Abstract
Objective
Inhibition and knockdown of GPR68 negatively affects glioblastoma cell survival in vitro by inducing ferroptosis. Herein, we aimed to demonstrate that inhibition of GPR68 reduces the survival of glioblastoma cells in vivo using two orthotopic larval xenograft models in Danio rerio, using GBM cell lines U87-MG and U138-MG. In vivo survival of the cancer cells was assessed in the setting of GPR68 inhibition or knockdown.
Results
In vitro, shRNA-mediated knockdown of GPR68 inhibition demonstrated potent cytotoxic effects against U87 and U138 glioblastoma cell lines. This effect was associated with increased intracellular lipid peroxidation, suggesting ferroptosis as the underlying mechanism of cell death. Translating these findings in vivo, we established a novel xenograft model in zebrafish by successfully grafting fluorescently labeled human glioblastoma cells, which were previously shown to overexpress GPR68. shRNA knockdown of GPR68 significantly reduced the viability of grafted GBM cells within this model. Additionally, treatment with ogremorphin (OGM), a highly specific small molecule inhibitor of GPR68, also reduced the viability of grafted GBM cells with limited toxicity to the developing zebrafish embryos. This study suggests that therapeutic targeting of GPR68 with small molecules like OGM represents a promising approach for the treatment of GBM.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-024-06900-x.
Keywords: Glioblastoma multiforme, Ogremorphin, Zebrafish, Xenograft, GPR68, OGR1
Introduction
Glioblastoma multiforme (GBM) is the deadliest and most prevalent primary brain tumor in adult patients [1]. The average survival for patients with GBM is 14 months with near universal recurrence [2]. Standard of care relies on the alkylating agent temozolomide (TMZ) and radiotherapy following maximal surgical resection [2]. However, recurrent GBM presents with nearly ubiquitous resistance to TMZ and radiotherapy [3–5]. Furthermore, GBM tumors have high molecular heterogeneity and plasticity, rendering many potentially therapeutic agents ineffective and presenting a significant barrier to identifying a universal therapeutic target [6, 7]. The severity of the disease and the dearth of therapeutic options underscores the critical need for the development of new therapeutic targets for GBM.
A hallmark of cancer is the Warburg effect in which cancer cells favor aerobic glycolysis, resulting in acidification of the extracellular tumor microenvironment (TME). The acidic TME promotes malignant clonal selection, metastasis, pro-oncogenic transcriptional responses, and immune escape [8–16]. Williams et al., developed a novel small molecule inhibitor of proton sensing G-protein coupled receptor GPR68, Ogremorphin (OGM), and demonstrated that GBMs utilize GPR68 to protect from ferroptosis in an ATF4-dependent manner. The authors suggest that GPR68 is a key sensor of these extracellular pH changes and a regulator of the tumor’s response. While significant in vitro data is presented detailing the ability of OGM to induce ferroptosis, Williams et al., lack in vivo efficacy studies [17].
Larval zebrafish xenograft models have been developed for many cancers including GBM. These models are useful for early proof of principle studies and early stages of drug development due to low cost, short study duration, and permissive dosing requirements. We established two xenograft models using Green CMFDA stained U87MG and U138MG injected intracranially in zebrafish. These orthotopic xenograft models were then used to demonstrate that OGM kills GBM in vivo. We further show genetically, using shRNAs, that this effect is due to on-target inhibition of GPR68 in the GBMs. Taken together, we show the GPR68 is an attractive therapeutic target for inhibiting GBM in vivo.
Main text
Methods
Cell staining
U87 and U138 cells were grown in DMEM with high glucose, GlutaMAX, and hepes containing 10% FBS, 100 units/ml penicillin, and 100 µg/mL streptomycin at 37 °C and 5% CO2 (ThermoFisher; catalogs: 10564011, 26140079, 15140122). Media was replaced with fresh media containing 2.5 µM CellTracker Green CMFDA Dye (ThermoFisher; catalog: C7025). Cells were then incubated for 45 min at 37 °C and 5% CO2. Samples were subsequently washed with Dulbecco Phosphate Buffered Saline (DPBS) and fresh media added. For imaging, cells were washed with DPBS and FluoroBrite DMEM with 10% FBS and penicillin/streptomycin was added (ThermoFisher; catalog: A1896701). Cells were then imaged and analyzed on a Lionheart FX (Agilent BioTek). Cells were grown in 6-well plates and treated with DMSO or OGM. Alternatively, cells were reverse transfected with 5 µgs of plasmid per well.
GPR68 shRNA knockdowns
shRNAs for knockdown were obtained from VectorBuilder. shRNA #1 targets 5ʹ-CCACCGTTGTCACAGACAATG-3ʹ (plasmid: VB221221-1234czj) and shRNA #2 targets 5ʹ-GAGCTGTACCATCGACCATAC-3ʹ (plasmid: VB221221-1235jft). For control we used the non-targeting shRNA 5ʹ-CCTAAGGTTAAGTCGCCCTCG-3ʹ (plasmid: VB010000-9259tcf). Plasmids were reverse transfected using lipofectamine 3000 (ThermoFisher; catalog: L3000001).
Liperfluo
Experiments were run as previously described [17] with the following two changes. shRNA samples were transfected with 20 µgs plasmid per 100 mm cell culture dish. 30,000 events were recorded instead of 10,000.
Cell viability assay
Cells where reverse transfected with 0.25 µgs plasmid per well in a 24-well plate. Cells were incubated for 3 days before being lysed in 1 × Passive Lysis Buffer and assayed with CellTiter-Glo on a GloMax-Multi Detection System (Promega; catalogs: E1941, G7570, and TM297).
qRT-PCR experiments
Cells were reverse transfected with 0.25 µgs plasmid per well in a 24-well plate. Cells were incubated for 3 days before being lysed in TRIzol (ThermoFisher; catalog: 15596026) and RNA extracted. cDNA was generated using the high-capacity cDNA reverse transcription kit with RNase Inhibitor (Applied Biosystems; catalog:4374966). Samples were run on a QuantStudio 5 (ThermoFisher) using the TaqMan primers: GAPDH (Hs02786624_g1) and GPR68 (Hs00268858_s1).
Xenograft experiments
Casper zebrafish were grown at 28.5 °C in E3 media. At 2 days post fertilization (dpf) embryos were briefly tricained (0.15 mg/ml) and then transferred to a fresh plate for injection. Fish were held with Dumont #5 Forceps (Fine Science Tools) and injected with a FemtoJet 4i (Eppendorf; catalog 5252000021). Micro-needles were made from capillaries (World Precision Instruments; catalog TW100F-4) using a P-97 Flaming/Brown micropipette puller (Sutter Instrument; Heat: 590, Pull: 30, Velocity: 70, Time: 90). Fish were injected posterior to the hypothalamus in the midbrain halfway between the midline and medial edge of the retina with 50 cells in 3 nl. For cell preparations, cells were stained as described above, trypsinized for 5 min, centrifuged, washed with DPBS, and re-centrifuged. Cells were quantified on a TC20 Automated Cell Counter (Bio-Rad) with trypan blue. Samples with < 90% cell viability were discarded. For all experiments cells were resuspended, diluted to the working concentration, and injected in FluoroBrite DMEM with 10% FBS and penicillin/streptomycin, except for the test of injection medias which also used E3 embryo media and PBS. Except for the 6 hpi DMSO/OGM treatment test, injected fish were allowed to recover for 24 h in fresh E3 media at 28.5 °C. At 24 hpi fish were evaluated for successful grafting on an EVOS fl Fluorescence Microscope (ThermoFisher). Un-injected, poorly injected, over-injected, and off-target injected samples were discarded.
A stock of 20 mM OGM was added to E3 media for a final concentration of 2 µM. shRNA-transfected cells were reverse-transfected for 24 h before staining and injection. For the DMSO/OGM studies samples were then randomly and blindly assigned to treatment groups. Fish were grown for an additional 48 h to 5 dpf at 28.5 °C and imaged on a Lionheart FX in tricaine. Sagittal images were analyzed using a plug over the entire head of the zebrafish while excluding the yolk. Samples with the migration of cells into the trunk were excluded from the analysis. Alternatively, fish were mounted in 2% low melting point agarose and z-stack images were taken on a Nikon Eclipse Ti2 spinning disk confocal.
Statistical analysis
All statistics were performed in R 4.3.1 and multiple t-test comparisons were analyzed post-hoc with Bonferroni correction. Graphs were made in RStudio 2023.09.0 + 463 and edited with Inkscape 1.3.2.
Results
hGPR68 shRNAs kill U87MG and U138MG cells without affecting dye uptake
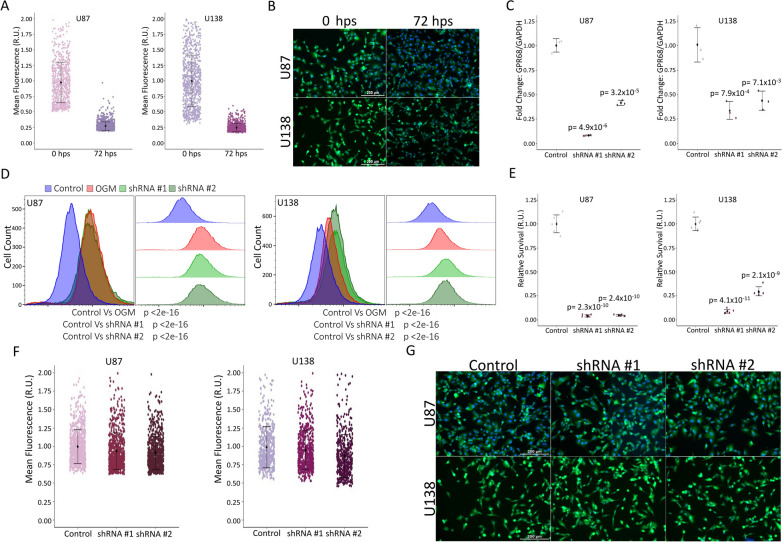
Prior to in vivo use, we optimized and characterized reagent efficacy and methodology. To visualize cancer cells upon transplantation, we used cell tracker dye CMFDA (excitation/emission: 492/517 nm), which is widely utilized to stain and monitor cells. It offers advantages such as bright fluorescence, good photostability, and the ability to transform into a highly fluorescent form once inside the cell's cytosol due to the cleavage of acetate groups by intracellular esterases, producing a bright fluorescein derivative (CMF) that becomes confined within the cell. These factors together allow for the persistence of signal and minimal effect on cell health. We evaluated the ability of both GBM cell lines (U87MG and U138MG) to take up and retain the dye for 72 h (Fig. 1A, B). Variability and total fluorescence decreased over time and the dye had no noticeable effect on cell viability (Fig. 1B) making the dye ideal for use in our xenograft experiments. Next, we validated the efficacies of two shRNAs against human GPR68, resulting in robust knockdown of the target (Fig. 1C). Furthermore, both shRNAs increase lipid peroxidation and cause comparable levels of cell death to OGM (Fig. 1D, E). However, the change in expression, lipid peroxidation, and cell death did not affect the uptake of the dye (Fig. 1F, G). These data establish the CMFDA dye is a suitable dye for labeling U87MG and U138MG cells and demonstrate that loss of GPR68 activity does not affect dye uptake or retention.
Fig. 1.
shRNA mediated ferroptosis does not inhibit dye uptake. A Stained U87 cells immediately after staining (n = 762) show more variability than at 72 h post stain (hps) (n = 1311). This pattern is recapitulated by U138 cells at 0 hps (n = 841) and 72 hps (n = 861). Data aggregated from n > 6 repeats. B Representative images of U87 and U138 cells at 0 hps and 72 hps (10 × magnification). C qRT-PCR shows significant knockdown of GPR68 mediated by shRNAs in U87 and U138 cells (n = 3 biological repeats). D Flow cytometry using Liperfluo demonstrates a significant increase in lipid peroxides with shRNA mediated knockdown of GPR68. This is consistent with the 2 µM OGM positive control in both U87 and U138 cells (n = 30,000 events). E Representative graph of CellTiter-Glo mediated assessment of cell survival 3 days post-transfection of U87s and U138s (n = 6 technical repeats; n = 3 biological repeats). F Stained shRNA #1 (n = 754) and shRNA #2 (n = 796) cells show no significant change in dye uptake from control shRNA treated U87 cells (n = 765). U138 cells recapitulate this data with shRNA #1 (n = 575) and shRNA #2 (n = 491) cells showing no significant change in dye uptake from control shRNA (n = 487). (Data aggregated from n > 6 repeats). G Representative images of control shRNA, shRNA #1, or shRNA #2 transfected U87 and U138 cells (10 × magnification). A, E, and F are relative units (R.U.). A Is normalized to 0 hps. C, E, and F are normalized to control shRNA. C, D, E Multiple two-tailed, equal variance, t-tests. A, C, E, and F mean ± SD C Bonferroni Correction of α-level of 0.05 is p < 0.025 and 0.01 is p < 0.005. D Bonferroni Correction of α-level of 0.001 is p < 0.00033. E Bonferroni Correction of α-level of 0.001 is p < 0.0005
U87 and U138 successfully graft when intracranially injected
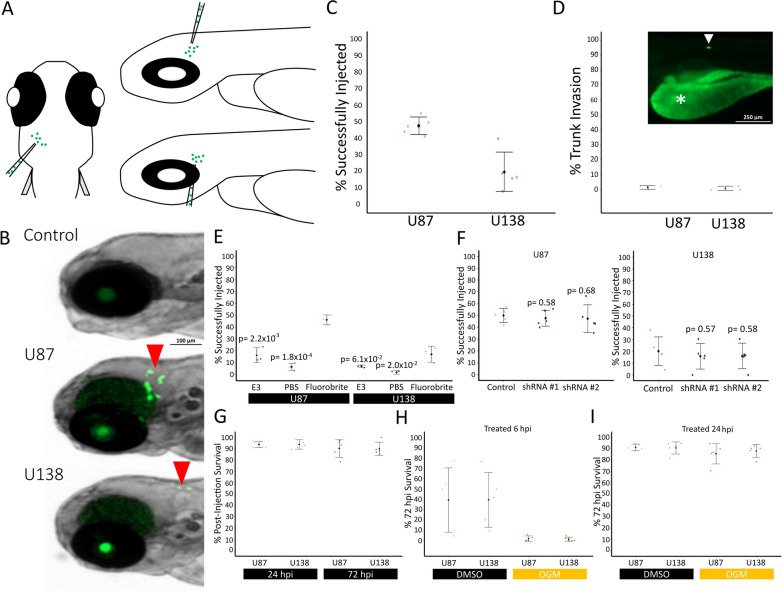
Numerous Zebrafish glioblastoma xenograft protocols vary broadly in methodology ranging from differences in the number of cells injected to injection sites (reviewed in [18]). To our knowledge, the vast majority of these use U87MG, while the U138MG line has not yet been used in zebrafish xenografts [18]. Early inhibition of GPR68 in zebrafish causes teratogenic effects such as a wavy notochord, reminiscent of a copper deficiency or knockout of lysyl oxidases in zebrafish embryos [19–21]. However, OGM treatment of later-stage embryos had normal morphology and did not result in neurotoxicity. Therefore, we decided to inject tumor cells into 2-day post fertilization (dpf) embryos. To avoid the use of 1-phenyl-2-thiourea (PTU), which can cause off-target toxicity and teratogenicity, to inhibit melanophore formation, we used the Casper line of zebrafish (which harbors mutations in mitfa and nacre), to prevent pigmentation of the zebrafish [22–29]. Furthermore, intercranial injections were chosen to provide a neural context for the GBM and to minimize noise from yolk autofluorescence in the GFP channel (Fig. 2A) [30].
Fig. 2.
U87 and U138 cells successfully xenograft in zebrafish in the forebrain-midbrain region. A Cartoon of injection strategy. B Representative samples were injected with stained cells 24 hpi. Red arrows indicate successfully xenografted Green CMFDA labeled cells (2.5× magnification). C Xenograft success rates at 72 h post injection (hpi) (U87: n = 246; U138: n = 285; Data aggregated from n = 5 repeats). D Migration of cells from brain into trunk tissues by 72 hpi (U87: n = 132; U138: n = 109; Data aggregated from n = 3 repeats; 4 × magnification). An example photo is trunk tissues invasion at 72 hpi. * Indicates yolk, arrow indicates U138 cell. E Xenograft success rates at 24 hpi of cells resuspended and injected in E3 (U87: n = 126; U138: n = 117), PBS (U87: n = 120; U138: n = 113), or FluoroBrite media (U87: n = 132; U138: n = 109) (Data aggregated from n = 3 repeats). F No difference in xenograft success rates between control (U87: n = 324; U138: n = 339), shRNA #1 (U87: n = 209; U138: n = 249), or shRNA #2 (U87: n = 239; U138: n = 220) transfected cells at 24 hpi was observed in U87 or U138 cells (Data aggregated from n = 5 repeats). G Injections of labeled cells did not affect acute (24 hpi) or short-term survival (72 hpi) of zebrafish (U87: n = 157; U138: n = 148; Data aggregated from n = 5 repeats). H Zebrafish injected with labeled cells and treated 6 hpi with DMSO (U87: n = 153; U138: n = 159) or OGM (U87: n = 162; U138: n = 175) show a significant decrease in survival. (Data aggregated from n = 5 repeats). I Treatment of zebrafish injected with labeled cells at 24 hpi with DMSO (U87: n = 151; U138: n = 151) or OGM (U87: n = 160; U138: n = 166) did not impact zebrafish survival. (Data aggregated from n = 5 repeats). C–I mean ± SD. E and F Multiple two-tailed, equal variance, t-tests; Bonferroni Correction of α-level of 0.05 is p < 0.025, 0.01 is p < 0.005, and 0.001 is p < 0.0005
Both U87MG and U138MG cells successfully grafted into the Casper embryos (Fig. 2B, C), showing minimal invasion beyond the brain tissue (Fig. 2D). The success rate of U87MG grafting was comparable to previous reports in wildtype zebrafish (Fig. 2B) [31, 32]. While various medias have been used for cell resuspension before injection [33–36], we found the use of cell culture media (Fluorobrite) provided maximal grafting success (Fig. 2E). Silencing hGPR68 with shRNA did not significantly affect the grafting efficiency of either cell line (Fig. 2F). Additionally, no significant decrease in survival was observed at 24- or 72-hpi, suggesting good tolerance of U87MG and U138MG grafted cells (Fig. 2G). However, treatment with DMSO or OGM at 6 hpi significantly decreased survival, but this effect was alleviated by extending the recovery period to 24 hpi (Fig. 2H, I). These data establish a robust U87MG and U138MG xenograft model using intercranial injection of labeled cells resuspended in cell media, followed by a 24-h recovery period before treatment with DMSO-containing compounds.
Inhibition of GPR68 kills glioblastoma in vivo
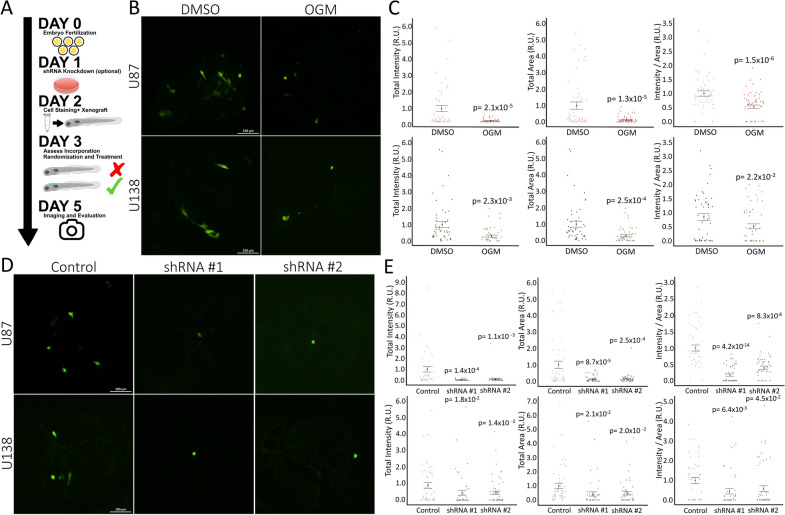
In our xenograft model, inhibiting GPR68 significantly reduced tumor burden for both U87MG and U138MG cells in the zebrafish brain after 48 h of treatment (Fig. 3A). OGM treatment had a significant effect on xenografts. It decreased the overall tumor cell area, indicating a reduction in the number and tumor volume. Secondly, it substantially lowered the total fluorescence intensity of the remaining cells (Fig. 3B, C). The decreased ratio of total fluorescence intensity to total tumor cell area (Fig. 3C) suggests that OGM did not just inhibit the growth of the tumor but induced cell death in the surviving population. Silencing hGPR68 with shRNA #1 and #2 in both U87MG and U138MG xenografts replicated these findings (Fig. 3D, E), further supporting the conclusion that inhibiting GPR68 specifically kills U87MG and U138MG cells in vivo without off-target toxicity.
Fig. 3.
Knockdown of GPR68 inhibit U87 and U138 Xenografts growth in zebrafish. A Schematic of xenograft testing in zebrafish. B Representative confocal (flattened z-stack) images of zebrafish head 72 hpi in 5 days post fertilization (dpf) zebrafish of DMSO or 2 µM OGM treated embryos (20× magnification). C The total fluorescent intensity and total area of Green CMFDA labeled cells in the zebrafish brain at 72 hpi treated with DMSO (U87: n = 50; U138: n = 62) or 2 µM OGM (U87: n = 51; U138: n = 54) is significantly decreased for U87 (Top) and U138 (bottom) cells. The ratio of intensity/area is also significantly decreased (Data aggregated from n = 5 repeats). D Representative confocal (flattened z-stack) images of zebrafish head 72 hpi in 5 dpf injected with control, shRNA #1, or shRNA #2 transfected cells (20 × magnification). E The total fluorescent intensity, total area, and ratio of intensity/area of Green CMFDA labeled cells in the zebrafish brain at 72 hpi transfected with shRNA #1 (U87: n = 55; U138: n = 51) or shRNA #2 (U87: n = 51; U138: n = 57) is significantly decreased for U87 (Top) and U138 (bottom) cells when compared to the control shRNA (U87: n = 50; U138: n = 50) (Data aggregated from n = 5 repeats). C and E mean ± SEM. C and E are relative units (R.U.). C Is normalized to DMSO control and E is normalized to control shRNA. C Two-tailed, equal variance, t-tests. E Multiple two-tailed, equal variance, t-tests; Bonferroni Correction of α-level of 0.05 is p < 0.025, 0.01 is p < 0.005, and 0.001 is p < 0.0005
Discussion
Zebrafish xenograft models are powerful in vivo tools, however, there is a lack of consistent methodology for establishing glioblastomas (reviewed in [18]). Here, we established that CellTracker Green CMFDA Dye is appropriate for tracking U87MG and U138MG cancer cell lines for up to 3 days. shRNA transfection did not affect the uptake of dye or the ability of the cells to graft into the zebrafish. Xenograft models most commonly use the glioblastoma cell line U87. Consistent with this, we found that U87MG cancer cells successfully grafted in about half of injected samples. Conversely, the U138MG cancer cell line was not a robust xenograft model, as it grafted at a low rate. Neither line invaded the trunk of the animal at an appreciable level when injected intracranially. However, the successful grafting of both cell lines was heavily dependent on the media in which they were suspended at the time of injection. Cells resuspended and injected in cell culture media grafted at a significantly higher rate than cells in PBS or E3 media. Furthermore, intercranial injections had minimal effects on sample survival. However, injected samples required a 24-h recovery period prior to drug treatment. Treatment 6 h after injection led to large amounts of variability in the DMSO controls and very low survival in the OGM-treated samples. This data suggests staining untreated or shRNA transfected glioblastoma cancer cells before resuspension in cell media and intercranial injection followed by a 24-h recovery period before treatment with small molecules is an appropriate and robust xenograft model in zebrafish.
Building on the extensive in vitro evidence of GPR68-mediated ferroptosis in GBM cells from Williams et al., we provide further support using both their small molecule inhibitor, OGM, and targeted shRNAs. First, we replicated their findings on cell survival and Liperfluo data, confirming ferroptosis induction by OGM and shRNA knockdown. Next, we established a robust U87MG and U138MG xenograft model in zebrafish and demonstrated OGM’s ability to inhibit tumor growth in vivo. This finding was further validated genetically through shRNA mediated GPR68 knockdown in the xenografts. Our data not only supports previous observations but also bridges a critical gap by demonstrating OGM’s efficacy in vivo. Importantly, Williams et al. found OGM to be non-toxic to healthy neural tissue in zebrafish. Together, these results strongly suggest OGM as a promising, non-toxic therapeutic approach for glioblastoma.
Limitations
Dye retention and dilution limits experimental length to 3 days. Furthermore, post-injection recovery limits small molecule treatments to 2 days. Long-term treatment with OGM may differ and/or increase toxicity.
Supplementary Information
Acknowledgements
We would like to acknowledge the University of Maryland School of Medicine Center for Innovative Biomedical Resources, Confocal Microscopy Core—Baltimore, Maryland. We would also like to thank Yi-Ju Chen at Johns Hopkins University for her help in de-bugging our RStudio scripts.
Authors' information
LRN, CCH, and CHW are authors on a previous manuscript published in Experimental Hematology & Oncology characterizing Ogremorphin, GPR68, ferroptosis, and glioblastoma cells including the cell lines U87 and U138.
Abbreviations
- dpf
Days post fertilization
- GBM
Glioblastoma multiforme
- hpi
Hours post injection
- hps
Hours post stain
- OGM
Ogremorphin
- PTU
1-Phenyl-2-thiourea
- TME
Tumor microenvironment
- TMZ
Temozolomide
Author contributions
LRN wrote the main manuscript text and prepared all figures. LRN and DTF generated data for the manuscript. CHW and CCH contributed insights and criticisms for the manuscript. LRN, CHW, and CCH conceptualized and planned experiments.
Funding
Our work was funded by NIGMS R01GM118557 to CCH. We also acknowledge the support of the University of Maryland, Baltimore, Institute for Clinical & Translational Research (ICTR) and the National Center for Advancing Translational Sciences (NCATS) Clinical Translational Science Award (CTSA) grant number 1UL1TR003098. Flow Cytometry Core is supported by the Maryland Department of Health’s Cigarette Restitution Fund Program and the National Cancer Institute—Cancer Center Support Grant (CCSG)—P30CA134274.
Availability of data and materials
All data generated or analyzed during this study are included in this published article as supplementary file: Data file. R-scripts used in generation of figures are included as R-script files.
Declarations
Ethics approval and consent to participate
Animal protocols all conform to the Guiding Principles in the Care and Use of Animals. The University of Maryland, Baltimore’s Committee on Use and Care of Animals approved all animal protocols.
Consent for publication
Not applicable.
Competing interests
CHW and CCH are inventors on an issued patent related to this manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–20. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–96. [DOI] [PubMed] [Google Scholar]
- 3.Brandes AA, Franceschi E, Tosoni A, Blatt V, Pession A, Tallini G, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008;26(13):2192–7. [DOI] [PubMed] [Google Scholar]
- 4.Cao VT, Jung TY, Jung S, Jin SG, Moon KS, Kim IY, et al. The correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomas. Neurosurgery. 2009;65(5):866–75. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 6.Lauko A, Lo A, Ahluwalia MS, Lathia JD. Cancer cell heterogeneity & plasticity in glioblastoma and brain tumors. Semin Cancer Biol. 2022;82:162–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science (1979). 2014;344(6190):1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boedtkjer E, Pedersen SF. The acidic tumor microenvironment as a driver of cancer. Annu Rev Physiol. 2020;82(1):103–26. [DOI] [PubMed] [Google Scholar]
- 9.Justus CR, Dong L, Yang LV. Acidic tumor microenvironment and pH-sensing G protein-coupled receptors. Front Physiol. 2013;4:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutoo S, Maeda T, Suzuki A, Kato Y. Adaptation to chronic acidic extracellular pH elicits a sustained increase in lung cancer cell invasion and metastasis. Clin Exp Metastasis. 2020;37(1):133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato Y, Ozawa S, Miyamoto C, Maehata Y, Suzuki A, Maeda T, et al. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013;13(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Worsley CM, Veale RB, Mayne ES. The acidic tumour microenvironment: Manipulating the immune response to elicit escape. Hum Immunol. 2022;83(5):399–408. [DOI] [PubMed] [Google Scholar]
- 13.Bailey KM, Wojtkowiak JW, Hashim AI, Gillies RJ. Targeting the metabolic microenvironment of tumors. Adv Pharmacol. 2012;65:63–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter A, Hendrikse A, Renan M, Abratt R. Does the tumor microenvironment influence radiation-induced apoptosis? Apoptosis. 2006;11(10):1727–35. [DOI] [PubMed] [Google Scholar]
- 15.Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci. 2019;20(4):840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo A, Yamamoto S, Nakaki R, Shimamura T, Hamakubo T, Sakai J, et al. Extracellular acidic pH activates the sterol regulatory element-binding protein 2 to promote tumor progression. Cell Rep. 2017;18(9):2228–42. [DOI] [PubMed] [Google Scholar]
- 17.Williams CH, Neitzel LR, Cornell J, Rea S, Mills I, Silver MS, et al. GPR68-ATF4 signaling is a novel prosurvival pathway in glioblastoma activated by acidic extracellular microenvironment. Exp Hematol Oncol. 2024;13(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pliakopanou A, Antonopoulos I, Darzenta N, Serifi I, Simos YV, Katsenos AP, et al. Glioblastoma research on zebrafish xenograft models: a systematic review. Clin Transl Oncol. 2023;26(2):311–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madsen EC, Gitlin JD. Zebrafish mutants calamity and catastrophe define critical pathways of gene-nutrient interactions in developmental copper metabolism. PLoS Genet. 2008;4(11): e1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gansner JM, Mendelsohn BA, Hultman KA, Johnson SL, Gitlin JD. Essential role of lysyl oxidases in notochord development. Dev Biol. 2007;307(2):202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendelsohn BA, Yin C, Johnson SL, Wilm TP, Solnica-Krezel L, Gitlin JD. Atp7a determines a hierarchy of copper metabolism essential for notochord development. Cell Metab. 2006;4(2):155–62. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Ptak D, Zhang L, Walls EK, Zhong W, Leung YF. Phenylthiourea specifically reduces zebrafish eye size. PLoS ONE. 2012;7(6): e40132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsalini OA, Rohr KB. Phenylthiourea disrupts thyroid function in developing zebrafish. Dev Genes Evol. 2003;212(12):593–8. [DOI] [PubMed] [Google Scholar]
- 24.Bohnsack BL, Gallina D, Kahana A. Phenothiourea sensitizes zebrafish cranial neural crest and extraocular muscle development to changes in retinoic acid and IGF signaling. PLoS ONE. 2011;6(8): e22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orger MB, Gahtan E, Muto A, Page-McCaw P, Smear MC, Baier H. Behavioral screening assays in zebrafish. Academic Press; 2004. p. 53–68. [DOI] [PubMed] [Google Scholar]
- 26.Parker MO, Brock AJ, Millington ME, Brennan CH. Behavioral phenotyping of Casper mutant and 1-pheny-2-thiourea treated adult zebrafish. Zebrafish. 2013;10(4):466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson J, von Hofsten J, Olsson PE. Generating transparent zebrafish: a refined method to improve detection of gene expression during embryonic development. Mar Biotechnol. 2001;3(6):0522–7. [DOI] [PubMed] [Google Scholar]
- 28.White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2(2):183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen XK, Kwan JSK, Chang RCC, Ma ACH. 1-phenyl 2-thiourea (PTU) activates autophagy in zebrafish embryos. Autophagy. 2021;17(5):1222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Y, Voshart D, Paridaen JTML, Oosterhof N, Liang D, Thiruvalluvan A, et al. CD146 increases stemness and aggressiveness in glioblastoma and activates YAP signaling. Cell Mol Life Sci. 2022;79(8):398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vargas-Patron LA, Agudelo-Dueñas N, Madrid-Wolff J, Venegas JA, González JM, Forero-Shelton M, et al. Xenotransplantation of Human glioblastoma in Zebrafish larvae: in vivo imaging and proliferation assessment. Biol Open. 2019;8(5):bio043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Shan A, Zhou Z, Li W, Xie L, Du B, et al. LncRNA TCONS_00004099-derived microRNA regulates oncogenesis through PTPRF in gliomas. Ann Transl Med. 2021;9(12):1023–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vittori M, Breznik B, Hrovat K, Kenig S, Lah TT. RECQ1 helicase silencing decreases the tumour growth rate of U87 glioblastoma cell xenografts in zebrafish embryos. Genes (Basel). 2017;8(9):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nešović M, Divac Rankov A, Podolski-Renić A, Nikolić I, Tasić G, Mancini A, et al. Src inhibitors pyrazolo[3,4-d]pyrimidines, Si306 and Pro-Si306, inhibit focal adhesion kinase and suppress human glioblastoma invasion in vitro and in vivo. Cancers (Basel). 2020;12(6):1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wehmas LC, Tanguay RL, Punnoose A, Greenwood JA. Developing a novel embryo-larval zebrafish xenograft assay to prioritize human glioblastoma therapeutics. Zebrafish. 2016;13(4):317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pudelko L, Edwards S, Balan M, Nyqvist D, Al-Saadi J, Dittmer J, et al. An orthotopic glioblastoma animal model suitable for high-throughput screenings. Neuro Oncol. 2018;20(11):1475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article as supplementary file: Data file. R-scripts used in generation of figures are included as R-script files.