Abstract
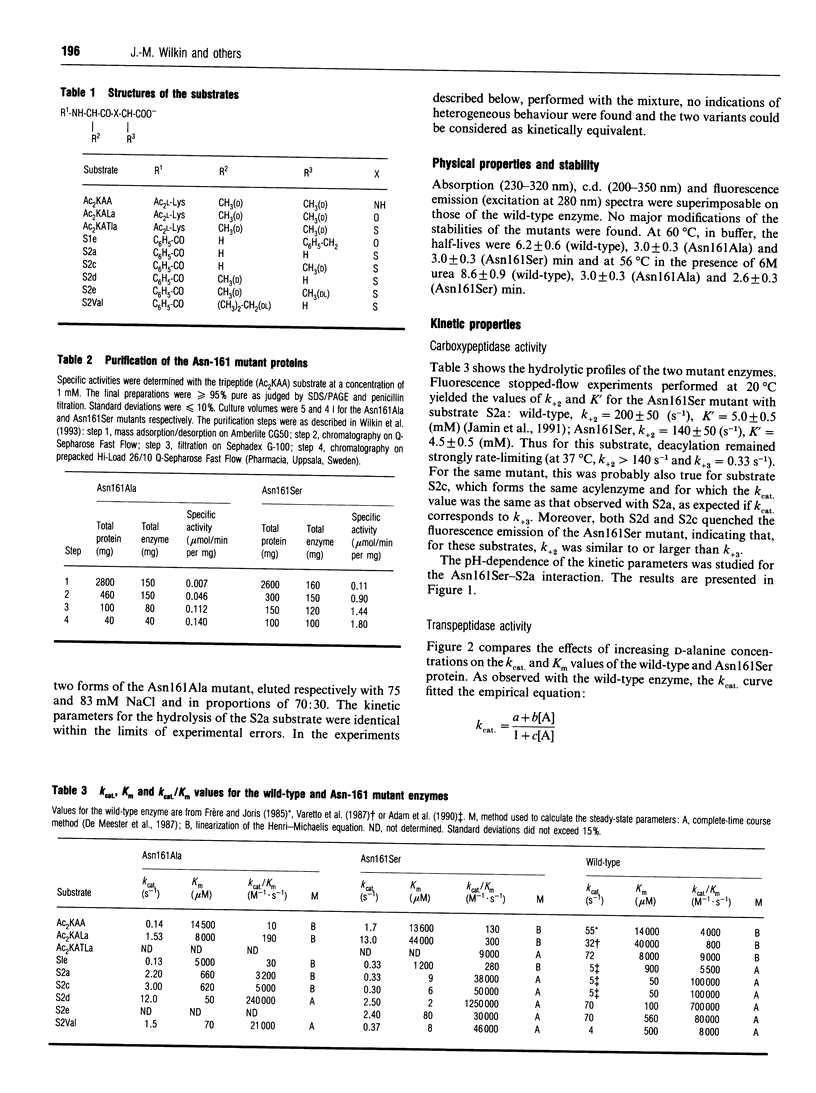
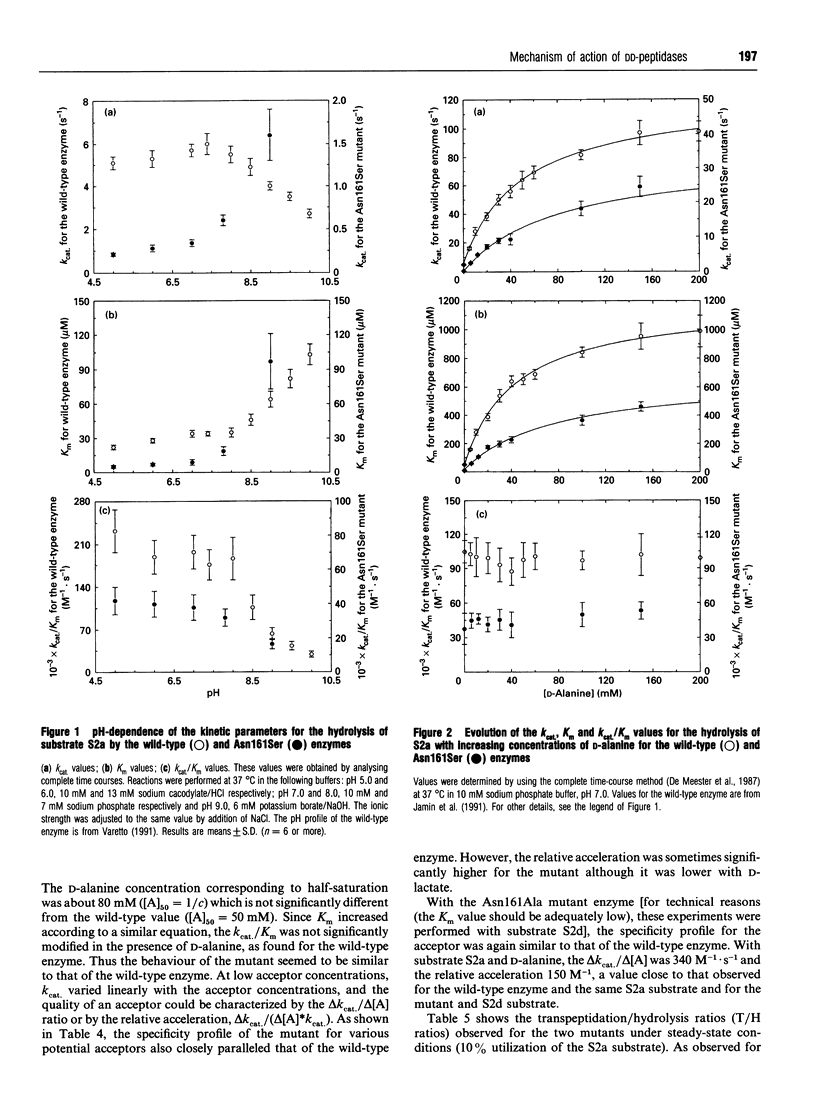
The role of residue Asn-161 in the interaction between the Streptomyces R61 DD-peptidase and various substrates or beta-lactam inactivators was probed by site-directed mutagenesis. The residue was successively replaced by serine and alanine. In the first case, acylation rates were mainly affected with the peptide and ester substrates but not with the thiol-ester substrates and beta-lactams. However, the deacylation rates were decreased 10-30-fold with the substrates yielding benzoylglycyl and benzoylalanyl adducts. The Asn161Ala mutant was more generally affected, although the acylation rates with cefuroxime and cefotaxime remained similar to those observed with the wild-type enzyme. Surprisingly, the deacylation rates of the benzoylglycyl and benzoylalanyl adducts were very close to those observed with the wild-type enzyme. The results also indicate that the interaction with the peptide substrate and the transpeptidation reaction were more sensitive to the mutations than the other reactions studied. The results are discussed and compared with those obtained with the Asn-132 mutants of a class A beta-lactamase.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam M., Damblon C., Plaitin B., Christiaens L., Frère J. M. Chromogenic depsipeptide substrates for beta-lactamases and penicillin-sensitive DD-peptidases. Biochem J. 1990 Sep 1;270(2):525–529. doi: 10.1042/bj2700525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester F., Joris B., Reckinger G., Bellefroid-Bourguignon C., Frère J. M., Waley S. G. Automated analysis of enzyme inactivation phenomena. Application to beta-lactamases and DD-peptidases. Biochem Pharmacol. 1987 Jul 15;36(14):2393–2403. doi: 10.1016/0006-2952(87)90609-5. [DOI] [PubMed] [Google Scholar]
- Frère J. M., Joris B. Penicillin-sensitive enzymes in peptidoglycan biosynthesis. Crit Rev Microbiol. 1985;11(4):299–396. doi: 10.3109/10408418409105906. [DOI] [PubMed] [Google Scholar]
- Herzberg O., Moult J. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 A resolution. Science. 1987 May 8;236(4802):694–701. doi: 10.1126/science.3107125. [DOI] [PubMed] [Google Scholar]
- Jacob F., Joris B., Dideberg O., Dusart J., Ghuysen J. M., Frère J. M. Engineering a novel beta-lactamase by a single point mutation. Protein Eng. 1990 Oct;4(1):79–86. doi: 10.1093/protein/4.1.79. [DOI] [PubMed] [Google Scholar]
- Jacob F., Joris B., Lepage S., Dusart J., Frère J. M. Role of the conserved amino acids of the 'SDN' loop (Ser130, Asp131 and Asn132) in a class A beta-lactamase studied by site-directed mutagenesis. Biochem J. 1990 Oct 15;271(2):399–406. doi: 10.1042/bj2710399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamin M., Adam M., Damblon C., Christiaens L., Frère J. M. Accumulation of acyl-enzyme in DD-peptidase-catalysed reactions with analogues of peptide substrates. Biochem J. 1991 Dec 1;280(Pt 2):499–506. doi: 10.1042/bj2800499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Ghuysen J. M., Dive G., Renard A., Dideberg O., Charlier P., Frère J. M., Kelly J. A., Boyington J. C., Moews P. C. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988 Mar 1;250(2):313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Ledent P., Dideberg O., Fonzé E., Lamotte-Brasseur J., Kelly J. A., Ghuysen J. M., Frère J. M. Comparison of the sequences of class A beta-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991 Nov;35(11):2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamotte-Brasseur J., Dive G., Dideberg O., Charlier P., Frère J. M., Ghuysen J. M. Mechanism of acyl transfer by the class A serine beta-lactamase of Streptomyces albus G. Biochem J. 1991 Oct 1;279(Pt 1):213–221. doi: 10.1042/bj2790213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moews P. C., Knox J. R., Dideberg O., Charlier P., Frère J. M. Beta-lactamase of Bacillus licheniformis 749/C at 2 A resolution. Proteins. 1990;7(2):156–171. doi: 10.1002/prot.340070205. [DOI] [PubMed] [Google Scholar]
- Strynadka N. C., Adachi H., Jensen S. E., Johns K., Sielecki A., Betzel C., Sutoh K., James M. N. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 A resolution. Nature. 1992 Oct 22;359(6397):700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- Varetto L., Frère J. M., Nguyen-Distèche M., Ghuysen J. M., Houssier C. The pH dependence of the active-site serine DD-peptidase of Streptomyces R61. Eur J Biochem. 1987 Feb 2;162(3):525–531. doi: 10.1111/j.1432-1033.1987.tb10671.x. [DOI] [PubMed] [Google Scholar]
- Wilkin J. M., Jamin M., Damblon C., Zhao G. H., Joris B., Duez C., Frère J. M. The mechanism of action of DD-peptidases: the role of tyrosine-159 in the Streptomyces R61 DD-peptidase. Biochem J. 1993 Apr 15;291(Pt 2):537–544. doi: 10.1042/bj2910537. [DOI] [PMC free article] [PubMed] [Google Scholar]


