Abstract
Chronic myeloid leukemia (CML) is associated with several breakpoint regions that result in different BCR::ABL1 fusion transcripts. These include the major breakpoint region (M-BCR), minor breakpoint region (m-BCR), and mu breakpoint region (u-BCR) corresponding to p210, p190, and p230 fusion transcripts, respectively. This patient is a 38-year-old female with a new diagnosis of CML in chronic phase. A novel p210 fusion transcript splice variant was detected with qualitative reverse transcription PCR and capillary electrophoresis. Subsequent FISH study was performed, which revealed 86.5% positive for the BCR::ABL1 fusion. Quantitative real-time polymerase chain reaction (PCR) showed a negative result for the p210 fusion transcript. The variant was further characterized by Sanger sequencing. This variant is in-frame and predicted to be functional. This case illustrates the need for a combination of different testing techniques to fully characterize the rare BCR::ABL1 fusion transcripts.
1. Introduction
CML is a myeloproliferative neoplasm that is most frequently (>95%) diagnosed in the chronic phase. The clinical findings in the chronic phase can range from asymptomatic to splenomegaly, fatigue, malaise, weight loss, night sweats, and anemia [1].
Complete blood counts in CML patients show leukocytosis, absolute basophilia, eosinophilia, and monocytosis. Peripheral blood smears in the chronic phase can demonstrate multiple findings, including increased myelocytes and metamyelocytes and less than 2% blasts [2]. Bone marrow specimens are usually hypercellular, with marked proliferation of granulocytes and expansion at the myelocyte stage. Dysplasia is not significant. Blasts are usually <5% of the marrow cells [3]. However, the morphological findings are nonspecific and diagnosis relies on detection of the BCR::ABL1 fusion transcript or chromosomal rearrangement.
The BCR::ABL1 fusion transcript results from the joining of the 5′-end of the BCR gene to the 3′-end of the ABL1 gene. This causes the promoter region of the BCR gene to activate the tyrosine kinase domain of the ABL1 gene. The two most common isoforms of the p210 fusion transcript (M-BCR) are between exon 14 on BCR and exon 2 on ABL1 (e14a2) and between exon 13 on BCR and exon 2 of ABL1 (e13a2). Other isoforms of p210 fusion transcript include e14a3 and e13a3 [4]. The isoforms associated with the p190 fusion transcript (m-BCR) are e1a2 and e1a3. In the p230 fusion transcript (u-BCR), the possible isoforms are e19a2 and e19a3 [1].
According to the European LeukemiaNet organization, a combination of different testing techniques would need to be done to fully characterize the isoforms. These include cytogenetics, interphase FISH, and quantitative real-time and qualitative reverse transcription PCR [5].
2. Case Presentation
This is a 38-year-old female with no significant past medical history or family history who presented to the emergency department for lightheadedness for 3 weeks. Peripheral blood smear shows red blood cells with polychromasia and anisopoikilocytosis. Leukocytosis with absolute neutrophilia, monocytosis, and basophilia were noted. A shift to immaturity was seen with the neutrophils, including myelocytes, promyelocytes, and rare blasts suspicious for CML.
3. Materials and Methods
Quantitative PCR was performed on the Cobas z480 analyzer with the Ipsogen BCR-ABL1 Mbcr IS-MMR kit per manufacturer's protocol. Qualitative PCR was performed using long-range reverse transcription PCR, followed by size characterization on the ABI 3130xl Genetic Analyzer using 3 different primer sets with different fluorophores targeting p190, p210, and p230 isoforms (Figure 1). The forward and reverse primer sequences for detecting the p210 fusion transcript on the ABI 3130xl Genetic Analyzer were [FAM] GTGCAGAGTGGAGGGAGAAC and ACTGTTGACTGG-CGTGATGT, respectively. An abnormal reverse transcription PCR product size using the p210 primer set prompted further investigation by Sanger sequencing. Interphase FISH was performed with Vysis probes labeling ABL1 (9q34) with Spectrum Orange and BCR (22q11.2) with Spectrum Green (Figure 2). 200 interphase cells in total were evaluated for each probe by two technologists independently.
Figure 1.
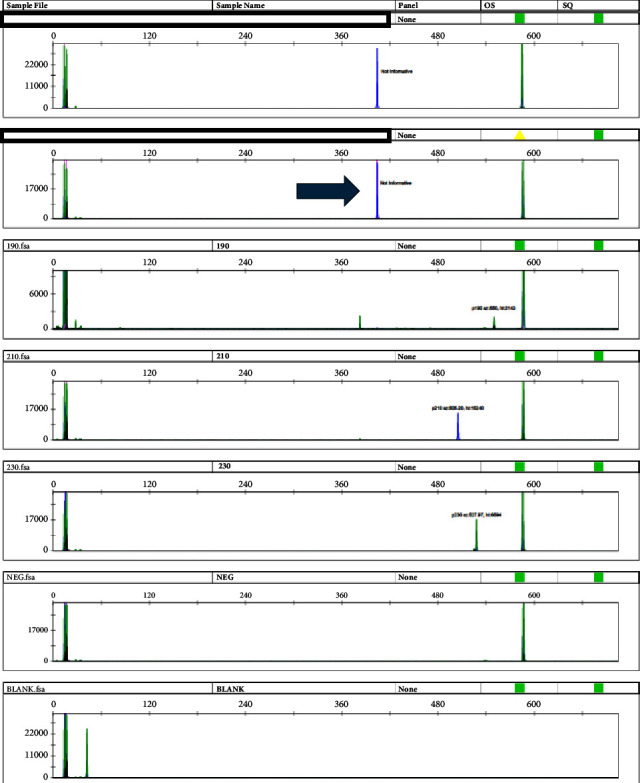
The qualitative reverse transcription PCR shows a positive p210 variant (arrow).
Figure 2.
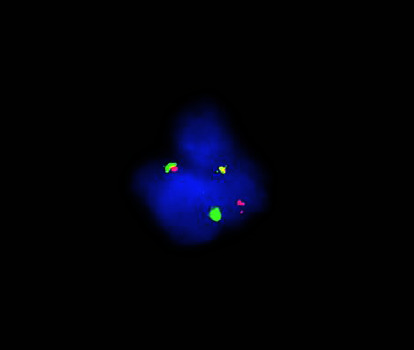
Break-apart FISH test using the BCR probe (green) and the ABL1 probe (orange) reveals a rearrangement with the two genes.
4. Results
A qualitative reverse transcription PCR test was done, which revealed a positive p210 splice variant. However, the splice variant size did not correspond to the typical isoforms (e14a2, e14a3, e13a2, or e13a3). A subsequent FISH study had confirmed the BCR::ABL1 rearrangement. The quantitative real-time PCR test used to monitor residual disease and targeting the most common fusion transcript isoforms (e14a2 and e13a2) was negative. Sanger sequencing was done to better characterize the fusion product, which revealed a novel isoform most closely related to the e13a2 isoform. The forward and reverse sequences had been run through a BLAST search, which revealed a missing portion of exon 13. In addition, a novel eight-nucleotide sequence, AACCCAAG, was inserted between the BCR and ABL1 sequences (Figure 3). It had also demonstrated that the forward primer site of the quantitative real-time PCR test was not included in this isoform explaining the negative result.
Figure 3.
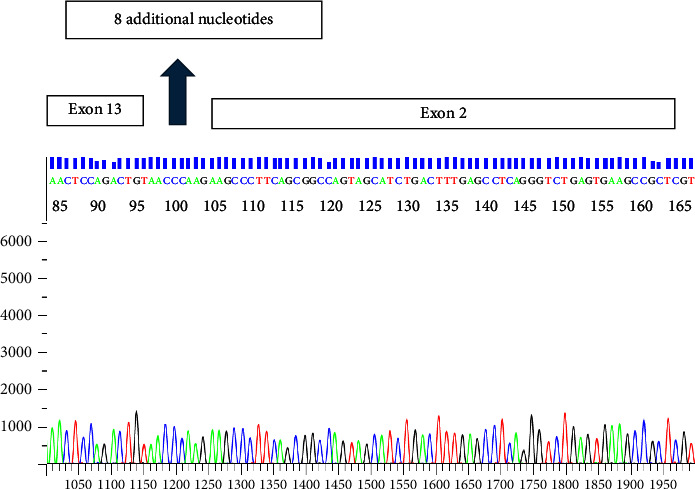
The sequence had been run through BLAST and ENSEMBL software programs. This portion of the electropherogram displays the site between exon 13 from the BCR gene and exon 2 of the ABL1 gene. The eight additional nucleotides–AACCCAAG–detected between the BCR and ABL1 are shown. The sequence AAGCCC is noted to be the first six nucleotides for the ABL1 gene.
5. Discussion
Variant BCR::ABL1 transcripts are seen in less than 5% of CML patients, most commonly due to alternative splicing of BCR and ABL1 exons. The most commonly involved isoforms are e6a2, e8a2, e13a3, and e14a3 for p210; e1a2 for p190; and e19a2 for p230 [6]. According to one study by Baccarani et al., the percentage of p190 fusion transcript isoforms is 0.93%, with e1a2 being 0.91% and e1a3 being 0.02%. The percentage of p230 e19a2 isoform is 0.31% [1].
Rare atypical breakpoints have been reported and can be organized into four categories. The first category contains BCR breakpoints located within the exons fused to ABL a2. The second category is characterized by BCR breakpoints located within the introns that are outside of the M-bcr, m-bcr, or μ-bcr fused to ABL a2. The third category features typical BCR breakpoints (M-bcr, m-bcr, or μ-bcr) fused to ABL breakpoints that are downstream of a2. The fourth category contains transcripts with intervening sequences between BCR and ABL a2, like the one described herein [7].
This patient had an in-frame fusion transcript that most closely resembles the e13a2 isoform. A partial deletion of exon 13 on BCR, insertion of eight additional nucleotides in exon 13 of BCR, and unaffected ABL1 exons 2 and 3 were noted. It has been shown in a previous study that the oncogenic properties of the BCR::ABL1 fusion product were not affected by the aforementioned changes on exon 13 along with the unaltered ABL1 exons [8]. In addition, the ABL1 SH3 domain is theorized to be involved with leukemogenesis by kinase domain (SH1) negative feedback and STAT5 signaling activation [8]. Due to the similarities between the changes seen on exon 13 of BCR in this patient and the unaffected ABL1 gene in both scenarios, it can be speculated that the isoform detected from this patient is functional. The patient subsequently received imatinib, which resulted in a complete cytogenetic and molecular (both p210 and p190) response.
Quantitative PCR has the best analytical measurement range for monitoring CML disease burden [7]. However, the failure to detect less common variants of BCR::ABL1 in quantitative real-time PCR testing can lead to false negative results. This is due to the PCR primers and probes being limited to their specific target regions. Therefore, according to the European LeukemiaNet organization, it is recommended to simultaneously do additional tests—FISH, cytogenetics, and qualitative PCR—for the diagnosis of CML in the chronic phase [5].
Cytogenetics can detect prognostically significant additional chromosomal aberrations (ACAs) but can miss complex rearrangements resulting in false negative results in up to 5% of the cases. Interphase FISH can detect all of the BCR::ABL1 rearrangements regardless of the breakpoint region but cannot characterize the isoform [5]. Qualitative reverse transcription PCR can identify and characterize the common and rare BCR::ABL1 transcript isoforms.
6. Conclusion
In summary, we present a patient with CML with a novel BCR::ABL1 fusion transcript, which is not detectable by quantitative real-time PCR. Diagnosis relied on detection by qualitative reverse transcription PCR and confirmation by FISH. This case highlights the need for a combination of different testing modalities for the diagnosis of CML as recommended by European LeukemiaNet organization.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethical Approval
Institutional Review Board approval was not required for this case report per institutional guidance as the information presented was obtained in the course of standard clinical care.
Consent
Written informed consent was not necessary for this case report per institutional guidance outlined in OHRP Exempt Categories 45 CFR 46.104–(HRP-312) Category 4, as the information presented was obtained in the course of standard clinical care (nonresearch).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
J.J. and L.J. wrote and edited the manuscript. D.D. performed and analyzed the Sanger sequence. M.S. interpreted and provided the FISH data. L.J. interpreted the qualitative and quantitative PCR results. J.G. interpreted the peripheral blood smear.
References
- 1.Baccarani M., Castagnetti F., Gugliotta G., et al. The proportion of different BCR-ABL1 transcript types in chronic myeloid leukemia. An international overview. Leukemia . 2019;33(5):1173–1183. doi: 10.1038/s41375-018-0341-4. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann V. S., Baccarani M., Hasford J., et al. The EUTOS population-based registry: incidence and clinical characteristics of 2904 CML patients in 20 European Countries. Leukemia . 2015;29(6):1336–1343. doi: 10.1038/leu.2015.73. [DOI] [PubMed] [Google Scholar]
- 3.Cortes J. E., Talpaz M., O’Brien S., et al. Staging of chronic myeloid leukemia in the imatinib era: an evaluation of the World Health Organization proposal. Cancer . 2006;106(6):1306–1315. doi: 10.1002/cncr.21756. [DOI] [PubMed] [Google Scholar]
- 4.Melo J. V. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype [editorial; comment] Blood . 1996;88(7):2375–2384. doi: 10.1182/blood.v88.7.2375.bloodjournal8872375. [DOI] [PubMed] [Google Scholar]
- 5.Cross N. C. P., Ernst T., Branford S., et al. European LeukemiaNet laboratory recommendations for the diagnosis and management of chronic myeloid leukemia. Leukemia . 2023;37(11):2150–2167. doi: 10.1038/s41375-023-02048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crampe M., Haslam K., Kelly J., Conneally E., Langabeer S. E. Characterization of a novel variant BCR-ABL1 fusion transcript in a patient with chronic myeloid leukemia: implications for molecular monitoring. Hematology/Oncology and Stem Cell Therapy . 2017;10(2):85–88. doi: 10.1016/j.hemonc.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Jinawath N., Norris-Kirby A., Smith B. D., et al. A rare e14a3 (b3a3) BCR-ABL fusion transcript in chronic myeloid leukemia: diagnostic challenges in clinical laboratory practice. Journal of Molecular Diagnostics . 2009;11(4):359–363. doi: 10.2353/jmoldx.2009.090008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu S., Hu Y., Fu Y., et al. Novel BCR-ABL1 fusion and leukemic mutations of SETBP1, PAX5, and TP53 detected by next generation sequencing in chronic myeloid leukemia. Cancer Biology & Therapy . 2016;17(10):1003–1009. doi: 10.1080/15384047.2016.1219821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


