Abstract
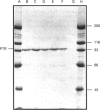
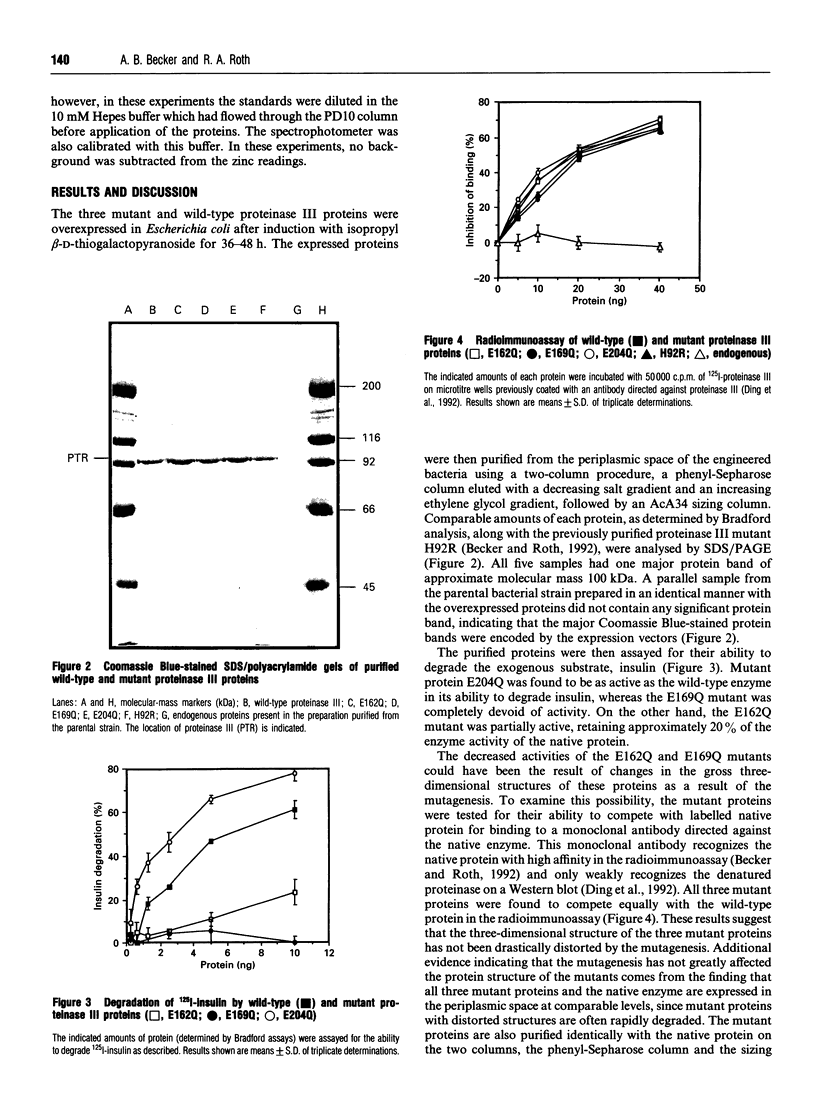
A novel active site has been identified in a family of zinc-dependent metalloendopeptidases that includes bacterial proteinase III, the human and Drosophila insulin-degrading enzymes, and the processing-enhancing protein subunit of the mitochondrial processing proteinase. None of these enzymes contains the conserved active site described in most other metalloendopeptidases, HEXXH; instead, all four contain an inversion of this motif, HXXEH. Prior mutagenesis studies of proteinase III indicate that the two histidines are essential for co-ordinating the zinc atom, while all three residues are required for enzyme activity. To identify the third zinc-binding residue in this protein family, three glutamates downstream from the active site were mutated to glutamine in proteinase III. The mutant proteins were expressed and their ability to degrade insulin was compared with the wild-type enzyme. The glutamate-204 mutant was as active as the wild-type protein, the glutamate-162 mutant retained 20% of the activity of the wild-type enzyme and the glutamate-169 mutant was completely devoid of insulin-degrading activity. The purified wild-type and glutamate-204 mutant enzymes were found to contain nearly stoichiometric levels of zinc by atomic absorption spectrophotometry, whereas the glutamate-162 mutant had a slight reduction in the level of zinc, and the glutamate-169 mutant retained less than 0.3 mol of zinc/mol of enzyme. These findings are consistent with glutamate-169 being the third zinc-binding residue in proteinase III.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Affholter J. A., Fried V. A., Roth R. A. Human insulin-degrading enzyme shares structural and functional homologies with E. coli protease III. Science. 1988 Dec 9;242(4884):1415–1418. doi: 10.1126/science.3059494. [DOI] [PubMed] [Google Scholar]
- Assoian R. K., Tager H. S. Peptide intermediates in the cellular metabolism of insulin. J Biol Chem. 1982 Aug 10;257(15):9078–9085. [PubMed] [Google Scholar]
- BEYER R. E. A study of insulin metabolism in an insulin tolerant strain of mice. Acta Endocrinol (Copenh) 1955 Aug;19(4):309–332. doi: 10.1530/acta.0.0190309. [DOI] [PubMed] [Google Scholar]
- Becker A. B., Roth R. A. An unusual active site identified in a family of zinc metalloendopeptidases. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3835–3839. doi: 10.1073/pnas.89.9.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. C., Lee Y. H. Extracellular autoprocessing of a metalloprotease from Streptomyces cacaoi. J Biol Chem. 1992 Feb 25;267(6):3952–3958. [PubMed] [Google Scholar]
- Cheng Y. S., Zipser D. Purification and characterization of protease III from Escherichia coli. J Biol Chem. 1979 Jun 10;254(11):4698–4706. [PubMed] [Google Scholar]
- Delporte C., Carvalho K. M., Leseney A. M., Winand J., Christophe J., Cohen P. A new metallo- endopeptidase from human neuroblastoma NB-OK-1 cells which inactivates atrial natriuretic peptide by selective cleavage at the Ser123-Phe124 bond. Biochem Biophys Res Commun. 1992 Jan 15;182(1):158–164. doi: 10.1016/s0006-291x(05)80125-1. [DOI] [PubMed] [Google Scholar]
- Devault A., Nault C., Zollinger M., Fournie-Zaluski M. C., Roques B. P., Crine P., Boileau G. Expression of neutral endopeptidase (enkephalinase) in heterologous COS-1 cells. Characterization of the recombinant enzyme and evidence for a glutamic acid residue at the active site. J Biol Chem. 1988 Mar 15;263(8):4033–4040. [PubMed] [Google Scholar]
- Devault A., Sales V., Nault C., Beaumont A., Roques B., Crine P., Boileau G. Exploration of the catalytic site of endopeptidase 24.11 by site-directed mutagenesis. Histidine residues 583 and 587 are essential for catalysis. FEBS Lett. 1988 Apr 11;231(1):54–58. doi: 10.1016/0014-5793(88)80701-4. [DOI] [PubMed] [Google Scholar]
- Ding L., Becker A. B., Suzuki A., Roth R. A. Comparison of the enzymatic and biochemical properties of human insulin-degrading enzyme and Escherichia coli protease III. J Biol Chem. 1992 Feb 5;267(4):2414–2420. [PubMed] [Google Scholar]
- Doherty J. J., 2nd, Kay D. G., Lai W. H., Posner B. I., Bergeron J. J. Selective degradation of insulin within rat liver endosomes. J Cell Biol. 1990 Jan;110(1):35–42. doi: 10.1083/jcb.110.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth W. C., Hamel F. G., Peavy D. E., Liepnieks J. J., Ryan M. P., Hermodson M. A., Frank B. H. Degradation products of insulin generated by hepatocytes and by insulin protease. J Biol Chem. 1988 Feb 5;263(4):1826–1833. [PubMed] [Google Scholar]
- Duckworth W. C. Insulin degradation: mechanisms, products, and significance. Endocr Rev. 1988 Aug;9(3):319–345. doi: 10.1210/edrv-9-3-319. [DOI] [PubMed] [Google Scholar]
- Ebrahim A., Hamel F. G., Bennett R. G., Duckworth W. C. Identification of the metal associated with the insulin degrading enzyme. Biochem Biophys Res Commun. 1991 Dec 31;181(3):1398–1406. doi: 10.1016/0006-291x(91)92094-z. [DOI] [PubMed] [Google Scholar]
- Fagan J. M., Waxman L. Purification of a protease in red blood cells that degrades oxidatively damaged haemoglobin. Biochem J. 1991 Aug 1;277(Pt 3):779–786. doi: 10.1042/bj2770779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faming Z., Kobe B., Stewart C. B., Rutter W. J., Goldsmith E. J. Structural evolution of an enzyme specificity. The structure of rat carboxypeptidase A2 at 1.9-A resolution. J Biol Chem. 1991 Dec 25;266(36):24606–24612. [PubMed] [Google Scholar]
- Finch P. W., Wilson R. E., Brown K., Hickson I. D., Emmerson P. T. Complete nucleotide sequence of the Escherichia coli ptr gene encoding protease III. Nucleic Acids Res. 1986 Oct 10;14(19):7695–7703. doi: 10.1093/nar/14.19.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehm B. D., Rosner M. R. Regulation of insulin, epidermal growth factor, and transforming growth factor-alpha levels by growth factor-degrading enzymes. Endocrinology. 1991 Mar;128(3):1603–1610. doi: 10.1210/endo-128-3-1603. [DOI] [PubMed] [Google Scholar]
- Hamel F. G., Mahoney M. J., Duckworth W. C. Degradation of intraendosomal insulin by insulin-degrading enzyme without acidification. Diabetes. 1991 Apr;40(4):436–443. doi: 10.2337/diab.40.4.436. [DOI] [PubMed] [Google Scholar]
- Hamel F. G., Posner B. I., Bergeron J. J., Frank B. H., Duckworth W. C. Isolation of insulin degradation products from endosomes derived from intact rat liver. J Biol Chem. 1988 May 15;263(14):6703–6708. [PubMed] [Google Scholar]
- Hari J., Shii K., Roth R. A. In vivo association of [125I]-insulin with a cytosolic insulin-degrading enzyme: detection by covalent cross-linking and immunoprecipitation with a monoclonal antibody. Endocrinology. 1987 Feb;120(2):829–831. doi: 10.1210/endo-120-2-829. [DOI] [PubMed] [Google Scholar]
- Houslay M. D., Siddle K. Molecular basis of insulin receptor function. Br Med Bull. 1989 Jan;45(1):264–284. doi: 10.1093/oxfordjournals.bmb.a072316. [DOI] [PubMed] [Google Scholar]
- Kayalar C., Wong W. T. Metalloendoprotease inhibitors which block the differentiation of L6 myoblasts inhibit insulin degradation by the endogenous insulin-degrading enzyme. J Biol Chem. 1989 May 25;264(15):8928–8934. [PubMed] [Google Scholar]
- Kirschner R. J., Goldberg A. L. A high molecular weight metalloendoprotease from the cytosol of mammalian cells. J Biol Chem. 1983 Jan 25;258(2):967–976. [PubMed] [Google Scholar]
- Kuo W. L., Gehm B. D., Rosner M. R. Cloning and expression of the cDNA for a Drosophila insulin-degrading enzyme. Mol Endocrinol. 1990 Oct;4(10):1580–1591. doi: 10.1210/mend-4-10-1580. [DOI] [PubMed] [Google Scholar]
- Kuo W. L., Gehm B. D., Rosner M. R. Regulation of insulin degradation: expression of an evolutionarily conserved insulin-degrading enzyme increases degradation via an intracellular pathway. Mol Endocrinol. 1991 Oct;5(10):1467–1476. doi: 10.1210/mend-5-10-1467. [DOI] [PubMed] [Google Scholar]
- Le Moual H., Devault A., Roques B. P., Crine P., Boileau G. Identification of glutamic acid 646 as a zinc-coordinating residue in endopeptidase-24.11. J Biol Chem. 1991 Aug 25;266(24):15670–15674. [PubMed] [Google Scholar]
- MIRSKY I. A. Insulinase, insulinase-inhibitors, and diabetes mellitus. Recent Prog Horm Res. 1957;13:429–471. [PubMed] [Google Scholar]
- Medina J. F., Wetterholm A., Rådmark O., Shapiro R., Haeggström J. Z., Vallee B. L., Samuelsson B. Leukotriene A4 hydrolase: determination of the three zinc-binding ligands by site-directed mutagenesis and zinc analysis. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7620–7624. doi: 10.1073/pnas.88.17.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D., Baumeister H., Buck F., Richter D. Atrial natriuretic peptide (ANP) is a high-affinity substrate for rat insulin-degrading enzyme. Eur J Biochem. 1991 Dec 5;202(2):285–292. doi: 10.1111/j.1432-1033.1991.tb16374.x. [DOI] [PubMed] [Google Scholar]
- Paluh J. L., Yanofsky C. High level production and rapid purification of the E. coli trp repressor. Nucleic Acids Res. 1986 Oct 24;14(20):7851–7860. doi: 10.1093/nar/14.20.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease R. J., Smith G. D., Peters T. J. Degradation of endocytosed insulin in rat liver is mediated by low-density vesicles. Biochem J. 1985 May 15;228(1):137–146. doi: 10.1042/bj2280137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J. Homologues of insulinase, a new superfamily of metalloendopeptidases. Biochem J. 1991 Apr 15;275(Pt 2):389–391. doi: 10.1042/bj2750389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick N. M., Maloy W. L., Guy H. R., Zasloff M. A novel endopeptidase from Xenopus that recognizes alpha-helical secondary structure. Cell. 1991 Aug 9;66(3):541–554. doi: 10.1016/0092-8674(81)90017-9. [DOI] [PubMed] [Google Scholar]
- Schneider H., Arretz M., Wachter E., Neupert W. Matrix processing peptidase of mitochondria. Structure-function relationships. J Biol Chem. 1990 Jun 15;265(17):9881–9887. [PubMed] [Google Scholar]
- Shii K., Roth R. A. Inhibition of insulin degradation by hepatoma cells after microinjection of monoclonal antibodies to a specific cytosolic protease. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4147–4151. doi: 10.1073/pnas.83.12.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbiah S., Harrison S. C. A method for multiple sequence alignment with gaps. J Mol Biol. 1989 Oct 20;209(4):539–548. doi: 10.1016/0022-2836(89)90592-5. [DOI] [PubMed] [Google Scholar]
- Thayer M. M., Flaherty K. M., McKay D. B. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-A resolution. J Biol Chem. 1991 Feb 15;266(5):2864–2871. doi: 10.2210/pdb1ezm/pdb. [DOI] [PubMed] [Google Scholar]
- Vallee B. L., Auld D. S. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990 Jun 19;29(24):5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- Williams F. G., Johnson D. E., Bauer G. E. [125I]-insulin metabolism by the rat liver in vivo: evidence that a neutral thiol-protease mediates rapid intracellular insulin degradation. Metabolism. 1990 Mar;39(3):231–241. doi: 10.1016/0026-0495(90)90041-a. [DOI] [PubMed] [Google Scholar]
- Witte C., Jensen R. E., Yaffe M. P., Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988 May;7(5):1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. J., Geli V., Oppliger W., Suda K., James P., Schatz G. The MAS-encoded processing protease of yeast mitochondria. Interaction of the purified enzyme with signal peptides and a purified precursor protein. J Biol Chem. 1991 Apr 5;266(10):6416–6423. [PubMed] [Google Scholar]