Abstract
Research organisms provide invaluable insights into human biology and diseases, serving as essential tools for functional experiments, disease modeling, and drug testing. However, evolutionary divergence between humans and research organisms hinders effective knowledge transfer across species. Here, we review state-of-the-art methods for computationally transferring knowledge across species, primarily focusing on methods that utilize transcriptome data and/or molecular networks. We introduce the term “agnology” to describe the functional equivalence of molecular components regardless of evolutionary origin, as this concept is becoming pervasive in integrative data-driven models where the role of evolutionary origin can become unclear. Our review addresses four key areas of information and knowledge transfer across species: (1) transferring disease and gene annotation knowledge, (2) identifying agnologous molecular components, (3) inferring equivalent perturbed genes or gene sets, and (4) identifying agnologous cell types. We conclude with an outlook on future directions and several key challenges that remain in cross-species knowledge transfer.
Introduction
The use of research organisms, also known as model species or model systems, is essential to biomedical research [1]. Leveraging their resemblance in anatomy, physiology, behavior and genetics to corresponding human conditions, research organisms help scientists investigate a wide range of biomedical phenomena and therapeutic treatments before they are applied to humans. For instance, the zebrafish (Danio rerio) is a well-established vertebrate research organism that has external and fast development in large numbers, transparent embryos, and allows for easy genetic manipulation and drug administration through compound exposure [2].
There are two main reasons that research organisms are critical to studying human phenotypes, processes, and diseases. First, it is often unethical to study biomedical processes or apply novel therapeutic treatments directly in humans. Although there are many unanswered questions on the ethics of using research organisms [3,4,5], the knowledge derived from these organisms has helped save countless human lives [2,6]. Secondly, genetic studies in humans typically have very high variability due to confounding effects such as population genetics and environmental factors [7,8,9,10], and these confounders can be controlled much more in laboratory research with research organisms.
However, using research organism data to explore human biology presents its own challenges. Evolutionary divergence often gives rise to similar yet different underlying biological processes between human and research organisms [11], thereby impeding the transfer of knowledge across species. Even orthologs (i.e. homologous genes across species), which have taken a privileged role in transferring functional annotations across species, may have significant functional changes across species [12,13,14,15,16,17]. Additionally, the complex genetics underlying phenotypes/processes shared across species may differ. This is because interactions among genes responsible for a phenotype could be rewired during the evolutionary process, potentially encompassing some species-specific genes [18]. Consequently, the effective and precise transfer of knowledge considering species-specific genes remains challenging. Finally, since no single research organism can fully recapitulate the entirety of a complex biological condition in humans [1], how do we predict which research organisms capture the different facets of the human biology of interest?
To address these issues, researchers have begun developing computational models that can robustly transfer knowledge between a variety of research organisms and humans to identify functionally similar genes or groups of genes across species regardless of their evolutionary origin. For functionally equivalent genes that may or may not be orthologous, we here coin the term “agnolog”. We define agnologs to be biological entities, processes, or responses — e.g., genes, gene sets, or even biological systems — that are “functionally equivalent” across species regardless of evolutionary origin. “Agno-” conveys the sense of being a data-driven observation that is noncommittal about the evolutionary origin (may they be homologous or convergently evolved) and compatible with any biological explanation. Recent discoveries of agnologs were evidenced by large-scale competitions such as sbvIMPROVER Species Translation Challenge [19] and the Critical Assessment of Protein Function Annotation Challenge (CAFA) [20,21,22,23].
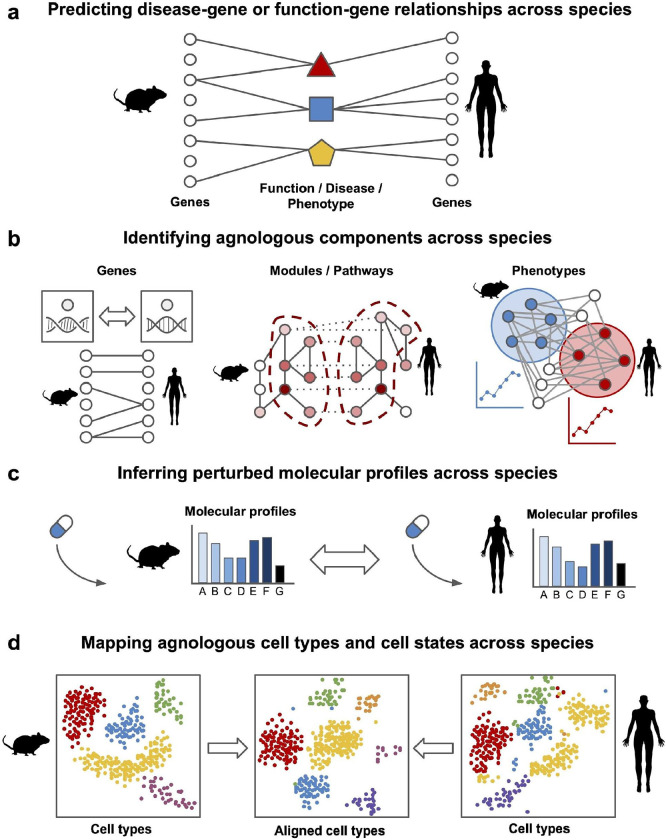
In this review, we present a suite of cross-species knowledge transfer approaches with a significantly broader scope than previous reviews [24,25,26,27]. We comprehensively lay out the landscape of recent and state-of-the-art data-driven strategies, including those that leverage AI and machine learning, to answer four classes of important questions that frequently arise when using research organisms to study biomedical questions and translating findings to humans (Figure 1): (1) How to predict disease-gene or function-gene relationships across species? (2) How to identify agnologous molecular components across species? (3) How to infer perturbed molecular profiles across species? (4) How to map agnologous cell types and cell states across species?
Figure 1: Schematic diagram of each section of this review article.
a. How to predict disease-gene or function-gene relationships across species? b. How to identify agnologous molecular components across species? c. How to infer perturbed molecular profiles across species? d. How to map agnologous cell types and cell states across species?
Instead of discussing methods through an algorithmic lens, we are taking a question-first perspective to provide inspiration and background both for computational researchers interested in developing new methods in this area and for experimental/wet-lab researchers interested in finding and using the best methods in this area. Although we have separated the methods into four sections, many of the approaches described across these sections share similarities in algorithmic design, data types, and output formats. Detailed information about the methods discussed can be found in Supplementary File 1, presented for researchers to easily find the methods and tools relevant to their application of interest. In addition, we have curated the benchmark datasets used in the discussed methods, summarized in Supplementary File 2 and Supplementary Note 1. These resources will enable researchers to test and improve upon existing computational methods in the field.
How to predict disease-gene or function-gene relationships across species
Comprehensive knowledge of the roles genes play in molecular functions, phenotypes, and diseases is fundamental to our understanding of the molecular underpinnings of biological, physiological, and pathological processes. However, the roles of less than half of all genes in the human genome have been experimentally characterized even in a single biological context. Knowledge of gene-function, gene-phenotype, and gene-disease relationships is significantly richer, though far from complete, in research organisms due to the availability of a variety of experimental techniques such as gene editing, knock-in, and knockout experiments [28,29]. How do we leverage this knowledge available across species to close massive annotation gaps, especially in light of the potential functional divergence of homologous genes and the presence of species-specific genes? Moreover, how do we use existing knowledge in human and traditional research organisms (e.g., mouse, frog, zebrafish, fly, worm) to fill annotation gaps in non-traditional research organisms (e.g., dog [30], python [31], gar [32], planaria [33]) that lack sufficient data. These needs have spurred the development of a number of computational methods for predicting disease-gene or function-gene relationships across multiple species (Figure 2).
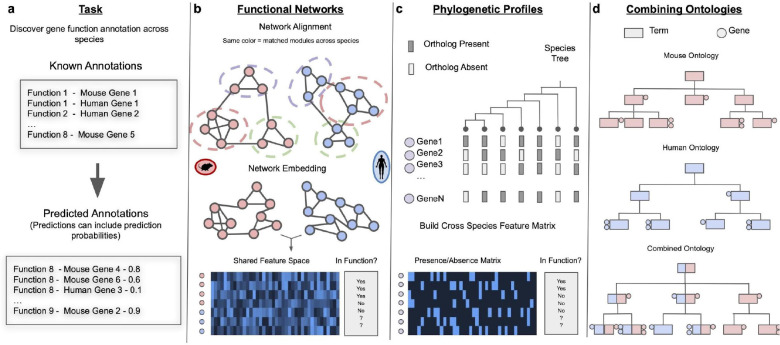
Figure 2: How to predict disease-gene or function-gene relationships across species.
(a) The objective is to predict gene-related functions or diseases across species by leveraging known annotations. (b) Network-based methods can annotate diseases or functions for genes across species by aligning networks or by embedding networks into a shared, low-dimensional feature space where a supervised learning model is then trained to propagate annotations. (c) Phylogenetic profile-based methods identify co-presence or co-absence of genes throughout evolutionary history, implying closely related functions among these genes. This relationship is then used to propagate annotations across species. (d) Disease and function annotations can be transferred by combining ontologies across species. Genes with similar functions or disease associations across species can then be identified through the ontology structure.
Utilizing molecular networks to predict functional gene annotations across species
Molecular interaction networks composed of experimentally verified or computationally predicted physical and/or functional relationships between pairs of genes (or their products) have become indispensable tools for predicting novel gene annotations based on the principle of “guilt-by-association”, which states that genes close to each other in the underlying molecular network will be involved in similar biological processes [34]. Interaction neighborhoods within these networks capture the “functional context” of genes, which is highly complementary to sequence homology information. Further, these networks typically contain and therefore enable making inferences about nearly all genes in the genome, including un(-der)studied and species-specific genes. Therefore, a number of network-based computational approaches have been developed to transfer and predict function, phenotype, and disease annotations across species, especially using machine learning (ML) approaches [35,36,37,38,39,40].
For instance, the Functional Knowledge Transfer (FKT) method [36] first finds “functionally-similar homologous gene pairs” that are in the same gene family and in similar network neighborhoods [41], and uses these pairs to propagate functional annotation across species. FKT significantly enhanced the prediction of gene-pathway associations, especially for biological processes lacking extensive experimental data in the target organism, enabling the transfer of functional insights from well-studied organisms to less-explored ones. For example, by transferring knowledge from other species, FKT successfully annotated the genes related to the regulation of exit from mitosis (GO:0007096) in zebrafish, which had no experimental annotation at the time of the study. One of the top genes predicted by FKT, cdh2, has then been experimentally confirmed related to cell cycle progression in zebrafish retina cells [42].
NetQuilt [37] addressed the challenges of transferring functional gene annotations across species by integrating multiple networks across species using IsoRank [43], which aligns networks considering both sequence similarity and network similarity. To predict annotations across species, NetQuilt utilized a deep learning (DL) model trained on node representations of annotated genes from one or multiple species to predict a fixed set of annotations for genes in the rest of the species.
GenePlexusZoo [38] simultaneously integrates networks of human and five research organisms (mouse, zebrafish, fly, worm, yeast) to generate a “functional representation” of genes from all these species that can be used for any pathway, phenotype, or disease prediction task. By training ML models on this joint multi-species network representation, GenePlexusZoo improves the performance of predicting gene annotations within a single species as well as facilitates knowledge-transfer across species.
Predicting candidate disease genes using networks
While, conceptually, the aforementioned methods can be used to also predict disease-gene relationships, other network-based methods have been developed exclusively for predicting disease associations based on cross-species information.
Katz measure [44], a well-established technique in social network link prediction [45], has been successfully applied to discover connections between diseases and genes across species [46]. The authors constructed a heterogeneous network that included gene-gene, gene-disease, and disease-disease links within humans and then added links between genes in humans and other species based on sequence homology. Then, novel disease gene candidates were identified as those in close proximity to disease nodes in this network (based on overall path lengths), estimated using the Katz measure. Further, “Katz features” that represent the number of paths of a certain length between a gene and a disease node, when incorporated into a ML framework, were shown to significantly enhance the performance of predicting gene-disease relationships [46].
DiseaseQUEST [47] predicts disease-gene candidates in research organisms by combining human genome-wide association studies (GWAS) [48] with gene networks that reflect pathways underlying tissue-specific physiology [49] and disease [50,51]. Specifically, in a target species, using homologs of GWAS disease genes as positive examples and gene interactions in an appropriate tissue network as features, diseaseQUEST trains an ML model to prioritize novel disease-gene candidates. The authors demonstrated this approach by prioritizing and experimentally validating genes related to Parkinson’s disease in the nematode Caenorhabditis elegans [47].
In addition, numerous methods have been developed for predicting disease-gene associations in single species, typically in humans. These methods can be adapted to predict disease-gene associations across species [39,52,53,54,55].
Discovering disease-related genes using phylogenetic profiles
Genes with similar functions tend to appear and disappear jointly during the evolutionary process. Most human disease genes have ancient origins [56,57,58,59], with many traceable to eukaryotic ancestors, some dating back to the evolution of multicellularity [56]. By identifying genes that co-evolve with known disease-related genes, we could propagate the disease annotations from the known genes to the co-evolved genes, thereby helping predict functions for genes that are not well characterized [60,61].
Maxwell and colleagues [62] used evolutionary profiles to examine the evolutionary distribution of human disease genes from the Online Mendelian Inheritance in Man (OMIM) database [63]. They revealed heterogeneity underlying the evolutionary origins of different classes of human disease genes. For example, genes related to inflammatory and immune diseases are of vertebrate origin, while genes associated with cardiovascular and hematological diseases originate from early metazoans.
Recent methods [64,65] enabled systematic screens of genome-wide co-occurring functional modules, leading to functional predictions for numerous previously uncharacterized genes. For example, using phylogenetic profiling, Dey et al. [65] identified understudied candidate genes linked to the actin-nucleating WASH complex and ciliary and centrosomal defects by scanning co-occurring gene modules across 177 species on the eukaryotic species tree. They further evaluated candidate functions of genes by identifying gene product co-localization and gene knockdown.
Bridging disease-gene association through phenotypic similarity
Extensive gene-phenotype associations in research organisms have been discovered through hypothesis-driven experiments and large-scale genetics screens [66,67,68]. These links can lead to the identification of genes in a research organism whose perturbation results in phenotypes similar to those observed in patients with a particular disease, thereby pointing to a viable model for the disease under observation.
However, comparing phenotypes across species is challenging due to evolutionary divergence and non-standard descriptions of phenotypes in different species. The continuous efforts in phenotypic ontology curation and the development of cross-species phenotype ontologies such as uPheno [69] and PhenomeNET [70] have now significantly mitigated this challenge, enabling researchers to use ontology-based semantic similarity measurements between phenotypes to explore suitable research organisms and identify new disease-gene and function-gene relationships. For instance, Exomiser [71,72] leveraged disease-to-gene relationships discovered through semantic similarity measures among integrated ontologies to assist in disease diagnosis.
Recent studies improve on semantic similarity by creating a latent embedding space based on the phenotype ontology combined with known disease-phenotype and disease-/phenotype-gene associations[73,74]. “Node embeddings” created this way contain a low-dimensional numerical representation of each entity, such as gene, phenotype, and disease that captures that entity’s relationships. Such embeddings naturally lend themselves as inputs into ML algorithms. In this study [73], the authors trained a supervised neural network model to predict gene-disease associations based on the node embeddings of genes and diseases, which performed better than an unsupervised approach based on the similarities between gene and disease embeddings.
How to identify agnologous molecular components across species
Complementary to inferring genes in each species that are associated with a particular function or disease, another critical task is to infer molecular components that are equivalent counterparts of each other across species. Termed “agnologs” above, these molecular counterparts reveal novel biology when they are inferred in a data-driven manner, in the context of a specific condition or perturbation, and regardless of their evolutionary origin (e.g., homology). A number of computational approaches have been developed to identify such agnologous components at the gene, pathway (gene set), and genomic level across species (Figure 3).
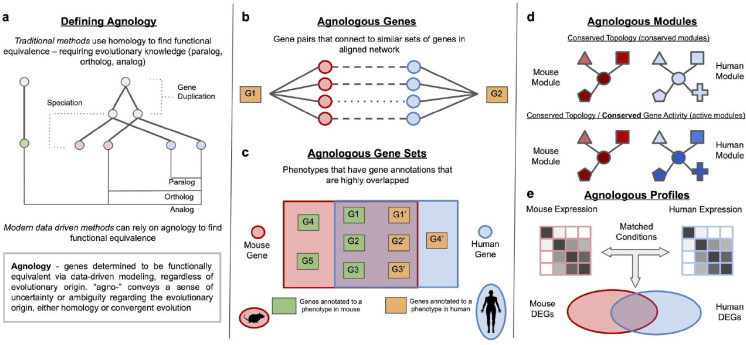
Figure 3: How to identify agnologous molecular components across species.
(a) Definition of agnology. (b) Agnologous gene pairs can be identified through genes that connect to similar sets of genes in aligned cross-species gene networks. (c) When prior knowledge about gene sets is known, agnologous gene sets are discovered through large overlaps between gene sets across species. (d) When prior knowledge is not available, agnologous gene sets can be discovered by finding sub-networks with similar topology and expression patterns across species. (e) Resemblance at the organismal level, i.e., agnologous profiles, can be examined by comparing genomic profiles across species.
Identifying agnologous gene pairs across species
An essential ingredient of finding agnologous genes is to go beyond solely using sequence similarity and homology because, in many organisms, proteins performing similar functions (e.g., playing similar roles in the same biological pathway) may not be the most similar in sequence [75]. Gene Analogue Finder implemented this notion by measuring functional similarity between a pair of genes based on the overlap between gene ontology (GO) terms associated with them [76]. Han et al. [15] used a comparable approach to calculate functional similarity between homologous human-mouse gene pairs based on the average pairwise similarity between human and mouse phenotype ontology terms annotated to those genes. This study identified several cases of functional divergence of orthologs that could be traced to changes in noncoding regulatory sequences of gene pairs with high protein sequence similarity. Nevertheless, the performance of such methods depends heavily on the completeness of gene annotations to terms in function and phenotype ontologies, with term overlap estimates becoming less meaningful for particular genes or entire species with sparse (i.e., highly incomplete) annotations [15].
Genome-scale gene interaction networks help overcome this limitation by providing a powerful alternative way to capture the “functional context” of each gene in terms of its local network neighborhood. Thus, two genes across species interacting with similar sets of genes in their respective molecular network neighborhoods are likely agnologs. Chikina et al. [41] realized this idea by first grouping the network neighbors of individual genes into meta-genes that correspond to Treefam families [77] and measuring the hypergeometric overlap between sets of meta-genes that are neighbors of a pair of genes across species in the respective species network. Intersecting meta-genes identified through this approach revealed specific biological processes underlying network similarities.
Other recent network-based approaches for finding agnologous genes take advantage of the idea of node embedding. Methods such as MUNK [78], MUNDO [79], and ETNA [80] create a joint network-based representation of genes by embedding the molecular networks in a pair of species individually and then align the two embeddings based on sequence orthologs. MUNK produces different joint embeddings based on the “source” species and the “target” species, where the joint embedding is as large as the number of genes in the smaller network. ETNA uses cross-training to align the two embeddings into a bidirectional compressed (low-dimensional) space. MUNK, MUNDO and ETNA use the similarity of embedding vectors of genes to estimate their functional similarity or to inform function or disease gene prediction.
Discovering agnologous gene sets across species
Going beyond gene pairs, it is also of interest to find functionally conserved gene sets across species because sets of genes could represent concepts like pathways and molecular mechanisms underlying conserved phenotypes. McGary et al. [81] introduced the concept of orthologous phenotypes, i.e. “phenologs”, defined as phenotypes that are associated with orthologs. Phenologs could include phenotype counterparts that are observably very different from each other while being influenced by conserved molecular functions (e.g., significantly overlapping sets of orthologous genes). Examples include a yeast model for angiogenesis defects, a worm model for breast cancer, mouse models of autism, and a plant model for human neural crest defects [81]. Phenologs serve as a valuable tool to quantitatively identify non-obvious research organism phenotypes for studying human diseases, along with disease gene candidates such as genes annotated to non-obvious phenotypes and which are not orthologous to any known disease genes.
Discovering agnologous gene modules across species
Phenolog approaches rely on prior knowledge of functional and phenotypic annotations of genes, which is often highly incomplete. Network-based methods have proven valuable in overcoming this limitation by helping to find conserved gene sets, usually called gene modules, while filling in knowledge gaps. This is because, in addition to capturing relationships between pairs of genes, networks also capture higher-level organization between groups of genes in the form of tight sub-networks. Consequently, similar sub-networks across species correspond to homologous functional modules or pathways. CoCoCoNet [82] takes such a network-based approach to test whether a given gene set in one species is involved in similar functions as homologous gene sets in another species. If a subset of genes accurately predicts the remaining genes in the gene set using neighbor voting in both species, that is taken as an indication that the gene set corresponds to a conserved functional module.
Molecular networks can further refine comparisons of differential gene expression across species to help identify functionally conserved “active modules” that comprise tightly connected sub-networks of conserved genes that respond similarly to a given condition (e.g., disease, perturbation). However, finding such modules is difficult because active modules in different species are often not conserved across species, while conserved modules are not necessarily active [83]. So, algorithms for finding conserved and active modules need to consider activity plus conservation at the same time.
The neXus algorithm [84] was developed to meet this need. neXus identifies modules using a greedy seed-and-extend algorithm. It begins with a pair of orthologous nodes as seeds and iteratively extends both sub-networks by incorporating neighboring orthologous gene pairs. ModuleBlast [85] works similarly to neXus with the added capability of distinguishing the resulting modules based on the direction of the expression change. This separation allows ModuleBlast to evaluate whether conserved active modules display expression patterns in the same or opposite way. Both neXus and ModuleBlast limit the identification of modules to fully conserved genes. xHeinz[83] relaxes this constraint by allowing users to define the proportion of conserved nodes, offering a more flexible approach that leads to including functionally conserved but non-homologous (even species-specific) genes in the final modules. xHeinz was further applied to understand the regulatory mechanism of interleukin-17-producing helper T (Th17) cell differentiation in humans and mice. The study revealed that key regulators of Th17 cells are conserved across species [83].
Measuring agnologous profiles between human and research organisms
Beyond genes and gene modules, evaluating the ability of a research organism to mimic specific human biological processes is crucial for functional studies and experimental design. However, such evaluations are subjective, with researchers often using vague terms such as “poorly” or “greatly” to indicate resemblances. Therefore, some recent methods have leveraged functional genomics data to draw quantitative conclusions about the biological resemblance between human and research organisms.
Congruence Analysis for Model Organisms (CAMO) [86] quantitatively measures the congruence between human and research organisms by comparing the distribution of differentially expressed gene (DEG) profiles under matching conditions. To improve the accuracy of identifying DEGs, CAMO infers differential posterior probabilities of genes based on p-values from conventional pipelines. The concordance level of differential gene expression across species was summarized as concordance (c-scores) and discordance scores (d-scores), which served as quantitative measures of congruence across species. CAMO was applied to reconcile studies [87,88] that reached contradicting conclusions about whether mice are a suitable model for studying human inflammatory diseases. By reanalyzing and comparing inflammatory expression data within and across species, the authors concluded that burn- and infection-induced inflammation in mice resembles human inflammation [86].
Comparing phenotypes provides another avenue to discover biological resemblance in research organisms. Cross-species phenotypic ontologies can link phenotypes related to human diseases to research organism phenotypes. Leveraging such connections, the Monarch Initiative [72] provides tools, such as the Phenotype Explorer, to prioritize research organisms based on phenotypic similarities, aiding in the discovery of relevant research organisms and phenotypes.
How to infer perturbed transcriptomes across species
Bulk and single-cell transcriptome profiling have emerged as preeminent technologies for nearly any organism to capture genome-wide molecular responses to a variety of developmental and physiological states as well as treatments, perturbations, and other conditions. Transcriptome profiling in research organisms has especially been valuable in studying perturbations that may be impractical or ethically infeasible in humans. Further, comparing transcriptomic profiles across species sheds light on conserved and distinct cellular states and gene responses, and makes way for context-specific knowledge transfer. However, directly comparing expression changes across species based on gene homology is challenging due to evolutionary divergence in gene expression programs. This section describes several methods that have been developed to utilize computational techniques to enable accurate comparisons of transcriptomic profiles across species (Figure 4).
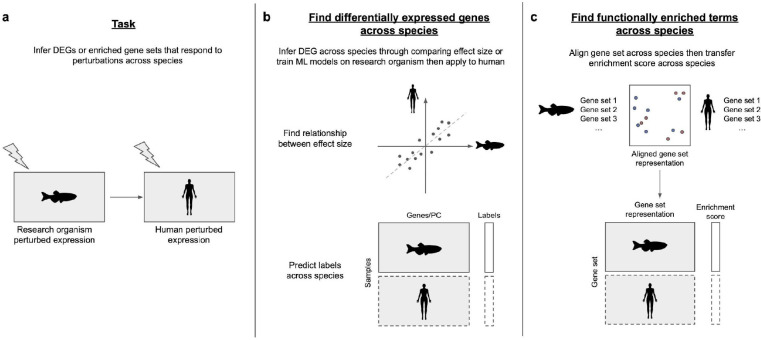
Figure 4: How to infer perturbed transcriptomes across species.
(a) The objective is to infer potential perturbed genes or gene sets in humans based on experimental perturbation results in research organisms. (b) To infer DEGs across species, FIT [89] uses linear models to describe relationships between the effect sizes of DEGs in humans and research organisms with matching conditions. Other approaches develop ML models based on research organisms to predict perturbed expression in humans. (c) To infer enriched gene sets across species, XGSEA [90] aligns gene set representations across species, train models in research organisms, and then infer enrichment scores of gene sets across species.
Determining differentially expressed genes across species
Identifying differentially expressed genes (DEGs) from transcriptome profiles has led to insights into the impact of numerous diseases, perturbations, or experimental conditions. To systematically capture the relationship of DEGs across species, a class of methods train ML models using carefully constructed cross-species dataset pairs (CSDPs) with matching conditions, offering curated examples of how expression changes correspond to phenotypic outcomes across species, which can be further used for model training [89,91].
Found In Translation (FIT) [89] is a linear regression model that is used to understand relationships of DEGs between human and mice for each orthologous gene pair. The authors built models based on 170 mouse-human CSDPs across 28 conditions, which were pairs of mouse and human experiments associated with the same disease or condition. The model was then used to predict DEGs in humans under conditions analogous to those captured in novel mouse data. FIT identified more human DEGs compared to simple transfers of DEGs from research organisms to human based on orthology. For instance, using protein immunostaining, the authors confirmed their prediction that the gene ILF3 is upregulated in the colon of patients with inflammatory bowel diseases (IBD) even though the gene was not differentially expressed in either human or mouse data [89].
The FIT approach is likely to be limited to diseases or drug perturbations with sufficient training data from both human and research organisms. In contrast, Brubaker et al. [91] proposed a semi-supervised method to predict DEGs across species that only requires phenotype information in research organisms. Initial supervised models were built to predict mouse phenotypes using mouse expression data, and then these models were iteratively augmented with high-confidence human samples to predict the phenotypes of the remaining human expression data. Finally, differential gene expression analyses were performed on human samples with distinct predicted phenotypes. Transcomp-R [92] was proposed as an alternative approach to predict DEGs across species when phenotype labels are available in only one species. Its prominent application involved finding mouse genes that are predictive of infliximab responses in chronic IBD patients when corresponding phenotype labels are not available in mice. After representing murine expression data in a low-dimensional space using principal component analysis (PCA) and projecting human expression data into the mouse PC space, Transcomp-R identified murine PCs (and corresponding genes) most associated with infliximab response by regressing the mouse PCs of the projected human data against the human phenotype labels. This approach identified the activated integrin pathway signaling in IBD patients with infliximab resistance. Single-cell sequencing on patient biopsies revealed over-expression of one of the top genes, ITGA1, in immune cells, which probably mediates infliximab resistance. The function of ITGA1 in immune cells was further experimentally validated by treating anti-ITGA1 on patients’ peripheral blood mononuclear cell samples [92].
Finally, other recent work [93,94] includes treating the biological condition such as a tissue or a disease as a “style” and then utilizing style transfer techniques to predict transcriptomic changes across conditions within the same species. Though promising, it is important to note that genetic differences across species are significantly larger and more complex than the condition changes observed within a single species. As a result, applying style transfer techniques directly to cross-species comparisons may face additional challenges and limitations that need to be carefully considered and addressed.
Determining functionally enriched terms across species
Gene set analysis (GSA) has been widely used to identify predefined gene sets (e.g. Gene Ontology biological processes) that are significantly enriched in a gene list of interest [95]. GSA provides a powerful approach to gaining insights into enriched functional terms associated with diseases, phenotypes, or perturbations. However, the discrepancy of genes across species makes the definition of functionally equivalent gene set across species difficult, so directly transferring enriched terms across species is impractical.
XGSEA [90] tackles the challenge of transferring enriched terms across species by using affine mapping, which projects genes from humans and research organisms into the same space. In this joint space, gene sets sharing more homologous genes will be closer to each other in the transformed space. With the transformed gene set representations, logistic regression models were built to characterize the relationship between the representation and enrichment metrics from the research organism, including p-values and enrichment scores of gene sets. Finally, the trained models are used to predict enrichment metrics for gene sets from humans. XGSEA successfully estimates enrichment metrics in human gene sets based on metrics of homologous gene sets in research organisms. For instance, XGSEA was able to train models based on zebrafish data and predict enriched pathways associated with melanomas in human patients [90].
How to map agnologous cell types and cell states across species
The advent of single-cell and single-nucleus sequencing techniques has opened up new avenues in biological research. This emerging frontier focuses on identifying and comparing cell types and states across different species to better understand the cellular diversity and species-specific cellular innovation arose from cell type evolution [96,97]. By transferring insights from extensively studied organisms to less-explored ones, these cross-species comparisons offer a powerful insight for cell type identification and discovery, thereby advancing our understanding of cellular biology across different species.
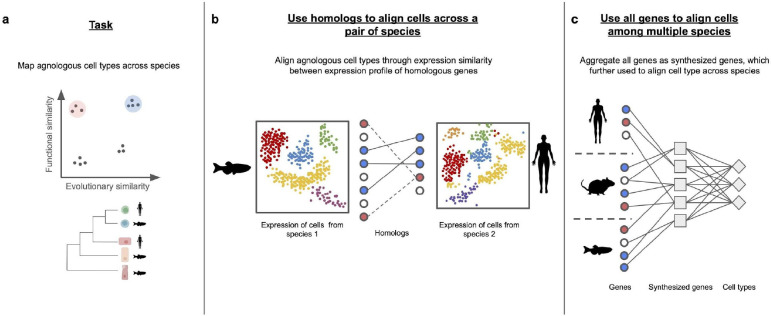
Cell mapping problems may be simply conceptualized as mapping transcriptionally similar cells across species (i.e., based on homologous genes having similar expression levels). However, cells of a particular type within a species are more alike than those of corresponding cell types across different species. [100,101]. Therefore, effectively mapping cells across species requires correcting for species-specific expression differences so that cells of homologous types are mixed in irrespective of species origin, while cells of different types remain distinct from each other. Additionally, methods must account for experiment-induced batch effects. Some tools have been developed to reconcile heterogeneous single-cell RNA-seq (scRNA-seq) data from multiple species [100,102,103,104,105]. These methods typically seek to project cells from multiple species into a unified space, facilitating cross-species comparison. As a result, the cell mapping problem can be viewed as a domain adaptation problem, i.e., aligning cells from different species in a unified space (Figure 5).
Figure 5: How to map agnologous cell types and cell states across species.
(a) The objective is to align analogous cells (represented by points shaded in red and blue patches) across species, given pairs of single-cell datasets or even multiple single-cell datasets from different species. (b) The majority of methods align cells with similar expression patterns in homologs across a pair of species. (c) SATURN [98] stands out as the only method capable of aligning single-cell datasets from multiple species simultaneously. It first aggregates genes into synthesized genes using PLM [99]. These synthesized genes are then utilized to align cells across multiple species.
Aligning cells across a pair of species based on homologs
Shafer [97] summarized various methods for cross-species scRNA-seq integration. Originally designed to mitigate batch effects introduced by experimental variations, these methods may not sufficiently correct for species differences, which are more pronounced than batch effects [101]. When comparing human cells to those of anciently polyploid species like the teleost fish, the abundance of gene duplicates results in fewer one-to-one gene matches, leading to a reduced signal for alignment. Moreover, orthology does not necessarily imply similar functions across species, and paralogs might exhibit more functional similarity than orthologs. For instance, when an ortholog acquires a loss-of-function mutation, its function might be compensated by the upregulation of a paralog, resulting in the paralog having a more similar function to the ortholog in the other species [106,107]. Therefore, incorporating one-to-many or many-to-many homology into the analyses could improve cross-species alignment performance by accounting for functional similarity between paralogs.
In a benchmark study conducted by Song et al. [108], SAMap [109] was identified as the only method that is capable of mapping divergent cell types. To account for the effect that expression patterns may be divergent in homologous cell types, SAMap uses the full gene homology information and incorporates neighboring cells within species into the calculation of similarity between cells across species. Consequently, cells with differing expression profiles remain closely associated if they are among the nearest cross-species neighbors to each other. Based on this analysis, SAMap identified general alignment between gene expression patterns and developmental relationships during embryogenesis in frogs and zebrafish. Interestingly, they also detected a group of secretory cell types that have similar expression patterns while having different developmental origins including arising from different germ layers [109].
Aligning cells among multiple species using all genes
Instead of using precalculated gene homology, a new framework SATURN [98] integrates cell atlases from divergent species by harnessing the power of protein language models (PLMs) to relate genes across species to each other and project all cells from multiple species into shared cell embedding space. PLMs can learn informative representations of protein sequences that can implicitly reflect similarity of function, protein structure, molecular characteristics [110], as well as evolutionary relationships [104] between proteins. Using a neural network, SATURN first projects scRNA-seq datasets to a joint space composed of “macrogenes” representing groups of genes across species that have similar protein embeddings inferred from the PLM. The neural network weights is used to define the importance of a gene to a macrogene. For each macrogene, SATURN learns the cross-species cell embedding as a non-linear combination of macrogenes, guided by an unsupervised objective function that simultaneously maximizes the distance between distinct cells within the same species and minimizes the distance between similar cells across species. This approach enables SATURN to integrate data from multiple species (and not just perform pairwise comparisons), including divergent species where gene homology relationships are hard to unravel. The output of SATURN also enables easy cross-species comparison, such as finding differentially-expressed macrogenes across species, which can then be traced back to actual genes based on the weight of connections to the macrogenes in the neural network. SATURN’s application to a multi-species dataset of frog and zebrafish embryogenesis shows success in aligning evolutionary-related cell types and revealing differentially expressed genes in macrophage/myeloid progenitors and ionocytes across these two remotely-related species [98].
Future perspective
The explosion of large-scale human biological data and accompanying analysis methods, while seen as a prospect of a golden age for human genetics, has also raised the question of whether it will diminish the significance of research organisms [7,111]. From the perspective of this review, the answer is certainly “No”. In fact, the ocean of new data will lead to an increasing number of candidate disease-related genes, dysregulated pathways, drugs, etc., that will require validation and functional characterization, which can be carried out in completely in vivo settings only in research organisms [7]. Therefore, the goal of increasingly better computational strategies to effectively gain biological and biomedical insights from research organisms will continue to remain significant. The landscape of current methods, summarized in this review, demonstrates great promise towards this goal. Instead of solely relying on sequence similarity and gene homology, these approaches utilize sophisticated inferential techniques and diverse datasets, paving a promising path for effectively transferring knowledge between human and research organisms. The next frontier in unlocking the full potential of research organisms and the ever-growing datasets involves addressing some key challenges that still remain.
Capturing the specific facets of complex diseases
Complex human diseases exhibit staggering genetic and phenotypic heterogeneity. Despite this complexity, much of the current focus is on identifying a single research organism that can fully recapitulate an entire disease. Consequently, a major need in the field is to develop methods that can identify optimal phenotypes, conditions, and genes for studying specific facets of complex diseases.
Several approaches have been proposed to dissect diseases into molecular components. Given that disrupted biological processes are often shared among diseases [112,113,114], even those seemingly unrelated [115], it is promising to dissect diseases into dysregulated processes [116] or modules [117,118]. Additionally, phenotype ontologies, such as Human Phenotype Ontology (HPO) [119], are widely used tools for phenotype-driven disease analysis, enabling the breakdown of diseases into related phenotypes [120,121].
By leveraging dissected processes, modules, or phenotypes, we can identify combinations of research organisms that maximally capture the multifaceted nature of complex diseases. The methods discussed in this review, particularly in the section “How to identify analogous molecular components across species”, shed light on approaches for relating these dissected components to suitable research organisms. However, there remains a critical need for future approaches that can utilize analogous molecular components of complex diseases to guide the strategic selection of the most appropriate research organisms for specific disease aspects.
From homology to agnology
Homology is widely used as the bridge to find functional similarities across species. However, the definition of homology does not include nor does it guarantee functional similarity. Several factors can lead to functional divergence between homologous genes. Examples include changes in non-coding regulatory sequences [15], reciprocal gene loss [122], and developmental system drift [123].
Moreover, homology restrains the power of predictive models to explore comprehensive functional relationships across divergent species. For instance, research organisms like the nematode C. elegans may not share a substantial number of gene orthologs with humans, resulting in a scarcity of genes available for model training and testing. This limited orthologous gene repertoire can hinder the ability of computational models to learn comprehensive functional relationships across species. Furthermore, orthologs alone can only explain a fraction of observed biological variations [97]. Therefore, it is crucial to incorporate species-/taxon-specific genes, i.e. agnologs, to construct more comprehensive models. Relying solely on homologous genes is limiting from the perspective of network biology, where species evolve through gain or loss of interactions, indicating that species-/taxon-specific genes can play a role in similar functional and regulatory programs [124].
While most methods discussed here suffer from the limitation of heavily relying on homology for knowledge transfer, approaches such as FKT [36], GenePlexusZoo [38] and SATURN [98] show how to include every gene in cross-species analyses, may they be homologous or not.
Networks in more species and more contexts
Molecular networks are one of the most widely applied data types in cross-species knowledge transfer. Regardless of their representation (e.g., edgelists, adjacency matrices, or node embeddings), networks capture interactions among neighboring genes and provide mechanistic understanding to computational models. However, network-based methods also have limitations.
Most networks generated using experimental approaches are for humans or popular research organisms like mice. There is an urgent need to expand the repertoire of networks beyond these species to include non-traditional research organisms. Current methods, such as STRING [125] and those described in recent literature [126], use orthology information to infer “interologs”, i.e., conserved interactions between pairs of proteins that have interacting homologs in another organism [127,128]. For example, STRING utilizes high-confidence networks in humans and data-rich research organisms to derive protein-protein interaction (PPI) networks for over 1,000 species. However, the assumption that paired orthologous genes have conserved interactions across species may not always hold true due to interaction rewiring during the course of evolution. Additionally, limiting the scope to orthologous genes cannot infer interactions involving taxon-specific genes. Other previously introduced methods like FKT/IMP [36,129] use functional analogs to transfer knowledge across species, but these methods require experimental functional genomics data as a prior to generate networks, limiting their application to popular research organisms.
Another important future step in generating species-specific networks is the development of context-specific networks, particularly those with tissue or cell-type specificity. Context specificity plays a crucial role in biomedicine since disease-gene associations frequently arise from disrupted interactions among tissue-specific and cell lineage-specific processes under particular environmental conditions [51,130]. Unfortunately, the nuanced interactions that vary across tissues may not be fully captured by experimentally generated large-scale networks such as PPI. Coexpression networks have the potential to capture tissue specificity more effectively [131], but they tend to be noisy [132]. To obtain robust signals, current studies often extract only a small fraction of information for downstream analysis, such as the top 0.5% of co-expressed gene pairs [133]. This approach, however, results in the loss of significant information. Limited efforts have been made to build tissue-specific networks by integrating multi-modal functional genomic data [47,51] or to contextualize network representations by incorporating tissue-specific expression [134,135,136].
Recent advances in sequence-based deep learning models could help expand the repertoire of species-specific and context-specific networks. For example, AlphaFold-Multimer [137] can predict protein interactions based on sequences. ExPecto [138] and ExPectoSC [139] can predict tissue-/cell- specific regulatory landscapes based on DNA sequences alone. Thanks to the advancement of genome projects for non-model species, such as the Vertebrate Genomes Project [140], genome sequence resources are much richer than other genomics types such as transcriptomes or epigenomes. Leveraging the power of these sequence-based models and transfer learning techniques poses a promising way to expand networks beyond popular research organisms. However, we need to integrate phylogenetic information into transfer learning to consider the taxonomic-specific logic of gene regulation or protein interactions.
Automated construction of ontologies and knowledge graphs
In this review, we have described the power of ontologies as a framework for transferring knowledge across species. Furthermore, combining various ontologies and annotations into knowledge graphs can provide new insights for translational biomedicine. The Monarch Knowledge Graph exemplifies this approach, integrating knowledge from 33 biomedical resources, including information from all major research organism databases [72]. Leveraging such knowledge graphs, we can employ advanced techniques like graph deep learning [141] to utilize information from research organisms and assist biomedical studies such as rare disease variant prioritization [142]. Integrating knowledge graphs with large language models can also help reduce “hallucinations” in AI-powered question-answering systems within the biomedical field [143].
Despite the power of ontologies and knowledge graphs in cross-species knowledge transfer, the generation of ontologies requires laborious curation by specialists. Manual curation may also introduce biases towards popular research areas. For instance, since zebrafish is a widely used research organism for developmental and embryogenic studies, GO terms annotated to zebrafish might be skewed towards these areas. Such differences in term annotation frequency could in turn bias downstream genomic analyses [144].
The capability of current large language models to effectively extract entities and relationships from the literature offers an efficient method for automatically generating ontologies and knowledge graphs. For example, SPIRE [145] can generate ontologies by processing text input and a user-provided schema that describes the desired structure of the ontology. While more effort is needed to maintain the quality and consistency of automatically generated ontologies, this approach holds great potential for significantly enriching ontologies across a wide spectrum of organisms, from well-studied to understudied research organisms and even non-model organisms.
Better benchmarking studies for cross-species scRNAseq analyses
Mapping single-cell and single-nuclei transcriptomics (scRNA-seq/snRNA-seq) data across different species provides valuable insights into cell type evolution and facilitates cell type and cell state annotation [96,97]. However, most existing cross-species cell mapping algorithms are limited to closely related species and do not account for full homology [97], while considering all homologous genes are crucial when comparing species involving gene duplication at their common ancestors. For example, due to a genome duplication in the ancestor of teleost fishes, considering full homology is essential for cross-species integration of cell types between human and biomedical fish models such as zebrafish [146]. To overcome this limitation, algorithms such as SAMap [109] and SATURN [98] have been developed. While recent benchmarking studies highlight properties of these methods that are important for effective cross-species integration of scRNA-seq data [108], most of these methods were originally designed for batch corrections. Furthermore, since SAMap applied additional restrictions on within-species manifolds, SAMap was not systematically compared to other methods in current benchmarking studies. Thus, it is essential to conduct more comprehensive benchmarking of these methods to evaluate their performance on different types of scRNA-seq datasets and species with varying degrees of divergence and data curation. Additionally, the development of new methods for downstream analyses is crucial to extract meaningful biological insights from the growing mountain of single-cell/nuclei data.
Extracting and curating knowledge from non-traditional research organisms
The majority of the organisms noted in this review are classic research organisms such as mouse, zebrafish, and the nematode worm, with non-traditional and emerging research organisms often being overlooked in biomedical studies. However, non-traditional research organisms have significant contributions to translational research due to their unique characteristics [147]. For instance, the axolotl (Ambystoma mexicanum), a neotenic salamander, shows remarkable regenerative abilities, making it a crucial research organism for regenerative medicine and developmental biology [148]. The spotted gar (Lepisosteus oculatus), a ray-finned fish with a slow evolutionary rate, has proven valuable in facilitating genomic comparison between teleost biomedical research organisms and the human genome [32]. The tunicate, Ciona intestinalis, is a close invertebrate relative of vertebrates. Its unique evolutionary position, simple body plan, and ease of manipulation make it highly suitable for studying embryonic development and morphogenesis [149]. However, challenges such as polyploidy, large genome sizes, genomic rearrangements, and taxon-specific cell types can make analyzing the genomic data from such species more difficult than classic research organisms. It is necessary to explore additional methods that can effectively transfer knowledge from and to non-traditional research organism data sets.
Conclusions
In this review, we have discussed the current state of computational methods for cross-species knowledge transfer. We emphasize that these methods surpass simple comparisons of molecular profiles across species and highlight the utilization of orthogonal information sources such as phenotypic ontologies and molecular networks to facilitate cross-species knowledge transfer.
Our review provides valuable resources and insights into the advancements and challenges of methods for cross-species knowledge transfer among various research organisms and human. The resources summarized in this review will facilitate biomedical studies using research organisms, including traditional and non-traditional research organisms, by leveraging knowledge from human or extensively studied model systems.
There are still needs and plenty of room for further improvements and refinements of existing approaches. More advanced computational approaches are needed to identify a range of research organisms for studying different aspects of diseases. Instead of relying solely on homology as a bridge, expanding analyses to agnologs can enhance the effectiveness of cross-species knowledge transfer. To gain a more comprehensive understanding of diseases, we need networks from more species, and also need to incorporate context specificity, especially cell- and tissue-specificity, into networks, despite the inherent difficulties involved. To fully unlock the power of ontology-based knowledge transfer, we need methods to automatically generate high-quality and robust ontologies as well as knowledge graphs. As a rapidly growing field, it is essential to establish more benchmarks for state-of-the-art cross-species cell mapping between humans and evolutionary divergent research organisms. Currently, most methods are focused on the few well-studied research organisms, but greater attention and analytical methods should be employed to extract biomedical insights from non-traditional, emerging research organisms.
We propose that with the application of comprehensive computational approaches, the field will gain more exciting insights from big data of research organisms, ultimately enhancing our understanding of human biology and diseases.
Acknowledgements
This work is supported by NIH R35 GM128765 and Simons Foundation 1017799 (to A.K.). zebrafish-human transfer research in the Braasch Lab has been supported by NIH R01OD011116. We thank all members in Krishnan and Braasch Labs for helpful discussion and feedback on the manuscript.
Footnotes
Ethics declaration
Competing interests
The authors declare no competing interests.
Contributor Information
Hao Yuan, Genetics and Genome Science Program; Ecology, Evolution, and Behavior Program, Michigan State University.
Christopher A. Mancuso, Department of Biostatistics & Informatics, University of Colorado Anschutz Medical Campus
Kayla Johnson, Department of Biomedical Informatics, University of Colorado Anschutz Medical Campus.
Ingo Braasch, Department of Integrative Biology; Genetics and Genome Science Program; Ecology, Evolution, and Behavior Program, Michigan State University.
Arjun Krishnan, Department of Biomedical Informatics, University of Colorado Anschutz Medical Campus.
References
- 1.Animal models are essential to biological research: issues and perspectives Barré-Sinoussi Françoise, Montagutelli Xavier Future Science OA (2015-November) https://doi.org/gftwtr DOI: 10.4155/fso.15.63 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Model organisms contribute to diagnosis and discovery in the undiagnosed diseases network: current state and a future vision Baldridge Dustin, Wangler Michael F, Bowman Angela N, Yamamoto Shinya, Schedl Tim, Pak Stephen C, Postlethwait John H, Shin Jimann, Solnica-Krezel Lilianna, … Westerfield Monte Orphanet Journal of Rare Diseases (2021-May-07) https://doi.org/gkjnhw DOI: 10.1186/s13023-021-01839-9 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Use of animals in experimental research: an ethical dilemma? Baumans V Gene Therapy (2004-September-29) https://doi.org/dsrjkb DOI: 10.1038/sj.gt.3302371 [DOI] [PubMed] [Google Scholar]
- 4.The ethics of animal research Festing Simon, Wilkinson Robin EMBO reports (2007-May-18) https://doi.org/bzr78f DOI: 10.1038/sj.embor.7400993 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harmonization of Animal Care and Use Guidance Demers Gilles, Griffin Gilly, Vroey Guy De, Haywood Joseph R, Zurlo Joanne, Bédard Marie Science (2006-May-05) https://doi.org/bh2jbk DOI: 10.1126/science.1124036 [DOI] [PubMed] [Google Scholar]
- 6.Model Organisms Facilitate Rare Disease Diagnosis and Therapeutic Research Wangler Michael F, Yamamoto Shinya, Chao Hsiao-Tuan, Posey Jennifer E, Westerfield Monte, Postlethwait John, Hieter Philip, Boycott Kym M, Campeau Philippe M, Bellen Hugo J Genetics (2017-August-31) https://doi.org/gbwb63 DOI: 10.1534/genetics.117.203067 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The future of model organisms in human disease research Aitman Timothy J, Boone Charles, Churchill Gary A, Hengartner Michael O, Mackay Trudy FC, Stemple Derek L Nature Reviews Genetics (2011-July-18) https://doi.org/b3ht5z DOI: 10.1038/nrg3047 [DOI] [PubMed] [Google Scholar]
- 8.Genetic Heterogeneity in Human Disease McClellan Jon, King Mary-Claire Cell (2010-April) https://doi.org/b9rhz5 DOI: 10.1016/j.cell.2010.03.032 [DOI] [PubMed] [Google Scholar]
- 9.Molecular Characterization Reveals Genetic Uniformity in Experimental Chicken Resources TADANO Ryo, KINOSHITA Keiji, MIZUTANI Makoto, ATSUMI Yuusuke, FUJIWARA Akira, SAITOU Toshiki, NAMIKAWA Takao, TSUDZUKI Masaoki Experimental Animals (2010) https://doi.org/fg9q89 DOI: 10.1538/expanim.59.511 [DOI] [PubMed] [Google Scholar]
- 10.Isogenic lines in fish – a critical review Franěk Roman, Baloch Abdul Rasheed, Kašpar Vojtěch, Saito Taiju, Fujimoto Takafumi, Arai Katsutoshi, Pšenička Martin Reviews in Aquaculture (2019-October-21) https://doi.org/ggddr9 DOI: 10.1111/raq.12389 [DOI] [Google Scholar]
- 11.Defining the functional divergence of orthologous genes between human and mouse in the context of miRNA regulation Cui Chunmei, Zhou Yuan, Cui Qinghua Briefings in Bioinformatics (2021-July-05) https://doi.org/gr86f6 DOI: 10.1093/bib/bbab253 [DOI] [PubMed] [Google Scholar]
- 12.Testing the Ortholog Conjecture with Comparative Functional Genomic Data from Mammals Nehrt Nathan L, Clark Wyatt T, Radivojac Predrag, Hahn Matthew W PLoS Computational Biology (2011-June-09) https://doi.org/cmg5m4 DOI: 10.1371/journal.pcbi.1002073 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The ortholog conjecture revisited: the value of orthologs and paralogs in function prediction Stamboulian Moses, Guerrero Rafael F, Hahn Matthew W, Radivojac Predrag Bioinformatics (2020-July-01) https://doi.org/gr96nw DOI: 10.1093/bioinformatics/btaa468 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution Nadimpalli Shilpa, Persikov Anton V, Singh Mona PLOS Genetics (2015-March-06) https://doi.org/f68jmg DOI: 10.1371/journal.pgen.1005011 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Divergence of Noncoding Regulatory Elements Explains Gene–Phenotype Differences between Human and Mouse Orthologous Genes Han Seong Kyu, Kim Donghyo, Lee Heetak, Kim Inhae, Kim Sanguk Molecular Biology and Evolution (2018-April-24) https://doi.org/gdd4g2 DOI: 10.1093/molbev/msy056 [DOI] [PubMed] [Google Scholar]
- 16.The origin and evolution of cell types Arendt Detlev, Musser Jacob M, Baker Clare VH, Bergman Aviv, Cepko Connie, Erwin Douglas H, Pavlicev Mihaela, Schlosser Gerhard, Widder Stefanie, Laubichler Manfred D, Wagner Günter P Nature Reviews Genetics (2016-November-07) https://doi.org/f9b62x DOI: 10.1038/nrg.2016.127 [DOI] [PubMed] [Google Scholar]
- 17.Tissue evolution: mechanical interplay of adhesion, pressure, and heterogeneity Büscher Tobias, Ganai Nirmalendu, Gompper Gerhard, Elgeti Jens New Journal of Physics (2020-March-01) https://doi.org/gqpwh8 DOI: 10.1088/1367-2630/ab74a5 [DOI] [Google Scholar]
- 18.Evolutionary rewiring of regulatory networks contributes to phenotypic differences between human and mouse orthologous genes Ha Doyeon, Kim Donghyo, Kim Inhae, Oh Youngchul, Kong JungHo, Han Seong Kyu, Kim Sanguk Nucleic Acids Research (2022-February-07) https://doi.org/gtsx5v DOI: 10.1093/nar/gkac050 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Understanding the limits of animal models as predictors of human biology: lessons learned from the sbv IMPROVER Species Translation Challenge Rhrissorrakrai Kahn, Belcastro Vincenzo, Bilal Erhan, Norel Raquel, Poussin Carine, Mathis Carole, Dulize Rémi HJ, Ivanov Nikolai V, Alexopoulos Leonidas, Jeremy Rice J, … Hoeng Julia Bioinformatics (2014-September-17) https://doi.org/f63qxc DOI: 10.1093/bioinformatics/btu611 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens Zhou Naihui, Jiang Yuxiang, Bergquist Timothy R, Lee Alexandra J, Kacsoh Balint Z, Crocker Alex W, Lewis Kimberley A, Georghiou George, Nguyen Huy N, Hamid Md Nafiz, … Friedberg Iddo Genome Biology (2019-November-19) https://doi.org/ggnxpz DOI: 10.1186/s13059-019-1835-8 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An expanded evaluation of protein function prediction methods shows an improvement in accuracy Jiang Yuxiang, Oron Tal Ronnen, Clark Wyatt T, Bankapur Asma R, D’Andrea Daniel, Lepore Rosalba, Funk Christopher S, Kahanda Indika, Verspoor Karin M, Ben-Hur Asa, … Radivojac Predrag Genome Biology (2016-September-07) https://doi.org/gg7qsc DOI: 10.1186/s13059-016-1037-6 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A large-scale evaluation of computational protein function prediction Radivojac Predrag, Clark Wyatt T, Oron Tal Ronnen, Schnoes Alexandra M, Wittkop Tobias, Sokolov Artem, Graim Kiley, Funk Christopher, Verspoor Karin, Ben-Hur Asa, … Friedberg Iddo Nature Methods (2013-January-27) https://doi.org/gjh8q2 DOI: 10.1038/nmeth.2340 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Community-Wide Evaluation of Computational Function Prediction Friedberg Iddo, Radivojac Predrag arXiv (2016) https://doi.org/gsc3dr DOI: 10.48550/arxiv.1601.01048 [DOI] [PubMed] [Google Scholar]
- 24.Translating preclinical models to humans Brubaker Douglas K, Lauffenburger Douglas A Science (2020-February-14) https://doi.org/ggk6dt DOI: 10.1126/science.aay8086 [DOI] [PubMed] [Google Scholar]
- 25.Applications of comparative evolution to human disease genetics McWhite Claire D, Liebeskind Benjamin J, Marcotte Edward M Current Opinion in Genetics & Development (2015-December) https://doi.org/gr86f5 DOI: 10.1016/j.gde.2015.08.004 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Transfer learning of clinical outcomes from preclinical molecular data, principles and perspectives Kowald Axel, Barrantes Israel, Möller Steffen, Palmer Daniel, Escobar Hugo Murua, Schwerk Anne, Georg Fuellen Briefings in Bioinformatics (2022-April-23) https://doi.org/gr86f7 DOI: 10.1093/bib/bbac133 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Systems biology approaches help to facilitate interpretation of cross-species comparisons Dougherty Bonnie V, Papin Jason A Current Opinion in Toxicology (2020-October) https://doi.org/gr86f4 DOI: 10.1016/j.cotox.2020.06.002 [DOI] [Google Scholar]
- 28.A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish Wu Roland S, Lam Ian I, Clay Hilary, Duong Daniel N, Deo Rahul C, Coughlin Shaun R Developmental Cell (2018-July) https://doi.org/gdv6c4 DOI: 10.1016/j.devcel.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 29.Loss of circadian rhythmicity in bdnf knockout zebrafish larvae D’Agostino Ylenia, Frigato Elena, Noviello Teresa MR, Toni Mattia, Frabetti Flavia, Cigliano Luisa, Ceccarelli Michele, Sordino Paolo, Cerulo Luigi, Bertolucci Cristiano, D’Aniello Salvatore iScience (2022-April) https://doi.org/gr8cpc DOI: 10.1016/j.isci.2022.104054 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leading the way: canine models of genomics and disease Shearin Abigail L, Ostrander Elaine A Disease Models & Mechanisms (2010-January-14) https://doi.org/dqcgd5 DOI: 10.1242/dmm.004358 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Burmese python genome reveals the molecular basis for extreme adaptation in snakes Castoe Todd A, de Koning APJason, Hall Kathryn T, Card Daren C, Schield Drew R, Fujita Matthew K, Ruggiero Robert P, Degner Jack F, Daza Juan M, Gu Wanjun, … Pollock David D Proceedings of the National Academy of Sciences (2013-December-02) https://doi.org/f5kfpx DOI: 10.1073/pnas.1314475110 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons Braasch Ingo, Gehrke Andrew R, Smith Jeramiah J, Kawasaki Kazuhiko, Manousaki Tereza, Pasquier Jeremy, Amores Angel, Desvignes Thomas, Batzel Peter, Catchen Julian, … Postlethwait John H Nature Genetics (2016-March-07) https://doi.org/f3rndn DOI: 10.1038/ng.3526 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms Grohme Markus Alexander, Schloissnig Siegfried, Rozanski Andrei, Pippel Martin, George Robert Young Sylke Winkler, Brandl Holger, Henry Ian, Dahl Andreas, Powell Sean, … Christian Jochen Rink Nature (2018-January-24) https://doi.org/gcv62s DOI: 10.1038/nature25473 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guilt-by-association goes global Oliver Stephen Nature (2000-February) https://doi.org/b6×4kj DOI: 10.1038/35001165 [DOI] [PubMed] [Google Scholar]
- 35.The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function Warde-Farley David, Donaldson Sylva L, Comes Ovi, Zuberi Khalid, Badrawi Rashad, Chao Pauline, Franz Max, Grouios Chris, Kazi Farzana, Lopes Christian Tannus, … Morris Quaid Nucleic Acids Research (2010-June-21) https://doi.org/bkds9d DOI: 10.1093/nar/gkq537 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Functional Knowledge Transfer for High-accuracy Prediction of Under-studied Biological Processes Park Christopher Y, Wong Aaron K, Greene Casey S, Rowland Jessica, Guan Yuanfang, Bongo Lars A, Burdine Rebecca D, Troyanskaya Olga G PLoS Computational Biology (2013-March-14) https://doi.org/f4qtp9 DOI: 10.1371/journal.pcbi.1002957 · This study introduced the Functional Knowledge Transfer (FKT) method, a network-based approach that utilizes agnologs to enhance gene annotation across species.
- 37.NetQuilt: deep multispecies network-based protein function prediction using homology-informed network similarity Barot Meet, Gligorijević Vladimir, Cho Kyunghyun, Bonneau Richard Bioinformatics (2021-February-12) https://doi.org/gk2rt6 DOI: 10.1093/bioinformatics/btab098 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Joint representation of molecular networks from multiple species improves gene classification Mancuso Christopher A, Johnson Kayla A, Liu Renming, Krishnan Arjun PLOS Computational Biology (2024-January-10) https://doi.org/gtsx8m DOI: 10.1371/journal.pcbi.1011773 · This study introduced GenePlexusZoo, a method to simultaneously integrate network information across more than two species to improve gene classification.
- 39.Supervised learning is an accurate method for network-based gene classification Liu Renming, Mancuso Christopher A, Yannakopoulos Anna, Johnson Kayla A, Krishnan Arjun Bioinformatics (2020-April-14) https://doi.org/gmvnfc DOI: 10.1093/bioinformatics/btaa150 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Network-based methods for human disease gene prediction Wang X, Gulbahce N, Yu H Briefings in Functional Genomics (2011-July-15) https://doi.org/ctnj62 DOI: 10.1093/bfgp/elr024 [DOI] [PubMed] [Google Scholar]
- 41. Accurate Quantification of Functional Analogy among Close Homologs Chikina Maria D, Troyanskaya Olga G PLoS Computational Biology (2011-February-03) https://doi.org/bh988k DOI: 10.1371/journal.pcbi.1001074 · This method uses functional gene networks and metagenes of neighboring genes in these networks to find agnologs across species.
- 42.Mutations in N-cadherin and a Stardust homolog, Nagie oko, affect cell-cycle exit in zebrafish retina Yamaguchi Masahiro, Imai Fumiyasu, Tonou-Fujimori Noriko, Masai Ichiro Mechanisms of Development (2010-May) https://doi.org/djzmf9 DOI: 10.1016/j.mod.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 43.Global alignment of multiple protein interaction networks with application to functional orthology detection Rohit Singh, Xu Jinbo, Berger Bonnie Proceedings of the National Academy of Sciences (2008-September-02) https://doi.org/cn7rgd DOI: 10.1073/pnas.0806627105 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A new status index derived from sociometric analysis Katz Leo Psychometrika (1953-March) https://doi.org/c6g25t DOI: 10.1007/bf02289026 [DOI] [Google Scholar]
- 45.The link prediction problem for social networks Liben-Nowell David, Kleinberg Jon Proceedings of the twelfth international conference on Information and knowledge management (2003-November-03) https://doi.org/c7ttqw DOI: 10.1145/956863.956972 [DOI] [Google Scholar]
- 46.Singh-Blom UMartin, Natarajan Nagarajan, Tewari Ambuj, Woods John O, Dhillon Inderjit S, Marcotte Edward M PLoS ONE (2013-May-01) https://doi.org/f24vnt DOI: 10.1371/journal.pone.0058977 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. An integrative tissue-network approach to identify and test human disease genes Yao Victoria, Kaletsky Rachel, Keyes William, Mor Danielle E, Wong Aaron K, Sohrabi Salman, Murphy Coleen T, Troyanskaya Olga G Nature Biotechnology (2018-October-22) https://doi.org/gfg4bd DOI: 10.1038/nbt.4246 · This study introduced DiseaseQUEST, a method to combine human GWAS-derived gene-disease associations and research organism functional networks to predict candidate disease-related genes in research organisms.
- 48.Large-Scale Discovery of Disease-Disease and Disease-Gene Associations Gligorijevic Djordje, Stojanovic Jelena, Djuric Nemanja, Radosavljevic Vladan, Grbovic Mihajlo, Kulathinal Rob J, Obradovic Zoran Scientific Reports (2016-August-31) https://doi.org/f8znpb DOI: 10.1038/srep32404 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Understanding Tissue-Specific Gene Regulation Sonawane Abhijeet Rajendra, Platig John, Fagny Maud, Chen Cho-Yi, Paulson Joseph Nathaniel, Lopes-Ramos Camila Miranda, DeMeo Dawn Lisa, Quackenbush John, Glass Kimberly, Kuijjer Marieke Lydia Cell Reports (2017-October) https://doi.org/ggcrpf DOI: 10.1016/j.celrep.2017.10.001 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hekselman Idan, Yeger-Lotem Esti Nature Reviews Genetics (2020-January-08) https://doi.org/ggkx9v DOI: 10.1038/s41576-019-0200-9 [DOI] [PubMed] [Google Scholar]
- 51.Understanding multicellular function and disease with human tissue-specific networks Greene Casey S, Krishnan Arjun, Wong Aaron K, Ricciotti Emanuela, Zelaya Rene A, Himmelstein Daniel S, Zhang Ran, Hartmann Boris M, Zaslavsky Elena, Sealfon Stuart C, … Troyanskaya Olga G Nature Genetics (2015-April-27) https://doi.org/f7dvkv DOI: 10.1038/ng.3259 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Network propagation: a universal amplifier of genetic associations Cowen Lenore, Ideker Trey, Raphael Benjamin J, Sharan Roded Nature Reviews Genetics (2017-June-12) https://doi.org/gbhkwn DOI: 10.1038/nrg.2017.38 [DOI] [PubMed] [Google Scholar]
- 53.Benchmarking network propagation methods for disease gene identification Picart-Armada Sergio, Barrett Steven J, Willé David R, Perera-Lluna Alexandre, Gutteridge Alex, Dessailly Benoit H PLOS Computational Biology (2019-September-03) https://doi.org/gtsx8k DOI: 10.1371/journal.pcbi.1007276 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krishnan Arjun, Zhang Ran, Yao Victoria, Theesfeld Chandra L, Wong Aaron K, Tadych Alicja, Volfovsky Natalia, Packer Alan, Lash Alex, Troyanskaya Olga G Nature Neuroscience (2016-August-01) https://doi.org/f889pv DOI: 10.1038/nn.4353 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.XGDAG: explainable gene–disease associations via graph neural networks Mastropietro Andrea, De Carlo Gianluca, Anagnostopoulos Aris Bioinformatics (2023-August-01) https://doi.org/gtsx8h DOI: 10.1093/bioinformatics/btad482 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An Ancient Evolutionary Origin of Genes Associated with Human Genetic Diseases Domazet-Loso T, Tautz D Molecular Biology and Evolution (2008-August-05) https://doi.org/bpbtgp DOI: 10.1093/molbev/msn214 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Similarly Strong Purifying Selection Acts on Human Disease Genes of All Evolutionary Ages Cai James J, Borenstein Elhanan, Chen Rong, Petrov Dmitri A Genome Biology and Evolution (2009-January-01) https://doi.org/bgbqrk DOI: 10.1093/gbe/evp013 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Genome-wide identification of genes likely to be involved in human genetic disease Lopez-Bigas N Nucleic Acids Research (2004-June-02) https://doi.org/bbzrfz DOI: 10.1093/nar/gkh605 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.On the Origins of Mendelian Disease Genes in Man: The Impact of Gene Duplication Dickerson JE, Robertson DL Molecular Biology and Evolution (2011-June-24) https://doi.org/czx744 DOI: 10.1093/molbev/msr111 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles Pellegrini Matteo, Marcotte Edward M, Thompson Michael J, Eisenberg David, Yeates Todd O Proceedings of the National Academy of Sciences (1999-April-13) https://doi.org/br9862 DOI: 10.1073/pnas.96.8.4285 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Detecting Protein Function and Protein-Protein Interactions from Genome Sequences Marcotte Edward M, Pellegrini Matteo, Ng Ho-Leung, Rice Danny W, Yeates Todd O, Eisenberg David Science (1999-July-30) https://doi.org/fxg2p3 DOI: 10.1126/science.285.5428.751 [DOI] [PubMed] [Google Scholar]
- 62.Evolutionary profiling reveals the heterogeneous origins of classes of human disease genes: implications for modeling disease genetics in animals Maxwell Evan K, Schnitzler Christine E, Havlak Paul, Putnam Nicholas H, Nguyen Anh-Dao, Moreland RTravis, Baxevanis Andreas D BMC Evolutionary Biology (2014-October-04) https://doi.org/f6qffg DOI: 10.1186/s12862-014-0212-1 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.OMIM.org: leveraging knowledge across phenotype–gene relationships Amberger Joanna S, Bocchini Carol A, Scott Alan F, Hamosh Ada Nucleic Acids Research (2018-November-16) https://doi.org/gjxh3j DOI: 10.1093/nar/gky1151 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling Tabach Yuval, Golan Tamar, Hernández-Hernández Abrahan, Messer Arielle R, Fukuda Tomoyuki, Kouznetsova Anna, Liu Jian-Guo, Lilienthal Ingrid, Levy Carmit, Ruvkun Gary Molecular Systems Biology (2013-January) https://doi.org/f2pc78 DOI: 10.1038/msb.2013.50 · This study uses phylogenetic profiles to enable systematic identifications of genome-wide functional modules that co-occur across eukaryotic phylogeny.
- 65.Systematic Discovery of Human Gene Function and Principles of Modular Organization through Phylogenetic Profiling Dey Gautam, Jaimovich Ariel, Collins Sean R, Seki Akiko, Meyer Tobias Cell Reports (2015-February) https://doi.org/ggvqsm DOI: 10.1016/j.celrep.2015.01.025 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.High-throughput mouse phenomics for characterizing mammalian gene function Brown Steve DM, Holmes Chris C, Mallon Ann-Marie, Meehan Terrence F, Smedley Damian, Wells Sara Nature Reviews Genetics (2018-April-06) https://doi.org/gr8fcd DOI: 10.1038/s41576-018-0005-2 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Progress towards completing the mutant mouse null resource Peterson Kevin A, Murray Stephen A Mammalian Genome (2021-October-26) https://doi.org/gr8fcc DOI: 10.1007/s00335-021-09905-0 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harmonizing model organism data in the Alliance of Genome Resources, Agapite Julie, Albou Laurent-Philippe, Aleksander Suzanne A, Alexander Micheal, Anagnostopoulos Anna V, Antonazzo Giulia, Argasinska Joanna, Arnaboldi Valerio, Attrill Helen, … Zytkovicz Mark Genetics (2022-February-25) https://doi.org/gr8fcg DOI: 10.1093/genetics/iyac022 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species Shefchek Kent A, Harris Nomi L, Gargano Michael, Matentzoglu Nicolas, Unni Deepak, Brush Matthew, Keith Daniel, Conlin Tom, Vasilevsky Nicole, Zhang Xingmin Aaron, … Osumi-Sutherland David Nucleic Acids Research (2019-November-08) https://doi.org/ghjzgs DOI: 10.1093/nar/gkz997 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.uPheno 2: Framework for standardised representation of phenotypes across species Matentzoglu Nico, Osumi-Sutherland David, Balhoff James P, Bello Susan, Bradford Yvonne, Cardmody Leigh, Grove Chris, Harris Midori A, Harris Nomi, Köhler Sebastian, … Haendel Melissa F1000 Research Limited (2019) https://doi.org/gr8fcj DOI: 10.7490/f1000research.1116540.1 [DOI] [Google Scholar]
- 71.Next-generation diagnostics and disease-gene discovery with the Exomiser Smedley Damian, Jacobsen Julius OB, Jäger Marten, Köhler Sebastian, Holtgrewe Manuel, Schubach Max, Siragusa Enrico, Zemojtel Tomasz, Buske Orion J, Washington Nicole L, … Robinson Peter N Nature Protocols (2015-November-12) https://doi.org/f73qtp DOI: 10.1038/nprot.2015.124 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.The Monarch Initiative in 2024: an analytic platform integrating phenotypes, genes and diseases across species Putman Tim E, Schaper Kevin, Matentzoglu Nicolas, Rubinetti Vincent P, Alquaddoomi Faisal S, Cox Corey, Caufield JHarry, Elsarboukh Glass, Gehrke Sarah, Hegde Harshad, … Munoz-Torres Monica C Nucleic Acids Research (2023-November-24) https://doi.org/gs6kmr DOI: 10.1093/nar/gkad1082 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Contribution of model organism phenotypes to the computational identification of human disease genes Alghamdi Sarah M, Schofield Paul N, Hoehndorf Robert Disease Models & Mechanisms (2022-July-01) https://doi.org/gr8fch DOI: 10.1242/dmm.049441 · This study uses phenotype ontologies to relate research organisms-specific and human-specific ontologies. By treating connected ontologies as graphs, they also proposed using graph embedding-based ML approaches to predict disease-gene associations across species.
- 74.Prioritizing genomic variants through neuro-symbolic, knowledge-enhanced learning Althagafi Azza, Zhapa-Camacho Fernando, Hoehndorf Robert Bioinformatics (2024-May-01) https://doi.org/gt6n4x DOI: 10.1093/bioinformatics/btae301 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Convergent Evolution of Enzyme Active Sites Is not a Rare Phenomenon Gherardini Pier Federico, Wass Mark N, Helmer-Citterich Manuela, Sternberg Michael JE Journal of Molecular Biology (2007-September) https://doi.org/cc7qn6 DOI: 10.1016/j.jmb.2007.06.017 [DOI] [PubMed] [Google Scholar]
- 76. Gene analogue finder: a GRID solution for finding functionally analogous gene products Tulipano Angelica, Donvito Giacinto, Licciulli Flavio, Maggi Giorgio, Gisel Andreas BMC Bioinformatics (2007-September-03) https://doi.org/dtn246 DOI: 10.1186/1471-2105-8-329 · This study introduced Gene Analogue Finder, a method to find agnologous gene pairs across species based on overlapping GO term annotations.
- 77.TreeFam: a curated database of phylogenetic trees of animal gene families Li H Nucleic Acids Research (2006-January-01) https://doi.org/fss2j9 DOI: 10.1093/nar/gkj118 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Functional protein representations from biological networks enable diverse cross-species inference Fan Jason, Cannistra Anthony, Fried Inbar, Lim Tim, Schaffner Thomas, Crovella Mark, Hescott Benjamin, Leiserson Mark DM Nucleic Acids Research (2019-March-08) https://doi.org/gmdn7p DOI: 10.1093/nar/gkz132 · This study introduces MUNK, a method that utilizes aligned PPI network representations to identify agnoglous gene pairs. Additionally, this work expands the concept of Phenologs, characterizing them by a significant overlap of agnoglous gene pairs.
- 79.MUNDO: protein function prediction embedded in a multispecies world Arsenescu Victor, Devkota Kapil, Erden Mert, Shpilker Polina, Werenski Matthew, Cowen Lenore J Bioinformatics Advances (2021-September-29) https://doi.org/gtsx8g DOI: 10.1093/bioadv/vbab025 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Joint embedding of biological networks for cross-species functional alignment Li Lechuan, Dannenfelser Ruth, Zhu Yu, Hejduk Nathaniel, Segarra Santiago, Yao Vicky Bioinformatics (2023-August-26) https://doi.org/gtsx8j DOI: 10.1093/bioinformatics/btad529 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Systematic discovery of nonobvious human disease models through orthologous phenotypes McGary Kriston L, Park Tae Joo, Woods John O, Cha Hye Ji, Wallingford John B, Marcotte Edward M Proceedings of the National Academy of Sciences (2010-March-22) https://doi.org/b8hnkn DOI: 10.1073/pnas.0910200107 · This study introduced the concept of Phenologs, which facilitates finding phenotype counterparts across species by looking at ortholog overlap between genes annotated to phenotypes.
- 82. CoCoCoNet: conserved and comparative co-expression across a diverse set of species Lee John, Shah Manthan, Ballouz Sara, Crow Megan, Gillis Jesse Nucleic Acids Research (2020-May-11) https://doi.org/ghjzdz DOI: 10.1093/nar/gkaa348 · This study introduced CoCoCoNet, a method that helps find agnoglous gene sets across species by comparing co-expression networks generated in different species.
- 83.xHeinz: an algorithm for mining cross-species network modules under a flexible conservation model El-Kebir Mohammed, Soueidan Hayssam, Hume Thomas, Beisser Daniela, Dittrich Marcus, Müller Tobias, Blin Guillaume, Heringa Jaap, Nikolski Macha, Wessels Lodewyk FA, Klau Gunnar W Bioinformatics (2015-May-27) https://doi.org/f7vck8 DOI: 10.1093/bioinformatics/btv316 [DOI] [PubMed] [Google Scholar]
- 84. A Scalable Approach for Discovering Conserved Active Subnetworks across Species Deshpande Raamesh, Sharma Shikha, Verfaillie Catherine M, Hu Wei-Shou, Myers Chad L PLoS Computational Biology (2010-December-09) https://doi.org/bfn9rk DOI: 10.1371/journal.pcbi.1001028 · This study introduced neXus, a method that integrates differentially expressed gene lists and network topology to find agnoglous gene modules across species.
- 85.ModuleBlast: identifying activated sub-networks within and across species Zinman Guy E, Naiman Shoshana, O’Dee Dawn M, Kumar Nishant, Nau Gerard J, Cohen Haim Y, B Ziv ar-Joseph Nucleic Acids Research (2014-November-26) https://doi.org/f66rt7 DOI: 10.1093/nar/gku1224 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Transcriptomic congruence analysis for evaluating model organisms Zong Wei, Rahman Tanbin, Zhu Li, Zeng Xiangrui, Zhang Yingjin, Zou Jian, Liu Song, Ren Zhao, Li Jingyi Jessica, Sibille Etienne, … Tseng George C Proceedings of the National Academy of Sciences (2023-February-02) https://doi.org/grqm9w DOI: 10.1073/pnas.2202584120 · This study introduced CAMO, a method to systematically quantify the similarity between perturbed transcriptomic profiles across species.
- 87.Genomic responses in mouse models poorly mimic human inflammatory diseases Seok Junhee, Warren HShaw, Cuenca Alex G, Mindrinos Michael N, Baker Henry V, Xu Weihong, Richards Daniel R, McDonald-Smith Grace P, Gao Hong, Hennessy Laura, … Proceedings of the National Academy of Sciences (2013-February-11) https://doi.org/f4p74d DOI: 10.1073/pnas.1222878110 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Genomic responses in mouse models greatly mimic human inflammatory diseases Takao Keizo, Miyakawa Tsuyoshi Proceedings of the National Academy of Sciences (2014-August-04) https://doi.org/f6zqzv DOI: 10.1073/pnas.1401965111 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Found In Translation: a machine learning model for mouse-to-human inference Normand Rachelly, Du Wenfei, Briller Mayan, Gaujoux Renaud, Starosvetsky Elina, Ziv-Kenet Amit, Shalev-Malul Gali, Tibshirani Robert J, Shen-Orr Shai S Nature Methods (2018-November-26) https://doi.org/gfkfvg DOI: 10.1038/s41592-018-0214-9 This study introduced FIT, a method that investigated the possibility of transferring DEG results across species using linear models.
- 90. XGSEA: CROSS-species gene set enrichment analysis via domain adaptation Cai Menglan, Hao Canh Nguyen, Hiroshi Mamitsuka, Li Limin Briefings in Bioinformatics (2021-January-30) https://doi.org/gr8hvz DOI: 10.1093/bib/bbaa406 This study introduced XGSEA, a method that transfers gene set enrichment results across species.
- 91.Computational translation of genomic responses from experimental model systems to humans Brubaker Douglas K, Proctor Elizabeth A, Haigis Kevin M, Lauffenburger Douglas A PLOS Computational Biology (2019-January-10) https://doi.org/gr8cpf DOI: 10.1371/journal.pcbi.1006286 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. An interspecies translation model implicates integrin signaling in infliximab-resistant inflammatory bowel disease Brubaker Douglas K, Kumar Manu P, Chiswick Evan L, Gregg Cecil, Starchenko Alina, Vega Paige N, Southard-Smith Austin N, Simmons Alan J, Scoville Elizabeth A, Coburn Lori A, … Lauffenburger Douglas A Science Signaling (2020-August-04) https://doi.org/gqbd3g DOI: 10.1126/scisignal.aay3258 · This study introduced Transcomp-R, a method that allows transferring knowledge across species when the data from each species was not measured using the same omics modality.
- 93.Style transfer with variational autoencoders is a promising approach to RNA-Seq data harmonization and analysis Russkikh Nikolai, Antonets Denis, Shtokalo Dmitry, Makarov Alexander, Vyatkin Yuri, Zakharov Alexey, Terentyev Evgeny Bioinformatics (2020-July-16) https://doi.org/gjrszt DOI: 10.1093/bioinformatics/btaa624 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.scGen predicts single-cell perturbation responses Lotfollahi Mohammad, Wolf FAlexander, Theis Fabian J Nature Methods (2019-July-29) https://doi.org/gf9t5z DOI: 10.1038/s41592-019-0494-8 [DOI] [PubMed] [Google Scholar]
- 95.Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles Subramanian Aravind, Tamayo Pablo, Mootha Vamsi K, Mukherjee Sayan, Ebert Benjamin L, Gillette Michael A, Paulovich Amanda, Pomeroy Scott L, Golub Todd R, Lander Eric S, Mesirov Jill P Proceedings of the National Academy of Sciences (2005-September-30) https://doi.org/d4qbh8 DOI: 10.1073/pnas.0506580102 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.The ancestral gene repertoire of animal stem cells Alié Alexandre, Hayashi Tetsutaro, Sugimura Itsuro, Manuel Michaël, Sugano Wakana, Mano Akira, Satoh Nori, Agata Kiyokazu, Funayama Noriko Proceedings of the National Academy of Sciences (2015-December-07) https://doi.org/gf3xjn DOI: 10.1073/pnas.1514789112 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cross-Species Analysis of Single-Cell Transcriptomic Data Shafer Maxwell ER Frontiers in Cell and Developmental Biology (2019-September-02) https://doi.org/gg2rpw DOI: 10.3389/fcell.2019.00175 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Toward universal cell embeddings: integrating single-cell RNA-seq datasets across species with SATURN Rosen Yanay, Brbić Maria, Roohani Yusuf, Swanson Kyle, Li Ziang, Leskovec Jure Nature Methods (2024-February-16) https://doi.org/gtsx8f DOI: 10.1038/s41592-024-02191-z This study introduced SATURN, a method that uses all genes, regardless of evolutionary relationships, to align single-cell transcriptomes across species. SATURN is also able to simultaneously integrate single-cell transcriptomes from more than two species.
- 99.Evolutionary-scale prediction of atomic-level protein structure with a language model Lin Zeming, Akin Halil, Rao Roshan, Hie Brian, Zhu Zhongkai, Lu Wenting, Smetanin Nikita, Verkuil Robert, Kabeli Ori, Shmueli Yaniv, … Rives Alexander Science (2023-March-17) https://doi.org/grzgts DOI: 10.1126/science.ade2574 [DOI] [PubMed] [Google Scholar]
- 100.Integrating single-cell transcriptomic data across different conditions, technologies, and species Butler Andrew, Hoffman Paul, Smibert Peter, Papalexi Efthymia, Satija Rahul Nature Biotechnology (2018-April-02) https://doi.org/gc87v6 DOI: 10.1038/nbt.4096 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benchmarking atlas-level data integration in single-cell genomics Luecken Malte D, Büttner M, Chaichoompu K, Danese A, Interlandi M, Mueller MF, Strobl DC, Zappia L, Dugas M, Colomé-Tatché M, Theis Fabian J Nature Methods (2021-December-23) https://doi.org/gnvvd5 DOI: 10.1038/s41592-021-01336-8 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors Haghverdi Laleh, Lun Aaron TL, Morgan Michael D, Marioni John C Nature Biotechnology (2018-April-02) https://doi.org/gc8754 DOI: 10.1038/nbt.4091 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Comprehensive Integration of Single-Cell Data Stuart Tim, Butler Andrew, Hoffman Paul, Hafemeister Christoph, Papalexi Efthymia, Mauck III William M, Hao Yuhan, Stoeckius Marlon, Smibert Peter, Satija Rahul Cell (2019-June) https://doi.org/gf3sxv DOI: 10.1016/j.cell.2019.05.031 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Efficient integration of heterogeneous single-cell transcriptomes using Scanorama Hie Brian, Bryson Bryan, Berger Bonnie Nature Biotechnology (2019-May-06) https://doi.org/gf9psf DOI: 10.1038/s41587-019-0113-3 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity Welch Joshua D, Kozareva Velina, Ferreira Ashley, Vanderburg Charles, Martin Carly, Macosko Evan Z Cell (2019-June) https://doi.org/gf3m3v DOI: 10.1016/j.cell.2019.05.006 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Variable paralog expression underlies phenotype variation Bailon-Zambrano Raisa, Sucharov Juliana, Mumme-Monheit Abigail, Murry Matthew, Stenzel Amanda, Pulvino Anthony T, Mitchell Jennyfer M, Colborn Kathryn L, Nichols James T eLife (2022-September-22) https://doi.org/gt6n5b DOI: 10.7554/elife.79247 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.OrthoDisease: tracking disease gene orthologs across 100 species Forslund K, Schreiber F, Thanintorn N, Sonnhammer ELL Briefings in Bioinformatics (2011-May-12) https://doi.org/fhwbjk DOI: 10.1093/bib/bbr024 [DOI] [PubMed] [Google Scholar]
- 108. Benchmarking strategies for cross-species integration of single-cell RNA sequencing data Song Yuyao, Miao Zhichao, Brazma Alvis, Papatheodorou Irene Nature Communications (2023-October-14) https://doi.org/gsv2bj DOI: 10.1038/s41467-023-41855-w · This is a comprehensive benchmarking study that compares methods to align single-cell transcriptomics across species.
- 109. Mapping single-cell atlases throughout Metazoa unravels cell type evolution Tarashansky Alexander J, Musser Jacob M, Khariton Margarita, Li Pengyang, Arendt Detlev, Quake Stephen R, Wang Bo eLife (2021-May-04) https://doi.org/gkczvh DOI: 10.7554/elife.66747 · This study introduced SAMap, a method that utilizes complex homology patterns to align single-cell transcriptomes across a pair of species.
- 110.Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences Rives Alexander, Meier Joshua, Sercu Tom, Goyal Siddharth, Lin Zeming, Liu Jason, Guo Demi, Ott Myle, Zitnick CLawrence, Ma Jerry, Fergus Rob Proceedings of the National Academy of Sciences (2021-April-05) https://doi.org/gmmft5 DOI: 10.1073/pnas.2016239118 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Modeling Human Disease Phenotype in Model Organisms Marian Ali J Circulation Research (2011-August-05) https://doi.org/b2mhvn DOI: 10.1161/circresaha.111.249409 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.A Pathway-Based View of Human Diseases and Disease Relationships Li Yong, Agarwal Pankaj PLoS ONE (2009-February-04) https://doi.org/d7483w DOI: 10.1371/journal.pone.0004346 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shared Biological Pathways Between Alzheimer’s Disease and Ischemic Stroke Cui Pan, Ma Xiaofeng, Li He, Lang Wenjing, Hao Junwei Frontiers in Neuroscience (2018-September-07) https://doi.org/gfbdpb DOI: 10.3389/fnins.2018.00605 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Unravelling the Shared Genetic Mechanisms Underlying 18 Autoimmune Diseases Using a Systems Approach Gokuladhas Sreemol, Schierding William, Golovina Evgeniia, Fadason Tayaza, O’Sullivan Justin Frontiers in Immunology (2021-August-13) https://doi.org/gt6n46 DOI: 10.3389/fimmu.2021.693142 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Molecular Processes Involved in the Shared Pathways between Cardiovascular Diseases and Diabetes Tokarek Julita, Budny Emilian, Saar Maciej, Stańczak Kamila, Wojtanowska Ewa, Młynarska Ewelina, Rysz Jacek, Franczyk Beata Biomedicines (2023-September-23) https://doi.org/gs3jxw DOI: 10.3390/biomedicines11102611 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mapping biological process relationships and disease perturbations within a pathway network Stoney Ruth, Robertson David L, Nenadic Goran, Schwartz Jean-Marc npj Systems Biology and Applications (2018-June-11) https://doi.org/gt6n4v DOI: 10.1038/s41540-018-0055-2 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.A network-based approach for isolating the chronic inflammation gene signatures underlying complex diseases towards finding new treatment opportunities Hickey Stephanie L, McKim Alexander, Mancuso Christopher A, Krishnan Arjun Frontiers in Pharmacology (2022-October-12) https://doi.org/gt6n47 DOI: 10.3389/fphar.2022.995459 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Network analysis reveals rare disease signatures across multiple levels of biological organization Buphamalai Pisanu, Kokotovic Tomislav, Nagy Vanja, Menche Jörg Nature Communications (2021-November-09) https://doi.org/gnpnsc DOI: 10.1038/s41467-021-26674-1 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.The Human Phenotype Ontology in 2024: phenotypes around the world Gargano Michael A, Matentzoglu Nicolas, Coleman Ben, Addo-Lartey Eunice B, Anagnostopoulos Anna V, Anderton Joel, Avillach Paul, Bagley Anita M, Bakštein Eduard, Balhoff James P, … Robinson Peter N Nucleic Acids Research (2023-November-11) https://doi.org/gt6qm8 DOI: 10.1093/nar/gkad1005 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Common genetic variation associated with Mendelian disease severity revealed through cryptic phenotype analysis Blair David R, Hoffmann Thomas J, Shieh Joseph T Nature Communications (2022-June-27) https://doi.org/gt6qm7 DOI: 10.1038/s41467-022-31030-y · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maassen Willem, Legger Geertje, Cinar Ovgu Kul, van Daele Paul, Gattorno Marco, Bader-Meunier Brigitte, Wouters Carine, Briggs Tracy, Johansson Lennart, van der Velde Joeri, … van Gijn Marielle Frontiers in Immunology (2023-September-12) https://doi.org/gt6qm9 DOI: 10.3389/fimmu.2023.1215869 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gene Loss and Evolutionary Rates Following Whole-Genome Duplication in Teleost Fishes Brunet Frédéric G, Roest Crollius Hugues, Paris Mathilde, Aury Jean-Marc, Gibert Patricia, Jaillon Olivier, Laudet Vincent, Robinson-Rechavi Marc Molecular Biology and Evolution (2006-June-29) https://doi.org/c4tn7b DOI: 10.1093/molbev/msl049 [DOI] [PubMed] [Google Scholar]
- 123.Developmental system drift and flexibility in evolutionary trajectories True John R, Haag Eric S Evolution and Development (2001-March) https://doi.org/fd5m8w DOI: 10.1046/j.1525-142x.2001.003002109.x [DOI] [PubMed] [Google Scholar]
- 124.Interactome evolution: insights from genome-wide analyses of protein–protein interactions Ghadie Mohamed A, Coulombe-Huntington Jasmin, Xia Yu Current Opinion in Structural Biology (2018-June) https://doi.org/gd7tzw DOI: 10.1016/j.sbi.2017.10.012 [DOI] [PubMed] [Google Scholar]
- 125.The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest Szklarczyk Damian, Kirsch Rebecca, Koutrouli Mikaela, Nastou Katerina, Mehryary Farrokh, Hachilif Radja, Gable Annika L, Fang Tao, Doncheva Nadezhda T, Pyysalo Sampo, … von Mering Christian Nucleic Acids Research (2022-November-12) https://doi.org/gs7sn3 DOI: 10.1093/nar/gkac1000 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.INTREPPPID - An Orthologue-Informed Quintuplet Network for Cross-Species Prediction of Protein-Protein Interaction Szymborski Joseph, Emad Amin Cold Spring Harbor Laboratory (2024-February-16) https://doi.org/gt6n44 DOI: 10.1101/2024.02.13.580150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Protein Interaction Mapping in C. elegans Using Proteins Involved in Vulval Development Walhout Albertha JM, Sordella Raffaella, Lu Xiaowei, Hartley James L, Temple Gary F, Brasch Michael A, Thierry-Mieg Nicolas, Vidal Marc Science (2000-January-07) https://doi.org/db3c99 DOI: 10.1126/science.287.5450.116 [DOI] [PubMed] [Google Scholar]
- 128.Annotation Transfer Between Genomes: Protein–Protein Interologs and Protein–DNA Regulogs Yu Haiyuan, Luscombe Nicholas M, Lu Hao Xin, Zhu Xiaowei, Xia Yu, Han Jing-Dong J, Bertin Nicolas, Chung Sambath, Vidal Marc, Gerstein Mark Genome Research (2004-June) https://doi.org/fn7gwg DOI: 10.1101/gr.1774904 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.IMP 2.0: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks Wong Aaron K, Krishnan Arjun, Yao Victoria, Tadych Alicja, Troyanskaya Olga G Nucleic Acids Research (2015-May-12) https://doi.org/f7nxbp DOI: 10.1093/nar/gkv486 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Machine learning methods to model multicellular complexity and tissue specificity Sealfon Rachel SG, Wong Aaron K, Troyanskaya Olga G Nature Reviews Materials (2021-July-15) https://doi.org/gr83px DOI: 10.1038/s41578-021-00339-3 [DOI] [Google Scholar]
- 131.Co-expression networks reveal the tissue-specific regulation of transcription and splicing Saha Ashis, Kim Yungil, Gewirtz Ariel DH, Jo Brian, Gao Chuan, McDowell Ian C, Engelhardt Barbara E, Battle Alexis Genome Research (2017-October-11) https://doi.org/gb2qx6 DOI: 10.1101/gr.216721.116 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Addressing noise in co-expression network construction Burns Joshua JR, Shealy Benjamin T, Greer Mitchell S, Hadish John A, McGowan Matthew T, Biggs Tyler, Smith Melissa C, Feltus FAlex, Ficklin Stephen P Briefings in Bioinformatics (2021-November-30) https://doi.org/gr83p2 DOI: 10.1093/bib/bbab495 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Applying differential network analysis to longitudinal gene expression in response to perturbations Xue Shuyue, Rogers Lavida RK, Zheng Minzhang, He Jin, Piermarocchi Carlo, Mias George I Frontiers in Genetics (2022-October-17) https://doi.org/gr83p3 DOI: 10.3389/fgene.2022.1026487 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.CONE: COntext-specific Network Embedding via Contextualized Graph Attention Liu Renming, Yuan Hao, Johnson Kayla A, Krishnan Arjun Cold Spring Harbor Laboratory (2023-October-24) https://doi.org/gt6n42 DOI: 10.1101/2023.10.21.563390 [DOI] [Google Scholar]
- 135.Contextual AI models for single-cell protein biology Li Michelle M, Huang Yepeng, Sumathipala Marissa, Liang Man Qing, Valdeolivas Alberto, Ananthakrishnan Ashwin N, Liao Katherine, Marbach Daniel, Zitnik Marinka Nature Methods (2024-July-22) https://doi.org/gt6n4w DOI: 10.1038/s41592-024-02341-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Predicting multicellular function through multi-layer tissue networks Zitnik Marinka, Leskovec Jure Bioinformatics (2017-July-12) https://doi.org/gbnk7p DOI: 10.1093/bioinformatics/btx252 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Protein complex prediction with AlphaFold-Multimer Evans Richard, O’Neill Michael, Pritzel Alexander, Antropova Natasha, Senior Andrew, Green Tim, Žídek Augustin, Bates Russ, Blackwell Sam, Yim Jason, … Hassabis Demis Cold Spring Harbor Laboratory (2021-October-04) https://doi.org/gm2vcp DOI: 10.1101/2021.10.04.463034 [DOI] [Google Scholar]
- 138.Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk Zhou Jian, Theesfeld Chandra L, Yao Kevin, Chen Kathleen M, Wong Aaron K, Troyanskaya Olga G Nature Genetics (2018-July-16) https://doi.org/gdvmqw DOI: 10.1038/s41588-018-0160-6 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Atlas of primary cell-type-specific sequence models of gene expression and variant effects Sokolova Ksenia, Theesfeld Chandra L, Wong Aaron K, Zhang Zijun, Dolinski Kara, Troyanskaya Olga G Cell Reports Methods (2023-September) https://doi.org/gt6n4t DOI: 10.1016/j.crmeth.2023.100580 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Towards complete and error-free genome assemblies of all vertebrate species Rhie Arang, McCarthy Shane A, Fedrigo Olivier, Damas Joana, Formenti Giulio, Koren Sergey, Uliano-Silva Marcela, Chow William, Fungtammasan Arkarachai, Kim Juwan, … Jarvis Erich D Nature (2021-April-28) https://doi.org/gjtrn9 DOI: 10.1038/s41586-021-03451-0 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Graph representation learning in biomedicine and healthcare Li Michelle M, Huang Kexin, Zitnik Marinka Nature Biomedical Engineering (2022-October-31) https://doi.org/gq533s DOI: 10.1038/s41551-022-00942-x · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.The effects of biological knowledge graph topology on embedding-based link prediction Bradshaw Michael S, Gaskell Alisa, Layer Ryan M Cold Spring Harbor Laboratory (2024-June-11) https://doi.org/gt6n45 DOI: 10.1101/2024.06.10.598277 [DOI] [Google Scholar]
- 143.Phenomics Assistant: An Interface for LLM-based Biomedical Knowledge Graph Exploration O’Neil Shawn T, Schaper Kevin, Elsarboukh Glass, Reese Justin T, Moxon Sierra AT, Harris Nomi L, Munoz-Torres Monica C, Robinson Peter N, Haendel Melissa A, Mungall Christopher J Cold Spring Harbor Laboratory (2024-February-02) https://doi.org/gt6n43 DOI: 10.1101/2024.01.31.578275 [DOI] [Google Scholar]
- 144.Gene Ontology: Pitfalls, Biases, and Remedies Gaudet Pascale, Dessimoz Christophe Methods in Molecular Biology (2016-November-04) https://doi.org/gf4d5v DOI: 10.1007/978-1-4939-3743-1_14 [DOI] [PubMed] [Google Scholar]
- 145.Structured prompt interrogation and recursive extraction of semantics (SPIRES): A method for populating knowledge bases using zero-shot learning Caufield JHarry, Hegde Harshad, Emonet Vincent, Harris Nomi L, Joachimiak Marcin P, Matentzoglu Nicolas, Kim HyeongSik, Moxon Sierra AT, Reese Justin T, Haendel Melissa A, … Mungall Christopher J arXiv (2023) https://doi.org/gt6n48 DOI: 10.48550/arxiv.2304.02711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.The zebrafish reference genome sequence and its relationship to the human genome Howe Kerstin, Clark Matthew D, Torroja Carlos F, Torrance James, Berthelot Camille, Muffato Matthieu, Collins John E, Humphray Sean, McLaren Karen, Matthews Lucy, … Stemple Derek L Nature (2013-April-17) https://doi.org/k9c DOI: 10.1038/nature12111 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Non-model model organisms Russell James J, Theriot Julie A, Sood Pranidhi, Marshall Wallace F, Landweber Laura F, Fritz-Laylin Lillian, Polka Jessica K, Oliferenko Snezhana, Gerbich Therese, Gladfelter Amy, … Brunet Anne BMC Biology (2017-June-29) https://doi.org/gfc837 DOI: 10.1186/s12915-017-0391-5 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.A chromosome-scale assembly of the axolotl genome Smith Jeramiah J, Timoshevskaya Nataliya, Timoshevskiy Vladimir A, Keinath Melissa C, Hardy Drew, Voss SRandal Genome Research (2019-January-24) https://doi.org/gf5xcq DOI: 10.1101/gr.241901.118 · [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.An organismal perspective on C. intestinalis development, origins and diversification Kourakis Matthew J, Smith William C eLife (2015-March-25) https://doi.org/gt6n49 DOI: 10.7554/elife.06024 · [DOI] [PMC free article] [PubMed] [Google Scholar]