blood. 2020;136:81–92. doi: 10.1182/blood.2019000106. [DOI] [PubMed] [Google Scholar]
- 43.Cogliati S, Scorrano L. A BID on mitochondria with MTCH2. Cell Res. 2010;20:863–865. doi: 10.1038/cr.2010.100. [DOI] [PubMed] [Google Scholar]
- 44.Zaltsman Y, Shachnai L, Yivgi-Ohana N, Schwarz M, Maryanovich M, Houtkooper RH, Vaz FM, De Leonardis F, Fiermonte G, Palmieri F, et al. MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat Cell Biol. 2010;12:553–562. doi: 10.1038/ncb2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou G, Liu J, Li X, Sang Y, Zhang Y, Gao L, Wang J, Yu Y, Ge W, Sun Z, Zhou X. Silica nanoparticles inducing the apoptosis via microRNA-450b-3p targeting MTCH2 in mice and spermatocyte cell. Environ Pollut. 2021;277:116771. doi: 10.1016/j.envpol.2021.116771. [DOI] [PubMed] [Google Scholar]
- 46.Ruggiero A, Aloni E, Korkotian E, Zaltsman Y, Oni-Biton E, Kuperman Y, Tsoory M, Shachnai L, Levin-Zaidman S, Brenner O, et al. Loss of forebrain MTCH2 decreases mitochondria motility and calcium handling and impairs hippocampal-dependent cognitive functions. Sci Rep. 2017;7:44401. doi: 10.1038/srep44401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buzaglo-Azriel L, Kuperman Y, Tsoory M, Zaltsman Y, Shachnai L, Zaidman SL, Bassat E, Michailovici I, Sarver A, Tzahor E, et al. Loss of muscle MTCH2 increases whole-body energy utilization and protects from diet-induced obesity. Cell Rep. 2016;14:1602–1610. doi: 10.1016/j.celrep.2016.01.046. [DOI] [PubMed] [Google Scholar]
- 48.Maryanovich M, Zaltsman Y, Ruggiero A, Goldman A, Shachnai L, Zaidman SL, Porat Z, Golan K, Lapidot T, Gross A. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat Commun. 2015;6:7901. doi: 10.1038/ncomms8901. [DOI] [PubMed] [Google Scholar]
- 49.Bahat A, Goldman A, Zaltsman Y, Khan DH, Halperin C, Amzallag E, Krupalnik V, Mullokandov M, Silberman A, Erez A, et al. MTCH2-mediated mitochondrial fusion drives exit from naïve pluripotency in embryonic stem cells. Nat Commun. 2018;9:5132. doi: 10.1038/s41467-018-07519-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y, Zhou L, Li H, Sun T, Wen X, Li X, Meng Y, Li Y, Liu M, Liu S, et al. Nuclear-encoded lncRNA MALAT1 epigenetically controls metabolic reprogramming in HCC cells through the mitophagy pathway. Mol Ther Nucleic Acids. 2021;23:264–276. doi: 10.1016/j.omtn.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang C, Svitkina TM. Ultrastructure and dynamics of the actin-myosin II cytoskeleton during mitochondrial fission. Nat Cell Biol. 2019;21:603–613. doi: 10.1038/s41556-019-0313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakamura S, Irie K, Tanaka H, Nishikawa K, Suzuki H, Saitoh Y, Tamura A, Tsukita S, Fujiyoshi Y. Morphologic determinant of tight junctions revealed by claudin-3 structures. Nat Commun. 2019;10:816. doi: 10.1038/s41467-019-08760-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim WS, Kim H, Joo MK, Choi BI, Yoo AY, Park JJ, Lee BJ, Kim SH, Chun HJ. High expression of claudin-4 is associated with synchronous tumors in patients with early gastric cancer. J Clin Med. 2022;11:3550. doi: 10.3390/jcm11123550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Souza WF, Fortunato-Miranda N, Robbs BK, De Araujo WM, De-Freitas-Junior JC, Bastos LG, Viola JP, Morgado-Díaz JA. Claudin-3 overexpression increases the malignant potential of colorectal cancer cells: Roles of ERK1/2 and PI3K-Akt as modulators of EGFR signaling. PloS One. 2013;8:e74994. doi: 10.1371/journal.pone.0074994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hashimoto I, Oshima T. Claudins and gastric cancer: An overview. Cancers (Basel) 2022;14:290. doi: 10.3390/cancers14020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang YH, Bao Y, Peng W, Goldberg M, Love K, Bumcrot DA, Cole G, Langer R, Anderson DG, Sawicki JA. Claudin-3 gene silencing with siRNA suppresses ovarian tumor growth and metastasis. Proc Natl Acad Sci USA. 2009;106:3426–3430. doi: 10.1073/pnas.0813348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He ZY, Wei XW, Luo M, Luo ST, Yang Y, Yu YY, Chen Y, Ma CC, Liang X, Guo FC, et al. Folate-linked lipoplexes for short hairpin RNA targeting claudin-3 delivery in ovarian cancer xenografts. J Control Release. 2013;172:679–689. doi: 10.1016/j.jconrel.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 58.Choi YL, Kim J, Kwon MJ, Choi JS, Kim TJ, Bae DS, Koh SS, In YH, Park YW, Kim SH, et al. Expression profile of tight junction protein claudin 3 and claudin 4 in ovarian serous adenocarcinoma with prognostic correlation. Histol Histopathol. 2007;22:1185–1195. doi: 10.14670/HH-22.1185. [DOI] [PubMed] [Google Scholar]
- 59.Lei N, Cheng Y, Wan J, Blasig R, Li A, Bai Y, Haseloff RF, Blasig IE, Zhu L, Qin Z. Claudin-3 inhibits tumor-induced lymphangiogenesis via regulating the PI3K signaling pathway in lymphatic endothelial cells. Sci Rep. 2022;12:17440. doi: 10.1038/s41598-022-22156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu J, Zhou T, Wang D, Hong W, Qian D, Meng X, Liu X. Pan-cancer analysis identifies AIMP2 as a potential biomarker for breast cancer. Curr Genomics. 2023;24:307–329. doi: 10.2174/0113892029255941231014142050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ku J, Kim R, Kim D, Kim D, Song S, Lee K, Lee N, Kim M, Yoon SS, Kwon NH, et al. Single-cell analysis of AIMP2 splice variants informs on drug sensitivity and prognosis in hematologic cancer. Commun Biol. 2020;3:630. doi: 10.1038/s42003-020-01353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in the present study may be requested from the corresponding author.