Abstract
The rapid advancement of DNA foundation language models has revolutionized the field of genomics, enabling the decoding of complex patterns and regulatory mechanisms within DNA sequences. However, the current evaluation of these models often relies on fine-tuning and limited datasets, which introduces biases and limits the assessment of their true potential. Here, we present a benchmarking study of three recent DNA foundation language models, including DNABERT-2, Nucleotide Transformer version-2 (NT-v2), and HyenaDNA, focusing on the quality of their zero-shot embeddings across a diverse range of genomic tasks and species through analyses of 57 real datasets. We found that DNABERT-2 exhibits the most consistent performance across human genome-related tasks, while NT-v2 excels in epigenetic modification detection. HyenaDNA stands out for its exceptional runtime scalability and ability to handle long input sequences. Importantly, we demonstrate that using mean token embedding consistently improves the performance of all three models compared to the default setting of sentence-level summary token embedding, with average AUC improvements ranging from 4.3% to 9.7% for different DNA foundation models. Furthermore, the performance differences between these models are significantly reduced when using mean token embedding. Our findings provide a framework for selecting and optimizing DNA language models, guiding researchers in applying these tools effectively in genomic studies.
Introduction
Led by the advances in Natural Language Processing (NLP) in recent years, foundation language models through self-supervised pre-training have been the paradigm of decoding information in sequences. By representing sequences as numerical embeddings, foundation language models can outperform previous methods in many downstream tasks such as sequence classification and sequence generation. As natural language-based foundation models like GPT-4 [1], Llama2 [2], and Mistral [3] have been proven successful, similar ideas have been extended to other domains by interpreting domain-specific languages with unique semantic rules, and examples include foundation models on programming codes, protein sequences and single-cell sequencing [4-7]. With the long-lasting interests in decoding DNA sequences to understand the epigenetic patterns, transcriptional regulations, and disease associations [8,9], DNA foundation language models have also emerged recently including DNABERT-2 [10], Nucleotide Transformer [11] and HyenaDNA [12]. These models are pre-trained on large genomic datasets such as the human reference genome [13], human whole-genome sequencing datasets like 1000 Genomes project datasets [14], and multi-species genome datasets [11]. After fine-tuning, they have shown promising results in DNA sequence classification tasks.
A critical aspect of DNA foundation models is the method used to generate sequence embeddings, with sentence-level summary token and mean token embeddings being two primary approaches. The comparative efficacy of these embedding methods in DNA sequence analysis remains understudied, despite their potential impact on model performance.
With the rapid evolution of DNA foundation models of various architectures and the wide range of genomic analysis tasks to be solved, there is a pressing need for effectively evaluating these models. However, most of the current evaluations on DNA foundation models are biased, as they are conducted after fine-tuning [10-12], which may introduce biases in model performance comparison. For instance, different models may have various levels of overfitting depending on which layers are selected to update during fine-tuning. The use of advanced parameter-efficient fine-tuning methods [15-16], further complicates this issue by introducing additional hyperparameters that could impact model fitting. Conversely, a recent work directly compared DNA foundation models based on their output embeddings [30], where the weights in all layers were frozen and a trainable convolutional neural network (CNN) was appended to the last layer. While this approach mitigates fine-tuning biases, the study scope was limited to several human genome analysis tasks and did not account for potential effects of CNN hyperparameters. Moreover, it did not investigate the impact of different embedding methods on model performance. Therefore, it is also important to expand current evaluation to more diverse settings and investigate the inherent qualities of the pre-trained models without the confounding factors introduced by fine-tuning.
In this study, we provide a comprehensive and unbiased evaluation of existing state-of-the-art DNA foundation language models. Our evaluation is focused on the zero-shot embeddings—specifically, the last hidden states of the pre-trained models. These embeddings are crucial as they reflect the models' understanding of DNA sequences and are strongly linked to performance in downstream fine-tuning tasks. To objectively assess the quality of these embeddings, we employ a supervised learning approach using efficient tree-based models, which enables a thorough hyperparameter search while minimizing inductive biases. We collect datasets from a wide variety of genomic tasks across multiple species, where DNA sequences are labelled with biological traits such as the association with specific methylation sites and chromatin regions. This diverse set of benchmarking datasets allows us to examine the performance and generalizability of DNA foundation models across different domains and species. Furthermore, we conduct a formal comparative analysis of sentence-level summary token and mean token embedding methods, evaluating their impact on model performance across various genomic tasks. We also explore how different output pooling methods affect the quality of zero-shot embeddings and examine the influence of sequence length on DNA foundation model efficiency. Our evaluation framework allows for a comparative analysis of current models, illustrates the factors influencing performance, and discusses the strengths and limitations of DNA foundation models in genomic applications. All the codes and datasets untilized in this study are available at https://github.com/ChongWuLab/dna_foundation_benchmark.
Methods
DNA foundation language models
To evaluate DNA foundation language models comprehensively, we identified the three most recent state-of-the-art DNA foundation language models, including DNABERT-2 [10], Nucleotide Transformer version-2 [11], and HyenaDNA [12]. These foundation models take DNA sequence as input, tokenize into sequence of tokens, and generate embeddings of fixed dimension for each token after passing multiple layers. In the following, we will briefly describe these three models.
DNABERT-2 [10]
has the network architecture similar to Bidirectional Encoder Representations from Transformers (BERT) [17], which usually contains a positional embedding layer added to input embeddings, and a series of encoders each consisting of a multi-head self-attention layer and a feedforward network. It is pre-trained using the masked language modelling approach [17] on genomes from 135 species, including the human reference genome. DNABERT-2 tokenizes DNA sequences by the Byte Pair Encoding (BPE) method, which is an iterative algorithm that searches for nucleotides combinations and builds the vocabulary at the same time; it makes no assumption on fixed words and grammars, so each input sequence is independently tokenized merely based on its pattern. It is worth noting that the number of tokens in the tokenized sequence is not fixed in DNABERT-2. DNABERT-2 modifies the architecture of BERT by using Attention with Linear Biases (ALiBi) instead of positional embedding layer. DNABERT-2 has about 117 million trainable parameters, the output embedding dimension is 768. There is no hard limit on the input sequence length, although the runtime is still quadratically increasing with sequence length.
Nucleotide Transformer Version 2 (NT-v2) [10]
is also based on the BERT architecture, and it is pre-trained using the masked language modelling approach on genomes from 850 species, including the human reference genome. To tokenize DNA sequence, NT-v2 employs the 6-mers tokenization method that uses a sliding window of size 6 and reads every 6 nucleotides; if there are leftover elements at the end of sequence, nucleotides will be tokenized individually into {A, T, C, G, N}. Therefore, the number of tokens produced by the tokenizer will be approximately 1/6 of DNA sequence length. NT-v2 modifies BERT by replacing the learned positional embeddings with the rotary embeddings, which rotates the embeddings output by each attention layer based on the token’s position, and the Swish activation without bias. These modifications reduce the number of model parameters in Nucleotide Transformer Version 1, and thus reduce the computation cost. The largest NT-v2 model has around 500 million trainable parameters, the output embedding dimension is 1,024, and the input sequence length limit is 12,000 nucleotides.
HyenaDNA [12]
differs from the architectures of DNABERT-2 and NT-v2 by eschewing the attention mechanism in favor of a decoder-based architecture. HyenaDNA is pre-trained exclusively on the human reference genome using a next nucleotide prediction approach. The key component of this model is the Hyena operators, which integrate long convolutions with implicit parameterization and data-controlled gating. Benefiting from this architecture, HyenaDNA can process extremely long DNA sequences with fewer model parameters than attention-based transformer. This enables a straightforward tokenization approach in HyenaDNA, where each nucleotide is treated as an individual token. HyenaDNA can also perform in-context learning such as soft-prompting [12, 31], and details can be found in its original article. The largest HyenaDNA model has around 30 million trainable parameters, the output embedding dimension is 256, and the input sequence length limit is one million nucleotides.
Model Configuration Selection
It is worth noting that both NT-v2 and HyenaDNA offer multiple pre-trained model configurations, varying in the number of parameters, output dimensions, and input length limitations, detailed in Supplementary Table 1. In this study, we selected the NT-v2-500M model in the group of NT-v2 pre-trained models, as it is the largest in size and is deemed optimal in the original study of Nucleotide Transformer. For HyenaDNA, the number of layers, number of parameters in each layer, and the output dimensions are the same for Hyena-160K, Hyena-360K and Hyena-1.6M, and the only difference comes from the input layer that adapts for different maximum input lengths. Therefore, we chose the smallest one (Hyena-160K) among them for computation efficiency, because the longest sequence length in our benchmarking datasets does not exceed 160K nucleotides.
Benchmarking Datasets
To unbiasedly evaluate the foundation models, we first collected 17 public datasets from four DNA sequence classification tasks. These tasks and datasets were selected to reflect a wide range of potential downstream use cases, ensuring they are both challenging and achievable. The four DNA sequence classification tasks are the following:
4mc sites detection in multiple species [18]:
We used six datasets containing DNA sequences from the following six species correspondingly: Escherichia coli (E. coli), Caenorhabditis elegans (C. elegans), Geobacter pickeringii (G. pickeringii), Geoalkalibacter subterraneus (G. subterraneus), Drosophila melanogaster (D. melanogaster), and Arabidopsis thaliana (A. thaliana). Each sub-dataset is dedicated to predicting whether a DNA sequence contains a DNA N4-methylcytosine region (4mC) or not. For each species, the dataset consists of DNA sequences with annotated 4mC sites, and all sequences are 41 base pairs long, including 20 base pairs upstream and downstream of the 4mC site.
DNase-I hypersensitive sites detection [19]:
The datasets used in the study consist of positive DNA sequences for the 280 Dnase I hypersensitive sites (DHS), and negative sequences for the 737 non-Dnase I hypersensitive sites. Identification of the DNA sequences containing DHS is crucial for detecting DNA regulatory regions, as DHS is indicative of genomic regulatory regions like promoters, enhancers, silencers, and suppressors. The sequence length ranges from 225 to 275 base pairs.
5mC and 6mA modifications detection [20]:
We used two datasets of human DNA samples from this study, where the tasks are the detection of 5-methylcytosine (5mC) and N6-methyladenosine (6mA) modifications in DNA sequences, respectively. The positive samples in these datasets are defined by the presence of either 5mC or 6mA and all sequences are 41 base pairs long. To account for potential bias and redundancy, sequences with high similarity were excluded, resulting in 4688 samples for the 5mC dataset and 36670 samples for the 6mA dataset.
Promoter identification in multiple species [21]:
We use 8 datasets from 4 distinct species including human (4 different cell lines of GM12878, NHEK, HeLa-S3, HUVEC), B. amyloliquefaciens, R. capsulatus, and Arabidopsis (TATA and non-TATA). The positive samples in these datasets are promoter sequences. Negative samples were generated by identifying genomic sequences with maximal similarity to each positive promoter sequence, while ensuring no overlap with known positive regions. These datasets, especially the human cell lines datasets, include DNA sequences with significant variation in their lengths, and the maximum length can exceed 2000 base pairs.
Besides, we also adopt the genomic analysis datasets used in comparison studies from the original articles of DNABERT-2, NT-v2 and HyenaDNA. These datasets involve genomic analysis tasks on either binary classification or multiple classification. Along with the datasets we collected, there are in total 57 datasets included in this study. Detailed descriptions of the names and sources of all 57 datasets can be found in Supplementary Text. The specific training size, testing size, and details of sequence lengths for all datasets used in our study can be found in Supplementary Table 2.
To facilitate a systematic analysis, we categorized the 57 datasets into four distinct classes based on the nature of their respective classification tasks: 1) Human Genome Sequence Region Classification: This category encompasses tasks such as identification of transcription factor binding sites, promoter regions, and other functional elements within the human genome. 2) Multi-Species Genome Sequence Region Classification: These tasks involve distinguishing genomic regions across different species, for example, differentiating between human and Caenorhabditis elegans (worm) genome sequences. 3) Human Genome Epigenetic Trait Classification: This group includes tasks related to identifying epigenetic modifications specific to the human genome, such as detection of N4-methylcytosine (4mC) sites. 4) Multi-Species Genome Epigenetic Trait Classification: These tasks focus on identifying and classifying epigenetic traits across multiple species' genomes. We present and analyze our findings separately for each of these four dataset categories, allowing for a comprehensive assessment of the models' capabilities and limitations in various genomic classification scenarios.
Evaluation methods
Supervised learning evaluation
We evaluated the inherent quality of zero-shot embeddings by the separation of different classes. This was examined by the performance of supervised learning classifiers on the zero-shot embeddings. For each dataset, we first generated zero-shot embeddings for all sequences, then split the samples into training and testing sets, and finally trained a classifier and reported its performance predicting the labels of each sequence in the test set from their zero-shot embeddings. We maintained the training and testing split of datasets from their original works if available; otherwise, we randomly split the samples into a ratio of 7:3 for training and testing. For supervised learning task, we intitially tested XGBoost [32] and random forest [33] as these tree-based models require minimal tuning, allowing us to focus on evaluating the quality of the DNA foundation model embeddings rather than optimizing classifier performance. In our experiments, we noticed that random forest consistently outperformed XGBoost across all evaluation metrics, and thus we report only the random forest results to maintain clarity and conciseness in our analysis.
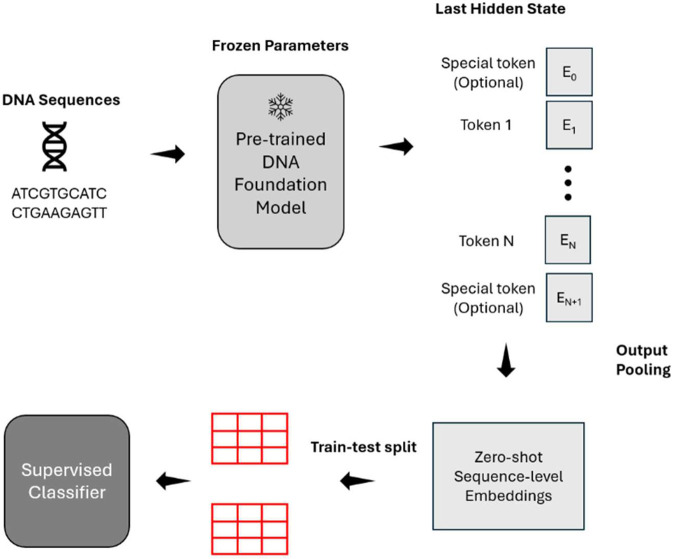
During training, we performed 5-fold cross-validation that divides the training set into five non-overlapping train-validation pairs for hyperparameters tuning, and then reported the testing performance on the test set. The hyperparameter grid is detailed in the Supplementary Table 3. We evaluated model performance on the test set using four metrics: Area Under the Curve (AUC), Matthews Correlation Coefficient (MCC) [29], F1 Score, and prediction accuracy. AUC serves as our primary measure of performance throughout this work, with the other metrics providing complementary information. To ensure rigorous comparison, we applied the DeLong’s test of AUC [34] to examine the statistical significance of differences in AUC values between models for each dataset. For the five datasets involving multi-class classification tasks, where DeLong’s test become less adaptable, we used classification accuracy as the primary metric instead. The detailed workflow of our method can be found in Figure 1.
Figure 1: Overview of DNA foundation models evaluation workflow.
DNA sequences are input into foundation models, generating token embeddings from the final layer. These embeddings undergo output pooling to produce high-dimensional representations of input sequences. A supervised classifier (random forest) is trained on these embeddings using labeled datasets. Model performance is evaluated on a held-out test set using multiple metrics, with AUC as the primary measure.
Benchmarking two pooling methods
We investigated the impact of output pooling methods on the quality of zero-shot embeddings in DNA sequence classification tasks. In foundation language models, output pooling methods refer to techniques for generating a single, fixed-dimensional embedding to represent an entire sequence [17]. We focused on two common output pooling methods in our evaluation: Sentence-level summary token method and mean pooling method.
Essentially, all the DNA foundation models in this study created additional special tokens during tokenization, alidning with common practice in natural language processing [23-25]. In the original BERT, the classify token “CLS” is appended to the start of every tokenized sequence. This CLS token serves as a sentence-level summary, capturing the context of the entire input sequence after processing through multiple self-attention layers. The embedding of the “CLS” token has been utilized for downstream fine-tuning and has proven effective [17]. In DNABERT-2 and NT-v2, the “CLS” token is created in the same way as in BERT. In HyenaDNA, the end of sequence token “EOS” is appended to the end of each tokenized sequence and has also been used as the output embedding to represent the whole sequence [12]. The mean pooling method, on the other hand, calculates the average of all the non-padding tokens in the tokenized sequence and uses it to represent the whole sequence. This approach provides an alternative representation that potentially captures information from all parts of the sequence equally.
In our study, we study the two pooling methods in two aspects: (1) we compare the performance across different DNA foundation models using the same pooling method; (2) we compare the performance across different pooling methods for the same DNA foundation model.
Runtime analysis
To evaluate computational efficiency, for each dataset, we measured the average time required for a single forward pass (i.e., an inference step) of an input batch for each model. Given the limited availability of GPUs and limited GPU memories, we conducted our experiments on CPUs. To ensure consistency across datasets, all experiments were performed on CPUs with an identical configuration of 20 cores, and the batch size was fixed at 256.
We also adjusted the tokenizer configurations in the models to roughly match sequence lengths; for example, the dataset of human genome promoter region classification [21] contained sequence length up to 2999 base pairs, so the 6-mers based tokenizer of NT-v2 took a maximum of 600 tokens and the single-nucleotide tokenizer of HyenaDNA took 2999 tokens. The tokenizer of DNABERT-2 generated an uncertain number of tokens, so we estimate it conservatively to ensure there is no truncated sequence causing information loss. This conservative approach may result in a slight inflation in DNABERT-2 runtime.
Results
Based on the four categories of DNA sequence classification tasks, we performed a thorough benchmark across multiple aspects.
Human genome sequence region classification
We evaluated the performance of the three DNA foundation models, DNABERT-2, NT-v2, and HyenaDNA, on a diverse set of human genome sequence region classification tasks. For the human genome sequence region classification tasks, as shown in Table 1, when using sentence-level summary token pooling method, all three models achieved AUC scores above 0.8 on the majority of tasks, indicating their ability to capture meaningful semantic information from human DNA sequences. These results show that zero-shot embeddings generated by these models are sufficiently informative for supervised learning models, even without fine-tuning. Among the three models, DNABERT-2 exhibited superior overall performance across multiple metrics, including AUC, MCC, F1 score, and accuracy (Supplementary Figure 1). Specifically, DNABERT-2 outperformed the NT-v2 and HyenaDNA by an average of 3.6% and 5.9% in AUC scores across all datasets. To assess the statistical significance of these performance differences, we conducted DeLong's test for AUC comparisons. DNABERT-2's superior performance was statistically significant (p < 0.01) in 11 out of the 24 tasks while NV-v2’s superior performance was statistically significant in 4 out of the 24 tasks (Table 1). When using the mean pooling method, the performance gap narrowed, but DNABERT-2 still maintained a lead in average AUC (Supplementary Table 3). Now DNABERT-2 has statistically significant (p < 0.01) highest AUC in 7 out of the 24 tasks, while NT-v2 has 4 and HyenaDNA has 1. Notably, DNABERT-2 excelled in promoter identification tasks for cell lines GM12878, HUVEC, Hela-S3, and NHEK. In these tasks, it achieved AUC scores of 0.964, 0.974, 0.971, and 0.912 respectively, which are comparable to those reported in the original studies where models were fully trained on the respective datasets [21]. This performance is particularly impressive given that our models were not fine-tuned for these specific tasks.
Table 1:
The AUC results for binary sequence classification tasks on human genome. The tasks include promoter region identification (multiple datasets), coding region detection, splice site donor and acceptor identification, enhancer identification (multiple datasets), transcription factor binding site identification (multiple datasets), and open chromatin region identification (multiple datasets). Using sentence-level summary token pooling method. Largest values row-wise are bolded. **DeLong Test significance < 0.01. Bolded value: DeLong Test significance < 0.05.
| Data | DNABERT-2 | NT-v2 | HyenaDNA |
|---|---|---|---|
| Promoter GM12878 | 0.964** | 0.878 | 0.884 |
| Promoter HUVEC | 0.974** | 0.912 | 0.906 |
| Promoter Hela-S3 | 0.971** | 0.909 | 0.9 |
| Promoter NHEK | 0.912** | 0.855 | 0.854 |
| Promoter NonTATA 251 bps | 0.861 | 0.834 | 0.853 |
| Promoter NonTATA 70 bps | 0.816 | 0.838** | 0.79 |
| Promoter TATA 70 bps | 0.809 | 0.872** | 0.732 |
| Promoter All 70 bps | 0.803 | 0.822** | 0.769 |
| Promoter NonTATA 300 bps | 0.938** | 0.91 | 0.818 |
| Promoter TATA 300 bps | 0.698 | 0.694 | 0.717 |
| Promoter All 300 bps | 0.897** | 0.875 | 0.797 |
| Coding | 0.915** | 0.863 | 0.885 |
| Donor | 0.823** | 0.636 | 0.626 |
| Acceptor | 0.793** | 0.632 | 0.67 |
| Enhancer | 0.863 | 0.879 | 0.833 |
| Enhancer Cohn | 0.792** | 0.728 | 0.733 |
| Enhancer Ensembl | 0.947 | 0.95** | 0.944 |
| TFBS Data 1 | 0.817 | 0.824 | 0.83 |
| TFBS Data 2 | 0.834 | 0.836 | 0.842 |
| TFBS Data 3 | 0.744 | 0.751 | 0.741 |
| TFBS Data 4 | 0.66 | 0.663 | 0.624 |
| TFBS Data 5 | 0.785 | 0.801 | 0.787 |
| Open chromatin region | 0.685** | 0.657 | 0.665 |
| DNase_I Hypersensitive | 0.815 | 0.806 | 0.787 |
Multispecies genome sequence region classification
To evaluate the cross-species generalizability of DNA foundation models, we assessed their performance on multispecies genome sequence region classification tasks. Using the summary token pooling method, DNABERT-2 demonstrates superior performance in terms of AUC (Table 2). DNABERT-2 achieved an average AUC of 0.860 across these tasks, outperforming NT-v2 (mean AUC: 0.802) and HyenaDNA (mean AUC: 0.731) by 7.2% and 17.6%, respectively. The performance advantage of DNABERT-2 was statistically significant (p < 0.01, DeLong's test) in 4 out of 6 tasks. Interestingly, when employing the mean pooling method, we observed a shift in relative performance. HyenaDNA's performance improved markedly, achieving a mean AUC of 0.857, compared to DNABERT-2's 0.866 and NT-v2's 0.856 (Supplementary Table 3). This improvement was particularly notable in the Arabidopsis promoter identification tasks, where HyenaDNA outperformed the other models by 1.0-1.5% in AUC. This observation suggests that HyenaDNA's capabilities are not substantially constrained by its pre-training exclusively on the human genome, and that mean pooling may be particularly effective in leveraging its learned representations across species. Furthermore, when using mean pooling, all models show only modest decreases in performance compared to similar human genome classification tasks. The average performance drop was only 0.9% for DNABERT-2, 0.7% for NT-v2, and 0.1% for HyenaDNA. These results suggest that DNA foundation models demonstrate great potential for effective application in analyzing genomic sequences across multiple species.
Table 2:
The AUC results for binary sequence classification tasks which have multi-species involved, including promoter region prediction (first four rows), human vs worm classification and mouse transcription factor binding site (TFBS) identification. The results for mouse TFBS are averaged over 5 independent datasets focusing on different TFBSs. Using sentence-level summary token pooling method. **DeLong Test significance < 0.01. Bolded value: DeLong Test significance < 0.05.
| Data | DNABERT-2 | NT-v2 | HyenaDNA |
|---|---|---|---|
| Promoter B_amyloliquefaciens | 0.856** | 0.797 | 0.688 |
| Promoter R_capsulatus | 0.661 | 0.668 | 0.602 |
| Promoter Arabidopsis NonTATA | 0.891** | 0.85 | 0.814 |
| Promoter Arabidopsis TATA | 0.903** | 0.855 | 0.82 |
| Human vs worm | 0.946** | 0.919 | 0.837 |
| Mouse TFBS | 0.700 | 0.722 | 0.624 |
Human epigenetic modification & Multispecies epigenetic modification prediction
We further investigated the performance of the models on human epigenetic modification prediction tasks, specifically the detection of 5-methylcytosine (5mC) and N6-methyladenosine (6mA) modifications. NT-v2 outperformed DNABERT-2 and HyenaDNA by an average of 4% and 14% in AUC using summary token pooling, and an average of 3.8% and 4.2% using mean pooling (Table 3, Supplementary Table 3). For epigenetic trait detection tasks on multi-species, NT-v2 is still leading clearly under mean pooling (Supplementary Table 3). It is also notable that, compared to the genome sequence region classification tasks, all models experienced a decline in AUCs in epigenetic modification prediction tasks under both pooling methods. This trend aligns with our expectation, as the information encoding epigenetic modifications in DNA sequences is likely to be more subtle and complex than the information distinguishing different genome regions.
Table 3:
The AUC results for each model on datasets which aim to detect the epigenetic modifications. Using sentence-level summary token pooling method. **DeLong Test significance < 0.01. Bolded value: DeLong Test significance < 0.05.
| Data | DNABERT-2 | NT-v2 | HyenaDNA |
|---|---|---|---|
| 5-methylcytosin (5mC) | 0.678 | 0.713 | 0.604 |
| N6-methyladenosine (6mA) | 0.731 | 0.752** | 0.681 |
It is also noteworthy that HyenaDNA achieved comparable accuracy to DNABERT-2 and NT-v2 in classification tasks with more than two classes (Table 5). HyenaDNA demonstrated comparable, and in some cases superior, performance to DNABERT-2 and NT-v2 in these multi-class classification tasks. For example, in the Regulatory Region Type classification task, HyenaDNA significantly outperformed the other models, achieving an accuracy of 70.2%, compared to 63.0% for DNABERT-2 and 55.5% for NT-v2. This observation suggests that HyenaDNA’s architecture may be particularly well-suited for capturing complex patterns and distinguishing between multiple classes.
Table 5:
The accuracy for all multi-class classification datasets in this study. Bolded value: row maximum.
| Data | DNABERT-2 | NT-v2 | HyenaDNA |
|---|---|---|---|
| Enhancer Strength | 0.515 | 0.471 | 0.485 |
| Splice Site Type, NT | 0.712 | 0.725 | 0.71 |
| Splice Site Type, DNABERT-2 | 0.608 | 0.607 | 0.607 |
| Covid Variants | 0.446 | 0.43 | 0.449 |
| Regulatory Region Type | 0.63 | 0.555 | 0.702 |
Pooling methods comparison
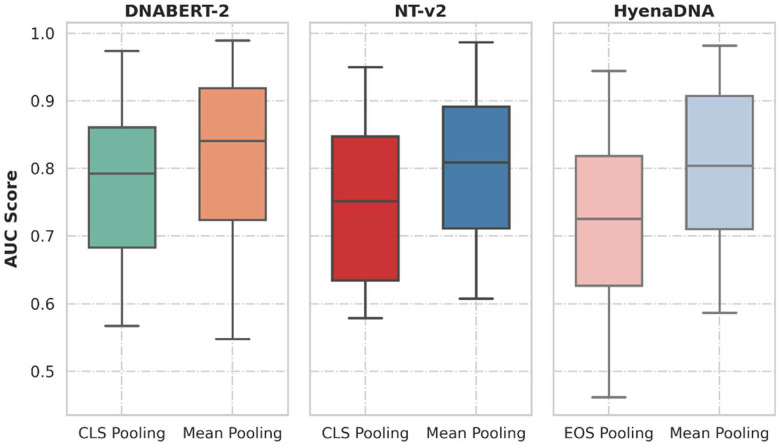
Our evaluation of the two pooling methods, sentence-level summary token ([CLS] or [EOS]) embedding and mean token embedding, revealed significant differences in the performance of DNA foundation models. Despite sentence-level summary token embedding being the default choice in most DNA foundation models, we observed that mean token embedding consistently improved the performance of DNABERT-2, NT-v2, and HyenaDNA across all task categories (Supplementary Tables 4-6). Figure 2 illustrates the improvement in AUC scores for all models when using mean token embedding. The average increase in AUC was 4.3% (interquartile range: 1.6%-6.1%) for DNABERT-2, 6.9% (interquartile range: 3.8%-8.4%) for NT-v2, and 9.7% (interquartile range: 6.3%-13.0%) for HyenaDNA across all tasks. This consistent enhancement indicates the superiority of mean token embedding over the currently favored summary-level token embedding.
Figure 2:
Boxplots comparing the AUC scores distribution over all datasets included in this study, on the choice of using mean output pooling or using summary-level token pooling.
The improved performance with mean token embedding suggests that this pooling method more effectively captures the overall information contained in the DNA sequences. By averaging the embeddings of all non-padding tokens, mean token embedding provides a more comprehensive representation of the entire sequence, as opposed to relying on a single summary token. This finding is particularly relevant for simple DNA sequence classification tasks, such as promoter identification and enhancer identification, where the discriminative features may be distributed throughout the sequence rather than concentrated in a specific region. For instance, in the promoter identification task for the GM12878 cell line, mean token embedding improved the AUC from 0.964 to 0.985 for DNABERT-2, a 2.2% increase (Supplementary Table 4). More strikingly, for the B.amyloliquefaciens genome, the improvement was from 0.688 to 0.862 for HyenaDNA, representing a 25.3% increase (Supplementary Table 6). These examples highlight how mean token embedding can capture distributed features more effectively across the entire sequence.
Moreover, the reduced performance differences among the models when using mean token embedding (Figure 2) imply that this pooling method helps to mitigate the architectural variations across the models. Specifically, the mean token embedding results in a generally lower difference of AUC scores across models. For example, in the coding region classification task, the range of AUC scores differences across models narrowed from 0.052 (0.915-0.863) with summary token pooling to 0.015 (0.944-0.929) with mean token pooling. Taking average over all datasets, the AUC scores differences decreased from 0.063 with summary token pooling to 0.032 with mean pooling. This observation underscores the importance of carefully selecting the pooling method when evaluating and comparing DNA foundation models, as it can significantly impact the assessment of their relative strengths and weaknesses.
Given the consistent improvement in performance across all models and task categories, we recommend using mean token embedding as the default pooling method for generating zero-shot embeddings in DNA sequence classification tasks, rather than the currently popular sentence-level summary token embedding. This recommendation may extend to the selection of pooling methods for fine-tuning, as the choice of pooling method during pre-training can influence the quality of the learned representations and, consequently, the performance of the fine-tuned models. However, further research is needed to validate this hypothesis in fine-tuning scenarios, and we leave such exciting topic to future research.
Runtime and user-friendliness analysis
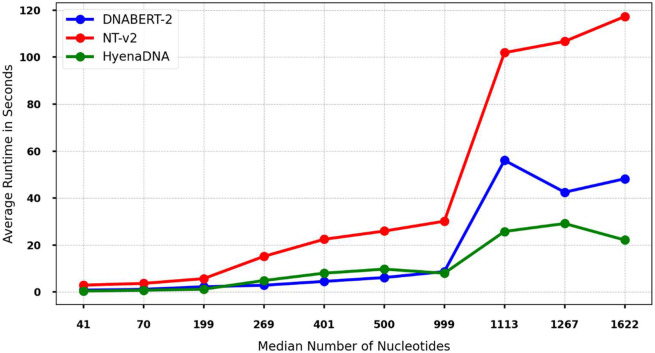
We conducted a runtime analysis by measuring the average time cost for a forward pass on each dataset using a fixed batch size of 256. Given the relatively small model sizes, inference was performed on CPUs to save our limited GPU resources. Figure 3 illustrates the average runtime with respect to the increase in median sequence length for each model. For the purpose of visualization, we selected a subset of datasets.
Figure 3:
The average runtime of passing a batch of 256 DNA sequences for selected datasets. X-axis: the median sequence length of the dataset. Y-axis: the average runtime in seconds. Runtime is recorded on the same set of CPUs with the same cores.
Our analysis revealed that HyenaDNA exhibits the most scalable runtime trend, corroborating its remarkable ability to process longer sequences efficiently. This scalability can be attributed to HyenaDNA's architecture, which leverages Hyena operators to integrate long convolutions with implicit parameterization and data-controlled gating. HyenaDNA demonstrated a scalable increase in runtime with sequence length, while NT-v2 and DNABERT-2 both exhibited a sharp runtime increase when the median sequence length exceeded 1000 base pairs. Specifically, we observed a 3.99 times increase in runtime for NT-v2 and a 4.92 times increase for DNABERT-2 when the median sequence length increased from 999 to 1,113 base pairs. In addition, NT-v2 consistently required the highest runtime among the three models, primarily due to its larger model size (500M parameters compared to 117M for DNABERT-2 and 30M for HyenaDNA). On average, NT-v2's runtime was 3.32 times that of DNABERT-2 and 4.37 times that of HyenaDNA across all sequence lengths. Despite these differences, we did not observe a obvious superiority in terms of runtime polynomial complexity between the models.
In addition to computational efficiency, we evaluated the user-friendliness of the DNA foundation models. All three models are implemented in PyTorch and have been integrated into the Hugging Face platform, enabling users to leverage various computationally efficient techniques, such as flash attention and mixed precision training. However, we encountered disparities in the ease of integration with parameter-efficient fine-tuning (PEFT) libraries. While DNABERT-2 and NT-v2 can be seamlessly integrated with PEFT and benefit from state-of-the-art algorithms for both classification and regression tasks, HyenaDNA currently lacks an efficient fine-tuning algorithm beyond updating the entire model, owing to its unique architecture.
Furthermore, we noticed that none of the models provides a straightforward way to switch from the default sentence-level summary token embedding to mean token embedding pooling. To perform the analysis using mean embedding pooling, we had to manually extract the padding tokens of sequences and calculate the mean embeddings. This limitation highlights the need for more flexible and user-friendly implementations of DNA foundation models, which would allow researchers to easily explore different pooling methods and adapt the models to their specific requirements.
Discussion
Our comprehensive study evaluates state-of-the-art DNA foundation models by focusing on the quality of their zero-shot embeddings in DNA sequence classification tasks. The analysis reveals that DNABERT-2 exhibits the most consistent performance across various datasets, demonstrating its robustness and reliability. HyenaDNA stands out for its exceptional runtime scalability and ability to handle extensive input lengths while maintaining competitive performance on human genome-related tasks and multi-class classification problems. In contrast, NT-v2 generates zero-shot embeddings that are most suitable for epigenetic modification detection tasks. Notably, all models show reduced embedding quality on non-human genome datasets compared to human datasets, and perform worse on epigenetic modification prediction tasks compared to DNA sequence classification tasks. Despite these challenges, for some simpler tasks such as promoter identification in human cell lines (e.g., GM12878 and HUVEC), the performance of zero-shot embeddings combined with random forest classifiers approaches that of task-specific models, highlighting the potential of DNA foundation models. This observation suggests possible synergies across different models, where their complementary strengths could be leveraged to improve overall performance across a wider range of genomic tasks.
Our comprehensive evaluation of DNA foundation models not only provides insights into their technical performance but also has important implications for biological and clinical research. For instance, in the task of predicting presence of N4-methylcytosine site in C.elegans DNA sequences, we observed that NT-v2 outperformed other models. This superior performance suggests that NT-v2 could be particularly valuable for studying epigenetic regulation in nematodes and potentially other organisms. Similarly, DNABERT-2's consistent performance across human genome-related tasks, such as promoter and enhancer identification, positions it as a powerful tool for understanding gene regulation in human diseases. The ability of these models to capture complex genomic patterns without task-specific training opens new avenues for discovering novel biomarkers and regulatory elements. For example, applying these models to cancer genomics could potentially identify previously unknown regulatory regions associated with tumor progression. Furthermore, HyenaDNA's capability to handle long input sequences efficiently makes it particularly suitable for analyzing large-scale genomic rearrangements or long-range interactions, which are often implicated in genetic disorders. By benchmarking these models across diverse genomic tasks, our study provides guidance for researchers to select the most appropriate model for their specific biological questions, potentially accelerating discoveries in fields ranging from developmental biology to personalized medicine.
A key strength of our approach lies in its broader and more diverse set of comparisons, which evaluates zero-shot embeddings from pre-trained models across a wide range of genomic tasks and species. This extensive dataset collection enables a more thorough and unbiased assessment of DNA foundation models' capabilities compared to existing studies. Moreover, our methodology introduces a novel evaluation criterion by utilizing efficient tree-based models for hyperparameter optimization, minimizing inductive biases, and offering unbiased tools and datasets to evaluate DNA foundation models. A significant contribution of our study is the comprehensive analysis of different pooling methods. We demonstrate that mean token embedding consistently outperforms the commonly used sentence-level summary token embedding across all models and tasks, with average AUC improvements ranging from 4.2% to 7.1%. This finding challenges the current default practice in DNA foundation models and provides a clear direction for improving model performance in genomic sequence analysis tasks.
Despite the comprehensive nature of our analysis, we acknowledge several limitations that warrant further exploration. Our current benchmarks are confined to zero-shot embeddings for sequence classification tasks and do not include regression-based tasks. Additionally, while fine-tuning may introduce biases in evaluating foundation models, there is still a need to investigate the fine-tuning potential of different models appropriately. For instance, NT-v2, with its larger size (500M parameters) compared to DNABERT-2 (117M) and HyenaDNA (30M), may exhibit more significant improvement when fine-tuned for specific applications. Another limitation is the scope of biological applications explored in our benchmarking. While we cover a range of genomic tasks, there are important areas that require further investigation. For instance, we have not extensively explored the potential of these models in identifying and classifying variants involved in human disease, particularly in non-coding regions. This application could have significant implications for understanding complex genetic disorders and advancing personalized medicine. Future studies should aim to incorporate a broader range of tasks, including regression, and develop more sophisticated benchmarking methods that unbiasedly account for the effects of fine-tuning. Additionally, direct comparisons with traditional genomic analysis methods would provide a more comprehensive understanding of the strengths and weaknesses of DNA foundation models. Exploring model ensembles that leverage the complementary strengths of different architectures may also prove fruitful in addressing the diverse challenges in genomic sequence analysis.
In conclusion, our study provides a comprehensive evaluation framework for DNA foundation models, offering insights into their strengths, limitations, and potential areas for improvement. The findings presented here can guide researchers in selecting appropriate models for specific genomic tasks and highlight promising directions for future development in this rapidly evolving field.
Supplementary Material
Table 4:
The AUC results for each model on epigenetic modification detection in multispecies genome. The results for yeast are averaged over 7 different datasets focusing on different epigenetic marks. Using sentence-level summary token pooling method. **DeLong Test significance < 0.01. Bolded value: DeLong Test significance < 0.05.
| Data | DNABERT-2 | NT-v2 | HyenaDNA |
|---|---|---|---|
| A.Thaliana 4mC | 0.59 | 0.6** | 0.557 |
| C.Elegans 4mC | 0.587 | 0.594 | 0.583 |
| D.Melanogaster 4mC | 0.604 | 0.611 | 0.57 |
| E.Coli 4mC | 0.567 | 0.579 | 0.579 |
| G.Pickeringii 4mC | 0.587 | 0.607 | 0.603 |
| G.Subterraneus 4mC | 0.588 | 0.581 | 0.577 |
| Yeast Epigenetic Marks | 0.734** | 0.643 | 0.665 |
Acknowledgements
This work is partially supported by grants from the National Institutes of Health R01CA263494, P30CA016672 and P50CA217674, and Cancer Prevention & Research Institute of Texas (CPRIT) grant RP230166. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and Cancer Prevention & Research Institute of Texas.
References
- [1].OpenAI et al. GPT-4 Technical Report. Preprint at 10.48550/arXiv.2303.08774 (2024). [DOI] [Google Scholar]
- [2].Touvron H. et al. Llama 2: Open Foundation and Fine-Tuned Chat Models. Preprint at 10.48550/arXiv.2307.09288 (2023). [DOI] [Google Scholar]
- [3].Jiang A. Q. et al. Mistral 7B. Preprint at 10.48550/arXiv.2310.06825 (2023). [DOI] [Google Scholar]
- [4].Chen M. et al. Evaluating Large Language Models Trained on Code. Preprint at 10.48550/arXiv.2107.03374 (2021). [DOI] [Google Scholar]
- [5].Madani A. et al. Large language models generate functional protein sequences across diverse families. Nat Biotechnol 41, 1099–1106 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cui H. et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat Methods 1–11 (2024) doi: 10.1038/s41592-024-02201-0. [DOI] [PubMed] [Google Scholar]
- [7].Lin Z. et al. Language models of protein sequences at the scale of evolution enable accurate structure prediction. Preprint at 10.1101/2022.07.20.500902 (2022). [DOI] [Google Scholar]
- [8].Gershman A. et al. Epigenetic patterns in a complete human genome. Science 376, eabj5089 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang G. et al. Understanding Transcription Factor Regulation by Integrating Gene Expression and DNase I Hypersensitive Sites. Biomed Res Int 2015, 757530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhou Z. et al. DNABERT-2: Efficient Foundation Model and Benchmark For Multi-Species Genome. Preprint at 10.48550/arXiv.2306.15006 (2024). [DOI] [Google Scholar]
- [11].Dalla-Torre H. et al. The Nucleotide Transformer: Building and Evaluating Robust Foundation Models for Human Genomics. Preprint at 10.1101/2023.01.11.523679 (2023). [DOI] [PubMed] [Google Scholar]
- [12].Nguyen E. et al. HyenaDNA: Long-Range Genomic Sequence Modeling at Single Nucleotide Resolution. Preprint at 10.48550/arXiv.2306.15794 (2023). [DOI] [Google Scholar]
- [13].Genome Reference Consortium. Genome reference consortium human build 38 (grch38). National Center for Biotechnology Information, 2013. URL https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.26/ [Google Scholar]
- [14].Byrska-Bishop M., Evani U. S., Zhao X., Basile A. O., Abel H. J., Regier A. A., Corvelo A., Clarke W. E., Musunuri R., Nagulapalli K., et al. , “High-coverage whole-genome sequencing of the expanded 1000 genomes project cohort including 602 trios,” Cell, vol. 185, no. 18, pp. 3426–3440, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hu E. J. et al. LoRA: Low-Rank Adaptation of Large Language Models. Preprint at 10.48550/arXiv.2106.09685 (2021). [DOI] [Google Scholar]
- [16].Liu H. et al. Few-Shot Parameter-Efficient Fine-Tuning is Better and Cheaper than In-Context Learning. Preprint at 10.48550/arXiv.2205.05638 (2022). [DOI] [Google Scholar]
- [17].Devlin J., Chang M.-W., Lee K. & Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. Preprint at 10.48550/arXiv.1810.04805 (2019). [DOI] [Google Scholar]
- [18].Xu H., Jia P. & Zhao Z. Deep4mC: systematic assessment and computational prediction for DNA N4-methylcytosine sites by deep learning. Briefings in Bioinformatics 22, bbaa099 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu B., Long R. & Chou K.-C. iDHS-EL: identifying DNase I hypersensitive sites by fusing three different modes of pseudo nucleotide composition into an ensemble learning framework. Bioinformatics 32, 2411–2418 (2016). [DOI] [PubMed] [Google Scholar]
- [20].Jin J. et al. iDNA-ABF: multi-scale deep biological language learning model for the interpretable prediction of DNA methylations. Genome Biology 23, 219 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang P., Zhang H. & Wu H. iPro-WAEL: a comprehensive and robust framework for identifying promoters in multiple species. Nucleic Acids Research 50, 10278–10289 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Grinsztajn L., Oyallon E. & Varoquaux G. Why do tree-based models still outperform deep learning on typical tabular data? Advances in Neural Information Processing Systems 35, 507–520 (2022). [Google Scholar]
- [23].Gillioz A., Casas J., Mugellini E. & Khaled O. A. Overview of the Transformer-based Models for NLP Tasks. in Annals of Computer Science and Information Systems vol. 21 179–183 (2020). [Google Scholar]
- [24].Zhang H. & Shafiq M. O. Survey of transformers and towards ensemble learning using transformers for natural language processing. Journal of Big Data 11, 25 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang X., Huang J. Y., Zhou W. & Chen M. Parameter-Efficient Tuning with Special Token Adaptation. Preprint at 10.48550/arXiv.2210.04382 (2023). [DOI] [Google Scholar]
- [26].Hubert L. & Arabie P. Comparing partitions. Journal of Classification 2, 193–218 (1985). [Google Scholar]
- [27].Vinh N. X., Epps J. & Bailey J. Information Theoretic Measures for Clusterings Comparison: Variants, Properties, Normalization and Correction for Chance. Journal of Machine Learning Research 11, 2837–2854 (2010). [Google Scholar]
- [28].Rousseeuw P. J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics 20, 53–65 (1987). [Google Scholar]
- [29].Matthews B. W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochimica et Biophysica Acta (BBA) - Protein Structure 405, 442–451 (1975). [DOI] [PubMed] [Google Scholar]
- [30].Marin F. I. et al. BEND: Benchmarking DNA Language Models on biologically meaningful tasks. Preprint at 10.48550/arXiv.2311.12570 (2024). [DOI] [Google Scholar]
- [31].Lester B., Al-Rfou R. & Constant N. The Power of Scale for Parameter-Efficient Prompt Tuning. Preprint at 10.48550/arXiv.2104.08691 (2021). [DOI] [Google Scholar]
- [32].Chen T. & Guestrin C. XGBoost: A Scalable Tree Boosting System. in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (Association for Computing Machinery, New York, NY, USA, 2016). doi: 10.1145/2939672.2939785. [DOI] [Google Scholar]
- [33].Breiman L. Random Forests. Machine Learning 45, 5–32 (2001). [Google Scholar]
- [34].DeLong Elizabeth R., et al. “Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach.” Biometrics, vol. 44, no. 3, 1988, pp. 837–45. JSTOR, 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.