Abstract
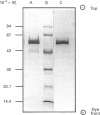
L(+)-Mandelate dehydrogenase was purified to homogeneity from the yeast Rhodotorula graminis KGX 39 by a combination of (NH4)2SO4 fractionation, ion-exchange and hydrophobic-interaction chromatography and gel filtration. The amino-acid composition and the N-terminal sequence of the enzyme were determined. Comprehensive details of the sequence determinations have been deposited as Supplementary Publication SUP 50172 (4 pages) at the British Library Document Supply Centre, Boston Spa, Wetherby, West Yorkshire LS23 7BQ, U.K., from whom copies can be obtained on the terms indicated in Biochem. J. (1993) 289, 9. The enzyme is a tetramer as judged by comparison of its subunit M(r) value of 59,100 and native M(r) of 239,900, estimated by SDS/PAGE and gel filtration respectively. There is one molecule of haem and approx. one molecule of non-covalently bound FMN per subunit. 2,6-Dichloroindophenol, cytochrome c and ferricyanide can all serve as electron acceptors. L(+)-Mandelate dehydrogenase is stereospecific for its substrate. D(-)-Mandelate and L(+)-hexahydromandelate are competitive inhibitors. The enzyme has maximum activity at pH 7.9 and it has a pI value of 4.4. HgCl2 and 4-chloromercuribenzoate are potent inhibitors, but there is no evidence that the enzyme is subject to feedback inhibition by potential metabolic effectors. The evidence suggests that L(+)-mandelate dehydrogenase from R. graminis is a flavocytochrome b which is very similar to, and probably (at least so far as the haem domain is concerned) homologous with, certain well-characterized yeast L(+)-lactate dehydrogenases, and that the chief difference between them is their mutually exclusive substrate specificities.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison N., O'Donnell M. J., Fewson C. A. Membrane-bound lactate dehydrogenases and mandelate dehydrogenases of Acinetobacter calcoaceticus. Purification and properties. Biochem J. 1985 Oct 15;231(2):407–416. doi: 10.1042/bj2310407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D. P., Kleanthous C., Keen J. N., Weinhold E., Fewson C. A. Mechanistic and active-site studies on D(--)-mandelate dehydrogenase from Rhodotorula graminis. Biochem J. 1992 Jan 1;281(Pt 1):211–218. doi: 10.1042/bj2810211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M. T., Gunn F. J., Chapman S. K., Reid G. A. Structural basis for the kinetic differences between flavocytochromes b2 from the yeasts Hansenula anomala and Saccharomyces cerevisiae. Biochem J. 1989 Nov 1;263(3):973–976. doi: 10.1042/bj2630973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers R. M., Fewson C. A. Purification and characterization of benzaldehyde dehydrogenase I from Acinetobacter calcoaceticus. Biochem J. 1989 Nov 1;263(3):913–919. doi: 10.1042/bj2630913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham D. R. Initial reactions involved in the dissimilation of mandelate by Rhodotorula graminis. J Bacteriol. 1984 Nov;160(2):778–780. doi: 10.1128/jb.160.2.778-780.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham D. R., McNamee C. G., Stewart D. B. Dissimilation of aromatic compounds in Rhodotorula graminis: biochemical characterization of pleiotropically negative mutants. J Bacteriol. 1984 Nov;160(2):771–777. doi: 10.1128/jb.160.2.771-777.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewson C. A. Microbial metabolism of mandelate: a microcosm of diversity. FEMS Microbiol Rev. 1988 Apr-Jun;4(2):85–110. doi: 10.1111/j.1574-6968.1988.tb02737.x. [DOI] [PubMed] [Google Scholar]
- Guiard B., Groudinsky O., Lederer F. Homology between bakers' yeast cytochrome b2 and liver microsomal cytochrome b5. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2539–2543. doi: 10.1073/pnas.71.6.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiard B., Lederer F. The "b5-like" domain from chicken-liver sulfite oxidase: a new case of common ancestral origin with liver cytochrome b5 and bakers' yeast cytochrome b2 core. Eur J Biochem. 1977 Mar 15;74(1):181–190. doi: 10.1111/j.1432-1033.1977.tb11379.x. [DOI] [PubMed] [Google Scholar]
- Guiard B. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: the yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2). EMBO J. 1985 Dec 1;4(12):3265–3272. doi: 10.1002/j.1460-2075.1985.tb04076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haumont P. Y., Thomas M. A., Labeyrie F., Lederer F. Amino-acid sequence of the cytochrome-b5-like heme-binding domain from Hansenula anomala flavocytochrome b2. Eur J Biochem. 1987 Dec 15;169(3):539–546. doi: 10.1111/j.1432-1033.1987.tb13642.x. [DOI] [PubMed] [Google Scholar]
- Hoey M. E., Allison N., Scott A. J., Fewson C. A. Purification and properties of L-mandelate dehydrogenase and comparison with other membrane-bound dehydrogenases from Acinetobacter calcoaceticus. Biochem J. 1987 Dec 15;248(3):871–876. doi: 10.1042/bj2480871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Labeyrie F., Baudras A., Lederer F. Flavocytochrome b 2 or L-lactate cytochrome c reductase from yeast. Methods Enzymol. 1978;53:238–256. doi: 10.1016/s0076-6879(78)53030-9. [DOI] [PubMed] [Google Scholar]
- Lederer F., Cortial S., Becam A. M., Haumont P. Y., Perez L. Complete amino acid sequence of flavocytochrome b2 from baker's yeast. Eur J Biochem. 1985 Oct 15;152(2):419–428. doi: 10.1111/j.1432-1033.1985.tb09213.x. [DOI] [PubMed] [Google Scholar]
- MacKintosh R. W., Fewson C. A. Benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase II from Acinetobacter calcoaceticus. Purification and preliminary characterization. Biochem J. 1988 Mar 15;250(3):743–751. doi: 10.1042/bj2500743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smékal O., Yasin M., Fewson C. A., Reid G. A., Chapman S. K. L-mandelate dehydrogenase from Rhodotorula graminis: comparisons with the L-lactate dehydrogenase (flavocytochrome b2) from Saccharomyces cerevisiae. Biochem J. 1993 Feb 15;290(Pt 1):103–107. doi: 10.1042/bj2900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou A. Y., Ransom S. C., Gerlt J. A., Buechter D. D., Babbitt P. C., Kenyon G. L. Mandelate pathway of Pseudomonas putida: sequence relationships involving mandelate racemase, (S)-mandelate dehydrogenase, and benzoylformate decarboxylase and expression of benzoylformate decarboxylase in Escherichia coli. Biochemistry. 1990 Oct 23;29(42):9856–9862. doi: 10.1021/bi00494a015. [DOI] [PubMed] [Google Scholar]
- Vanni P., Giachetti E., Pinzauti G., McFadden B. A. Comparative structure, function and regulation of isocitrate lyase, an important assimilatory enzyme. Comp Biochem Physiol B. 1990;95(3):431–458. doi: 10.1016/0305-0491(90)90002-b. [DOI] [PubMed] [Google Scholar]