Summary
Heterozygous mutations in SLC40A1, encoding a multi-pass membrane protein of the major facilitator superfamily known as ferroportin 1 (FPN1), are responsible for two distinct hereditary iron-overload diseases: ferroportin disease, which is associated with reduced FPN1 activity (i.e., decrease in cellular iron export), and SLC40A1-related hemochromatosis, which is associated with abnormally high FPN1 activity (i.e., resistance to hepcidin). Here, we report three SLC40A1 missense variants with opposite functional consequences. In cultured cells, the p.Arg40Gln and p.Ser47Phe substitutions partially reduced the ability of FPN1 to export iron and also partially reduced its sensitivity to hepcidin. The p.Ala350Val substitution had more profound effects, resulting in low FPN1 iron egress and weak FPN1/hepcidin interaction. Structural analyses helped to differentiate the first two substitutions, which are predicted to cause local instabilities, and the third, which is thought to prevent critical rigid-body movements that are essential to the iron transport cycle. The phenotypic traits observed in a total of 12 affected individuals are highly suggestive of ferroportin disease. Our findings dismantle the classical dualism of FPN1 loss versus gain of function, highlight some specific and unexpected functions of FPN1 transmembrane helices in the molecular mechanism of iron export and its regulation by hepcidin, and extend the spectrum of rare genetic variants that may cause ferroportin disease.
Keywords: Ferroportin disease, SLC40A1-related hemochromatosis, MFS transporters, Hepcidin, Pathogenicity of missense variants
Heterozygous mutations in the SLC40A1 gene are responsible for two iron-overload diseases, ferroportin disease and SLC40A1-related hemochromatosis, depending on whether they result from a loss or gain of function of the FPN1 protein. We dismantle this apparent dichotomy by providing new genetic, clinical, functional, and structural data.
Introduction
The solute-carrier family 40 member 1 (HUGO Gene Nomenclature Committee: SLC40A1; UniProt: Q9NP59), also referred to as ferroportin 1 (FPN1), is the sole iron export protein found in mammals.1 It is expressed in the plasma membrane of cells highly specialized in iron metabolism, including macrophages, hepatocytes, duodenal enterocytes, placenta syncytiotrophoblasts, and erythrocytes.2,3 FPN1 activity is predominantly regulated by the hepatic hyposideremic hormone hepcidin, which, depending on the cell type, induces internalization and degradation of FPN1 and/or blocks the FPN1-related iron export mechanism.4,5
More than 65 SLC40A1 heterozygous pathogenic variants have now been reported in affected individuals of different ethnic groups.6 These mutations fall into two functional categories, loss of function and gain of function (LoF and GoF, respectively), and are responsible for two distinct human diseases.7,8,9 Ferroportin disease (FD), which is the most frequently reported in the literature, is caused by germline mutations that reduce the ability of human FPN1 (HsFPN1) to export iron through variable molecular mechanisms.10,11 The disease phenotype is characterized by iron accumulation in reticuloendothelial macrophages, correlating with elevated serum ferritin and low to normal transferrin saturation. Over time, iron can escape into the bloodstream, resulting in increased transferrin saturation and iron accumulation in hepatocytes. Clinical manifestations appear to be rare judging from the reports available in the literature.6,7,8,9 SLC40A1-related hemochromatosis (HC) is caused by germline mutations that more or less protect HsFPN1 from the effects of hepcidin downregulation.12,13 This leads to excessive iron export to the bloodstream, increased transferrin saturation, and progressive iron accumulation in parenchymal cells (primarily hepatocytes). When observed, hyperferritinemia correlates with the degree of iron overload and its progression. Clinical symptoms are similar to those seen in affected individuals with HFE-related HC.
FPN1 belongs to the major facilitator superfamily (MFS), which is the largest family of secondary active transporters.14 MFS proteins share a common architecture consisting of two bundles of six transmembrane (TM) helices (N-domain: TM1–TM6; C-domain: TM7–TM12), related by 2-fold pseudosymmetry. Each bundle can be further subdivided into two inverted-topology repeats of three TMs, where the first helices (i.e., TMs 1, 4, 7, and 10; in blue in Figure 1) are referred to as the core helices or the gate helices, since they form the central cavity and are also thought to play an active role in the formation of thin gates on both sides of the lipid bilayer. Structural plasticity of these helices is actually often observed during substrate loading and release, in tight connection with the second helices (i.e., TMs 2, 5, 8, and 11; in green), which are uniquely involved in the interface between the N- and C-domains, while the third helices (i.e., TMs 3, 6, 9, and 12; in red) appear less conformationally diverse.15,16,17,18 This gave rise to the “clamp-and-switch” transport cycle model that involves (at least) five conformational states (outward-facing [OF], outward-occluded, occluded, inward-occluded, inward-facing [IF]) and is modulated by MFS substrates and gating residues.17,18
Figure 1.
Ribbon representation of the 3D structures of human FPN1 in complex with hepcidin (pink) in the OF conformation (PDB: 6WBV)
TM helices are colored to highlight the four inverted repeats of three helices.
So far, the molecular and physiopathological bases of FD and SLC40A1-related HC have largely been regarded as antithetic. Nevertheless, we have previously shown that the non-clinical (i.e., not described in an affected individual, nor in the Genome Aggregation Database [gnomAD]) p.Trp42Ala mutant displays mixed behavior in vitro, with a very substantial effect on the ability of HsFPN1 to export iron out of the cell, but also a partial resistance to hepcidin.19 The hypothesis of a close link between the effect of hepcidin and the ability of HsFPN1 to export iron also emerged from two studies investigating the p.Asp181Val and p.Arg178Gln pathogenic variants.20,21 The present study, which combines original genetic, clinical, cellular, and structural data, provides further evidence that LoF mutations in the SLC40A1 gene cause FD, regardless of the sensitivity of the HsFPN1 protein to hepcidin.
Subjects and methods
Genetics studies
The complete coding sequence of SLC40A1 and intron/exon boundaries was investigated by next-generation sequencing (conditions are available upon request) in the probands, while family members were only assessed for the p.Ser47Phe variant by Sanger sequencing. All probands were negative for genotypes known to cause HC (HFE, HJV, HAMP, TFR2 genes) or hereditary hyperferritinemia (FTL gene). They were also negative for the few BMP6 missense variations (p.Pro95Leu, p.Leu96Pro, and p.Gln113Glu) that have been associated with mild to moderate late-onset iron overload in French affected individuals.22,23
FPN1 plasmid constructs
The wild-type (WT) FPN1-V5 and FPN1-V5/CD8 (cluster of differentiation 8) bicistronic plasmid constructs were generated as previously described.10,11 The full-length human SLC40A1 cDNA (GenBank: NM_0414585.5) was inserted into the pcDNA5-FRT-TO vector (Thermo Fisher Scientific) to produce isogenic stably transfected tetracycline-inducible HEK293 cells (Flp-In 293 T-Rex). All FPN1 mutations were introduced in the different vectors by using the QuikChange Site-Directed Mutagenesis Kit, according to the manufacturer’s instructions (Agilent Technologies). Sequencing analyses were performed to check the integrity of all plasmid constructs (full-length SLC40A1 cDNA sequenced after each site-directed mutagenesis).
Culture and transfection of HEK293T cells
HEK293T cells, from the American Type Culture Collection, were incubated at 37°C in a 5% CO2 humidified atmosphere and propagated in DMEM (Lonza) supplemented with 10% fetal bovine serum. Cells were transiently transfected using JetPEI (Polyplus), according to the manufacturer’s instructions, and a 2:1 transfection reagent (μL)/plasmid DNA ratio (μg).
Generation of stable HEK293T cells exhibiting tetracycline-inducible expression of FPN1
Flp-In 293 T-Rex cells were transfected with the different pcDNA5-FRT-TO plasmid constructs and the pOG44 vector using the TransIT2020 cationic lipid (Mirus Bio), according to the manufacturer’s instructions (Thermo Fisher Scientific). Stably transfected cells were selected in DMEM (Lonza) supplemented with 10% fetal bovine serum, 150 μg/mL blasticidin (Thermo Fisher Scientific), and 300 μg/mL hygromycin (Thermo Fisher Scientific) for 3 weeks. The resulting clones were propagated and maintained in the same media. SLC40A1 cDNA integration was verified by PCR amplification and Sanger sequencing, while FPN1 expression was assessed by western blotting.
55Fe release measurements
55Fe loading of human apotransferrin was performed as previously described.24 Briefly, HEK293T cells (1 × 105 cells/well in 12-well plates) were grown for 24 h in supplemented DMEM (Lonza), before being incubated with 20 μg/mL 55Fe-transferrin for 24 h and transiently transfected with WT or mutated FPN1-V5 plasmid constructs. At 15 h post-transfection, cells were washed once with PBS and cultured in Pro293a-CDM serum-free medium (BioWhittaker) for up to 36 h. 55Fe exported into the supernatant was collected, mixed with liquid scintillation fluid (Ultima Gold MV, Packard Bioscience), and counted for 10 min in a Tri-Carb 1600 CA scintillation counter (Packard). The percentage of 55Fe export was calculated using the following formula: (55Fe in the supernatant at endpoint/cellular 55Fe at time zero) × 100.
Intracellular 55Fe measurements
HEK293T cells were transfected with WT or mutated pcDNA3.1_FPN1-V5 constructs for 24 h, before being cultured in Pro293a-CDM serum-free medium (Lonza) and preloaded with 20 μg/mL 55Fe-transferrin for 16 h. Each pcDNA3.1_FPN-V5 construct was co-delivered with the pSV-β-galactosidase (β-gal) vector (Promega). Cells were harvested with trypsin, mixed with liquid scintillation fluid (Ultima Gold MV, PerkinElmer) and counted for 2 min in a Tri-Carb 1600 CA scintillation counter (PerkinElmer). 55Fe radioactivity was normalized on total protein content and β-gal activity.
Human hepcidin-25 synthesis and secretion by HEK293 T-Rex cells
The native human hepcidin-25 was recovered from the supernatant of HEK293 T-Rex cells as previously described.10
Flow cytometry analysis
Flow cytometry experiments were done as previously described.11 Briefly, HEK293T cells (1.75 × 105 cells/well in 12-well plates) transfected with the pIRES_FPN1-V5_CD8 constructs were treated (or not) with 4.3 nM native human hepcidin-25 (24 h after transfection) for 16 h. Cells were harvested with trypsin, pelleted (500 g, 5 min, 4°C), and resuspended in PBS (pH 7.4) containing EDTA (Lonza) and 10% fetal bovine serum, before being incubated for 20 min at 4°C with anti-V5-FITC (fluorescein isothiocyanate; Thermo Fisher Scientific) and anti-CD8-APC (Miltenyi Biotec). Stained cells were pelleted (500 g, 5 min, 4°C) and resuspended in 400 μL PBS-EDTA. Cells were analyzed using a BD Accuri C6 flow cytometer (BD Biosciences) and FlowLogic software (Miltenyi Biotec).
Hepcidin-binding analysis
Flp-In T-Rex cells (4 × 105 cells/well in 6-well plates), induced with 1 μg/mL tetracyclin for 24 h, or transiently transfected HEK293T (3.5 × 105 cells/well in 6-well plates) were treated with 10 μg/mL C terminus biotinylated hepcidin-25 (Bachem) for 30 min at 37°C. Proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer (Boston BioProducts) supplemented with protease inhibitor cocktail (Roche). The total protein concentration was determined using the bicinchoninic assay (BCA, Pierce). Protein lysates, 600 μg (in 300 μL), were co-immunoprecipitated using the Dynabeads MyOne Streptavidin T1, according to the manufacturer’s instructions (Thermo Fisher Scientific). Western blot analysis was performed with horseradish peroxidase (HRP)-conjugated anti-V5 monoclonal antibody (Thermo Fisher Scientific).
Detection of FPN1 ubiquitination
Flp-In T-Rex cells (4 × 105 cells/well in 6-well plates), induced with 1 μg/mL tetracycline for 24 h, or transiently transfected HEK293T (3.5 × 105 cells/well in 6-well plates) were treated with 4.3 nM native humanhepcidin-25 for 30–120 min. Proteins were extracted using RIPA buffer (Boston BioProducts) supplemented with protease inhibitor cocktail (Roche). The total protein concentration was determined using the BCA (Pierce). FPN1 proteins were purified from 500 μL total protein lysates (normalized: 500–1,000 μg in the different experiments) using the V5-tagged Protein Purification Kit version 2 (MBL International Corporation), according to the manufacturer’s instructions. Western blot analysis was performed with HRP-conjugated anti-V5 monoclonal antibody (Thermo Fisher Scientific), or primary anti-mono-/polyubiquitin monoclonal antibody FK2 (Enzo Life Sciences) followed by HRP-coupled secondary antibody.
3D structure modeling and analysis
Three-dimensional (3D) structures were extracted from the PDB (https://rcsb.org/) and manipulated using Chimera 1.13.1.25 We also considered a model of the global organization of mammalian ferroportin in the IF state26; this one was based on the superimposition of the N- and C-lobes of the OF 3D structures of mammalian ferroportin proteins on the IF 3D structures of Bdellovibrio bacteriovorus (Bb) Fpn.
Statistics
Data are presented as scatter dot plots and means. Comparisons used a one-tailed Student’s t test.
Study approval
Informed consent for molecular studies and clinical data publication was obtained from all affected individuals and family members, in accordance with the Declaration of Helsinki. In line with French ethical guidelines, the Clinical Research Ethics Committee of the University Hospital of Brest approved the study.
Results
Genetic and phenotypic data
Three heterozygous missense variations were identified in SLC40A1 (NM_014585.5): c.119G>A (p.Arg40Gln), c.140C>T (p.Ser47Phe), and c.1049C>T (p.Ala350Val). They are absent (c.140C>T, c.1049C>T) or present at extreme low frequencies (c.119G>A; <0.00001) in gnomAD (version 2.1.1). The p.Ser47Phe and p.Ala350Val substitutions have been previously reported in a few affected individuals with iron overload.27,28,29
The p.Ser47Phe variation was observed in 9 affected individuals belonging to a family from northeastern France (15 members investigated: 10 positive for the p.Ser47Phe variant, but only 9 with phenotypic data available; Figure S1) and 3 unrelated affected individuals, whereas the p.Arg40Gln and p.Ala350Val variations were observed in single individuals (Table 1). Together, the three missense variations were found in 9 males and 3 females, ages 20–75 years at diagnosis, who displayed significant hyperferritinemia (males: 889–2,000 μg/L; females: 439–922 μg/L). Ten affected individuals (7 males and 3 females) had transferrin saturation values above 45%, the threshold usually used for early detection of HC.30 All values, however, remained below 75%, the threshold for the appearance of toxic free iron species in plasma.31 Hepatic iron concentration (HIC) was evaluated using Gandon’s MRI method in 8 of the 12 considered affected individuals with values ranging from slightly increased (<100 μmol/g) to highly increased (≥230 μmol/g). No information was available on iron levels in the spleen.
Table 1.
Description of the study sample: SLC40A1 missense variations, family relationships, and biological and clinical data
| Variant | Gender | Family relationships | Age, yearsa | TS, % | SF, μg/L | ASAT, IU/L | ALAT, IU/L | GGT, IU/L | ALP, IU/L | RBC, tera/L | Hb, g/dL | Ht, % | MCV, fL | HIC, μmol/g | Clinical observations | Alcohol consumptionb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p.Arg40Gln | M | index case | 68 | 55 | 2,000 | 230 | ||||||||||
| p.Ser47Phe | M | index case (III.6) | 49 | 66 | 2,000 | 25 | 5.0 | 16.0 | 46.0 | 90.0 | 300 | daily | ||||
| p.Ser47Phe | M | brother (III.9) | 40 | 67 | 1,769 | 22 | 35 | 340 | daily | |||||||
| p.Ser47Phe | M | father (II.5) | 71 | 50 | 944 | 96.0 | fatigue | occasional | ||||||||
| p.Ser47Phe | M | uncle (II.4) | 75 | 58 | 1,166 | |||||||||||
| p.Ser47Phe | F | first cousin (III.4) | 58 | 67 | 439 | 128 | ||||||||||
| p.Ser47Phe | F | first cousin (III.5) | 51 | 52 | 922 | 77 | ||||||||||
| p.Ser47Phe | M | uncle (II.2) | 65 | 59 | 1,000 | 20 | 18 | 14.8 | 96.9 | 80 | cataract | occasional | ||||
| p.Ser47Phe | F | first cousin (III.3) | 53 | 14 | 36 | 15 | 16 | 29 | 84 | 4.5 | 12.7 | 38.2 | 84.0 | coronaropathy, high blood pressure | abstinent | |
| p.Ser47Phe | M | son of first cousin (IV.1) | 22 | 25 | 182 | |||||||||||
| p.Ser47Phe | F | index case | 50 | 52 | 609 | 16 | 18 | 21 | 4.0 | 12.7 | 37.0 | 92.4 | 100 | occasional | ||
| p.Ser47Phe | M | index case | 70 | 50 | 944 | 21 | 12 | 13 | 96.0 | fatigue | occasional | |||||
| p.Ser47Phe | M | index case | 33 | 24 | 889 | 77 | 36 | 14 | 93.0 | abstinent | ||||||
| p.Ala350Val | M | index case | 20 | 15 | 1,413 | 78 |
Age at diagnosis.
Occasional: less than 1 drink per day for women and 2 drinks per day for men. Daily: at least than 1 drink per day for women and 2 drinks per day for men. Alkaline phosphatase, normal range 44–147 IU/L; ALT, alanine aminotransferase (normal range 5–50 IU/L); ASAT, aspartate aminotransferase (normal range 5–50 IU/L); GGT, γ-glutamyl transferase (normal range 5–55 IU/L); Hb, hemoglobin (normal range 12.0–18.0 g/dL); HIC, hepatic iron concentration (normal value <36 μmol/g); Ht, hematocrit (normal range 37%–52%); MCV, mean corpuscular volume (normal range 80–95 fL); RBC, red blood cells (normal range 4.0–5.7 × 1012/L); SF, serum ferritin (normal value ≤200 μg/L in females, ≤300 in males); TS, transferrin saturation (normal value ≤50% in females, ≤60% in males).
Biochemical analysis of the three identified SLC40A1 amino acid changes
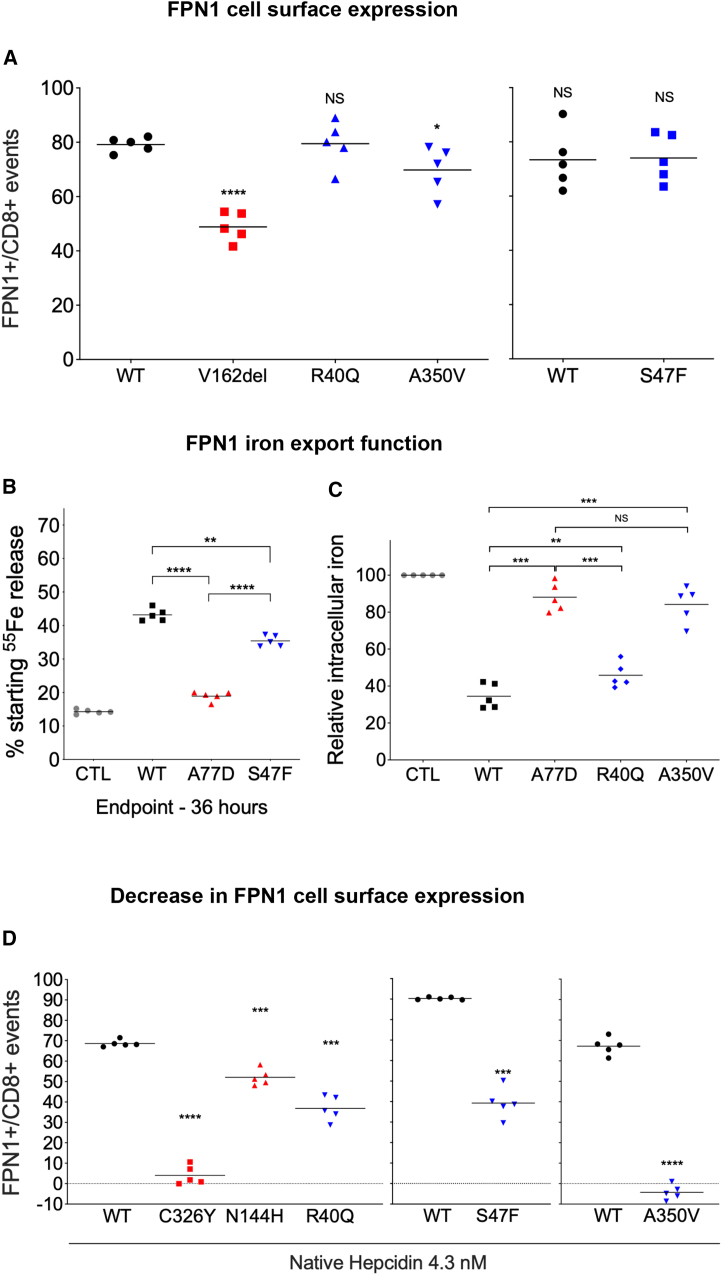
A bicistronic construct was used to evaluate the concurrent plasma membrane expression of human FPN1 (conjugated with a V5 epitope) and CD8 in transiently transfected HEK293T cells on flow cytometry, as previously reported.11 The p.Val162del single amino acid deletion, which is known to reduce cell surface localization of HsFPN1,32 was used as negative control (Figure 2A). The level of expression of HsFPN1-Val162del on the surface of CD8-expressing cells was indeed markedly decreased in comparison with WT HsFPN1 (p < 0.001). A more moderate reduction was observed with the p.Ala350Val mutant (p < 0.05), whereas no difference was observed with the p.Arg40Gln and p.Ser47Phe mutants.
Figure 2.
Evaluation of FPN1 (WT versus mutants) cell surface expression, iron export ability, and resistance to hepcidin
(A) HEK293T cells were transiently transfected with the bicistronic pIRES2 plasmid encoding both full-length human FPN1-V5 and CD8. After 36 h, cells were double stained for CD8 (APC) and the FPN1-V5 fusion protein (FITC) and analyzed by two-color flow cytometry. Data are presented as percentages of FPN1+ over CD8+ events. NS, not significant.
(B) HEK293T cells were grown in 20 μg/mL 55Fe-transferrin for 24 h before being washed and transiently transfected with WT or mutated SLC40A1-V5 expression plasmids. After 15 h, cells were washed and then serum starved. The 55Fe exported into the supernatant was collected at 36 h. Data are presented as percentages of cellular radioactivity at time zero.
(C) HEK293T cells were transfected with pcDNA3.1-FPN1-V5-His vectors, grown for 24 h, and then fed with 20 μg/mL 55Fe-transferrin for 16 h. Cells were then washed and counted. Counts per minute were normalized by total protein and β-gal activity.
(D) HEK293T cells were transiently transfected with the bicistronic pIRES2 plasmid encoding both full-length human FPN1-V5 and CD8 24 h before being treated with 4.3 nM native hepcidin for 16 h. Cells were then double stained for CD8 (APC) and the FPN1-V5 fusion protein (FITC) and analyzed by two-color flow cytometry. Data are presented as percentages of FPN1+ over CD8+ events. Each point represents the average value (from triplicate) of five independent experiments. p values were calculated by a Student’s t test. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
The in vitro activity of the HsFPN1 mutants was assessed using radioactive iron isotopes in experiments measuring either the export of iron (Figure 2B) or its cellular retention (Figure 2C) according to previously validated protocols.11,33 As shown in Figure 2B, HEK293T cells transiently transfected with a pcDNA3.1 vector encoding a WT HsFPN1-V5 fusion protein displayed a 3-fold increase in iron release than cells transfected with the corresponding empty vector (commercial pcDNA3.1-V5-His; CTL). The p.Ser47Phe mutant was not able to export 55Fe in amounts that are comparable with WT HsFPN1 (p < 0.01), but it was more active than the well-known p.Ala77Asp iron export defective mutant (p < 0.0001). In the inverse experiment, cells transiently transfected with WT HsFPN1 displayed 2- to 3-fold lower iron accumulation than cells not overexpressing HsFPN1 (CTL). Similar to the p.Ser47Phe mutant, the p.Arg40Gln mutant was not able to export 55Fe in amounts comparable with WT HsFPN1 (p < 0.001), but it was more active than the LoF p.Ala77Asp mutant (p < 0.001). The p.Ala350Val mutant was almost completely unable to export iron, as evidenced by a large difference with WT HsFPN1 (p < 0.001) but not with p.Ala77Asp (p = 0.285).
To investigate whether the HsFPN1 mutants could modify their response to hepcidin, transiently transfected HEK293T cells were cultured for 16 h with conditioned media derived from HEK293 T-Rex cells stably expressing the full-length human HAMP cDNA. Two known HsFPN1 GoF mutants served as positive controls: p.Asn144His, which shows partial resistance to hepcidin inhibition, and p.Cys326Tyr, which abolishes hepcidin binding to HsFPN1 and is responsible for complete resistance.12,34 As expected, the addition of hepcidin to cells expressing WT HsFPN1 resulted in the disappearance of the iron exporter from the plasma membrane. The p.Arg40Gly and p.Ser47Phe mutants showed intermediate sensitivity (between the p.Asn144His and p.Cys326Tyr controls), whereas the p.Ala350Val was found unresponsive (Figure 2D).
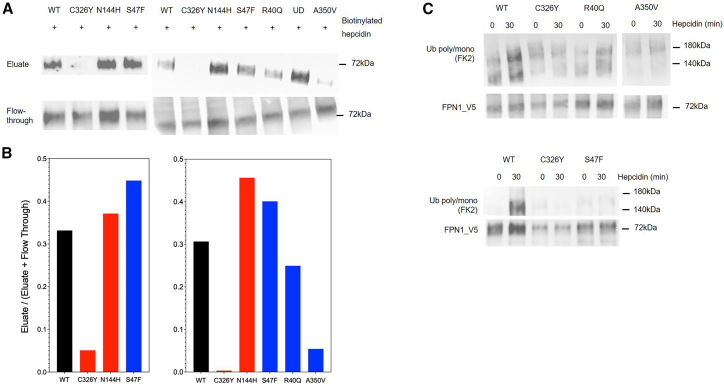
Fernandes and collaborators demonstrated that the cysteine-to-serine (p.Cys326Ser) and cysteine-to-threonine (p.Cys326Tyr) changes at position 326 of human HsFPN1 prevent hepcidin binding, whereas the p.Asn144Asp and p.Asn144Thr missense mutations do not.34 We obtained comparable results using stably transfected HEK293 T-Rex cells expressing, upon induction by tetracycline, either the WT HsFPN1-V5 fusion protein or the HsFPN1-C326Y-V5 and HsFPN1-N144H-V5 mutants (Figure S2). We reproduced these findings in transiently transfected HEK293T cells, where we further tested the p.Arg40Gln, p.Ser47Phe, and p.Ala350Val mutants. As shown in Figure 3A, two HsFPN1 mutants co-purified with hepcidin in amounts similar to that of WT HsFPN1 (data are representative of two independent experiments; see Figure S3). Only the interaction between the p.Ala350Val mutant and hepcidin proved tenuous.
Figure 3.
Evaluation of FPN1 binding to hepcidin (WT versus mutants) and underlying ubiquitination
(A) HEK293T cells transiently transfected with pcDNA3.1_FPN1-V5 constructs (encoding either FPN1-WT, FPN1-Cys326Tyr, FPN1-Asn144His, FPN1-Arg40Gln, FPN1-Ser47Phe, or FPN1-Ala350Val), were treated with a C-ter biotinylated hepcidin-25 for 30 min before being washed and lysed. The FPN1-hepcidin complexes were purified on streptavidin beads and revealed by western blot analysis with V5 antibody (Eluate); the fraction of FPN1-V5 not bound to hepcidin in each lysate is shown for comparison (flowthrough).
(B) Densitometry scans of the binding to hepcidin experiments; data are expressed as the ratio between eluate and the sum of eluate and flowthrough.
(C) Transiently transfected HEK293T cells were treated with 4.3 nM native hepcidine for 30 min. Cell lysates were immunoprecipitated with anti-V5 and blotted with anti-poly-/monoubiquitin antibody (FK2). The membrane was reblotted with the anti-V5 antibody to confirm FPN1-V5 protein expression.
Ubiquitination is a necessary condition for internalization and degradation of HsFPN1 after exposure to hepcidin in various cells, including the HEK293 cell line.35 We finally studied the ubiquitination of the iron transporter before and after hepcidin treatment in stably transfected HEK293 T-Rex cells and in transiently transfected HEK293T cells. Cells overexpressing different HsFPN1-V5 fusion proteins were treated with human native hepcidin-25 and, after V5-tagged protein purification, the ubiquitination was revealed by western blotting with an anti-poly-/monoubiquitin (FK2) antibody. As described previously,35 we detected ubiquitination after 30 min of hepcidin treatment on the WT protein. The ubiquitinated species of human FPN1 migrated in SDS-PAGE with an apparent mass of between 140 and 180 kDa, which was indicative of the addition of about 10 ubiquitin molecules (Figure S4). Compared to WT HsFPN1, the ubiquitination of the p.Cys326Tyr, p.Ser47Phe, and p.Ala350Val mutants was very low to undetectable, while that of the p.Arg40Gln mutant appeared weaker (Figure 3B; replicas are presented in Figure S3).
3D structure-guided prediction of the effect of the three identified SLC40A1 amino acid changes
Several 3D structures of the Fpn MFS transporter have been captured in different organisms, highlighting residues that are directly involved in metal binding and the formation of intra- and extracellular gates.11,36,37,38,39 In mammalian proteins (human: HsFPN1; primate Philippine Tarsier: TsFpn), which so far have only been solved in OF states, transition-metal ion-binding sites have been identified in both the N- and C-domains. There are two essential residues in each site: Asp39 and His43 on TM1 and Cys326 and His507 on TM7 and TM11 (highlighted in yellow in Figure 1).36,39 The second site is different from the first one in that it exploits a structural feature that is not found in all MFS proteins18: a non-helical stretch of six residues in the center of TM7 that separates the helix into a longer TM7a and a shorter TM7b. This uncommon feature has been posited to be important in both iron and hepcidin binding.36,39,40
In an OF structure of HsFPN1 (PDB: 6WBV), solved in the presence of cobalt (an iron mimetic) and hepcidin,36 Arg40 and Ser47 emerge in the close vicinity of TM1-Asp39 and His43 (Figure 4). One hydrogen bond is formed between the side chain of Ser47 and the main-chain oxygen atom of His43, which may stabilize the N-domain metal-binding site. What is remarkable, and has not been described before, is that Ser47 also forms a hydrogen bond with the main chain oxygen atom of hepcidin Pro5. Ser47 thus forms a second interaction network with Asp39 and His43 in the center of TM1 (Figure 4A), which echoes the one that has been already described in detail (in all-atom molecular dynamic simulations of HsFPN1 with Fe2+) between Asp325 (TM7b), Cys326 (TM7b), and His507 (TM11) in the C-domain.36
Figure 4.
Close-up views of the metal- and hepcidin-binding sites within the HsFPN1 N- and C-lobes in an OF conformation
(A and B) HsFPN1 in an OF conformation. Ribbon (A) and surface (B) representations of the human FPN1 3D structures are shown in complex with hepcidin (pink ribbon) (OF conformation; PDB: 6WBV). TM helices are colored as in Figure 1. Amino acids discussed in the text are shown with atomic details. The cobalt (iron mimetic) ions are represented as pink spheres, while a water molecule is shown in red in the ion-binding site of the C-lobe.
(C) Mammalian FPN1 in an IF conformation. Crude assembly of the N- and C-lobes of Philippine Tarsier TsFPN1 (as extracted from its experimental 3D structure in the OF conformation: PDB: 6VYH), after superimposition on the N- and C-lobes of BbFPN in the IF conformation (PDB: 6BTX).26 The cobalt (iron mimetic) ions are represented as green spheres.
(D and E) Close-up views of Ala350 in a central pivot position between conformations. (D) Ribbon representations of the 3D structures of human FPN1 in complex with hepcidin in OF conformation (PDB: 6WBV). (E) Bdellovibrio bacteriovorus (Bb) Fpn in the OF (PDB: 5AYM, top right) and IF conformations (PDB: 6BTX, bottom right) conformations, respectively. The atoms of the two amino acids from TM5 and TM8 in close contact in the two conformations are depicted as spheres.
Arg40 is not in position to directly interact with Asp39, His43, or the metal ion. However, as shown in Figure 4B, the side chains of Ser47 (in red), but also Arg40 (in blue), are accessible in the deepest buried surface area of the central cavity, in which hepcidin is inserted. Arg40 may thus impact hepcidin binding through its participation in the electrostatic surface potential of the pocket. Moreover, the importance of Arg40 must be considered in the light of the IF conformation of plasma membrane MFS transporters, in which the extracellular gate is mainly achieved by the extracellular halves of TM1 and TM7, as well as by the flanking helices TM2 and TM8.15,16,17,18 Based on the possible arrangement of HsFPN1 and TsFPN1 N- and C-domains in the IF state, predicted on the basis of the experimental 3D structure of a bacterial homolog (BbFpn) in an identical state, we previously suggested that Asp325 (TM7b) forms an inter-lobe salt bridge with Arg40 (TM1) when not interacting with Cys326 (TM7b) (Figure 4C). This interaction may be a key feature of the extracellular gate in mammalians, playing a role in the conformational switch when iron sits in the core of the transporter awaiting translocation.26
The importance of Ala350 in TM8 must be assessed by considering the role of this helix in the interface it makes with the N-domain through contact with TM5 (green helices in Figure 1). In the OF conformation of HsFPN1, TM5 is bent, and close contacts are observed only in the N- and C-terminal parts of TM5 and TM8, respectively. Ile186, in the middle of TM5, is the last amino acid of this secondary structure in contact with TM8 Ala350 and, one helical turn before, Gly247 (Figure 4D). A similar situation is observed in BbFpn OF conformation, while contacts between the two helices are inverted in the IF state (Figure 4E), the only amino acids staying in contact in the two conformations being TM5 Val167 (human counterpart: Ile186) and TM8-Ala288 (human counterpart: Ala350). This invariant contact point thus acts like a gear to facilitate the rotation of the two interfacial helices. This could be a critical structural feature of the iron transport cycle, implying that the effects of the p.Ala350Val variant on FPN1 movements within the lipid bilayer could be very important. This may explain both the low iron export seen in Figure 2 and the minimal binding to hepcidin seen in Figure 3.
Discussion
Clinical, genetic, and functional data published in the literature for more than 20 years have led a majority of authors to consider that FD is caused by LoF mutations, whereas SLC40A1-related HC is caused by GoF mutations. The challenge for physicians and scientists has essentially been to distinguish LoF and GoF mutations from neutral missense variants identified in affected individuals with phenocopies (i.e., secondary, not clearly defined causes of iron overload).6,7,29 The hypothesis of a third category of pathogenic variants, accumulating LoF and GoF characteristics, has been raised recently, given the proximity between HsFPN1 amino acids involved in iron or hepcidin binding, the involvement of other residues in conformational changes also governing the iron export function and its regulation by hepcidin, and some functional studies suggesting that intact iron export by HsFPN1 is necessary for hepcidin-induced downregulation.13,19,20,21,36 We present here some original data that strongly support this hypothesis.
Our functional analyses (Figures 2 and 3) reveal that the two missense variations, p.Arg40Gln and p.Ser47Phe of TM1, are fairly similar. They bothI (1) reduce the ability of HsFPN1 to export iron out of the cell, although they do not cause HsFPN1 mislocalization as we have previously reported for numerous missense variations altering the intracellular gate,10,11 and (2) reduce the sensitivity of HsFPN1 to hepcidin, although they do not obviously alter the SLC40A1-hepcidin interaction as has been previously reported by Fernandes et al. for other missense variations located in the TM2 or TM4 helices of the N-domain.34 The functional consequences of the p.Ala350Val mutant are more significant, since it does not reach the cell surface correctly and is much more limited in terms of iron egress, while exhibiting strong resistance to hepcidin in a way that suggests a major interaction defect between the two proteins.
Our structural analyses, however, make it possible to distinguish between different molecular mechanisms. The first involves a cross-section of HsFPN1, parallel to the membrane plane, in which interactions with iron and/or hepcidin are obviously concentrated between the N- and C-domains. Here, the Ser47Phe novel missense variation is very likely to cause local instability with disrupting a previously unreported H-bond interaction network that associates Ser47 with both key iron coordinating residues Asp39 and His43, and also the N terminus moiety of hepcidin (Pro5). That the primary binding site of hepcidin, at the Cys326 position and flanking positions in the TM7b and TM11 helices of the C-domain,36 is not altered may explain that we did not observe a marked decrease in the HsFPN1-hepcidin interaction in Figure 3A. It is worth noting that the double mutant Asp39Ala-His43Ala in TsFpn has been documented as defective in iron transport and insensitive to hepcidin.39 The other molecular mechanisms are associated with the extracellular gate architecture of HsFPN1 and its conformational dynamics. Here, the p.Arg40Gln mutant is thought to prevent an inter-domain salt bridge between TM1 and TM7b in the IF state before iron binding, which could be a key element for stabilizing HsFPN1 in this conformation.26 This salt bridge, which is supposed to be formed between Arg40 in TM1 and Asp325 in TM7b and standing between the critical Asp39 and His43 positions, could also play an essential role in the local conformation of the primary iron-binding site within the central cavity. Arg40 also emerges from deeply buried positions of HsFPN1 that participate in the formation of the hepcidin-binding sites within the N- and C-domains. This suggests that TM1 plays a particular role in the conformational changes of the N-lobe that must happen as a result of hepcidin binding, and especially those which must facilitate the access of the large intracellular loop between TM6 and TM7 for ubiquitination.35 Such a role has been proposed by Billesbølle et al. for the flanking TM2 helix where the Tyr64 residue has been identified as interacting with the hepcidin Phe4 and Ile6 residues.36 It is also worth noting that the p.Tyr64Asn SLC40A1-related HC causing mutation has been consistently reported to reduce FPN1 ubiquitination and prevent its intracellular degradation, without altering hepcidin binding.13,34 Consistent with the results obtained in vitro, the Ala350Val mutant is predicted to have a more pronounced structural impact. This mutant would hinder a critical interaction between TM8 and TM5, at a point where the two TM helices rotate relatively to each other, allowing critical rigid-body movements that are part of the clamp-and-switch MFS transport cycle model.16,17,41 Two consequences can then be anticipated: (1) reduced dynamics of HsFPN1 within the plasma membrane, with a lower capacity to bind and export iron, and (2) minimal hepcidin binding to HsFPN1. This latter assumption is also supported from data produced by Billesbølle et al., who reported that hepcidin binding to the C-domain of HsFPN1 is coupled to iron binding, with an 80-fold increase in hepcidin affinity in the presence of iron, and further showed that the purified mutant proteins p.Asp325Asn, p.Cys326Ser, and p.His507Arg (where the C-domain iron-binding site is disrupted) bind iron weakly. The p.Ala350Val amino acid change might also alter the overall polar and hydrophobic properties of the C-domain hepcidin-binding site, as expected for the N-domain p.Arg40Gln and p.Ser47Phe substitutions (Figure S5).
A key difference between the various types of HC and FD is that iron overload does not develop in the same way. In HC, it appears in the blood compartment (reflected by an increase in transferrin saturation), without being immediately associated with iron accumulation in organs (which may correspond to a second stage in the progression of the disease), whereas in FD, it begins at the cellular level (reflected by high levels of serum ferritin), without being immediately associated with an increase in plasma iron. In the present study, hyperferritinemia was observed in all affected individuals. It was associated with low to slightly elevated transferrin saturation levels (Table 1). Transferrin saturation values were above 45% in 10 (of 12) affected individuals, the threshold commonly used for early detection of HC,30 but below 75%, the threshold corresponding to the accumulation of toxic non-transferrin bound iron in plasma. Free, unbound plasma iron has a strong propensity to penetrate various organs and induce tissue iron overload.31,42,43 As a matter of fact, transferrin saturation is generally well above the normal range in HFE-related HC-affected individuals with hyperferritinemia.44,45,46 Studying the spectra of disease-causing mutations in the SLC40A1 gene, we recently showed that the median transferrin saturation value was 33% in index cases with a LoF pathogenic variation, compared with 91% in index cases with a GoF pathogenic variation. The median values for serum ferritin were greater than 2,000 μg/L in both groups. We also documented a trend toward increased plasma iron in FD-affected individuals with the highest serum ferritin values.6 This observation, which highlights a certain degree of phenotypic heterogeneity, had previously been attributed to an exacerbation of FD, with iron escaping into the bloodstream and being more likely to cause mixed iron overload and tissue injury.8 Here, three men displayed serum ferritin concentrations >1,500 μg/L and transferrin saturations ranging from 55% to 67%. They also had strikingly increased HICs (≥230 μmol/g). A 53-year-old woman and her 22-year-old son had normal iron indices. This suggests incomplete penetrance.
The observed phenotypes do not appear to be the result of a balance between LoF and GoF properties. Rather, they are consistent with the unique iron export function of HsFPN1, which is considered a permanent action in different specialized cells of iron metabolism (primarily the macrophages) to fulfill the daily needs of erythropoiesis (among others), and the fact that hepcidin synthesis is closely related to circulating iron levels.2,47 Basically, the less HsFPN1 is able to export iron, the less the liver is expected to produce hepcidin to regulate the concentration of iron in plasma and its subsequent distribution in various preferred tissues. It also important to note that hepcidin binding to HsFPN1 is iron dependent.36 Therefore, it seems appropriate to ascribe the phenotypic expression of the ambivalent SLC40A1 missense mutations to the permanent state of HsPN1 inactivation and to consider the GoF situation as secondary to this state. A similar hypothesis was put forward by Praschberger and collaborators when they studied the p.Asp181Val missense variation in an English family whose affected individuals presented typical FD phenotypes.20 Further observations are, however, warranted to draw a more definitive conclusion on this point. A longitudinal assessment of serum hepcidin concentration in affected individuals with LoF, GoF, and ambivalent SLC40A1 mutations could prove particularly interesting; so far this information has only been collected in a 63-year-old woman with the p.Arg178Gln pathogenic variation.21
In conclusion, while FD is typically considered a SLC40A1 LoF disease and HC4 a SLC40A1 GoF disease, our results situate the p.Arg40Gln, p.Ser47Phe, and p.Ala350Val missense variations in an intermediate set of FD disease-causing mutations with both LoF and GoF properties. By bringing in vitro and in silico analyses into the clinical interpretation of rare missense variants, our results also provide mechanistic insights into FPN1-related iron export and hepcidin responsiveness. Further clarification of the role of FPN1 helices TM1, TM7, TM5, and TM8 will improve our knowledge of the structural and molecular biology of the MFS proteins, which are associated with various genetic and non-genetic diseases in adults and children,48 while continuing to improve our ability to classify single amino acid variants of unknown significance.
Data and code availability
The data supporting the present study have not yet been deposited in a public repository but are available from the corresponding author on request.
Acknowledgments
This work was supported by grants to G.L.G. from the French Hospital Clinical Research Program (Progamme Hospitalier de Recherche Clinique 2009; Brest University Hospital UF0857), the Gaétan Saleün Association and the GR-Ex Laboratory of Excellence (reference ANR-11-LABX-0051). The GR-Ex label is funded by the IdEx “Investissements d’avenir” program of the French National Research Agency (reference ANR-18-IDEX-0001). We thank Dr. Philippe Campeau for his advice.
Declaration of interests
The authors declare no competing interests.
Published Online: July 22, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100335.
Web resources
PDB, https://rcsb.org/.
Supplemental information
References
- 1.Donovan A., Lima C.A., Pinkus J.L., Pinkus G.S., Zon L.I., Robine S., Andrews N.C. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metabol. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Drakesmith H., Nemeth E., Ganz T. Ironing out Ferroportin. Cell Metabol. 2015;22:777–787. doi: 10.1016/j.cmet.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson G.J., Frazer D.M. Current understanding of iron homeostasis. Am. J. Clin. Nutr. 2017;106:1559S–1566S. doi: 10.3945/ajcn.117.155804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nemeth E., Tuttle M.S., Powelson J., Vaughn M.B., Donovan A., Ward D.M., Ganz T., Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 5.Zhang D.-L., Ghosh M.C., Ollivierre H., Li Y., Rouault T.A. Ferroportin deficiency in erythroid cells causes serum iron deficiency and promotes hemolysis due to oxidative stress. Blood. 2018;132:2078–2087. doi: 10.1182/blood-2018-04-842997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uguen K., Ka C., Collod-Béroud G., Le Tertre M., Guellec J., Férec C., Béroud C., Callebaut I., Le Gac G. The Spectra of Disease-Causing Mutations in the Ferroportin 1 (SLC40A1) Encoding Gene and Related Iron Overload Phenotypes (Hemochromatosis Type 4 and Ferroportin Disease) Hum. Mutat. 2023;2023 doi: 10.1155/2023/5162256. [DOI] [Google Scholar]
- 7.Vlasveld L.T., Janssen R., Bardou-Jacquet E., Venselaar H., Hamdi-Roze H., Drakesmith H., Swinkels D.W. Twenty Years of Ferroportin Disease: A Review or An Update of Published Clinical, Biochemical, Molecular, and Functional Features. Pharmaceuticals. 2019;12 doi: 10.3390/ph12030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piperno A., Pelucchi S., Mariani R. Inherited iron overload disorders. Transl. Gastroenterol. Hepatol. 2020;5:25. doi: 10.21037/tgh.2019.11.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pietrangelo A. Ferroportin disease: pathogenesis, diagnosis and treatment. Haematologica. 2017;102:1972–1984. doi: 10.3324/haematol.2017.170720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ka C., Guellec J., Perpermans X., Kannengiesser C., Ged C., Wuyts W., Cassiman D., de Ledinghen V., Varet B., de Kerguenec C., et al. The SLC40A1 R178Q mutation is a recurrent cause of hemochromatosis and is associated with a novel pathogenic mechanism. Haematologica 2018. 103(11):1796-1805doi:10.3324/haematol.2018.189845. [DOI] [PMC free article] [PubMed]
- 11.Guellec J., Elbahnsi A., Tertre M.L., Uguen K., Gourlaouen I., Férec C., Ka C., Callebaut I., Gac G.L. Molecular model of the ferroportin intracellular gate and implications for the human iron transport cycle and hemochromatosis type 4A. FASEB J. 2019;33:14625–14635. doi: 10.1096/fj.201901857R. [DOI] [PubMed] [Google Scholar]
- 12.Drakesmith H., Schimanski L.M., Ormerod E., Merryweather-Clarke A.T., Viprakasit V., Edwards J.P., Sweetland E., Bastin J.M., Cowley D., Chinthammitr Y., et al. Resistance to hepcidin is conferred by hemochromatosis-associated mutations of ferroportin. Blood. 2005;106:1092–1097. doi: 10.1182/blood-2005-02-0561. [DOI] [PubMed] [Google Scholar]
- 13.Aschemeyer S., Qiao B., Stefanova D., Valore E.V., Sek A.C., Ruwe T.A., Vieth K.R., Jung G., Casu C., Rivella S., et al. Structure-function analysis of ferroportin defines the binding site and an alternative mechanism of action of hepcidin. Blood. 2018;131:899–910. doi: 10.1182/blood-2017-05-786590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marger M.D., Saier M.H. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 15.Yan N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu. Rev. Biophys. 2015;44:257–283. doi: 10.1146/annurev-biophys-060414-033901. [DOI] [PubMed] [Google Scholar]
- 16.Fowler P.W., Orwick-Rydmark M., Radestock S., Solcan N., Dijkman P.M., Lyons J.A., Kwok J., Caffrey M., Watts A., Forrest L.R., Newstead S. Gating topology of the proton-coupled oligopeptide symporters. Structure. 2015;23:290–301. doi: 10.1016/j.str.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quistgaard E.M., Löw C., Guettou F., Nordlund P. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat. Rev. Mol. Cell Biol. 2016;17:123–132. doi: 10.1038/nrm.2015.25. [DOI] [PubMed] [Google Scholar]
- 18.Drew D., North R.A., Nagarathinam K., Tanabe M. Structures and General Transport Mechanisms by the Major Facilitator Superfamily (MFS) Chem. Rev. 2021;121:5289–5335. doi: 10.1021/acs.chemrev.0c00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Gac G., Ka C., Joubrel R., Gourlaouen I., Lehn P., Mornon J.-P., Férec C., Callebaut I. Structure-function analysis of the human ferroportin iron exporter (SLC40A1): effect of hemochromatosis type 4 disease mutations and identification of critical residues. Hum. Mutat. 2013;34:1371–1380. doi: 10.1002/humu.22369. [DOI] [PubMed] [Google Scholar]
- 20.Praschberger R., Schranz M., Griffiths W.J.H., Baumgartner N., Hermann M., Lomas D.J., Pietrangelo A., Cox T.M., Vogel W., Zoller H. Impact of D181V and A69T on the function of ferroportin as an iron export pump and hepcidin receptor. Biochim. Biophys. Acta. 2014;1842:1406–1412. doi: 10.1016/j.bbadis.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Viveiros A., Panzer M., Baumgartner N., Schaefer B., Finkenstedt A., Henninger B., Theurl I., Nachbaur K., Weiss G., Haubner R., et al. Reduced iron export associated with hepcidin resistance can explain the iron overload spectrum in ferroportin disease. Liver Int. 2020;40:1941–1951. doi: 10.1111/liv.14539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daher R., Kannengiesser C., Houamel D., Lefebvre T., Bardou-Jacquet E., Ducrot N., de Kerguenec C., Jouanolle A.-M., Robreau A.-M., Oudin C., et al. Heterozygous Mutations in BMP6 Pro-peptide Lead to Inappropriate Hepcidin Synthesis and Moderate Iron Overload in Humans. Gastroenterology. 2016;150:672–683.e4. doi: 10.1053/j.gastro.2015.10.049. [DOI] [PubMed] [Google Scholar]
- 23.Le Gac G., Gourlaouen I., Ka C., Férec C. The p.Leu96Pro Missense Mutation in the BMP6 Gene Is Repeatedly Associated With Hyperferritinemia in Patients of French Origin. Gastroenterology. 2016;151:769–770. doi: 10.1053/j.gastro.2016.03.054. [DOI] [PubMed] [Google Scholar]
- 24.Ka C., Le Gac G., Dupradeau F.-Y., Rochette J., Férec C. The Q283P amino-acid change in HFE leads to structural and functional consequences similar to those described for the mutated 282Y HFE protein. Hum. Genet. 2005;117:467–475. doi: 10.1007/s00439-005-1307-y. [DOI] [PubMed] [Google Scholar]
- 25.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Le Tertre M., Elbahnsi A., Ka C., Callebaut I., Le Gac G. Insights into the Role of the Discontinuous TM7 Helix of Human Ferroportin through the Prism of the Asp325 Residue. Int. J. Mol. Sci. 2021;22:6412. doi: 10.3390/ijms22126412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos P.C.J.L., Cançado R.D., Pereira A.C., Schettert I.T., Soares R.A.G., Pagliusi R.A., Hirata R.D.C., Hirata M.H., Teixeira A.C., Figueiredo M.S., et al. Hereditary hemochromatosis: mutations in genes involved in iron homeostasis in Brazilian patients. Blood Cells Mol. Dis. 2011;46:302–307. doi: 10.1016/j.bcmd.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Détivaud L., Island M.-L., Jouanolle A.-M., Ropert M., Bardou-Jacquet E., Le Lan C., Mosser A., Leroyer P., Deugnier Y., David V., et al. Ferroportin Diseases: Functional Studies, a Link Between Genetic and Clinical Phenotype. Hum. Mutat. 2013;34:1529–1536. doi: 10.1002/humu.22396. [DOI] [PubMed] [Google Scholar]
- 29.Landemaine A., Hamdi-Roze H., Cunat S., Loustaud-Ratti V., Causse X., Si Ahmed S.N., Drénou B., Bureau C., Pelletier G., De Kerguenec C., et al. A simple clinical score to promote and enhance ferroportin disease screening. J. Hepatol. 2022;76:568–576. doi: 10.1016/j.jhep.2021.10.022. [DOI] [PubMed] [Google Scholar]
- 30.Zoller H., Schaefer B., Vanclooster A., Griffiths B., Bardou-Jacquet E., Corradini E., Porto G., Ryan J., Cornberg M., European Association for the Study of the Liver Corrigendum to “EASL Clinical Practice Guidelines on haemochromatosis” [J Hepatol 2022 (77) 479-502] J. Hepatol. 2023;79:1341. doi: 10.1016/j.jhep.2023.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Le Lan C., Loréal O., Cohen T., Ropert M., Glickstein H., Lainé F., Pouchard M., Deugnier Y., Le Treut A., Breuer W., et al. Redox active plasma iron in C282Y/C282Y hemochromatosis. Blood. 2005;105:4527–4531. doi: 10.1182/blood-2004-09-3468. [DOI] [PubMed] [Google Scholar]
- 32.Schimanski L.M., Drakesmith H., Merryweather-Clarke A.T., Viprakasit V., Edwards J.P., Sweetland E., Bastin J.M., Cowley D., Chinthammitr Y., Robson K.J.H., Townsend A.R.M. In vitro functional analysis of human ferroportin (FPN) and hemochromatosis-associated FPN mutations. Blood. 2005;105:4096–4102. doi: 10.1182/blood-2004-11-4502. [DOI] [PubMed] [Google Scholar]
- 33.Létocart E., Le Gac G., Majore S., Ka C., Radio F.C., Gourlaouen I., De Bernardo C., Férec C., Grammatico P. A novel missense mutation in SLC40A1 results in resistance to hepcidin and confirms the existence of two ferroportin-associated iron overload diseases. Br. J. Haematol. 2009;147:379–385. doi: 10.1111/j.1365-2141.2009.07834.x. [DOI] [PubMed] [Google Scholar]
- 34.Fernandes A., Preza G.C., Phung Y., De Domenico I., Kaplan J., Ganz T., Nemeth E. The molecular basis of hepcidin-resistant hereditary hemochromatosis. Blood. 2009;114:437–443. doi: 10.1182/blood-2008-03-146134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiao B., Sugianto P., Fung E., Del-Castillo-Rueda A., Moran-Jimenez M.-J., Ganz T., Nemeth E. Hepcidin-induced endocytosis of ferroportin is dependent on ferroportin ubiquitination. Cell Metabol. 2012;15:918–924. doi: 10.1016/j.cmet.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Billesbølle C.B., Azumaya C.M., Kretsch R.C., Powers A.S., Gonen S., Schneider S., Arvedson T., Dror R.O., Cheng Y., Manglik A. Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms. Nature. 2020;586:807–811. doi: 10.1038/s41586-020-2668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tortosa V., Bonaccorsi di Patti M.C., Iacovelli F., Pasquadibisceglie A., Falconi M., Musci G., Polticelli F. Dynamical Behavior of the Human Ferroportin Homologue from Bdellovibrio bacteriovorus: Insight into the Ligand Recognition Mechanism. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21186785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi R., Kato H.E., Font J., Deshpande C.N., Wada M., Ito K., Ishitani R., Jormakka M., Nureki O. Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin. Nat. Commun. 2015;6:8545. doi: 10.1038/ncomms9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan Y., Ren Z., Gao S., Shen J., Wang L., Xu Z., Yu Y., Bachina P., Zhang H., Fan X., et al. Structural basis of ion transport and inhibition in ferroportin. Nat. Commun. 2020;11:5686. doi: 10.1038/s41467-020-19458-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deshpande C.N., Ruwe T.A., Shawki A., Xin V., Vieth K.R., Valore E.V., Qiao B., Ganz T., Nemeth E., Mackenzie B., Jormakka M. Calcium is an essential cofactor for metal efflux by the ferroportin transporter family. Nat. Commun. 2018;9:3075. doi: 10.1038/s41467-018-05446-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Latorraca N.R., Fastman N.M., Venkatakrishnan A.J., Frommer W.B., Dror R.O., Feng L. Mechanism of Substrate Translocation in an Alternating Access Transporter. Cell. 2017;169:96–107.e12. doi: 10.1016/j.cell.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brissot P., Ropert M., Le Lan C., Loréal O. Non-transferrin bound iron: a key role in iron overload and iron toxicity. Biochim. Biophys. Acta. 2012;1820:403–410. doi: 10.1016/j.bbagen.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 43.Cabantchik Z.I., Breuer W., Zanninelli G., Cianciulli P. LPI-labile plasma iron in iron overload. Best Pract. Res. Clin. Haematol. 2005;18:277–287. doi: 10.1016/j.beha.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Adams P.C., Reboussin D.M., Barton J.C., McLaren C.E., Eckfeldt J.H., McLaren G.D., Dawkins F.W., Acton R.T., Harris E.L., Gordeuk V.R., et al. Hemochromatosis and iron-overload screening in a racially diverse population. N. Engl. J. Med. 2005;352:1769–1778. doi: 10.1056/NEJMoa041534. [DOI] [PubMed] [Google Scholar]
- 45.Le Gac G., Scotet V., Gourlaouen I., L’Hostis C., Merour M.-C., Karim Z., Deugnier Y., Bardou-Jacquet E., Lefebvre T., Assari S., Ferec C. Prevalence of HFE-related haemochromatosis and secondary causes of hyperferritinaemia and their association with iron overload in 1059 French patients treated by venesection. Aliment. Pharmacol. Ther. 2022;55:1016–1027. doi: 10.1111/apt.16775. [DOI] [PubMed] [Google Scholar]
- 46.Allen K.J., Gurrin L.C., Constantine C.C., Osborne N.J., Delatycki M.B., Nicoll A.J., McLaren C.E., Bahlo M., Nisselle A.E., Vulpe C.D., et al. Iron-Overload–Related Disease in HFE Hereditary Hemochromatosis. N. Engl. J. Med. 2008;358:221–230. doi: 10.1056/NEJMoa073286. [DOI] [PubMed] [Google Scholar]
- 47.Hentze M.W., Muckenthaler M.U., Galy B., Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 48.Bai X., Moraes T.F., Reithmeier R.A.F. Structural biology of solute carrier (SLC) membrane transport proteins. Mol. Membr. Biol. 2017;34:1–32. doi: 10.1080/09687688.2018.1448123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the present study have not yet been deposited in a public repository but are available from the corresponding author on request.