Abstract
Background
Glioblastoma (GBM) is a malignant astrocytic tumor and its progression involves the regulation of vascular endothelial growth factor-A (VEGFA). However, the mechanism of VEGFA in regulating GBM progression remains unclear.
Methods
VEGFA mRNA expression was analyzed by quantitative real-time polymerase chain reaction. Protein expression of VEGFA, cluster of differentiation 9 (CD9), CD81, and transforming growth factor-β1 (TGF-β1) was detected by western blotting assay. Flow cytometry assay was conducted to assess cell proliferation, cell apoptosis and myeloid-derived suppressor cell (MDSC) differentiation. TUNEL cell apoptosis detection kit was utilized to analyze cell apoptosis of tumors. Angiogenic capacity was investigated by tube formation assay. Transwell assay was used to assess cell migration and invasion. The effect of VEGFA on tumor formation was determined by a xenograft mouse model assay. Immunohistochemistry assay was used to analyze positive expression rate of VEGFA in tumor tissues. TGF-β1 level was detected by enzyme-linked immunosorbent assay.
Results
VEGFA expression was upregulated in GBM tissues, GBM cells, and exosomes from GBM patients and GBM cells. VEGFA silencing led to decreased cell proliferation, tube formation, migration and invasion and increased cell apoptosis. Moreover, VEGFA knockdown also delayed tumor formation. VEGFA promoted MDSC differentiation and TGF-β1 secretion by MDSCs by being packaged into exosomes. In addition, TGF-β1 knockdown displayed similar effects with VEGFA silencing on GBM cell phenotypes, and MDSCs attenuated VEGFA knockdown-induced effects by secreting TGF-β1 in A172 and U251 cells.
Conclusion
VEGFA contributed to tumor property of GBM cells by promoting MDSC differentiation and TGF-β1 secretion by MDSCs, providing potential targets for GBM treatment.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-024-12803-8.
Keywords: GBM, VEGFA, MDSC, TGF-β1, Exosomes
Highlights
VEGFA expression was upregulated in GBM tissues and cells;
VEGFA knockdown inhibited proliferation, tube formation, migration and invasion and induced apoptosis of A172 and U251 cells;
VEGFA promoted MDSC differentiation and TGF-β1 secretion by MDSCs by being packaged into exosomes;
MDSCs attenuated VEGFA knockdown-induced effects by producing TGF-β1 in A172 and U251 cells.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-024-12803-8.
Introduction
Glioblastoma, also known as glioblastoma multiforme (GBM), is an aggressive type of brain tumor that originates from the brain’s glial cells [1]. It tends to occur more frequently in older individuals, with a median age of diagnosis around 64 years [2]. The exact cause of glioblastoma is not yet fully understood. Some risk factors that have been identified include exposure to ionizing radiation and certain hereditary conditions such as neurofibromatosis type 1 and Li-Fraumeni syndrome [3, 4]. However, it is important to note that most cases of GBM occur sporadically without any known risk factors. The symptoms of glioblastoma can vary depending on the location and size of the tumor. Common signs and symptoms may include headaches, seizures, cognitive changes, papilledema, personality changes, and motor deficits [5]. Due to its aggressive nature, GBM tends to progress rapidly and can infiltrate surrounding brain tissue, making complete surgical removal challenging. Treatment for GBM often involves a combination of surgery, radiation therapy, and chemotherapy, and research into potential new treatments and therapies for GBM is ongoing [6, 7]. Although treatment options are available, it remains a complex disease with a poor prognosis. Continued research and advancements in medical knowledge are crucial in improving outcomes for patients with GBM.
Peripheral blood mononuclear cells (PBMCs) are a type of immune cells in blood, which can participate in immune reactions and produce various immune factors. In certain diseases such as cancer, infection, and inflammation, PBMCs may exhibit functional abnormalities, leading to inflammation and the release of inflammatory factors and chemokines [8, 9]. These factors can further promote the differentiation, proliferation, and functional activation of myeloid-derived suppressor cells (MDSCs), resulting in enhanced immunosuppressive effects [10]. MDSCs are a type of immune cells that play a crucial role in tumor development and progression [11]. These cells are characterized by their ability to suppress the immune response against tumors, thereby promoting tumor growth and survival. MDSCs are composed of two major subsets, granulocytic MDSCs (gMDSCs) and monocytic MDSCs (mMDSCs) [12]. In particular, MDSCs have been found to cause reversible T cell dysfunction [13], enabling the tumors to grow and spread. Furthermore, the interaction between MDSCs and tumor cells can enhance the tumor’s resistance to chemotherapy and radiation therapy [14]. Exosomes are small vesicles released by cells that contain a variety of biological molecules, including proteins, RNA, and DNA [15, 16]. They have been increasingly recognized as important players in the development and progression of GBM. These exosomes can be detected in the blood of GBM patients [17], making them potential biomarkers for diagnosis and prognosis. Additionally, exosomes can transfer cancer effectors to GBM cells, promoting tumorigenesis and metastasis [18].
Vascular endothelial growth factor-A (VEGFA) is a highly specific factor that regulates vascular growth and vascular permeability, playing a crucial role in angiogenesis, the process of forming new blood vessels [19]. The VEGFA gene is located on the short arm of chromosome 6 in humans, and the protein is expressed in various cells including endothelial cells. It is widely recognized as a potent pro-angiogenic factor that is highly expressed in various human tumor tissues and has significant implications in tumor metastasis and disease progression [20, 21]. Bevacizumab antagonizes VEGFA, and Food and Drug Administration has approved bevacizumab for recurrent GBM [22]. Previous studies have revealed that VEGFA contributes to GBM progression [23, 24], however, how VEGFA regulates GBM remains unclear. Further research and clinical trials regarding the mechanism of GBM progression will continue to refine our understanding of the role of VEGFA in GBM and potentially lead to more effective treatments for this devastating disease.
Based on the above evidence, we hypothesized that VEGFA contributed to GBM development by inducing MDSC differentiation by being packaged into exosomes, and the hypothesis was verified by a series of experiments with the hope of seeking attractive targets for novel therapeutic interventions.
Materials and methods
Patient samples
Consented patients who underwent surgical treatment at No. 215 Hospital of Shaanxi Nuclear Industry provided a total of 64 brain tissues, including GBM tissues and normal brain tissues, as well as serum. GBM patients not underwent chemotherapy or radiotherapy and diagnosed blindly by senior pathologists. Control patients had no any congenital diseases and hospitalized owing to car accidents-caused craniocerebral trauma. Tissue collection was approved from the Ethics Committee of No. 215 Hospital of Shaanxi Nuclear Industry. The collected tissues were kept at -80℃, while serum was stored at 4℃. Participants signed the written informed consent.
Exosome isolation, identification and treatment
Total exosome isolation kits (#4478360 and #4478359; Invitrogen) were used to isolate exosomes from human serum and cells according to supplier’s instructions. Serum and cell supernatant were centrifuged at 2000 × g, and obtained supernatant was re-suspended in Reagent. Samples were centrifuged after incubation at 4 °C. Then, supernatant was removed, and 1 × phosphate buffered saline (PBS; Yuanye Bio-technology, Shanghai, China) was added to dissolve the pellet (exosomes). Subsequently, exosomes were observed under a JEM-1010 transmission electron microscope (TEM; JEOL, Tokyo, Japan) to analyze their morphology as previously reported [25]. Nanoparticle tracking analysis (NTA) was performed to measure the distribution of exosome size. Additionally, protein markers of exosomes, CD9 and CD81, were detected via western blotting assay.
Cell culture and treatment
GBM cells including A172 and U251, human astrocytes (NHA) and human umbilical vein endothelial cells (HUVECs) were provided by Sbjbio@ life Science (Nanjing, China) and cultured in DMEM. Jining Shiye Biotech (Shanghai, China) provided human peripheral blood mononuclear cells (PBMCs), which were isolated from peripheral blood and maintained in complete culture medium of PBMCs. During cell culture at 37℃with 5% CO2, medium was supplemented with 10% fetal bovine serum (Sbjbio@ life Science). PBMCs were also incubated with PBS (Yuanye Bio-technology) or exosomes isolated from A172 and U251 cell supernatants expressing si-VEGFA or si-NC.
In addition, exosomes were treated with PKH67 linker (Umibio, Shanghai, China) for green fluorescent cell labeling and then mixed using a vortex oscillator, followed by incubation with PBS. Exosomes were isolated according to the method described above. PBMCs were incubated with the isolated exosomes and observed under an IX71 fluorescence microscope (Olympus, Tokyo, Japan) to analyze whether the exosomes were taken up by PBMCs.
Cell transfection
Small interfering RNA (siRNA) and small hairpin RNA (shRNA) of VEGFA (Accession: NM_001025366.3) as well as siRNA of TGF-β1 (Accession: NM_000660.7) were designed through the online designer https://rnaidesigner.thermofisher.com/rnaiexpress/. Transfection of siRNA and shRNA was conducted using InvitroRNA™ reagent (#IVG1101-10; Yingweiwo Biotechnology, Shanghai, China).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Brain tissues were grinded using pestle and mortar and diluted PBS (Yuanye Bio-technology). Then, these tissues, cells and exosomes were placed into tubes and mixed with Trizol reagent (TaKaRa, Dalian, China). The tubes were centrifuged after 30 s of oscillating on the vortex shaker. The supernatants were collected and then RNA was isolated using chloroform (Sigma, St. Louis, MO, USA), isopropyl (Sigma), and ethyl alcohol (Sigma). cDNA synthesis is performed on an ice box using High Fidelity PCR Master Mix (Sangon Biotech, Shanghai, China). Subsequently, cDNA was used for quantification analysis according to the guidebook of TaqMan Fast qPCR mixture (Sangon Biotech). IQ5 thermocycler (Bio-Rad, Hercules, CA, USA) was used for quantification reaction. Primer sequences are listed in Table 1.
Table 1.
Primer sequences used for RT-PCR.
| Name | Primers for RT-PCR (5’-3’) | |
|---|---|---|
| VEGFA | Forward | AAGGGGCAAAAACGAAAGCG |
| Reverse | GCTCCAGGGCATTAGACAGC | |
| TGFβ1 | Forward | TGATGTCACCGGAGTTGTGC |
| Reverse | GTGAACCCGTTGATGTCCACT | |
| GAPDH | Forward | ATCACTGCCACCCAGAAGAC |
| Reverse | CCGTTCAGCTCAGGGATGAC |
Western blotting assay
Part of the brain tissues were removed and cut into small pieces. After grinding, these tissues were treated with an appropriate amount of PBS (Yuanye Bio-technology). The supernatant, cells and isolated exosomes were transferred to a clean tube and then treated with 1 mL of lysis buffer and 10 µL of phenylmethylsulfonyl fluorid (Sigma). The sample was placed on ice prepared in advance and centrifuged at 12,000 r/min for 10 min to collect the supernatant. The required sample volume was calculated and protein bands were separated using separating gels and stacking gels (Thermo Fisher, Waltham, MA, USA). The target band region was found according to the protein Marker, and the gels were cut. The membranes were transferred with the prepared transfer buffer and incubated with 5% skimmed milk solution at room temperature on an incubation shaker for 2 h, followed by incubation with primary antibodies, including VEGFA (#AF5131; 1:1000; Affinity, Nanjing, China), CD9 (#AF5139; 1:1000; Affinity), CD81 (#AF5131; 1:1000; Affinity), TGF-β1 (#AF1027; 1:1000; Affinity), and β-actin (#AF7018; 1:1000; Affinity), and secondary antibody (1:5000; Affinity). An appropriate amount of BeyoECL Plus working solution was added to the membrane according to the size of the membrane and protein bands (Beyotime, Shanghai, China) were visualized with a chemical luminescence imaging instrument (Gel Doc™ XR+, Bio-Rad).
Flow cytometry analysis
The assay used to analyze cell cycle distribution was conducted on A172 and U251 cells in the logarithmic growth phase. GBM cells with various treatments were harvested and fixed using ethanol, followed by incubation with RNase A and propidium iodide (PI) in accordance with the guidebook of the DNA content quantitation kit (Solarbio, Beijing, China). At last, an AttuneTM NxT flow cytometer (Thermo Fisher) was used to analyze the stained cells.
Tube formation assay
The tube formation assay is a widely used technique to study angiogenesis, the process by which new blood vessels are formed. Growth factor-reduced Matrigel (Abwbio, Shanghai, China) was prepared according to the manufacturer’s instructions in advance. A suitable volume of HUVEC suspension was added to culture plates, ensuring an even distribution of cells. After cell culture at 37℃ with 5% CO2, cell treatments were performed and cell medium was replaced using GBM-conditioned medium. Cells were cultured for 16 h, and the formation of tube-like structures (tubule length and number of branching points) was assessed using an IX71 microscope (Olympus) and Image J software (NIH, Bethesda, MD, USA).
Transwell assays
The Transwell inserts were rinsed using PBS (Yuanye Bio-technology) to wet the membrane surface. GBM cells were counted using an automated cell counter and their concentration was adjusted to the desired density in culture medium. The upper chambers were used to culture cells. The lower chambers were filled with culture medium added with 10% fetal bovine serum (Sbjbio@ life Science). The cells were allowed to attach to the membrane for 24 h, and the inserts were carefully removed from the plate at the end of the incubation period. The cells were fixed using 4% paraformaldehyde and stained using 0.1% crystal violet. Finally, migrated cells were analyzed under an IX71 microscope (Olympus). The cell invasion was performed according to the above method with the transwell inserts coated with Matrigel (Abwbio).
Xenograft mouse model assay
Hunan Slyke Jingda Experimental Animal Co., LTD (Changsha, China) provided male BALB/c nude mice (N = 12, 5 weeks of age), which were housed under pathogen-free conditions. A172 cells stably expressing sh-VEGFA or sh-NC were adjusted to 5 × 106 cells/0.2 mL PBS and then subcutaneously injected into nude mouse flanks. The mice were monitored regularly for tumor growth. All mice were sacrificed using pentobarbital sodium (40 mg/kg) when the tumor reaches a certain size. The formed tumors were harvested for further analysis. The Animal Care and Use Committee of No. 215 Hospital of Shaanxi Nuclear Industry approved the study.
Immunohistochemistry (IHC) assay
The formed tumors were fixed using appropriate fixatives, such as formalin, and then processed into paraffin blocks by dehydration, clearing, and embedding. The tissue sections were cut from the paraffin blocks at appropriate thicknesses using a microtome and deparaffinized in xylene, followed by rehydration through a graded series of alcohol. Antigens were retried by subjecting the tissue sections to appropriate antigen retrieval methods, and non-specific binding sites were blocked using a blocking solution. The tissue sections were incubated with primary antibody against VEGFA (#AF5131; 1:100; Affinity), and the immunohistochemical staining was developed. The tissue sections were counterstained to visualize the tissue morphology and mounted using coverslips. The stained sections were analyzed using a TCS SP8 confocal microscopy (Leica Microsystems, Mannheim, Germany).
TUNEL assay
Cell apoptosis of tumor tissues was analyzed using the TUNEL Apoptosis Detection Kit (Yeasen Biotech, Shanghai, China). The tumor tissues were cut into small tumor sections and immersed into xylene and ethyl alcohol. The tumor tissues were treated with Proteinase K for 20 min and Equilibration Buffer for 30 min, followed by incubation with TdT buffer. The slide was immersed in a dyeing tank containing PI solution for 5 min. Samples were immediately analyzed under an IX71 fluorescence microscope (Olympus).
Statistical analysis
Statistical analysis was performed using GraphPad Prism, and results were shown as means ± standard deviations (SD). Significant differences were compared using Wilcoxon signed-rank test, Student’s t-tests or analysis of variance. P < 0.05 indicated statistical significance.
Results
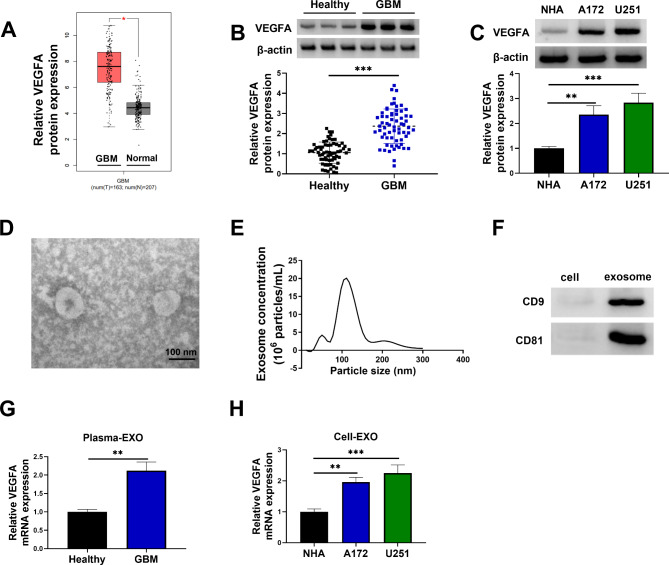
VEGFA expression was upregulated in GBM tissues and cells
The Gene Expression Profiling Interactive Analysis (GEPIA) online dataset was used to analyze VEGFA expression in GBM tissues and normal brain tissues. The results showed that VEGFA expression was upregulated in GBM tissues compared with normal tissues (Fig. 1A). Subsequently, the data revealed that VEGFA protein expression was upregulated in clinical GBM tissues when compared with healthy brain tissues (Fig. 1B). Moreover, GBM cell lines (A172 and U251) showed high VEGFA protein expression in comparison with human astrocytes (NHA), as shown in Fig. 1C. The study then isolated exosomes from serum of GBM patients, and the isolated vesicles were identified through TEM and NTA. As shown in Fig. 1D, the isolated vesicles were approximately 100 nm in diameter and had similar morphology with exosomes. NTA assay showed that the diameter of the most particles was approximately 110 nm (Fig. 1E). The study also found that exosomes marker proteins CD9 and CD81 were expressed in the isolated vesicles (Fig. 1F). qRT-PCR analysis showed that VEGFA expression was upregulated in the isolated exosomes from GBM patients when compared with the isolated exosomes from healthy volunteers (Fig. 1G). Comparatively, VEGFA mRNA expression was significantly increased in the exosomes from A172 and U251 cell supernatants in comparison with exosomes from NHA cell supernatant (Fig. 1H). These data demonstrated that VEGFA expression was upregulated in GBM tissues and cells.
Fig. 1.
VEGFA expression was upregulated in GBM tissues and cells. (A) The GEPIA online dataset was used to analyze VEGFA expression in GBM tissues and normal brain tissues. (B) VEGFA protein expression was analyzed by western blotting assay in GBM tissues (N = 64) and normal brain tissues (N = 64). (C) Western blotting assay was used to determine VEGFA protein expression in NHA, A172 and U251 cells. (D-F) TEM, NTA and western blotting assay were used to identify the isolated vesicles. (G and H) qRT-PCR was used to detect VEGFA mRNA expression in the isolated exosomes from serum of GBM patients and normal volunteers and supernatant of NHA, A172 and U251 cells. *P < 0.05, **P < 0.01 and ***P < 0.001
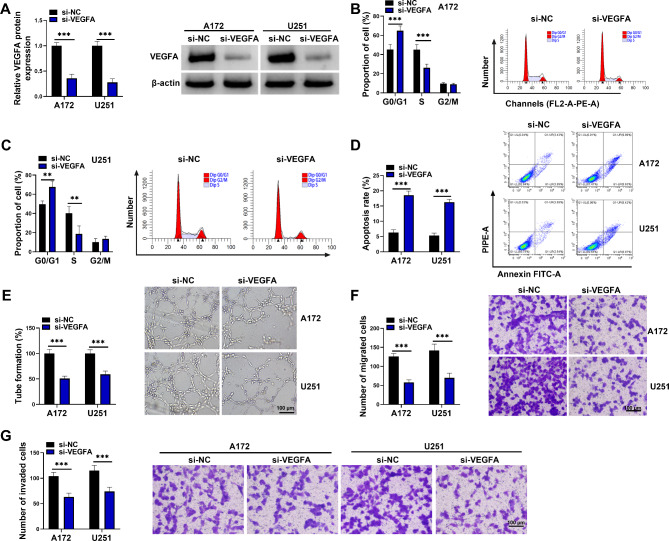
VEGFA knockdown inhibited proliferation, tube formation, migration and invasion and induced apoptosis of A172 and U251 cells
The study then analyzed the effects of VEGFA silencing on A172 and U251 cell phenotypes, including cell proliferation, tube formation, migration, invasion and apoptosis. The data first showed low VEGFA protein expression after transfection with siRNA of VEGFA (Fig. 2A), indicating the successful knockdown of VEGFA in A172 and U251 cells. Subsequently, the results showed that VEGFA silencing increased the proportion of G0/G1 phase cells and decreased the proportion of S phase cells (Fig. 2B and C), suggesting the inhibitory effect of VEGFA silencing on cell proliferation. As shown in Fig. 2D and E, VEGFA absence induced cell apoptosis and repressed tube formation. Comparatively, the migration and invasion were inhibited after VEGFA knockdown (Fig. 2F and G). The above data demonstrated that VEGFA knockdown inhibited tumor property of GBM cells.
Fig. 2.
VEGFA knockdown inhibited proliferation, tube formation, migration and invasion and induced apoptosis of A172 and U251 cells. A172 and U251 cells were divided into si-NC group and si-VEGFA group. (A) VEGFA protein expression was analyzed by western blotting assay in A172 and U251 cells. (B-D) Flow cytometry assay was used to assess cell cycle and apoptosis of A172 and U251 cells. (E) Tube formation assay was performed to assess the effect of A172 and U251 cells on angiogenic capacity of HUVECs. (F and G) Transwell assays were used to assess the migration and invasion of A172 and U251 cells. **P < 0.01 and ***P < 0.001
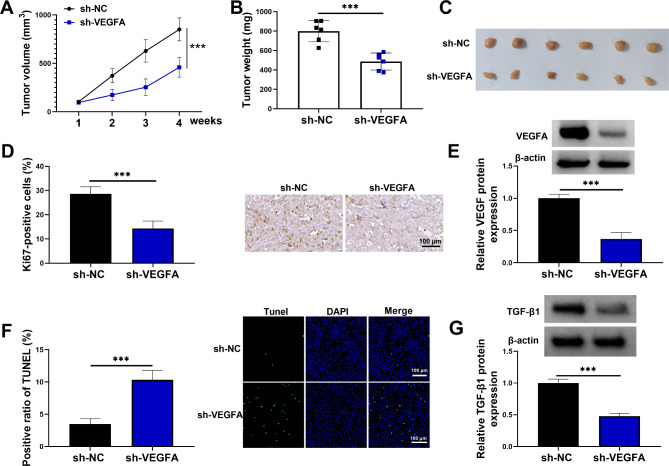
VEGFA silencing delayed tumor formation in vivo
The study continued to analyze the effect of VEGFA silencing on tumor formation in vivo. The results showed that the tumors formed from VEGFA-deficient A172 cells grew more slowly and had lower weight than those derived from A172 cells transfected with sh-NC (Fig. 3A-C). Subsequently, the data revealed that the positive expression rate of Ki67 and the protein expression of VEGFA was lower in the formed tumors from the sh-VEGFA group than those from the sh-NC group (Fig. 3D and E). Comparatively, cell apoptosis of the formed tumors from the sh-VEGFA group was induced when compared with those from the sh-NC group (Fig. 3F). Further, the results showed that TGF-β1 protein expression was downregulated in the formed tumors from the sh-VEGFA group than those from the sh-NC group (Fig. 3G). These data demonstrated that VEGFA knockdown inhibited tumor property of A172 cells.
Fig. 3.
VEGFA silencing delayed tumor formation in vivo. A172 cells stably expressing sh-VEGFA or sh-NC were injected into the left flank of the mice, and the formed tumors were harvested after 4 weeks. (A) The volume of the formed tumors was measured every 7 days for 4 cycles. (B) The effect of VEGFA knockdown on tumor weight. (C) The tumor images in the sh-VEGFA group and sh-NC group. (D) IHC assay was used to analyze Ki67 protein expression in the formed tumors in the sh-VEGFA group and sh-NC group. (E) Western blotting assay was used to detect VEGFA protein expression in the formed tumors from the sh-VEGFA group and sh-NC group. (F) TUNEL cell apoptosis detection kit was used to analyze cell apoptosis in the formed tumors from the sh-VEGFA group and sh-NC group. (G) TGF-β1 protein expression was detected by western blotting assay in the resulting tumors from A172 cells transfected with sh-NC or sh-VEGFA. ***P < 0.001
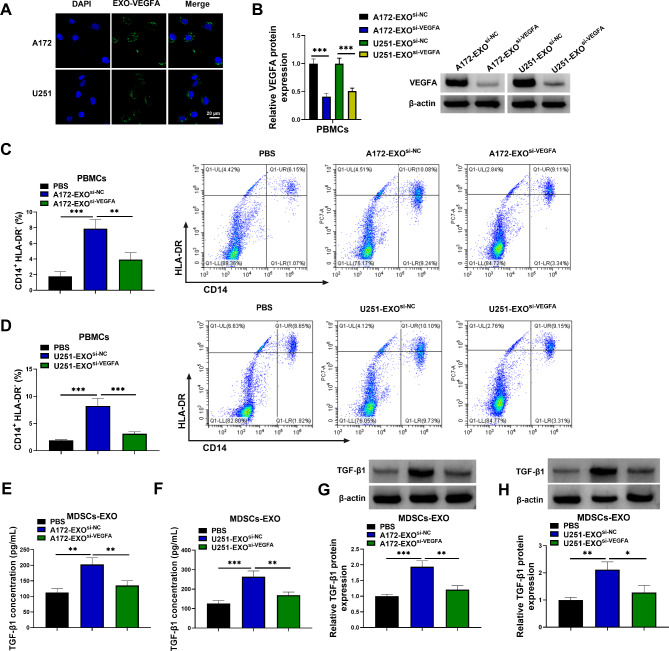
VEGFA promoted MDSC differentiation and TGF-β1 secretion by MDSCs by being packaged into exosomes
The study then incubated PBMCs with exosomes isolated from A172 and U251 cell supernatants expressing si-VEGFA or si-NC and then investigated the consequent effects on MDSC differentiation and TGF-β1 secretion. As shown in Fig. 4A, the exosomes were taken up by PBMCs, as the green fluorescent dye was observed in the PBMCs. Moreover, the result showed that VEGFA protein expression was lower in the PBMCs incubated with exosomes isolated from A172 and U251 cell supernatants expressing si-VEGFA than in those isolated from A172 and U251 cell supernatants expressing si-NC (Fig. 4B). These data demonstrated that VEGFA contained in the exosomes was taken up by PBMCs. Subsequently, exosomes from A172 and U251 cell supernatants significantly promoted differentiation of PBMCs into CD14+HLA-DR−-MDSCs compared to control group, but the incubation with exosomes from A172 and U251 cell supernatants expressing si-VEGFA inhibited the differentiation (Fig. 4C and D). Further, the data showed that exosomes from A172 and U251 cells promoted TGF-β1 secretion by MDSCs, but the incubation with exosomes from A172 and U251 cell supernatants expressing si-VEGFA inhibited TGF-β1 production, as revealed by ELISAs and western blotting assay (Fig. 4E-H). The above results suggested that VEGFA contributed to MDSC differentiation and TGF-β1 secretion by MDSCs by being packaged into exosomes.
Fig. 4.
VEGFA contributed to MDSC differentiation and TGF-β1 secretion by MDSCs by being packaged into exosomes. (A) Exosomes were treated with PKH67 linker for green fluorescent cell labeling and then mixed. Exosomes were isolated, and PBMCs were incubated with the isolated exosomes and observed under a fluorescence microscope. (B) VEGFA protein expression was analyzed by western blotting assay in PBMCs incubated with exosomes isolated from A172 and U251 cells expressing si-VEGFA or si-NC. (C and D) Flow cytometry was used to analyze CD14+HLA-DR−MDSCs. (E-H) The exosomes isolated from both A172 cells and U251 cells expressing si-VEGFA or si-NC were incubated with MDSCs, and TGF-β1 level was assessed by ELISAs and Western blotting assay in the exosomes from MDSCs. *P < 0.05, **P < 0.01 and ***P < 0.001
TGF-β1 knockdown inhibited proliferation, tube formation, migration and invasion and induced apoptosis of A172 and U251 cells
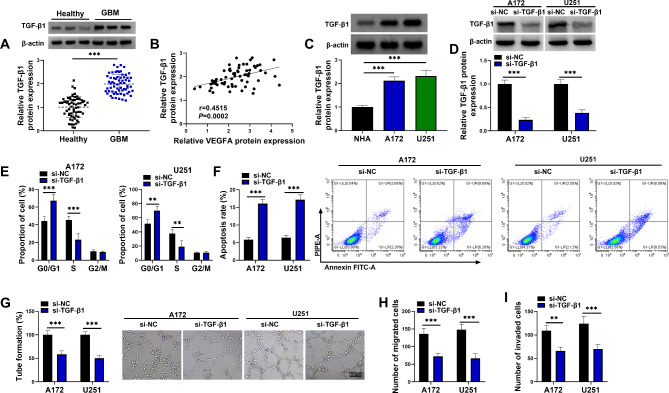
The results showed that TGF-β1 protein expression was upregulated and was positively correlated with VEGFA expression in GBM tissues (Fig. 5A and B). Moreover, its protein expression was higher in both A172 and U251 cells than in NHA cells (Fig. 5C). The study also analyzed the effects of TGF-β1 silencing on tumor property of A172 and U251 cells. The data showed low TGF-β1 protein expression after transfection with siRNA of TGF-β1 (Fig. 5D). Subsequently, the results showed that TGF-β1 silencing increased the proportion of G0/G1 phase cells and decreased the proportion of S phase cells (Fig. 5E), suggesting the inhibitory effect of TGF-β1 silencing on cell proliferation. As shown in Fig. 5F and G, TGF-β1 knockdown induced cell apoptosis and repressed tube formation. Comparatively, the migration and invasion were inhibited after TGF-β1 knockdown (Fig. 5H and I). The above data demonstrated that TGF-β1 knockdown inhibited tumor property of GBM cells.
Fig. 5.
TGF-β1 knockdown inhibited proliferation, tube formation, migration and invasion and induced apoptosis of A172 and U251 cells. (A) TGF-β1 protein expression was analyzed by western blotting assay in GBM tissues (N = 64) and normal brain tissues (N = 64). (B) Spearman correlation analysis was performed for correlation identification of TGF-β1 protein expression and VEGFA protein expression in GBM tissues. (C) TGF-β1 protein expression was detected by western blotting assay in NHA cells, A172 cells and U251 cells. (D-I) A172 and U251 cells were divided into si-NC group and si-TGF-β1 group. (D) TGF-β1 protein expression was analyzed by western blotting assay in A172 and U251 cells. (E and F) Flow cytometry assay was used to assess cell cycle and apoptosis of A172 and U251 cells. (G) Tube formation assay was performed to assess the effect of A172 and U251 cells on angiogenic capacity of HUVECs. (H and I) Transwell assays were used to assess the migration and invasion of A172 and U251 cells. **P < 0.01 and ***P < 0.001
MDSCs attenuated the effects of VEGFA knockdown in A172 and U251 cells by producing TGF-β1
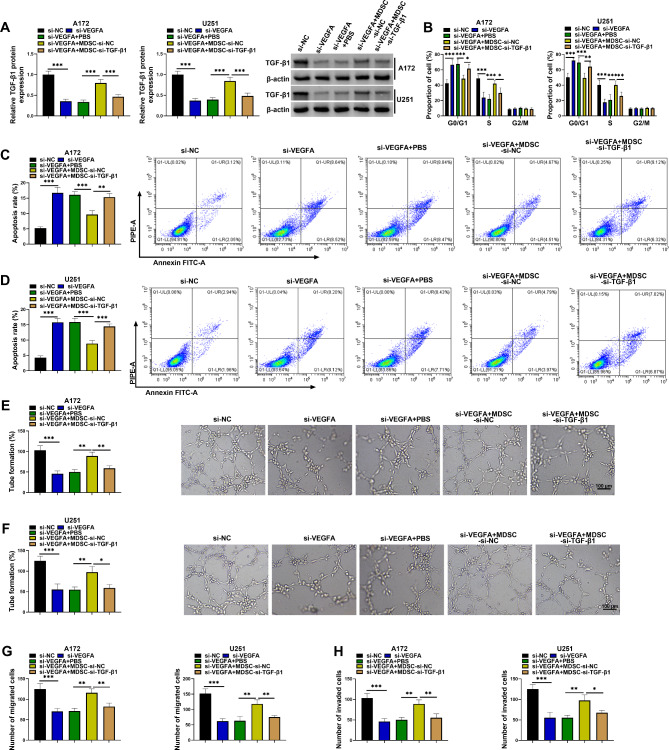
The study further analyzed the effects of MDSCs with low TGF-β1 expression on A172 and U251 cell phenotypes. To achieve this, both U251 and A172 cells were transfected with si-NC, si-VEGFA, and/or cocultured with MDSCs transfected with si-TGF-β1 or si-NC. The data first showed that MDSCs attenuated the inhibitory effect of VEGFA knockdown on TGF-β1 protein expression, whereas the effect was rescued by TGF-β1-deficient MDSCs (Fig. 6A). Subsequently, the results showed that VEGFA silencing-induced inhibitory effect on cell proliferation and promoting effect on cell apoptosis were relieved by MDSCs, however, these effects were rescued by TGF-β1-deficient MDSCs (Fig. 6B-D). Comparatively, MDSCs treatment also relieved VEGFA absence-induced effects on tube formation and cell migration and invasion, but these effects were counteracted by MDSCs lowly expressing TGF-β1 (Fig. 6E-H). These data revealed that MDSCs rescued VEGFA knockdown-induced effects by increasing TGF-β1 expression in A172 and U251 cells.
Fig. 6.
MDSCs attenuated VEGFA knockdown-induced effects by producing TGF-β1 in A172 and U251 cells. A172 and U251 cells were divided into si-NC group, si-VEGFA group, si-VEGFA + PBS group, si-VEGFA + MDSC-si-NC group and si-VEGFA + MDSC-TGF-β1 group. (A) TGF-β1 protein expression was analyzed by western blotting assay in A172 and U251 cells. (B-D) Flow cytometry assay was used to assess cell cycle and apoptosis of A172 and U251 cells. (E and F) Tube formation assay was performed to assess the effect of A172 and U251 cells on angiogenic capacity of HUVECs. (G and H) Transwell assays were used to assess the migration and invasion of A172 and U251 cells. *P < 0.05, **P < 0.01 and ***P < 0.001
Discussion
Vascular endothelial growth factor (VEGF) plays a crucial role in promoting angiogenesis, the formation of new blood vessels, which is essential for tumor growth and metastasis. VEGF is a key molecule involved in the development and progression of GBM, a highly aggressive type of brain cancer, primarily through its effects on angiogenesis, invasion, and tumorigenesis. Targeting VEGF signaling pathways has become an important strategy in the treatment and management of GBM, as it aims to inhibit tumor growth and reduce the risk of recurrence [26]. The further investigation related to the role of VEGFA in the development of GBM can provide a basis for the development of new therapies that can help improve treatment outcomes and survival in patients with GBM. The present work analyzed the role of VEGFA in tumor property of GBM cells and its working mechanism. The results showed that VEGFA promoted GBM cell malignancy by promoting MDSC differentiation and TGF-β1 secretion by MDSCs.
VEGFA exhibited a high expression in various cancers and promoted angiogenesis, proliferation and invasion of tumor cells [27]. In GBM, single nucleotide polymorphism rs2010963 of VEGFA increased risk of vascular events [28]. VEGFA induced angiogenesis and contributed to lower grade glioma to progress into GBM [29]. Another paper revealed that macrophage-derived VEGFA participated in the regulation of α-lactose to glioma angiogenesis [30]. Moreover, VEGFA expression in serum was associated with response to bevacizumab treatment, and GBM patients with high VEGFA expression had positive response to anti-angiogenic therapy [31]. Our data showed that VEGFA was highly expressed in GBM tissues. In addition, GBM cells had a high VEGFA expression. VEGFA silencing inhibited GBM cell proliferation, tube formation and tumor formation. Meanwhile, loss-of-function assays revealed that VEGFA knockdown induced cell apoptosis and repressed cell motility. Exosomes are small membrane-bound structures that are released by cells into the extracellular space. These vesicles contain various biomolecules, including proteins, nucleic acids (such as DNA and RNA), lipids, and metabolites. They can transfer their cargo to neighboring or distant cells, thereby influencing cell behavior and function [15]. This transfer of information can occur in both healthy and diseased states. Our results showed that VEGFA expression was upregulated in exosomes isolated from GBM patients and GBM cells, and VEGFA promoted the differentiation of PBMCs to MDSCs. Further, the exosomes with low VEGFA expression was taken up by PBMCs. These results demonstrated that VEGFA might function by promoting MDSCs differentiation.
TGF-β1 is a cytokine that plays a crucial role in tumor development and progression. It is a multifunctional molecule involved in various cellular processes, including cell growth, apoptosis, differentiation, and immune regulation [32]. In cancer, TGF-β1 has been found to have a dual role. Initially, it acts as a tumor suppressor by inhibiting cell proliferation and inducing apoptosis in early stages of tumor development [33]. However, as the tumor progresses, TGF-β1 can switch its function and promote tumor growth and metastasis [34]. Our data showed that PBMCs incubated with exosomes from GBM cells could produce more TGF-β1, while PBMCs incubated with exosomes from GBM cells with low VEGFA expression could produce less TGF-β1. Functional assays revealed the inhibitory effect of TGF-β1 silencing on tumor property of GBM cells. Further, MDSCs attenuated VEGFA knockdown-induced effects by increasing TGF-β1 expression in A172 and U251 cells. One way by which TGF-β1 promotes tumor development is through its ability to induce immune evasion or immune escape. The immune system has mechanisms to recognize and eliminate cancer cells, but tumors can develop strategies to evade immune detection and destruction. TGF-β1 plays a key role in this process by suppressing the immune response against the tumor [35]. It stimulates the production of growth factors that promote cell proliferation, leading to uncontrolled cell growth [36]. Additionally, TGF-β1 promotes the epithelial-to-mesenchymal transition (EMT), a process that allows cancer cells to acquire migratory and invasive properties, further enhancing tumor progression [37]. These data suggest that VEGFA regulates GBM cell malignancy by regulating TGF-β1 in MDSCs.
Taken together, VEGFA was secreted in the form of exosomes outside the GBM cells, which was subsequently taken up by PBMCs to induce differentiation of MDSCs and TGF-β1 secretion by MDSCs, thereby regulating the tumor properties of GBM cells. However, both in vivo and in vitro assays do not fully replicate the tumor microenvironment necessary for tumor growth, and an orthotopic murine model, which involves the intracranial injection of GBM cells, should be established to validate these results more effectively. Our results suggest that the tumor properties of GBM cells may be inhibited by interfering with the expression of VEGFA or its receptor, or by adjusting exosomes secretion and uptake, which may provide a new strategy for the treatment of GBM.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
Shangjun Chen designed and performed the research; Yufeng Ren, Xiao Gao and Xuechao Yang analyzed the data; Yanlong Tian wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by Xianyang key research and development Project [approval number: S2022-ZDYF-SF-1283] and No. 215 Hospital of Shaanxi Nuclear Industry scientific research project [approval number: 215KYJJ-202209].
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The present study was approved by the ethical review committee of the No. 215 Hospital of Shaanxi Nuclear Industry. Written informed consent was obtained from all enrolled patients.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Le Rhun E, Devos P, Houillier C, Cartalat S, Chinot O, Di Stefano AL, Lepage C, Reyns N, Dubois F, Weller M. Romiplostim for temozolomide-induced thrombocytopenia in glioblastoma: the PLATUM trial. Neurology. 2019;93(19):e1799–806. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: primary brain and other Central Nervous System tumors diagnosed in the United States in 2009–2013. Neurooncology. 2016;18(suppl5):v1–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher JL, Schwartzbaum JA, Wrensch M, Wiemels JL. Epidemiology of brain tumors. Neurol Clin. 2007;25(4):867–90. vii. 10.1016/j.ncl.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 4.Wrensch M, Lee M, Miike R, Newman B, Barger G, Davis R, Wiencke J, Neuhaus J. Familial and personal medical history of cancer and nervous system conditions among adults with glioma and controls. Am J Epidemiol. 1997;145(7):581–93. 10.1093/oxfordjournals.aje.a009154 [DOI] [PubMed] [Google Scholar]
- 5.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–50. 10.1001/jama.2013.280319 [DOI] [PubMed] [Google Scholar]
- 6.Rong L, Li N, Zhang Z. Emerging therapies for glioblastoma: current state and future directions. J Experimental Clin cancer Research: CR. 2022;41(1):142. 10.1186/s13046-022-02349-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang B, Wang X, Dong D, Pan Y, Wu J, Liu J. Existing Drug Repurposing for Glioblastoma to Discover candidate drugs as a New a Approach. Lett Drug Des Discovery. 2022;19(1):31–43. 10.2174/1570180818666210509141735 [DOI] [Google Scholar]
- 8.Bhardwaj RG, Al-Khabbaz A, Karched M. Cytokine induction of peripheral blood mononuclear cells by biofilms and biofilm supernatants of Granulicatella and Abiotrophia spp. Microb Pathog. 2018;114:90–4. 10.1016/j.micpath.2017.11.037 [DOI] [PubMed] [Google Scholar]
- 9.Zare Moayedi M, Ahmmadpour F, Rashidi M, Ahmadzadeh A, Salmasi A, Mohammadzadeh G. The association between mRNA expression of resistin, TNF- α, IL-6, IL-8, and ER-α in peripheral blood mononuclear cells and breast cancer. Turk J Med Sci. 2021;51(3):1345–53. 10.3906/sag-2008-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mace TA, Ameen Z, Collins A, Wojcik S, Mair M, Young GS, Fuchs JR, Eubank TD, Frankel WL, Bekaii-Saab T, et al. Pancreatic cancer-associated stellate cells promote differentiation of myeloid-derived suppressor cells in a STAT3-dependent manner. Cancer Res. 2013;73(10):3007–18. 10.1158/0008-5472.CAN-12-4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groth C, Hu X, Weber R, Fleming V, Altevogt P, Utikal J, Umansky V. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br J Cancer. 2019;120(1):16–25. 10.1038/s41416-018-0333-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Wang X, Zhang R, Wang X, Fu H, Yang W. MDSCs interactions with other immune cells and their role in maternal-fetal tolerance. Int Rev Immunol. 2022;41(5):534–51. 10.1080/08830185.2021.1938566 [DOI] [PubMed] [Google Scholar]
- 13.Raychaudhuri B, Rayman P, Huang P, Grabowski M, Hambardzumyan D, Finke JH, Vogelbaum MA. Myeloid derived suppressor cell infiltration of murine and human gliomas is associated with reduction of tumor infiltrating lymphocytes. J Neurooncol. 2015;122(2):293–301. 10.1007/s11060-015-1720-6 [DOI] [PubMed] [Google Scholar]
- 14.Lakshmanachetty S, Cruz-Cruz J, Hoffmeyer E, Cole AP, Mitra SS. New insights into the multifaceted role of myeloid-derived suppressor cells (MDSCs) in high-Grade gliomas: from metabolic reprograming, immunosuppression, and therapeutic resistance to current strategies for Targeting MDSCs. Cells 2021, 10(4). [DOI] [PMC free article] [PubMed]
- 15.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science (New York, NY) 2020, 367(6478). [DOI] [PMC free article] [PubMed]
- 16.Dong Y, Zhang L, Chang H. Regulation of exosomes-mediated circNR4A1 on Chemoresistance and Biological effects of oral squamous cell carcinoma cells. Lett Drug Des Discovery. 2023;20(7):921–9. 10.2174/1570180819666220610140616 [DOI] [Google Scholar]
- 17.Puigdelloses M, González-Huárriz M, García-Moure M, Martínez-Vélez N, Esparragosa Vázquez I, Bruna J, Zandio B, Agirre A, Marigil M, Petrirena G, et al. RNU6-1 in circulating exosomes differentiates GBM from non-neoplastic brain lesions and PCNSL but not from brain metastases. Neuro-oncology Adv. 2020;2(1):vdaa010. 10.1093/noajnl/vdaa010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kros JM, Mustafa DM, Dekker LJ, Sillevis Smitt PA, Luider TM, Zheng PP. Circulating glioma biomarkers. Neurooncology. 2015;17(3):343–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273(2):114–27. 10.1111/joim.12019 [DOI] [PubMed] [Google Scholar]
- 20.Andreozzi M, Quagliata L, Gsponer JR, Ruiz C, Vuaroqueaux V, Eppenberger-Castori S, Tornillo L, Terracciano LM. VEGFA gene locus analysis across 80 human tumour types reveals gene amplification in several neoplastic entities. Angiogenesis. 2014;17(3):519–27. 10.1007/s10456-013-9396-z [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto K, Ema M. Roles of VEGF-A signalling in development, regeneration, and tumours. J BioChem. 2014;156(1):1–10. 10.1093/jb/mvu031 [DOI] [PubMed] [Google Scholar]
- 22.Ramezani S, Vousooghi N, Joghataei MT, Chabok SY. The role of kinase signaling in resistance to Bevacizumab Therapy for Glioblastoma Multiforme. Cancer Biother Radiopharm. 2019;34(6):345–54. [DOI] [PubMed] [Google Scholar]
- 23.Yang B, Liu D, Ren YQ, Sun YQ, Zhang JP, Wang XG, Wu YQ, Wang SL, Guo SH, Guo G. FRAT1 promotes the angiogenic properties of human glioblastoma cells via VEGFA. Mol Med Rep 2022, 25(3). [DOI] [PubMed]
- 24.Long N, Xu X, Lin H, Lv Y, Zou S, Cao H, Chen X, Zhao Y, Qi X, Yang H, et al. Circular RNA circPOSTN promotes neovascularization by regulating miR-219a-2-3p/STC1 axis and stimulating the secretion of VEGFA in glioblastoma. Cell Death Discovery. 2022;8(1):349. 10.1038/s41420-022-01136-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Zhang H, Bai M, Ning T, Ge S, Deng T, Liu R, Zhang L, Ying G, Ba Y. Exosomes serve as nanoparticles to deliver anti-mir-214 to reverse chemoresistance to cisplatin in gastric Cancer. Mol Therapy: J Am Soc Gene Therapy. 2018;26(3):774–83. 10.1016/j.ymthe.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulte JD, Aghi MK, Taylor JW. Anti-angiogenic therapies in the management of glioblastoma. Chin Clin Oncol. 2021;10(4):37. 10.21037/cco.2020.03.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuzziol CI, Castanhole-Nunes MMU, Pavarino ÉC, Goloni-Bertollo EM. MicroRNAs as regulators of VEGFA and NFE2L2 in cancer. Gene. 2020;759:144994. 10.1016/j.gene.2020.144994 [DOI] [PubMed] [Google Scholar]
- 28.Di Stefano AL, Labussiere M, Lombardi G, Eoli M, Bianchessi D, Pasqualetti F, Farina P, Cuzzubbo S, Gallego-Perez-Larraya J, Boisselier B et al. VEGFA SNP rs2010963 is associated with vascular toxicity in recurrent glioblastomas and longer response to bevacizumab. Journal of neuro-oncology 2015, 121(3):499–504. [DOI] [PubMed]
- 29.Biterge-Sut B. A comprehensive analysis of the angiogenesis-related genes in glioblastoma multiforme vs. brain lower grade glioma. Arq Neuropsiquiatr. 2020;78(1):34–8. 10.1590/0004-282x20190131 [DOI] [PubMed] [Google Scholar]
- 30.Ni X, Wu W, Sun X, Ma J, Yu Z, He X, Cheng J, Xu P, Liu H, Shang T, et al. Interrogating glioma-M2 macrophage interactions identifies Gal-9/Tim-3 as a viable target against PTEN-null glioblastoma. Sci Adv. 2022;8(27):eabl5165. 10.1126/sciadv.abl5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-Romero N, Palacín-Aliana I, Madurga R, Carrión-Navarro J, Esteban-Rubio S, Jiménez B, Collazo A, Pérez-Rodríguez F, Ortiz de Mendivil A, Fernández-Carballal C, et al. Bevacizumab dose adjustment to improve clinical outcomes of glioblastoma. BMC Med. 2020;18(1):142. 10.1186/s12916-020-01610-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaminska B, Kocyk M, Kijewska M. TGF beta signaling and its role in glioma pathogenesis. Adv Exp Med Biol. 2013;986:171–87. 10.1007/978-94-007-4719-7_9 [DOI] [PubMed] [Google Scholar]
- 33.Li Q, Gao H, Zhou S, Liao Y. LncRNA PlncRNA-1 overexpression inhibits the growth of breast cancer by upregulating TGF-β1 and downregulating PHGDH. Breast cancer (Tokyo Japan). 2018;25(5):619–25. 10.1007/s12282-018-0858-4 [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Zheng W, Zhang L, Gu Y, Zhu L, Huang Y. LncRNA FBXO18-AS promotes gastric cancer progression by TGF-β1/Smad signaling. Eur J Histochemistry: EJH 2023, 67(2). [DOI] [PMC free article] [PubMed]
- 35.Sheng J, Chen W, Zhu HJ. The immune suppressive function of transforming growth factor-β (TGF-β) in human diseases. Growth Factors (Chur Switzerland). 2015;33(2):92–101. 10.3109/08977194.2015.1010645 [DOI] [PubMed] [Google Scholar]
- 36.Iglesias-de la Cruz MC, Ziyadeh FN, Isono M, Kouahou M, Han DC, Kalluri R, Mundel P, Chen S. Effects of high glucose and TGF-beta1 on the expression of collagen IV and vascular endothelial growth factor in mouse podocytes. Kidney Int. 2002;62(3):901–13. 10.1046/j.1523-1755.2002.00528.x [DOI] [PubMed] [Google Scholar]
- 37.Song Y, Chen Y, Li Y, Lyu X, Cui J, Cheng Y, Zhao L, Zhao G. Metformin inhibits TGF-β1-induced epithelial-to-mesenchymal transition-like process and stem-like properties in GBM via AKT/mTOR/ZEB1 pathway. Oncotarget. 2018;9(6):7023–35. 10.18632/oncotarget.23317 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.