Abstract
Fibrosis in the lung is thought to be driven by epithelial cell dysfunction and aberrant cell-cell interactions. Unveiling the molecular mechanisms of cellular plasticity and cell-cell interactions is imperative to elucidating lung regenerative capacity and aberrant repair in pulmonary fibrosis. By mining publicly available RNA-Seq data sets, we identified loss of CCAAT enhancer-binding protein alpha (CEBPA) as a candidate contributor to idiopathic pulmonary fibrosis (IPF). We used conditional KO mice, scRNA-Seq, lung organoids, small-molecule inhibition, and potentially novel gene manipulation methods to investigate the role of CEBPA in lung fibrosis and repair. Long-term (6 months or more) of Cebpa loss in AT2 cells caused spontaneous fibrosis and increased susceptibility to bleomycin-induced fibrosis. Cebpa knockout (KO) in these mice significantly decreased AT2 cell numbers in the lung and reduced expression of surfactant homeostasis genes, while increasing inflammatory cell recruitment as well as upregulating S100a8/a9 in AT2 cells. In vivo treatment with an S100A8/A9 inhibitor alleviated experimental lung fibrosis. Restoring CEBPA expression in lung organoids ex vivo and during experimental lung fibrosis in vivo rescued CEBPA deficiency–mediated phenotypes. Our study establishes a direct mechanistic link between CEBPA repression, impaired AT2 cell identity, disrupted tissue homeostasis, and lung fibrosis.
Keywords: Pulmonology, Stem cells
Keywords: Adult stem cells, Fibrosis, Pulmonary surfactants
<strong>CEBPA deficiency causes loss of AT2 cells and induces profibrotic response, which provides new possibilities for lung fibrosis treatment by restoring insufficient CEBPA expression.</strong>
Introduction
Fibrosis is the end-stage pathological outcome of many chronic diseases, including IPF. It not only involves the accumulation of excess extracellular matrix components but also likely shares intrinsic mechanisms through which structural cells activate the mesenchymal compartment by creating a fibrotic niche (1). Healthy type 2 alveolar epithelial (AT2) cells play central roles in maintaining lung homeostasis, including secreting surfactant, transporting sodium and fluids, and performing immunomodulation as well as serving as adult tissue stem cells (2) in lung tissue maintenance, repair, and regeneration. The concept that AT2 cell failure contributes to pulmonary fibrosis has been tested in multiple animal studies (3), with persistent injury signals from AT2 cells leading to elaboration of profibrogenic mediators that activate a pathologic mesenchymal response. To date, tremendous progress has been made in understanding the mechanisms underlying epithelial cell injury (4) and fibroblast activation (5) in lung fibrosis. However, challenges remain in fully understanding AT2 cell plasticity in driving lung fibrosis and intrinsic and extrinsic mechanisms leading to AT2 cell failure.
Recent findings (6–9) emphasize an important concept that healthy epithelial cells engage in continuous interactions with neighboring mesenchyme to maintain homeostasis. Moreover, the loss of these homeostatic interactions in fibrotic diseases such as IPF has only begun to be studied. Recently, single-cell RNA-Seq (scRNA-Seq) identified loss of normal epithelial cell identities and gain of abnormal transitional states of differentiation in IPF (10–14). Animal studies (11, 15) and lung organoid studies (15, 16) have also demonstrated that transitional cell states are linked to experimental lung fibrosis. Such transitional cell states are likely a common property of AT2 and other epithelial cells in the development of lung fibrosis and other chronic lung diseases. However, it remains unclear how AT2 cells lose their identities and to what extent such mechanisms contribute to fibrosis initiation and progression.
The biology of CEBPA has been studied for over 20 years. CEBPA is known to play critical roles in cellular proliferation (17), lipogenesis (18–20), lung development (21, 22), tumor suppression (23), and epithelial cell homeostasis (24). Given the significant roles of CEBPA in liver fibrosis (25), COPD, and lung fibrosis (26), we aim to understand in depth how the loss of CEBPA contributes to the depletion of AT2 cells and the promotion of lung fibrosis. Using publicly available scRNA-Seq data from patients with IPF, we confirmed that CEBPA is among the top downregulated genes in alveolar epithelial cells from the lungs of patients with IPF, highlighting a potentially important role of CEBPA in maintaining lung homeostasis. Therefore, we aimed to investigate the connections between loss of CEBPA expression in human IPF and its roles in the mouse models, with the goal of validating CEBPA as a valuable therapeutic target for restoring epithelial identity and fibrosis resolution.
Specifically, we took advantage of an aged mouse injury model to confirm low Cebpa expression in experimental lung fibrosis. We then tested whether deleting Cebpa leads to spontaneous lung fibrosis, as well as whether its loss disrupts fibrosis resolution after bleomycin exposure. Using scRNA-Seq, we obtained a better understanding of how Cebpa deletion disrupts tissue homeostasis. Lineage-tracing mouse and human lung organoid experiments confirmed the role of CEBPA in maintaining AT2 cell identity and tissue homeostasis, as well as initiating and resolving fibrosis. We then employed pharmacological intervention, AAV-mediated overexpression, and nongenome editing CRISPR gene activation to demonstrate the benefit of targeting CEBPA and downstream pathways in alleviating experimental lung fibrosis, supporting CEBPA as a key regulator of tissue homeostasis and repair in the lung.
Results
CEBPA is downregulated in human IPF lung samples.
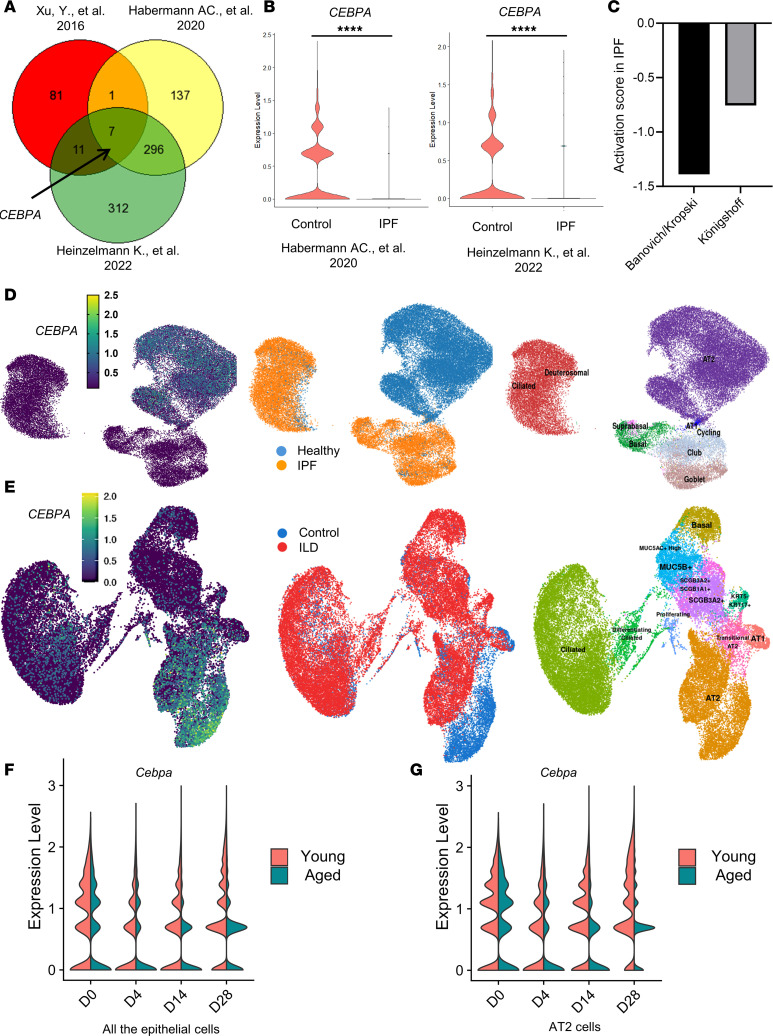
We began by querying 3 publicly available scRNA-Seq data sets to identify genes significantly downregulated in lung samples from patients with and without IPF. CEBPA was the most consistently downregulated gene in IPF epithelial cells (Figure 1A). CEBPA expression levels were significantly lower in IPF versus control epithelial cells in all available data sets (10, 14, 27) (Figure 1B). In addition, an upstream regulator analysis based on differentially expressed genes from 2 of these data sets predicted that CEBPA is negatively regulated in IPF (Figure 1C). Uniform Manifold Approximation and Projection (UMAP) representation of epithelial cells derived from available scRNA-Seq data sets (14, 27) demonstrated that CEBPA is enriched in AT2 cells and its expression is decreased in IPF lungs (Figure 1, D and E).
Figure 1. CEBPA is downregulated in human IPF and in experimental lung fibrosis.
(A) In this Venn diagram, each circle represents genes significantly downregulated in human IPF versus control lung samples from each of 3 independent studies. (B) Violin plot of CEBPA expression in epithelial cells from the lungs of patients with and without IPF, from 2 of the human scRNA-Seq data sets analyzed (14, 27). (C) Activation score of Upstream Regulator Analysis (from QIAGEN Ingenuity Pathway Analysis) for the data from the 2 human scRNA-Seq studies in B (14, 27). (D and E) Uniform manifold approximation projection (UMAP) plots of human lung epithelial cells from the 2 scRNA-Seq data sets generated via http://ipfcellatlas.com (F and G) Violin plot of Cebpa expression in the epithelial cells (F) or AT2 cells (G) at different time points (day 0 [D0], D4, D14, D28) after bleomycin injury in the lung of young and aged mice from reanalysis of GSE157995. Data were analyzed using an unpaired, 2-tailed Student’s t test. ****P < 0.0001.
Cebpa is downregulated in experimental lung fibrosis.
After establishing that CEBPA expression levels are consistently lower in the epithelial cells from human IPF versus control lung samples, we aimed to investigate Cebpa expression in an experimental lung fibrosis mice model. We exposed young (~2-month-old) and aged mice (~18-month-old) to bleomycin to simulate self-resolving (28) and persistent (29, 30) lung fibrosis, respectively. At baseline, the number of AT2 cells (represented by pulmonary-associated surfactant protein C–expressing [Sftpc-expressing] cells) did not differ between the young and aged mice. However, at day 30 after bleomycin injury, the number of AT2 cells in aged mice was significantly lower than in young mice (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.175290DS1). This finding demonstrates that AT2 cell numbers decline substantially in the bleomycin-induced persistent lung fibrosis aged mice model.
We then compared Cebpa expression levels in sorted epithelial cells from young and aged mice over the course of the experiment. In young mice, the majority of epithelial cells (CD326+) are Sftpc cells (Sftpc _GFP+), demonstrated by flow cytometry (Supplemental Figure 1C). Cebpa expression significantly but transiently declined after bleomycin injury, with gradual recovery by day 30 (Supplemental Figure 1D). By contrast, in aged mice, Cebpa expression remained significantly repressed at day 30 after bleomycin injury (Supplemental Figure 1D). We used immunostaining to confirm differences in lung tissue Sftpc and Cebpa expression at select time points (Supplemental Figure 1E). Strikingly, the transient loss of Cebpa expression in young mice and sustained loss in aged mice directly mirrored the transient versus sustained loss of AT2 cells and transient versus persistent fibrosis in young and aged mice. These data have also been validated by scRNA-Seq reanalysis from comparison of young and aged mice in the bleomycin-induced fibrosis model at different time points (day 0, day 4, day 14, day 28). UMAP (Supplemental Figure 1F) and violin plots have shown that Cebpa expression from both epithelial cells (Figure 1F) and AT2 cells (Figure 1G) was decreased after bleomycin injury in both young and aged mice and remained expressed to a lower degree in older mice than young mice at a later time (day 28) after bleomycin injury. These data consistently demonstrate that Cebpa is preferentially expressed in AT2 cells and that levels of both Cebpa and AT2 cells decrease in aged mice with persistent experimental lung fibrosis.
Loss of Cebpa in AT2 cells promotes lung fibrosis.
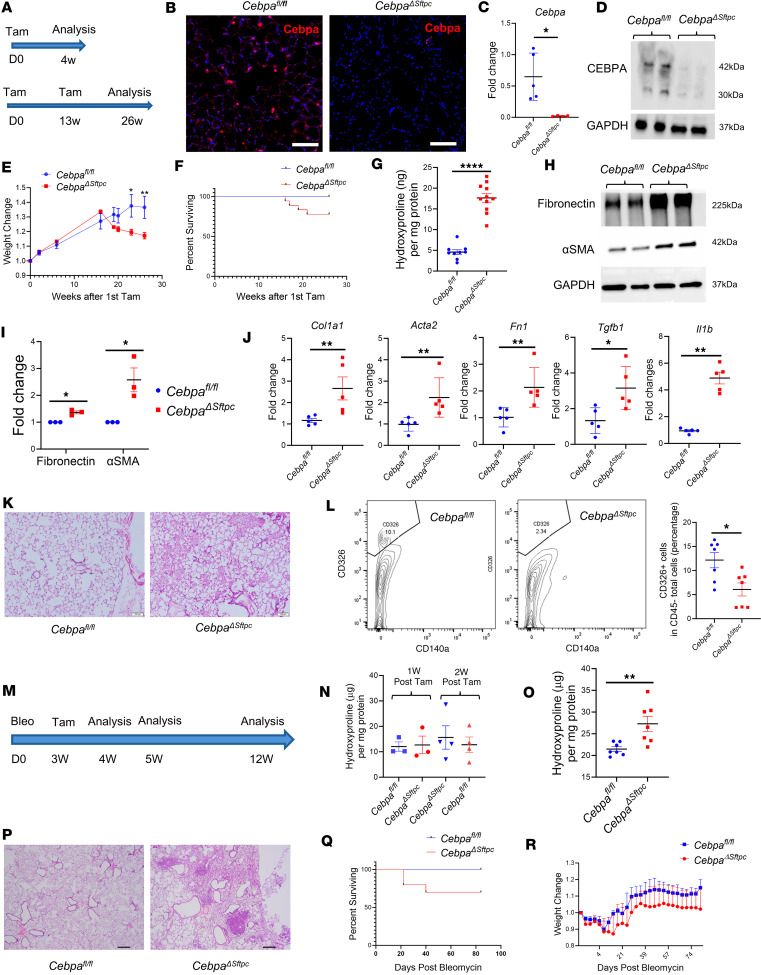
Based on the association between low CEBPA levels and IPF in humans and in the mice models, we hypothesized that Cebpa loss could initiate lung fibrosis. To test this hypothesis, we generated CebpaΔSftpc mice in which we conditionally deleted Cebpa in AT2 cells via tamoxifen-induced Cre recombinase activation (Figure 2A). At 4 weeks after tamoxifen treatment, Cebpa expression in CebpaΔSftpc mice was significantly diminished compared with Cebpa-floxed mice (Cebpafl/fl), as confirmed by immunostaining, quantitative PCR (qPCR), and Western blot (Figure 2, B–D). However, levels of collagen, a marker of fibrosis that can be measured with a hydroxyproline assay, were only slightly increased in the lungs of CebpaΔSftpc versus Cebpafl/fl mice (Supplemental Figure 2A), and no difference in lung tissue histology was evaluated by H&E staining and mean linear intercept measurement (Supplemental Figure 2, B and C).
Figure 2. Loss of Cebpa in AT2 cells promotes lung fibrotic response, impairs fibrosis resolution and lung repair.
(A) Schematic showing tamoxifen treatment and analysis timeline. (B–D) Representative immunostaining images (n = 3) of Cebpa expression, qPCR of Cebpa transcripts, and representative Western blot (n = 3) showing Cebpa expression from CebpaΔSftpc and Cebpafl/fl mice lungs, 4 weeks after tamoxifen treatment. Scale bar: 50 μM. (E and F) Weight change of CebpaΔSftpc (n = 14) and Cebpafl/fl (n = 12) mice, and survival curve of CebpaΔSftpc (n = 18) and Cebpafl/fl (n = 10) mice after first tamoxifen treatment, over a 26-week period. (G) Hydroxyproline assay showing collagen deposition from CebpaΔSftpc (n = 11) and Cebpafl/fl (n = 9) mice lungs 26 weeks after first tamoxifen treatment. (H and I) Representative Western blot and their quantification show fibronectin and α-SMA expression from CebpaΔSftpc and Cebpafl/fl mice lungs 26 weeks after tamoxifen treatment. (J) qPCR for profibrotic gene transcripts from CebpaΔSftpc and Cebpafl/fl mice lungs 26 weeks after first tamoxifen treatment. (K) Representative H&E staining showing lung sections from CebpaΔSftpc (n = 3) and Cebpafl/fl (n = 3) mice 26 weeks after first tamoxifen treatment. Scale bar: 100 μM. (L) Flow cytometry analysis of CD326+ cells from CebpaΔSftpc (n = 7) and Cebpafl/fl (n = 7) mice lungs 26 weeks after first tamoxifen treatment. (M) Schematic showing timeline for bleomycin treatment, tamoxifen treatment, and analysis. (N) Hydroxyproline assay from CebpaΔSftpc and Cebpafl/fl mice lungs at 4 and 5 weeks after bleomycin injury. (O) Hydroxyproline assay from CebpaΔSftpc and Cebpafl/fl mice lungs at 12 weeks after bleomycin injury. (P) Representative H&E staining showing lung sections from CebpaΔSftpc (n = 3) and Cebpafl/fl (n = 3) mice at 12 weeks after bleomycin injury. Scale bar: 200 μM. (Q and R) Survival curve and weight change of CebpaΔSftpc (n = 10) and Cebpafl/fl (n = 7) mice over a 12-week postbleomycin injury period. Data were analyzed using a Mann-Whitney U test. *P < 0.05, **P < 0.01, ****P < 0.0001.
To investigate the long-term effects of Cebpa deletion, CebpaΔSftpc mice were also compared with Cebpafl/fl mice at 26 weeks after first tamoxifen treatment (Figure 2A). CebpaΔSftpc mice displayed significant weight loss (Figure 2E) and increased mortality (Figure 2F) beginning at 20 weeks after tamoxifen treatment. Hydroxyproline assay (Figure 2G) and Western blot (Figure 2, H and I) of whole lung samples demonstrated that, compared with Cebpafl/fl mice, CebpaΔSftpc mice displayed increased tissue content of total collagen, fibronectin, and α-SMA. qPCR (Figure 2J) also showed increased transcripts of the profibrotic genes Col1a, Fn1, Acta2, Tgfb1, and Il1b in lung samples from CebpaΔSftpc mice. Moreover, H&E staining (Figure 2K) showed significantly increased architectural distortion in whole lung samples from CebpaΔSftpc mice. Flow cytometry analysis indicated that the percentage of epithelial cells in samples from CebpaΔSftpc mice also decreased (Figure 2L).
Next, we combined bleomycin-induced lung fibrosis with Cebpa deletion and hypothesized that mice with the Cebpa deletion would be particularly susceptible to developing lung fibrosis. When bleomycin was administered 4 months after tamoxifen administration (Supplemental Figure 2D), CebpaΔSftpc mice lost more weight than Cebpafl/fl mice (Supplemental Figure 2E), and half of CebpaΔSftpc mice died before the 21-day endpoint (Supplemental Figure 2F). In addition, both the H&E staining (Supplemental Figure 2G) and hydroxyproline assay (Supplemental Figure 2H) revealed that CebpaΔSftpc mice exhibited greater architectural distortion and higher total collagen content than Cebpafl/fl mice.
Then, to determine if Cebpa plays a role in resolving lung fibrosis, we reversed the order of treatments, deleting Cebpa (via tamoxifen administration) 3 weeks after administering a bleomycin injury to initiate lung fibrosis (Figure 2M). At 4 and 5 weeks after bleomycin injury (1 and 2 weeks after tamoxifen treatment), the hydroxyproline assay did not show an obvious difference in collagen deposition between CebpaΔSftpc and Cebpafl/fl mice (Figure 2N). However, 12 weeks after bleomycin injury, we observed increased collagen deposition and more fibrotic patches (Figure 2, O and P), as well as increased mortality (Figure 2Q) in CebpaΔSftpc mice, but without significant weight differences (Figure 2R). This indicates that conditional deletion of Cebpa after bleomycin injury compromised the ability to resolve the fibrosis initiated by bleomycin injury and restore homeostasis to the same extent as Cebpafl/fl mice. Taken together, these data demonstrate that Cebpa loss in AT2 cells promotes lung fibrogenesis and impairs fibrosis resolution in mouse models.
Single-cell transcriptome analysis reveals that Cebpa loss disrupts lung tissue homeostasis.
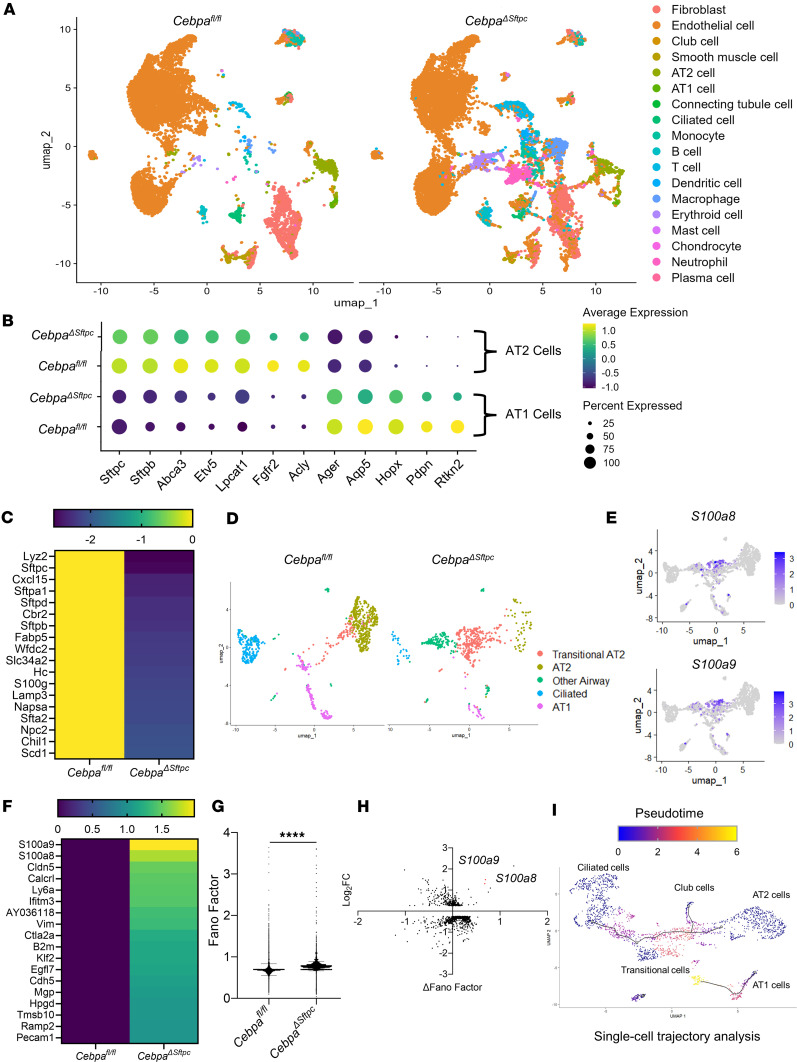
To better understand how loss of Cebpa in the AT2 cells contributes to fibrosis progression, single-cell suspensions of whole lungs were generated for scRNA-Seq from CebpaΔSftpc (n = 3) and Cebpafl/fl (n = 3) mice, which are 6 months after the first tamoxifen treatment. After filtering, normalization, and quality control, a total of 25,094 cells (11,721 Cebpafl/fl cells and 13,373 CebpaΔSftpc cells) were used for downstream analysis. Unsupervised clustering analysis revealed 18 distinct clusters in both CebpaΔSftpc and Cebpafl/fl mice, including all major known epithelial, mesenchymal, and leukocyte lineages (Figure 3A and Supplemental Figure 3A). However, significantly more immune cells were present in CebpaΔSftpc mice, including neutrophils, erythroid cells, macrophages, B cells, T cells, and DCs. In contrast, there were fewer epithelial and endothelia cells in CebpaΔSftpc mice (Figure 3A and Supplemental Figure 3B), suggesting the presence of inflammation and impaired tissue homeostasis in the CebpaΔSftpc mice.
Figure 3. Single-cell transcriptome analysis reveals disrupted epithelial homeostasis.
(A) UMAP plot of lung samples from CebpaΔSftpc (n = 3) and Cebpafl/fl (n = 3) mice showing major cellular clusters, each representing a different cell type. (B) Dot plot for AT1 and AT2 cells obtained from CebpaΔSftpc versus Cebpafl/fl mice showing (a) percentage of cells expressing a marker gene, proportional to dot size, and (b) average expression level of that gene based on unique molecular identifier (UMI) counts, accounting for differences in total UMI counts and cell-type frequencies. (C) Heatmap showing top downregulated genes in AT2 cells from CebpaΔSftpc versus Cebpafl/fl mice. (D) UMAP plot of lung epithelial cells from CebpaΔSftpc versus Cebpafl/fl mice. (E) UMAP plot of S100a8/a9 expression in the lung epithelial cells. (F) Heatmap showing top upregulated genes in AT2 cells from CebpaΔSftpc versus Cebpafl/fl mice. (G) Graph of transcriptional noise (represented by the Fano factor) in epithelial cells from CebpaΔSftpc versus Cebpafl/fl mice. (H) Transcriptional noise plotted against log2 fold change for genes differentially expressed between CebpaΔSftpc and Cebpafl/fl mouse epithelial cells. (I) UMAP plot of pseudotime single-cell trajectory analysis in epithelial cells from CebpaΔSftpc versus Cebpafl/fl mice. Data were analyzed using an unpaired, 2-tailed Student’s t test. ****P < 0.0001.
Markers and cell numbers for both AT2 and AT1 cells were significantly lower in CebpaΔSftpc versus Cebpafl/fl mice (Figure 3B). Pathway analysis (Supplemental Figure 3C) and heatmap analysis (Figure 3C) indicated that the most downregulated genes in AT2 cells in CebpaΔSftpc versus Cebpafl/fl mice were linked to lipid metabolism, surfactant homeostasis, and inflammation. Unsupervised clustering analysis of all the epithelial cells from the lungs of CebpaΔSftpc mice revealed fewer normal epithelial cells — including AT2 cells, AT1 cells, ciliated cells, and other airway cells — than in Cebpafl/fl mice (Figure 3D). In CebpaΔSftpc mice, a unique population of transitional AT2 cells emerged, characterized by high expression of S100a8/a9 but lower expression of Sftpc compared with Cebpafl/fl mice (Figure 3E and Supplemental Figure 3D). In addition, S100a8/9 were the genes with the greatest overexpression in AT2 cells from the lungs of CebpaΔSftpc versus Cebpafl/fl mice (Figure 3F).
Next, we quantified transcriptional noise in single cells following previous work (31, 32) as an indicator of the variability in gene expression. Transcriptional noise is often associated with loss of transcriptional control with aging, leading to a loss of cell identity. Here we observed an increase in transcriptional noise in epithelial cells from the lungs of CebpaΔSftpc versus Cebpafl/fl mice (Figure 3G). More specifically, both noise and expression levels for the S100a8/a9 increased in samples from CebpaΔSftpc mice (Figure 3H). Pseudotime trajectory analysis revealed that AT2 cells tended to become less distinct and more transitional following Cebpa KO (Figure 3I), suggesting that Cebpa deficiency causes a loss of epithelial identity, consistent with other studies (11, 13, 15).
Cebpa is essential to maintain AT2 cells.
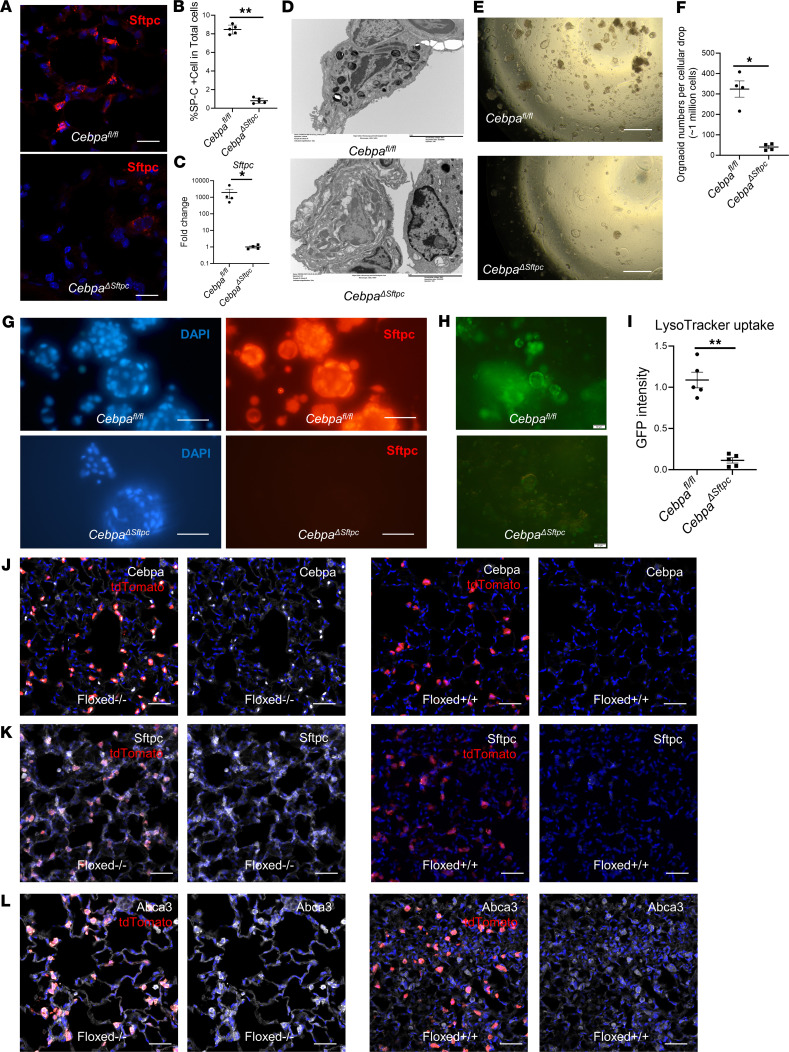
We have shown that, in mouse models, Cebpa loss in AT2 cells promotes lung fibrosis and impairs its resolution, while also reducing AT2 cell–associated transcripts. Thus, we hypothesized that Cebpa is critical for maintaining AT2 cell identity. To test this hypothesis, we performed immunostaining and gene expression analyses 6 months after Cebpa deletion in CebpaΔSftpc mice, a time point associated with significant fibrosis. Sftpc expression and the number of Sftpc+ cells were significantly reduced in CebpaΔSftpc versus Cebpafl/fl mice (Figure 4, A and B), indicating a loss of AT2 cells in CebpaΔSftpc mice. qPCR (Figure 4C) also showed significantly decreased transcripts of Sftpc from CebpaΔSftpc versus Cebpafl/fl mice. Transmission electron microscopy confirmed that deleting Cebpa in AT2 cells led to loss of lamellar bodies, a defining feature of AT2 cells (Figure 4D).
Figure 4. Loss of Cebpa induces loss of AT2 cell identity.
(A) Representative images of immunostaining of Sftpc, a surfactant and marker of AT2 cells, in lung samples taken from CebpaΔSftpc (n = 5) and Cebpafl/fl (n = 5) mice. Scale bar: 20 μM. (B) Quantification of Sftpc+ cells as a percentage of total cells from CebpaΔSftpc (n = 5) and Cebpafl/fl (n = 5) mice. (C) qPCR for Sftpc transcripts from CebpaΔSftpc (n = 4) and Cebpafl/fl mice (n = 4). (D) Representative transmission electron microscopy images showing lamellar bodies, a defining feature of AT2 cells, in lung samples from CebpaΔSftpc (n = 4) and Cebpafl/fl (n = 4) mice. Scale bar: 5 μM. (E) Representative bright-field images of lung organoid formation from samples taken from CebpaΔSftpc (n = 4) and Cebpafl/fl (n = 4) mice. Scale bar: 200 μM. (F) Quantification of organoid formation from CebpaΔSftpc and Cebpafl/fl mice in E. (G) Representative immunostaining images showing Sftpc expression in the lung organoids from CebpaΔSftpc and Cebpafl/fl mice. Scale bar: 50 μM. (H) Representative immunofluorescence images showing LysoTracker uptake in lung organoids from CebpaΔSftpc and Cebpafl/fl mice. Organoids were stained with LysoTracker, which selectively accumulates in lamellar bodies in AT2 cells. Scale bar: 50 μM. (I) Quantification of LysoTracker uptake (i.e., GFP intensity) from H. (J–L) Representative immunostaining images of Cebpa, Sftpc, and Abca3 in lung samples from tdTomato Sftpc Cre lineage-tracing mice with Cebpafi+/fl+ (n = 3) and Cebpafl–/fl– (n = 3) mice. Each protein of interest fluoresces white, while lineage-tracing tdTomato cells fluoresce red. Scale bar: 50 μm. Data were analyzed using a Mann–Whitney U test. *P < 0.05, **P < 0.01.
To determine whether Cebpa plays a role in AT2 cell stemness, we created organoids from lung cells gathered from Cebpafl/fl and CebpaΔSftpc mice 6 months after Cebpa deletion, according to established protocols (33, 34). Organoid formation requires stemness potential of the AT2 cells, as does lung regeneration. Cebpa deletion in AT2 cells significantly reduced organoid formation (Figure 4, E and F), indicating that stemness potential in the lung requires Cebpa. Immunostaining confirmed abundant expression of Sftpc in Cebpafl/fl organoids compared with CebpaΔSftpc organoids (Figure 4G). Next, we investigated LysoTracker uptake to determine how many mature AT2 cells were present. We observed significantly reduced uptake of LysoTracker, which stains acidic compartments in live cells and selectively accumulates in lamellar bodies (35), in CebpaΔSftpc versus Cebpafl/fl organoids (Figure 4, H and I), indicting that fewer mature AT2 cells are present in the CebpaΔSftpc organoids.
To further evaluate the role of Cebpa in determining the identity of AT2 cells, we generated lineage-tracing mice (tdTomato+/+ Sftpc-Cre+/+) to track the fate of AT2 cells in the presence (Cebpa-floxed–/– [Cebpafl-/fl-]) and absence of Cebpa (Cebpa-floxed+/+ [Cebpafl+/fl+]). We hypothesized that mice with a Cebpa deletion (6 months after first tamoxifen) would disrupt the intact AT2 identity of those lineage-tracing tdTomato+ cells in their lungs. Immunostaining showed significantly reduced expression of Cebpa, Sftpc, and another AT2 cell marker, ATP Binding Cassette Subfamily A Member 3 (Abca3), in tdTomato+ cells from Cebpa-deleted mice (Figure 4, J–L). Taken together, our results show that (a) Cebpa is essential to maintain AT2 cell identity and function and (b) its deletion leads to lung fibrosis. These findings align with those from previous studies (12, 36) showing that AT2 cell dysfunction contributes to lung fibrosis initiation and progression.
S100A8/A9 inhibitor paquinimod alleviates lung fibrosis.
Chronic inflammation prevents AT2-AT1 cell differentiation, leading to the accumulation of aberrant AT2 cells and impaired alveolar regeneration (15). Thus, we hypothesized that creating a fibrotic environment for lung organoids would decrease markers of healthy, mature AT2 cells (such as Cebpa). When we treated lung organoids from Sftpc-GFP mice with recombinant proteins S100A8/A9, Cebpa and Sftpc expression were reduced (Supplemental Figure 4), similar to effects observed under treatment with TGF-β1 (Supplemental Figure 4).
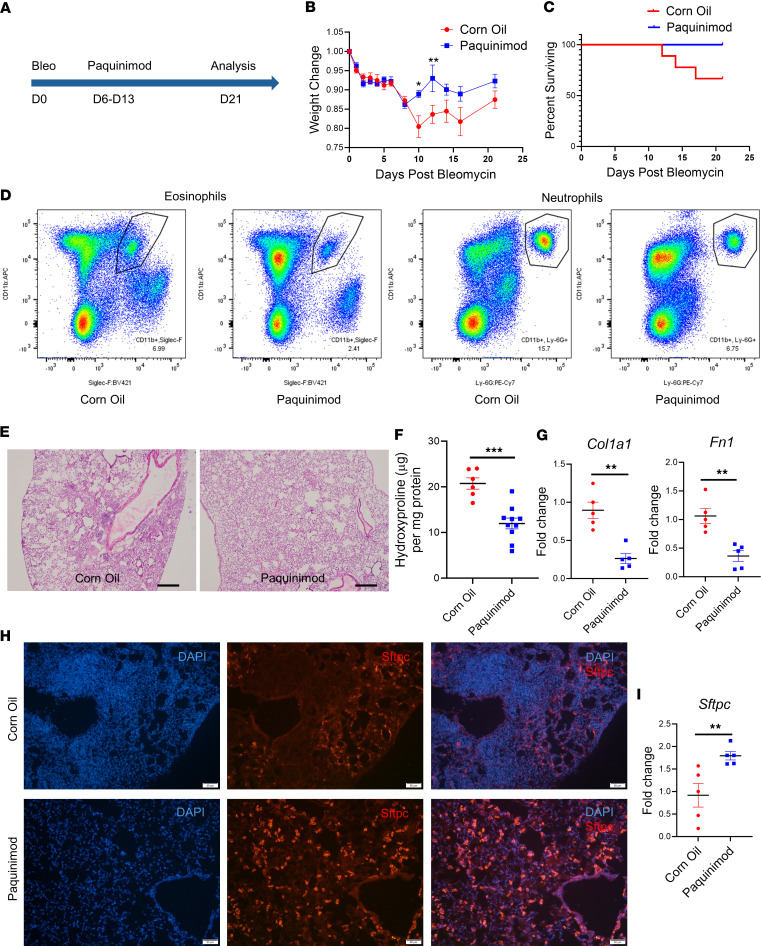
Persistent inflammation can also trigger fibrosis (37). In the previous experiments, we showed that (a) Cebpa loss induces persistent inflammation with prominent S100A8/A9 expression and (b) the S100A8/A9 proteins themselves can contribute to Cebpa repression. Therefore, we hypothesized that S100A8/A9 are mediators of fibrosis. To test our hypothesis, we employed paquinimod, an orally active quinoline-3-carboxamide class immunomodulator that targets S100A8/A9 via direct binding, blocking their interaction with their receptors (38). Paquinimod has been found to be effective in other nonlung fibrosis conditions (39–41). We treated 68-week-old aged mice i.p. with paquinimod (10 mg/kg) starting 6 days after bleomycin injection (daily for 7 days) and then harvested the mice 21 days after bleomycin injury (Figure 5A). This scheme allowed us to determine the effect of blocking S100A8/A9 in a model of persistent lung fibrosis. After 4 days, weight significantly improved in paquinimod-treated versus control mice (Figure 5B). Survival was also higher in paquinimod-treated mice (Figure 5C). Flow cytometry analysis showed that paquinimod treatment reduced neutrophil (CD45+CD11b+Ly6G+) and eosinophil (CD45+CD11b+Siglec-F+) accumulation in the lungs (Figure 5D). Histologically, paquinimod-treated mice exhibited less lung fibrosis and total collagen content than control mice, as assessed by H&E staining (Figure 5E) and the hydroxyproline assay (Figure 5F). Finally, paquinimod treatment ameliorated the increase in profibrotic gene expression (Figure 5G) and the decrease in AT2 cells (shown by Sftpc expression) observed in control mice (Figure 5, H and I). These results indicate that blocking the inflammatory signals from S100A8/A9 can alleviate lung fibrosis in a mouse model.
Figure 5. S100A8/9 inhibitor paquinimod alleviates experimental lung fibrosis.
(A) Schematic showing timeline of paquinimod treatment, which inhibits the inflammatory S100A8/9 proteins, in the CebpaΔSftpc mouse model of fibrosis. (B) Weight of mice treated with paquinimod (n = 10) or corn oil (n = 9), as depicted in A. (C) Survival curve for mice from B. (D) Flow cytometry analysis performed 21 days after bleomycin injury, showing eosinophils and neutrophils from the lungs of mice treated with paquinimod (n = 3) or corn oil (n = 3). (E) Representative H&E staining images showing lung sections taken 21 days after bleomycin injury from mice treated with paquinimod (n = 3) or corn oil (n = 3). (F) Hydroxyproline assay performed 21 days after bleomycin injury in lung samples from mice in E. (G) qPCR performed 21 days after bleomycin injury for profibrotic gene transcripts in mice treated with paquinimod (n = 5) or corn oil (n = 5). (H) Representative immunostaining images of Sftpc expression from the lungs of mice harvested 21 days after bleomycin injury and after treatment with paquinimod (n = 3) or corn oil (n = 3). (I) qPCR for Sftpc expression performed 21 days after bleomycin injury in mice from H. Data were analyzed using a Mann–Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001.
Restoring Cebpa expression alleviates lung fibrosis.
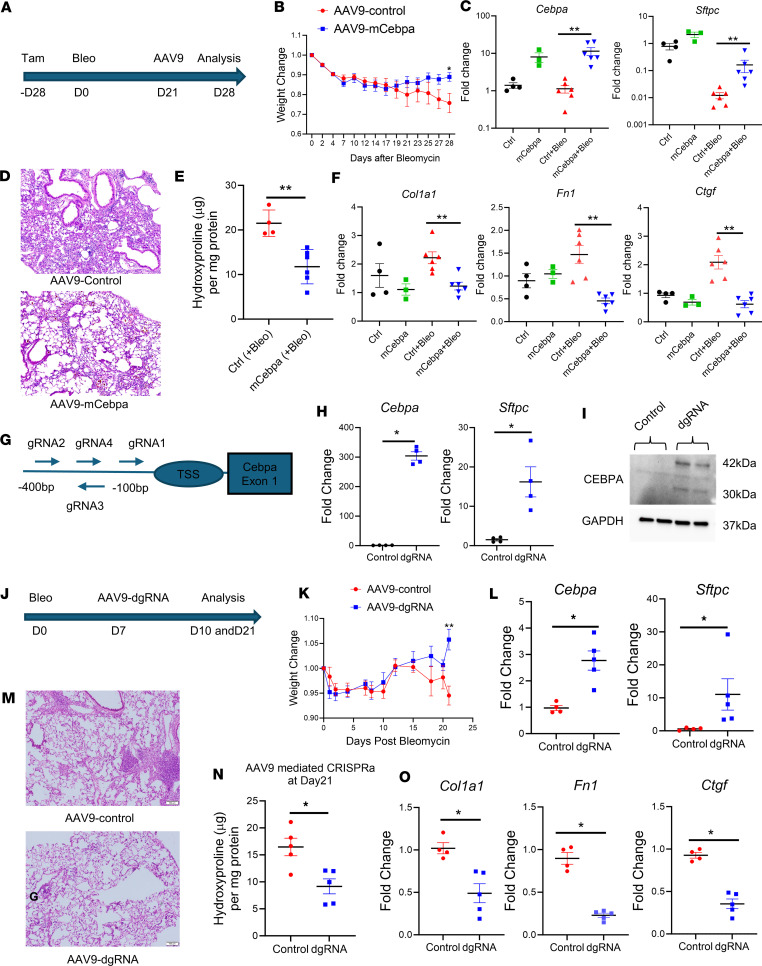
Because Cebpa KO induces long-lasting lung fibrosis, we aimed to determine if restoring Cebpa could alleviate lung fibrosis. First, we administered tamoxifen to delete Cebpa in CebpaΔSftpc mice, and 4 weeks later, we administered a single dose of bleomycin intratracheally to induce experimental lung fibrosis. Twenty-one days after bleomycin injury, mice received a single intratracheal dose of 5 × 1010 genome copies of AAV9-mCebpa or AAV9-control. We then performed analyses 28 days after bleomycin injury (Figure 6A). After day 21 after bleomycin injury, mice treated with AAV9-mCebpa (those with restored Cebpa expression) exhibited less weight loss than control mice (Figure 6B). At 28 days after bleomycin injury, Sftpc and Cebpa expression from sorted lung epithelial cells was restored by AAV9-mCebpa treatment, both with and without bleomycin treatment (Figure 6C). Architectural distortion (Figure 6D) and total collagen in the lungs of mice treated with AAV9-mCebpa was also significantly lower (Figure 6E). Bleomycin increased profibrotic gene transcripts in whole lung tissue from control mice, but this increase was attenuated by treatment with AAV9-mCebpa (Figure 6F).
Figure 6. Rescue of Cebpa expression alleviates lung fibrosis.
(A) Schematic timeline of AAV9 treatment to alleviate lung fibrosis. (B) Weight of CebpaΔSftpc mice treated with AAV9-mCebpa (n = 6) or AAV9-control (n = 5). (C) qPCR for Cebpa and Sftpc transcripts performed 28 days after bleomycin injury on sorted epithelial cells from CebpaΔSftpc mice lungs treated with AAV9-mCebpa or AAV9-control. (D) Representative H&E staining showing 28 days after bleomycin injury from CebpaΔSftpc mice lung treated with AAV9-dgRNA (n = 3) or AAV9-control (n = 3). (E) Hydroxyproline assay performed 28 days after bleomycin injury from CebpaΔSftpc mice lungs treated with AAV9-mCebpa (n = 6) or AAV9-control (n = 5). (F) qPCR for profibrotic gene transcripts performed 28 days after bleomycin injury from CebpaΔSftpc mice lungs treated with AAV9-mCebpa or AAV9-control. (G) Schematic showing how endogenous Cebpa expression was enhanced using an inactivated CRISPR-Cas9 system. (H) qPCR for Cebpa and Sftpc transcripts in cultured epithelial cells from Cas9 mice and then transfected with the dgRNA1 plasmid (n = 4). (I) Representative Western blot showing Cebpa activation in cultured epithelial cells from Cas9 mice treated with the dgRNA1 plasmid (n = 3). (J) Schematic timeline showing endogenous Cebpa reactivation in the 52- to 60-week-old Cas9 mice with bleomycin induced lung fibrosis. (K) Weight change of Cas9 mice after bleomycin injury and after treatment with AAV9-dgRNA (n = 5) or AAV9-control (n = 5). (L) qPCR of Cebpa and Sftpc transcripts performed 10 days after bleomycin injury on sorted epithelial cells from Cas9 mice treated with AAV9-dgRNA or AAV9-control. (M and N) Representative H&E staining (n = 3) and hydroxyproline assay showing 21 days after bleomycin injury from Cas9 mice lungs treated with AAV9-dgRNA or AAV9-control. (O) qPCR for profibrotic gene transcripts performed 21 days after bleomycin injury from Cas9 mice lungs treated with AAV9-dgRNA (n = 5) or AAV9-control (n = 4). Data were analyzed using a Mann-Whitney U test. *P < 0.05, **P < 0.01.
To explore the relevance of targeting Cebpa to treat human lung fibrosis, we generated organoids from human lung tissue (Supplemental Figure 5A) and treated them with TGF-β1 (10 ng/mL) to induce a fibrotic phenotype with or without Cebpa overexpression (Supplemental Figure 5B). Overexpression of Cebpa significantly upregulated Sftpc expression in the lung organoids derived from sorted mouse lung epithelial cells (Supplemental Figure 5C). For human lung organoids, representative bright-field images showed mixed cellular populations, including epithelial spheres and fibroblasts present in the lung organoids (Supplemental Figure 5D). The goal of using mixed cell culture is to recapitulate the effects of Cebpa gene delivery in vivo, since AAV might infect both epithelial cells and fibroblasts in the lung. Delivery of Cebpa to these human lung organoids via AAV9-mCebpa treatment increased SFTPC expression and decreased profibrotic gene expression (Supplemental Figure 5E), which help us understand the overall effects of this approach, specifically the restoration of AT2 gene expression and antifibrotic effects simultaneously.
We also tested the ability to enhance endogenous Cebpa expression using an inactivated CRISPR-Cas9 system. In this system, sgRNAs target sequences within −400 and +100 bp of the transcriptional start site (TSS, in particular between −100 and +50 bp) to enhance transcriptional activation (42–44). Following the protocol published by Liao et al. (45), we created an AAV-dgRNA_Cebpa construct and identified 4 target sequences near the Cebpa TSS (Figure 6G), which were inserted into plasmids. To test the effectiveness of these 4 gRNAs, we cultured sorted lung CD326+ cells from Cas9 mice and transfected them with the 4 individual AAV-dgRNA_Cebpa plasmids in vitro and compared their Cebpa expression (Supplemental Figure 6). The gRNA1 plasmid significantly increased the expression of Cebpa (Figure 6, H and I) as well as Sftpc expression (Figure 6H). To study the effect of endogenous Cebpa reactivation in a mouse model of fibrosis, a single dose of 5 × 1011 genomic copies of AAV9-dgRNA_Cebpa or AAV9-control was injected intratracheally into the lungs of Cas9 mice 7 days after administering bleomycin (Figure 6J). Mice that received AAV9-dgRNA_Cebpa lost less weight than control mice at day 21 (Figure 6K). At day 10 after bleomycin injury, AAV9-dgRNA_Cebpa treatment significantly upregulated both Cebpa and Sftpc expression in sorted epithelial cells (Figure 6L). At day 21 after bleomycin injury, H&E staining (Figure 6M) and the hydroxyproline assay (Figure 6N) further demonstrated that treatment with AAV9-dgRNA_Cebpa versus the control vector reduced total collagen content. AAV9-dgRNA_Cebpa treatment also reduced profibrotic gene expression in whole-lung tissue samples (Figure 6O). These results indicate that restoring endogenous Cebpa expression in the lung is able to accelerate fibrosis resolution and alleviates fibrotic response in a mouse model as well as a human lung organoid model of fibrosis.
Discussion
In this study, we set out to identify a candidate gene involved in fibrosis, confirm its contribution to the disease process, and investigate the molecular mechanisms responsible. By comparing unbiased gene expression of epithelial cell populations from human IPF versus control lung samples, we identified loss of CEBPA as a candidate regulator of epithelial dysfunction in AT2 cells from IPF samples. Next, we used mouse models of lung fibrosis to establish Cebpa’s role in epithelial identity, tissue homeostasis, and repair in the lung. Deleting Cebpa in AT2 cells was sufficient to initiate mild, spontaneous fibrosis in mice, and deletion significantly amplified and prolonged the fibrotic response to bleomycin injury. Cebpa KO in mouse AT2 cells also resulted in the loss of surfactant proteins and lamellar bodies and increased inflammation levels, demonstrating that Cebpa deficiency causes AT2 cells to lose key markers of their identity. Lastly, restoring Cebpa expression via AAV-mediated gene overexpression or CRISPR activation was sufficient to protect AT2 cells and diminish lung fibrosis in mouse models, as well as human lung organoids.
These findings are consistent with a growing body of literature showing epithelial alterations in scRNA-Seq studies from patients with IPF (10, 14, 27, 46). Recent studies have shown that transitional AT2 cells or indeterminate epithelial cells mediate pulmonary fibrosis, displaying aberrant activation of the TGF-β, ER stress, and TP53 signaling pathways (11). Our findings that Cebpa KO causes AT2 cell surfactant protein loss, lamellar body loss, and increased inflammation demonstrates that AT2 cells can lose their identity due to Cebpa deficiencies. We report that Cebpa is crucial to maintaining AT2 cell identity, which is consistent with its role in epithelial maintenance by preventing epithelial-to-mesenchymal transition (24). CEBPA has been known as a master regulator of adipogenesis, which control lipids generation (18, 19). CEBPA has been also studied mainly in the embryonic lung (21, 22). While many factors contribute to lung development and maturation, their losses do not necessarily lead to lung fibrosis. The uniqueness of lower CEBPA expression in the IPF highlights its association with fibrosis phenotype. Previous work demonstrated that specific deletion of Cebpa in the lung epithelium blocks the production of surfactant lipids and proteins necessary for lung function, which cause lung epithelium immaturity and respiratory failure (22). Mice that survived postnatally from embryonic deletion of Cebpa developed COPD and fibrosis phenotypes characterized by histology and inflammatory indicators (26). Our findings linking Cebpa loss to a fibrotic phenotype in the lungs of adult mice build on these earlier findings, highlighting the pathobiological relevance of Cebpa deficiency in the AT2 cells from human IPF lungs.
Prior studies have shown that bleomycin-induced lung fibrosis spontaneously resolves in young mice and, thus, fails to fully recapitulate IPF phenotypes in humans, including aberrant epithelial remodeling and progressive fibrosis (28, 29). Thus, there remains an unmet need for a preclinical model with features that more closely resemble the human IPF. By contrast, the persistent pulmonary fibrosis model we present in this study has strong clinical relevance, as evidenced by the CEBPA deficiency we documented in AT2 cells from patients with IPF. Fibrosis is not maintained by bleomycin-induced lung injury but is induced and sustained by Cebpa KO in AT2 cells, in agreement with the role for AT2 cell dysfunction in mediating lung fibrosis (12, 36).
It is well known that persistent inflammation increases with age, contributing to organ dysfunction and fibrosis (47). However, the molecular mechanisms linking age, inflammation, and fibrosis remain mysterious. Aging is often associated with cellular “fate drifts” and ambiguous cell-type identities (48, 49). Aging can also result in dysregulated transcriptional programs that lead to transcriptional noise, which may contribute to inflammation. Transcriptional noise serves as an indicator for the variability in gene expression and is often associated with the loss of transcriptional control, particularly with aging, leading to a loss of cell-type specificity. Aging involves stochastic changes rather than a precisely orchestrated alteration in gene activity. For example, scRNA-Seq studies have shown that lung cells (50) and pancreatic cells (49) become transcriptionally noisy with age. Increased transcriptional noise and deregulated epigenetic control were found in the lungs of aged mice. Individual endocrine cells in older individuals were more likely to express irrelevant hormone genes, mirroring our findings that the loss of Cebpa indicates a loss of transcriptional control in AT2 cells, resulting in increased expression of erratic genes such as S100a8 and S100a9. Therefore, progressive fibrosis could result from transcriptional noise and erratic inflammation disrupting AT2 cells homeostasis. Consistent with this hypothesis, we showed that AT2 cells from mouse lungs harboring Cebpa KO displayed significantly increased transcriptional noise across lung epithelial populations, leading to disrupted tissue homeostasis and aberrant inflammation. These aberrant AT2 cells were characterized by increased expression of S100a8/a9, which are also upregulated during aging (51) and play a crucial role in fibrosis onset and the subsequent formation of a feed-forward fibrotic loop that promotes fibrosis (51–53). Furthermore, we have shown that the S100A8/A9 inhibitor paquinimod effectively alleviated fibrotic response and restored AT2 cell populations in a bleomycin-induced lung fibrosis aged mice model. We also acknowledged that the likely effects of paquinimod might not only result from regulating epithelial cells but also from blocking the proinflammatory and profibrotic effects of monocytes. This represents a limitation of this experiment. In short, our study presents a mechanism by which Cebpa deficiency in lung AT2 cells links aging, disrupted homeostasis, and fibrosis.
Our experiments also suggest that increasing CEBPA might be protective or therapeutic in IPF. However, CEBPA is a transcription factor that is considered to be an undruggable target. Increasing gene expression by delivery and overexpression from plasmids via AAVs is a potential strategy for targeting this undruggable pathway. We also note that intratracheal instillation of AAVs might infect multiple cell types including macrophages and fibroblasts in the lung. Here, we also used a CRISPR activation system to enhance Cebpa, which involves the use of a modified version of the CRISPR-Cas9 system. In CRISPR activation, a deactivated version of the Cas9 enzyme (called dCas9) is fused with transcriptional activator domains and is guided to a specific DNA sequence by a gRNA complementary to the target gene’s sequence (54). Once the dCas9-gRNA complex binds to the promoter of the target gene, the transcriptional activator domain recruits transcriptional machinery, leading to increased target gene expression. It has the advantages of bypassing the size limitation of AAV gene delivery (45), avoiding the need for genome editing/insertion (55), limiting the risk of aberrant overexpression, and ensuring a physiological range of expression levels (56) depending on basal gene expression levels and the gene’s epigenetic status. Clearly, the safe and efficient delivery of a CRISPR activation system in vivo, as performed here, remains a major challenge to the widespread clinical success of any CRISPR-Cas9 therapeutics (57). Our study provides an initial proof of concept that insufficient gene expression can be restored by CRISPR activation in vivo to alleviate a model of chronic lung disease. Future research may uncover the epigenetic mechanism of CEBPA repression in human pulmonary fibrosis, potentially leading to additional strategies for restoring gene regulation.
In conclusion, our results demonstrate that CEBPA plays a critical role in AT2 cell maintenance and tissue homeostasis in the mouse lung. We speculate that lower levels of CEBPA expression in the lungs of patients with IPF are a central feature of chronic lung injury characterized by AT2 cell loss, persistent inflammation, and disrupted tissue homeostasis. Our work provides mechanistic insights into the regulation of AT2 cell identity by Cebpa and by Cebpa deficiency–induced inflammation, characterized by increased S100a8/a9 expression. Moreover, we have shown that restoring Cebpa expression was sufficient to alleviate fibrosis (Supplemental Figure 7). Future research will be necessary to uncover the epigenetic mechanism responsible for CEBPA repression in human IPF and to design a more precise gene restoration method based on that specific mechanism. In the future, such an approach could be harnessed to rescue expression of CEBPA or other regulators of aberrant epithelial cell types in patients with IPF and other chronic lung diseases, to restore tissue homeostasis and promote lung repair.
Methods
Sex as a biological variable.
Sex was not considered as a biological variable. Both male and female mice were included in this study.
Human specimens.
Human lung samples were used to generate lung organoids. Lung tissues were collected from patients whose “healthy” lungs were deemed unsuitable for transplantation due to size incompatibility or were unsuitable for lung transplantation.
Mouse models.
Sftpc-CreERT2 mice (58), Cebpa “floxed” allele mice (59), and tdTomato Cre mice (60) were purchased from The Jackson Laboratory. All mice analyzed had mixed genetic backgrounds and were age matched unless mentioned specifically. The mice used for experiments were 3 months old unless otherwise indicated. Cebpa conditional KO in the AT2 cells was conducted in the Sftpc-CreER+ Cebpafl/fl mice (referred to as CebpaΔSftpc mice) and their control Cebpafl/fl mice. Additional lineage tracing was carried out in Sftpc-CreER+ tdTomato+ mice, which have tdTomato labeling with (Cebpafl+/fl+) or without (Cebpafl−/fl−) the deletion of Cebpa in the AT2 cells after tamoxifen treatment. Cas9 mice were purchased from The Jackson Laboratory (Rosa26-Cas9 knockin on B6J, stock no. 026179). These CRISPR/Cas9-knockin mice constitutively express CRISPR-associated protein 9 (cas9) endonuclease and EGFP in a widespread fashion under the direction of a CAG promoter, which can be used with designed gRNAs for manipulating gene expression in vivo or ex vivo.
Mouse experimental fibrosis experiments.
Cebpa deletion was induced with 5 injections of 75 mg tamoxifen/kg body weight, daily. A second round of weekly tamoxifen injections was given if analyses were performed more than 3 months after the initial round of tamoxifen injection. To induce experimental fibrosis, bleomycin (0.5 U/kg in 50 μL saline) was administered to the mice intratracheally, as previously described (29). Mice were then randomized to receive 10 mg/kg/day paquinimod (or corn oil) i.p. for 7 days to inhibit the effect of S100a8/a9. Mice were also randomized to receive a single intratracheal dose of AAV9-mCebpa, AAV9-dgRNA_Cebpa, or AAV9-empty control. Lungs were collected at designated time points.
Lung harvest, FACS, and analysis.
Mice were anaesthetized with 90–120 mg/kg ketamine combined with 10 mg/kg xylazine, after which their lungs were perfused with ice-cold PBS. The lungs were then immediately harvested, minced in 10 cm petri dishes, and incubated in digestive solution: DMEM (Thermo Fisher Scientific, 11965092), 0.2 mg/mL Liberase DL (MilliporeSigma, 5401119001), and 100 U/mL DNase I (MilliporeSigma, 4536282001). Tissue was digested at 37°C for 40 minutes. Digestive solution was inactivated with 1× volume DMEM (Thermo Fisher Scientific, 11965092) containing 10% FBS (Thermo Fisher Scientific, A5256701). Cell and tissue suspensions were subjected to a 40 μm filter and centrifuged at 300g for 5 minutes. Cell pellets were then resuspended in RBC lysis buffer (BioLegend, 420301) for 90 seconds and diluted in a 4× volume of PBS. Cells were centrifuged at 300g for 5 minutes and resuspended in 200 μL MojoSort Buffer (BioLegend, 480017). Single-cell suspensions were then incubated with PerCpCy5.5 anti–mouse CD45 (BioLegend, 1:200, 157612), PE anti–mouse CD326 (BioLegend; 1:200, 118206), and DAPI (Thermo Fisher Scientific, 1:1,000, 62248) antibodies for 30 minutes on ice. To isolate and analyze CD326+ cells, cells were sorted by FACS using a BD FACS Aria II (BD Biosciences). The following selection strategy was used: debris exclusion (forward scatter area [FSC-A] by side scatter A [SSC-A]), doublet exclusion (SSC width [SSC-W] by SSC height [SSC-H] and FSC-W by FSC-H), dead cell exclusion (DAPI–), CD45 cell removal. Epithelial cells were sorted as DAPI–CD45–CD326+ cells. For RNA analysis, CD326+ cells were sorted directly into RLT lysis buffer (Qiagen, 74004). To analyze eosinophils and neutrophils, additional antibodies for APC anti-CD11b (BioLegend, 1:200, 101211), BV421 anti-mouse CD170 (Siglec-F, BioLegend, 1:200, 155509), and PE/Cyanine7 anti–mouse Ly-6G (BioLegend, 1:200, 127617) were included in the antibody incubation. Population analysis was performed using FlowJo FACS analysis software (BD Biosciences).
Lung organoid culture and primary cell culture.
Briefly, fresh lung tissue was cut in small pieces (1–2 mm2) and incubated in digestive solution (100 U/mL DNase I plus 0.2 mg/mL Liberase at 35 minutes for mouse lung, 2 mg/mL collagenase I at 90 minutes for human lung). After 70 μm filter and cells were embedded into a Matrigel (Corning, 356231) dome, which was incubated at 37°C for 30 minutes to allow time for polymerization. Once embedded, the explants were fed with AT2 medium following an established protocol (34). The medium was changed every other day. For mouse lung epithelial cells organoids, DAPI–CD45–CD326+ cells will be used for Matrigel culture. For monolayer epithelial cell culture, cells were cultured in Epithelial Cell Medium (Cell Biologics, M6621) with 5% CO2 and 95% air in a humidified incubator. Transient transfection was performed using a Lipofectamine 3000 kit (Thermo Fisher Scientific, L3000015) according to the manufacturer’s instructions.
CRISPR activation in vitro and in vivo.
Following the protocol published by Liao et al. (45), we constructed sgRNAs that identified 4 target sequences near the Cebpa TSS (Figure 6G). These predesigned, 14-bp gRNA spacers (5′–3′; GCGCAGGAGTCAGT; GGGCTCCCTAGTGT; CTGCAAGGCGAACC; ACAGCGCCGCCGGG) were inserted into plasmid AAV-U6-dgRNA-CAG-MPH, which utilizes a modified version of the Synergistic Activation Mediator (SAM) system. This plasmid was originally produced by Juan Belmonte (Addgene plasmid 106259) and expresses the transcription activator MS2:P65:HSF1 (MPH) from the CAG promoter and one U6-driven dgRNA in an AAV backbone. For the in vitro CRISPR activation experiment, AAV-U6-dgRNA-CAG-MPH plasmids carrying dgRNA_Cebpa (AAV-dgRNA_Cebpa) were transiently transfected into sorted CD326+Cas9+ cells using a Lipofectamine 3000 kit (Invitrogen) according to the manufacturer’s instructions. For the in vivo CRISPR activation experiment, AAV9 plasmids expressing AAV-dgRNA_Cebpa or AAV-control were produced by the University of Pennsylvania Vector Core. AAV9 was delivered to the mouse lung intratracheally, following an established protocol for bleomycin delivery (29).
Hydroxyproline assay for measuring collagen.
Collagen content in the lung was measured using a hydroxyproline assay kit (Abcam, ab222941) according to the manufacturer’s instructions, as previously described (61). Frozen lung tissue was homogenized in sterile water at a ratio of 10 mg tissue to 100 μL water and hydrolyzed in 12M HCl in a pressure-tight, Teflon-capped vial at 120°C for 3 hours, followed by filtration through a 45 μm Spin-X Centrifuge Tube filter (Corning, 8162). Next, a 10 μL sample was dried in a Speed-Vac overnight, followed by incubation with 100 μL Chloramine T reagent for 5 minutes at room temperature and then incubation with 100 μL DMAB for 90 minutes at 60°C. The absorbance of oxidized hydroxyproline was measured at 560 nm. Hydroxyproline concentrations were calculated from a standard curve generated using known concentrations of trans-4-hydroxyl-L-proline. The total amount of protein isolated from the weighed tissues was determined by using a protein assay kit (Bio-Rad). Hydroxyproline content data were expressed as μg collagen per mg total lung protein (μg/mg).
RNA extraction and quantitative PCR.
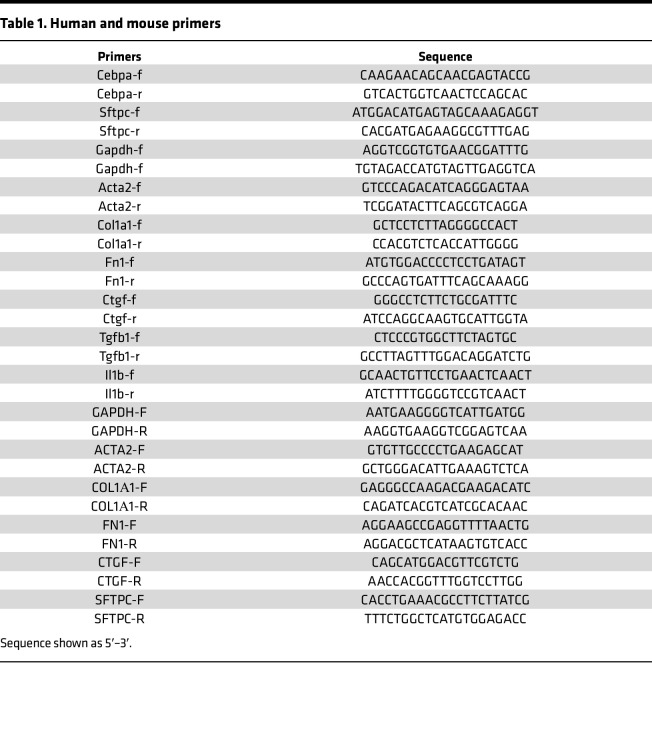
Total RNA was isolated from samples using the RNeasy Plus Mini kit (Qiagen, 74134) or RNeasy Micro Kit (Qiagen, 74004) depending on the RNA quantities. cDNA was synthesized with SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific, 18090050). Quantitative PCR (qPCR) amplification was carried out using the FastStart Essential DNA Green Master (Roche, 06402712001) for SYBR Green I-based real-time PCR on the Lightcycler 96 Real-Time PCR System (Roche), according to the manufacturer’s instructions. qPCR was performed by incubating plates at 95°C for 10 minutes and then cycling 40 times at 95°C for 10 seconds, 60°C for 10 seconds, and 72°C for 10 seconds. Ct values within each experiment were normalized against GAPDH/Gapdh levels, and fold change was calculated for all conditions relative to a single, randomly selected, control result. Both human and mice primers are listed in Table 1.
Table 1. Human and mouse primers.
Tissue preparation, H&E staining, and immunostaining.
Lungs were inflated with 4% formaldehyde solution (Thermo Fisher Scientific, 28908) and continually fixed in 4% formaldehyde at 4°C for 24 hours. Then the lungs were cryoprotected in 30% sucrose and embedded in OCT (Fisher HealthCare, 4585). H&E staining followed the standard H&E protocol. Briefly, slides were washed with water to remove the OCT. The nuclei were stained with hematoxylin (Abcam, ab150678) for 2 minutes, and the cytoplasm was stained with eosin (MilliporeSigma, HT110280) for 3 minutes. Slices were sealed with neutral resin after the dehydration and clearing steps.
Tissue sections (10 μm) from each block were cut in a cryostat at –21°C and mounted onto Fisherbrand Tissue Path Superfros Plus Gold slides (Thermo Fisher Scientific). Slides were permeabilized in 0.25 % Triton X-100 (Sigma-Aldrich, X-100), blocked with 1% BSA for 1 hour, and incubated overnight with CEBPA antibody (dilution 1:100, Cell Signaling Technology, 8178S), SFTPC antibody (dilution 1:100, Santa Cruz Animal Health, PA5-71680), or ABCA3 antibody (dilution 1:100, Thermo Fisher Scientific, PA5-103632) at 4°C overnight in PBS with 1% BSA, followed by incubation with a mixture of fluorescence-conjugated secondary antibody (dilution 1:500, Thermo Fisher Scientific) and DAPI (dilution 1:1,000, Thermo Fisher Scientific) at room temperature for 1 hour. All images were captured using a Zeiss LSM 780 confocal microscope or an Olympus CKX53 microscope.
Western blotting.
Cells were harvested into RIPA Lysis Buffer (Thermo Fisher Scientific, 89900) with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, 78440). Lysates were then quantitated using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225), and equal amounts of protein were subjected to 4%–15% Mini-PROTEAN TGX Gel (Bio-Rad, 15 μL [4568086] or 50 μL [4561084]). Proteins were transferred from the gel to PVDF membranes with Trans-Blot Turbo Transfer System (Bio-Rad). After blocking for 1 hour at room temperature with 5% nonfat dry milk (Bio-Rad, Bloting-Grade Blocker, 1706404) in TBST blocking buffer, PVDF membranes were probed with GAPDH (Cell Signaling Technology, 2218S), Cebpa (Cell Signaling Technology, 8178S), α-SMA (Sigma-Aldrich, F3777), and Fibronectin (Santa Cruz Biotechnology Inc., sc-81767) antibodies at 4°C overnight followed by incubation with HRP-conjugated goat anti-rabbit antibody (Promega, W4011) for 1 hour at room temperature. Bands were detected using Super Signal West Pico Plus (Thermo Fisher Scientific, 34580) and visualized using a Bio-Rad ChemiDoc Imaging system (Bio-Rad). Quantification was performed via densitometry. Expression of a specific antibody relative to GAPDH was computed using the ImageLab software provided by Bio-Rad.
Transmission electron microscopy.
Lung tissue was placed into fixative containing 4% paraformaldehyde and 1% glutaraldehyde in 0.1M phosphate buffered solution, pH 7.2 (PBS). Then the fixed tissue was washed with PBS, stained with 1% osmium tetroxide, washed in H2O, stained with 2% uranyl acetate, washed in H2O, dehydrated through a graded series of ethanol and acetone, and embedded in Embed 812 resin. Following a 24-hour polymerization at 60°C, 0.1 μM ultrathin sections were prepared and poststained with lead citrate. Micrographs were acquired using a JEOL 1400 Plus transmission electron microscope (JEOL Inc.) at 80 kV equipped with a Gatan Orius camera (Gatan Inc.).
scRNA-Seq and analysis.
In our mouse fibrosis experiments involving Cebpa-KO mice, 26 weeks after the first tamoxifen injections, the lungs of CebpaΔSftpc (n = 3) and Cebpafl/fl (n = 3) mice were harvested and disassociated into single-cell suspensions as described earlier. Cell suspensions were brought to the Genome Analysis Core at Mayo Clinic for processing. Cell viability and counts were determined using a Countess II Automated Cell Counter. Single cells, reagents, and a single Gel Bead containing barcoded oligonucleotides were encapsulated into a nanoliter-sized Gel Bead-in-Emulsion using the 10X Genomics GemCode platform (10X Genomics). cDNA libraries were prepared with a Chromium Single Cell 3′ GEM Library & Gel Bead Kit (10X Genomics). All cDNA libraries were sequenced on an Illumina HiSeq 4000 instrument.
scRNA-Seq data were aligned and quantified using the 10X Genomics Cell Ranger Software Suite (v6.1.1) against the mouse reference genome mm10 from raw count data. Data were processed and analyzed with the Seurat R package V4.0 (62) to perform integrated analyses of single cells. Genes expressed in < 3 cells and cells that expressed < 200 genes and > 20% mitochondria genes were excluded from downstream analysis in each sample. One Seurat object was created from filtered count matrices from pooled CebpaF/F and CebpaΔSftpc cells. Each data set was SCTransform-normalized and the top 3,000 highly variable genes (HVGs) across cells were selected.
Principal component analysis (PCA) and UMAP were performed on the integrated object to identify cell clusters. UMAP was performed using the first 30 PCA dimensions. Cell populations in whole lung tissue were annotated using lung cells from the Mouse Cell Atlas 3.0 as a reference (63) using Seurat. Epithelial cells were subset and reanalyzed with a clustering resolution of 0.1, and cell-type annotations were assigned manually to each cluster based on expression of canonical marker genes.
Differential expression testing was performed using MAST (64). Pseuodotime trajectories were constructed using the Monocle 3 R package (65). Transcriptional noise was measured by calculating the Fano factor for each gene using SCTransform-normalized count values across all epithelial cells. Pathway analysis was performed using DAVID Gene Ontology analysis (https://david.ncifcrf.gov/home.jsp).
For publicly available scRNA-Seq analysis, downregulate gene lists are extracted from original papers and are included in Supplemental Table 1. Venn diagrams were prepared using the webtool Venny (https://bioinfogp.cnb.csic.es/tools/venny/index.html). Violin plots of CEBPA expression were constructed using the Monocle 3 R package. The upstream regulator analysis was performed using Ingenuity Upstream Regulator Analysis in IPA (QIAGEN) and are available in Supplemental Table 2. UMAP was generated directly from (http://www.ipfcellatlas.com/).
Statistics.
Data are presented as means ± SEM. Differences between conditions were examined with GraphPad Prism 9 using an unpaired, 2-tailed Student’s t test with Welch’s correction or a nonparametric Mann–Whitney U test for experiments with n < 10. P <0.05 was considered significant.
Study approval.
All experiments with human tissue samples were performed under protocols approved by IRB at Mayo Clinic. All mouse experiments were conducted under protocols approved by the IACUCs of Mayo Clinic and the University of Minnesota.
Data availability.
RNA-Seq raw data have been deposited to Gene Expression Omnibus (GEO) database with the accession no. GSE264629. Values for graphs in the figures and supplemental figures are provided in the Supporting Data Values file.
Author contributions
QT contributed funding acquisition, conceptualization, data acquisition and analysis, methodology, and manuscript writing and editing. JHW contributed data acquisition, methodology and data analysis, and manuscript editing. SS contributed methodology, data acquisition, and analysis. SRD contributed data acquisition and analysis as well as manuscript editing. JAM contributed methodology, data acquisition, and analysis. KMC contributed methodology, data acquisition, and analysis. YL contributed methodology and data acquisition. YSP contributed funding acquisition and intellectual input. DJT contributed funding acquisition, conceptualization, intellectual input, and manuscript writing and editing.
Supplementary Material
Acknowledgments
We thank Tao Mao for his bioinformatic support, Sean McBride for his mouse breeding service, the Mayo Microscopy and Cell Analysis Core for experimental and technical support, and Kristin Harper, of Harper Health & Science Communications LLC, for providing editorial support in accordance with Good Publication Practice guidelines.
This work was supported by funding for JHW from the NIH T32 grant HL105355, NIH R01 grant HL092961 awarded to DJT, and NIH R01 grant HL153026 awarded to QT.
Version 1. 07/16/2024
In-Press Preview
Version 2. 08/22/2024
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2024, Tan et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2024;9(16):e175290.https://doi.org/10.1172/jci.insight.175290.
Contributor Information
Qi Tan, Email: qtan@umn.edu.
Jack H. Wellmerling, Email: Wellmerling.Jack@mayo.edu.
Shengren Song, Email: song0683@umn.edu.
Sara R. Dresler, Email: Dresler.sara@mayo.edu.
Jeffrey A. Meridew, Email: Meridew.Jeffrey@mayo.edu.
Kyoung M. Choi, Email: Choi.Kyoung@mayo.edu.
Yong Li, Email: Li.Yong2@mayo.edu.
Y.S. Prakash, Email: prakash.ys@mayo.edu.
Daniel J. Tschumperlin, Email: Tschumperlin.Daniel@mayo.edu.
References
- 1.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18(7):1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123(7):3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang J, et al. Hyaluronan and TLR4 promote surfactant-protein-C-positive alveolar progenitor cell renewal and prevent severe pulmonary fibrosis in mice. Nat Med. 2016;22(11):1285–1293. doi: 10.1038/nm.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selman M, Pardo A. Role of epithelial cells in idiopathic pulmonary fibrosis: from innocent targets to serial killers. Proc Am Thorac Soc. 2006;3(4):364–372. doi: 10.1513/pats.200601-003TK. [DOI] [PubMed] [Google Scholar]
- 5.Phan SH. Noncanonical Wnt signaling promotes myofibroblast differentiation in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2002;65(5):489–499. doi: 10.1165/rcmb.2020-0499OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman HA. Epithelial-mesenchymal interactions in pulmonary fibrosis. Annu Rev Physiol. 2011;73:413–435. doi: 10.1146/annurev-physiol-012110-142225. [DOI] [PubMed] [Google Scholar]
- 7.Horowitz JC, Thannickal VJ. Epithelial-mesenchymal interactions in pulmonary fibrosis. Semin Respir Crit Care Med. 2006;27(6):600–612. doi: 10.1055/s-2006-957332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hewlett JC, et al. Idiopathic pulmonary fibrosis: epithelial-mesenchymal interactions and emerging therapeutic targets. Matrix Biol. 2018;71–72:112–127. doi: 10.1016/j.matbio.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan Q, et al. Nascent lung organoids reveal epithelium- and bone morphogenetic protein-mediated suppression of fibroblast activation. Am J Respir Cell Mol Biol. 2019;61(5):607–619. doi: 10.1165/rcmb.2018-0390OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1(20):e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi Y, et al. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat Cell Biol. 2020;22(8):934–946. doi: 10.1038/s41556-020-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katzen J, Beers MF. Contributions of alveolar epithelial cell quality control to pulmonary fibrosis. J Clin Invest. 2020;130(10):5088–5099. doi: 10.1172/JCI139519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strunz M, et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat Commun. 2020;11(1):3559. doi: 10.1038/s41467-020-17358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Habermann AC, et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv. 2020;6(28):eaba1972. doi: 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi J, et al. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell. 2020;27(3):366–382. doi: 10.1016/j.stem.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kathiriya JJ, et al. Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5+ basal cells. Nat Cell Biol. 2022;24(1):10–23. doi: 10.1038/s41556-021-00809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, et al. C/EBPα arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell. 2001;8(4):817–828. doi: 10.1016/S1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 18.Rosen ED, et al. C/EBPα induces adipogenesis through PPARγ: a unified pathway. Genes Dev. 2002;16(1):22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefterova MI, et al. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22(21):2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mason RJ, et al. Keratinocyte growth factor and the transcription factors C/EBPα, C/EBPδ, and SREBP-1c regulate fatty acid synthesis in alveolar type II cells. J Clin Invest. 2003;112(2):244–255. doi: 10.1172/JCI16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martis PC, et al. C/EBPα is required for lung maturation at birth. Development. 2006;133(6):1155–1164. doi: 10.1242/dev.02273. [DOI] [PubMed] [Google Scholar]
- 22.Basseres DS, et al. Respiratory failure due to differentiation arrest and expansion of alveolar cells following lung-specific loss of the transcription factor C/EBPα in mice. Mol Cell Biol. 2006;26(3):1109–1123. doi: 10.1128/MCB.26.3.1109-1123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato A, et al. CCAAT/enhancer-binding protein-α suppresses lung tumor development in mice through the p38α MAP kinase pathway. PLoS One. 2013;8(2):e57013. doi: 10.1371/journal.pone.0057013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lourenco AR, et al. C/EBPɑ is crucial determinant of epithelial maintenance by preventing epithelial-to-mesenchymal transition. Nat Commun. 2020;11(1):785. doi: 10.1038/s41467-020-14556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao LL, et al. C/EBP-α ameliorates CCl(4)-induced liver fibrosis in mice through promoting apoptosis of hepatic stellate cells with little apoptotic effect on hepatocytes in vitro and in vivo. Apoptosis. 2012;17(5):492–502. doi: 10.1007/s10495-012-0700-y. [DOI] [PubMed] [Google Scholar]
- 26.Didon L, et al. Lung-specific inactivation of CCAAT/enhancer binding protein α causes a pathological pattern characteristic of COPD. Eur Respir J. 2010;35(1):186–197. doi: 10.1183/09031936.00185008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinzelmann K, et al. Single-cell RNA sequencing identifies G-protein coupled receptor 87 as a basal cell marker expressed in distal honeycomb cysts in idiopathic pulmonary fibrosis. Eur Respir J. 2022;59(6):2102373. doi: 10.1183/13993003.02373-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan Q, et al. Spontaneous lung fibrosis resolution reveals novel antifibrotic regulators. Am J Respir Cell Mol Biol. 2021;64(4):453–464. doi: 10.1165/rcmb.2020-0396OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporarello N, et al. PGC1α repression in IPF fibroblasts drives a pathologic metabolic, secretory and fibrogenic state. Thorax. 2019;74(8):749–760. doi: 10.1136/thoraxjnl-2019-213064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pham TX, et al. Transcriptional analysis of lung fibroblasts identifies PIM1 signaling as a driver of aging-associated persistent fibrosis. JCI Insight. 2022;7(6):e153672. doi: 10.1172/jci.insight.153672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blake WJ M KA, et al. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez A, Golding I. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013;342(6163):1188–1193. doi: 10.1126/science.1242975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sachs N, et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38(4):e100300. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katsura H, et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell. 2020;27(6):890–904. doi: 10.1016/j.stem.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Velden JL, et al. LysoTracker is a marker of differentiated alveolar type II cells. Respir Res. 2013;14(1):123. doi: 10.1186/1465-9921-14-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers RC, Mercer PF. Mechanisms of alveolar epithelial injury, repair, and fibrosis. Ann Am Thorac Soc. 2015;12(suppl 1):S16–S20. doi: 10.1513/AnnalsATS.201410-448MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214(2):199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Q, et al. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell Host Microbe. 2021;29(2):222–235. doi: 10.1016/j.chom.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fransen Pettersson N, et al. The immunomodulatory quinoline-3-carboxamide paquinimod reverses established fibrosis in a novel mouse model for liver fibrosis. PLoS One. 2018;13(9):e0203228. doi: 10.1371/journal.pone.0203228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stenstrom M, et al. Paquinimod reduces skin fibrosis in tight skin 1 mice, an experimental model of systemic sclerosis. J Dermatol Sci. 2016;83(1):52–59. doi: 10.1016/j.jdermsci.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 41.Hesselstrand R, et al. An open-label study to evaluate biomarkers and safety in systemic sclerosis patients treated with paquinimod. Arthritis Res Ther. 2021;23(1):204. doi: 10.1186/s13075-021-02573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159(3):647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kearns NA, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141(1):219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao HK, et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell. 2017;171(7):1495–1507. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams TS, et al. Single-cell RNA-Seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv. 2020;6(28):eaba1983. doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanada F, et al. Source of chronic inflammation in aging. Front Cardiovasc Med. 2018;5:12. doi: 10.3389/fcvm.2018.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Jimenez CP, et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science. 2017;355(6332):1433–1436. doi: 10.1126/science.aah4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enge M, et al. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell. 2017;171(2):321–330. doi: 10.1016/j.cell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angelidis I, et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat Commun. 2019;10(1):963. doi: 10.1038/s41467-019-08831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S, et al. S100A8/A9 in inflammation. Front Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leclerc E, et al. Binding of S100 proteins to RAGE: an update. Biochim Biophys Acta. 2009;1793(6):993–1007. doi: 10.1016/j.bbamcr.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 53.Araki K, et al. The heterodimer S100A8/A9 is a potent therapeutic target for idiopathic pulmonary fibrosis. J Mol Med (Berl) 2021;99(1):131–145. doi: 10.1007/s00109-020-02001-x. [DOI] [PubMed] [Google Scholar]
- 54.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12(4):326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9(1):1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoger E, et al. CRISPR-mediated activation of endogenous gene expression in the postnatal heart. Circ Res. 2020;126(1):6–24. doi: 10.1161/CIRCRESAHA.118.314522. [DOI] [PubMed] [Google Scholar]
- 57.Behr M, et al. In vivo delivery of CRISPR-Cas9 therapeutics: progress and challenges. Acta Pharm Sin B. 2021;11(8):2150–2171. doi: 10.1016/j.apsb.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108(52):E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang P, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBPα. Immunity. 2004;21(6):853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 60.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haak AJ, et al. Selective YAP/TAZ inhibition in fibroblasts via dopamine receptor D1 agonism reverses fibrosis. Sci Transl Med. 2019;11(516):eaau6296. doi: 10.1126/scitranslmed.aau6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang R, et al. Construction of a cross-species cell landscape at single-cell level. Nucleic Acids Res. 2023;51(2):501–516. doi: 10.1093/nar/gkac633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finak G, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao J, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq raw data have been deposited to Gene Expression Omnibus (GEO) database with the accession no. GSE264629. Values for graphs in the figures and supplemental figures are provided in the Supporting Data Values file.