Abstract
Advances have led to a greater understanding of the genetics of Alzheimer’s Disease (AD). However, the gap between the predicted and observed genetic heritability estimates when using single nucleotide polymorphisms (SNPs) and small indel data remains. Large genomic rearrangements, known as structural variants (SVs), have the potential to account for this missing genetic heritability. By leveraging data from two ongoing cohort studies of aging and dementia, the Religious Orders Study and Rush Memory and Aging Project (ROS/MAP), we performed genome-wide association analysis testing around 20,000 common SVs from 1,088 participants with whole genome sequencing (WGS) data. A range of Alzheimer’s Disease and Related Disorders (AD/ADRD) clinical and pathologic traits were examined. Given the limited sample size, no genome-wide significant association was found, but we mapped SVs across 81 AD risk loci and discovered 22 SVs in linkage disequilibrium (LD) with GWAS lead variants and directly associated with AD/ADRD phenotypes (nominal P < 0.05). The strongest association was a deletion of an Alu element in the 3’UTR of the TMEM106B gene. This SV was in high LD with the respective AD GWAS locus and was associated with multiple AD/ADRD phenotypes, including tangle density, TDP-43, and cognitive resilience. The deletion of this element was also linked to lower TMEM106B protein abundance. We also found a 22 kb deletion associated with depression in ROSMAP and bearing similar association patterns as AD GWAS SNPs at the IQCK locus. In addition, genome-wide scans allowed the identification of 7 SVs, with no LD with SNPs and nominally associated with AD/ADRD traits. This result suggests potentially new ADRD risk loci not discoverable using SNP data. Among these findings, we highlight a 5.6 kb duplication of coding regions of the gene C1orf186 at chromosome 1 associated with indices of cognitive impairment, decline, and resilience. While further replication in independent datasets is needed to validate these findings, our results support the potential roles of common structural variations in the pathogenesis of AD/ADRD.
BACKGROUND
Alzheimer’s disease (AD) is a complex disorder heavily influenced by genetics. While twin studies have estimated the broad-sense heritability of AD between 60–80% (Gatz et al. 2006), SNP-based heritability varies across different studies, ranging from 5–30% (Escott-Price and Hardy 2022; Wang et al. 2021). Epistatic interactions, unbalanced age-matched controls, and the large proportion of “proxy” cases are among the reasons that could explain such a gap between the expected and observed heritability (Escott-Price and Hardy 2022). Large genomic rearrangements, known as structural variants (SVs), can also account for some missing genetic heritability. This class of variants, often not discoverable using genotyping assays, is recently garnering attention with the popularization of whole-genome sequencing data which allows detection of such variants at a large scale with considerable confidence (Collins et al. 2020; Sudmant et al. 2015).
Although a few SVs have already been linked to AD, with the most well-known being rare duplications of the APP gene causally linked to the early onset form of the disease (Blom et al. 2008; Hooli et al. 2012; Kasuga et al. 2009; Rovelet-Lecrux et al. 2006; Sleegers et al. 2006), our knowledge of the impact of common SVs in late-onset AD (LOAD) is mostly limited to a few copy number variants (CNVs), which are usually detected using SNP arrays or PCR assays and identified in small sample-size studies that have weak replication (Wang et al. 2022). For other neurodegenerative diseases, the role of SVs is usually clearer. For example, in frontotemporal dementia (FTD), repeat expansions in C9orf72 and the MAPT locus inversion are linked to disease risk (Baker et al. 1999; DeJesus-Hernandez et al. 2011). The same inversion is also a major genetic risk factor for progressive supranuclear palsy (PSP) (Baker et al. 1999), and it is associated with Parkinson’s disease (PD) (Zabetian et al. 2007). Additionally, a recent study found a common deletion in the gene TPCN1 associated with Lewy body dementia (LBD) in a locus also associated with AD GWAS (Bellenguez et al. 2022; Kaivola et al. 2023). These findings highlight the fact that mapping SVs across multiple diseases and related clinical and neuropathological traits can be beneficial to understanding their role in complex diseases.
To investigate the role of SVs in AD/ADRD traits, we leveraged data from two ongoing cohort studies of aging and dementia: the Religious Orders Study and the Rush Memory and Aging Project (ROS/MAP). These cohorts benefit from having a comprehensive set of phenotypes measured from the same set of individuals. We previously reported that SVs have an impact on many molecular phenotypes in the human brain, including over 300 SVs that were also in LD with GWAS traits (Vialle et al. 2022). Here we extend our findings in many ways by effectively linking SVs to an extensive list of AD/ADRD phenotypes measured for the same individuals. Over 20 thousand common SVs mapped in 1,088 samples with whole genome sequencing (WGS) data were tested for association with multiple indices for AD/ADRD, including clinical diagnoses of AD dementia and mild cognitive impairment (MCI); multiple measurements of cognition (e.g., cognitive decline and resilience); diagnosis of major depressive disorder (MDD) and depressive symptoms; indices of motor function; neuropathological evaluations of -amyloid load, neurofibrillary tangles density, Lewy Bodies and TDP-43; and indices of cerebrovascular diseases (CVD) such as cerebral amyloid angiopathy (CAA), gross and microinfarcts, atherosclerosis, and arteriolosclerosis.
RESULTS
Characteristics of the samples
Genotyped structural variant calls were obtained on 1,088 non-Latino white subjects from the ROS/MAP cohort studies (David A. Bennett et al. 2018). In both studies, participants underwent annual clinical evaluations and donated their brains at death. The mean (SD) age at enrollment and age at death across the participants used in this study was 80.9 (6.8) and 89.0 (6.4) years, respectively, with an average (SD) follow-up period of 7.2 (4.9) years. Of all participants, 43.7% had a diagnosis of Alzheimer’s dementia at death, and nearly two-thirds had pathologic AD confirmed post-mortem. A decline in cognition was observed, with the Mini-Mental State Examination (MMSE) score decreasing from 28 (IQR 26–29) at baseline to 25 (IQR 15–28) proximate to death. TDP-43 pathology extending beyond the amygdala was observed in just over a third of brains, and 13% presented Lewy bodies in nigra and/or cortex regions. Cerebrovascular diseases, including macroscopic infarcts and microinfarcts, were observed in more than a third and a quarter, respectively, and moderate to severe amyloid angiopathy, atherosclerosis, and arteriolosclerosis were observed in about a third of the brains. Detailed characteristics of each sample are presented in Supplementary Table 1.
Genome-wide SV association scans with clinical and neuropathologic phenotypes
Genome-wide association scans of structural variants were performed on a range of clinical and neuropathologic phenotypes covering multiple clinical and pathologic variables related to aging and dementia. Twenty four phenotypes were analyzed, including clinical diagnosis of AD, MCI, and MDD; depressive symptomatology; measurements of cognitive function, motor function, frailty and parkinsonian scores; neuropathologies, such as -amyloid, tangles, TDP-43, Lewy Bodies, and multiple cerebrovascular diseases indices. SV discovering and genotyping pipelines were obtained from combined ROS/MAP samples as previously described (Vialle et al. 2022). SVs were classified into different classes of variation, including deletions (DEL), insertions (INS), duplications (DUP), inversions (INV), complex rearrangements (CPX), and three classes of mobile element insertions (MEI), Alu, SVA, and LINE1. A total of 72,348 SVs were initially mapped. For the association analysis, variants with Hardy-Weinberg equilibrium (HWE) P-value lower than 10−6 and minor allele frequency (MAF) lower than 1% were removed. Single variant association tests were applied across 16 quantitative and eight binary traits (Supplementary Table 2) for about 20 thousand SVs with minor allele count (MAC) greater than 10 in each cohort. Scans were performed using the tool SAIGEgds (Scalable and Accurate Implementation of GEneralized mixed model) (Zheng and Davis 2021; Zhou et al. 2018). All tests were controlled by age at death, sex, years of education, five genetic principal components, and the genetic correlation matrix (modeled as a random effect). A meta-analysis was performed combining results from both ROS and MAP cohorts. Genomic inflation () ranged from 0.59 to 1.08, indicating a reasonable control of population stratification and relatedness (Table 1). None SVs reached genome-wide significance (P < 5 × 10−8) in any of the samples and phenotypes tested at the current sample size. Complete summary stats for each phenotype are provided in Additional Data (see Data availability).
Table 1.
Genomic inflation of genome wide association tests in each cohort and meta-analysis
| Phenotype | ROS () | MAP () | Meta-Analysis (ROS/MAP) () |
|---|---|---|---|
| Cognition | |||
| Alzheimer’s Dementia | 1.01 | 1.01 | 0.99 |
| Mild cognitive impairment | 1.01 | 0.99 | 0.98 |
| Global cognitive function | 1.00 | 0.73 | 0.89 |
| Cognitive decline | 1.02 | 1.02 | 0.97 |
| Cognitive resilience | 1.02 | 1.01 | 1.03 |
| Depression | |||
| Depressive symptoms (CES-D) | 0.67 | 1.03 | 1.01 |
| Major Depressive Disorder | 1.08 | 1.02 | 1.03 |
| Motor | |||
| Frailty | 0.59 | 0.99 | 1.02 |
| Motor functions | 0.90 | 1.01 | 0.94 |
| Global parkinsonian score | 0.91 | 0.85 | 1.00 |
| Pathology | |||
| NIA-Reagan diagnosis of AD | 1.01 | 1.03 | 0.99 |
| Global AD pathology | 0.99 | 1.00 | 0.99 |
| Beta-Amyloid | 1.01 | 1.00 | 1.01 |
| Tangle density | 1.03 | 0.99 | 0.99 |
| Diffuse plaques | 1.01 | 0.91 | 0.99 |
| Neuritic plaques | 1.01 | 1.02 | 1.00 |
| Neurofibrillary tangle | 1.01 | 0.86 | 0.92 |
| TDP-43 | 1.01 | 0.99 | 1.01 |
| Lewy Body disease | 0.99 | 1.00 | 1.00 |
| Vascular Pathology | |||
| Arteriolosclerosis | 1.03 | 0.96 | 0.99 |
| Cerebral Atherosclerosis | 1.03 | 0.99 | 1.00 |
| Cerebral amyloid angiopathy | 1.00 | 0.99 | 1.00 |
| Cerebral Infarctions (Gross) | 1.01 | 1.01 | 1.00 |
| Cerebral Infarctions (Micro) | 1.00 | 1.00 | 1.00 |
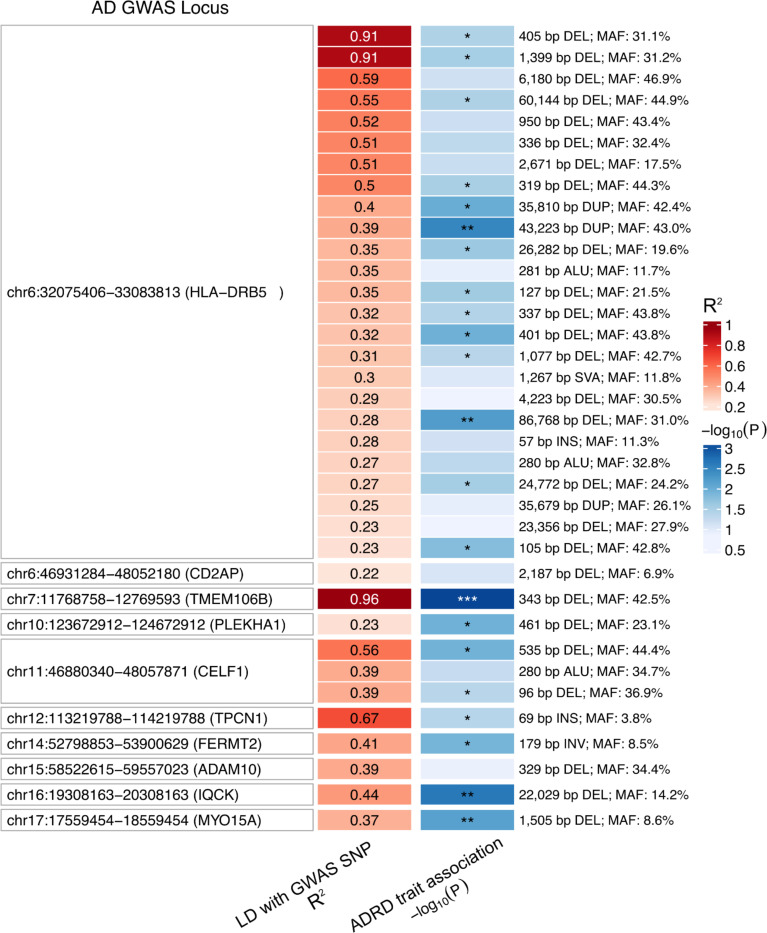
SVs in AD GWAS loci
To investigate the impact of SVs in known AD GWAS loci, we examined 81 genome-wide significant loci collectively identified in six previous studies that did not examine SVs (Andrews et al. 2023). We first mapped the presence of SVs (discovered in ROS/MAP) in each locus. On average, 10 SVs were identified by locus across all 81 loci (Supplementary Figure S1). As expected, complex genomic regions, such as the HLA locus, harbored a considerably higher number of SVs (75), followed by the ABCA7 (47), the TMEM121 (35), and IDUA (32) loci. 36 SVs were in LD with the lead variant in 10 of the 81 loci, with R2 ranging from 0.217 to 0.964. Among these, 22 SVs were nominally associated (P < 0.05) with at least one of the 24 AD/ADRD phenotypes tested (Figure 1 and Supplementary Table 3).
Figure 1. Structural variants in LD with AD GWAS variants.
Heatmap shows 36 SVs in LD (R2 > 0.2) with 10 AD GWAS loci lead SNPs. The best association of each SV with ROS/MAP AD/ADRD phenotypes is highlighted in the blue colored column (shown as −log10(nominal P); *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
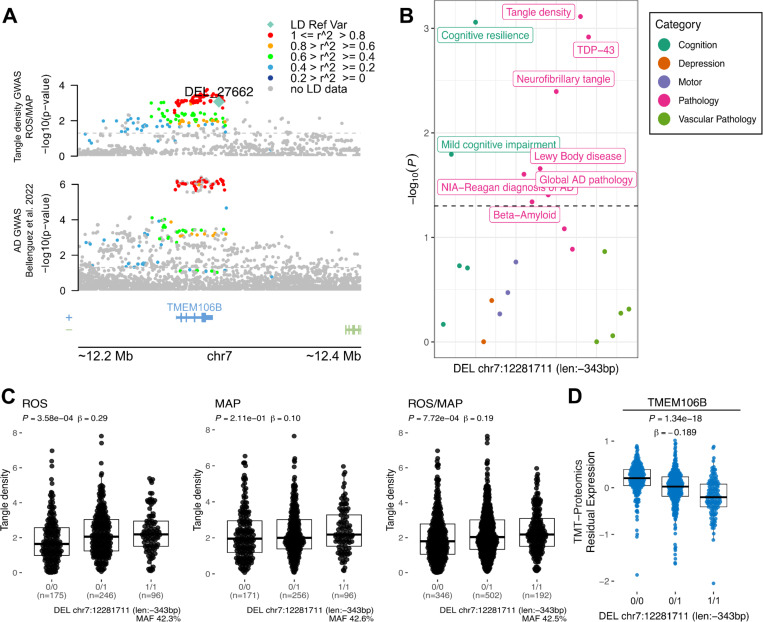
The SV with the strongest results (P = 7.72 × 10−4) was a 343 bp deletion (Figure 2C), which deletes an Alu element at the 3’UTR of TMEM106B (Rodney et al. 2024), and was in high LD with the lead variant at the TMEM106B locus (rs5011436; R2 = 0.96) (Figure 2A). In ROS/MAP participants, this SV was associated with multiple AD/ADRD phenotypes, including tangle density, cognitive resilience, TDP-43, and others, illustrating the pleiotropy among SVs (Figure 2B). We further collected information on SV-xQTL and found that the deletion was also associated with lower protein abundance of TMEM106B (Figure 2D).
Figure 2. 343 bp deletion associated with tangle density at TMEM106B locus.
A, locus zoom plot for the TMEM106B locus (chr7:11,768,758–12,769,593) showing a 200 Kbp window around a 343 bp deletion with high LD with the lead SNP rs5011436. On the top, the y-axis shows the nominal P values (as −log10) for the association tests with tangle density in ROS/MAP participants. On the bottom, the y-axis shows the AD GWAS results from Bellenguez et al. (2022). The SV is plotted in a diamond shape, while SNPs are plotted in circles. Points are colored by the LD (R2) to the SV. The dashed line represents nominal P = 0.05. B, shows the nominal P values (as −log10) for the association of the SV with all AD/ADRD traits tested. Dots are colored by phenotype category. The dashed line represents nominal P = 0.05. C, Boxplots showing the tangle density measures by the deletion alleles (ROS, MAP, ROS/MAP). D, Boxplot shows the SV-pQTL between the deletion and protein expression levels of TMEM106B measured from DLPFC brain tissues of 663 ROS/MAP participants.
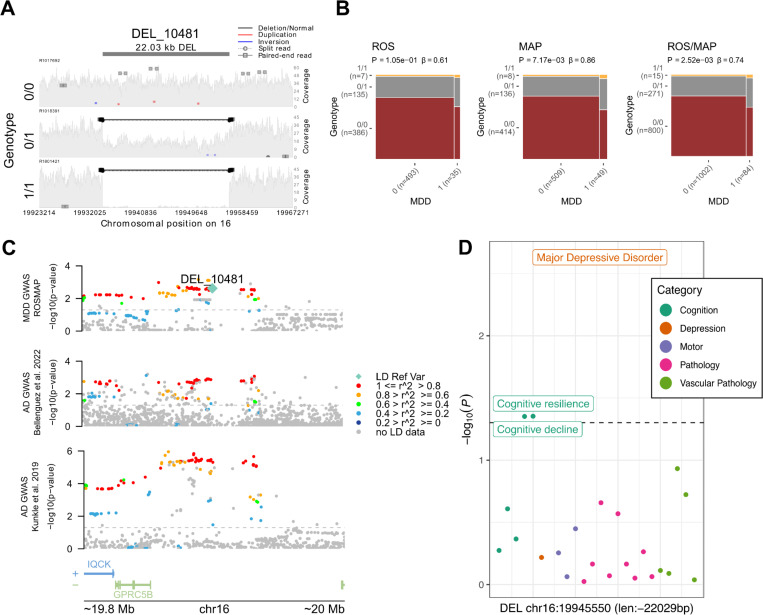
Another four associations were found with a relaxed P < 0.01. That included a 22,029 bp long deletion (Figure 3A) associated with diagnosis of MDD in ROS/MAP (P = 0.0025) and in LD (R2 = 0.44) with the lead variant in the IQCK locus (chr16:19,308,163–20,308,163)(Figure 3B). Two SVs at the HLA locus, an 86,768 bp deletion (R2 = 0.27) and a 43,223 bp duplication (R2 = 0.39) associated with cognitive resilience (P = 0.002) and MDD (P = 0.003), respectively. And one 1,505 bp deletion at the MYO15A locus (R2 = 0.37), associated with TDP-43 (P = 0.007). Although the AD association at the IQCK locus was identified only in one paper (Kunkle et al. 2019), there are still nominally significant associations at more powered AD GWAS (Figure 3C). In ROS/MAP, apart from depression status, cognitive resilience and cognitive decline also reached a nominal significant threshold (Figure 3D). Depression is a well-known risk factor for AD, associated with an increased likelihood of developing dementia (Bellou et al. 2017; Dafsari and Jessen 2020; Harerimana et al. 2022; Holmquist, Nordström, and Nordström 2020), and this association could represent a genetic link between depression and AD. The SVs at the HLA locus overlap the genes HLA-DRB1, HLA-DRB5, and HLA-DRB6, characterizing the class II sub-region haplotypes. However, their association and LD patterns are overly complex to disentangle the causal variants from the haplotypes.
Figure 3. 22 Kbp deletion associated with major depressive disorder at IQCK locus.
A, mapping of sequencing reads at the locus from three representative individuals corresponding to the possible genotypes: deletion not present (0/0), heterozygous deletion (0/1), and homozygous deletion (1/1). B, mosaic plots showing the proportion of individuals with MDD and respective deletion alleles for ROS, MAP, and ROS/MAP. C, locus zoom plot for the AD GWAS IQCK locus (chr16:19,308,163–20,308,163) showing a 200 Kbp window around the deletion in LD with the lead SNP rs7185636. The scatter plot on the top shows the nominal P values (as −log10) for the association tests with MDD status in ROS/MAP participants. The scatter plots in the middle and bottom show AD GWAS results from Bellenguez et al. (2022) and Kunkle et al. (2019), respectively. The 22 Kbp deletion is plotted in a diamond shape, while SNPs are plotted in circles. Points are colored by the LD (R2) to the SV, as measured in ROS/MAP. The dashed line represents nominal P = 0.05. D, shows the nominal P values (as −log10) for the association of the 22 Kbp deletion with all AD/ADRD traits tested. Dots are colored by phenotype category. The dashed line represents nominal P = 0.05.
Genome-wide association SV reveals potentially novel ADRD risk loci
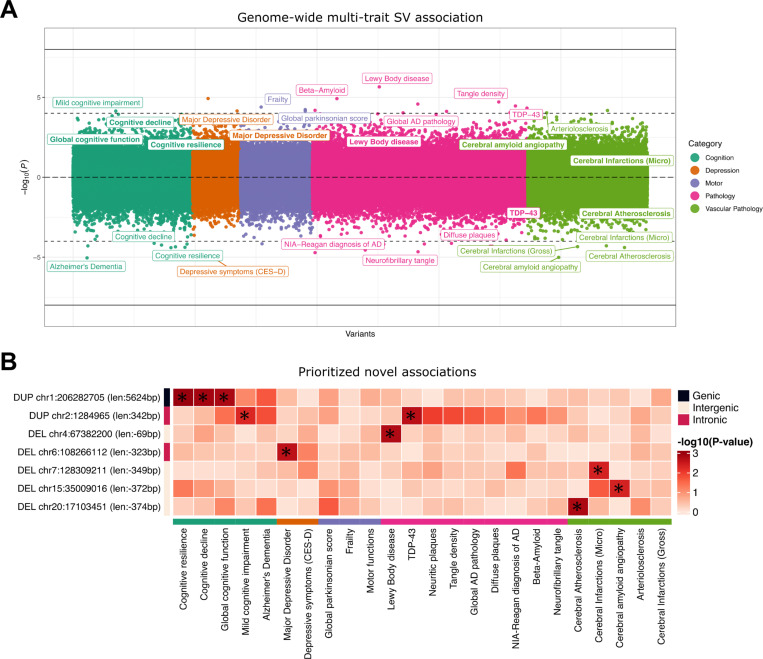
As an exploratory analysis, we performed a meta-analysis combining association results from 529 ROS participants and 559 individuals from MAP. We were able to map suggestive associations that could represent potential novel risk loci by focusing on SVs with no LD with SNPs and a relaxed P-value threshold of P < 5 × 10−3 with at least one ADRD phenotype. To verify the robustness of the variant call and avoid false positives, we selected only SVs also mapped in gnomAD (with 80% reciprocal overlap). That resulted in 9 SV-phenotype associations comprising 7 SVs (Table 2; Figure 4), four mapped to intergenic regions, two intronic, and one with overlaps to coding regions. SV lengths ranged from 69 bp to 5.6 kb, with MAFs from 5.2% to 42.4%.
Table 2 -.
SVs associated with AD/ADRD traits with no LD with SNPs
| Phenotype | SV | MAF | ROS | MAP | Meta-Analysis (ROS/MAP) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Beta (SE) | P | Beta (SE) | P | Beta (SE) | P | Gene | |||
| Cognitive resilience | DUP_267 chr1:206282705 (len:5,624bp) | 34.3% | 0.01 (0.01) | 1.27 × 10−1 | 0.02 (0.01) | 2.41 × 10−3 | 0.01 (0.00) | 1.02 × 10−3 | C1orf186 |
| Cognitive decline | DUP_267 chr1:206282705 (len:5,624bp) | 34.3% | 0.02 (0.01) | 3.90 × 10−2 | 0.02 (0.01) | 1.11 × 10−2 | 0.02 (0.01) | 1.28 × 10−3 | C1orf186 |
| Global cognition | DUP_267 chr1:206282705 (len:5,624bp) | 34.3% | 0.26 (0.12) | 2.96 × 10−2 | 0.29 (0.12) | 1.71 × 10−2 | 0.27 (0.08) | 1.51 × 10−3 | C1orf186 |
| Cerebral Atherosclerosis | DEL_17353 chr20:17103451 (len:-374bp) | 8.5% | −0.20 (0.09) | 2.41 × 10−2 | −0.22 (0.10) | 2.20 × 10−2 | −0.21 (0.07) | 1.58 × 10−3 | RNU6–27P |
| Lewy Body disease | DEL_21899 chr4:67382200 (len:-69bp) | 28.3% | 0.49 (0.22) | 2.44 × 10−2 | 0.46 (0.20) | 2.42 × 10−2 | 0.46 (0.15) | 1.73 × 10−3 | RPS23P3 |
| TDP-43 | DUP_2394 chr2:1284965 (len:342bp) | 42.4% | −0.70 (0.27) | 8.19 × 10−3 | −0.41 (0.22) | 6.40 × 10−2 | −0.53 (0.17) | 2.19 × 10−3 | SNTG2 |
| MDD | DEL_26626 chr6:108266112 (len:-323bp) | 28.2% | 0.78 (0.36) | 2.85 × 10−2 | 0.71 (0.32) | 2.81 × 10−2 | 0.76 (0.24) | 2.31 × 10−3 | SEC63 |
| Micro-chronic cerebral infarctions | DEL_28995 chr7:128309211 (len:-349bp) | 8.6% | −0.10 (0.25) | 6.98 × 10−1 | 0.95 (0.26) | 2.87 × 10−4 | 0.40 (0.18) | 4.29 × 10−3 | RP11–274B21.5 |
| Cerebral amyloid angiopathy | DEL_9454 chr15:35009016 (len:-372bp) | 5.2% | 0.36 (0.14) | 8.63 × 10−3 | 0.19 (0.13) | 1.27 × 10−1 | 0.26 (0.09) | 4.40 × 10−3 | GJD2 |
Figure 4. Chicago plot showing the direction of associations with results of the SV genome-wide scan in 1,088 ROS/MAP participants across 24 AD/ADRD phenotypes.
A, Phenotypes are colored by common related categories. Solid-line represents the genome-wide significance of P < 5 × 10−8. The dashed line represents a suggestive threshold of P < 10−4. Phenotype labels are shown only for associations below the suggestive threshold. Prioritized associations (PMETA < 5 × 10−5 and no LD with SNPs) are highlighted in bold. B, List of 7 prioritized SV-trait associations under the conditions of PMETA < 5 × 10−3 for at least one ADRD phenotype, no LD with SNPs in their respective loci, and supporting evidence of the SV call in gnomAD. Bonferroni adjusted P-values are displayed in the heatmap. Asterisks indicate adj. P < 0.05.
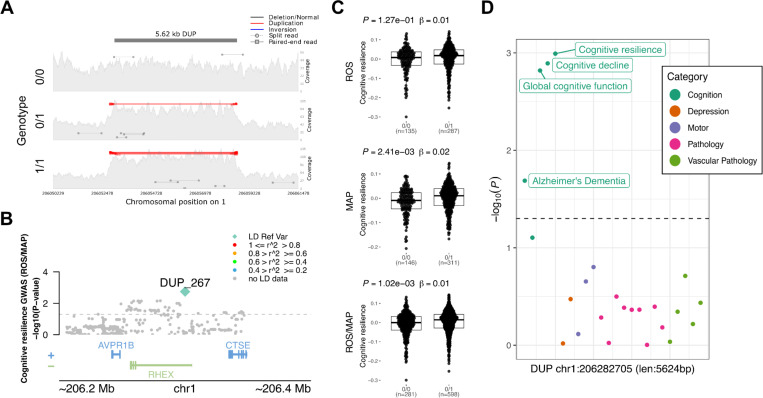
The most promising finding was a 5.6 kb duplication at the 1q31.1 locus, which completely overlaps a non-canonical exon and parts of the 5’UTR of the gene C1orf186 (which codes for the protein RHEX)(Figure 5A-B). This duplication was associated with better measures of cognitive resilience (PROS = 1.27 × 10−1, PMAP = 2.41 × 10−3, PMETA = 1.02 × 10−3) (Figure 5C), and also slopes of cognitive decline and global cognition proximal to death (Figure 5D and Table 2). MAF in both studies was around 34%, with no homozygous carriers with measures of resilience available. Given that C1orf186 is not expressed in brain tissues, no QTL mapping was measured between the duplication with the expression of C1orf186 in ROS/MAP brains. Other suggestive findings included two SVs overlapped intronic regions, a 342 bp duplication in an intronic region of the gene SNTG2 at chr2 associated with TDP-43 and several other AD neuropathologies, and a 323 bp deletion in an intronic region of the gene SEC63 at chr6 associated with MDD. Finally, four other SVs were mapped to intergenic regions, a 374 bp deletion at chr20 associated with cerebral atherosclerosis; a 69 bp deletion at chr4 associated with the presence of Lewy bodies; a 349 bp deletion at chromosome 7 overlapping associated with micro-chronic cerebral infarctions; and a 372 bp deletion at chr15 associated with cerebral amyloid angiopathy.
Figure 5. 5.6 Kbp duplication at the 1q31.1 locus associated with cognitive resilience.
A, mapping of sequencing reads at the locus from three representative individuals corresponding to the possible genotypes: duplication not present (0/0), heterozygous allele (0/1), and homozygous allele (1/1). B, locus zoom plot showing 200 Kbp window around the SV. The y-axis shows the nominal P values (as −log10) for the association tests with cognition in ROS/MAP participants. The SV is plotted in a diamond shape, while SNPs are plotted in circles. Points are colored by the LD (R2) to the SV. C, Boxplots showing the cognitive resilience measures by the duplication alleles (ROS, MAP, ROS/MAP). D, shows the nominal P values (as −log10) for the association of the SV with all other AD/ADRD traits tested. Dots are colored by phenotype category. The dashed line represents nominal P = 0.05.
DISCUSSION
GWAS have enabled the identification of dozens of common SNVs and small indels contributing to AD/ADRD traits (Bellenguez et al. 2022; Chia et al. 2021; Farrell et al. 2022; Howard et al. 2019; Nalls et al. 2019; Sherva et al. 2023; Zhang et al. 2022; Yan et al. 2021). The contribution of SVs, by contrast, lags. Previous studies were restricted mainly to either CNVs, as in autism (Marshall et al. 2008); rare SVs, as in schizophrenia(Halvorsen et al. 2020; Sebat, Levy, and McCarthy 2009; Walsh et al. 2008); or repeat expansions, as in amyotrophic lateral sclerosis (ALS), FTD, and Huntington’s disease. Here, by leveraging the deep ROS/MAP phenotypic data, we performed for the first time a comprehensive analysis of the role of SVs in AD/ADRD traits by linking common SVs alleles to multiple clinical and neuropathological traits.
We investigated the impact of SVs in known AD GWAS loci. By mapping the presence of SVs in each of the 81 loci, we identified 26 SVs in moderate to high LD with GWAS lead SNPs and directly associated (nominal P < 0.05) with AD/ADRD phenotypes in ROS/MAP. The most significant SV was a 343 bp deletion at the 3’UTR of TMEM106B, associated with multiple phenotypes, including cognitive resilience, tangle density, and TDP-43, and in high LD with the lead variant at the locus. This SV has already been reported as being the likely causal variant at the locus, with reported risks for frontotemporal lobar dementia with TDP-43 inclusions (FTLD-TDP) (Chemparathy et al. 2023) and neurodegeneration (Salazar et al. 2023). The proposed mechanism involved is via an aging-related negative feedback loop mediated by the presence of the AluYb8 that dysregulates TDP-43 due to increasing demethylation of TMEM160B (Salazar et al. 2023). TMEM106B variants have also been reported to affect cell fraction in brain tissues from ROS/MAP, specifically for a subpopulation of excitatory neurons (Fujita et al. 2024; Green et al. 2023).
Our integrated results identified suggestive associations with the potential to be novel ADRD risk loci. We highlight seven SVs with sizes ranging from 69 bp to 5,624 bp. Not surprisingly, most of these were found in non-coding regions, two intronic and four intergenic. Interestingly, a large duplication overlapped the UTR and the coding exonic regions of the gene C1orf186. This duplication was consistently associated with cognition-related phenotypes, including cognitive decline and resilience, and seemed to provide a protective effect to AD. The C1orf186 gene encodes for the RHEX protein, which is involved in the erythropoietin-mediated signaling pathway and positive regulation of erythrocyte differentiation (Verma et al. 2014), and although it is not expressed in brain tissues, that does not exclude the possibility of the duplication affecting expression in other tissues.
Recently, a study analyzed associations of common SVs with PD (Billingsley et al. 2023), finding three deletions reaching genome-wide significance. Another study focused on non-Alzheimer’s dementia, including almost 6 thousand individuals, identified a novel deletion associated with LBD and replicated known loci associated with FTD and ALS (Kaivola et al. 2023). Unfortunately, our study could not test these findings due to differences in variant calling methods. Still, besides those highlighted results, many other potentially interesting findings can still be explored from the genome-wide scans, including SVs in moderate to high LD with SNVs in their loci, where SVs having equal or higher significance with the traits tested might suggest potential causal effects. Although the association tests were restricted to the ROS/MAP samples due to the specificity of phenotypic data, the meta-analysis findings still provide a better selection of candidate variants with consistent effects between two cohort design studies.
While our results represent a step forward in understanding the effects of common genetic variation in AD/ADRD traits, important limitations must be noted: 1) the power for association discovery is constrained by the current sample size; 2) the replication of associations in independent samples is limited to available AD-related phenotypes and might not capture the same nuances from ROS/MAP; 3) SV calling is restricted to deletions, insertions, inversions, and duplication and is still prone to falsely discovered variants and low sensitivity (especially for insertions); 4) the suggestive associations do not represent suggestive causal effects on the traits, especially when LD is present, which would require a more precise fine-mapping analysis; 5) analyses were restricted to germline common autosomal structural variation; 6) since the individuals in this study have a European genetic background, these associations might not transfer to ancestrally diverse population-based data. These limitations could be overcome with additional sample size and deeper sequencing data (e.g., long-reads). Most importantly, these results should be interpreted as suggestive associations, and a more comprehensive replication in an independent sample data set is crucial for the validity of the findings.
METHODS
Study participants
The study uses data from 529 participants from the Religious Orders Study (ROS) and 559 participants from the Rush Memory and Aging Project (MAP), two longitudinal cohort studies. ROS and MAP enroll participants free of known dementia. Participants agree to annual clinical evaluations and to donate their brains at death. ROS was initiated in 1994 and enrolls older Catholic priests, nuns, and brothers from nearly 40 groups located in 12 U.S. states (D. A. Bennett et al. 2006; David A. Bennett et al. 2018). By the time the samples were sent for sequencing (end of 2017), 1,437 individuals had completed their baseline assessment. The MAP was established in 1997 and recruits older men and women from retirement communities and individual householders in the greater Chicago area (D. A. Bennett et al. 2006; David A. Bennett et al. 2018, 2012, 2005). As of the end of 2017, 1,967 participants had completed their baseline evaluation.
ROS and MAP are studies conducted by the same team of investigators sharing a common core of measures and procedures, allowing direct comparison between variables and efficient data merging for combined analyses. The follow-up rate for surviving participants surpasses 90%. Both studies were approved by a Rush University Medical Center Institutional Review Board. Each participant provided written informed consent at enrollment and signed the Uniform Anatomical Gift Act.
Alzheimer’s dementia and cognitive function
Standardized cognitive and clinical assessments are conducted annually by examiners unaware of previous data. The clinical diagnosis for dementia follows the directives provided by the joint working group of the National Institute of Neurological and Communicative Disorders and Stroke and the AD and Related Disorders Association, as described (David A. Bennett, Schneider, Aggarwal, et al. 2006; McKhann et al. 1984). Mild cognitive impairment (MCI) was defined as individuals assessed by the neuropsychologist as cognitively impaired but not diagnosed as having dementia by the examining physician, as previously outlined (D. A. Bennett et al. 2002). Persons without dementia or MCI were designated as having no cognitive impairment (NCI) as described (D. A. Bennett et al. 2002). A final consensus cognitive diagnosis is determined by a neurologist with proficiency in dementia after reviewing select clinical information after death without knowledge of any postmortem data (Julie A. Schneider et al. 2007). For the current association analysis, two binary statuses were used: Alzheimer’s dementia vs no dementia (MCI + NCI) and with cognitive impairment (AD + MCI) vs no cognitive impairment (NCI).
Quantitative measurements of cognitive function were measured yearly. Cognitive evaluations comprise 19 cognitive performance tests that are common to both studies. A Mini-Mental State Examination is employed for descriptive reasons, whereas the Complex Ideational Material from the Boston Diagnostic Aphasia Examination is solely utilized for diagnostic classification. The remaining 17 tests are merged into a comprehensive metric of global cognition. The scores for each test were converted to a composite score (Robert S. Wilson et al. 2015). For the association analysis, measurements were considered proximal to death.
Estimated slopes for global cognition were also included as a measure of cognitive decline. The random slope of global cognition is calculated to the individualized projected pace of change in the global cognition variable across time. This projection is generated through a linear mixed-effects model, with global cognition serving as the longitudinal outcome. The model adjusts for age at baseline, sex, and years of education (De Jager et al. 2012; Oveisgharan et al. 2023). Additionally, as a measure of cognitive resilience, another random slope of global cognition is calculated, controlling for demographics and neuropathologies. The linear mixed-effects model generates the projection using global cognition as the outcome while adjusting for age at baseline, sex, and years of education as demographics, and global AD pathology burden, -amyloid, PHF tau tangles, gross chronic cerebral infarctions, chronic microinfarctions, Lewy body disease, TDP-43, hippocampal sclerosis, cerebral amyloid angiopathy, cerebral atherosclerosis, and arteriolosclerosis as pathology factors (Boyle et al. 2021).
Parkinsonism, frailty and motor function
A global Parkinsonian summary score was also tested. The global Parkinsonian summary score is a composite measure of Parkinsonian signs. It is calculated as the average of four separate domains based on a 26-item modified version of the motor portion of the United Parkinson’s Disease Rating Scale (mUPDRS). These domains include bradykinesia, gait, rigidity, and tremor, and they are administered by a trained nurse clinician. These measures are highly reliable and reproducible in both men and women across various cohorts and have been modified to be more applicable to individuals without Parkinson’s disease and easier for non-physicians to administer and score (Aron S. Buchman et al. 2012).
In addition, a total of two motor-related indices were evaluated. A measure of frailty and a composite measure of global motor function. Each of the indices is described below. Frailty is defined as multiple system weaknesses. A continuous composite measure of frailty is based on four components: grip strength, timed walking, body composition (BMI), and fatigue. The raw scores for each component are converted into z-scores using the mean and standard deviation values from all participants at baseline (Aron S. Buchman et al. 2007; A. S. Buchman et al. 2009). The global motor function combines multiple motor tests, including the Purdue Pegboard Test, finger-tapping test, the time and number of steps to cover a distance of 8 feet, the time and number of steps for 360-degree turn, leg and toe stand, grip strength, and pinch strength. Each test’s performance score is converted to a score based on the mean score of all participants at baseline, and then the scores are averaged together to create the composite measure. This measure provides a comprehensive assessment of motor and gait function in individuals (Aron S. Buchman et al. 2009).
Depression and depressive symptoms
Two measures of depression were evaluated: a binary status of clinical diagnosis of major depressive disorder (MDD) and a quantitative score of depressive symptoms. The clinical diagnosis of major depressive disorder was made by an examining physician at each evaluation. Diagnosis was based on criteria of the Diagnostic and Statistical Manual of Mental Disorders (DSM-III-R), clinical interview with the participant, and a review of responses to questions adapted from the Diagnostic Interview Schedule (David A. Bennett et al. 2004). A binary variable was tested classifying probably or highly probable vs. possible or not present MDD into the presence or absence of MDD. Depressive symptoms were assessed using a modified 10-item version of the Center for Epidemiologic Studies Depression Scale (CES-D) (Kohout et al. 1993; Robert S. Wilson et al. 2002, 2014). Participants were asked whether they experienced each of the ten symptoms frequently in the past week. An overall score was obtained by aggregating the number of symptoms reported.
Neuropathological evaluations
Systematic assessment of various neurodegenerative and cerebrovascular conditions, including pathological diagnosis of Alzheimer’s disease, Lewy bodies, LATE, hippocampal sclerosis, chronic macroscopic infarcts and microinfarcts, cerebral amyloid angiopathy, atherosclerosis, and arteriolosclerosis were performed as previously reported (Boyle et al. 2019, 2021). The assessments were conducted by examiners who were blinded to all clinical data. A summary of each variable used in the current study is described below:
NIA-Reagan diagnosis of AD:
The NIA-Reagan diagnosis of Alzheimer’s disease was measured based on a set of consensus recommendations for diagnosing the disease after death, taking in account the presence of both neurofibrillary tangles (Braak) and neuritic plaques (CERAD). This criteria was modified because the neuropathological evaluation is carried out without knowledge of the patient’s clinical information, including a dementia diagnosis. Thus, the level of Alzheimer’s disease pathology is determined by a neuropathologist (Bennett et al. 2006). In our association analysis, we utilized a dichotomized variable where individuals with an intermediate or high likelihood fulfill the criteria for a pathological diagnosis of Alzheimer’s disease.
Burden of neuritic and diffuse plaques and neurofibrillary tangles:
Neuric and diffuse plaques were determined through microscopic examination of silver-stained slides from five specific brain regions. The index count in each region is scaled by dividing it by its corresponding standard deviation. The scaled regional measures are then averaged to obtain a summary measure for the plaque burden. The five regions examined are the midfrontal cortex, midtemporal cortex, inferior parietal cortex, entorhinal cortex, and mid-hippocampus CA1. In addition, a global score of AD pathology was also tested as quantitative measure derived from the counts of three silver-stained measures of AD pathologies: neuritic plaques, diffuse plaques, and neurofibrillary tangles. Measures were made in same five regions, and each regional count was scaled by dividing by the corresponding standard deviation. The average of the three measured was used (D. A. Bennett et al. 2003; David A. Bennett, Schneider, Tang, et al. 2006).
-amyloid load:
-amyloid protein is identified by molecularly specific immunohistochemistry and quantified by image analysis in 8 brain regions. The percent area of the cortex occupied by -amyloid is calculated, and a mean score is determined from 4 or more regions (R. S. Wilson et al. 2007).
PHFtau tangle density:
Neuronal tangles are identified with phosphorylated Tau protein antibodies (AT8) and quantified in 8 brain regions using systematic sampling by stereology to determine cortical density. A mean tangle score is then calculated from 4 or more regions (R. S. Wilson et al. 2007).
TDP-43:
TDP-43 cytoplasmic inclusions in neurons and glia are assessed for each of the eight brain regions and scored based on four stages of TDP-43 distribution (ranging from none to involvement of all eight regions). A dichotomized presence or absence is used in these analyses (Nag et al. 2017).
Lewy Body disease (LBD):
A pathologic diagnosis of LBD is determined based on four stages of distribution of -synuclein in the brain. Brain tissue samples from multiple regions are evaluated with -synuclein immunostaining. The McKeith criteria were modified to assess the presence of LBD in different categories: not present, nigral-predominant, limbic-type, and neocortical-type. A dichotomized version of this variable is used, referring to Lewy bodies present or absent (J. A. Schneider et al. 2012).
Arteriolosclerosis:
Arteriolosclerosis refers to histological changes observed in small brain vessels during aging, including intimal deterioration, smooth muscle degeneration, and fibrohyalinotic thickening that narrows the vascular lumen. The severity of arteriolosclerosis is evaluated as none, mild, moderate, or severe (Aron S. Buchman et al. 2011).
Cerebral Atherosclerosis:
The severity of large vessel cerebral atherosclerosis is visually assessed by examining several arteries and their proximal branches in the circle of Willis. The rating was based on the extent of involvement, including the number of arteries affected and the degree of occlusion. A semiquantitative scale is used: none or possible, mild, moderate, and severe (Arvanitakis et al. 2017).
Cerebral Amyloid Angiopathy:
A semiquantitative summary of cerebral amyloid angiopathy (CAA) pathology in 4 neocortical regions is calculated using paraffin-embedded sections that were immunostained for -amyloid using one of three monoclonal anti-human antibodies. Meningeal and parenchymal vessels are assessed for -amyloid deposition and scored from 0 to 4 based on the extent of circumferential deposition for each region. The CAA score for each region is the maximum of the meningeal and parenchymal CAA scores, which are then averaged across regions to summarize as a continuous measure of CAA pathology (Boyle et al. 2015).
Cerebral Infarctions - Gross-Chronic:
Neuropathologic evaluations are currently performed at Rush to determine the presence of one or more gross chronic cerebral infarctions. The evaluations are blinded to clinical data and reviewed by a board-certified neuropathologist. The examination documents the age (acute/subacute/chronic), size, and location (side and region) of infarcts visible to the naked eye on fixed slabs. All visible and suspected macroscopic infarcts are dissected for histologic confirmation. A value is one or more gross chronic infarctions vs none (Arvanitakis et al. 2011; J. A. Schneider et al. 2005).
Cerebral Infarctions - Micro-Chronic:
Neuropathologic evaluations are performed at Rush to determine the presence of one or more chronic microinfarcts, which are chronic microscopic infarctions. The evaluations are blinded to clinical data and reviewed by a board-certified neuropathologist. At least nine regions in one hemisphere are examined for microinfarcts on 6μm paraffin-embedded sections stained with hematoxylin/eosin. The examination includes six cortical regions (midfrontal, middle temporal, entorhinal, hippocampal, inferior parietal, and anterior cingulate cortices), two subcortical regions (anterior basal ganglia, thalamus), and midbrain. Age (acute/subacute/chronic) and location (side and region) of microinfarcts are recorded. A value of 0 indicates no chronic microinfarcts, while a value of 1 indicates the presence of one or more chronic microinfarcts (Arvanitakis et al. 2011).
WGS data and variant calling
Whole-genome sequencing (WGS) data were previously generated from DNA samples from blood or cortex tissues (De Jager et al. 2018). Briefly, libraries were sequenced on an Illumina HiSeq X sequencer using 2 × 150 bp cycles. Single nucleotide variant and small indel discovery and genotyping were performed utilizing an NYGC automated pipeline, which included alignment to the GRCh37 human reference using the Burrows-Wheeler Aligner and processing using the GATK best-practices workflow. The workflow included marking duplicate reads, local realignment around indels, and using Genome Analysis Toolkit (GATK) base quality score recalibration.
Structural variant discovery and genotyping were also previously described. A combination of seven different software tools, including DELLY (Rausch et al. 2012), LUMPY (Layer et al. 2014), Manta (X. Chen et al. 2016), BreakDancer (K. Chen et al. 2009), CNVnator (Abyzov et al. 2011), BreakSeq (Abyzov et al. 2015), and MELT (Gardner et al. 2017), was applied to identify SVs in each sample. The variants were then merged at the individual level using SURVIVOR (Jeffares et al. 2017) and genotyped at the study level using smoove (GitHub - Brentp/smoove). ROS and MAP were jointly genotyped, resulting in a final set of 72,348 SVs mapped in 1,106 individuals after quality control. Details about the specific SV pipeline steps are described in the original publication (Vialle et al. 2022).
Linkage disequilibrium with GWAS variants
Linkage disequilibrium between SNPs and SVs was also previously generated (Vialle et al. 2022). A joint call set with 8,566,510 SNPs and 72,348 SVs was used to calculate LD in terms of R2 for all SVs using PLINK and considering a window of 5 Mb.
Single variant association analysis of SVs and AD/ADRD traits
We performed genome-wide association analysis testing for each common SV with 16 quantitative and eight binary AD/ADRD phenotypes. The variables measuring -amyloid tangle density, neurofibrillary tangle burden, neuritic and diffuse plaque burden were squared root transformed before the association analysis.
Analysis was performed using SAIGEgds (Scalable and Accurate Implementation of GEneralized mixed model) (Zheng and Davis 2021; Zhou et al. 2018). The method uses the saddlepoint approximation to calibrate the distribution of score test statistics and state-of-the-art optimization strategies to reduce computational costs. SAIGE can analyze large-scale data while controlling for unbalanced case-control ratios and sample relatedness, making it applicable to GWAS for thousands of phenotypes by large biobanks. The method was tested on UK Biobank data and demonstrated its efficiency in analyzing extensive sample data. The SAIGE method involves two main steps: (1) fitting the null logistic mixed model to estimate model parameters using the average information restricted maximum likelihood algorithm, and (2) testing for associations between each genetic variant and phenotype using the saddlepoint approximation method on score test statistics. Several optimization strategies have been applied to make fitting the null logistic mixed model practical for large data sets, such as using the raw genotypes as input and the preconditioned conjugate gradient method to solve linear systems iteratively. These optimizations make SAIGE computationally efficient and applicable to GWAS for thousands of phenotypes by large biobanks.
Here, we performed separate genome-wide scans using 529 participants from ROS and 559 participants from MAP. All tests were controlled by age at death, sex, years of education, five genetic principal components, and the genetic correlation matrix (modeled as a random effect). To combine consistent genetic effects across both studies, a meta-analysis was conducted using METASOFT v.2.0.1 (Han and Eskin 2011). Effect sizes and standard errors of each SV–trait pair were used as input. We carried out a random-effects meta-analysis using the RE2 model, optimized to detect associations under heterogeneity.
Proteomics data
In our analysis, we utilized ROS/MAP proteomics data to link the impact of SVs with protein abundances. The methods for data generation were previously published in detail (Seifar et al. 2024; Higginbotham et al. 2022; Wingo et al. 2020; Yu et al. 2020). Briefly, frozen DLPFC tissue samples underwent homogenization, followed by the quantification of protein concentrations. Isobaric TMT peptide labels were subsequently introduced and fractioned by high pH. These fractions were then subjected to liquid chromatography-mass spectrometry, and the resulting spectra were cross-referenced with the UniProt database. After quality control to eliminate technical confounders, 10,030 proteins from 971 individuals were available for downstream analysis (Seifar et al. 2024).
Supplementary Material
Acknowledgments
We thank the participants of ROS/MAP cohorts for their essential contributions and gift to these projects, as well the investigators and staff at the Rush Alzheimer’s Disease Center. This work has been supported the following National Institute on Aging (NIA) grants: P30AG10161 (DAB), P30AG72975 (JAS), R01AG15819 (DAB), R01AG17917 (DAB), U01AG46152 (PLD, DAB), U01AG61356 (PLD, DAB), R01AG22018 (LLL), U01AG079847 (CG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Code availability
All relevant code used in this study has been provided in a single repository on GitHub (https://github.com/RushAlz/ADRD_SV_GWAS).
Data availability
All SV site frequency data and complete nominal summary statistics for all phenotypes tested are publicly available on GitHub (https://github.com/RushAlz/ADRD_SV_GWAS). ROS/MAP resources, including individual-level genotyping and phenotypic data can be requested at https://www.radc.rush.edu.
REFERENCES
- Abyzov Alexej, Li Shantao, Kim Daniel Rhee, Mohiyuddin Marghoob, Stütz Adrian M., Parrish Nicholas F., Mu Xinmeng Jasmine, et al. 2015. “Analysis of Deletion Breakpoints from 1,092 Humans Reveals Details of Mutation Mechanisms.” Nature Communications 6 (June):7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov Alexej, Urban Alexander E., Snyder Michael, and Gerstein Mark. 2011. “CNVnator: An Approach to Discover, Genotype, and Characterize Typical and Atypical CNVs from Family and Population Genome Sequencing.” Genome Research 21 (6): 974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews Shea J., Renton Alan E., Fulton-Howard Brian, Podlesny-Drabiniok Anna, Marcora Edoardo, and Goate Alison M. 2023. “The Complex Genetic Architecture of Alzheimer’s Disease: Novel Insights and Future Directions.” EBioMedicine 90 (104511): 104511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitakis Zoe, Capuano Ana W., Leurgans Sue E., Buchman Aron S., Bennett David A., and Schneider Julie A. 2017. “The Relationship of Cerebral Vessel Pathology to Brain Microinfarcts.” Brain Pathology 27 (1): 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitakis Zoe, Leurgans Sue E., Barnes Lisa L., Bennett David A., and Schneider Julie A. 2011. “Microinfarct Pathology, Dementia, and Cognitive Systems.” Stroke; a Journal of Cerebral Circulation 42 (3): 722–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M., Litvan I., Houlden H., Adamson J., Dickson D., Perez-Tur J., Hardy J., Lynch T., Bigio E., and Hutton M. 1999. “Association of an Extended Haplotype in the Tau Gene with Progressive Supranuclear Palsy.” Human Molecular Genetics 8 (4): 711–15. [DOI] [PubMed] [Google Scholar]
- Bellenguez Céline, Küçükali Fahri, Jansen Iris E., Kleineidam Luca, Moreno-Grau Sonia, Amin Najaf, Naj Adam C., et al. 2022. “New Insights into the Genetic Etiology of Alzheimer’s Disease and Related Dementias.” Nature Genetics 54 (4): 412–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellou Vanesa, Belbasis Lazaros, Tzoulaki Ioanna, Middleton Lefkos T., Ioannidis John P. A., and Evangelou Evangelos. 2017. “Systematic Evaluation of the Associations between Environmental Risk Factors and Dementia: An Umbrella Review of Systematic Reviews and Meta-Analyses.” Alzheimer’s & Dementia: The Journal of the Alzheimer’s Association 13 (4): 406–18. [DOI] [PubMed] [Google Scholar]
- Bennett D. A., Schneider J. A., Arvanitakis Z., Kelly J. F., Aggarwal N. T., Shah R. C., and Wilson R. S. 2006. “Neuropathology of Older Persons without Cognitive Impairment from Two Community-Based Studies.” Neurology 66 (12): 1837–44. [DOI] [PubMed] [Google Scholar]
- Bennett David A., Buchman Aron S., Boyle Patricia A., Barnes Lisa L., Wilson Robert S., and Schneider Julie A. 2018. “Religious Orders Study and Rush Memory and Aging Project.” Journal of Alzheimer’s Disease: JAD 64 (s1): S161–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett David A., Schneider Julie A., Aggarwal Neelum T., Arvanitakis Zoe, Shah Raj C., Kelly Jeremiah F., Fox Jacob H., et al. 2006. “Decision Rules Guiding the Clinical Diagnosis of Alzheimer’s Disease in Two Community-Based Cohort Studies Compared to Standard Practice in a Clinic-Based Cohort Study.” Neuroepidemiology 27 (3): 169–76. [DOI] [PubMed] [Google Scholar]
- Bennett David A., Schneider Julie A., Buchman Aron S., Barnes Lisa L., Boyle Patricia A., and Wilson Robert S. 2012. “Overview and Findings from the Rush Memory and Aging Project.” Current Alzheimer Research 9 (6): 646–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett David A., Schneider Julie A., Buchman Aron S., de Leon Carlos Mendes, Bienias Julia L., and Wilson Robert S. 2005. “The Rush Memory and Aging Project: Study Design and Baseline Characteristics of the Study Cohort.” Neuroepidemiology 25 (4): 163–75. [DOI] [PubMed] [Google Scholar]
- Bennett David A., Schneider Julie A., Tang Yuxiao, Arnold Steven E., and Wilson Robert S. 2006. “The Effect of Social Networks on the Relation between Alzheimer’s Disease Pathology and Level of Cognitive Function in Old People: A Longitudinal Cohort Study.” Lancet Neurology 5 (5): 406–12. [DOI] [PubMed] [Google Scholar]
- Bennett David A., Wilson Robert S., Schneider Julie A., Bienias Julia L., and Arnold Steven E. 2004. “Cerebral Infarctions and the Relationship of Depression Symptoms to Level of Cognitive Functioning in Older Persons.” The American Journal of Geriatric Psychiatry: Official Journal of the American Association for Geriatric Psychiatry 12 (2): 211–19. [PubMed] [Google Scholar]
- Bennett D. A., Wilson R. S., Schneider J. A., Evans D. A., Aggarwal N. T., Arnold S. E., Cochran E. J., Berry-Kravis E., and Bienias J. L. 2003. “Apolipoprotein E epsilon4 Allele, AD Pathology, and the Clinical Expression of Alzheimer’s Disease.” Neurology 60 (2): 246–52. [DOI] [PubMed] [Google Scholar]
- Bennett D. A., Wilson R. S., Schneider J. A., Evans D. A., Beckett L. A., Aggarwal N. T., Barnes L. L., Fox J. H., and Bach J. 2002. “Natural History of Mild Cognitive Impairment in Older Persons.” Neurology 59 (2): 198–205. [DOI] [PubMed] [Google Scholar]
- Billingsley Kimberley J., Ding Jinhui, Jerez Pilar Alvarez, Illarionova Anastasia, Levine Kristin, Grenn Francis P., Makarious Mary B., et al. 2023. “Genome-Wide Analysis of Structural Variants in Parkinson Disease.” Annals of Neurology 93 (5): 1012–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom Elin S., Viswanathan Jayashree, Kilander Lena, Helisalmi Seppo, Soininen Hilkka, Lannfelt Lars, Ingelsson Martin, Glaser Anna, and Hiltunen Mikko. 2008. “Low Prevalence of APP Duplications in Swedish and Finnish Patients with Early-Onset Alzheimer’s Disease.” European Journal of Human Genetics: EJHG 16 (2): 171–75. [DOI] [PubMed] [Google Scholar]
- Boyle Patricia A., Wang Tianhao, Yu Lei, Wilson Robert S., Dawe Robert, Arfanakis Konstantinos, Schneider Julie A., and Bennett David A. 2021. “To What Degree Is Late Life Cognitive Decline Driven by Age-Related Neuropathologies?” Brain: A Journal of Neurology 144 (7): 2166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle Patricia A., Yu Lei, Leurgans Sue E., Wilson Robert S., Brookmeyer Ron, Schneider Julie A., and Bennett David A. 2019. “Attributable Risk of Alzheimer’s Dementia Attributed to Age-related Neuropathologies.” Annals of Neurology 85 (1): 114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle Patricia A., Yu Lei, Nag Sukriti, Leurgans Sue, Wilson Robert S., Bennett David A., and Schneider Julie A. 2015. “Cerebral Amyloid Angiopathy and Cognitive Outcomes in Community-Based Older Persons.” Neurology 85 (22): 1930–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman Aron S., Boyle Patricia A., Wilson Robert S., Fleischman Debra A., Leurgans Sue, and Bennett David A. 2009. “Association between Late-Life Social Activity and Motor Decline in Older Adults.” Archives of Internal Medicine 169 (12): 1139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman Aron S., Boyle Patricia A., Wilson Robert S., Tang Yuxiao, and Bennett David A. 2007. “Frailty Is Associated with Incident Alzheimer’s Disease and Cognitive Decline in the Elderly.” Psychosomatic Medicine 69 (5): 483–89. [DOI] [PubMed] [Google Scholar]
- Buchman Aron S., Leurgans Sue E., Nag Sukriti, Bennett David A., and Schneider Julie A. 2011. “Cerebrovascular Disease Pathology and Parkinsonian Signs in Old Age.” Stroke; a Journal of Cerebral Circulation 42 (11): 3183–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman Aron S., Shulman Joshua M., Nag Sukriti, Leurgans Sue E., Arnold Steven E., Morris Martha C., Schneider Julie A., and Bennett David A. 2012. “Nigral Pathology and Parkinsonian Signs in Elders without Parkinson Disease.” Annals of Neurology 71 (2): 258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. S., Wilson R. S., Bienias J. L., and Bennett D. A. 2009. “Change in Frailty and Risk of Death in Older Persons.” Experimental Aging Research 35 (1): 61–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemparathy Augustine, Le Guen Yann, Zeng Yi, Gorzynski John, Jensen Tanner, Yang Chengran, Kasireddy Nandita, et al. 2023. “A 3’UTR Insertion Is a Candidate Causal Variant at the TMEM106Blocus Associated with Increased Risk for FTLD-TDP.” bioRxiv. 10.1101/2023.07.06.23292312. [DOI] [PMC free article] [PubMed]
- Chen Ken, Wallis John W., McLellan Michael D., Larson David E., Kalicki Joelle M., Pohl Craig S., McGrath Sean D., et al. 2009. “BreakDancer: An Algorithm for High-Resolution Mapping of Genomic Structural Variation.” Nature Methods 6 (9): 677–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Xiaoyu, Schulz-Trieglaff Ole, Shaw Richard, Barnes Bret, Schlesinger Felix, Källberg Morten, Cox Anthony J., Kruglyak Semyon, and Saunders Christopher T. 2016. “Manta: Rapid Detection of Structural Variants and Indels for Germline and Cancer Sequencing Applications.” Bioinformatics 32 (8): 1220–22. [DOI] [PubMed] [Google Scholar]
- Chia Ruth, Sabir Marya S., Bandres-Ciga Sara, Saez-Atienzar Sara, Reynolds Regina H., Gustavsson Emil, Walton Ronald L., et al. 2021. “Genome Sequencing Analysis Identifies New Loci Associated with Lewy Body Dementia and Provides Insights into Its Genetic Architecture.” Nature Genetics 53 (3): 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins Ryan L., Brand Harrison, Karczewski Konrad J., Zhao Xuefang, Alföldi Jessica, Francioli Laurent C., Khera Amit V., et al. 2020. “A Structural Variation Reference for Medical and Population Genetics.” Nature 581 (7809): 444–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafsari Forugh S., and Jessen Frank. 2020. “Depression-an Underrecognized Target for Prevention of Dementia in Alzheimer’s Disease.” Translational Psychiatry 10 (1): 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager De, Philip L., Ma Yiyi, McCabe Cristin, Xu Jishu, Vardarajan Badri N., Felsky Daniel, Klein Hans-Ulrich, et al. 2018. “A Multi-Omic Atlas of the Human Frontal Cortex for Aging and Alzheimer’s Disease Research.” Scientific Data 5 (August):180142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager De, Philip L., Shulman Joshua M., Chibnik Lori B., Keenan Brendan T., Raj Towfique, Wilson Robert S., Yu Lei, et al. 2012. “A Genome-Wide Scan for Common Variants Affecting the Rate of Age-Related Cognitive Decline.” Neurobiology of Aging 33 (5): 1017.e1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez Mariely, Mackenzie Ian R., Boeve Bradley F., Boxer Adam L., Baker Matt, Rutherford Nicola J., Nicholson Alexandra M., et al. 2011. “Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS.” Neuron 72 (2): 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escott-Price Valentina, and Hardy John. 2022. “Genome-Wide Association Studies for Alzheimer’s Disease: Bigger Is Not Always Better.” Brain Communications 4 (3): fcac125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell Kurt, Kim Soongho, Han Natalia, Iida Megan A., Gonzalez Elias M., Otero-Garcia Marcos, Walker Jamie M., et al. 2022. “Genome-Wide Association Study and Functional Validation Implicates JADE1 in Tauopathy.” Acta Neuropathologica 143 (1): 33–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Masashi, Gao Zongmei, Zeng Lu, McCabe Cristin, White Charles C., Ng Bernard, Green Gilad Sahar, et al. 2024. “Cell Subtype-Specific Effects of Genetic Variation in the Alzheimer’s Disease Brain.” Nature Genetics 56 (4): 605–14. [DOI] [PubMed] [Google Scholar]
- Gardner Eugene J., Lam Vincent K., Harris Daniel N., Chuang Nelson T., Scott Emma C., Pittard W. Stephen, Mills Ryan E., 1000 Genomes Project Consortium, and Devine Scott E. 2017. “The Mobile Element Locator Tool (MELT): Population-Scale Mobile Element Discovery and Biology.” Genome Research 27 (11): 1916–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz Margaret, Reynolds Chandra A., Fratiglioni Laura, Johansson Boo, Mortimer James A., Berg Stig, Fiske Amy, and Pedersen Nancy L. 2006. “Role of Genes and Environments for Explaining Alzheimer Disease.” Archives of General Psychiatry 63 (2): 168–74. [DOI] [PubMed] [Google Scholar]
- “GitHub - Brentp/smoove: Structural Variant Calling and Genotyping with Existing Tools, But, Smoothly.” n.d. GitHub. Accessed August 9, 2024. https://github.com/brentp/smoove. [Google Scholar]
- Green Gilad Sahar, Fujita Masashi, Yang Hyun-Sik, Taga Mariko, McCabe Cristin, Cain Anael, White Charles C., et al. 2023. “Cellular Dynamics across Aged Human Brains Uncover a Multicellular Cascade Leading to Alzheimer’s Disease.” bioRxiv. 10.1101/2023.03.07.531493. [DOI]
- Halvorsen Matthew, Huh Ruth, Oskolkov Nikolay, Wen Jia, Netotea Sergiu, Giusti-Rodriguez Paola, Karlsson Robert, et al. 2020. “Increased Burden of Ultra-Rare Structural Variants Localizing to Boundaries of Topologically Associated Domains in Schizophrenia.” Nature Communications 11 (1): 1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Buhm, and Eskin Eleazar. 2011. “Random-Effects Model Aimed at Discovering Associations in Meta-Analysis of Genome-Wide Association Studies.” The American Journal of Human Genetics 88 (5): 586–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harerimana Nadia V., Liu Yue, Gerasimov Ekaterina S., Duong Duc, Beach Thomas G., Reiman Eric M., Schneider Julie A., et al. 2022. “Genetic Evidence Supporting a Causal Role of Depression in Alzheimer’s Disease.” Biological Psychiatry 92 (1): 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham Lenora, Carter E. Kathleen, Dammer Eric B., Haque Rafi U., Johnson Erik C. B., Duong Duc M., Yin Luming, et al. 2022. “Unbiased Classification of the Human Brain Proteome Resolves Distinct Clinical and Pathophysiological Subtypes of Cognitive Impairment.” bioRxiv. 10.1101/2022.07.22.501017. [DOI] [PMC free article] [PubMed]
- Holmquist Sofie, Nordström Anna, and Nordström Peter. 2020. “The Association of Depression with Subsequent Dementia Diagnosis: A Swedish Nationwide Cohort Study from 1964 to 2016.” PLoS Medicine 17 (1): e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooli B. V., Mohapatra G., Mattheisen M., Parrado A. R., Roehr J. T., Shen Y., Gusella J. F., et al. 2012. “Role of Common and Rare APP DNA Sequence Variants in Alzheimer Disease.” Neurology 78 (16): 1250–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard David M., Adams Mark J., Clarke Toni-Kim, Hafferty Jonathan D., Gibson Jude, Shirali Masoud, Coleman Jonathan R. I., et al. 2019. “Genome-Wide Meta-Analysis of Depression Identifies 102 Independent Variants and Highlights the Importance of the Prefrontal Brain Regions.” Nature Neuroscience 22 (3): 343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares Daniel C., Jolly Clemency, Hoti Mimoza, Speed Doug, Shaw Liam, Rallis Charalampos, Balloux Francois, Dessimoz Christophe, Bähler Jürg, and Sedlazeck Fritz J. 2017. “Transient Structural Variations Have Strong Effects on Quantitative Traits and Reproductive Isolation in Fission Yeast.” Nature Communications 8 (January):14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaivola Karri, Chia Ruth, Ding Jinhui, Rasheed Memoona, Fujita Masashi, Menon Vilas, Walton Ronald L., et al. 2023. “Genome-Wide Structural Variant Analysis Identifies Risk Loci for Non-Alzheimer’s Dementias.” Cell Genomics 3 (6): 100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga K., Shimohata T., Nishimura A., Shiga A., Mizuguchi T., Tokunaga J., Ohno T., et al. 2009. “Identification of Independent APP Locus Duplication in Japanese Patients with Early-Onset Alzheimer Disease.” Journal of Neurology, Neurosurgery, and Psychiatry 80 (9): 1050–52. [DOI] [PubMed] [Google Scholar]
- Kohout F. J., Berkman L. F., Evans D. A., and Cornoni-Huntley J. 1993. “Two Shorter Forms of the CES-D (Center for Epidemiological Studies Depression) Depression Symptoms Index.” Journal of Aging and Health 5 (2): 179–93. [DOI] [PubMed] [Google Scholar]
- Kunkle Brian W., Grenier-Boley Benjamin, Sims Rebecca, Bis Joshua C., Damotte Vincent, Naj Adam C., Boland Anne, et al. 2019. “Genetic Meta-Analysis of Diagnosed Alzheimer’s Disease Identifies New Risk Loci and Implicates , Tau, Immunity and Lipid Processing.” Nature Genetics 51 (3): 414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer Ryan M., Chiang Colby, Quinlan Aaron R., and Hall Ira M. 2014. “LUMPY: A Probabilistic Framework for Structural Variant Discovery.” Genome Biology 15 (6): R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall Christian R., Noor Abdul, Vincent John B., Lionel Anath C., Feuk Lars, Skaug Jennifer, Shago Mary, et al. 2008. “Structural Variation of Chromosomes in Autism Spectrum Disorder.” American Journal of Human Genetics 82 (2): 477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G., Drachman D., Folstein M., Katzman R., Price D., and Stadlan E. M. 1984. “Clinical Diagnosis of Alzheimer’s Disease: Report of the NINCDS-ADRDA Work Group under the Auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease.” Neurology 34 (7): 939–44. [DOI] [PubMed] [Google Scholar]
- Nag Sukriti, Yu Lei, Wilson Robert S., Chen Er-Yun, Bennett David A., and Schneider Julie A. 2017. “TDP-43 Pathology and Memory Impairment in Elders without Pathologic Diagnoses of AD or FTLD.” Neurology 88 (7): 653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls Mike A., Blauwendraat Cornelis, Vallerga Costanza L., Heilbron Karl, Bandres-Ciga Sara, Chang Diana, Tan Manuela, et al. 2019. “Identification of Novel Risk Loci, Causal Insights, and Heritable Risk for Parkinson’s Disease: A Meta-Analysis of Genome-Wide Association Studies.” Lancet Neurology 18 (12): 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oveisgharan Shahram, Yang Jingyun, Yu Lei, Burba Dominika, Bang Woojeong, Tasaki Shinya, Grodstein Fran, et al. 2023. “Estrogen Receptor Genes, Cognitive Decline, and Alzheimer Disease.” Neurology 100 (14): e1474–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch Tobias, Zichner Thomas, Schlattl Andreas, Stütz Adrian M., Benes Vladimir, and Korbel Jan O. 2012. “DELLY: Structural Variant Discovery by Integrated Paired-End and Split-Read Analysis.” Bioinformatics 28 (18): i333–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodney Alana, Karanjeet Kul, Benzow Kellie, and Koob Michael D. 2024. “A Common Alu Insertion in the 3’UTR of TMEM106B Is Associated with Risk of Dementia.” Alzheimer’s & Dementia: The Journal of the Alzheimer’s Association 20 (7): 5071–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovelet-Lecrux Anne, Hannequin Didier, Raux Gregory, Le Meur Nathalie, Laquerrière Annie, Vital Anne, Dumanchin Cécile, et al. 2006. “APP Locus Duplication Causes Autosomal Dominant Early-Onset Alzheimer Disease with Cerebral Amyloid Angiopathy.” Nature Genetics 38 (1): 24–26. [DOI] [PubMed] [Google Scholar]
- Salazar Alex, Tesi Niccolò, Knoop Lydian, Pijnenburg Yolande, Sven van der Lee, Wijesekera Sanduni, Krizova Jana, et al. 2023. “An AluYb8 Retrotransposon Characterises a Risk Haplotype of TMEM106B Associated in Neurodegeneration.” bioRxiv. 10.1101/2023.07.16.23292721. [DOI]
- Schneider J. A., Arvanitakis Z., Yu L., Boyle P. A., Leurgans S. E., and Bennett D. A. 2012. “Cognitive Impairment, Decline and Fluctuations in Older Community-Dwelling Subjects with Lewy Bodies.” Brain: A Journal of Neurology 135 (Pt 10): 3005–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. A., Bienias J. L., Wilson R. S., Berry-Kravis E., Evans D. A., and Bennett D. A. 2005. “The Apolipoprotein E epsilon4 Allele Increases the Odds of Chronic Cerebral Infarction [corrected] Detected at Autopsy in Older Persons.” Stroke; a Journal of Cerebral Circulation 36 (5): 954–59. [DOI] [PubMed] [Google Scholar]
- Schneider Julie A., Arvanitakis Zoe, Bang Woojeong, and Bennett David A. 2007. “Mixed Brain Pathologies Account for Most Dementia Cases in Community-Dwelling Older Persons.” Neurology 69 (24): 2197–2204. [DOI] [PubMed] [Google Scholar]
- Sebat Jonathan, Levy Deborah L., and McCarthy Shane E. 2009. “Rare Structural Variants in Schizophrenia: One Disorder, Multiple Mutations; One Mutation, Multiple Disorders.” Trends in Genetics: TIG 25 (12): 528–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifar Fatemeh, Fox Edward J., Shantaraman Anantharaman, Liu Yue, Dammer Eric B., Modeste Erica, Duong Duc M., et al. 2024. “Large-Scale Deep Proteomic Analysis in Alzheimer’s Disease Brain Regions Across Race and Ethnicity.” bioRxiv : The Preprint Server for Biology, April. 10.1101/2024.04.22.590547. [DOI] [PMC free article] [PubMed]
- Sherva Richard, Zhang Rui, Sahelijo Nathan, Jun Gyungah, Anglin Tori, Chanfreau Catherine, Cho Kelly, et al. 2023. “African Ancestry GWAS of Dementia in a Large Military Cohort Identifies Significant Risk Loci.” Molecular Psychiatry 28 (3): 1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers Kristel, Brouwers Nathalie, Gijselinck Ilse, Theuns Jessie, Goossens Dirk, Wauters Jan, Del-Favero Jurgen, Cruts Marc, van Duijn Cornelia M., and Van Broeckhoven Christine. 2006. “APP Duplication Is Sufficient to Cause Early Onset Alzheimer’s Dementia with Cerebral Amyloid Angiopathy.” Brain: A Journal of Neurology 129 (Pt 11): 2977–83. [DOI] [PubMed] [Google Scholar]
- Sudmant Peter H., Rausch Tobias, Gardner Eugene J., Handsaker Robert E., Abyzov Alexej, Huddleston John, Zhang Yan, et al. 2015. “An Integrated Map of Structural Variation in 2,504 Human Genomes.” Nature 526 (7571): 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma Rakesh, Su Su, McCrann Donald J., Green Jennifer M., Leu Karen, Young Peter R., Schatz Peter J., Silva Jeffrey C., Stokes Matthew P., and Wojchowski Don M. 2014. “RHEX, a Novel Regulator of Human Erythroid Progenitor Cell Expansion and Erythroblast Development.” The Journal of Experimental Medicine 211 (9): 1715–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialle Ricardo A., de Paiva Lopes Katia, Bennett David A., Crary John F., and Raj Towfique. 2022. “Integrating Whole-Genome Sequencing with Multi-Omic Data Reveals the Impact of Structural Variants on Gene Regulation in the Human Brain.” Nature Neuroscience 25 (4): 504–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh Tom, McClellan Jon M., McCarthy Shane E., Addington Anjené M., Pierce Sarah B., Cooper Greg M., Nord Alex S., et al. 2008. “Rare Structural Variants Disrupt Multiple Genes in Neurodevelopmental Pathways in Schizophrenia.” Science 320 (5875): 539–43. [DOI] [PubMed] [Google Scholar]
- Wang Hui, Bennett David A., De Jager Philip L., Zhang Qing-Ye, and Zhang Hong-Yu. 2021. “Genome-Wide Epistasis Analysis for Alzheimer’s Disease and Implications for Genetic Risk Prediction.” Alzheimer’s Research & Therapy 13 (1): 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Hui, Wang Li-San, Schellenberg Gerard, and Lee Wan-Ping. 2022. “The Role of Structural Variations in Alzheimer’s Disease and Other Neurodegenerative Diseases.” Frontiers in Aging Neuroscience 14:1073905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Robert S., Barnes L. L., Mendes de Leon C. F., Aggarwal N. T., Schneider J. S., Bach J., Pilat J., et al. 2002. “Depressive Symptoms, Cognitive Decline, and Risk of AD in Older Persons.” Neurology 59 (3): 364–70. [DOI] [PubMed] [Google Scholar]
- Wilson Robert S., Boyle Patricia A., Yang Jingyun, James Bryan D., and Bennett David A. 2015. “Early Life Instruction in Foreign Language and Music and Incidence of Mild Cognitive Impairment.” Neuropsychology 29 (2): 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Robert S., Capuano Ana W., Boyle Patricia A., Hoganson George M., Hizel Loren P., Shah Raj C., Nag Sukriti, Schneider Julie A., Arnold Steven E., and Bennett David A. 2014. “Clinical-Pathologic Study of Depressive Symptoms and Cognitive Decline in Old Age.” Neurology 83 (8): 702–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. S., Arnold S. E., Schneider J. A., Tang Y., and Bennett D. A. 2007. “The Relationship between Cerebral Alzheimer’s Disease Pathology and Odour Identification in Old Age.” Journal of Neurology, Neurosurgery, and Psychiatry 78 (1): 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingo Aliza P., Fan Wen, Duong Duc M., Gerasimov Ekaterina S., Dammer Eric B., Liu Yue, Harerimana Nadia V., et al. 2020. “Shared Proteomic Effects of Cerebral Atherosclerosis and Alzheimer’s Disease on the Human Brain.” Nature Neuroscience 23 (6): 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Qi, Nho Kwangsik, Del-Aguila Jorge L., Wang Xingbin, Risacher Shannon L., Kang-Hsien Fan, Snitz Beth E., et al. 2021. “Genome-Wide Association Study of Brain Amyloid Deposition as Measured by Pittsburgh Compound-B (PiB)-PET Imaging.” Molecular Psychiatry 26 (1): 309–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Lei, Tasaki Shinya, Schneider Julie A., Arfanakis Konstantinos, Duong Duc M., Wingo Aliza P., Wingo Thomas S., et al. 2020. “Cortical Proteins Associated with Cognitive Resilience in Community-Dwelling Older Persons.” JAMA Psychiatry (Chicago, Ill.) 77 (11): 1172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabetian Cyrus P., Hutter Carolyn M., Factor Stewart A., Nutt John G., Higgins Donald S., Griffith Alida, Roberts John W., et al. 2007. “Association Analysis of MAPT H1 Haplotype and Subhaplotypes in Parkinson’s Disease.” Annals of Neurology 62 (2): 137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Sai, Cooper-Knock Johnathan, Weimer Annika K., Shi Minyi, Moll Tobias, Marshall Jack N. G., Harvey Calum, et al. 2022. “Genome-Wide Identification of the Genetic Basis of Amyotrophic Lateral Sclerosis.” Neuron 110 (6): 992–1008.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Xiuwen, and Davis J. Wade. 2021. “SAIGEgds-an Efficient Statistical Tool for Large-Scale PheWAS with Mixed Models.” Bioinformatics 37 (5): 728–30. [DOI] [PubMed] [Google Scholar]
- Zhou Wei, Nielsen Jonas B., Fritsche Lars G., Dey Rounak, Gabrielsen Maiken E., Wolford Brooke N., LeFaive Jonathon, et al. 2018. “Efficiently Controlling for Case-Control Imbalance and Sample Relatedness in Large-Scale Genetic Association Studies.” Nature Genetics 50 (9): 1335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All SV site frequency data and complete nominal summary statistics for all phenotypes tested are publicly available on GitHub (https://github.com/RushAlz/ADRD_SV_GWAS). ROS/MAP resources, including individual-level genotyping and phenotypic data can be requested at https://www.radc.rush.edu.