Abstract
Ginsenoside Compound K (GCK) is the main metabolite of natural protopanaxadiol ginsenosides with diverse pharmacological effects. Gut microbiota contributes to the biotransformation of GCK, while the effect of gut microbiota on the pharmacokinetics of GCK in vivo remains unclear. To illustrate the role of gut microbiota in GCK metabolism in vivo, a systematic investigation of the pharmacokinetics of GCK in specific pathogen free (SPF) and pseudo-germ-free (pseudo-GF) rats were conducted. Pseudo-GF rats were treated with non-absorbable antibiotics. Liquid chromatography tandem mass spectrometry (LC–MS/MS) was validated for the quantification of GCK in rat plasma. Compared with SPF rats, the plasma concentration of GCK significantly increased after the gut microbiota depleted. The results showed that GCK absorption slowed down, Tmax delayed by 3.5 h, AUC0-11 increased by 1.3 times, CLz/F decreased by 0.6 times in pseudo-GF rats, and Cmax was 1.6 times higher than that of normal rats. The data indicated that gut microbiota played an important role in the pharmacokinetics of GCK in vivo.
Introduction
Ginsenoside Compound K (GCK), a protopanaxadiol (PPD)-type saponin, is the main metabolite of natural ginsenosides from Panax species biotransformed by gut microbiota in vivo [1]. Comparing with the parent saponins, recent studies find that GCK shows stronger and broader pharmacological effects such as hepatoprotective [2, 3], anti-atherosclerosis [4, 5], anti-diabetes [6, 7], neuroprotection [8], anti-inflammatory [9, 10] and anti-tumor activities [11]. Currently, GCK tablets are being tested in patients with rheumatoid arthritis (ClinicalTrials.gov Identifier: NCT03755258). However, the fate of GCK pertinent to pharmacokinetics in vivo is not well-known.
Evidences from several animal and clinical studies suggest that GCK is safe and well-tolerated [12, 13]. However, relative poor bioavailability and great individual variations of pharmacokinetics and pharmacodynamics limit the development and application of GCK [14]. Gene polymorphisms only partially explain the individual pharmacokinetics variation of GCK [15]. A clinical trial demonstrates that the pharmacokinetics of GCK is influenced by diet, but the mechanism remains unclear [14]. Gut microbiota plays an important role in drug metabolism [16]. Some studies have shown that probiotics and prebiotics improve the metabolism of the parent saponins of GCK [17, 18]. Moreover, in vitro studies also demonstrate that GCK can be biotransformed by gut microbiota [19]. However, the effects of gut microbiota on the pharmacokinetics of GCK are not well investigated.
In this study, the antibiotics including ampicillin, vancomycin, neomycin sulfate and metronidazole were used to construct a pseudo-germ-free (GF) rat model. A Liquid chromatography tandem mass spectrometry (LC-MS/MS) was validated for the quantification of GCK in rat plasma. Compared with the specific pathogen free (SPF) group, the area under the concentration-time curve (AUC) and the CLz/F of GCK were obviously increased in the pseudo-GF group. Our results implied that gut microbiota played an indispensable role in the metabolism of GCK in vivo.
Materials and methods
Chemicals and reagents
Ampicillin, vancomycin, neomycin sulfate and metronidazole were purchased from BBI Life Sciences Co., LTD. (Shanghai, China). GCK (purity ≥ 98.0%) was purchased from Baoji Hebest Biotechnology Co., LTD. (Shanxi, China). HPLC grade acetonitrile (ACN) was provided by Merck (Darmstadt, Germany).
Ethics statement
Male Sprague-Dawley (SD) rats (8 weeks old, 180–220 g) were purchased from Hunan SJA Laboratory (Hunan, China) (Certificate Number: SCXK-2019-0004). The rats were reared in a controlled environment with relative humidity (60 ± 5%), temperature (20–24°C), and maintained light and darkness for 12 hours each day. All animal works were approved by the experimental animal welfare ethical Review Institution of Central South University (Hunan, China. Approved number: CSU-2022-0003).
Construction of a pseudo-germ-free rat model
SD rats were randomly divided into two groups: normal control group (SPF group, n = 7) and pseudo-GF group (n = 7). To deplete the gut microbiota, a solution of ampicillin (50 mg/kg), vancomycin (25 mg/kg), neomycin sulfate (50 mg/kg), and metronidazole (50 mg/kg) were orally administered. Fresh antibiotic concoction with fed ampicillin in water (0.25 g/L) were prepared every day for a week. The feces were collected three days after withdrawal, and then the gut bacterial DNA was extracted with a stool DNA kit (Omega, D4015-01) and measured by Nonodrop 3000 spectrophotometer (Shimadzu, Japan) [20].
Instruments and condition
The mass spectrometer analysis was performed on QTRAP 6500+ low mass spectrometers. The Phenomenex LUNA C18 (2) Reversed Phase (Phenomenex, 150 × 2.0 mm, i.d., 5 μm) was used for the chromatographic separation with a mobile phase of acetonitrile (A) and the aqueous phase of 2 mM ammonium acetate (B) at a flow rate of 0.3 mL/min. The gradient elution procedure was set at 0−2 min: 20%A, 2−3 min: 20−65%A, 3−4 min: 65−75%A, 4−5 min: 75−90%A, 5−6 min: 90−100%A, and the re-equilibration time was set for 5 mins with 20%A after gradient elution. The tailing factors under the selected chromatographic conditions were calculated as 0.993 and 0.996 for GCK and PPD, respectively, while the theoretical plates were measured as 1.13 × 104. The injection volume was 10 μL with the temperature of column at 40°C. The mass spectrometer parameters were optimized as follow, Curtain Gas (CUR), 35; collision gas, 9;, ionspray voltage,-4500 V; temperature, 450°C; curtain gas, 35 psi; ion source gas 1, 50 psi; ion source gas 2, 55 psi. Both GCK and PPD were analyzed in multiple reaction monitoring (MRM) mode, shown as Fig 1.
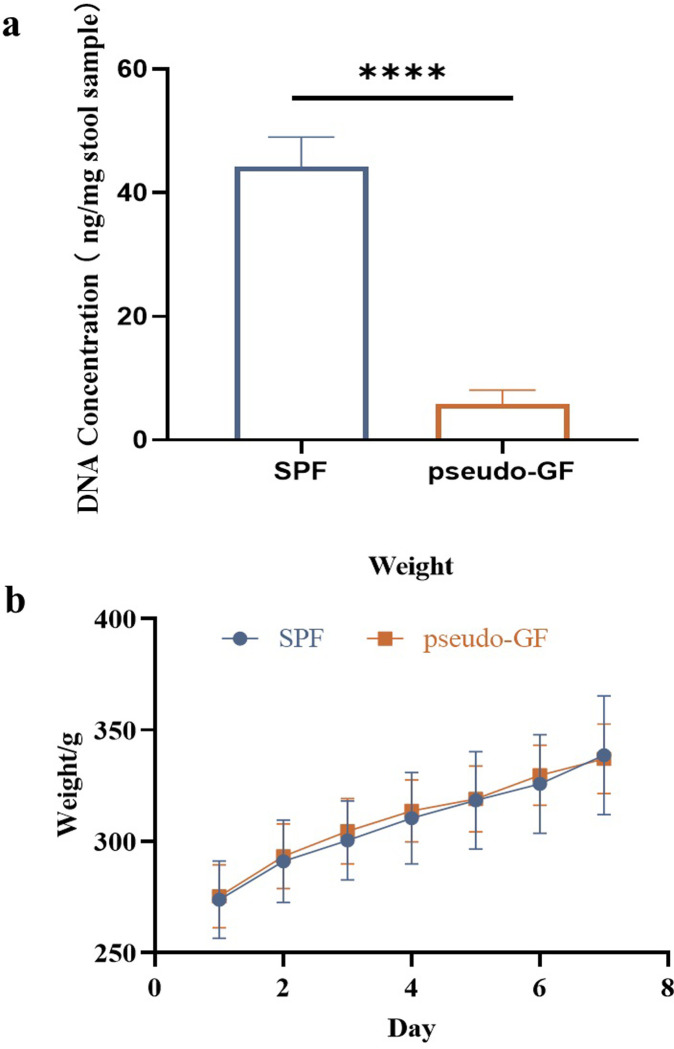
Fig 1. Validation of pseudo-GF model in rats.
(a) Body weights of rats in both normal and pseudo-GF rats after oral antibiotics (n = 5); (b) Concentration of gut microbial DNA in feces (data were expressed as mean ± SD, n = 7); SPF, normal control group; pseudo-GF, pseudo-germ-free group, ****p < 0.0001.
Experimental protocol
After fasted for 12 h, all animals were once orally administrated with a medium dose of GCK (22.5 mg/kg) [13] in 0.9% normal saline. Blood samples were collected from the tail vein at 0 h, 0.5 h, 1 h, 1.5 h, 2.5 h, 3 h, 3.5 h, 4 h, 5 h, 6 h, 7 h, 9.5 h, and 11 h after oral administration. All blood samples were immediately centrifuged at 4000 rpm for 20 min at 4°C. Approximate 100 μL of supernatant was collected and stored at -80°C for analysis. Non-compartmental model analysis was operated on DAS 2.0 software of the Chinese Pharmacological Society, and pharmacokinetic parameters were calculated accordingly.
Blood sample preparation
The blood samples (50 μL) were mixed with 150 μL of digoxin solution (Internal standard (IS), 3.33 ng/mL), subsequently vortex-mixed and centrifuged at 1.4 × 104 rpm (4°C) to remove the precipitated proteins for 10 min, respectively. Finally, 100 μL of supernatant was subjected to LC-MS/ MS analysis.
Calibration standards and quality controls (QC)
The primary stock solutions of GCK (1.38 mg/mL), and PPD (1.14 mg/mL) were prepared by dissolving them in methanol, respectively. The working solution was diluted with methanol. The standard calibration curve and QC samples were prepared by spiking 5 μL of working solution with 45 μL of blank rat plasma. The solution of IS was diluted to 3.33 ng/mL using acetonitrile. Calibration standards and QC were validated according to the 2018 FDA Guidelines.
Validation of method
Selectivity
Each blank plasma sample from six rats spiked with/without IS or IS and analytes were analyzed to assess the selectivity. The peak areas near all the analytes in the blank plasma must be less than 20% of all the analytes, respectively, which were less than 5% of blank plasma spiked with IS and analytes. Additionally, the chromatographic separation between IS and analytes must be achieved without interference on the quantification of all the analytes.
Linearity and quantification
The calibration curves were constructed in a weighted (1/X2) linear least squares regression model, and rectified by analyte/IS peak area ratios versus the standards. The correlation coefficient (R2) of calibration curves were qualified more than 0.99. The low limits of quantification (LLOQ) were defined as the lowest concentration for the calibration curve with signal-to-noise ratios (S/N) about 10 at least. Six LLOQ samples were repeatedly analyzed with the concentration acceptance due to accuracy within 85%–115%.
Precision and accuracy
Precision was evaluated through relative standard deviation (RSD), while accuracy was assessed using QC samples. The LQC, MQC, and HQC samples were quantified to calculate the intra- and inter-day precision and accuracy. The inter-day precision and accuracy were evaluated by determining QC samples in three successive days. Generally, the variability should be within 15%.
Extraction recovery
The LQC, MQC, and HQC plasma samples with six replicates at each group were used to evaluate extraction recovery which was assessed by calculating the ratios between peak areas of QC samples and/with post-extracted blank samples spiked with analytes and IS. The matrix effect was assessed by calculating peak areas ratios of post-extracted samples and/with spiked with IS and analytes. The variability was feasible to be less than 20%.
Stability
The LQC, MQC, and HQC samples were stored at –80°C for thirty days. The employed samples were tested to assess long-term stability. Six replicates of all QC samples were frozen at –80°C and thawed at 25°C for three cycles, which were then determined to assess the freeze/thaw cycle stability. The variabilities of precision and accuracy should be within15%.
Statistical analysis
Pharmacokinetics parameters including AUC, terminal elimination half-life (T1/2), mean retention time (MRT), apparent distribution volume (Vz/ F), and clearance rate (CL) were calculated using non-compartmental model analysis (DAS 2.0 software, Chinese Pharmacological Association, China). Other parameters, such as maximum plasma concentration (Cmax) and time to maximum plasma concentration (Tmax), were obtained directly from the plasma concentration-time diagram. All data were expressed as mean ± SD. Significant differences were marked by the symbols ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
Result
Pseudo-germ-free rat model construction
After oral administration with antibiotic cocktails, the concentration of microbial DNA significantly decreased in the pseudo-GF rats. Meanwhile, the body weight showed no significant difference between the two groups (Fig 1). Therefore, the results indicated that the gut microbiota was extremely disrupted in the pseudo-GF rats treated with antibiotic cocktails.
Method validation
Selectivity
The analytical method exhibited good selectivity for the detection of GCK in plasma samples. The typical MRM chromatograms of GCK, protopanaxadiol (PPD) and IS in blank rat plasma were shown in Figs 2 and 3. No significant matrix interference was found around the chromatographic retention time of analytes and IS in six different blank plasma samples.
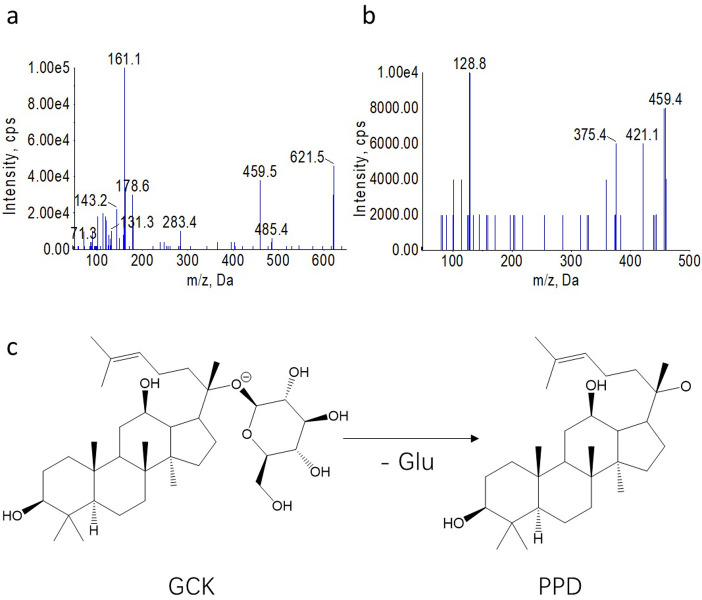
Fig 2.
Mass spectra and fragmentation pathways of GCK (a), PPD (b) in negative ion mode. (c) The MRM transitions of GCK and PPD.
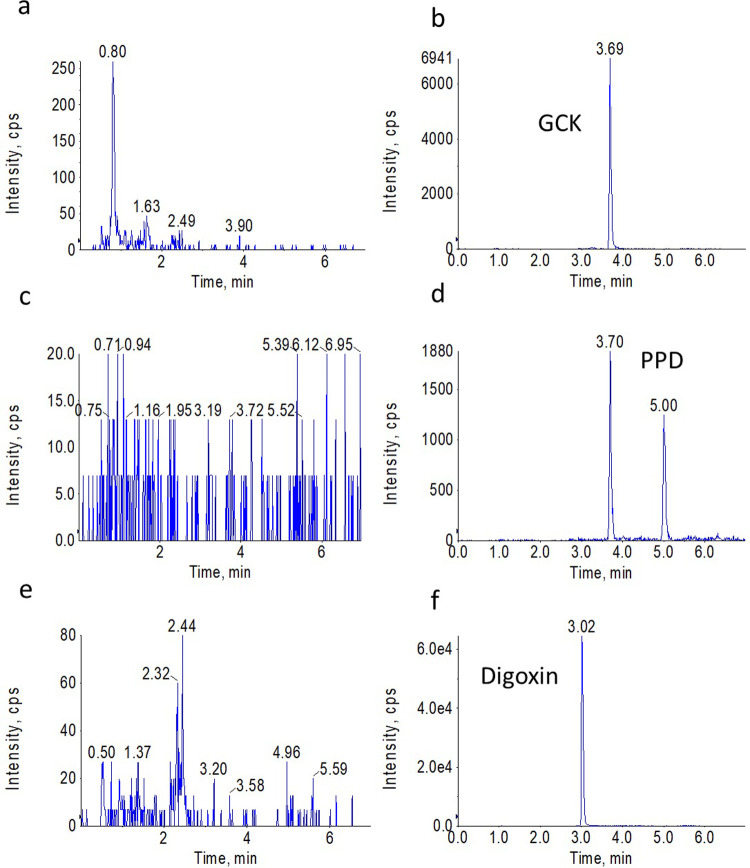
Fig 3. Typical MRM chromatograms of blank rat plasma and/or spiked with analytes and IS.
(a), (c), and (e) were the chromatograms of blank plasma; (b), (d), and (f) were the chromatograms of GCK, PPD and digoxin, respectively.
Linearity
The linearity range, calibration curves, correlation coefficients and LLOQ of the GCK and PPD in plasma samples were shown in Table 1. Both regression equations showed good linearity (R2 ≥ 0.99), and the LLOQ of GCK and PPD were 5.39 and 4.45 ng/mL, respectively. All QC samples were within the standard range (RSD ≤ 15%). The developed method was sensitive for analyzing the GCK and PPD in plasma samples.
Table 1. Linearity range, correlation coefficients (r), calibration curves and LLOQ of GCK and PPD in plasma samples.
| Compounds | Linear range (ng/ml) | Correlation coefficients (r) | Calibration curves | LLOQ (ng/ml) |
|---|---|---|---|---|
| GCK | 5.39–690 | 0.9965 | y = 0.000191x+0.00078 | 5.39 |
| PPD | 4.45–570 | 0.9938 | y = 0.000224x+0.000474 | 4.45 |
Precision and accuracy
The results of inter- and intra- day precision and accuracy of GCK and PPD were listed in Table 2. The precision of GCK and PPD in QC samples ranged from 3.34% to 9.25%, and 3.73% to 9.09%, respectively. The inter- and intra-day accuracy of GCK and PPD in QC samples were ranged from 97.2% to 104.6%, and 92.2% to 99.6%, respectively. The results indicated that the method was validated with good precision and accuracy.
Table 2. Precision and accuracy of GCK and PPD.
| Compounds | Conc. added (ng/mL)a | Intra-day (n = 6) | Inter-day (n = 18) | |||
|---|---|---|---|---|---|---|
| Accuracy (%) | Precision (%) | Accuracy (%) | Precision (%) | |||
| GCK | 5.39 | 97.59 | 6.17 | 97.17 | 7.21 | |
| 10.8 | 99.80 | 9.25 | 102.4 | 7.86 | ||
| 86.3 | 104.6 | 3.34 | 103.6 | 6.65 | ||
| 552 | 102.4 | 4.22 | 98.82 | 7.55 | ||
| PPD | 4.45 | 96.29 | 8.50 | 99.56 | 9.09 | |
| 8.91 | 92.18 | 4.16 | 99.28 | 9.04 | ||
| 71.3 | 99.60 | 4.41 | 95.47 | 6.22 | ||
| 456 | 95.61 | 3.73 | 94.17 | 5.67 | ||
aconc. is the abbreviation of concentration.
Extraction recovery
The results of extraction recovery were listed in Table 3. The mean of extraction recovery of GCK at three levels of QC samples was 77.80%, 65.22%, and 72.85%, respectively. Deviation at three levels was within 15%. It indicated that a high and reproducible extraction recovery of GCK in this method.
Table 3. Extraction recovery of GCK in rat plasma (mean ± SDa).
| Compounds | Conc. (ng/mL)b | Recovery (%) (n = 6) | RSD (%)c |
|---|---|---|---|
| GCK | 10.8 | 77.80±11.21 | 14.41 |
| 86.3 | 65.22±8.25 | 12.65 | |
| 552 | 72.85±10.66 | 14.64 | |
| PPD | 8.91 | 77.75±8.44 | 10.85 |
| 71.3 | 81.61±4.53 | 5.55 | |
| 456 | 87.75±4.20 | 4.78 |
a SD: standard deviation.
b conc. is the abbreviation of concentration.
c RSD, relative standard deviation (%) = standard deviation/ mean.
Stability
Comprehensive analysis of freeze/thaw stability and long-term stability, the precision of GCK and PPD in three levels of QC samples were ranged from 2.88%–10.65%, and 3.21%–9.29%, respectively. The accuracy of GCK and PPD in three levels of QC samples were ranged from 95.56%–106.47%, and 89.65%–101.93%, respectively (Table 4). The results suggested that all analytes were stable after storing at –80°C for 30 days or 3 freeze-thaw cycles.
Table 4. Freeze/thaw stability and long-term stability of GCK in rat plasma.
| Compounds | conc. (ng/mL)a | Freeze/thaw stability (n = 6) | long-term stability (n = 6) | ||
|---|---|---|---|---|---|
| Accuracy (%) | Precision (RSD %) | Accuracy (%) | Precision (RSD %)b | ||
| GCK | 10.8 | 102.76 | 9.07 | 95.56 | 10.65 |
| 86.3 | 106.47 | 3.37 | 103.32 | 3.57 | |
| 552 | 104.11 | 4.15 | 103.91 | 2.88 | |
| PPD | 8.91 | 92.44 | 6.64 | 96.17 | 9.29 |
| 71.3 | 93.50 | 5.51 | 97.83 | 8.86 | |
| 456 | 89.65 | 3.21 | 101.93 | 4.13 | |
a conc. is the abbreviation of concentration.
Pharmacokinetics of GCK
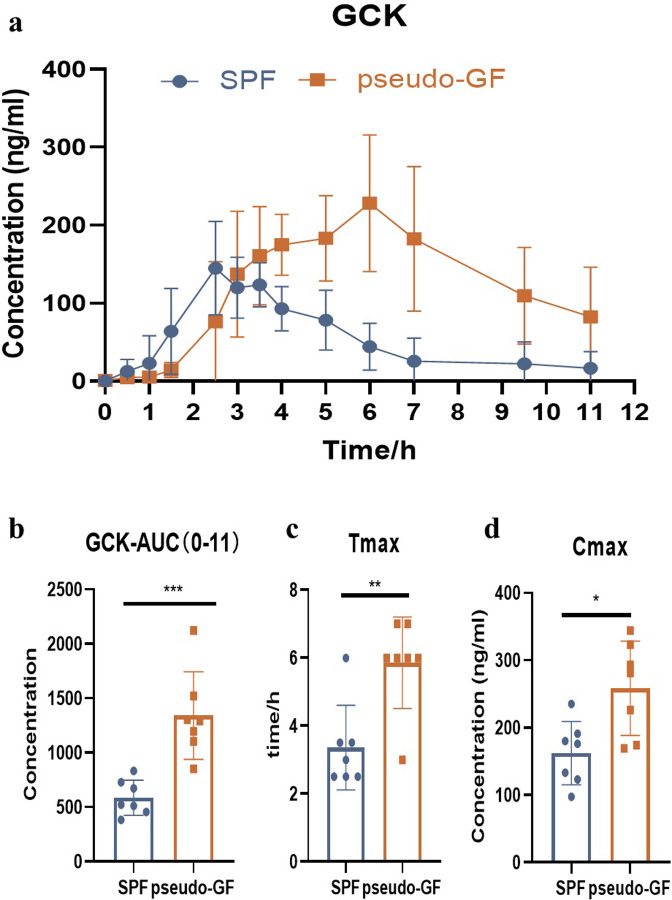
To further confirm whether the pharmacokinetics of GCK was changed after that the gut microbiota was disrupted in vivo. The chromatograms of GCK were shown in Fig 4. Fig 5 showed the concentration-time curves of GCK in SPF and pseudo-GF rats, and Table 5 summarized the pharmacokinetics parameters. Compared with SPF rats, GCK absorption in pseudo-GF rats were slowed down, Tmax were delayed by 3.5 h, AUC 0–11 in pseudo-GF rats were increased by 1.3 times, CLz/F were decreased by 0.6 times (P <0.01), and Cmax was 1.6 times higher than that of SPF rats. However, the predominant metabolite, PPD, was not detected in the two groups in this study.
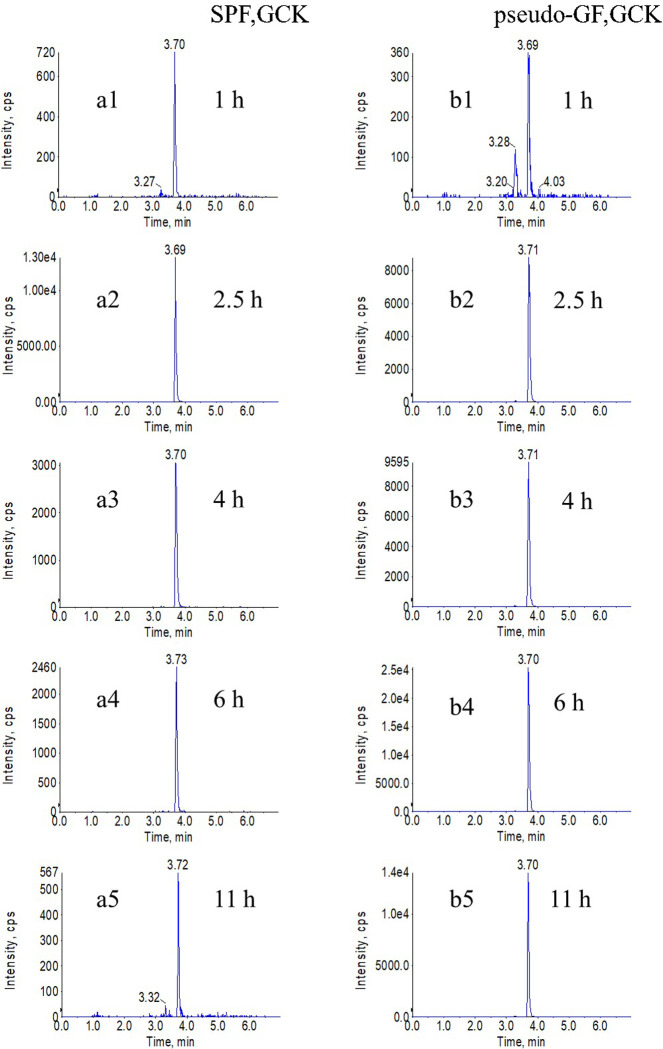
Fig 4. Typical MRM Chromatograms of GCK at checkpoints in the plasma of SPF group and pseudo-GF group.
(a1), (a2), (a3), (a4), and (a5) were typical MRM chromatograms of GCK at 1 h, 2.5 h, 4 h, 6 h, and 11 h in SPF group, respectively. (b1), (b2), (b3), (b4), and (b5) were typical MRM chromatograms of GCK at 1 h, 2.5 h, 4 h, 6 h, 11 h in pseudo-GF group, respectively.
Fig 5. The mean plasma concentration–time profiles of GCK in rat plasma after intragastric administration of GCK (22.5 mg/kg); each point and bar represent the mean ± SD (n = 7), *p < 0.05, **p < 0.01 and ***p < 0.001.
Table 5. Summary pharmacokinetic parameters of ginsenoside CK.
| Parameters | SPF group | pseudo-GF group | P value |
|---|---|---|---|
| Cmax (ng/L) | 162.21±46.93 | 258.57±69.97 | < 0.05 |
| Tmax (h) | 3.36±1.25 | 5.86±1.35 | < 0.01 |
| T1/2 (h) | 2.20±1.69 | 3.93±1.65 | 0.077 |
| MRT(0–11) (h) | 4.16±1.11 | 6.05±0.42 | < 0.01 |
| AUC(0-∞) (ng/L*h) | 678.95±284.40 | 2299.05±1626.69 | < 0.05 |
| AUC (0–11) (ng/L*h) | 586.32±161.33 | 1340.82±401.50 | 0.001 |
| CLz/F (L/h/kg) | 37941.58±13973.39 | 13561.93±6948.47 | < 0.01 |
| Vz/F (L/kg) | 96540.94±44906.34 | 63166.94±15378.70 | 0.088 |
Cmax: maximum plasma concentration; Tmax: time of maximum plasma concentration; T1/2: half-life of elimination; MRT: mean residence time; AUC: area under the plasma concentration vs time curve; CLz/F: clearance rate; Vz/F: apparent volume of distribution.
Discussion
The fates of many natural products are related with gut microbiota in vivo. The antibiotics alter the characteristics and function of gut microbiota [21]. In order to better understand the impact of gut microbiota on GCK metabolism and its pharmacokinetics, we constructed a pseudo-sterile rat model on which a mixture of antibiotics was selected to disrupt a broad spectrum of both G+ and G- bacteria [22]. The result of the microbial DNA concentration displayed that the combination of broad-spectrum antibiotics efficiently destroyed the gut microbiota.
To analyze the metabolism of GCK in vivo, the different check-point concentrations of the parent drug and PPD in plasma were measured in both groups after a single oral administration. Compared with the SPF group, the concentration of GCK was significantly higher, but the Tmax was delayed. Meanwhile, the exposed time was longer after oral administration, and the AUC increased by approximately twice in the pseudo-GF group. These results indicated that the pharmacokinetics of GCK greatly changed with the depletion of gut microbiota in vivo.
In view of our data, gut microbiota played an important role in the metabolism of GCK in vivo. Moreover, when the bacterial community was out-of-balance, the biotransformation of GCK was severely altered due to the catalyzing enzyme secreted by gut microbiota. Besides, the host metabolism of GCK may be affected by the dysbiosis of gut microbiota. Additionally, the data showed that the individual variation of GCK pharmacokinetics was observed in both groups. Herein, the diversity of gut microbiota may lead to significant variations in the metabolic behavior of GCK.
In a clinical trial, the T1/2 of PPD in the human plasma sample reaches more than 19 hours, while the Tmax and Cmax of PPD are approximate 25 hours and 4.2 ng/mL after a single dose of oral GCK [14]. Our method was more sensitive than the employed methods for the detection of GCK and PPD in plasma samples. However, PPD was not detected in our study due to the possibility of poor generation and absorption of PPD after oral administration of GCK in rats. Therefore, the pharmacokinetics parameters of PPD were unable to be calculated for evaluating its pharmacokinetics in this study. Nevertheless, the validated method was greatly applied for the pharmacokinetics of GCK in vivo.
Conclusion
The pharmacokinetics of GCK was significantly altered along with the depletion of gut microbiota in rats. Though GCK is able to be metabolized to generate PPD, which was not detected in rat plasma from both normal and pseudo-GF groups after single oral administration of GCK. The pharmacokinetics of GCK presented individual variation due to potential binary metabolism of host and gut microbiota.
Acknowledgments
The authors would like to great acknowledge Mr. Wang Yicheng and Dr. Gao Yongchao of the Institute of Clinical Pharmacology, Central South University, for their technical assistance in technical methods and statistical analysis.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was supported by the National Natural Scientific Foundation of China (82074000, 81903784), the Hunan Provincial Natural Science Foundation of China (2023JJ30459, 2024JJ5585, 2024JJ5529, 2024JJ9533), the Scientific Research Program of Furong laboratory (2023SK2083), the Scientific Research Project of Department of Education of Hunan Province (20K136).
References
- 1.Yang X.D., et al., A review of biotransformation and pharmacology of ginsenoside compound K. Fitoterapia, 2015. 100: p. 208–20. doi: 10.1016/j.fitote.2014.11.019 [DOI] [PubMed] [Google Scholar]
- 2.Zhou L., et al., Ginsenoside compound K alleviates sodium valproate-induced hepatotoxicity in rats via antioxidant effect, regulation of peroxisome pathway and iron homeostasis. Toxicol Appl Pharmacol, 2020. 386: p. 114829. doi: 10.1016/j.taap.2019.114829 [DOI] [PubMed] [Google Scholar]
- 3.Chen X.J., et al., Ameliorative effects of Compound K and ginsenoside Rh1 on non-alcoholic fatty liver disease in rats. Sci Rep, 2017. 7: p. 41144. doi: 10.1038/srep41144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou L., et al., Compound K Attenuates the Development of Atherosclerosis in ApoE(-/-) Mice via LXRalpha Activation. Int J Mol Sci, 2016. 17(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu S., et al., Ginsenoside compound K protects human umbilical vein endothelial cells against oxidized low-density lipoprotein-induced injury via inhibition of nuclear factor-kappaB, p38, and JNK MAPK pathways. J Ginseng Res, 2019. 43(1): p. 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei S., et al., Ginsenoside Compound K suppresses the hepatic gluconeogenesis via activating adenosine-5’monophosphate kinase: A study in vitro and in vivo. Life Sci, 2015. 139: p. 8–15. doi: 10.1016/j.lfs.2015.07.032 [DOI] [PubMed] [Google Scholar]
- 7.Han GC K.S., Sung JH, Chung SH., Compound K enhances insulin secretion with beneficial metabolic effects in db/db mice. J Agric Food Chem., 2007. [DOI] [PubMed] [Google Scholar]
- 8.Oh J.M., et al., Ginsenoside Compound K Induces Adult Hippocampal Proliferation and Survival of Newly Generated Cells in Young and Elderly Mice. Biomolecules, 2020. 10(3). doi: 10.3390/biom10030484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J.O., et al., Compound K, a ginsenoside metabolite, plays an antiinflammatory role in macrophages by targeting the AKT1-mediated signaling pathway. J Ginseng Res, 2019. 43(1): p. 154–160. doi: 10.1016/j.jgr.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J.W., et al., Compound K ameliorates airway inflammation and mucus secretion through the regulation of PKC signaling in vitro and in vivo. J Ginseng Res, 2022. 46(3): p. 496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L., et al., Targeting PLA2G16, a lipid metabolism gene, by Ginsenoside Compound K to suppress the malignant progression of colorectal cancer. J Adv Res, 2022. 36: p. 265–276. doi: 10.1016/j.jare.2021.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A. and Lee H.J., Ginsenoside Compound K: Insights into Recent Studies on Pharmacokinetics and Health-Promoting Activities. Biomolecules, 2020. 10(7). doi: 10.3390/biom10071028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L., et al., Single- and Multiple-Dose Trials to Determine the Pharmacokinetics, Safety, Tolerability, and Sex Effect of Oral Ginsenoside Compound K in Healthy Chinese Volunteers. Front Pharmacol, 2017. 8: p. 965. doi: 10.3389/fphar.2017.00965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L., et al., Food and Sex-Related Impacts on the Pharmacokinetics of a Single-Dose of Ginsenoside Compound K in Healthy Subjects. Front Pharmacol, 2017. 8: p. 636. doi: 10.3389/fphar.2017.00636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou L., et al., Impact of NR1I2, adenosine triphosphate-binding cassette transporters genetic polymorphisms on the pharmacokinetics of ginsenoside compound K in healthy Chinese volunteers. J Ginseng Res, 2019. 43(3): p. 460–474. doi: 10.1016/j.jgr.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Javdan B., et al., Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell, 2020. 181(7): p. 1661–1679 e22. doi: 10.1016/j.cell.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang A., et al., Gut microbiota in the pharmacokinetics and colonic deglycosylation metabolism of ginsenoside Rb1 in rats: Contrary effects of antimicrobials treatment and restraint stress. Chem Biol Interact, 2016. 258: p. 187–96. doi: 10.1016/j.cbi.2016.09.005 [DOI] [PubMed] [Google Scholar]
- 18.Kim D.H., Gut microbiota-mediated pharmacokinetics of ginseng saponins. J Ginseng Res, 2018. 42(3): p. 255–263. doi: 10.1016/j.jgr.2017.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y.P., et al., Bioconversion variation of ginsenoside CK mediated by human gut microbiota from healthy volunteers and colorectal cancer patients. Chin Med, 2021. 16(1): p. 28. doi: 10.1186/s13020-021-00436-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuno T., et al., Effect of Intestinal Flora on Protein Expression of Drug-Metabolizing Enzymes and Transporters in the Liver and Kidney of Germ-Free and Antibiotics-Treated Mice. Mol Pharm, 2016. 13(8): p. 2691–701. doi: 10.1021/acs.molpharmaceut.6b00259 [DOI] [PubMed] [Google Scholar]
- 21.Ge X., et al., Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. J Transl Med, 2017. 15(1): p. 13. doi: 10.1186/s12967-016-1105-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill D.A., et al., Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol, 2010. 3(2): p. 148–58. doi: 10.1038/mi.2009.132 [DOI] [PMC free article] [PubMed] [Google Scholar]