Abstract
The myocyte enhancer factor 2B (MEF2B) transcription factor is frequently mutated in germinal center (GC)-derived B-cell lymphomas. Its ammino (N)-terminal mutations drive lymphomagenesis by escaping interaction with transcriptional repressors, while the function of carboxy (C)-terminal mutations remains to be elucidated. Here, we show that MEF2B C-tail is physiologically phosphorylated at specific residues and phosphorylation at serine (S)324 is impaired by lymphoma-associated mutations. Lack of phosphorylation at S324 enhances the interaction of MEF2B with the SWI/SNF chromatin remodeling complex, leading to higher transcriptional activity. In addition, these mutants show an increased protein stability due to impaired interaction with the CUL3/KLHL12 ubiquitin complex. Mice expressing a phosphorylation-deficient lymphoma-associated MEF2B mutant display GC enlargement and develop GC-derived lymphomas, when crossed with Bcl2 transgenic mice. These results unveil converging mechanisms of action for a diverse spectrum of MEF2B mutations, all leading to its dysregulation and GC B-cell lymphomagenesis.
Subject terms: B-cell lymphoma, B-cell lymphoma
N-terminal mutations in the transcription factor Myocyte Enhancer Factor 2B (MEF2B) are reported to drive lymphomagenesis. Here, the authors show that lymphoma-associated C-terminal mutations in MEF2B impair its phosphorylation, leading to increased stability and transcriptional activity to promote B-cell lymphomagenesis.
Introduction
Diffuse large B cell lymphoma (DLBCL) and follicular lymphoma (FL), are the most common forms of mature B cell malignancies1. Although the majority of patients (about 60%) are curable with current R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone) immunochemotherapy, the remaining are not cured and about ~20% of DLBCL patients are subject to early relapses with poor outcomes2,3. DLBCL and FL originate from the germinal centers (GCs), histological structures that form in the secondary lymphoid organs and where B cells undergo multiple rounds of immunoglobulin (Ig) gene editing, in the dark zone (DZ), followed by affinity-driven selection, in the light zone (LZ), in order to generate memory B cells and antibody secreting plasma cells4,5.
In the last two decades, numerous efforts have contributed to dissecting the heterogeneity of GC-derived lymphomas and the complex networks that are hijacked during malignant transformation. Two major DLBCL subtypes were identified based on gene expression profiling: the Germinal Center B cell-like (GCB) DLBCL, displaying transcriptional similarities to normal GC B cells; and the Activated B Cell-like (ABC) DLBCL, expressing genes that are induced by B cell receptor (BCR) signaling and activation and associated with a more unfavorable prognosis6,7. The analysis of mutations affecting the DLBCL coding genome has allowed a genetic classification showing several subgroups of prognostic significance8–14. Recently, the identification in DLBCL of a pervasive mutational mechanism, which targets the non-coding regulatory domains affecting the transcriptional regulation of multiple genes, including established oncogenes (such as BCL6, BCL2, and CXCR4), has added a further layer of complexity15.
Among the genes that are genetically targeted during lymphomagenesis the transcription factor MEF2B is somatically mutated in 11% of FL and 10% of DLBCL, with a prevalence in the EZB-DLBCL genetic subgroup8–12,14,16. MEF2B belongs to the MEF2 (myocyte enhancer factor 2) family of transcription factors that includes three additional members MEF2A, MEF2C, and MEF2D. MEF2 family members were initially identified as transcriptional modulators of myocyte differentiation and muscle development17–19. The N-terminus portion of MEF2 family members includes highly similar MADS and MEF2 domains that are involved in DNA binding and protein dimerization, while the C-terminus is divergent among MEF2 proteins and it is believed to play a role in transcriptional activation19–21. In mature B cells, MEF2B is exclusively expressed at the GC stage with two isoforms, isoform A (isoA) and isoform B (isoB), which differ in their C-terminal regions from amino acid (aa) 257 to the end22,23. MEF2B contributes to GC physiology by promoting BCL6 expression and regulating an array of genes involved in cell cycle, DNA replication and repair, apoptosis, differentiation, and GC B cell confinement22–24. In vivo Mef2b deletion in GC B cells reduces GC formation and deletion of all Mef2 family members completely abrogates GC formation, indicating a functional redundancy between MEF2B and other MEF2 members23.
By analyzing lymphoma-associated MEF2B mutations, we previously identified a group of highly recurrent MEF2B N-terminal mutations which lead to escape from multiple layers of negative regulation by impairing interaction with the HUCA (HIRA/UBN1/CABIN1/ASF1a) complex and class IIa histone deacetylases (HDACs)22,23. The most frequent lymphoma-associated N-terminal MEF2B mutant (D83V)8,10,13,22,25 shows hypomorphic features, including reduced DNA binding and protein stability, yet it acts dominantly due to its deregulated constitutive activity23,25,26. Mice conditionally expressing Mef2bD83V in GC B cells displayed enlarged GCs due to LZ expansion and, when crossed with Bcl2 transgenic mice, to mimic a common genetic lesion co-occurring with MEF2B mutations in human lymphoma, developed tumors recapitulating the phenotypic features of human FL and DLBCL23.
The C-terminus half of MEF2B is also targeted by genetic alterations, accounting for one-third of MEF2B-mutated cases and including nonsense, frameshift, and missense mutations that are scattered and rarely recurrent. The C-terminal mutations seem to affect the C-tail and its post-translational modifications22,23. However, the extent and function of post-translational modifications and their connections to the C-terminal mutations remain largely unexplored.
In this study we examine the function and consequence of the C-terminal mutations and their contribution to lymphomagenesis.
Results
Lymphoma-associated mutations targeting the C-terminus of MEF2B
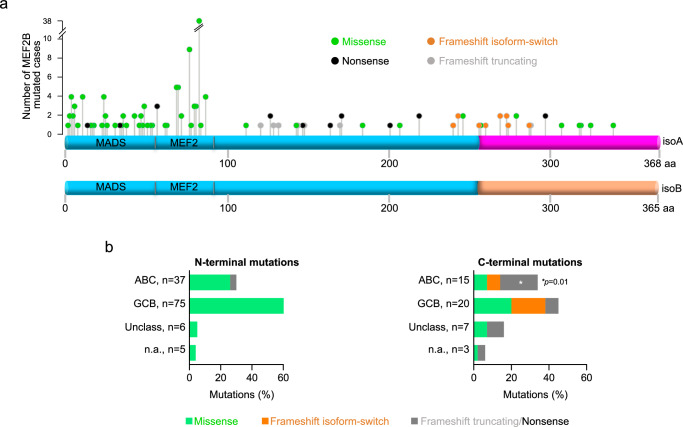
The MEF2B mutational landscape in DLBCL and FL displays a bimodal distribution pattern with highly recurrent missense mutations targeting the N-terminal domain of the MEF2B protein (aa1-88), and scattered mutations of different types, including missense, frameshift, and nonsense, in the C-terminus (Fig. 1a)8–12,16. Previously, we have functionally characterized N-terminus mutants and reported that mutations targeting the C-terminus appeared to interfere with the expression of MEF2B isoform A (isoA) in favor of isoform B (isoB) or to eliminate the C-tail of the protein22,23. Based on the analysis of the currently available expanded collection of sequencing data (1,412 DLBCL and 142 FL primary cases)8–12,16, we confirm here that MEF2B C-terminal mutations represent ~30% of the total and are heterogeneous for position and type. A sizable fraction of them (n = 11/45; 24.4% of MEF2B C-terminal mutants, hereafter referred to as isoform-switch) affect the isoform balance by encoding mutant proteins that resemble the wild-type (WT) isoB, which, although less abundant (20% of the MEF2B transcripts), displays higher transcriptional activity and stability compared to isoA (Fig. 1a and Supplementary Fig. 1)22,23. A second group of mutations (n = 18/45, 40%) lead to C-tail truncated forms of the MEF2B protein by introducing frameshifts that result in early termination (G121fs, R129fs, P132fs, Y144fs, P148fs, F170fs, and A288fs), or by generating stop codons (R127X, L147X, Q164X, R171X, Y201X, R219X, and R297X) (Fig. 1a and Supplementary Fig. 1). Lastly, the C-terminal region is also targeted by a fraction of missense mutations (n = 16/45, 35.5%), the function of which remains to be explored (Fig. 1a).
Fig. 1. Lymphoma-associated C-terminal mutations target the C-tail region of MEF2B isoform A.
a Distribution of MEF2B missense (green), nonsense (black), and frameshift mutations that are predicted to generate truncated proteins (truncating, gray) or proteins with an isoform B tail (isoform-switch, orange) in primary cases of DLBCL (147/1,412; 10.4%) and FL (16/142; 11.3%). The y-axis indicates the number of MEF2B mutated cases reported in8–12,16. The bottom schemes represent MEF2B isoform-A (isoA) and -B (isoB) proteins, respectively. The mutations are aligned with the targeted amino acid in isoA. The distinct C-tails of isoA and isoB are displayed in purple and orange respectively. b Percentage of N-terminal and C-terminal MEF2B mutations in primary cases, grouped according to the Cell-Of-Origin classification (ABC, Activated B Cell-like; GCB, Germinal Center B cell-like; Unclass, unclassified). The *p value was measured by Fisher’s Exact test (two-tailed). Source data are provided as a Source Data file.
Although more frequent in GCB-DLBCL, MEF2B mutations are detected in both DLBCL subtypes8–12,16 (Fig.1b). Nonetheless, the C-terminal mutations appear to segregate by type in the DLBCL subgroups with truncating mutations (both frameshift and stop codon) significantly enriched in the ABC-DLBCL, and missense and isoform-switch mutations mostly associated with GCB-DLBCL cases (Fig. 1b).
Overall, these alterations appear to replace (isoform-switch mutations) or eliminate (truncating mutations) the C-tail of MEF2B isoA protein. These observations, together with our previously reported data showing that phosphorylation of MEF2B C-tail negatively modulates MEF2B transcriptional activity22, suggest a critical functional role of the MEF2B C-terminal region.
MEF2B is phosphorylated in GC B cells
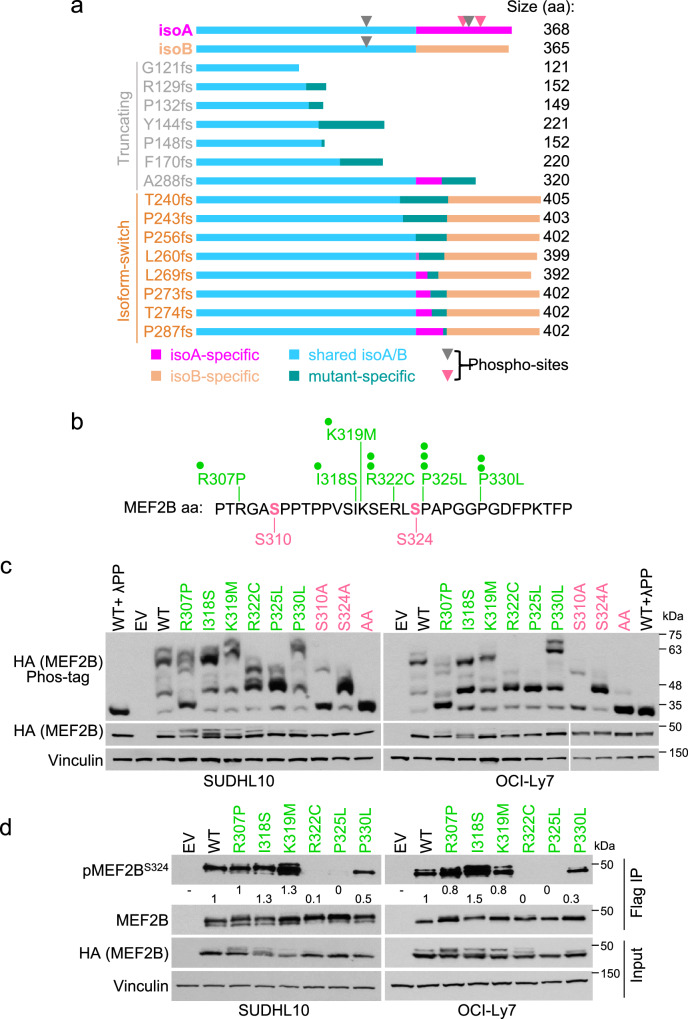
Our previous observation that MEF2B can be phosphorylated22, was confirmed and expanded to normal and malignant GC B cells using a phos-tag gel approach (Fig. 2a). In order to characterize the full pattern of MEF2B phosphorylation and map the phosphorylation sites, we performed Flag and HA sequential immunoprecipitation (IP) followed by mass spectrometry (MS) analysis in a DLBCL cell line (SUDHL10) that was engineered to express Flag-HA double-tagged MEF2B isoA. We identified recurrent phosphorylation at T196 (present in both isoA and isoB), and at the isoA-specific aa S310, T313, and S324 (Supplementary Fig. 2a), which were all confirmed in normal GC B cells (Fig. 2b). Considering the fraction of phosphorylated/unphosphorylated peptides, we estimated that phosphorylation at S310 and S324 accounted for the large majority of phosphorylated peptides (>95%) (Fig. 2b). We then engineered SUDHL10 cells to express Flag-HA double-tagged phosphorylation-deficient isoA mutants (serine or threonine residues were mutated to alanine) and analyzed changes in MEF2B phosphorylation pattern by phos-tag gel. Consistent with the MS results, the S310A and S324A mutants displayed dramatic changes in the phosphorylation pattern compared to MEF2B WT-isoA, whereas expression of T196A and T313A mutants seemed not to alter the overall phosphorylation features (Fig. 2c). These results were confirmed by phos-tag analysis in multiple DLBCL cell lines that were engineered to express the S310A and S324A mutants (Supplementary Fig. 2b).
Fig. 2. MEF2B isoform A is mainly phosphorylated at S310 and S324 in normal and malignant GC B cells.
a Phos-tag gel analysis of MEF2B phosphorylation in DLBCL cell lines and normal GC B cells isolated from human tonsil. Controls (MEF2B and Actin) were resolved on a Tris-Glycine gel. Actin was used as loading control; lambda phosphatase (λPP)-treated lysate was used as negative control for phosphorylation detection. b Schematic representation of MEF2B phosphorylation sites as identified by mass spectrometry (MS) analysis upon Flag and HA sequential immunoprecipitation (IP) in SUDHL10 cells expressing Flag-HA MEF2B WT-isoA and upon MEF2B IP in normal GC B cells isolated from human tonsil. Percentages associated to each phosphorylation site represent the fraction of phosphorylated peptides detected in normal GC B cells. c Phos-tag analysis of SUDHL10 DLBCL cell line that was engineered to express Flag-HA MEF2B WT-isoA or phosphorylation-deficient mutants T196A, S310A, T313A, S324A and S310A + S324A (AA). Controls (MEF2B and Vinculin) were resolved on a Tris-Glycine gel. Vinculin was used as loading control; λPP treated lysate was used as negative control for phosphorylation detection. d Flag (MEF2B) IP followed by immunoblotting (IB) to detect S324- or S310-phosphorylated MEF2B in SUDHL10 and OCI-Ly7 cells expressing Flag-HA MEF2B WT-isoA or phosphorylation-deficient mutants. Vinculin was used as loading control; λPP-treated lysate was used as negative control for phosphorylation detection. e MEF2B IP followed by IB to detect S324- or S310-phosphorylated MEF2B in nuclear extracts of SUDHL10 and OCI-Ly7 DLBCL cells, and normal GC B cells isolated from human tonsil. Lamin B1 was used as a nuclear fraction control; IgG IP was used as negative control. Asterisk, non-specific band. The experiments were repeated twice in (a) and (d); three times in (e); six times in (c) with similar results. Source data are provided as a Source Data file.
Consistently, antibodies generated to specifically recognize MEF2B phosphorylation at S310 and S324 detected the phosphorylated WT protein upon IP (Supplementary Fig. 2c), but not the S310A or S324A phosphorylation-deficient MEF2B mutants (Fig. 2d). As expected, treatment of the protein lysate with lambda-phosphatase completely abrogated the detection (Fig. 2d). Using the phospho-specific antibodies, we confirmed that endogenous MEF2B protein is phosphorylated at S310 and S324 in both malignant and normal GC B cells (Fig. 2e). Phosphorylation at S310 and S324 occurs independently, as shown by IP followed by MS analysis or immunoblotting with phospho-specific antibodies in cell lines expressing the S310A or S324A mutants (Fig. 2d and Supplementary Fig. 2d).
In conclusion, MEF2B is abundantly phosphorylated at S310 and S324 in GC B cells.
MEF2B C-terminal mutations impair S324 phosphorylation
To further investigate the relationship between C-terminal frameshift or missense mutations and phosphorylation, we compared the predicted MEF2B mutant proteins to MEF2B isoA C-tail, which includes S310 and S324. All frameshift truncated proteins were predicted to lack the region of interest with the minimum common deletion starting at position 297 of isoA (Fig. 3a). In addition, frameshift mutations that lead to an isoform-switch, replaced the isoA tail with the isoB sequence and therefore lacked the S310 and S324 phosphorylation sites (Fig. 3a). Thus, frameshift mutations resulted in truncated mutants or in isoB-like proteins, both of which lacked the isoA-specific C-terminal phosphorylation region. The majority (12/14) of the truncated mutants displayed low or no protein expression, when exogenously expressed in DLBCL cell lines (Supplementary Fig. 3 and “Discussion”).
Fig. 3. MEF2B C-terminal missense and frameshift mutations impair S324 phosphorylation.
a The scheme shows the amino acid alignment of MEF2B WT-isoA with MEF2B WT-isoB and the predicted truncated and isoform-switch mutants. The identified phosphorylation sites are marked by triangles (rose red, sites with phosphorylation >95%; gray, phosphorylation <1%). b Schematic representation of the phosphorylation region in MEF2B isoA aa sequence. Phosphorylated aa are labeled in rose red. Amino acid changes found in lymphoma cases are reported on the top using green dots, each representing one patient8–12,16,27. c Phos-tag analysis of MEF2B phosphorylation in SUDHL10 and OCI-Ly7 DLBCL cell lines that were engineered to express Flag-HA MEF2B WT-isoA, lymphoma-associated missense mutants (R307P, I318S, K319M, R322C, P325L and P330L) or phosphorylation-deficient mutants (S310A, S324A and S310A + S324A, labeled as AA). Controls (HA and Vinculin) were resolved on a Tris-Glycine gel. Vinculin was used as loading control; λPP-treated lysate was used as a negative control for phosphorylation detection. d Flag (MEF2B) IP followed by IB with phospho-specific pMEF2BS324 antibody in SUDHL10 and OCI-Ly7 cells that were engineered to express Flag-HA MEF2B WT-isoA or lymphoma-associated missense mutants (R307P, I318S, K319M, R322C, P325L and P330L). Vinculin was used as loading control. Quantifications were normalized to the immunoprecipitated MEF2B (HA). Fold changes were calculated relative to the MEF2B WT sample. The experiments displayed in (c) and (d) were repeated 4 times with similar results. Source data are provided as a Source Data file.
Several missense mutations appeared to target the phosphorylation region, in particular the aa surrounding the S324 site (Fig. 3b)8–12,16,27. In order to test the effects of these missense mutations on S324 phosphorylation, we generated SUDHL10 and OCI-Ly7 cell lines expressing double-tagged MEF2B missense mutant proteins and analyzed their phosphorylation profile by phos-tag assay in comparison to the MEF2B WT-isoA and the phospho-deficient mutants (S310A and S324A). The results showed that R322C and P325L mutants display a phosphorylation pattern similar to S324A mutant, suggesting impaired phosphorylation at S324 (Fig. 3c). No mutants displayed a pattern resembling the S310A mutant, suggesting that missense mutations are targeting phosphorylation at S324, but not at S310 (Fig. 3c). The cell lines expressing the MEF2B mutants were analyzed by immunoblotting using the pMEF2BS324 antibody, upon Flag IP, confirming abrogation (R322C and P325L) or reduction (P330L) of S324 phosphorylation by MEF2B mutants (Fig. 3d). In order to exclude that the impaired detection was due to reduced affinity of the pMEF2BS324 antibody because of the mutations affecting residues in the phosphorylation motif, we performed MS analysis in SUDHL10-Flag-HA-MEF2B-R322C cells and confirmed the lack of phosphorylation at S324 (Supplementary Fig. 2d).
Taken together, our data show that MEF2B C-terminal mutations eliminate, replace, or target the C-tail region that is phosphorylated at S324, suggesting a relevant role for S324 phosphorylation in the modulation of MEF2B function.
Enhanced binding to SWI/SNF complex and transcriptional activity of mutants
In order to investigate whether MEF2B phosphorylation at S324 affects MEF2B protein complex formation, we performed MS analysis upon MEF2B immunoprecipitation in SUDHL10 cells expressing MEF2B WT-isoA, WT-isoB, or P325L. The list of interactors was filtered for common contaminants and for proteins with low abundance (intensity values in all the analyzed samples less than 1E + 04). The results confirmed the interaction of MEF2B WT-isoA with previously reported interactors, including the HUCA complex, HDAC4, HDAC5, and HDAC723,28. A subset of interactors (653/1,996; 33%) displayed affinity changes that were consistent in both MEF2B WT-isoB and P325L compared to MEF2B WT-isoA (Supplementary Data 1).
Among the interactors that displayed higher binding affinity to MEF2B WT-isoB and P325L, we identified the switch/sucrose non-fermentable (SWI/SNF) chromatin remodeling complex29,30, as a notable candidate among the protein complexes for which multiple subunits were detected (Supplementary Data 1). The SWI/SNF complex is involved in chromatin unwinding and therefore it may contribute to promote MEF2B transcriptional activity. Additional IP assays followed by MS analysis in normal GC B cells and in engineered DLBCL cell lines expressing MEF2B WT-isoA or P325L confirmed the enhanced interaction between MEF2B phospho-deficient mutant and multiple subunits of the SWI/SNF chromatin remodeling complex, including the SMARCA4 ATPase subunit (Supplementary Data 2). To further validate the interaction between MEF2B and SMARCA4, we performed co-IP for the endogenous proteins and identified their interaction in the nucleus of both malignant and normal GC B cells (Fig. 4a). Nuclear/cytoplasm fractionation in normal GC B cells and in DLBCL cell lines confirmed that MEF2B and SMARCA4 are detected exclusively in the nucleus (Supplementary Fig. 4a). In addition, protein proximity labeling in SUDHL10 cells engineered to express MEF2B fused with a biotinylating enzyme (MEF2B-TurBoID)31 confirmed the interaction of MEF2B/SMARCA4 in GC B cells (Supplementary Fig. 4b).
Fig. 4. MEF2B interacts with the SWI/SNF complex.
a Detection of MEF2B/SMARCA4 interaction by MEF2B immunoprecipitation (IP) followed by immunoblotting (IB) using nuclear extracts of SUDHL10 cells or normal GC B cells isolated from human tonsil. Lamin B1 was used as a nuclear fraction control. Normal GC B cell samples are identical to those used in Fig. 2e. The experiment was repeated twice. b Heat maps showing the read density and distance plots displaying the distribution of MEF2B or SMARCA4 bound regions in relationship to each other closest peak, as detected in normal GC B cells by Cut&Run-seq analysis. In the heat maps, data is normalized to number of reads per bin/sum of all reads per bin (in millions). c Genomic binding profile of MEF2B and SMARCA4 at BCL6, CCND3, CXCR4, CD83, CD86 and CD79A loci, as detected by Cut&Run-seq analysis in normal GC B cells. Significantly bound regions that were recurrently detected in both biological replicates (2 independent donors) are displayed as bars under the read profiles. MEF2B/SMARCA4 overlapping bound regions are displayed as black bars at the bottom (MEF2B/SMARCA4 overlap). Genomic coordinates based on hg38. BPM, bins per million map reads. Source data are provided as a Source Data file.
In order to investigate the transcriptional program modulated by MEF2B in cooperation with the SWI/SNF complex, we analyzed the chromatin binding profiles of MEF2B and SMARCA4 in normal human GC B cells by Cut&Run followed by sequencing. MEF2B binding profile identified by Cut&Run was largely overlapping (~55% peaks) with our previous characterization by ChIP-seq (~66% of ChIP-seq peaks were identified by Cut&Run). Of note, co-occupancy of MEF2B and SMARCA4 was detected in ~74% of SMARCA4 and ~50% of MEF2B-bound regions (Fig. 4b), which were annotated as promoters (50%; identified as −2Kb/+1Kb from the TSS), intragenic (20%) and intergenic (30%) regions. Overall, we identified 5,661 genes that were associated with the MEF2B/SMARCA4 co-bound regions and were expressed in mature B cells (Supplementary Data 3). Pathway enrichment analysis for these genes using the Hallmark, KEGG and Biocarta databases32, largely recapitulated the overall MEF2B program, including genes involved in cell cycle, DNA repair, apoptosis, and GC confinement (Supplementary Data 4)23. The same analysis on the genes associated with MEF2B in the absence of SMARCA4 did not reveal enrichment for additional pathways (Supplementary Data 4). SMARCA4 co-localizes with MEF2B in previously identified MEF2B-bound regions of the BCL6 locus both in the promoter and in the upstream super-enhancer (Fig. 4c)22,24. In addition, the SMARCA4/MEF2B co-bound targets included several DZ (i.e., CXCR4, CCND3) and LZ (i.e., CD83, CD86) markers, as well as genes involved in signaling pathways that are relevant for the affinity-based selection occurring in the LZ, including B cell receptor, NF-κB, and MAPK pathways (Fig. 4c and Supplementary Data 4).
Consistent with the MS analysis, co-IP in SUDHL10 cells expressing the phospho-deficient mutants (S324A and the lymphoma-associated R322C, P325L, and P330L) and in OCI-Ly7 isogenic clones edited to endogenously express S324A or P325L showed an increase in the amount of SMARCA4 that was immunoprecipitated by MEF2B mutants compared to the MEF2B WT-isoA, suggesting that the lack of phosphorylation promotes the interaction between MEF2B proteins and SMARCA4 (Fig. 5a, b and Supplementary Fig. 4c, d). As expected, the phospho-mimic S324E mutant did not show enhanced binding to SMARCA4 in both the overexpression and endogenous cell systems (Fig. 5a, b and Supplementary Fig. 4c, d). In addition, the double mutant S324E/P325L (EL), in which the phospho-mimic S324E was combined with the phospho-deficient P325L mutant, behaved like the single S324E mutant (Fig. 5a and Supplementary Fig. 4c). The enhanced interaction with the SWI/SNF chromatin remodeling complex may contribute to the increased transcriptional activity that is associated with the phospho-deficient mutants, as shown by luciferase reporter assay using an artificial reporter including canonical MEF2B binding sites (3xMEF2) (Fig. 5c).
Fig. 5. Phosphorylation-deficient MEF2B mutants display enhanced binding affinity to SWI/SNF complex and increased transcriptional activity.
a Detection of MEF2B/SMARCA4 interaction by Flag (MEF2B) immunoprecipitation (IP) followed by immunoblotting (IB) from SUDHL10 cells expressing Flag-HA-MEF2B-WT-isoA or missense mutants (EL, S324E + P325L). Lamin B1, loading control. Quantifications were normalized to the immunoprecipitated MEF2B (HA). Fold changes relative to MEF2B-WT. b MEF2B IP followed by IB from isogenic OCI-Ly7 control (Neutral) and MEF2B missense mutant clones. IgG, IP negative control. Quantifications were normalized to the immunoprecipitated MEF2B. Fold changes relative to the neutral clones. c Luciferase activity driven by synthetic wild-type (3xMEF2-WT) or mutated (3xMEF2-Mut) MEF2 binding sites, as measured in HEK293T cells transfected with empty vector (EV) or plasmids expressing MEF2B-WT-isoA (n = 15, 5 experiments) or the missense mutants S324A (n = 15, 5 experiments), S324E (n = 6, 2 experiments), P325L (n = 15, 5 experiments), EL (n = 6, 2 experiments), R322C (n = 9, 3 experiments) and P330L (n = 9, 3 experiments). Data are shown as average fold change ± SD relative to MEF2B-WT-isoA of indicated independent experiments, each performed in triplicates. Significance assessed by two-tailed Mann-Whitney test. d Flag (MEF2B) IP followed by IB from SUDHL10 cells expressing Flag-HA-MEF2B-WT-isoA, MEF2B-WT-isoB or isoform-switch mutants. Lamin B1, loading control. Quantifications were normalized to the immunoprecipitated MEF2B (HA). Fold changes relative to MEF2B-WT-isoA. e MEF2B IP followed by IB from isogenic OCI-Ly7 control (Neutral) and MEF2B isoform-switch mutant clones. IgG, IP negative control. Quantifications were normalized to the immunoprecipitated MEF2B. Fold changes relative to the neutral clones. f Luciferase activity of synthetic WT or mutated 3xMEF2-luc reporter in HEK293T cells transfected with EV or plasmids expressing MEF2B-WT-isoA (n = 18, 6 experiments), -isoB (n = 18, 6 experiments) or isoform-switch mutants T240fs (n = 9, 3 experiments), P243fs (n = 9, 3 experiments), P256fs (n = 9, 3 experiments), L260fs (n = 9, 3 experiments), L269fs (n = 9, 3 experiments), P273fs (n = 6, 2 experiments), T274fs (n = 9, 3 experiments) and P287fs (n = 9, 3 experiments). Data shown as average ± SD relative to MEF2B-WT-isoA of indicated independent experiments, each performed in triplicates (two-tailed Mann-Whitney test). The experiments reported in (a) and (d) were repeated three times with similar results. Source data are provided as a Source Data file.
Consistently, both the WT-isoB protein and the isoform-switch mutants expressing iso-B-like C-tails, which lack the S324 phosphorylation site, displayed enhanced interaction with SMARCA4 both in the over-expression and endogenous cell systems (Fig. 5d, e and Supplementary Fig. 4e, f). In addition, similar to the missense mutants, all these proteins showed an increased transcriptional activity compared to MEF2B WT-isoA (Fig. 5f). Collectively, these data show that MEF2B phosphorylation at S324 contributes to the dissociation between MEF2B and the SWI/SNF complex. C-terminal mutants, which cannot be phosphorylated at S324, display an enhanced interaction with SMARCA4 and increased transcriptional activity.
Stabilization of mutants by dissociation from the CUL3/KLHL12 complex
Among the interactors displaying lower affinity for MEF2B WT-isoB and P325L mutant, our analysis identified several subunits of the CUL3 ubiquitin E3 ligase complex, including Broad-Complex, Tramtrack and Bric a brac domain (BTB) adaptor proteins (Supplementary Data 1). In order to refine this analysis and identify proteins interacting with both MEF2B and CUL3, we performed MS analysis upon sequential immunoprecipitations of Flag (MEF2B) and CUL3 in the SUDHL10-Flag-HA-MEF2B WT-isoA cells. The results confirmed and expanded the list of MEF2B interactors that belong to the CUL3 ubiquitin E3 ligase complex (Supplementary Data 5). Of note, we identified several adaptor proteins the most abundant of which was the KLHL12 BTB-containing protein (Supplementary Data 5)33,34. These results suggest that KLHL12 may act as the adaptor protein, which recruits MEF2B in the CUL3 E3 ligase ubiquitin complex for ubiquitin-mediated degradation.
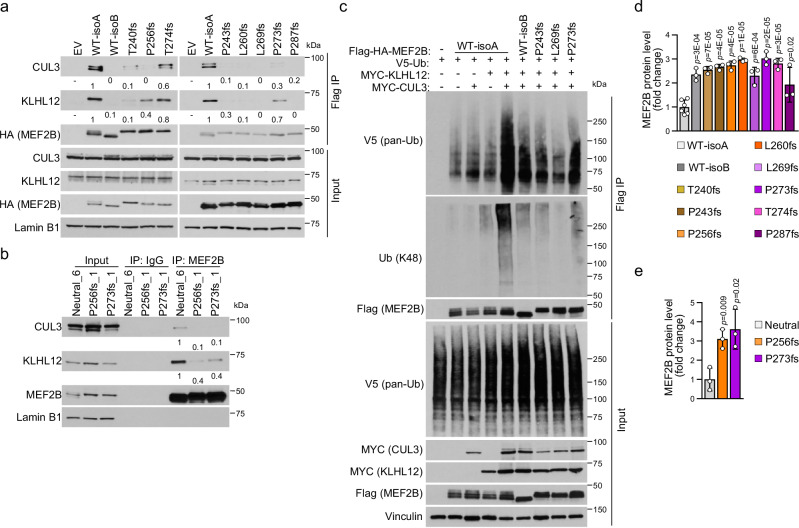
The native interaction between MEF2B, KLHL12 and CUL3 was confirmed in nuclear extracts by co-IP in both malignant and normal GC B cells (Fig. 6a). Nuclear/cytoplasm fractionation in normal GC B cells and in DLBCL cell lines confirmed expression of CUL3 and KLHL12 in both the cytoplasm and nucleus (Supplementary Fig. 5a). Protein proximity labeling in SUDHL10 cells expressing MEF2B-TurBoID protein confirmed the interaction of MEF2B/KLHL12/CUL3 (Supplementary Fig. 5b).
Fig. 6. Stabilization of MEF2B C-terminal missense mutants by dissociation from the CUL3/KLHL12 complex.
a Detection of MEF2B/CUL3/KLHL12 interaction by MEF2B immunoprecipitation (IP) followed by immunoblotting (IB) from SUDHL10 and normal GC B cells. Lamin B1, nuclear fraction control. Asterisks, non-specific bands. GC B cell samples are same as in Fig. 2e. b Flag (MEF2B) IP followed by IB in SUDHL10 cells expressing Flag-HA-MEF2B-WT-isoA or missense mutants. Lamin B1, loading control. Quantifications were normalized to the immunoprecipitated MEF2B (HA). Fold changes relative to MEF2B-WT. Samples in the left panel are same as in Fig. 5a. c MEF2B IP followed by IB using nuclear extracts of isogenic OCI-Ly7 control (Neutral) and MEF2B missense mutant clones. IgG, IP negative control. Quantifications were normalized to the immunoprecipitated MEF2B. Fold changes were calculated relative to the neutral clones. Samples are same as in Fig. 5b. d Ubiquitination assay upon Flag (MEF2B) IP followed by IB of HEK293T cells co-transfected with expression vectors for Flag-HA-MEF2B-WT-isoA or missense mutants, and V5-Ub, Myc-KLHL12 and Myc-CUL3. Total ubiquitin and K48 ubiquitin chain type were detected. The samples derive from the same experiment but different gels for Ub(K48) in the IP samples and V5 (pan-Ub) in the input samples were processed in parallel. e MEF2B relative expression level, upon CHX treatment (12 h) in SUDHL10 cells expressing Flag-HA-MEF2B-WT-isoA (n = 13) or the missense mutants S324A (n = 6), S324E (n = 3), P325L (n = 8), EL (n = 3), R322C (n = 3) and P330L (n = 3). Numbers (n) refer to independent experiments. Complete CHX pulse-chase experiment in Supplementary Fig. 7b. Average fold change ± SD relative to MEF2B-WT-isoA (two-tailed unpaired T test). f MEF2B relative expression level, upon CHX treatment (12 h) in isogenic OCI-Ly7 control (Neutral; n = 6) and MEF2B-P325L mutant (n = 6) clones. Complete CHX pulse-chase experiment in Supplementary Fig. 7c. Average fold change ± SD relative to neutral clones of two independent experiments (3 neutral compared to 3 P325L clones in each experiment; two-tailed unpaired T test). The experiment in (a), (b), and (d) were repeated three times with similar results. Source data are provided as a Source Data file.
In order to map the interaction of MEF2B and KLHL12, we performed co-IP assays on cells co-transfected with full-length or deletion mutants of both MEF2B and KLHL12 proteins (Supplementary Fig. 5c). The results showed that MEF2B full-length binds to both the N-terminus and the C-terminus of KLHL12, with the N-terminus of MEF2B being associated with the N-terminus of KLHL12. Although the MEF2B C-terminus by itself did not show binding to KLHL12 in this assay, the fact that (i) we detected binding between KLHL12 C-terminus and MEF2B full-length, (ii) the MEF2BP325L mutant displayed reduced binding affinity to KLHL12, and (iii) the canonical binding of KLHL12 to approximately ten amino acids of its substrates occurs through the C-terminus35–37, prompted us to test an oligopeptide surrounding the S324 phosphorylation region. Indeed, the binding of KLHL12 C-terminus in the MEF2B C-terminus was efficiently detected using this oligopeptide (M7 in Supplementary Fig. 5c). These results suggest that the interaction between KLHL12 and MEF2B requires the MEF2B N-terminus to retain the proper configuration of the C-terminus, and the ~270 aa C-terminus is a highly dynamic entity to be stabilized by interaction of merely ten residues with the KLHL12 Kelch domain. Thus, we mapped the interaction of MEF2B and KLHL12 to both the MEF2B N-terminal domain and the C-terminal phosphorylation region containing S324 (aa304-333), which displayed binding to KLHL12 N-terminus (BTB-BACK domain) and C-terminus (Kelch domain), respectively (Supplementary Fig. 5c).
In order to confirm that MEF2B is recruited by KLHL12 into the CUL3 E3 ligase complex for ubiquitin-mediated degradation, we co-expressed MEF2B with the KLHL12-AMF mutant (A60E, M61E and F62A) that cannot interact with CUL338. The results showed that KLHL12-AMF mutant was unable to bind CUL3 and, when co-expressed with MEF2B, failed to mediate MEF2B degradation (Supplementary Fig. 5d, e).
Co-IP in SUDHL10 cells expressing the phospho-deficient mutants (S324A or the lymphoma-associated R322C, P325L and P330L) and in OCI-Ly7 isogenic clones edited to endogenously express S324A or P325L confirmed the impaired interaction with KLHL12 and CUL3, while the phospho-mimic S324E mutant behaved similarly to the MEF2B WT-isoA protein (Fig. 6b, c and Supplementary Fig. 6a, b). MEF2B ubiquitination, detected both in SUDHL10 cells and in transient transfection assays, was reduced in the phospho-deficient mutants consistent with the impaired interaction with the KLHL12/CUL3 complex (Fig. 6d and Supplementary Fig. 6c, d). Accordingly, we detected poly-ubiquitin chains linked to K48, a known signal for protein degradation by the proteasome39(Fig. 6d). Ubiquitination not associated with protein degradation (i.e., K63-linked) was not detected (Supplementary Fig. 6e). Consistently, a significant increase of MEF2B protein stability and half-life was detected in SUDHL10 cells expressing the S324A, R322C, P325L or P330L mutants and in the OCI-Ly7 isogenic clones edited to endogenously express P325L (Fig. 6e, f and Supplementary Fig. 7). We note that although the S324E/P325L double mutant displays a protein stability similar to the WT (Fig. 6e), it remains deficient in its binding to KLHL12 and CUL3 (Fig. 6b), suggesting that alternative adaptor proteins may be involved in restoring the protein degradation of this artificial mutant.
The impaired interaction and ubiquitin-mediated degradation was not limited to the missense mutants affecting MEF2B phosphorylation at S324. In line with the fact that MEF2B WT-isoB lacks the S324 phosphorylation site, both the WT-isoB protein and the isoform-switch mutants displayed impaired interaction with KLHL12 and CUL3 both in the over-expression and endogenous cell contexts (Fig. 7a, b and Supplementary Fig. 8a, b). Similar to the missense mutants, the isoform-switch mutants showed a reduced ubiquitination, and extended half-life compared to MEF2B WT-isoA (Fig. 7c–e and Supplementary Fig. 8c–e).
Fig. 7. Stabilization of MEF2B C-terminal isoform-switch mutants by dissociation from the CUL3/KLHL12 complex.
a Detection of MEF2B/CUL3/KLHL12 interaction by Flag (MEF2B) immunoprecipitation (IP) using nuclear extracts of SUDHL10 cells expressing Flag-HA-MEF2B-WT-isoA, -isoB or isoform-switch mutants. Lamin B1 was used as loading control. Quantifications were normalized to the immunoprecipitated MEF2B (HA). Fold changes were calculated relative to the MEF2B WT-isoA sample. b MEF2B IP followed by immunoblotting (IB) using nuclear extracts of isogenic OCI-Ly7 clones including controls (Neutral) and cells expressing MEF2B isoform-switch (P256fs and P273fs) mutants. IgG IP was used as negative control. Quantifications were normalized to the immunoprecipitated MEF2B. Fold changes were calculated relative to the neutral clones. Samples are the same as in Fig. 5e. c Ubiquitination assay upon Flag (MEF2B) IP followed by IB using total extracts of HEK293T cells co-transfected with expression vectors for Flag-HA-MEF2B-WT-isoA, -isoB or isoform-switch mutants (P243fs, L269fs, and P273fs), and V5-Ub, Myc-KLHL12 and Myc-CUL3. Total ubiquitin and K48 ubiquitin chain type were detected. The samples derive from the same experiment but different gels for Ub(K48), in the IP samples, and V5 (pan-Ub), in the input samples, were processed in parallel. d Relative MEF2B protein levels, as measured upon CHX treatment for 12 h in SUDHL10 cells expressing Flag-HA-MEF2B-WT-isoA (n = 6) or isoform-switch mutants (n = 3 each). Numbers (n) refer to independent experiments. The complete CHX pulse-chase experiment is reported in Supplementary Fig. 8d. Data shown as average fold change ± SD compared to MEF2B-WT-isoA (two-tailed unpaired T test). e MEF2B relative expression level, as measured upon CHX treatment for 12 h in isogenic OCI-Ly7 control (Neutral; n = 3) and MEF2B isoform-switch mutant (n = 3 each) clones. Complete CHX pulse-chase experiment in Supplementary Fig. 8e. Data shown as average fold change ± SD relative to neutral clones (two-tailed unpaired T test). The experiments in (a) and (c) were repeated three times with similar results. Source data are provided as a Source Data file.
In conclusion, these results show that MEF2B phosphorylation at S324 favors the interaction with CUL3/KLHL12, while both lymphoma-associated missense and isoform-switch frameshift mutants escape CUL3/KLHL12 complex-mediated ubiquitination and degradation.
Mef2bP297L lymphoma-associated mutant enhances GC formation in mice
In order to understand the role of MEF2B phosphorylation at S324 and its lymphoma-associated impairment in GC development and lymphomagenesis, we generated a mouse model conditionally expressing the Mef2bP297L mutant, the orthologous of human MEF2BP325L (Fig. 8a). Toward this end, the coding region of Mef2b exon 2 and part of intron 2 were replaced by a cassette including the mouse Mef2b WT coding sequence (CDS; refer to NM_001045484.2) flanked by loxP sites and followed by the mutated Mef2b CDS (encoding Mef2bP297L mutant) linked, by a P2A self-cleavage sequence, to EGFP (Supplementary Fig. 9a). Upon crossing with GC-specific Cγ1-cre mice40, the expression of WT Mef2b in GC B cells is replaced by the Mef2bP297L mutant, which is co-transcribed with EGFP. Expression of the mutated transcripts in GC B cells was confirmed by RT-PCR followed by Sanger sequencing (Supplementary Fig. 9b). As expected, EGFP was detected exclusively in GC B cells of heterozygous (HET; Mef2bP297L/+;Cγ1-Cretg/+) and homozygous (HOMO; Mef2bP297L/P297L;Cγ1-Cretg/+) mutant mice, proportionally to allelic dosage, but not in WT (Mef2b+/+;Cγ1-Cretg/+) mice (Supplementary Fig. 9c-e).
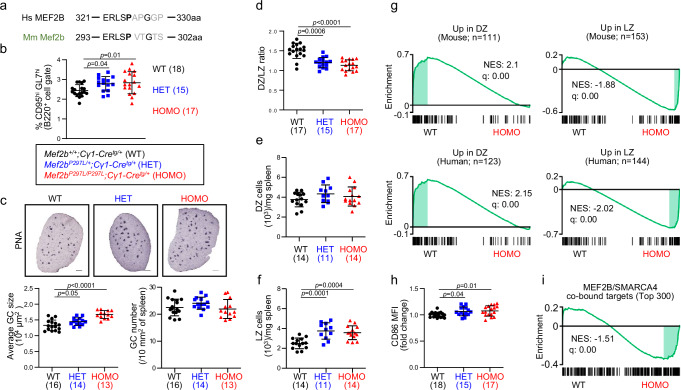
Fig. 8. The Mef2bP297L lymphoma-associated mutant enhances GC formation in mice.
a Amino acid sequence alignment of human MEF2B S324 phosphorylation region with the orthologous mouse Mef2b sequence. Human P325 and the conserved murine P297 aa are displayed in bold. b Percentage of splenic GC B cells (B220+/GL7hi/CD95hi) in Mef2b+/+;Cγ1-Cretg/+ (WT), Mef2bP297L/+;Cγ1-Cretg/+ (HET) and Mef2bP297L/P297L;Cγ1-Cretg/+ (HOMO) mice, as measured by flow cytometry 10 days post-SRBC immunization. c Top, immunohistochemistry for the GC marker PNA on representative spleen sections. Scale bars, 500 μm. Bottom, average GC size and numbers, as measured by quantification of PNA staining on spleen tissue sections in a subset of the mice displayed in (b). d Ratios between dark zone (DZ: CXCR4hi/CD86lo) and light zone (LZ: CXCR4lo/CD86hi) GC B cells (B220+/GL7hi/CD95hi) from WT, HET and HOMO mice. e DZ and f LZ B cell numbers normalized by spleen weight in a subset of the mice displayed in (d). g Gene set enrichment analysis (GSEA) in WT versus HOMO GC B cell RNA-seq data using previously reported mouse and human DZ versus LZ gene signatures41. NES, normalized enrichment score. h CD86 mean fluorescence intensity (MFI) in GC B cells (B220+/GL7hi/CD95hi) measured by flow cytometry analysis and displayed as fold change relative to WT mice. i GSEA in WT versus HOMO GC B cell RNA-seq data using the top 300 genes displaying co-binding of MEF2B and SMARCA4 in their regulatory regions as identified by Cut&Run-seq analysis in human GC B cells isolated from tonsil tissue. The enrichment is measured by ranking all the assessed transcripts in the RNA-seq profiles and evaluating any distribution bias along the ranked gene list. NES, normalized enrichment score. Dot plots display average ± SD for the number of animals reported in parentheses. The p values in dot plots were determined by one-way ANOVA with Dunnett’s Multiple Comparison Test. Source data are provided as a Source Data file.
Analysis of splenic B cells 10 days after sheep red blood cells (SRBC) immunization showed a significant increase of GC B cells in the mutant (HET and HOMO) mice compared to WT mice (Fig. 8b and Supplementary Fig. 9f, g), while no changes were detected in non-GC B cell populations, including follicular B cells, marginal zone B cells and plasma cells (Supplementary Fig. 9h–j). The GC B cell increase was due to an enlargement of the GC structures, not to an increased number of GCs, as shown by quantification of GC areas and numbers upon immunohistochemical staining of peanut agglutinin (PNA) (Fig. 8c). Analysis of the DZ and LZ GC B cell subpopulations showed a significant reduction in the DZ/LZ ratio, due to LZ expansion in the mutant mice (Fig. 8d–f and Supplementary Fig. 9f, k). Class-switch recombination was normal in the mutant mice, while their ability to generate high-affinity antibodies appeared slightly reduced (Supplementary Fig. 9l, m). No significant gender effect was detected since both the male and female cohorts showed statistically significant and consistent differences in GC size, DZ/LZ ratios, and LZ cell number, although the female cohort failed to reach significance regarding the increased percentage of GC B cells.
In order to characterize the transcriptional changes driven by expression of Mef2bP297L, we performed gene expression profiling of GC B cells sorted from SRBC-immunized WT and mutant (HET and HOMO) mice (at least 3 mice/genotype). We identified 137 genes that were significantly differentially expressed between WT and HOMO mice with a fold change >1.2 (65 up- and 72 down-regulated in HOMO compared to WT; q < 0.05; Supplementary Data 6). This gene signature was consistently enriched in the expression profiles of HET mice (Supplementary Fig. 10a). Consistent with the LZ expansion phenotype, Gene Set Enrichment Analysis (GSEA) in WT vs HET or HOMO expression profiles using previously reported DZ and LZ signatures both in mouse and human41 showed significant enrichment for the LZ signatures in the mutant mice (Fig. 8g and Supplementary Fig. 10b). In particular, CD86, a marker of the LZ compartment, was significantly upregulated in the GC B cells of mutant mice (Supplementary Data 6), which was consistent with increased protein levels, as detected by cytofluorimetric analysis (Fig. 8h). Consistent with the LZ expansion, the plasma cell master regulator Prdm142,43 was among the genes that were downregulated in mutant mice (Supplementary Data 6).
In order to evaluate the effects of the enhanced interaction between Smarca4 and the phospho-deficient Mef2b mutant in vivo, we performed GSEA in the mouse transcriptomic data using the targets that we identified as co-bound by SMARCA4 and MEF2B in human GC B cells. The results showed a significant enrichment for SMARCA4/MEF2B targets in the genes up-regulated in the mutant mice, consistent with their enhanced transcriptional expression when Mef2b cannot be phosphorylated (Fig. 8i). Consistent with the pathway enrichment analysis for SMARCA4/MEF2B targets in human GC B cells (Supplementary Data 4), shared targets are enriched for genes associated with the LZ compartment also in the murine GC B cells.
A few targets which displayed MEF2B/SMARCA4 co-binding in normal GC B cells (Supplementary Data 3) and were transcriptionally affected in the murine mutant GC B cells were tested for changes in expression in the DLBCL OCI-Ly7 isogenic clones expressing endogenous P325L. Consistent with the observations in mouse GC B cells, we detected transcriptional up-regulation of CD86, CD180, MAP3K8, PAX5 and RASGRP3 in the OCI-Ly7 isogenic clones expressing endogenous P325L when compared to neutral control clones (Supplementary Fig. 10c). These results suggest that the enhanced transcriptional effects of MEF2B phospho-deficient mutant are detectable both in normal and malignant GC B cells.
Overall, these data indicate that the expression of Mef2bP297L in GC B cells pathologically alters the GC development by enforcing LZ expansion, which, in multiple instances, has been shown to represent a phenotype preceding malignant transformation23,44,45.
Mef2bP297L mutant promotes GC B cell lymphomagenesis in mice
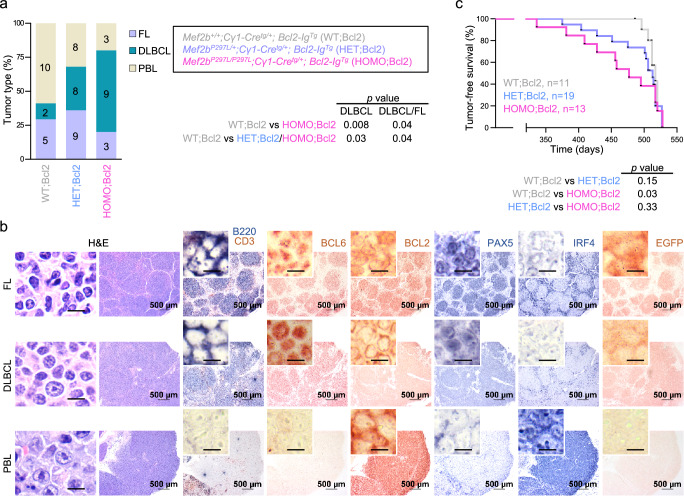
To investigate the role of Mef2bP297L in tumor development, mice were chronically immunized with SRBC (every 2 months, for six times in total) and monitored for signs of disease over a time frame of 17–18 months. The expression of Mef2bP297L mutant alone did not promote lymphoma development and had no impact on the tumor-free survival (Supplementary Fig. 11a, b). Since MEF2B mutations (including the phospho-deficient mutants) significantly co-occur with BCL2 re-arrangements in human DLBCL and FL8,13,16,46, the Mef2b mutant mice were also crossed with Bcl2-Ig transgenic mice47. We observed that Bcl2-Ig transgenic mice (Mef2b+/+;Cγ1-Cretg/+; Bcl2-IgTg) developed FL (4/19, 21%), but also more commonly plasmablastic lymphoma (PBL, 8/19, 42%), and in a few cases, multiple concomitant tumors (Fig. 9a and Supplementary Fig. 11a), as determined based on morphological features and analysis of T cell (Cd3), B cell (B220 and Pax5), GC (Bcl6) and plasma cell (Irf4) markers in the tumor tissue (Fig. 9b). In addition, expression of exogenous (human) BCL2 was confirmed in the tumors from Bcl2-Ig transgenic and compound mice (Fig. 9b). Tumors were clonal, as shown by the presence of clonally rearranged immunoglobulin loci (Supplementary Fig. 11c).
Fig. 9. Expression of Mef2bP297L mutant promotes GC B cell lymphomagenesis in mice.
a Distribution of B cell lymphomas across genotypes. Colored bars indicate different diagnoses, namely follicular lymphoma (FL), diffuse large B cell lymphoma (DLBCL), and plasmablastic lymphoma (PBL). The numbers in the bars refer to the number of tumors (note that some mice bear more than one tumor type). The p values were determined by Fisher’s Exact test (two-tailed). b H&E staining and immunohistochemical analysis of selected markers in representative tumors from mice diagnosed with FL, DLBCL, or PBL. EGFP is the tracking marker of Mef2bP297L. High magnification scale bar, 10 μm. These immunohistochemical analyses were performed for all tumors. c Tumor-free survival curve. The p values were determined by Gehan-Breslow-Wilcoxon Test (two-tailed) across genotypes. Source data are provided as a Source Data file.
PBLs originate from B cells committed to the plasma cell lineage, and as expected, do not express GC B cell markers, including Bcl6 and Mef2b (Fig. 9b). This also applies to the mutant Mef2bP297L, which is under the control of the endogenous promoter and regulatory regions. As expected, Mef2bP297L (EGFP) expression was detected in FL and DLBCL, but not in PBL tumors (Fig. 9b). Thus, the PBL tumors are not driven by Mef2b and do not reflect its function for lymphomagenesis. In contrast, the compound mice displayed a significantly higher incidence of DLBCL (32% in HET; Bcl2 and 60% in HOMO; Bcl2 vs 12% in WT; Bcl2; Fig. 9a). Accordingly, when considering GC tumors only, the compound HET; Bcl2 mice showed a shorter tumor-free survival that reached statistical significance for the HOMO; Bcl2 mice (Fig. 9c). No differences were observed regarding the tumor types and their histological features when comparing the Mef2bP297L with the previously published Mef2bD83V cohorts23.
Although expression of the mutant Mef2bP297L did not enhance overall lymphomagenesis, the increased incidence of Mef2b-expressing tumors and the associated reduction in tumor-free-survival suggest that expression of Mef2bP297L mutant, in combination with Bcl2-deregulation, promotes malignant transformation of GC B cells.
Discussion
The lymphoma-associated MEF2B mutational pattern is unusually heterogeneous, including missense, frameshift, and nonsense mutations, which are unevenly distributed along the MEF2B gene. We have previously demonstrated that the large majority of mutations targeting the N-terminus act through a convergent mechanism impairing MEF2B interaction with negative modulators (HUCA complex and class IIa histone deacetylases) and leading to its deregulated constitutive activity22,23. In this study we expanded this view demonstrating that multiple and distinct (by type and location) C-terminal mutations impair phosphorylation at S324, which negatively regulates MEF2B transcriptional activity and protein stability. Of note, the mutations impairing MEF2B phosphorylation at S324 never directly affect this amino acid, rather they disrupt the phosphorylation motif. We speculate that the lack of mutations affecting S324 is related to the fact that not all nucleotides have the same probability to be targeted by mutations. Based on these results, about 70% of MEF2B mutated cases are predicted to express a stable and/or deregulated mutant MEF2B protein.
Our data show that MEF2B interacts with the SWI/SNF chromatin remodeling complex, and that this interaction is regulated by S324 phosphorylation. This observation places MEF2B as a sequence-specific recruiter of chromatin modifiers that act as positive (SWI/SNF) or negative (HDACs) modulator of transcription. We cannot exclude that MEF2B may also recruit the SWI/SNF complex when it functions as a mediator, not directly binding to DNA28. The chromatin modifications downstream the MEF2B-mediated SWI/SNF (or HDACs) recruitment may have an impact in exposing or closing surrounding regions and therefore promote or impair the binding and function of additional transcription factors.
A key feature of MEF2B biology is the formation of complexes and this process appears to be hijacked by all types of functionally characterized mutations. Notably, it appears that different complexes, and therefore functions, are altered by distinct mutations. Indeed, the N-terminal mutants, which are impaired in their interaction with negative modulators of transcription22,23, are not affected in their complex formation with the SWI/SNF chromatin remodeling complex. On the other side, interaction between the C-terminal mutants and the HUCA complex and class IIa histone deacetylases is retained. N- and C-terminal mutants show also differences in protein stability, with a reduction observed in the N-terminal mutants and, conversely, an increased stability for the C-terminal mutants that are impaired in their interaction with the CUL3/KLHL12 E3 ligase complex. Of note, all these features are combined to generate MEF2B mutant proteins that are less stable but insensitive to negative modulation of their transcriptional activity (N-terminal mutants) or more stable and with an enhanced ability to promote transcription (C-terminal mutants). Regardless of their specific aberrant features, all these mutants appear to contribute to the pathogenesis of lymphoma without significant differences. These observations point to the presence of multiple mechanisms converging on de-regulation of the MEF2B oncogenic activity, with implications for the distinct approaches to be used if therapeutic targeting of MEF2B will be considered.
An aspect of extreme importance will be the identification of the structure of the full-length MEF2B protein. The N-terminal domain of MEF2B protein, spanning residues 1–91, is structurally ordered, as shown by the three-dimensional model of the full-length MEF2B generated by AlphaFold48 and by the crystal structures that have already been reported49–51. Conversely, there is no crystal structure for the MEF2B C-terminus, which is predicated to be disordered, most likely due to the prevalence of proline residues that impose a flexible loop conformation as opposed to stable ß-strand or α-helix topology. More importantly, the abundant presence of proline residues in the C-terminus suggests that this region is involved in protein-protein interactions52. The identification herein of a direct interaction between KLHL12 and MEF2B at both the N- and C-terminus provides an essential element to make the full-length MEF2B protein ordered and amenable to structural studies.
While most C-terminal mutations, including missense and frameshift ones, could be functionally characterized, a fraction of C-terminal mutants remain of difficult interpretation since they generate truncated forms of the protein, most of which appear to be unstable when expressed in DLBCL cell lines (no data are available from primary specimens). This type of mutants raises the possibility that haplo-insufficiency may contribute to lymphomagenesis, as previously suggested25. However, this hypothesis is not supported by the fact that (i) Mef2b deletion in mouse GC B cells induces a compensatory response from other Mef2 family members, suggesting that GC B cells require MEF2 activity; (ii) hemizygous expression of Mef2b in mouse GC B cells induces a decrease of DZ GC B cells, rather than a pre-neoplastic LZ expansion23. Thus, the experimental evidence in mouse models clearly suggests that Mef2b is essential for GC B development and maintenance, while its loss or hemizygous expression has a negative impact. A possible explanation for the C-terminus truncating mutations is suggested by their significant enrichment in the DLBCL-ABC subtype, the cell-of-origin of which is to be found in late GC B cells committed to post-GC differentiation6,53. These cells down-regulate the GC program, including expression of MEF2B, therefore in some cases, MEF2B truncating mutations may occur at a stage in which MEF2B function is not required.
GC lymphomagenesis is a complex multi-factorial process and modeling the effects of candidate oncogenes and onco-suppressors in vivo requires a long latency and often the presence of other pro-oncogenic (i.e., anti-apoptotic) alterations45,54–56. Indeed, the oncogenic potential of Mef2b mutants is best revealed in combination with Bcl2 deregulation for both the phospho-deficient mutant and the previously described N-terminal D83V mutant23. We note that the Bcl2-Ig transgenic model used here47 develop a significantly high fraction of PBL. However, our data clearly show that expression of a phosphorylation-defective Mef2b mutant, in combination with Bcl2 deregulation, promotes GC B cell malignant transformation toward FL and DLBCL, but not PBL, suggesting that Mef2b expression is required for tumor initiation and maintenance. Accordingly, BCL2 rearrangements co-occur significantly with MEF2B phospho-deficient mutations in human DLBCL and FL, while the much less frequent mutations targeting SMARCA4 are mutually exclusive with those affecting MEF2B, although this does not reach statistical significance due to the low number of cases (Supplementary Table 1).
Overall, these results unveil converging mechanisms of action for a diverse spectrum of MEF2B mutations, all leading to its dysregulation and GC B-cell lymphomagenesis. These assorted mechanisms provide multiple opportunities for the development of targeted therapeutic approaches.
Methods
This research complies with all ethical regulations. All in vivo experiments and procedures conformed to ethical principles and guidelines revised and approved by the Institutional Animal Care and Use Committee at Columbia University. This study includes the use of anonymized human tonsil tissue collected as residual material after diagnosis and therefore exempt from consent, as confirmed by the Columbia University Human Research Protection Office & Institutional Review Board. No material was obtained/analyzed through interaction/intervention/patient enrollment. Additional human data used in this study were previously published. As such the study represents “secondary research using data or biospecimens not collected specifically for this study” and has been defined as exempt human subject research in accordance with 45 CFR 46.
Mouse models and strains
Mice expressing the Cre recombinase under the control of the Cγ1 promoter and Bcl2-Ig transgenic mice were previously described40,47. All mouse strains were backcrossed into C57BL/6 background. Immunological responses were evaluated in immune-competent mice at ~3 months of age. Animals assigned to the tumor cohorts were monitored twice a week for tumor incidence and survival over a period of 17–18 months and were killed for analysis when visibly ill or at the end of the study. Although our approved protocol allows tumor sizes up to 20 mm, we sacrificed animals with masses of 10 mm or smaller. Both females and males were included in the experiments. Mice were housed in a dedicated pathogen-free environment with controlled temperature, humidity, and dark/light cycle.
Generation of Mef2bP297L conditional mouse model
The conditional Mef2bP297L C57BL/6 mouse was generated by Cyagen (Santa Clara, CA, USA). Briefly, the coding region of exon 2 plus partial intron 2 of Mef2b was replaced with the WT Mef2b coding DNA sequence (CDS; NCBI reference sequence: NM_001045484.2) flanked with two loxP sites and followed by a sequence encoding rabbit beta globin polyA signal. Downstream of the polyA cassette was inserted the Mef2b coding sequence carrying a C > T (nucleotide 890 of CDS) mutation (P297L amino-acid change) in exon 9. The mutated Mef2b coding region was linked to a sequence encoding enhanced green fluorescent protein (EGFP) by a 2 A sequence of porcine teschovirus-1 (P2A). In the targeting vector, the neomycin cassette was flanked by self-deletion anchor (SDA) sites. C57BL/6 ES cells were used for gene targeting, and correct homologous recombination was confirmed by Southern blotting and PCR followed by sequencing. Then, the targeted mouse ES cells were injected into blastocysts derived from C57BL/6 mice to generate chimeras. The following primers were used for genotyping and amplifying the fragment with the Mef2bP297L mutation (Supplementary Fig. 9a):
F1: 5’-CTTACCATCAGGCTCCACAGATT; R1: 5’-CTTCAACATCTCCTGCTTGCTTTA; F2: 5’-CCACATTGCCTTGCATTTATTGAG; R2: 5’-AAGACCATTTGACTTTGGCCTG; R3: 5’-CCCATGGTGGCTATAACTTCGTATAAT.
Mouse immunization and analysis
In order to induce GC responses, both female and male 3-month-old mice were immunized by intraperitoneal injection of 1 × 109 sheep red blood cells (SRBCs, Cocalico Biologicals) or 100 μg of 4-hydroxy-3-nitrophenyl-acetyl (NP) conjugated to keyhole limpet hemocyanin (KLH, BioSearch) in complete Freund’s Adjuvant (Sigma-Aldrich) and analyzed on day 10–12 post-immunization. To generate larger amounts of GC B cells (i.e., for gene expression profile studies), we performed two sequential injections of SRBCs (day 0, 1 × 108 SRBCs; day 5, 1 × 109 SRBC) and collected cells at day 12. This protocol yielded about three to four folds more GC B cells (~12–15% of the B cell fraction). For sorting GC B cells, B cells were first isolated from spleen by B cell isolation kit (130-090-862, Miltenyi Biotec), following manufacturer’s instructions. Then, the B cells were subjected to staining for the B cell and GC markers B220 (561086, BD Biosciences), GL7 (144613, BioLegend), and PNA (L32460, Thermo Fisher Scientific) and sorting on an Influx instrument (BD Biosciences). Only EGFP+ GC B cells were sorted from Mef2bP297L KI mice.
Spleens and other lymphoid tissues were isolated and divided into fragments, which were processed for histological staining and flow cytometry analysis. The mononuclear cells were isolated by crushing the tissue through 40 µm cell strainers in ice-cold 1Xphosphate-buffered saline (PBS) + 0.5% bovine serum albumin (BSA) followed by red blood cell lysis.
For flow cytometry analysis, mononuclear cell suspensions were stained for 20 min on ice with antigen-specific fluorochrome-conjugated antibodies. Samples were acquired on an Attune (Thermo Fisher Scientific) or Canto (BD Biosciences) instruments. FlowJo Software (V10.10.0, TreeStar) was used for data analyses and plot rendering. To calculate the absolute numbers of cells within splenic B cell subsets, spleen fragments were weighted, and erythrocyte-depleted mononuclear cell suspension were counted by Trypan blue exclusion using the Countess Automated Cell Counter (Thermo Fisher Scientific). The total number of counted splenic B cells was normalized according to the weight of the specimen and then multiplied by the fraction of each subpopulation, as identified by the cytofluorimetric analyses.
Mouse analyses were performed on sub-cohorts including 2–4 mice/genotype, which were obtained from 2–4 litters born at the same time.
Immunohistochemistry
3-μm-thick formalin-fixed and paraffin-embedded (FFPE) sections were used for histological staining, performed based on established protocols23. The GC number, GC size, and spleen areas were measured by Aperio ImageScope software (Leica).
The tumor diagnosis in mice was performed by histopathological analysis of tissue sections according to the following criteria: i) follicular lymphoma (FL) was diagnosed based on the presence of nodular proliferation of B220+ (553086, BD Biosciences)/BCL6+ (5650, Cell Signaling Technology) B cells with centrocytic morphology (characterized by angulated/cleaved nuclei and inconspicuous nucleoli) and admixed centroblastic cells (round-ovoid nuclei and prominent nucleoli) detectable to varying degrees; ii) diffuse large B cell lymphoma (DLBCL) was diagnosed based on diffuse proliferation of medium to large B220+/PAX5+ (sc-1974, Santa Cruz) B cells with round-ovoid nuclei, vesicular chromatin and distinct nucleoli; iii) plasmablastic lymphoma (PBL) was diagnosed in cases showing sheets of large cells with prominent nucleoli and exhibiting expression of IRF4 (sc-6059, Santa Cruz) and absent B (B220, PAX5), GC (BCL6) or T cell (CD3) markers.
Southern blotting
High molecular weight genomic DNA was extracted from frozen tissues by DNeasy Blood & Tissue Kit (Qiagen) and 5 μg were digested with the FastDigestion EcoRI (Thermo fisher scientific) for 3 h at 37 °C. The digested DNA was resolved on 0.8% agarose gel overnight followed by transfer to nitrocellulose membrane according to standard procedures. Hybridization was performed at 37 °C overnight using a 32P-labeled JH4 specific probe, as previously reported57.
Expression vectors
The plasmids pCMV-HA-MEF2B WT-isoA, pCDNA3-Flag-HA-MEF2B WT-isoA, pCDNA3-Flag-HA-MEF2B WT-isoB, pCCL-Flag-HA-MEF2B WT-isoA, pCCL-Flag-HA-MEF2B WT-isoB were previously reported22,23. pCCL-V5-TurboID was generated by subcloning the previously reported sequence encoding the V5-tagged TurboID31 into the pCCL vector. The pCCL-V5-TurboID-MEF2B WT-isoA was generated by subcloning MEF2B into the pCCL-V5-TurboID construct. pCDNA3-Flag-HA-MEF2B and pCMV-HA-MEF2B mutants were generated by site-direct mutagenesis. pCCL-Flag-HA-MEF2B mutants were constructed by subcloning from the pCDNA3-Flag-HA-MEF2B mutants. pCDNA3-EGFP were generated by subcloning EGFP cDNA into pCDNA3 vector (Invitrogen). pCDNA3-EGFP-MEF2B WT-isoA, pCDNA3-EGFP-MEF2B isoA fragments, pCDNA3-EGFP-MEF2B WT-isoB and pCDNA3-EGFP-MEF2B isoB were generated by subcloning into the pCDNA3-EGFP plasmid fragments obtained by PCR amplification from pCDNA3-Flag-HA-MEF2B WT-isoA and pCDNA3-Flag-HA-MEF2B WT-isoB. CUL3 cDNA (NM_003590.5) was amplified and inserted into pCDNA3 vector. Ubiquitin cDNA (NM_021009.7) together with V5 tag were amplified and inserted into pCDNA3 vector. KLHL12 cDNA (NM_001303051.2) was amplified and inserted into the pCMV-HA vector (Clontech). pCMV-Myc-KLHL12 and pCDNA3-Myc-CUL3 were generated by subcloning into the pCMV-Myc vector (Clontech) PCR amplicons obtained from pCMV-HA-KLHL12 and pCDNA3-Flag-CUL3. pCMV-HA-KLHL12-AMF and pCMV-Myc-KLHL12-AMF were generated by site-direct mutagenesis. pCMV-HA-SMARCA4 was generated by subcloning into pCMV-HA vector the PCR-amplified insert from pQCXIH BRGI-Flag (Addgene: #19148). All the plasmids were confirmed by Sanger sequencing.
Human normal GC B cell isolation
Human normal GC B cells were obtained from tonsils at Columbia University Medical Center. Specimens were exempt from informed consent being residual material after diagnosis and fully anonymized. The GC B cell isolation was performed as previously described58. Briefly, the tonsillar mononuclear cells (MNCs) were isolated by Ficoll-Isopaque centrifugation upon smashing the tonsil tissues. Then, GC B cells were purified by magnetic cell separation with anti-CD77-FITC antibody (BioLegend) and magnetic anti-FITC beads (Miltenyi Biotec), according to the manufacturer’s instructions (Miltenyi Biotec). The purity of GC B cells was confirmed by flow cytometry analysis with staining of anti-CD38-PE antibodies (BD Biosciences). The samples with at least 90% of GC B cell purity were used for downstream experiments.
Cell lines
HEK293T cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 100 mg/ml penicillin, and streptomycin. The DLBCL cell lines BJAB, DB, FARAGE, HBL1, HLY1, Karpas 422, OCI-LY1, OCI-LY3, OCI-LY7, OCI-LY8, RCK8, RIVA, SUDHL2, SUDHL4, SUDHL6, SUDHL7, SUDHL10 (ATCC), SUDHL1059, TMD8, U2932, VAL, WSU-DLCL2 and WSU-NHL were cultured in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% FCS, 100 mg/ml penicillin and streptomycin. Cell lines were maintained at 37 °C in humidified incubators in the presence of 5% CO2. All the cell lines tested negative for mycoplasma contamination. DB and SUDHL4 cell lines carry MEF2B mutations leading to the expression of the MEF2BD83V protein13. All remaining cell lines are wild-type for MEF2B.
Cell transfection and lentiviral transduction
For transient expression in HEK293T cells, polyethylenimine (PEI) (Polysciences) was used as previously described22. Briefly, DNA and PEI with amount ratio of 1:6 were incubated in 150 mM NaCl for 15 min at room temperature. DNA-PEI particles were gently added to pre-seeded HEK293T cells. After 6-h incubation at 37 °C, the transfection medium was removed and replaced with fresh DMEM medium. At 48 h post-transfection, cells were harvested for downstream analysis. Lentiviral transduction was done to establish stable cell lines as performed previously23. Viral supernatants were obtained by co-transfection of HEK293T cells with lentiviral vectors with EGFP expression, a lentivirus packaging vector (Δ8.9), and a plasmid encoding vesicular stomatitis virus envelope glycoprotein (VSVg). The indicated DLBCL cell lines were infected with the viral supernatants for two rounds, and then the infected cells were subjected to EGFP+ cell sorting on a SH800 Cell Sorter (Sony).
Generation of cell lines carrying endogenous mutations
The Alt-R CRISPR-Cas9 System (Integrated DNA Technologies) was used to edit OCI-Ly7 cells by introducing missense (S324A, S324E, and P325L) and isoform-switch (P256fs and P273fs) mutations in the MEF2B locus, as well as irrelevant mutations in intron 1 of PPP1R12C gene (neutral control), as reported previously15. Briefly, a ribonuclear protein (RNP) complex including specific crRNA and homology-directed repair (HDR) donor template, a tracerRNA labeled with ATTO 550 and Alt-R SpCas9 Nuclease V3 was formed according to the manufacturer’s protocol. The Neon NxT Electroporation System (Thermo Fisher Scientific) was used to deliver the RNP complex into OCI-Ly7 cells (electroporation condition: 1700 V, 20 ms and 1 pulse; buffer R). After 24 h, the top 70% of ATTO550-positive cells were sorted on an Influx Cell Sorter (BD Biosciences) and single-cell plated. Recovered single-cell clones were selected and analyzed by Sanger sequencing upon genomic DNA isolation and PCR amplification for the specific mutated region. In brief, clone pellets were resuspended in 100 µL of lysis buffer (10 mM Tris-HCL pH 7.5; 10 mM NaCl; 10 mM EDTA; 0.5% Sarcosyl) containing 0.5 mg/mL Proteinase K and incubated at 55 °C overnight. Genomic DNA was precipitated by adding 100 µL of cold EtOH/75 mM NaCl mixture followed by centrifugation at 956 g for 20 min at 4 °C. Finally, precipitated DNA was washed twice with cold 70% EtOH and centrifuged at 956 g at 4 °C for 10 min, air dried for 15 min and resuspended in molecular grade water for downstream analyses. PCR amplification of the targeted regions was performed using Taq polymerase and touch-down PCR conditions. Correct editing of recovered clones was assessed by using Mutation Surveyor (v.4.0.8).
The sequences of crRNAs and HDR donor templates are as following:
crRNA for MEF2B P325L: CAAGTCTGAGCGCCTCTCTC
HDR donor template for MEF2B P325L:
AGGGGAAGGTCTTAGGAAAGTCGCCGGGGCCCCCGGGGGCAAGAGAGAGGCGCTCAGACTTGATGCTGACTGGGGGGGTCGGCGG
crRNA for MEF2B S324A and S324E: CAAGTCTGAGCGCCTCTCTC
HDR donor template for MEF2B S324A:
GGAAGGTCTTAGGAAAGTCGCCGGGGCCCCCGGGGGCTGGAGCGAGGCGCTCAGACTTGATGCTGACTGGGGGGGTCGGCGGG
HDR donor template for MEF2B S324E:
GGAAGGTCTTAGGAAAGTCGCCGGGGCCCCCGGGGGCTGGTTCGAGGCGCTCAGACTTGATGCTGACTGGGGGGGTCGGCGGG
crRNA for MEF2B P256fs: GGTTAGGGGATGTACCTGGG
HDR donor template for MEF2B P256fs:
CGCTTCCCAGGTGGGGCAGGGCAGGTTAGGGGATGTACCTGGGGGCCTCCGGGGAGGAAGGGGAAGCTCCCCAGTGGGGGT
crRNA for MEF2B P273fs: CTTGTTGCAGCCCCCCACCC
HDR donor template for MEF2B P273fs:
CCTGGGAGACCCTCCACCGCCCCCTGGCTTGTTGCAGCCCCCACCCTGGCCCCCTGGCAGCCCTCGAGGGGTGATGGGCCCCC
crRNA for PPP1R12C intron mutation: CCAGCGAGTGAAGACGGCAT
HDR donor template for PPP1R12C intron mutation:
GAAGGCCATCCTAAGAAACGAGAGATGGCACAGGCCCCAGAAGGAGAAGGAAAAGGGAACCCAGCGAGTGAAGACGCCATGCGGTTGGGTGAGGGAGGAGAGATGCCCGGAGAGGACCCAGACACGGGGAGGATCCGCTCAGAGGACATC
The sequences of primers used for PCR amplification of the edited regions are as following:
S324A, S324E, P325L Forward: CTGGCTTCCTTCCCTTCCTC
S324A, S324E, P325L Reverse: CGTCACTGTTGGGTCTTCTC
P256fs Forward: CCTCTCTTCTCACCAAGCCA
P256fs Reverse: CACTTACCTGGGCTGGGAG
P273fs Forward: GTACATCCCCTAACCTGCCC
P273fs Reverse: CAAAATAGCTGCCCATCCGT
PPP1R12C Forward: GGGTGTCCAGGCAAAGAAAG
PPP1R12C Reverse: CCTTCAGGTTCCGTCTTCCT
Luciferase reporter assay
The luciferase reporter constructs (3xMEF2 WT-Luc and 3xMEF2 Mutant-Luc) were generated by cloning into the pGL4.26 vector (Promega) in our lab as previously described23. HEK293T cells were transiently transfected by PEI (Polysciences) with pCMV-HA-MEF2B WT or mutant plasmids along with the pGL4.26–3xMEF2 reporter construct and a renilla luciferase control reporter (pGL4.54-renilla). At 48 h post transfection, cells were harvested and lysed. Luciferase activity was measured using the Dual-Luciferase Reporter Assay Kit (E1960, Promega), according to the manufacturer’s instructions in a Glomax plate reader (Promega).
Immunoprecipitation and immunoblotting
Whole-cell protein extracts were isolated using the following immunoprecipitation (IP) buffer: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.2 mM EDTA pH 8.0, 0.05% NP40, 0.2% TritonX-100, 30 mM beta-glycerophosphate, 0.5 mM PMSF, 50 mM sodium fluoride, 1 mM sodium orthovanadate and protease inhibitor cocktail (Sigma-Aldrich). For nuclear extraction, cells were lysed with cytoplasmic extraction buffer (10 mM HEPES pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 0.5% NP40, and protease inhibitor cocktail) for 10 min on ice. Upon centrifugation for 10 min at 500 × g, the supernatant was transferred to a new tube as the cytoplasmic fraction and subjected to further centrifugation at 20000 × g for 10 min. The nuclei pellet was washed with cytoplasmic extraction buffer three times (500 × g for 10 min), and then nuclear extraction buffer (10 mM HEPES, 1.5 mM MgCl2, 420 mM NaCl, 1 mM DTT, 20% Glycerol and protease inhibitor cocktail) was added to lyse for 30 min on ice. The nuclear fraction was obtained with centrifugation at 20000 × g for 15 min.
Lysates were incubated overnight at 4 °C with anti-Flag (A2220, Sigma-Aldrich) or anti-HA affinity beads (F2426, Sigma-Aldrich) or indicated antibodies. In the latter case lysates were supplemented with Protein G beads (17-0618-01, Cytiva) for 2 h at 4 °C. Beads were washed 5 times in a modified IP buffer containing 400 mM NaCl. Immunocomplexes were eluted with 2.5 mg/ml MEF2B, Flag, or HA peptides (Synthesized by Genemed Synthesis, Inc.; San Antonio, TX, USA) or in 100 mM Glycine (pH 2.5) buffer. If proteins were eluted by 100 mM Glycine, neutralization was further performed by adding 20% (volume of Glycine solution) 500 mM Tris-HCl (pH 8). Eluted samples were concentrated by Vivaspin Protein Concentrators (Cytiva) or Amicon® Ultra Centrifugal Filters (Millipore), if elution volume was greater than 30 ul. The protein lysates or IP eluents were resolved on Tris-Glycine mini gels (Thermo Fisher Scientific), and subsequently transferred on a 0.45 µm nitrocellulose membrane. Membranes were blocked for 1 h in TBS (20 mM Tris, pH7.4 and 150 mM Nacl) with 0.05% Tween-20 and 5% non-fat dry milk. The membranes were incubated overnight at 4 °C with the primary antibodies against MEF2B (custom23 and NBP3-03920, Novus Biologicals; 1:1000 dilution), pMEF2BS310 (custom; 1:1000 dilution), pMEF2BS324 (custom; 1:1000 dilution), Actin (A5441, Sigma-Aldrich; 1:3000 dilution), Vinculin (SAB4200080, Sigma-Aldrich; 1:2000 dilution), Lamin B1 (12586, Cell Signaling Technology; 1:1000 dilution), HA (3724, Cell Signaling Technology; 1:1000 dilution), Myc (sc-40, Santa Cruz; 1:1000 dilution), Flag (P2983, Sigma-Aldrich; 1:1000 dilution), EGFP (2956, Cell Signaling Technology; 1:1000 dilution), Ubiquitinylated proteins (04-263, Sigma-Aldrich; 1:1000 dilution), K48-linkage Specific Polyubiquitin (8081, Cell Signaling Technology; 1:1000 dilution), K63-linkage Specific Polyubiquitin (5621, Cell Signaling Technology; 1:1000 dilution), V5 (13202, Cell Signaling Technology; 1:1000 dilution), SMARCA4 (49360, Cell Signaling Technology; 1:1000 dilution), CUL3 (A301-109A, Bethyl Laboratories; 1:1000 dilution) and KLHL12 (9406, Cell Signaling Technology; 1:1000 dilution). After washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (NA931 and NA934, Cytiva; 1:5000 dilution) or Trueblot secondary antibodies (18-8816-33 and 18-8817-33, Rockland; 1:5000 dilution) for 1 h at room temperature. Detection was performed by ECL or Dura Reagent (Thermo Fisher Scientific) followed by film exposure. ImageJ software was used to quantitate band density.
TurboID-mediated biotinylated protein pull-down
SUDHL10 cells stably expressing V5-TurBoID or V5-TurBoID-MEF2B WT-isoA were treated with 50 μM biotin (B4639, Sigma-Aldrich) for 24 h. Then, the cells were subjected to nuclear fraction isolation, as described in the Immunoblotting section. The nuclear extracts were incubated with Streptavidin Magnetic Beads (88817, Thermo Fisher Scientific) at 4 °C overnight. Subsequently, the beads were subjected to three rounds of washing (1st round: 2% SDS; 2nd round: 50 mM pH 7.5 HEPES, 0.5 M NaCl, 1 mM EDTA, 0.1% Deoxycholic acid and 1% Triton x100; 3rd round: 10 mM pH 7.5 Tris-HCl, 0.25 M LiCl, 1 mM EDTA, 0.1% Deoxycholic acid and 1% NP40). Finally, the beads were boiled before proceeding to electrophoresis on Tris-Glycine gels followed by immunoblotting.
Phos-tag gel analysis
Whole cell lysates were extracted by EDTA- and phosphate-free lysis buffer (50 mM Tris-Hcl pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, EDTA-free cOmplete protease inhibitor and phosSTOP phosphatase inhibitor). The samples were loaded into the 7.5% SuperSep Phos-tag™ gels (192-18001, Wako Chemicals) and electrophoresis was performed at 100 V on ice using freshly prepared Tris-MOPS buffer (50 mM Tris, 50 mM MOPS, 0.5% SDS and 5 mM sodium bisulfite). After electrophoresis, the gel was soaked three times in NuPAGE transfer buffer (Thermo Fisher Scientific) for 10 min/each. Protein transfer to 0.45 µm PVDF membrane (Millipore) was performed using the wet-tank method (Bio-Rad) at 150 mA for 16 h in the cold room. Antibody incubation and signal detection were performed using standard immunoblotting procedures.
Protein half-life measurement
SUDHL10 cells stably expressing N-terminal double-tagged Flag-HA-MEF2B WT or mutants were treated with 100 μg/ml cycloheximide (Sigma-Aldrich) at different time points and collected at the same time. To confirm proteasome-mediated degradation, 5 μM MG132 (Cayman Chemicals) was added in combination with cycloheximide treatments for 12 or 24 h. Cells lysates were resolved by Tris-Glycine gels and analyzed by immunoblotting with anti-HA and anti-vinculin antibodies. MEF2B half-life was quantified using the exponential regression function of densitometry values measured using the ImageJ processing software.
Ubiquitination assay
HEK293T cells were transfected with indicated plasmids by PEI, and then cultured for 48 h. 4 h before cell harvest, 10 μM MG132 was added into the medium for ubiquitin accumulation. For ubiquitin detection in DLBCL cells, SUDHL10 cells stably expressing N-terminal double-tagged Flag-HA EV or Flag-HA-MEF2B isoA-WT were treated with 5 μM MG132 for ~16 h. Both the HEK293T and SUDHL10 cells were subjected to lysis and then denatured at 95 °C for 15 min using a modified IP buffer containing 2 μM NEM (N-Ethylmaleimide; Sigma-Aldrich) and 1% SDS. Subsequently, sonication was performed to shear genomic DNA (Bioruptor, Diagenode). The SDS in the cell lysate was quenched by adding 10% Triton-100 and diluted to 0.1% SDS with IP buffer containing 2 μM NEM. Flag IP was performed overnight, and the beads were boiled before electrophoresis on Tris-Glycine gels followed by immunoblotting.
RNA extraction, cDNA synthesis, and RT-PCR
Total RNA was extracted using the Trizol Reagent (Thermo Fisher Scientific), RNeasy Kit (Qiagen), or the Nucleospin RNA XS RNA Isolation Kit (Macherey-Nagel), according to the manufacturer’s instructions. cDNA synthesis was performed with 1–3 µg of total RNA using the SuperScript II First-Strand Synthesis System (Thermo Fisher Scientific). PCR products were analyzed by electrophoresis on agarose gel. Quantitative retrotranscription q(RT)-PCR was performed using SYBR Green PCR Master Mix (Thermo Fisher Scientific) on a 7500 Fast Real-Time PCR System (Thermo Fisher Scientific). The primer sequences are as following:
Exogenous Flag-HA-MEF2B:
Forward: GATCCAAGCCACCATGTACC; Reverse: CAGAGCACGCTCAGCTCATA
CD86:
Forward: TTGATAATGGGATGAATGGAAGGA; Reverse: CGTAGGACATCTGTAGGCTAAG
CD180:
Forward: CACCTCCTGGGATCAGATGT; Reverse: TGGTAGAGTGTCAGGGATTTCA
MAP3K8:
Forward: GGCCGCAGATGCAATCTTCTTA; Reverse: TGGCTTTGCAGATACTGCGTT
PAX5:
Forward: GTCCCAGCTTCCAGTCACAG; Reverse: CGGAGACTCCTGAATACCTTCG
RASGRP3:
Forward: GGGAAAAGCCTGTCTGCTGTT; Reverse: GCTCCAGAAAAGTGAGGTGCT
GAPDH:
Forward: CATTTCCTGGTATGACAACGAA; Reverse: AGGGGTCTACATGGCAACTG
RNA-seq
Total RNA extracted from sorted murine GC B cells were quantified and subject to quality controls using a Bioanalyzer (Agilent). ~200 ng RNA was used for library preparation and sequencing by Azenta Life Sciences (South Plainfield, NJ, USA). RNA-seq reads were mapped to the Mus musculus (mm10/GRCm38) genome reference using the hisat2 prebuilt genome index60. Genome-mapped reads were aligned to exons on the mm10 (or GRCh38) transcriptome reference61 based on the information in the genomic BAM files, using featureCounts62 to produce abundance tables. We sanitized the transcriptome references (exon only) by removing read-through genes, anti-sense elements, miRNA, and rRNA. The count tables were subsequently normalized to produce transcript per million (TPM) tables. Differentially expressed genes were determined using DESeq263 with the following filters: FDR < 0.05 (after Benjamini-Hochberg correction) and absolute fold change (FC) > 1.2.
Cut and run
Nuclei were isolated from GC B cells freshly purified from human tonsil using a nuclear extraction buffer (20 mM pH 7.9 HEPES, 10 mM KCl, 0.1% Triton X-100, 20% glycerol, and 1 mM MnCl2). Cut&Run sample preparation was performed using the CUTANA ChIC/CUT&RUN Kit version 3.1 (14-1048, EpiCypher), following the manufacturer’s instructions. Briefly, activated ConA Beads (EpiCypher) were added to the nuclei samples followed by incubation with 0.5 μg of IgG (13-0042, EpiCypher), SMARCA4 (49360, Cell Signaling Technology), MEF2B (custom), H3K27Ac (C15410196, Diagenode) and H3K4me3 (13-0041, EpiCypher) antibodies to capture protein/DNA complexes. Subsequently, pAG-MNase (EpiCypher), added to all samples, was activated by CaCl2 to cleave target chromatin. DNA purification was performed using CUTANA™ DNA Purification Kit to capture DNA fragments >50 bp. Library preparation was performed with 5 ng of CUT&RUN enriched DNA (or the total sample volume if DNA output was less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit version 1.0 (14-1001, EpiCypher) following the manufacturer’s instructions.
After quantification, using QubitTM fluorometer with 1X dsDNA HS Assay Kit (Invitrogen), as per manufacturer’s instructions, and analysis of the library fragment size, using the High Sensitivity DNA Kit (Bioanalyzer), libraries were pooled and sequenced using HiSeq 3000/4000 Illumina paired-end sequencing. Paired-end samples were aligned to the GRCh38 genome using hisat264 with the following options: hisat2, no mixed, no discordant, phred33, minins 10, and maxins 700. After alignment, the BAM files were converted to BED files using bedtools bamtobed with the bedpe option. The BED files were filtered to keep properly paired fragments <1000 bp in length. MACS3 was used to find peaks in the filtered BED files using the matching IgG samples as controls with the following options: macs3 callpeak, format BEDPE, and keep dup all g hs q 0.05.
Sample preparation for mass spectrometry (MS) analysis
The protein samples for MS analysis were purified from normal GC B cells and SUDHL10 cells stably expressing the N-terminal double-tagged Flag-HA-MEF2B WT or mutants according to the IP procedure. The IP eluate for interactor identification was separated on 4–12% Tris-Glycine gel (Thermo Fisher Scientific) for Coomassie blue (Thermo Fisher Scientific) staining, and all the bands were excised. For protein modification analysis, the IP eluates were subject to electrophoresis using 10% Tris-Glycine gel (Thermo Fisher Scientific) and ~42 KDa bands were excised. The excised protein gel slices were used for in-gel digestion as previously described65, with minor modifications. Gel slices were washed with 1:1 Acetonitrile and 100 mM ammonium bicarbonate for 30 min then dehydrated with 100% acetonitrile for 10 min until shrunk. The excess acetonitrile was then removed, and the slices dried in a speed vacuum at room temperature for 10 min. Gel slices were reduced with 5 mM DTT for 30 min at 56 °C in an air thermostat, cooled down to room temperature, and alkylated with 11 mM IAA for 30 min with no light. Gel slices were then washed with 100 mM of ammonium bicarbonate and 100% acetonitrile for 10 min each. Excess acetonitrile was removed and dried in a speed-vacuum for 10 min at room temperature and the gel slices were re-hydrated in a solution of 25 ng/μl trypsin in 50 mM ammonium bicarbonate for 30 min on ice and digested overnight at 37 °C in an air thermostat. Digested peptides were collected and further extracted from gel slices in an extraction buffer (1:2 ratio by volume of 5% formic acid:acetonitrile) at high speed, shaking in an air thermostat. The supernatants from both extractions were combined and dried in a speed vacuum. Peptides were dissolved in 3% acetonitrile/0.1% formic acid.
Liquid chromatography with tandem mass spectrometry (LC-MS/MS) for interactor identification
Peptides were separated within 80 min at a flow rate of 400 nl/min on a reversed-phase C18 column with an integrated Captive Spray Emitter (25 cm × 75 µm, 1.6 µm, IonOpticks). Mobile phases A and B were using 0.1% formic acid in water and 0.1% formic acid in ACN. The fraction of B was linearly increased from 2 to 23% within 70 min, followed by an increase to 35% within 10 min and a further increase to 80% before re-equilibration. The timsTOF Pro was operated in PASEF mode66 with the following settings: Mass Range 100 to 1700 m/z, 1/K0 range 0.6 to 1.6, Ramp time 100 ms, lock duty cycle to 100%, Capillary Voltage 1600 V, Dry Gas 3 l/min, Dry Temp 200 °C, PASEF settings: 10 MS/MS Frames (1.16 s duty cycle), charge range 0–5, the active exclusion for 0.4 min, Target intensity 20,000, Intensity threshold 2500, CID collision energy 59 eV. A polygon filter was applied to the m/z and ion mobility plane to select features most likely representing peptide precursors rather than singly charged background ions.
LC-MS/MS data analysis for interactor identification
Acquired PASEF raw files were analyzed using the MaxQuant environment v. 2.0.3.0 and Andromeda for database searches at default settings67. The default is used for the first search tolerance and main search tolerance (20 ppm and 4.5 ppm, respectively). MaxQuant was set up to search with the reference human proteome database (UP000005640) downloaded from UniProt. MaxQuant performed the search trypsin digestion with up to 2 missed cleavages. Peptide, site, and protein false discovery rates (FDR) were all set to 1% with a minimum of 1 peptide needed for identification. The following modifications were used for protein identification and quantification: Carbamidomethylation of cysteine residues (+57.021 Da) was set as static modifications, while the oxidation of methionine residues (+15.995 Da), deamidation (+0.984) on asparagine and glutamine were set as a variable modification. Results obtained from MaxQuant were used to identify MEF2B complex formation. Moreover, the common protein contaminants including keratins, trypsin, serum albumin, and actin were excluded as previously reported68.
LC-MS/MS analysis for MEF2B phosphorylation
The dried peptides were dissolved in 10 µl of 3% acetonitrile/0.1% formic acid and 500 ng analyzed on an Orbitrap Fusion mass spectrometer coupled to a Dionex Ultimate 3000 (Thermo Fisher Scientific) using the PRM method69. Peptides were separated on an EASY-Spray C18 25 cm column (Thermo Fisher Scientific). Peptides elution and separation were achieved at a non-linear flow rate of 300 nl/min using a gradient of 5%-30% of buffer B (0.1% v/v formic acid, 100% acetonitrile) for 100 min with a temperature of the column maintained at 40 °C during the entire experiment. The Thermo Scientific Orbitrap Fusion Tribrid mass spectrometer was used for peptide tandem mass spectroscopy (MS/MS). MS data were acquired with combined two scan events corresponding to a full scan and a tMS2 method targeting the specific MEF2B-unmodified and phospho modified peptides. The target value for the full scan MS spectra was 1 × 106 ions in the 450–900 m/z range with a maximum injection time of 60 ms and resolution of 120,000 at 200 m/z. The tMS2 method were acquired with an isolation width of 1.6 m/z, a resolution of 30,000 on an Orbitrap, a target AGC value of 1.0 × 106, fragmented by higher-energy C-trap dissociation with a normalized collision energy of 27 eV, and a maximum injection time of 110 ms.
Data analysis was performed using Thermo Xcalibur v4.5.445.18 and the area under the curve of selected precursor ions summed to determine the abundance of the respective MEF2B-unmodified and modified phospho peptides.
Enzyme-linked immunosorbent assay (ELISA)
ELISA for NP-specific antibodies using plates coated with NP5 or NP25 in BSA (Biosearch Technologies; n5050) was done as previously described70. Peripheral blood was obtained before immunization and 14d after immunization. The levels of serum IgG were measured by ELISA using unlabeled anti-mouse Ig antibodies (Southern Biotech) as capture reagent, Alkaline phosphatase (AP)-labeled anti-mouse lg subclass-specific antibodies (Southern Biotech) as developing reagents, and 4-methyl umbelliferyl phosphate as AP substrate.
Statistics
GraphPad Prism v.9.0 software was used for statistical analyses. One-way analysis of variance (ANOVA) paired with Dunnett’s Multiple Comparison Test, Mann-Whitney test, unpaired two-tailed T-test, or one-sample T-test were used to assess statistically significant differences between groups. Fisher’s Exact test was used to assess significance of MEF2B mutation distribution and tumor incidence. Detailed information of the statistical test, number of replicates, and number of animals (defined as n) used in each experiment, as well as the definition of center and dispersion are reported in the figures and in the legends. Significance was associated to a p ≤ 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Source data
Acknowledgements
We would like to thank Clara Reglero Gomez and Hui Zhou for help with protein purification, and Bangmin Zhang for technical support. We are grateful to Shan Zha, Wei Gu, and Zhiguo Zhang for suggestions and discussion. These studies were supported by NIH grant R35CA-210105 (to RD-F) and funded in part through the NIH/NCI Cancer Center Support Grant P30CA013696. This research used the resources of the Herbert Irving Comprehensive Cancer Center (HICCC); the Flow Core Facility supported in part by the Office of the Director, National Institutes of Health under award S10OD020056; Proteomics and Macromolecular Crystallography Shared Resource; the HICCC Transgenic Mouse Facility; the Digital and Computational Pathology Laboratory in the Department of Pathology & Cell Biology at Columbia University. C.C. is supported by a Lymphoma Research Foundation fellowship. L.P. is on leave from the University of Perugia.
Author contributions
L.P., R.D.F., and K.B. designed and supervised the study; C.Y. designed and performed experiments; C.Y. and K.B. analyzed data; A.B.H. performed computational analyses; Q.S., A.T., C.C., and S.K. performed experiments; T.M. assisted with mouse husbandry and immunizations; R.K.S. performed mass-spectrometry experiments and data analysis; S.H. performed mouse pathology analyses; F.F. performed protein structural analysis; C.Y., L.P., R.D.F. and K.B. wrote the paper. All authors read, reviewed, and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The datasets generated during the current study are available in the GEO repository as super-series GSE234684 or separately under accession numbers GSE234657 (RNA-seq data of mouse GC B cells) and GSE234656 (Cut&Run data of human normal GC B cells) and in the ProteomeXchange Consortium via the PRIDE repository with dataset identifier PXD043998 (mass spectrometry proteomics data). Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Riccardo Dalla-Favera, Katia Basso.
Contributor Information
Riccardo Dalla-Favera, Email: rd10@cumc.columbia.edu.
Katia Basso, Email: kb451@cumc.columbia.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51644-8.
References
- 1.Swerdlow SH, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (IARC Publications 2017).
- 2.Sehn, L. H. & Salles, G. Diffuse large B-cell lymphoma. N. Engl. J. Med.384, 842–858 (2021). 10.1056/NEJMra2027612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casulo, C. et al. Early relapse of follicular lymphoma after rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone defines patients at high risk for death: an analysis from the National Lymphocare Study. J. Clin. Oncol.33, 2516–2522 (2015). 10.1200/JCO.2014.59.7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basso, K. & Dalla-Favera, R. Germinal centers and B cell lymphomagenesis. Nat. Rev. Immunol.15, 172–184 (2015). 10.1038/nri3814 [DOI] [PubMed] [Google Scholar]
- 5.Victora, G. D. & Nussenzweig, M. C. Germinal centers. Annu. Rev. Immunol.40, 413–442 (2022). [DOI] [PubMed]
- 6.Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature403, 503–511 (2000). 10.1038/35000501 [DOI] [PubMed] [Google Scholar]
- 7.Rosenwald, A. et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med.346, 1937–1947 (2002). 10.1056/NEJMoa012914 [DOI] [PubMed] [Google Scholar]
- 8.Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet.43, 830–837 (2011). 10.1038/ng.892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapuy, B. et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med.24, 679–690 (2018). 10.1038/s41591-018-0016-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ennishi, D. et al. Double-hit gene expression signature defines a distinct subgroup of germinal center b-cell-like diffuse large B-Cell lymphoma. J. Clin. Oncol.37, 190–201 (2019). 10.1200/JCO.18.01583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubschmann, D. et al. Mutational mechanisms shaping the coding and noncoding genome of germinal center derived B-cell lymphomas. Leukemia35, 2002–2016 (2021). 10.1038/s41375-021-01251-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz, R. et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med378, 1396–1407 (2018). 10.1056/NEJMoa1801445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature476, 298–303 (2011). 10.1038/nature10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright, G. W. et al. A probabilistic classification tool for genetic subtypes of diffuse large B Cell lymphoma with therapeutic implications. Cancer Cell37, 551–568.e514 (2020). 10.1016/j.ccell.2020.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bal, E. et al. Super-enhancer hypermutation alters oncogene expression in B cell lymphoma. Nature607, 808–815 (2022). 10.1038/s41586-022-04906-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasqualucci, L. et al. Genetics of follicular lymphoma transformation. Cell Rep.6, 130–140 (2014). 10.1016/j.celrep.2013.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gossett, L. A., Kelvin, D. J., Sternberg, E. A. & Olson, E. N. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol. Cell Biol.9, 5022–5033 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black, B. L. & Olson, E. N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol.14, 167–196 (1998). 10.1146/annurev.cellbio.14.1.167 [DOI] [PubMed] [Google Scholar]
- 19.Potthoff, M. J. & Olson, E. N. MEF2: a central regulator of diverse developmental programs. Development134, 4131–4140 (2007). 10.1242/dev.008367 [DOI] [PubMed] [Google Scholar]
- 20.Molkentin, J. D., Black, B. L., Martin, J. F. & Olson, E. N. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell Biol.16, 2627–2636 (1996). 10.1128/MCB.16.6.2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molkentin, J. D. et al. MEF2B is a potent transactivator expressed in early myogenic lineages. Mol. Cell Biol.16, 3814–3824 (1996). 10.1128/MCB.16.7.3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ying, C. Y. et al. MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat. Immunol.14, 1084–1092 (2013). 10.1038/ni.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brescia, P. et al. MEF2B instructs germinal center development and acts as an oncogene in B cell lymphomagenesis. Cancer Cell34, 453–465.e459 (2018). 10.1016/j.ccell.2018.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan, R. J. et al. Detection of enhancer-associated rearrangements reveals mechanisms of oncogene dysregulation in B-cell lymphoma. Cancer Discov.5, 1058–1071 (2015). 10.1158/2159-8290.CD-15-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pon, J. R. et al. MEF2B mutations in non-Hodgkin lymphoma dysregulate cell migration by decreasing MEF2B target gene activation. Nat. Commun.6, 7953 (2015). 10.1038/ncomms8953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei, X. et al. The cancer mutation D83V induces an alpha-helix to beta-strand conformation switch in MEF2B. J. Mol. Biol.430, 1157–1172 (2018). 10.1016/j.jmb.2018.02.012 [DOI] [PubMed] [Google Scholar]
- 27.Reddy, A. et al. Genetic and functional drivers of diffuse large B Cell lymphoma. Cell171, 481–494.e415 (2017). 10.1016/j.cell.2017.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu, C. S. et al. Unique immune cell coactivators specify locus control region function and cell stage. Mol. Cell80, 845–861.e810 (2020). 10.1016/j.molcel.2020.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clapier, C. R., Iwasa, J., Cairns, B. R. & Peterson, C. L. Mechanisms of action and regulation of ATP-dependent chromatin-remodeling complexes. Nat. Rev. Mol. Cell Biol.18, 407–422 (2017). 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Centore, R. C., Sandoval, G. J., Soares, L. M. M., Kadoch, C. & Chan, H. M. Mammalian SWI/SNF chromatin remodeling complexes: emerging mechanisms and therapeutic strategies. Trends. Genet.36, 936–950 (2020). 10.1016/j.tig.2020.07.011 [DOI] [PubMed] [Google Scholar]
- 31.Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol.36, 880–887 (2018). 10.1038/nbt.4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc.4, 44–57 (2009). 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 33.Angers, S. et al. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting dishevelled for degradation. Nat. Cell Biol.8, 348–357 (2006). 10.1038/ncb1381 [DOI] [PubMed] [Google Scholar]
- 34.Rondou, P., Haegeman, G., Vanhoenacker, P. & Van Craenenbroeck, K. BTB Protein KLHL12 targets the dopamine D4 receptor for ubiquitination by a Cul3-based E3 ligase. J. Biol. Chem.283, 11083–11096 (2008). 10.1074/jbc.M708473200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao, B. et al. Structural elucidation of peptide binding to KLHL-12, a substrate specific adapter protein in a Cul3-Ring E3 ligase complex. Biochemistry59, 964–969 (2020). 10.1021/acs.biochem.9b01073 [DOI] [PubMed] [Google Scholar]
- 36.Canning, P. et al. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J. Biol. Chem.288, 7803–7814 (2013). 10.1074/jbc.M112.437996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen, Z. et al. Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12. Open Biol.10, 200041 (2020). 10.1098/rsob.200041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akopian, D., McGourty, C. A. & Rape, M. Co-adaptor driven assembly of a CUL3 E3 ligase complex. Mol. Cell82, 585–597 e511 (2022). 10.1016/j.molcel.2022.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolla, S., Ye, M., Mark, K. G. & Rape, M. Assembly and function of branched ubiquitin chains. Trends Biochem. Sci.47, 759–771 (2022). 10.1016/j.tibs.2022.04.003 [DOI] [PubMed] [Google Scholar]
- 40.Casola, S. et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. USA103, 7396–7401 (2006). 10.1073/pnas.0602353103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Victora, G. D. et al. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood120, 2240–2248 (2012). 10.1182/blood-2012-03-415380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaffer, A. L. et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity17, 51–62 (2002). 10.1016/S1074-7613(02)00335-7 [DOI] [PubMed] [Google Scholar]
- 43.Shapiro-Shelef, M., Lin, K. I., Savitsky, D., Liao, J. & Calame, K. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J. Exp. Med.202, 1471–1476 (2005). 10.1084/jem.20051611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beguelin, W. et al. Mutant EZH2 induces a pre-malignant lymphoma niche by reprogramming the immune response. Cancer Cell37, 655–673.e611 (2020). 10.1016/j.ccell.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, J. et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat. Med.21, 1190–1198 (2015). 10.1038/nm.3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okosun, J. et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet.46, 176–181 (2014). 10.1038/ng.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDonnell, T. J. et al. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell57, 79–88 (1989). 10.1016/0092-8674(89)90174-8 [DOI] [PubMed] [Google Scholar]
- 48.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han, A. et al. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature422, 730–734 (2003). 10.1038/nature01555 [DOI] [PubMed] [Google Scholar]
- 50.Lei, X. et al. Crystal structure of apo MEF2B reveals new insights in DNA binding and cofactor interaction. Biochemistry57, 4047–4051 (2018). 10.1021/acs.biochem.8b00439 [DOI] [PubMed] [Google Scholar]
- 51.Lei, X. et al. Crystal structures of ternary complexes of MEF2 and NKX2-5 bound to DNA reveal a disease related protein-protein interaction interface. J. Mol. Biol.432, 5499–5508 (2020). 10.1016/j.jmb.2020.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morgan, A. A. & Rubenstein, E. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One8, e53785 (2013). 10.1371/journal.pone.0053785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holmes, A. B. et al. Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. J. Exp. Med.217, e20200483 (2020). [DOI] [PMC free article] [PubMed]
- 54.Beguelin, W. et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell23, 677–692 (2013). 10.1016/j.ccr.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flumann, R. et al. An autochthonous mouse model of Myd88- and BCL2-driven diffuse large B-cell lymphoma reveals actionable molecular vulnerabilities. Blood Cancer Discov.2, 70–91 (2021). 10.1158/2643-3230.BCD-19-0059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pasqualucci, L. & Klein, U. NF-kappaB mutations in germinal center b-cell lymphomas: relation to NF-kappaB Function in normal B cells. Biomedicines10, 2450 (2022). [DOI] [PMC free article] [PubMed]
- 57.Zhang, J. et al. The CREBBP acetyltransferase is a haploinsufficient tumor suppressor in B-cell lymphoma. Cancer Discov.7, 322–337 (2017). 10.1158/2159-8290.CD-16-1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klein, U. et al. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl Acad. Sci. USA100, 2639–2644 (2003). 10.1073/pnas.0437996100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Epstein, A. L. et al. Biology of the human malignant lymphomas. IV. Functional characterization of ten diffuse histiocytic lymphoma cell lines. Cancer42, 2379–2391 (1978). [DOI] [PubMed] [Google Scholar]
- 60.Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc.11, 1650–1667 (2016). 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harrow, J. et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol.7, 1–9 (2006). 10.1186/gb-2006-7-s1-s4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics30, 923–930 (2014). 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 63.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol.37, 907–915 (2019). 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc.1, 2856–2860 (2006). 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- 66.Meier, F. et al. Online Parallel Accumulation-Serial Fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell Proteom.17, 2534–2545 (2018). 10.1074/mcp.TIR118.000900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res.10, 1794–1805 (2011). 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- 68.Hodge, K., Have, S. T., Hutton, L. & Lamond, A. I. Cleaning up the masses: exclusion lists to reduce contamination with HPLC-MS/MS. J. Proteom.88, 92–103 (2013). 10.1016/j.jprot.2013.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gallien, S., Kim, S. Y. & Domon, B. Large-scale targeted proteomics using internal standard triggered-parallel reaction monitoring (IS-PRM). Mol. Cell Proteom.14, 1630–1644 (2015). 10.1074/mcp.O114.043968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye, B. H. et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet.16, 161–170 (1997). 10.1038/ng0697-161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
The datasets generated during the current study are available in the GEO repository as super-series GSE234684 or separately under accession numbers GSE234657 (RNA-seq data of mouse GC B cells) and GSE234656 (Cut&Run data of human normal GC B cells) and in the ProteomeXchange Consortium via the PRIDE repository with dataset identifier PXD043998 (mass spectrometry proteomics data). Source data are provided with this paper.