Abstract
Protein solubility plays a crucial role in various biotechnological, industrial, and biomedical applications. With the reduction in sequencing and gene synthesis costs, the adoption of high-throughput experimental screening coupled with tailored bioinformatic prediction has witnessed a rapidly growing trend for the development of novel functional enzymes of interest (EOI). High protein solubility rates are essential in this process and accurate prediction of solubility is a challenging task. As deep learning technology continues to evolve, attention-based protein language models (PLMs) can extract intrinsic information from protein sequences to a greater extent. Leveraging these models along with the increasing availability of protein solubility data inferred from structural database like the Protein Data Bank holds great potential to enhance the prediction of protein solubility. In this study, we curated an Updated Escherichia coli protein Solubility DataSet (UESolDS) and employed a combination of multiple PLMs and classification layers to predict protein solubility. The resulting best-performing model, named Protein Language Model-based protein Solubility prediction model (PLM_Sol), demonstrated significant improvements over previous reported models, achieving a notable 6.4% increase in accuracy, 9.0% increase in F1_score, and 11.1% increase in Matthews correlation coefficient score on the independent test set. Moreover, additional evaluation utilizing our in-house synthesized protein resource as test data, encompassing diverse types of enzymes, also showcased the good performance of PLM_Sol. Overall, PLM_Sol exhibited consistent and promising performance across both independent test set and experimental set, thereby making it well suited for facilitating large-scale EOI studies. PLM_Sol is available as a standalone program and as an easy-to-use model at https://zenodo.org/doi/10.5281/zenodo.10675340.
Keywords: protein solubility prediction, protein language models, enzymes of interest
Introduction
Proper folding of proteins to maintain enough solubility and homeostasis is essential for nearly every protein-based biological process. Unsatisfied solubility or aggregation can impede protein-based drug development, such as antibody production. The low solubility of antibodies may limit their shelf-life and potentially induce adverse immune responses [1–3]. Apart from antibodies, more and more enzymes of interest (EOI) are being discovered with an increasing speed due to the decreasing cost of sequencing and gene synthesis as well as continuous improvement of high-throughput functional screening platforms [4–6]. In these large-scale EOI screening studies, enhancing the accuracy of protein solubility prediction can improve the success rate of protein purification and facilitate the downstream biophysical or biochemical characterization. Common hosts such as bacterial cells, insect cells, yeast cells, plant cells, and mammalian cells are often used for recombinant protein expression [7]. Among these options, bacterial cells, typically Escherichia coli, provide the advantages of easy genetic manipulation and cost-effectiveness, therefore serving as one of the major platforms for recombinant protein production [8]. Improving the accuracy of protein solubility prediction in E. coli thus has great potential to reduce experimental cost and increase the success rate of novel EOI discovery.
Protein solubility in E. coli is a complex issue influenced by numerous factors at different levels. Firstly, regarding the sequence level, several attributes have been identified as pivotal determinants of solubility, encompassing the composition of specific amino acids (Asn, Thr, Tyr), the frequency of tripeptides [9], and the ratio of charged amino acids on the protein surface [10, 11]. Secondly, during the protein expression process, the ineffective translation of the mRNA and the manifestation of protein toxicity to E. coli may impede normal growth. In addition, there is evidence suggesting that protein instability can lead to aggregation, thereby inducing the formation of inclusion bodies [12–14]. Thirdly, at the experimental level, factors such as the selection of expression strains, appropriate fusion tags, culture temperature, and the pH value of the protein lysis buffer collectively contribute to the protein solubility [15]. Notwithstanding the abundance of available data, extracting essential features responsible for insolubility remains difficult, rendering the prediction of protein solubility a challenging task.
Over the past three decades, extensive researches have been dedicated to investigate the correlation between protein sequence and solubility, culminating in the formulation of numerous predictive models. Early studies employed statistical methods with a small dataset to extract essential protein sequence features for solubility classification [16]. In addition, SWI [17] employed the arithmetic mean of sequence composition scoring to predict protein’s solubility. Another booster for protein solubility predictor is the emergency of machine learning (ML), which shifted the focus toward the utilization of feature engineering and supervised ML algorithms, such as linear regression, support vector machines, and gradient boosting machines [18]. Some of the models leveraging traditional ML algorithms include PROSO [19], SOLpro [20], PROSO II [21], SCM [22], Protein-Sol [23], PaRSnIP [24], and SoluProt [25]. Recently, with the fast development of deep learning (DL), a transition from conventional ML to DL algorithms has been observed [26]. Researchers generated several DL models, such as DeepSol [27], SKADE [28], EPSOL [29], DSResSol [30], and DeepSoluE [31], which employ Convolutional Neural Networks (CNNs), bidirectional Gated Recurrent Unit (biGRU), or Long Short-Term Memory (LSTM)-based approaches for protein solubility prediction.
With the continuous development of natural language processing, attention-based algorithms deepen the understanding of relationships among tokens [32, 33]. The protein sequences, serving as the language of proteins, have catalyzed the development of diverse protein language models (PLMs), such as ProteinBERT [34], Evolutionary Scale Modeling (ESM) [35, 36], and ProtTrans [37]. These models excel in general contextual embeddings of protein sequences by training transformer models on large protein databases, such as UniRef [38] and BFD (Big Fantastic Database) [39]. By training with a labeled dataset, these models can be fine-tuned for predicting specific protein properties. For example, NetSolP utilized ESM1b and employed multilayer perceptron (MLP) for protein solubility classification [40].
Despite the significant advancements achieved by the aforementioned models, there still exists room for improvement. First of all, high-quality training data are critical for the success of any model. However, the most widely utilized dataset to date, originating from PROSO II [21] in 2012, has become outdated and suffers from ambiguous annotations. Another frequently used dataset, provided by SoluProt [25] in 2021, comprises only 10,912 sequences. Secondly, the accessibility and usability of models are also critical. For example, some of the reported models are out of service [19, 21, 22], while others require the inclusion of protein secondary structure information as inputs which is time consuming and compute intensive [24, 27, 29, 30]. Thirdly, model architectures play a crucial role in DL-based classification tasks. With the expanding of UniRef50D [38] database, ESM2 updated multiparameter versions of models [36], showcasing improvements in predictive capabilities and overall effectiveness in capturing intricate features of protein sequences. In terms of classification layers, the aforementioned NetSolP [40] applied only a simple MLP. Thus, the incorporation of updated protein language encoders and different classifier layers holds great potential to further improve prediction performance.
Given the aforementioned limitations, we attempted to improve the performance of protein solubility prediction by focusing on two key aspects: generating high-quality datasets, and developing a robust and easy-to-use model. For dataset generation, we integrated existing protein solubility data from TargetTrack [41], DNASU [42], eSOL [43], and Protein Data Bank (PDB) [44], making diligent efforts to improve the accuracy and comprehensiveness of the dataset. We designated this compiled dataset as the Updated E. coli protein Solubility DataSet (UESolDS). For model design, we trained a series of architectures by integrating multiple PLMs with diverse classifiers. Three pretrained PLMs, namely proteinBERT, ESM2, and ProtTrans, were employed alongside different classification layers, comprising MLP, Light Attention (LA) [45], and the biLSTM_TextCNN [46]. Subsequently, we systematically evaluated their performance on an independent test set. In comparison to previously reported models, the combination of ProtT5-XL-UniRef50 (ProtT5) with biLSTM_TextCNN demonstrated superior performance, denoting as the Protein Language Model-based protein Solubility prediction model (PLM_Sol). Finally, we performed an experimental test by assessing the solubility of 216 understudied proteins belonging to three distinct family types, and PLM_Sol consistently exhibited good performance.
Materials and Methods
UESolDS source
TargetTrack The TargetTrack database compiles experimental results curated by >100 investigators across 35 centers from 2000 to 2015, focusing on investigating the expression levels and structures of 350,000 proteins [41]. We applied a comparable data filtering analysis approach as that employed in SoluProt [25]. Utilizing experimental protocols provided by each contributor in the TargetTrack database, we extracted 17 datasets specifically focusing on protein expression in E. coli (Supplementary Table S1). The TargetTrack database contained numerous entries marked as “work stopped” or “other” due to uncertain final statuses, and these data were consequently excluded [21]. Subsequently, based on the experimental status of proteins, they were categorized into insoluble (Insol) and soluble (Sol). Insol proteins were those labeled with the tags “tested”, “selected”, “cloned”, and “expression tested”. Sol proteins were those labeled with “expressed”, “soluble”, “purified” and “in PDB” and had subsequent structural analysis data.
DNASU The DNASU is a global plasmid repository. Data obtained from DNASU originated from the Protein Structure Initiative: Biology (PSI:Biology) [42]. The entries using the common vectors (“pET21_NESG”, “pET15_NESG”) for recombinant protein expression were retrieved. Then, they were categorized based on the following tags: Insol proteins were labeled as “Tested_Not_Soluble” while Sol proteins were labeled as “Protein_Soluble”.
eSOL eSOL is a database offering solubility information for E. coli proteins through the protein synthesis using recombinant elements cell-free expression system [43]. The eSOL database assigns a solubility score to Sol proteins, whereas those lacking solubility scores are categorized as Insol proteins.
PDB Protein sequences with E. coli as expression host from the PDB database (March 2023 version) were extracted and annotated as Sol [44].
UESolDS data cleaning
The aforementioned datasets contained a total of 210,304 entries. Six steps were implemented sequentially for data cleaning to compile the aforementioned UESolDS.
(1) 19,576 membrane proteins predicted by TransMembrane helix Hidden Markov Model (TMHMM) [47] were filtered out due to their commonly insolubility upon overexpression [21, 25, 48].
(2) The following His tag fragments in proteins were excluded due to an uneven distribution of these tags between Insol and Sol proteins revealed by NetSolP [40]: “MGSDKIHHHHHH”, “MGSSHHHHHH”, “MHHHHHHS”, “MRGSHHHHHH”, “MAHHHHHH”, “MGHHHHHH”, “MGGSHHHHHH”, “HHHHHHH” and “AHHHHHHH”.
(3) 1,454 sequences containing special characters “X|x”, “U|u”, “Z|z”, “*”, “.”, “+” and “−” were thoroughly removed.
(4) 232 sequences with lengths <25 aa or >2,500 aa were filtered out.
(5) During the data categorization process, we identified data contamination, as some Insol proteins exhibit high similarity with Sol proteins. To enhance the dataset quality, we searched for matches of Insol protein sequences in Sol by using BLAST blastp [49]. The overlap of 23,933 Insol entries exhibiting an identity >75% and coverage >70% was removed.
(6) Clustering above filtered sequences using MMseqs [50] with 25% identity and coverage >70% threshold.
Finally, there were 31,581 entries categorized as Insol and 46,450 entries categorized as Sol, resulting in the creation of the UESolDS.
Independent test set generation
For an unbiased evaluation of model performance, we first aggregated the training sets of the previously reported models slated for evaluation in this study. Next, sequences in UESolDS with an identity of ≥25% to the integrated training sets were excluded. Subsequently, we randomly selected 2,000 sequences from the remaining Sol/Insol data to form the independent test set.
Model architecture and training process
ProteinBERT ProteinBERT has six encoder layers and is trained on the UniRef90 dataset, along with protein gene ontology annotation information. The inputs of proteinBERT are the protein sequences and the outputs are fixed-dimension vectors. Then, the vectors are fed into a vanilla MLP layer to obtain solubility probabilities. For the training process, ProteinBERT underwent a process where all layers of the pretrained model were initially frozen, except for the classification layers, which were trained for up to 10 epochs. Afterward, all layers were unfrozen and trained additional epochs until the test loss did not decline for 3 epochs. To optimize the learning process, the dynamic learning rate adjustment technique ReduceLROnPlateau was employed. The loss was calculated using binary cross-entropy. The training process was executed on a single Graphics Processing Unit (GPU) (Tesla P100-PCIE-12GB), with the learning rate and batch size set to 0.001 and 80, respectively.
ProtTrans To employ ProtTrans as the encoder, six models were trained by combining two architectures from ProtTrans with three classifiers, respectively. The first architecture, ProtBert_BFD, based on BERT with a total of 30 encoder layers, is trained on the BFD dataset. The second one is ProtT5, which utilized an architecture of T5 with a total of 24 encoder layers. It is pretrained on the BFD dataset and fine-tuned on UniRef50. The inputs of these PLM models are the protein sequences, and the outputs are matrices whose shape is n × L, where n represents the embedding dimensions and L is the maximum protein sequence length. Following the ProtBert_BFD and ProtT5 architectures, three classification modules were introduced and tested, respectively. The first classification module is a vanilla MLP. The second is the LA architecture, as described in a previous study [45], which shows excellent performance in protein localization classification. Thirdly, the biLSTM_TextCNN architecture, known for its effectiveness in sentiment classification [46], utilizes a bidirectional LSTM (biLSTM) to convert the information from the encoder into corresponding matrices. Following the classification process, a Text-attentional CNN (TextCNN) is applied, featuring three parallel one-dimensional convolution (Conv-1D) layers for crucial feature extraction. Next, the max pooling is executed along the direction of Conv-1D. The resulting three vectors obtained are concatenated and feed into fully connected layers to generate probabilities. For the training process, Bio-embedding software [51] was used to extract embedding information from ProtBert_BFD and ProtT5. In the model training phase, all layers of the pretrained model remained frozen. The classification layers were trained for 15 epochs. The learning rate was initially set to 0.001 and adjusted by the optimizer AdamW [52] with a weight decay of 0.001. Binary cross-entropy was utilized to calculate the loss. The training process was conducted on a single GPU (Tesla P100-PCIE-12GB) with a batch size of 72.
ESM2 To employ ESM2 as the encoder, six models were trained by combining two architectures from ESM2 with three classifiers, respectively. As for ESM2-based models, two implementations, esm2_t30_150M_UR50D (ESM2_30) and esm2_t33_650M_UR50D (ESM2_33), were selected. After the protein sequences are input into ESM2, the outputs matrices’ shape was L × n. L is the maximum protein sequence length and n is the embedding dimension. The classification modules utilized the same settings as the classification modules used above ProtTrans. For the training process, fine-tuning involved unfreezing the embedding norm before layers, embedding norm after layers, and Roberta Head for masked language modeling layers in the pretrained model. Training consisted of 15 epochs with a learning rate of 0.001 and a batch size of 119. The optimizer utilized AdamW with a weight decay of 0.0001. Cross-entropy was used to calculate the loss. A total of eight Tesla P100-PCIE-12GB GPUs were used for this process.
Experimental test set generation procedure
To better understand PLM_Sol’s performance on the understudied EOI, we selected various types of in-house EOI, consisting of tandem repeat proteins, DNA transposases, and deaminases (unpublished data). These EOI were intended to be synthesized for the development of novel gene editing tools, without any prior solubility prediction steps. To minimize redundancy, we clustered these EOI at 25% sequence identity, yielding 216 entries including 155 tandem repeat proteins, 30 DNA transposases, and 31 deaminases. Then, we performed in vitro recombinant protein expression assay in E. coli using the following procedure.
The pET28a vector with an N-terminal His tag and SUMO tag was applied for protein expression in E. coli Rosetta (DE3) cells. Protein expression was induced by adding 0.1 mM Isopropyl ß-D-1-thiogalactopyranoside (IPTG) when the OD600 reached 0.6–0.8, followed by incubation at 16 °C for 18 h. Cell pellets were sonicated (50 W, 3 s on/3 s off on ice for 3 min) after being resuspended in binding buffer (50 mM Tris–HCl, 500 mM NaCl, pH 7.5). Supernatant was extracted and analyzed via SDS-PAGE gels, where protein bands with overexpression strips at the right molecular weight were identified as Sol hits based on the Coomassie blue staining gel results.
Evaluation metrics
The performance metrics, including accuracy, area under the curve (AUC), area under the precision–recall curve (AUPR), precision, sensitivity, specificity, F1_score, confusion matrix. and Matthews correlation coefficient (MCC) were calculated using the scikit-learn packages [53].
Results
Dataset organization and analysis
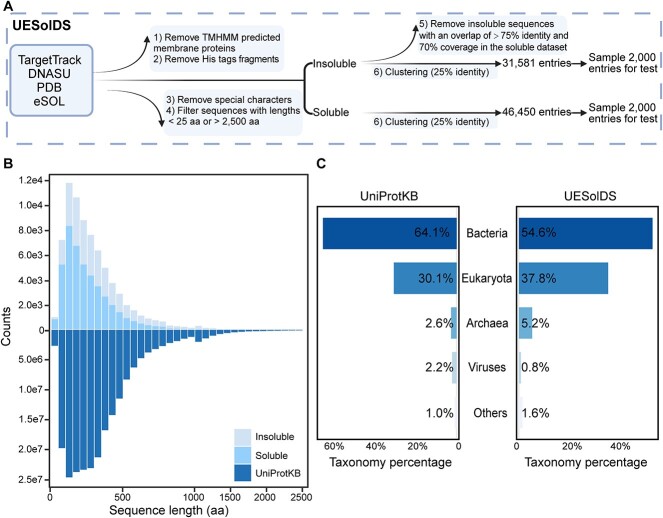
The previously reported datasets are outdated, with ambiguous annotation [21] or limited coverage [25]. To generate a more comprehensive and precise dataset, we reconstructed the dataset on recombinant protein solubility in E. coli by integrating and updating related databases, including TargetTrack, DNASU, eSOL, and PDB. We executed a six-step filtration process, encompassing the removal of membrane proteins, His tag fragments, special characters, sequences within certain length ranges, and ambiguous Insol sequences (Methods, Fig. 1A). This refined dataset was denoted as the UESolDS with a total of 78,031 entries. In order to establish an independent test set, we conducted a sequence identity analysis between UESolDS and the integrated training datasets of seven previously reported models, including Protein_sol [23], SKADE [28], SWI [17], Soluprot [25], EPSOL [29], NetSolP [40], and DeepSoluE [31]. Sequences with >25% identity to any of the previously reported training sets were removed. Subsequently, a random selection of 2,000 Sol and 2,000 Insol entries from the remaining dataset were compiled to form the independent test set (Fig. 1A).
Figure 1.
Generation and analysis of UESolDS. (A) Data processing pipeline of UESolDS. (B) Distribution of sequence lengths in the UniProtKB (release 2024_01) and UESolDS. (C) Taxonomy distribution of sequences from the UniProKB and UESolDS.
To elucidate the characteristics of UESolDS, we conducted an analysis of the sequence length and species distribution. The results revealed a concentration of protein lengths within the range of 25–500 amino acids in UESolDS (Fig. 1B). It is worth noting that UESolDS showed a very similar distribution pattern with UniProtKB [54] database (Fig. 1B), indicating a good coverage and representation of the existing protein space. The statistical analysis of the species distribution showed that bacteria constituted the majority with ~54.6%, followed by eukaryotes with ~37.8%, archaea with ~5.2%, viruses with 1.6%, and other sequences with 0.8%, which were annotated as unclassified (Fig. 1C). In addition, we compared UESolDS with UniProtKB, revealing a similar distribution in species composition, with bacteria, eukaryotes, and archaea ranking as the top three categories (Fig. 1C). These findings collectively emphasized a broad diversity of our refined dataset, thereby establishing a solid foundation for improving the model’s generalization capabilities.
Model construction and performance evaluation
With the rapidly evolving development of PLMs, the utilization of updated PLMs like ESM2 [36], coupled with different classification layers, holds the potential to improve the extraction of protein solubility features with higher accuracy. Therefore, we constructed models based on the following two parts: an encoder responsible for generating protein sequence embeddings from the PLMs, and a classification module for selecting and categorizing the crucial features. In this work, three PLMs have been tested, including ProteinBERT [34], ProtTrans [37], and ESM2 (Table 1). In addition, three classification architectures have been employed, namely MLP, LA [45], and biLSTM_TextCNN [46]. A total of 13 models were developed and trained (Methods).
Table 1.
Characteristics of the PLM embeddings used in this study
| Embeddings | Language models | Layers | Parameters | Training databases |
|---|---|---|---|---|
| ProteinBERT | Modified BERT | 6 | 16M | Uniref90 (106M seqs) |
| esm2_t30_150M_UR50D | BERT | 30 | 150M | UniRef50D (2021_04) (50M seqs) |
| esm2_t33_650M_UR50D | BERT | 33 | 650M | UniRef50D (2021_04) (50M seqs) |
| ProtT5-XL | T5 | 24 | 3B | BFD100 (2B seqs) + UniRef50 (45M seqs) |
| ProtBert | BERT | 30 | 420 M | BFD100 (2B seqs) |
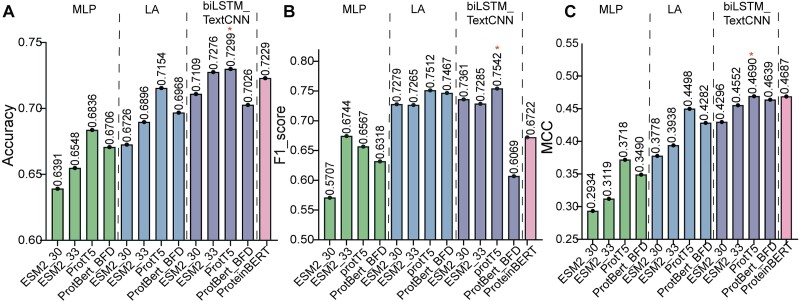
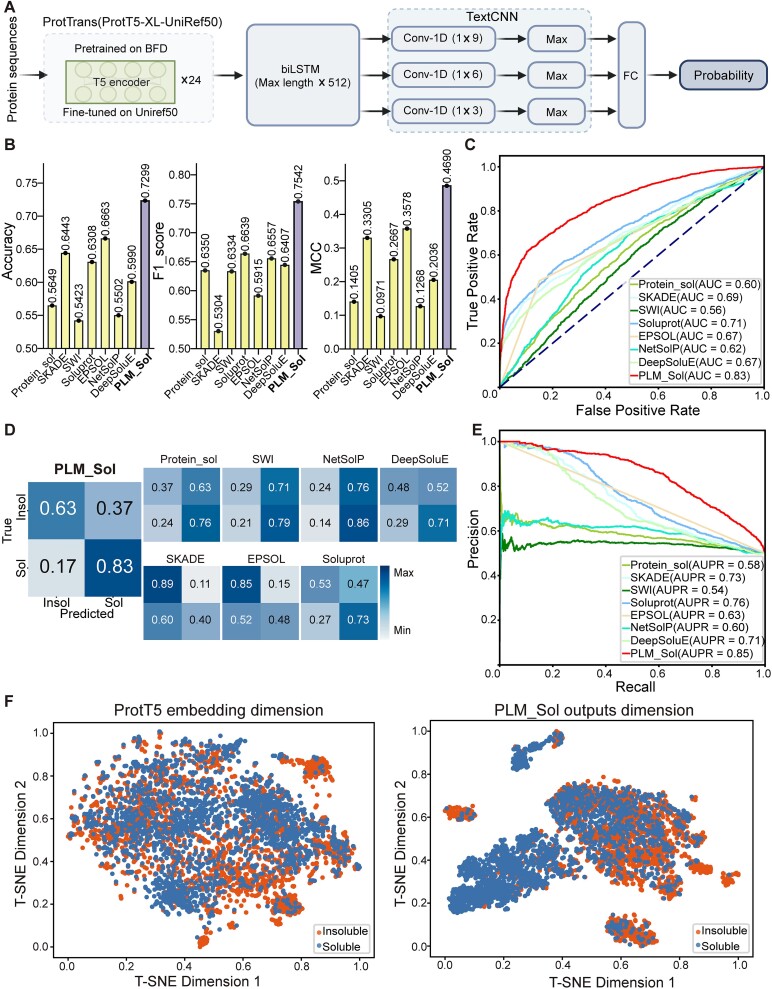
We then evaluated the performance of the 13 in-house trained models on the independent test set. Among these, the combination of ProtT5 with biLSTM_TextCNN demonstrated the best performance, achieving the highest accuracy of 0.7299, top F1_score of 0.7542, and maximum MCC of 0.4690 (Fig. 2A, B, C). Hence, the combination of ProtT5 with biLSTM_TextCNN was selected as our protein solubility classification model, designated as the PLM_Sol (Fig. 3A).
Figure 2.
Performance of 13 in-house models on independent test set. (A) Comparison of accuracy among 13 in-house models. The training results of different PLMs combined with MLP, LA, and BiLSTM_TextCNN classifiers are separated with dashed lines. (B) Comparison of F1_score among 13 in-house models. The results are presented in the same order as in (A). (C) Comparison of MCC among 13 in-house models. The results are presented in the same order as in (A).
Figure 3.
Performance of PLM_Sol on independent test set. (A) Model architecture of PLM_Sol; FC, fully connected layer. (B) Comparison of accuracy, F1_score, and MCC between PLM_Sol and previously reported models. (C) ROC curve for PLM_Sol and previously reported models. (D) Confusion matrices depicting PLM_Sol predictions in comparison with previously reported models. (E) Precision–recall curve for PLM_Sol and previously reported models. (F) Dimensionality reduction using t-SNE for ProtT5 embeddings and PLM_Sol FC layers vector values on independent test set. Each point represents a sequence.
Subsequently, we compared PLM_Sol with seven previously reported models, and PLM_Sol exhibited enhancement over the previously best-performing software across various metrics. For example, PLM_Sol showed a 6.4% increase in accuracy over EPSOL, a 9.0% increase in F1_score over Soluprot, and an 11.1% increase in MCC score over EPSOL on the test set (Fig. 3B, Table 2). PLM_Sol also showed the highest AUC and AUPR score among the models evaluated based on the receiver operating characteristic (ROC) and precision–recall curve (Fig. 3C, E). Next, we analyzed the confusion matrix of each model and observed distinctive prediction outcomes among the previously reported tools (Fig. 3D). For example, Protein_sol, SWI, and NetSolP preferred to predict proteins as Sol, while SKADE and EPSOL preferred to predict proteins as Insol. Conversely, PLM_Sol showed more accurate predictions power.
Table 2.
Comparison of solubility prediction performance of PLM_Sol with existing models on independent test set.
| Models | T | AUC | Accuracy | F1_score | MCC | Precision | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|
| Protein_sol | 0.5 | 0.5985 | 0.5649 | 0.6350 | 0.1405 | 0.5470 | 0.7569 | 0.3729 |
| SKADE | 0.5 | 0.6882 | 0.6443 | 0.5304 | 0.3305 | 0.7811 | 0.4015 | 0.8874 |
| SWI | 0.5 | 0.5597 | 0.5423 | 0.6334 | 0.0971 | 0.5284 | 0.7905 | 0.2938 |
| Soluprot | 0.5 | 0.7126 | 0.6308 | 0.6639 | 0.2667 | 0.6095 | 0.7290 | 0.5325 |
| EPSOL | — | 0.6664 | 0.6663 | 0.5915 | 0.3578 | 0.7630 | 0.4830 | 0.8499 |
| NetSolP | 0.5 | 0.6183 | 0.5502 | 0.6557 | 0.1268 | 0.5312 | 0.8564 | 0.2437 |
| DeepSoluE | 0.4 | 0.6660 | 0.5990 | 0.6407 | 0.2036 | 0.5804 | 0.7150 | 0.4830 |
| PLM_Sol | 0.5 | 0.8342 | 0.7299 | 0.7542 | 0.4690 | 0.6919 | 0.8289 | 0.6308 |
T represents the threshold value of the models. The bolded string indicates the highest score within the respective column.
To visually showcase the classification performance of PLM_Sol, we applied t-SNE [55] to the test set using ProtT5 embeddings and the vector values from PLM_Sol’s FC layer. Compared with the ProtT5 embeddings, the data processed by the PLM_Sol classification layer effectively separated Sol and Insol proteins data, suggesting a robust classification performance (Fig. 3F).
Solubility prediction for diverse types of EOI
To further validate the generalization of PLM_Sol, we conducted an evaluation of protein solubility prediction for 216 in-house EOI (Methods). These diverse protein types exhibit significant variability in solubility, and some of them are reported to be poorly soluble [56, 57], providing valuable material for the assessment of the model’s generalizability. We utilized the in vitro recombination protein expression assay to evaluate the solubility of EOI, resulting in 134 Insol proteins and 82 Sol proteins (Supplementary Table S2).
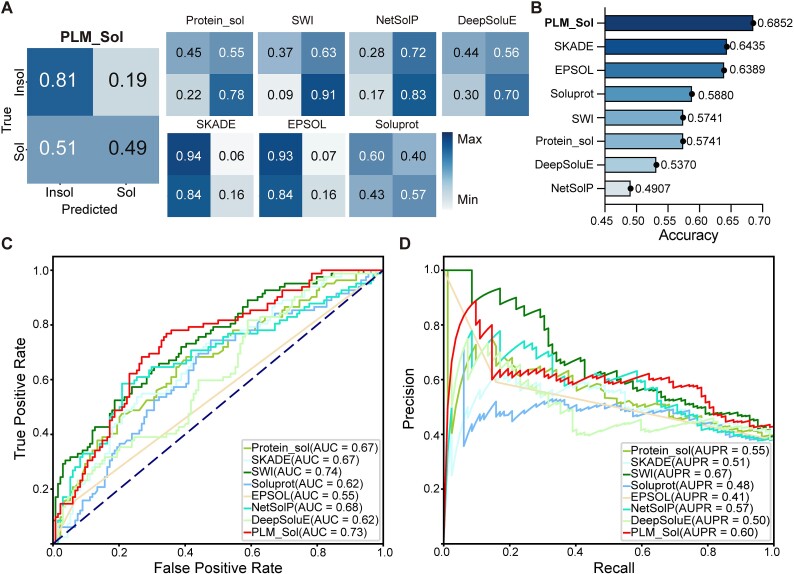
Next, PLM_Sol along with seven previously reported models were employed to predict the solubility of the aforementioned in-house EOI, followed by an assessment of their overall performance. We set the threshold to 0.5, which represents the average of the optimal thresholds in our two test sets and is also adopted by many previously reported models [17, 23, 25, 28, 40]. We then calculated the confusion matrix and accuracy. The confusion matrix was utilized to showcase the comparison between predicted and experimentally observed outcomes (Fig. 4A). The predictive preferences in our experimental test set were consistent with those in the independent test set for some reported models. It is noteworthy that PLM_Sol demonstrated a higher proportion of correct predictions, suggesting a better performance in predicting protein solubility for EOI screening (Fig. 4B). In addition, we plotted the ROC and precision–recall curves to illustrate PLM_Sol’s performance on the experimental test set (Fig. 4C, D). Due to the limited size of the test set, the curves were not smooth. Although PLM_Sol is not the best on every metrics of ROC and precision–recall curves, it performed well overall.
Figure 4.
Performance of PLM_Sol on experimental test set. (A) Confusion matrices depicting PLM_Sol predictions in comparison with previously reported models. (B) Accuracy for PLM_Sol and previously reported models. (C) ROC curve for PLM_Sol and previously reported models. (D) Precision–recall curve for PLM_Sol and previously reported models.
Discussion
Deep learning relies on two fundamental pillars: data and model [58–60]. However, in the development of solubility prediction tasks, most models have predominantly focused on model optimization, overlooking the quality of the datasets [27–29]. For instance, a commonly used dataset from PROSO II [21] showed data contamination, as we identified 9.8% of Insol proteins that showed strong similarity (>90% identity and >70% coverage) to those present in the Sol dataset. This can be attributed to the ambiguity of the annotations for Insol proteins in the TargetTrack database, which serves as the dominant source of Insol data. In this study, a more comprehensive and accurate database UESolDS was generated by integrating the existing datasets, followed by a more stringent blastp parameters (>75% identity and >70% coverage) to eliminate overlapping Insol instances within the Sol category. Furthermore, given the inherent complexity of protein insolubility states, more in-depth investigation and detailed annotation of insoluble protein experimental information may further improve the quality of the datasets. This, in turn, holds potential to further enhance the classification capability of DL models.
For model construction, we utilized the refined databases and employed PLMs incorporating additional classification layers for enhanced solubility prediction. After evaluation on an independent test set, we identified the best-performing model, named PLM_Sol. Compared to the state-of-the-art models, PLM_Sol exhibited a 6.4% increase in accuracy on the test set. In addition to the independent test set, we also validated the efficiency of PLM_Sol using our in-house experimental data, which showed improved prediction performance with an increase in accuracy of ~4.2%. Furthermore, our models could benefit from further optimization. The evaluation of the experimental test set shows that there is still room to improve the performance of PLM_sol. In the fast-paced world of large language models, the utilization of parameter-efficient fine-tuning modules, such as LoRA [61], Adapter Tuning [62], IA3 [63], and Prompt Tuning [64], may enhance the model performance. Alternatively, employing the protein structure features derived from AlphaFold2 [39] or ESMFold [36] may provide the capability to identify key structural features to better separate Sol and Insol proteins. Improvement based on these continuously emerging algorithms, combined with updated high-quality datasets, would ensure a better prediction accuracy.
All in all, PLM_Sol is easy to use and provides accurate predictions of protein solubility based solely on protein sequences as input. The integration of PLM_Sol into a high-throughput EOI screening pipeline offers the potential to avoid Insol gene synthesis, thereby improving the success rate and facilitating the scale-up screen process. Furthermore, the ongoing accumulation of experimental results from the above design, coupled with the continuously updated UESolDS, holds the prospect of iteratively refining PLM_Sol, thereby continuously enhancing its overall predictive performance.
Key Points
We curated an Updated E. coli protein Solubility DataSet (UESolDS) by incorporating newly available PDB data and consolidating three solubility datasets to explore protein solubility.
For the prediction of protein solubility, we benchmarked a combination of diverse PLMs along with various classification architectures. An independent test dataset was compiled for evaluation purposes. The ProtT5 combined with biLSTM_TextCNN demonstrating the best performance, designated as PLM_Sol, exhibited substantial improvements over previously reported models with a notable increase of 6.4% in accuracy, 9.0% in F1_score, and 11.1% in MCC score.
To further evaluate the performance of PLM_Sol, we performed the experimental testing by assessing the solubility of 216 understudied proteins across three distinct families. PLM_Sol also exhibited good performance.
Supplementary Material
Acknowledgements
We thank Duan Liu for his assistance in project management. We thank Computer Network Information Center, Chinese Academy of Sciences, for kindly providing the computing clusters for training the deep learning model. We also thank Prof. Yong E. Zhang and Dr. Daqi Yu for their insightful comments and constructive suggestions.
Contributor Information
Xuechun Zhang, Key Laboratory of Organ Regeneration and Reconstruction, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; University of Chinese Academy of Sciences, No. 1 Yanqihu East Rd, Huairou District, Beijing 101408, China.
Xiaoxuan Hu, Key Laboratory of Organ Regeneration and Reconstruction, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; University of Chinese Academy of Sciences, No. 1 Yanqihu East Rd, Huairou District, Beijing 101408, China.
Tongtong Zhang, Key Laboratory of Organ Regeneration and Reconstruction, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; University of Chinese Academy of Sciences, No. 1 Yanqihu East Rd, Huairou District, Beijing 101408, China.
Ling Yang, Key Laboratory of Organ Regeneration and Reconstruction, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; University of Chinese Academy of Sciences, No. 1 Yanqihu East Rd, Huairou District, Beijing 101408, China.
Chunhong Liu, Key Laboratory of Organ Regeneration and Reconstruction, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; University of Chinese Academy of Sciences, No. 1 Yanqihu East Rd, Huairou District, Beijing 101408, China.
Ning Xu, Key Laboratory of Organ Regeneration and Reconstruction, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; University of Chinese Academy of Sciences, No. 1 Yanqihu East Rd, Huairou District, Beijing 101408, China.
Haoyi Wang, Key Laboratory of Organ Regeneration and Reconstruction, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; University of Chinese Academy of Sciences, No. 1 Yanqihu East Rd, Huairou District, Beijing 101408, China; Beijing Institute for Stem Cell and Regenerative Medicine, A 3 Datun Road, Chaoyang District, Beijing 100100, China.
Wen Sun, Key Laboratory of Organ Regeneration and Reconstruction, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, 1 Beichen West Road, Chaoyang District, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, A 3 Datun Road, Chaoyang District, Beijing 100100, China.
Conflict of interest: None declared.
Funding
This research was supported by the Ministry of Agriculture and Rural Affairs of China, Biological Breeding-Major Projects [2023ZD0407401 to H.W.], the Strategic Priority Research Program of the Chinese Academy of Sciences [XDA16010503 to H.W.], Beijing Institute for Stem Cell and Regenerative Medicine [2022FH122, 2023FH105 to H.W., 2023FH106 to W.S.], National Natural Science Foundation of China [32001062 to W.S.], Initiative Scientific Research Program, Institute of Zoology, Chinese Academy of Sciences [2023IOZ0204 to H.W.], and Chinese Academy of Sciences [ZDBS-LY-SM005 to H.W.].
Data availability
The data underlying this article are available at https://zenodo.org/doi/10.5281/zenodo.10675340. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Author contributions
W.S. and H.Y.W. supervised the project. H.Y.W., X.C.Z., X.X.H., and W.S. conceived and designed the study. X.C.Z., T.T.Z., Y.L., C.H.L., and N.X. conducted the expression experiments. X.C.Z. and X.X.H. performed the computational analysis. H.Y.W., X.C.Z., X.X.H., and W.S. wrote the manuscript.
References
- 1. Jain K, Salamat-Miller N, Taylor K. Freeze–thaw characterization process to minimize aggregation and enable drug product manufacturing of protein based therapeutics. Sci Rep 2021;11:11332. 10.1038/s41598-021-90772-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ratanji KD, Dearman RJ, Kimber I. et al. Editor’s highlight: subvisible aggregates of immunogenic proteins promote a Th1-type response. Toxicol Sci 2016;153:258–70. 10.1093/toxsci/kfw121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hermeling S, Crommelin DJ, Schellekens H. et al. Structure-immunogenicity relationships of therapeutic proteins. Pharm Res 2004;21:897–903. 10.1023/B:PHAM.0000029275.41323.a6. [DOI] [PubMed] [Google Scholar]
- 4. Jia B, Han X, Kim KH. et al. Discovery and mining of enzymes from the human gut microbiome. Trends Biotechnol 2022;40:240–54. 10.1016/j.tibtech.2021.06.008. [DOI] [PubMed] [Google Scholar]
- 5. Xiang G, Li Y, Sun J. et al. Evolutionary mining and functional characterization of TnpB nucleases identify efficient miniature genome editors. Nat Biotechnol 2023;42:1–13. [DOI] [PubMed] [Google Scholar]
- 6. Huang J, Lin Q, Fei H. et al. Discovery of deaminase functions by structure-based protein clustering. Cell 2023;186(15):3182–3195.e14. 10.1016/j.cell.2023.05.041. [DOI] [PubMed] [Google Scholar]
- 7. Tripathi NK, Shrivastava A. Recent developments in bioprocessing of recombinant proteins: expression hosts and process development. Front Bioeng Biotechnol 2019;7:420. 10.3389/fbioe.2019.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shih YP, Kung WM, Chen JC. et al. High-throughput screening of soluble recombinant proteins. Protein Sci 2002;11:1714–9. 10.1110/ps.0205202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Idicula-Thomas S, Balaji PV. Understanding the relationship between the primary structure of proteins and its propensity to be soluble on overexpression in Escherichia coli. Protein Sci 2005;14:582–92. 10.1110/ps.041009005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carballo-Amador MA, McKenzie EA, Dickson AJ. et al. Surface patches on recombinant erythropoietin predict protein solubility: engineering proteins to minimise aggregation. BMC Biotechnol 2019;19:1–10. 10.1186/s12896-019-0520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sankar K, KrystekSR, Jr, Carl SM. et al. AggScore: prediction of aggregation-prone regions in proteins based on the distribution of surface patches. Proteins Struct Funct Bioinf 2018;86:1147–56. 10.1002/prot.25594. [DOI] [PubMed] [Google Scholar]
- 12. Tartaglia GG, Pechmann S, Dobson CM. et al. A relationship between mRNA expression levels and protein solubility in E. coli. J Mol Biol 2009;388:381–9. 10.1016/j.jmb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 13. Ventura S. Sequence determinants of protein aggregation: tools to increase protein solubility. Microb Cell Fact 2005;4:1–8. 10.1186/1475-2859-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem 2017;86:27–68. 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 15. Costa S, Almeida A, Castro A. et al. Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Front Microbiol 2014;5:63. 10.3389/fmicb.2014.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilkinson DL, Harrison RG. Predicting the solubility of recombinant proteins in Escherichia coli. Bio/technology 1991;9:443–8. [DOI] [PubMed] [Google Scholar]
- 17. Bhandari BK, Gardner PP, Lim CS. Solubility-weighted index: fast and accurate prediction of protein solubility. Bioinformatics 2020;36:4691–8. 10.1093/bioinformatics/btaa578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sarker IH. Machine learning: algorithms, real-world applications and research directions. SN Comput Sci 2021;2:160. 10.1007/s42979-021-00592-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smialowski P, Martin-Galiano AJ, Mikolajka A. et al. Protein solubility: sequence based prediction and experimental verification. Bioinformatics 2007;23:2536–42. 10.1093/bioinformatics/btl623. [DOI] [PubMed] [Google Scholar]
- 20. Magnan CN, Randall A, Baldi P. SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics 2009;25:2200–7. 10.1093/bioinformatics/btp386. [DOI] [PubMed] [Google Scholar]
- 21. Smialowski P, Doose G, Torkler P. et al. PROSO II–a new method for protein solubility prediction. FEBS J 2012;279:2192–200. 10.1111/j.1742-4658.2012.08603.x. [DOI] [PubMed] [Google Scholar]
- 22. Huang H-L, Charoenkwan P, Kao T-F. et al. Prediction and analysis of protein solubility using a novel scoring card method with dipeptide composition. BMC Bioinformatics. 2012;13:S3. 10.1186/1471-2105-13-S17-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hebditch M, Carballo-Amador MA, Charonis S. et al. Protein–sol: a web tool for predicting protein solubility from sequence. Bioinformatics 2017;33:3098–100. 10.1093/bioinformatics/btx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rawi R, Mall R, Kunji K. et al. PaRSnIP: sequence-based protein solubility prediction using gradient boosting machine. Bioinformatics 2018;34:1092–8. 10.1093/bioinformatics/btx662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hon J, Marusiak M, Martinek T. et al. SoluProt: prediction of soluble protein expression in Escherichia coli. Bioinformatics 2021;37:23–8. 10.1093/bioinformatics/btaa1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chauhan NK, Singh K. A review on conventional machine learning vs deep learning. 2018 International Conference on Computing, Power and Communication Technologies (GUCON). 2018, pp. 347–52. 10.1109/GUCON.2018.8675097. [DOI] [Google Scholar]
- 27. Khurana S, Rawi R, Kunji K. et al. DeepSol: a deep learning framework for sequence-based protein solubility prediction. Bioinformatics 2018;34:2605–13. 10.1093/bioinformatics/bty166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raimondi D, Orlando G, Fariselli P. et al. Insight into the protein solubility driving forces with neural attention. PLoS Comput Biol 2020;16:e1007722. 10.1371/journal.pcbi.1007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu X, Yu L. EPSOL: sequence-based protein solubility prediction using multidimensional embedding. Bioinformatics 2021;37:4314–20. 10.1093/bioinformatics/btab463. [DOI] [PubMed] [Google Scholar]
- 30. Madani M, Lin K, Tarakanova A. DSResSol: a sequence-based solubility predictor created with dilated squeeze excitation residual networks. Int J Mol Sci 2021;22:13555. 10.3390/ijms222413555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang C, Zou Q. Prediction of protein solubility based on sequence physicochemical patterns and distributed representation information with DeepSoluE. BMC Biol 2023;21:1–11. 10.1186/s12915-023-01510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vaswani A, Shazeer N, Parmar N. et al. Attention is all you need. Adv Neural Inf Process Syst 2017;30. [Google Scholar]
- 33. Devlin J, Chang M-W, Lee K. et al. Bert: pre-training of deep bidirectional transformers for language understanding. arXiv preprint. arXiv:1810.04805. 2018. [Google Scholar]
- 34. Brandes N, Ofer D, Peleg Y. et al. ProteinBERT: a universal deep-learning model of protein sequence and function. Bioinformatics 2022;38:2102–10. 10.1093/bioinformatics/btac020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rives A, Meier J, Sercu T. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc Natl Acad Sci 2021;118:e2016239118. 10.1073/pnas.2016239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin Z, Akin H, Rao R. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023;379:1123–30. 10.1126/science.ade2574. [DOI] [PubMed] [Google Scholar]
- 37. Elnaggar A, Heinzinger M, Dallago C. et al. Prottrans: toward understanding the language of life through self-supervised learning. IEEE Trans Pattern Anal Mach Intell 2021;44:7112–27. 10.1109/TPAMI.2021.3095381. [DOI] [PubMed] [Google Scholar]
- 38. Suzek BE, Wang Y, Huang H. et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015;31:926–32. 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jumper J, Evans R, Pritzel A. et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–9. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thumuluri V, Martiny H-M, Almagro Armenteros JJ. et al. NetSolP: predicting protein solubility in Escherichia coli using language models. Bioinformatics 2022;38:941–6. 10.1093/bioinformatics/btab801. [DOI] [PubMed] [Google Scholar]
- 41. Berman HM, Gabanyi MJ, Kouranov A. et al. Protein structure initiative–targettrack 2000-2017–all data files. Zenodo 2017;10:vbab035. [Google Scholar]
- 42. Seiler CY, Park JG, Sharma A. et al. DNASU plasmid and PSI: biology-materials repositories: resources to accelerate biological research. Nucleic Acids Res 2014;42:D1253–60. 10.1093/nar/gkt1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kitagawa M, Ara T, Arifuzzaman M. et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 2005;12:291–9. 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 44. Berman HM, Westbrook J, Feng Z. et al. The protein data bank. Nucleic Acids Res 2000;28:235–42. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stärk H, Dallago C, Heinzinger M. et al. Light attention predicts protein location from the language of life. Bioinform Adv 2021;1:1–8. 10.1093/bioadv/vbab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jiang X, Song C, Xu Y. et al. Research on sentiment classification for netizens based on the BERT-BiLSTM-TextCNN model. PeerJ Comput Sci 2022;8:e1005. 10.7717/peerj-cs.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krogh A, Larsson B, Von Heijne G. et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001;305:567–80. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 48. Grisshammer R, Tate C. Preface: overexpression of integral membrane proteins. Biochim Biophys Acta 2003;1610:1–2. 10.1016/S0005-2736(02)00706-X. [DOI] [Google Scholar]
- 49. Boratyn GM, Camacho C, Cooper PS. et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res 2013;41:W29–33. 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Steinegger M, Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 2017;35:1026–8. 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 51. Dallago C, Schütze K, Heinzinger M. et al. Learned embeddings from deep learning to visualize and predict protein sets. Curr Protoc 2021;1:1–26. 10.1002/cpz1.113. [DOI] [PubMed] [Google Scholar]
- 52. Loshchilov I, Hutter F. Decoupled weight decay regularization. The International Conference on Learning Representations 2018;1–8. [Google Scholar]
- 53. Pedregosa F, Varoquaux G, Gramfort A. et al. Scikit-learn: machine learning in python. J Mach Learn Res 2011;12:2825–30. [Google Scholar]
- 54. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res 2023;51:D523–31. 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res 2008;9:2579–605. [Google Scholar]
- 56. Dyda F, Chandler M, Hickman AB. The emerging diversity of transpososome architectures. Q Rev Biophys 2012;45:493–521. 10.1017/S0033583512000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hickman AB, Perez ZN, Zhou L. et al. Molecular architecture of a eukaryotic DNA transposase. Nat Struct Mol Biol 2005;12:715–21. 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- 58. Shrestha A, Mahmood A. Review of deep learning algorithms and architectures. IEEE access 2019;7:53040–65. 10.1109/ACCESS.2019.2912200. [DOI] [Google Scholar]
- 59. Sun C, Shrivastava A, Singh S. et al. Proceedings of the IEEE international conference on computer vision, 2017;843–52.
- 60. Dodge S, Karam L, 2016 eighth international conference on quality of multimedia experience (QoMEX). IEEE, 2016; 1–6. [Google Scholar]
- 61. Hu EJ, Shen Y, Wallis P. et al. LoRA: low-rank adaptation of large language models. The International Conference on Learning Representations 2022;1–13. [Google Scholar]
- 62. Houlsby N, Giurgiu A, Jastrzebski S. et al. International conference on machine learning. PMLR, 2019;2790–9. [Google Scholar]
- 63. Liu H, Tam D, Muqeeth M. et al. Few-shot parameter-efficient fine-tuning is better and cheaper than in-context learning. Adv Neural Inf Proc Syst 2022;35:1950–65. [Google Scholar]
- 64. Lester B, Al-Rfou R, Constant N. The power of scale for parameter-efficient prompt tuning. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Punta Cana, Dominican Republic, 2021, pp. 3045–59. 10.18653/v1/2021.emnlp-main.243. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available at https://zenodo.org/doi/10.5281/zenodo.10675340. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.